Abstract
Metabolons, multiprotein complexes consisting of sequential enzymes of a metabolic pathway, are proposed to be biosynthetic “hotspots” within the cell. However, experimental demonstration of their presence and functions has remained challenging. We used metabolomics and in situ three-dimensional submicrometer chemical imaging of single cells by gas cluster ion beam secondary ion mass spectrometry (GCIB-SIMS) to directly visualize de novo purine biosynthesis by a multienzyme complex, the purinosome. We found that purinosomes comprise nine enzymes that act synergistically, channeling the pathway intermediates to synthesize purine nucleotides, increasing the pathway flux, and influencing the adenosine monophosphate/guanosine monophosphate ratio. Our work also highlights the application of high-resolution GCIB-SIMS for multiplexed biomolecular analysis at the level of single cells.
De novo purine biosynthesis (DNPB) is a highly conserved, energy-intensive pathway that coordinates with the purine nucleotide salvage process to maintain purine levels to support cellular proliferation, survival, and metabolic adaptation under varying nutritional supply and evolving environmental demands (Fig. 1A) (1). The de novo pathway is carried out by six enzymes in higher organisms, including humans, commencing with phosphoribosyl pyrophosphate (PRPP), which is converted to inosine monophosphate (IMP) in 10 sequential steps (fig. S1A). IMP is the branchpoint intermediate that is converted to either adenosine 5′-monophosphate (AMP) or guanosine 5′-monophosphate (GMP) by four enzymes. By contrast, the free purine bases hypoxanthine, adenine, and guanine can be combined with PRPP to regenerate their respective mononucleotides by the action of salvage enzymes—hypoxanthine guanine phosphoribosyltransferase (HGPRT) and adenine phosphoribosyltransferase (APRT)—in a single step (Fig. 1A) (2–5).
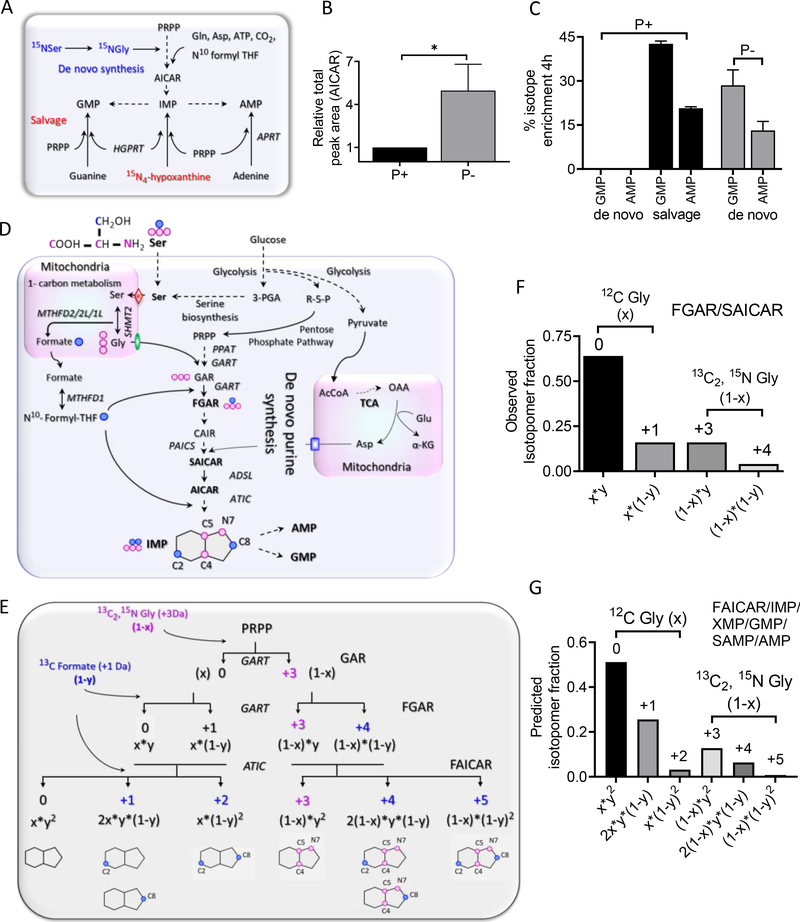
Fig. 1. A theoretical model to interrogate DNPB in HeLa cells upon pathway upregulation.
(A) Purine nucleotides can be regenerated by salvage synthesis (conversion of free purine base back to the nucleotides) or a 10-step de novo purine biosynthesis (DNPB) starting with PRPP. [15N4]Hypoxanthine (Hypo) incorporation is used to monitor salvage; [15N]Serderived Gly ([15N]Gly) incorporation is used to monitor DNPB flux. (B) Abundance of the DNPB pathway intermediate AICAR for purine-rich (P+) or purine-depleted (P−) media conditions. *P < 0.02 (two-tailed t test). (C) [15N]Ser incorporation in the end products AMP and GMP. Data in (B) and (C) are means ± SD of three independent experiments. (D) The metabolic interdependence of the cytosolic and mitochondrial metabolism to support DNPB and the flow of stable isotope–labeled Gly and formate, generated in mitochondria from labeled [13C3,15N]Ser, into the DNPB pathway intermediates and the end-product nucleotides. Pink circles denote 13C and 15N atoms of the labeled Ser backbone (produces [13C2,15N]Gly, +3 Da); blue circles represent the 13C at its side-chain β position ([13C]formate, +1 Da). Each pink and blue circle denotes incorporation of one labeled atom. Red diamond, SFXN1 (mitochondrial Ser transporter); green oval, SLC25A38 (mitochondrial Gly transporter); blue rectangle, SLC25A12/SLC25A13 (mitochondrial glutamate/aspartate transporter). See fig. S1 caption for acronyms not defined in text. (E) The classical DNPB pathway model and the isotopomers of intermediates and end nucleotides, with the labeled positions indicated on the product purine ring. Labeled Gly [x, unlabeled fraction; (1 – x), labeled fraction] and labeled formate [y, unlabeled fraction; (1 – y), labeled fraction] generated from [13C3,15N]Ser enter the DNPB pathway at three different steps (curved arrows); the first two are catalyzed by the enzyme GART and the third is catalyzed by the enzyme ATIC. Note that the +3, +4, and +5 isotopomer species in the end nucleotides show no interference from the preexisting or unlabeled nucleotide pools that are generated in parallel. (F) Example showing the use of the model to calculate (1 – x) and (1 – y) from the observed FGAR and SAICAR isotopomers. The values were calculated for each individual experiment. (G) The isotopomer distribution in FAICAR is predicted using the values of (1 – x) and (1 – y) and the observed SAICAR isotopomer distribution. All the end-product nucleotides (IMP, AMP, GMP) generated via DNPB in the time course of the experiment are expected to have the same isotopic distribution as FAICAR (fig. S1, A to C).
We previously reported the partial colocalization of multiple DNPB enzymes observed as cytosolic punctate structures, heterogeneous in their size, number, and composition, by fluorescence microscopy (2, 6–8). A substantial fraction of these dynamic structures, called purinosomes, show proximity to mitochondria (9), whereas others reside on microtubules and show directed movement toward mitochondria (10). The mitochondria-associated purinosomes are proposed to constitute the active DNPB metabolon, but the dynamic and fragile nature of such multienzyme complexes (purinosomes and all metabolons in general) (11, 12) has impeded their in vitro reconstitution. Moreover, artifacts introduced by transient overexpression and limitations of fluorescence imaging–based methods have made it challenging to determine the complete enzymatic and ancillary protein composition, to elucidate their relative stoichiometry in the complex, and to ascertain the functional state of the enzymes in purinosomes.
With the purinosome as a potential precedent, we sought support for the hypothesis that cellular metabolic pathways might generally organize in space and time to confer different properties on the collective versus the individual enzymes. Although this study is restricted to HeLa cells, we envision the extension of this methodology to a variety of cells in normal and disease states, leading to insights into how the function of metabolic pathways might be altered to sustain cellular processes.
A model to probe the de novo purine biosynthesis pathway
The two alternative routes of purine synthesis, salvage and de novo, were probed by following the incorporation of labeled hypoxanthine or glycine (Gly), respectively, in the intermediates and end nucleotides of the two pathways (Fig. 1A). Upon purine depletion, where purinosome formation has previously been observed, HeLa cells showed significant de novo synthesis, indicated by higher abundance of the DNPB intermediate 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) (Fig. 1B) as well as [15N]Gly incorporation in the products, AMP and GMP (hereafter AMP/GMP) (Fig. 1C). When cultured in purine-rich (P+) media, the cells instead seemed to only carry out salvage synthesis, quantified by [15N4]hypoxanthine incorporation in AMP/GMP (Fig. 1C). Thus, we used the purine-depleted HeLa cells, which show high DNPB flux, as the model system to probe the metabolic consequence of purinosome formation.
We developed a model to mathematically predict the distribution of isotopic labels that would be expected in the absence of the DNPB metabolon, and then tested the predictions by performing an in vivo isotope incorporation experiment. When cells are grown in isotopically labeled [13C3,15N]Ser (label on backbone and side-chain carbons as well as the backbone amine), labeled carbon and nitrogen can enter into various metabolites within the pathway (Fig. 1D). Cells take up labeled Ser from the media, which is then internalized by mitochondria through the mitochondrial Ser transporter SFXN1 and is acted upon by a suite of enzymes [serine hydroxymethyltransferase 2 (SHMT2) and different isoforms of methylenetetrahydrofolate dehydrogenase/cyclohydrolase (MTHFD)] that constitute the mitochondrial one-carbon metabolism, producing Gly and formate (13, 14). These products exit the mitochondria and are incorporated in different cytosolic pathways, including the DNPB pathway, by the action of the enzymes GART (trifunctional phosphoribosylglycinamide formyltransferase) and ATIC (bifunctional 5-aminoimidazole-4-carboxamide nucleotide formyltransferase/IMP cyclohydroxylase) (Fig. 1D) (15).
We considered a null hypothesis model assuming each of the 10 steps of the pathway, starting with PRPP and leading up to the formation of IMP, to be independent of each other; all the intermediates to be completely equilibrated with their respective cytosolic pools; and a homogeneous distribution of all the enzymes (i.e., no purinosome formation and localization proximal to mitochondria), coenzymes, substrates, and cofactors in the cytosol. The model allows prediction of the isotopomer distribution in DNPB pathway intermediates and end products assuming uniform isotopically labeled Gly (1 – x) and formate (1 – y) incorporation across the pathway (Fig. 1E). If x and (1 – x) are the respective fractions of unlabeled and labeled Gly, and y and (1 – y) are the respective fractions of unlabeled and labeled formate generated from the mitochondrial metabolism of [13C3,15N]Ser, the incorporation of labeled Gly and formate into the DNPB pathway can be mathematically described (Fig. 1E). It follows that the observed isotopomer distribution in DNPB pathway intermediates that show one Gly and one formate incorporation [i.e., phosphoribosyl-N-formylglycinamide (FGAR), phosphoribosyl aminoimidazole succinocarboxamide (SAICAR), and AICAR] can be used to calculate the source Gly (1 – x) and formate (1 – y) isotope enrichment (Fig. 1F), as well as to predict the expected isotopomer distribution in 5-formamidoimidazole-4-carboxamide ribonucleotide (FAICAR) and all the downstream nucleotides (Fig. 1G). [13C3,15N]Ser is particularly useful in generating unique isotopomer species arising from unlabeled and labeled Gly with no overlap between them and allowing for calculation of the newly generated pools of each intermediate and end product (see supplementary materials).
Channeled de novo purine biosynthesis by purinosomes adjacent to mitochondria
Our model describes DNPB as a simple diffusive relay of reactions (Fig. 1E), and the following testable predictions arise from it: (i) IMP and all the downstream nucleotides must have the same isotopomer distribution of +3, +4, and +5 species, because there is no further isotope incorporation after FAICAR formation; and (ii) labeled Gly and formate enrichment in the DNPB intermediates and end nucleotides generated during the course of the experiment should be the same (fig. S1, B and C). To test these predictions, we performed isotopic incorporation experiments to probe the mitochondriadependent generation of isotopically labeled Gly and formate and their incorporation in purines by supplying a limited concentration of [13C3,15N]Ser (30 μM) for 4 hours, followed by high-resolution quantitative liquid chromatography–mass spectrometry (LC/MS) of cellular metabolite extracts (fig. S2, A to C).
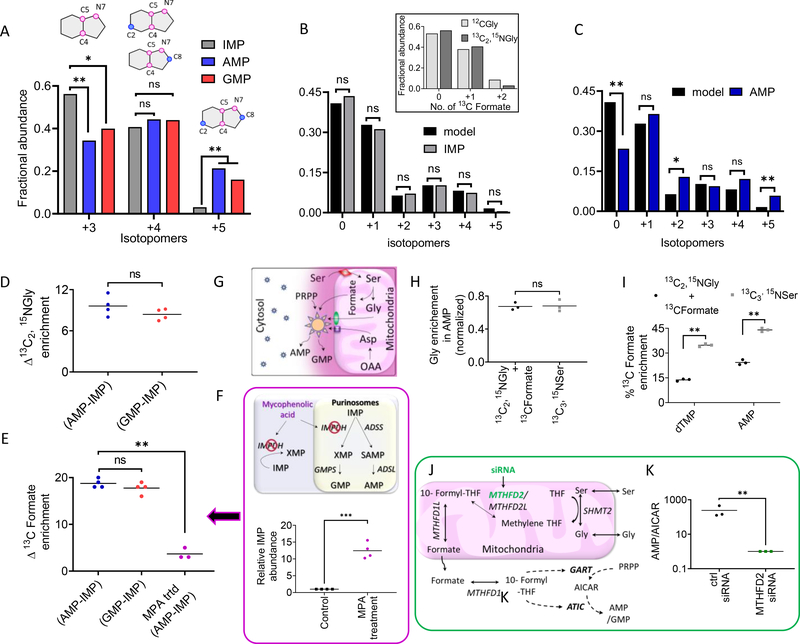
In contrast to the prediction, the fractional abundances of the two isotopomers (+3 and +5) in IMP were significantly different from those in AMP and GMP (Fig. 2A and fig. S2F), respectively, signifying that IMP and AMP/GMP have distinct isotopomeric distributions. To understand the source of this difference, using the observed FGAR and SAICAR isotopomer distribution in each independent experiment, we computed the complete isotopomer distribution in IMP as described by the model and compared it with the observed isotopomer distribution for IMP and all the nucleotides downstream of it, namely xanthosine monophosphate (XMP), succinyl-AMP (SAMP), AMP, and GMP (pathway steps shown in fig. S1A). Whereas the observed distribution for IMP and XMP matched the model-predicted isotopomer distribution (Fig. 2B and fig. S2D), AMP, SAMP, and GMP all showed a significantly different isotopomer distribution relative to the prediction (Fig. 2C and fig. S2, E and F). Consequently, IMP and its precursor substrates FGAR and SAICAR appear to have been synthesized from the same cytosolic pool of substrates (Gly and formate) and to have different isotopic enrichment relative to the substrate pool used for AMP and GMP synthesis. Moreover, no pathway intermediates with the isotopic enrichment seen in AMP/GMP were detected.
Fig. 2. HeLa cells show highly channeled DNPB by enzymes proximal to mitochondria.
(A) Fractional distribution of de novo synthesized +3, +4, and +5 isotopomers in IMP, AMP, and GMP. The +3 isotopomer has a significantly lower fractional contribution in AMP and GMP relative to IMP, but that of the +5 isotopomer is significantly higher than IMP. A paired two-tailed t test was performed to analyze the ratio of isotopic abundance of each isotopomer. Data from four independent replicates were combined to assess the significance of the isotopomeric differences between IMP and AMP/GMP. *P < 0.05, **P < 0.005; ns, not significant. (B and C) The isotopomer distribution calculated according to the model described in Fig. 1G matched the observed distribution in IMP (B) but not AMP (C) [or GMP (fig. S2F)]. Inset in (B) shows overlay of the observed fractional abundances of IMP isotopomers arising from 0, 1, or 2 13C incorporations (derived from formyl-THF) in purine ring containing either unlabeled Gly or [13C2,15N]Gly for one representative experiment. The values were computed as described in fig. S1C. (D and E) There is a ~10% difference in [13C2,15N]Gly (D) and a ~15 to 20% difference in [13C]formate enrichment (E) between newly synthesized AMP/GMP and IMP. (F) [13C]Formate enrichment difference between AMP and IMP is significantly lowered upon mycophenolic acid (MPA) treatment [magenta symbols in (E)]. (G) Localization of a “complete” functional purinosome (which shows channeling, shown as large sun symbol) near mitochondria, the site of production of isotopically labeled Gly and formate, leads to higher isotope enrichment in the purine nucleotides, AMP and GMP, produced by metabolic channeling. Free enzymes and incomplete purinosomes residing away from the mitochondria (small sun symbols) show lower isotope enrichment because of their limited accessibility to labeled substrates. Other symbols and acronyms are the same as in Fig. 1A and fig. S1A. (H and I) Comparison of Gly and formate enrichment, respectively, in AMP [and GMP (fig. S2H)] when molar equivalents of [13C2,15N]Gly + [13C]formate were added to the media instead of [13C3,15N]Ser. As a result of the regeneration of labeled Ser from Gly and formate (fig. S2G) and further Ser metabolism in the mitochondria, channeled synthesis of AMP/GMP was still supported. (I) Consistent with the lower formate uptake, indicated by lower isotope enrichment in deoxythymidine monophosphate (dTMP), enrichment in AMP was lower with Gly + formate than with Ser. (J) DNPB enzymes GART and ATIC use 10-formyl-THF, and the cytosolic availability of this cofactor depends on the mitochondrial one-carbon metabolism that generates formate from Ser. (K) Knockdown of the mitochondrial one-carbon metabolism enzyme MTHFD2 by siRNA treatment led to a significant reduction in the de novo synthesized AMP and GMP flux but significant AICAR accumulation. For each experiment, three or four biological replicates were performed with one or two technical replicates; (A), (B), and (C) and fig. S2, D to F, correspond to the same representative experiment. In (B) and (C), data from four independent experiments were used for statistical analysis. *P < 0.05, **P < 0.005. In (D), (E), (H), (I), and (K), the mean and individual data points are plotted. *P < 0.05, **P < 0.005 (paired two-tailed t tests).
Next, using the observed isotopomer distribution in AMP and GMP, we computed the percentage isotopic enrichment in the source substrates of the channeled pathway. The newly generated channeled DNPB products, AMP/GMP, had higher isotope enrichment than IMP, synthesized by the unchanneled pathway, with respect to both Gly and formate. The newly synthesized AMP/GMP showed ~10% higher Gly enrichment (Fig. 2D) and 15 to 20% higher formate enrichment relative to IMP (Fig. 2E). We conclude that the synthesis of AMP and GMP is accomplished in a highly channeled manner, preventing pathway intermediates from equilibrating with their bulk cytosolic pools. The higher isotope enrichment in AMP/GMP relative to IMP indicates the physical proximity of an “active” DNPB metabolon or plausible direct association of the enzymes with the mitochondrial metabolite transporters.
To test this explanation, we poisoned cells with mycophenolic acid (MPA), a specific high-affinity inhibitor of IMP dehydrogenase (IMPDH), an enzyme involved in the synthesis of guanine nucleotides (16). IMPDH inhibition is expected to cause accumulation and leakage of IMP from purinosomes, leading to intermixing of IMP produced from the two independent pathways. As expected, treatment of cells with the inhibitor resulted in significant accumulation of IMP (by a factor of ~12) (Fig. 2F) as a result of forced release of IMP synthesized by the channeled purinosomes. Consistent with our hypothesis, upon MPA treatment the observed labeled formate enrichment in AMP and IMP was similar (Fig. 2E and fig. S2I). Likewise, when the concentration of isotope-labeled Ser (120 μM) was increased by a factor of 4, it led to homogeneous spread of labeled formate across the cytosolic volume. Under these conditions, GMP and AMP (synthesized by channeled pathway) and the intermediates XMP and IMP (synthesized by unchanneled pathway) showed similar formate isotope enrichment (fig. S2, C and M).
These observations provide a rationale for the interpretation that the mitochondria-proximal purinosome must represent the “active” DNPB metabolon. This proximity results in the preferential capture of Ser-derived Gly and formate by “active” purinosomes for channeled DNPB, in which the equilibration of the purinosome-synthesized intermediates with their respective bulk cytosolic pools is limited. The detected IMP, on the other hand, must arise from a second diffusive substrate pathway or incomplete purinosomes. Consequently, the “active” DNPB metabolon represents the assembly of nine enzymes localized proximal to mitochondria, capable of catalyzing the conversion of PRPP to AMP and GMP in a sequence of 14 highly channeled steps (Fig. 2G). This may also rationalize the previously reported directed migration of purinosomes along the microtubule to facilitate the access of mitochondrially generated metabolites, Gly, Asp, and formate.
Instead of labeled Ser, when molar equivalents of labeled Gly and formate are supplemented in the media, action of the enzymes MTHFD1 and SHMT1 regenerated cytosolic labeled Ser (17) (fig. S2G).Under these conditions, we still observed channeled AMP/GMP synthesis, as reflected by the similar isotope incorporation in AMP and GMP (Fig. 2, H and I, and fig. S2H). The percentage of Gly incorporation in the end nucleotides AMP/GMP was normalized by the observed Gly enrichment in reduced glutathione to account for the differences in the uptake of Ser and Gly (Fig. 2H and fig. S2H).
Next, we examined whether the channeled DNPB exhibited an increase in the pathway efficiency, a hallmark of metabolons. We estimated the difference in the efficiencies of the DNPB channeled versus unchanneled pathways operating in parallel. The ratio of total newly synthesized AMP and GMP was higher than that of the total IMP produced during our experiment by a factor of ~7, highlighting the effectiveness of the mitochondria-associated multienzyme assembly in achieving greater pathway flux (fig. S2J). The role of mitochondrial metabolism in supporting the channeled pathway was further tested by knocking down the mitochondrial formate production pathway, and consequently 10-formyltetrahydrofolate (10-formyl-THF) production, by targeting MTHFD2 (one of the enzymes involved in mitochondrial one-carbon metabolism) (18) (Fig. 2J). Upon small interfering RNA (siRNA)–mediated knockdown of MTHFD2 (fig. S2K), there was a disruption in the pathway efficiency by a factor of ~100, reflected in the accumulation of the intermediate AICAR and a decrease in the production of the end product (AMP) relative to the control (Fig. 2K).
IMP lies at a branchpoint step, and its partitioning into the two possible downstream nucleotides AMP or GMP presents an intriguing scenario. The kinetic parameters for the individual enzymes and their overall availability are expected to favor guanine nucleotide synthesis over the adenine nucleotide by at least a factor of ~25 (7). On the other hand, the abundance of adenine nucleotides is higher than that of guanine nucleotides by a factor of ~10, and the adenine nucleotide content in the human genome is higher (~30%) than the guanine nucleotide content (~20%) (19). The channeled pathway favors the synthesis of adenine nucleotides over guanine nucleotides (fig. S2L). We hypothesize that cells may achieve preference for adenine nucleotides by regulating the composition of purinosomes to favor the enzymes adenylosuccinate synthetase (ADSS) and bifunctional adenylosuccinate lyase over IMPDH and GMP synthetase, by modulating the orientation of the branchpoint enzymes in the purinosome, or by localizing purinosomes close to the mitochondrial site of Asp production, thus increasing the availability of Asp for ADSS.
Application of high-resolution GCIB-SIMS imaging to probe biochemistry at the single-cell level
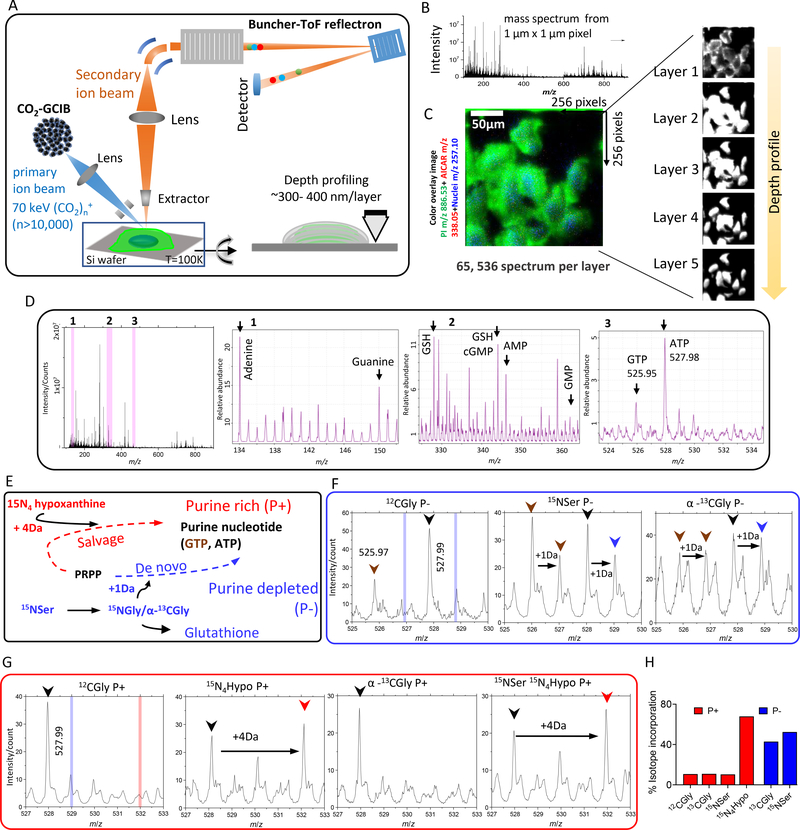
Mass spectrometry imaging (MSI) has emerged as a powerful tool to spatially locate the endogenous and exogenous compounds in various biological systems (20–23). We used GCIB-SIMS, which permits high-mass ion detection with low chemical damage, to study the cytoplasmic distribution of intact molecular ions of purine biosynthetic pathway intermediates and end nucleotides in a frozen hydrated monolayer of HeLa cells (fig. S3) (24–28). Cryo–scanning electron microscopy (cryo-SEM) images of frozen hydrated HeLa cells demonstrated that the cell size and morphology remain unperturbed after sample preparation (fig. S5, A to C). For multilayer in situ chemical profiling, we used high-voltage GCIB-SIMS with a (CO2)n+ (n > 10,000) cluster size and a focus spot 1 μm in diameter, generating an array of mass spectra [mass/charge ratio (m/z) 90 to 900] corresponding to each 1 μm×1 μm × ~300 to 400 nm voxel and covering a total lateral field of view of ~256 μm × 256 μm in each layer (Fig. 3, A to D).
Fig. 3. Identification of unique molecular ions of purine nucleotides in the intracellular pool by in situ GCIB-SIMS.
(A) Schematic of GCIB-SIMS imaging of HeLa cells. Imaging uses a finely focused 70-keV (CO2)n+ (n > 10,000) cluster beam to interrogate frozen hydrated HeLa cells three-dimensionally at 1-μm spatial resolution. Coupled with a buncher-ToF and direct-current beam setup, maximum spatial resolution and mass resolution can be retained. A pixel-by-pixel analysis was performed across a lateral field of view of 256 μm × 256 μm. (B) Mass spectra in the m/z range 0 to 900 were recorded for each pixel. (C) A composite two-dimensional colored image was generated combining the signal across all the layers PI (38:4; green) at m/z 886.53, phosphate-sugar backbone at m/z 257.10 (blue) from nucleotides, and 15N-enriched DNPB intermediate AICAR (red). Combination of mass spectral analysis and the spatial distribution of specific cellular signals demonstrates the reliability of the method for in situ biochemical studies. (D) Complete negative ion spectra from frozen hydrated HeLa cells with the unique peak assignments for the metabolites relevant to the study. The ordinate axis represents absolute intensity for each ion. Zoom-in view of the m/z ranges marked as 1, 2, and 3 and highlighted in pink bar are presented to show the peaks corresponding to (1) adenine and guanine base, (2) reduced glutathione (GSH), AMP, and GMP, and (3) salt adducts of ATP and GTP (spectra of all the standard compounds can be found in fig. S3). The intensities are relative to the highest abundant molecular ion. (E) Stable isotope enrichment under purine-rich (P+) and purine-depleted (P−) conditions via salvage (red) or de novo synthesis (blue) pathways, respectively. Isotope tracer experiments were leveraged to specifically label the purine base ring using either [15N4]hypoxanthine, imparting +4 Da mass increment, or [15N]Ser (metabolized to [15N]Gly) or [13C]Gly, imparting +1 Da mass increment. (F and G) Select SIMS ion spectra from P− (F) and P+ (G) HeLa cells for unlabeled (black arrowhead), 15N4-labeled (red arrowhead), or [13C,15N]ATP (blue arrowhead) and GTP (brown arrowhead) are shown along with the expected peak positions corresponding to +4 Da (vertical red bar) or +1 Da (vertical blue bar) mass increment, respectively. The intensities are relative to the highest abundant molecular ion. (H) The isotope enrichment profile was similar to that obtained from the high-resolution LC/MS spectrum of metabolite extracts of similarly grown cells.
The validity of the GCIB-SIMS images was confirmed by monitoring the deprotonated molecular ion [M-H]− of phosphoinositol lipid [PI 38:4, m/z 885.53, known to localize in the cell membrane (29)] (fig. S5G). Next, we confirmed that the cellular metabolites were localized within the cellular boundary and that the lateral and depth distribution of metabolites was also preserved (fig. S5D). The analysis confirmed that the spectrum obtained from each pixel of a lateral layer remained unaffected by the analysis performed on the adjacent pixels. Subsequently, a depth profile through the cell during a continuous layered scan showed that deeper layers remained undisturbed while the upper layers were being analyzed (Fig. 3C).
A GCIB-SIMS scan of cells produces a set of complex mass spectra because the ionizable metabolites yield several ionic species, including the pseudo-molecular ion, salt adducts, and fragment ions; hence, interpretation can be challenging (20, 28) (Fig. 3D and fig. S3). Before further analysis of the purine biosynthetic pathway, unique peaks were identified that primarily constituted the metabolites of interest, with minimum interference from other compounds. The ionization of pure standard compounds, including all purine nucleotides and the intermediates SAICAR and AICAR, were studied to optimize SIMS measurement that would yield intact deprotonated ions as a characteristic ion (fig. S3). Isotope tracer experiments were leveraged to validate the peak assignments by following the incorporation of isotope-labeled hypoxanthine, Ser, and Gly in the cells grown under P+ or P− conditions, respectively (Fig. 3, E to H). Before MSI, cells were supplemented with stable isotope–labeled [15N]Ser/[13C]Gly (+1 Da isotopomer generation via the DNPB pathway) (Fig. 3F) or [15N4]hypoxanthine (+3.988 Da isotopomer generation via purine salvage pathway) (Fig. 3G and fig. S5, H to L). Isotope label incorporation continued for 12 to 14 hours to ensure sufficient enrichment allowing detection of all isotopomers by GCIB-SIMS. The isotope incorporation percentage measured by SIMS was in agreement with that obtained in bulk analysis by high-resolution LC/MS of cell extracts after 12 hours of incorporation (Fig. 3H).
In situ GCIB-SIMS imaging captures the metabolon in action
As estimated from fluorescence imaging, purinosomes are roughly spheroid multiprotein assemblies with a diameter range of 0.2 to 0.9 μm (2), thus making 3D molecular scanning GCIB with a focal diameter of 1 μm particularly suitable to capture active functional purinosomes. The metabolic channeling observed for the DNPB pathway, if arising as a result of active DNPB by purinosomes, would lead to higher local concentration of the pathway intermediates and end nucleotides close to the enzyme complexes acting as biosynthetic hotspots. We exploited this feature to identify and characterize purinosomes. HeLa cells were grown in purine-depleted media and supplemented with [15N]Ser (see supplementary materials for experimental details) for isotope enrichment for 12 to 14 hours before performing MSI. Because of the mitochondria compartment–specific conversion of Ser to Gly, we expected the mitochondria-associated DNPB metabolon to show spatially confined higher concentrations of isotopically labeled intermediates and end-product nucleotides. Bulk metabolomic estimations by LC/MS showed that AICAR is efficiently channeled and accumulates only under limited formyl-THF availability (figs. S5K and S6A). Therefore, the cytoplasmic loci with high concentrations of the 15N-labeled DNPB intermediate AICAR were used as the reporter of the active purinosomes in the flash-frozen HeLa cells.
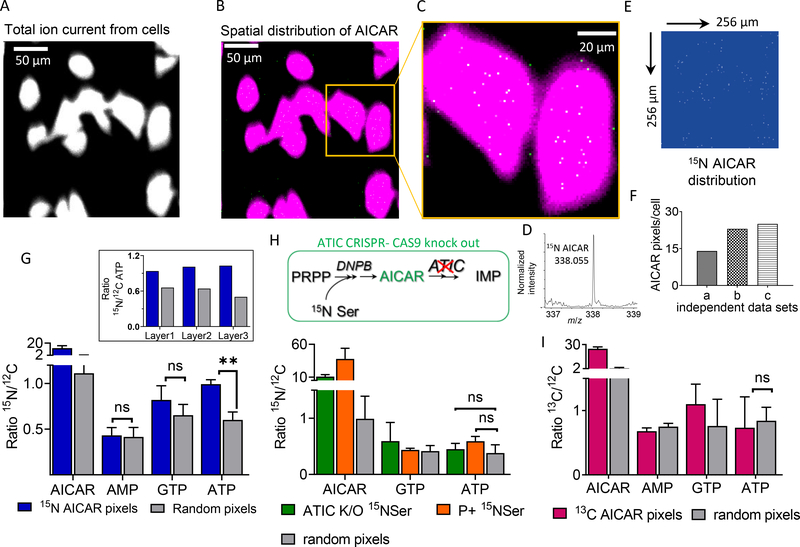
The total ion spectrum image was used to define the cellular boundary in each layer (Fig. 4A). From the total ion spectrum of cells, the peak corresponding to labeled AICAR (m/z 338.05, Δm/z 0.01) was selected to obtain its spatial distribution in each layer (Fig. 4, B to D). Each layer was analyzed independently, and pixels with less than 30% of the highest intensity per pixel and/or that appeared outside the defined cell boundary were discarded from the analysis. AICAR showed a nonhomogeneous distribution with distinct, isolated higher-concentration voxels (Fig. 4E and fig. S7, A and B), with ≥3 AICAR ions per voxel. This result suggests a higher abundance of AICAR molecules per voxel by a factor of 300 to 1000 relative to the abundance expected for a homogeneous distribution throughout the cellular volume (fig. S6B). We observed an average density of 10 to 30 15N-enriched AICAR pixels per cell (Fig. 4F), although we expect this to be an underestimation of the number of active purinosomes per cell because of technical limitations.
Fig. 4. Combining GCIB-SIMS and isotope label incorporation to identify the loci of channeled DNPB in single cells.
(A) Representative GCIB-SIMS image of P− HeLa cells grown on a Si substrate, with a field of view of 256 μm × 256 μm, after allowing [15N]Ser enrichment for 14 hours. Image was generated using cumulative total ion current in negative ion mode. (B) Spatial distribution of [15N]AICAR pixels in each analyzed layer was generated using Δm/z = 0.01, centered at m/z 338.055. (C) Zoomed-in view of the area of interest [yellow box in (B)] shows an overlay of the pixels with high [15N]AICAR abundance (white) and cell image generated using total ion current (magenta). (D) Zoomed-in region of mass spectrum showing [15N]AICAR peak from the selected pixels within the cell boundary with AICAR signal/noise ratio above 30%. (E) Spatial distribution of the labeled AICAR pixels after applying the signal cutoff. (F) Total number of [15N]AICAR pixels per cell obtained from three independent biological replicates. (G) Comparison of level of 15N enrichment in the DNPB intermediate AICAR and the end-product nucleotides AMP, GTP, and ATP for the [15N]AICAR pixels and equivalent numbers of random pixels selected from across the cell. Error bar corresponds to the variation observed for each layer analyzed for a sample. The ratio of total signal intensity for the unique peak corresponding to the isotope-enriched (15N-labeled) and the respective unlabeled (12C) molecule of interest was determined for each layer using Δm/z = 0.02, centered at the expected exact mass corresponding to the molecular ions of interest. **P < 0.005. Inset: In each layer analyzed, the ratio of [15N]ATP/[12C]ATP in the [15N]AICAR pixels was consistently found to be higher than that in the random pixels selected from the whole cell. (H) In the HeLa cells with ATIC CRISPR-Cas9 knockout (ATIC K/O; deficient in ATIC, one of the de novo pathway enzymes) and in cells grown in purine-rich media, selective 15N enrichment in ATP in the pixels corresponding to [15N]AICAR was not observed upon [15N]Ser supplementation. (I) P− HeLa cells under high Gly concentration were used as a negative control where limited 10-formyl-THF results in diminished purinosome-mediated synthesis and thus leads to low ATP enrichment. [13C]AICAR pixels in the negative control showed no selective labeled ATP enrichment. MSI and statistical analysis were performed on at least three independent biological replicates.
To analyze the chemical composition of the labeled AICAR pixels, we generated cumulative mass spectra of all such pixels in the top two or three layers from all the cells in a single field of view. Next, we analyzed the isotope enrichment of the downstream pathway metabolites AMP, ATP, and GTP. In each layer, [15N]AICAR pixels showed an elevated 15N/12C ratio for the downstream end-product nucleotide ATP relative to random cellular pixels (Fig. 4G and fig. S7, D and F). The higher 15N/12C ratio of ATP in the labeled AICAR pixels was consistently seen in each layer analyzed and across all replicate experiments performed (Fig. 4G, inset, and fig. S7, E and G). This trend confirms that the enrichment observed in P− HeLa cells was a specific signal arising as a result of an active, channeled DNPB pathway.
We suspect that a similar correlation was not observed for the peak corresponding to the molecular ion from GTP because of the contribution of deoxy-GTP to the same peak in the cytosolic signal. Similarly, 15N label enrichment in AMP could not be observed because of low mass resolution, resulting in overlap of the AMP +1 peak with unlabeled IMP and thus interfering with the precise estimation of [15N]AMP. Also, such correlation between labeled AICAR and ATP was not observed in any of the control experiments—namely, ATIC CRISPR knockout HeLa cells (lacking the enzyme to catalyze the conversion of DNPB intermediate AICAR to IMP) (30) (Fig. 4H and fig. S7H), P+ [15N]Ser (cells with no observable DNPB flux) (Figs. 1C and 4H), and P– [13C]Gly (with leaky and inefficient channeled DNPB) (Fig. 4I and fig. S5, H to L). Together, our results are consistent with the hypothesis that purine production is localized to biosynthetic “hotspots” congruent with the “active” purinosome metabolon within the cell. GCIB-SIMS allows selective identification and analysis of the mitochondria-associated active purinosomes and shows that the levels of the isotopically labeled metabolites AICAR and ATP are statistically above those of the purinosome’s surroundings.
We have shown that the DNPB pathway is carried out by a metabolon that consists of at least nine enzymes that act synergistically to increase the pathway flux by a factor of ~7 and to preferentially partition a key intermediate, IMP, into AMP by a factor of 3 to 4 over GMP. On the basis of our findings, we propose a functional definition of the purinosomes as the “active” DNPB metabolon, located proximal to the mitochondria (Fig. 2G). We envision that a better understanding of the importance of the purinosome metabolon for human health and its role in aggressive cancers with high purine demand may reveal therapeutically important metabolic vulnerabilities. Our work demonstrates the application of mass spectrometry–based techniques to investigate and quantify metabolic channeling in pathways where enzyme coclustering has been observed (31) and highlights the usefulness of high-energy GCIB-SIMS imaging to explore single-cell biochemistry.
Supplementary Material
ACKNOWLEDGMENTS
HeLa ATIC CRISPR-CAS9 knockout cells were a gift from the Zikanova lab, Charles University and General University Hospital, Prague, Czech Republic. V.P. and S.J.B. thank A. Patterson and P. Smith (Huck Institutes of Life Sciences, Penn State) for running LC/MS. H.T. and V.P. acknowledge technical assistance from P. Blenkinsopp and E. Mengusoglu (Ionoptika, UK) for image processing and single-pixel analysis. V.P. thanks S.J.B.’s lab members A. M. Pedley and L.-N. Zou for very useful discussions and inputs.
Funding: Supported by NIH grant GM024129 (S.J.B.), the Huck Institutes of Life Sciences (V.P.), and the Materials Research Institute and the Institutes of Energy and the Environment at Penn State (H.T.).
Footnotes
Competing interests: The authors declare no conflict of interest.
Data and materials availability: The extracted peak areas for the metabolites analyzed in this study can be found in the accessory files. The GCIB-SIMS imaging data are available from Penn State ScholarSphere (32).
SUPPLEMENTARY MATERIALS
science.sciencemag.org/content/368/6488/283/suppl/DC1 Materials and Methods
REFERENCES AND NOTES
- 1.Villa E, Ali ES, Sahu U, Ben-Sahra I, Cancers 11, 688 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedley AM, Benkovic SJ, Trends Biochem. Sci. 42, 141–154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchanan JM, Hartman SC, in Advances in Enzymology and Related Areas of Molecular Biology, Vol. 21, Nord FF, Ed. (Wiley, 1959), pp. 199–261. [Google Scholar]
- 4.Hartman SC, Buchanan JM, Annu. Rev. Biochem. 28, 365–410 (1959). [DOI] [PubMed] [Google Scholar]
- 5.Greenberg GR, Jaenicke L, in CIBA Foundation Symposium on the Chemistry and Biology Of Purines, Wolstenholme GEW, O’Connor CM, Eds. (Wiley, 1957), pp. 204–232. [Google Scholar]
- 6.An S, Kumar R, Sheets ED, Benkovic SJ, Science 320, 103–106 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Zhao H et al. , J. Biol. Chem. 290, 6705–6713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan CY et al. , Proc. Natl. Acad. Sci. U.S.A. 112, 1368–1373 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.French JB et al. , Science 351, 733–737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan CY et al. , Proc. Natl. Acad. Sci. U.S.A. 115, 13009–13014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyoung M, Russell SJ, Kohnhorst CL, Esemoto NN, An S, Biochemistry 54, 870–880 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng Y et al. , J. Biol. Chem. 287, 36201–36207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang M, Vousden KH, Nat. Rev. Cancer 16, 650–662 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Kory N et al. , Science 362, eaat9528 (2018).30442778 [Google Scholar]
- 15.Ducker GS, Rabinowitz JD, Cell Metab. 25, 27–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sintchak MD et al. , Cell 85, 921–930 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD, Cell Rep. 7, 1248–1258 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Nilsson R et al. , Nat. Commun. 5, 3128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venter JC et al. , Science 291, 1304–1351 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Winograd N, Annu. Rev. Anal. Chem. 11, 29–48 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Fearn S, Mater. Sci. Technol. 31, 148–161 (2015). [Google Scholar]
- 22.Boxer SG, Kraft ML, Weber PK, Annu. Rev. Biophys. 38, 53–74 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Fletcher JS, Vickerman JC, Winograd N, Curr. Opin. Chem. Biol. 15, 733–740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian H et al. , Angew. Chem. Int. Ed. 58, 3156–3161 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Angerer TB, Blenkinsopp P, Fletcher JS, Int. J. Mass Spectrom. 377, 591–598 (2015). [Google Scholar]
- 26.Rabbani S, Barber AM, Fletcher JS, Lockyer NP, Vickerman JC, Anal. Chem. 83, 3793–3800 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Fletcher JS et al. , Anal. Chem. 80, 9058–9064 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Passarelli MK, Ewing AG, Curr. Opin. Chem. Biol. 17, 854–859 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian H et al. , Anal. Chem. 89, 4611–4619 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baresova V et al. , Mol. Genet. Metab. 119, 270–277 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Sweetlove LJ, Fernie AR, Nat. Commun. 9, 2136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pareek V et al. , Penn State ScholarSphere (2020); 10.26207/8rm3-g809. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.