Elevated APOC2 expression correlates with MLL-rearrangement and worse prognosis in AML. APOC2 promotes AML growth through CD36-dependent lipid metabolism, and blocking APOC2 or CD36 slows down AML progression in mice.
Abstract
Acute myeloid leukemia (AML) is a devastating hematologic malignancy that affects the hematopoietic stem cells. The 5-year overall survival (OS) of patients with AML is less than 30%, highlighting the urgent need to identify new therapeutic targets. Here, we analyze gene expression datasets for genes that are differentially overexpressed in AML cells compared with healthy hematopoietic cells. We report that apolipoprotein C2 (APOC2) mRNA is significantly overexpressed in AML, particularly in patients with mixed-lineage leukemia rearrangements. By multivariate analysis, high APOC2 expression in leukemia blasts is significantly associated with decreased OS (HR: 2.51; 95% CI, 1.03–6.07; P = 0.04). APOC2 is a small secreted apolipoprotein that constitutes chylomicrons, very-low-density lipoproteins, and high-density lipoproteins with other apolipoproteins. APOC2 activates lipoprotein lipase and contributes to lipid metabolism. By gain and loss of function approaches in cultured AML cells, we demonstrate that APOC2 promotes leukemia growth via CD36-mediated LYN–ERK signaling activation. Knockdown or pharmacological inhibition of either APOC2 or CD36 reduces cell proliferation, induces apoptosis in vitro, and delays leukemia progression in mice. Altogether, this study establishes APOC2–CD36 axis as a potential therapeutic target in AML.
Significance:
The majority of patients with AML die within five years of diagnosis. We reveal that lipid transporter APOC2 is elevated in AML and promotes leukemic cell metabolism and growth via CD36, and provide preclinical evidence that targeting this pathway may be beneficial in AML.
This article is highlighted in the In This Issue feature, p. 127
Introduction
Acute myeloid leukemia (AML) is a devastating hematologic malignancy that occurs in patients of all ages, but is more common in older adults (1, 2). Certain cytogenetic and molecular genetic mutations have been implicated in the pathogenesis of AML, resulting in abnormal proliferation and differentiation of what are called myeloid blasts. Even with treatment, 5-year survival is only 35% to 40% among younger patients and 5% to 15% among older patients (3), highlighting the urgent need for new therapeutic targets. To screen for genes that are differentially upregulated in AML, we compared gene expression profiles from multiple public datasets on Oncomine. We found that apolipoprotein C2 (APOC2) was significantly upregulated in AML cells compared with that in healthy hematopoietic cells.
The APOC2 gene clusters with other apolipoprotein genes on chromosome 19 (q13), a known translocation hotspot in AML and a gene-rich region that includes the AKT2, TGFB1, and KMT2D genes (4). APOC2 is expressed mainly in the liver, and then secreted into the plasma where it binds lipids and lipoprotein lipase. APOC2 also participates in hydrolysis of very-low-density lipoproteins (VLDL) and high-density lipoproteins (HDL; ref. 5), thus facilitating energy delivery and storage (6). In addition to its hepatic expression, APOC2 is also expressed in macrophages to enhance the delivery of lipids and energy to these highly metabolic myeloid cells (7). APOC2 is known for its role in lipid metabolism, acting as a physiologic activator of lipoprotein lipase (LPL) by guiding lipoproteins to the active LPL site (8). APOC2 deficiency has been reported to cause severe hypertriglyceridemia and contribute to cardiovascular diseases (9); to our knowledge, no study to date has shown a role of APOC2 in cancer.
Myeloid leukemia cells are expected to exhibit higher energy requirements than their healthy myeloid counterparts to satisfy their increased proliferation potential. Thus, we hypothesized that the upregulation of APOC2 plays a role in maintaining the metabolic needs of leukemic cells. Here, we characterize APOC2 upregulation in AML and analyze its association with patients' clinical and molecular characteristics. We also demonstrate the functional and mechanistic role of APOC2 in AML cells and in AML xenograft murine models. Our study establishes APOC2 as a novel therapeutic target in AML acting via the CD36–ERK signaling pathway.
Results
APOC2 Is Hypomethylated and Upregulated in AML and Is Associated with Poor Clinical Outcomes
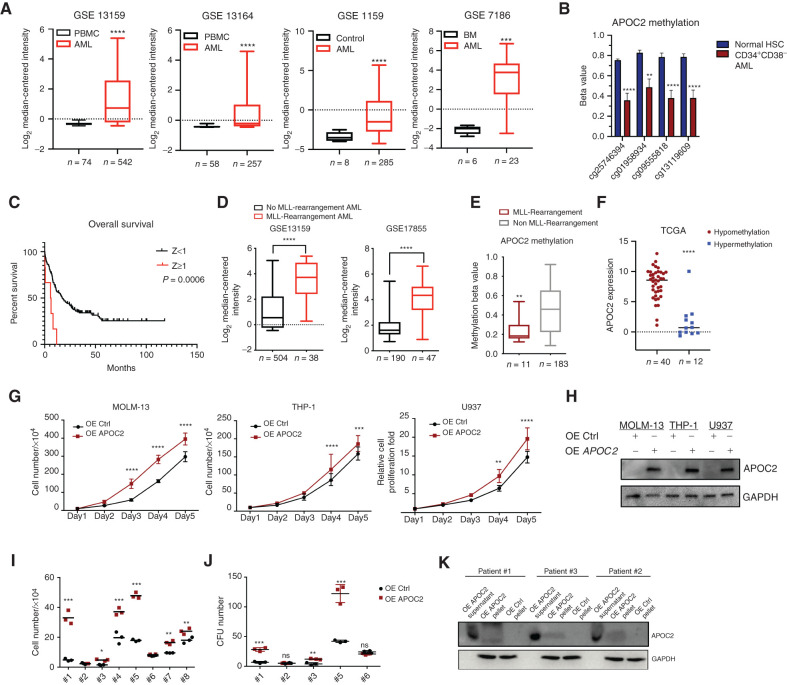
We found that APOC2 was consistently expressed at a significantly higher level in samples from patients with AML than in control samples. In the GSE13159 dataset, APOC2 mRNA was significantly overexpressed in 542 AML patient samples compared with 74 healthy peripheral blood mononuclear cells (PBMC) samples (2.9-fold, P < 0.0001). In the GSE13164 dataset, APOC2 mRNA was significantly higher in 257 AML samples compared with 58 healthy PBMC samples (1.7-fold, P < 0.0001). Consistently, APOC2 mRNA was upregulated in 285 AML samples compared with eight control samples in the GSE1159 dataset (6.0-fold, P < 0.0001). In the GSE7186 dataset, APOC2 exhibited a higher mRNA level in 23 AML patient samples compared with six normal bone marrow (BM) samples (32.0-fold, P = 0.0003; Fig. 1A).
Figure 1.
APOC2 is upregulated in AML and associated with poor clinical outcomes and promotes leukemia growth in vitro. A, The relative expression of APOC2 in: 542 AML cases compared with 74 healthy donors in the GSE13159 dataset; 257 AML cases compared with 58 healthy donors in the GSE13164 dataset; 285 AML cases compared with eight healthy donors in the GSE1159 dataset; and 23 AML cases compared with six healthy donors in the GSE7186 dataset (compared by Mann–Whitney tests). B, Comparisons of APOC2 methylation beta values between normal HSC and CD34+CD38−cells (LSCs) from patients with AML (unpaired t tests). C, Patients in TCGA dataset dichotomized into high (Z ≥ 1) or low (Z < 1) groups according to their APOC2 mRNA expression Z-score (RNA Seq V2 RSEM). Patients with high APOC2 levels had significantly shorter OS than patients with low APOC2 expression (median OS 6 vs. 17.4 months, P = 0.0006). D, The relative expression of APOC2 comparison between patients with and without MLL-rearrangements from GSE13159 and GSE17855 (compared by Mann–Whitney tests). E, APOC2 methylation beta value comparison between patients with and without MLL–rearrangements (unpaired t-test, P = 0.0035). F, Expression of APOC2 in AML patients with APOC2 hypomethylation (β ≤ 0.2) and hypermethylation (β ≥ 0.8). G, Cell proliferation assays in MOLM-13, THP-1, and U937 cells transduced with control or APOC2 overexpression lentiviral particles (compared by two-way ANOVA). H, Western blot analysis of APOC2 overexpression in AML cell lines. I, Viability assays for the growth of AML patient primary blasts transduced with either APOC2 or control lentiviral particles (compared by unpaired t tests). J, Colony formation unit (CFU) assays in AML patient primary blasts transduced with either APOC2 or a control vector (compared by unpaired t tests). K, Western blot analysis of APOC2 overexpression in primary patient samples showing APOC2 bands in the supernatant and cell pellet of blasts transduced with APOC2 lentiviral particles compared with control transduced blasts. (****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant).
To address whether APOC2 upregulation in AML is mediated by epigenetic mechanisms, we compared APOC2 methylation patterns between patients with AML and healthy controls from the GSE63409 dataset (10). We identified CpG sites within the APOC2 gene that were differentially methylated in AML leukemia stem cells (LSC, CD34+CD38−cells) relative to hematopoietic stem cells (HSC; Supplementary Table S1). The CpG sites corresponding to methylation probes cg25746394, cg01958934, cg09555818, and cg13119609 showed significantly less methylation in LSCs compared with the same loci in HSCs (Fig. 1B, cg25746394, P < 0.0001; cg01958934, P = 0.0016; cg09555818, P < 0.0001; cg13119609, P < 0.0001).
Next, we assessed APOC2 expression according to the French-American-British (FAB) classification of AML and observed higher levels of APOC2 in the M3 and M5 FAB subtypes compared with other subtypes (Supplementary Fig. S1A and S1B, one-way ANOVA, P < 0.0001). We dichotomized patients in The Cancer Genome Atlas (TCGA) dataset into high (Z ≥ 1) and low (Z < 1) mRNA expression groups based on their APOC2 mRNA expression Z-score (RNA Seq V2 RSEM). Patients with high APOC2 expression had a significantly higher percentage of BM blasts (median: 89.5% vs. 72%, P = 0.0099) and had a lower percentage of peripheral blood blasts (PB median: 0% vs. 40%, P = 0.0033; Supplementary Table S2) than patients with low APOC2. Five of eight patients in the APOC2 high mRNA expression group were reported to have no PB blasts compared with 17 of 165 patients in the low APOC2 group (P = 0.0009 by Fisher exact test).
For survival analysis, after excluding patients with t(15;17), we observed that patients with high APOC2 mRNA expression (Z ≥ 1) had significantly shorter overall survival (OS) than patients with low APOC2 mRNA expression (median OS: 6 vs. 17.4 months, P = 0.0006; Fig. 1C). Multivariate survival analysis using the Cox proportional hazards model showed that high APOC2 expression (Z ≥ 1) was significantly associated with decreased overall survival (n = 153 patients, excluding patients with FAB M3 classification and 4 patients without cytogenetic risk information; HR: 2.51; 95% CI, 1.03–6.07; P = 0.042) after adjusting for age, transplant status, cytogenetic risk, and FLT3 and p53 mutation status (Supplementary Table S3).
APOC2 Overexpression Is Associated with MLL Rearrangements
Because AML is a heterogeneous disease with distinct cytogenetic and molecular subtypes, it is crucial to determine how APOC2 expression levels vary according to cytogenetic and mutational status across patients. Our analysis of AML datasets showed that APOC2 is significantly upregulated in t(11q23)/MLL-rearranged AML. APOC2 mRNA levels were higher in 38 patients with MLL-rearrangements compared with 504 patients without MLL-rearrangements in the GSE13159 dataset (Fig. 1D, 9.0-fold, P < 0.0001). APOC2 mRNA levels were also higher in 47 patients with MLL-rearrangements compared with 190 patients without MLL-rearrangements in the GSE17855 dataset (Fig. 1D, 6.6-fold, P < 0.0001). Similar findings were observed in three other datasets (Supplementary Fig. S1C, GSE13164, 5.4-fold, P < 0.0001; TCGA, 9.2-fold, P = 0.0007; GSE1159, 5.1-fold, P = 0.0006). When we dichotomized patients in the TCGA dataset into high (Z ≥ 1) and low (Z < 1) expression groups according to APOC2 mRNA expression Z-scores (RNASeq V2 RSEM), we observed that high APOC2 expression (Z ≥ 1) was significantly associated with MLL-rearrangements (Supplementary Table S4, Fisher exact test, P = 0.003). Patients with the FLT3 mutations exhibited higher APOC2 mRNA expression than patients with wild-type FLT3 (FLT3-WT; Supplementary Fig. S1D, TCGA data; FLT3-ITD vs. FLT3-WT 6-fold, P = 0.013 and FLT3 point-mutation vs. FLT3-WT, 14-fold, P = 0.035). Similar observation was found in the GSE1159 dataset (Supplementary Fig. S1D). However, the frequency of FLT3 mutations in patients with high APOC2 (Z ≥ 1) was not significantly different than that in patients with low APOC2 (Z < 1; 25% vs. 28.5%, Supplementary Table S4). In addition, APOC2 levels were higher in patients with t(15;17) compared with patients without t(15;17), (Supplementary Fig. S1E, TCGA, 2.0-fold, P < 0.0001; GSE1159, 2.7-fold, P = 0.004). There was no significant association between high APOC2 (Z ≥ 1) and mutations in IDH1, IDH2, RUNX1, DNMT3A, or NPM1 (Fisher exact test, Supplementary Table S4).
APOC2 upregulation in the MLL-rearrangement AML was not limited to rearrangements that involved chromosome 19. This suggested a potential mechanism of epigenetic regulation of APOC2 expression in the MLL-rearrangement AML. To address this, we compared the methylation levels of APOC2 between patients with MLL-rearrangements and those without in TCGA dataset. We found that patients with MLL-rearrangements had significantly lower APOC2 methylation β values compared with patients without MLL-rearrangements (Fig. 1E, 2.0-fold, P = 0.0038). In TCGA data, APOC2 expression level is significantly higher in the hypomethylation group compared with the hypermethylation group (Fig. 1F, P < 0.0001). To further establish the epigenetic regulation of APOC2 expression, we treated four AML cell lines, MV4-11 and MOLM-13 (with MLL-rearrangements), and U937 and NB4 (without MLL-rearrangements) with increasing concentrations of the hypomethylating agent 5-azacytidine (5-Aza). APOC2 mRNA expression significantly increased in U937 and NB4, but not in MV4-11 and MOLM-13 (Supplementary Fig. S2A–S2C). Methylation-specific PCR (MSP) was performed to detect methylation level in the APOC2 promoter. Amplified bands for NB4 and U937 cells were stronger than bands for MOLM-13 and MV4-11 cells, indicating higher methylation level of APOC2 promoter in NB4 and U937 cells. In accordance with the mRNA expression level, NB4 and U937 showed decreased methylation by increasing 5-Aza concentration (Supplementary Fig. S2D). This further established that the presence of MLL-rearrangements protects CpG clusters from methylation within the APOC2 gene.
APOC2 Ectopic Expression Promotes Leukemia Growth In Vitro
To investigate the functional role of APOC2 in leukemic cells, we first assessed the level of APOC2 mRNA in a panel of AML cells by qPCR (Supplementary Fig. S3). We selected cell lines with low or medium levels of APOC2 expression to conduct gain-of-function studies. We established an ectopic lentiviral expression system to generate stable cells with APOC2 overexpression (OE APOC2). In three different AML cell lines (MOLM-13, THP-1, and U937), APOC2 overexpression caused slight, but statistically significant, increases in cell proliferation compared with controls (OE Ctrl) four and five days after puromycin selection (Fig. 1G, MOLM-13, 33%–74%, P < 0.0001; THP-1, 17%–34%, P = 0.002; U937, 33%–50%, P = 0.003). Overexpression of APOC2 was verified by Western blot analysis (Fig. 1H). In primary blasts from patients with AML, overexpression of APOC2 significantly increased cell growth in six of eight primary blast samples (Fig. 1I, means: #1: 4.87 × 104 vs. 33.03 × 104, P = 0.0005; #3: 1.54 × 104 vs. 4.78 × 104, P = 0.001; #4: 19.73 × 104 vs. 37.07 × 104, P = 0.003; #5: 17.90 × 104 vs. 47.83 × 104, P < 0.0001; #7: 9.56 × 104 vs. 16.47 × 104, P = 0.0008; #8: 18 × 104 vs. 24 × 104, P = 0.021). AML blast colony-forming cell (CFC) assays showed that APOC2 overexpression promoted colony-forming ability compared with controls in 3 of 5 patient samples (Fig. 1J, means: #1: 6.67 vs. 28.33, P = 0.0004; #3: 4 vs. 12, P = 0.0034; #5: 42 vs. 122.30, P = 0.0008). Overexpressed APOC2 protein levels were detected inboth cell lysates and cell culture supernatant (Fig. 1K). Patients' samples information are listed in Supplementary Table S5.
APOC2 Knockdown Induces Apoptotic Cell Death in Leukemia Cells
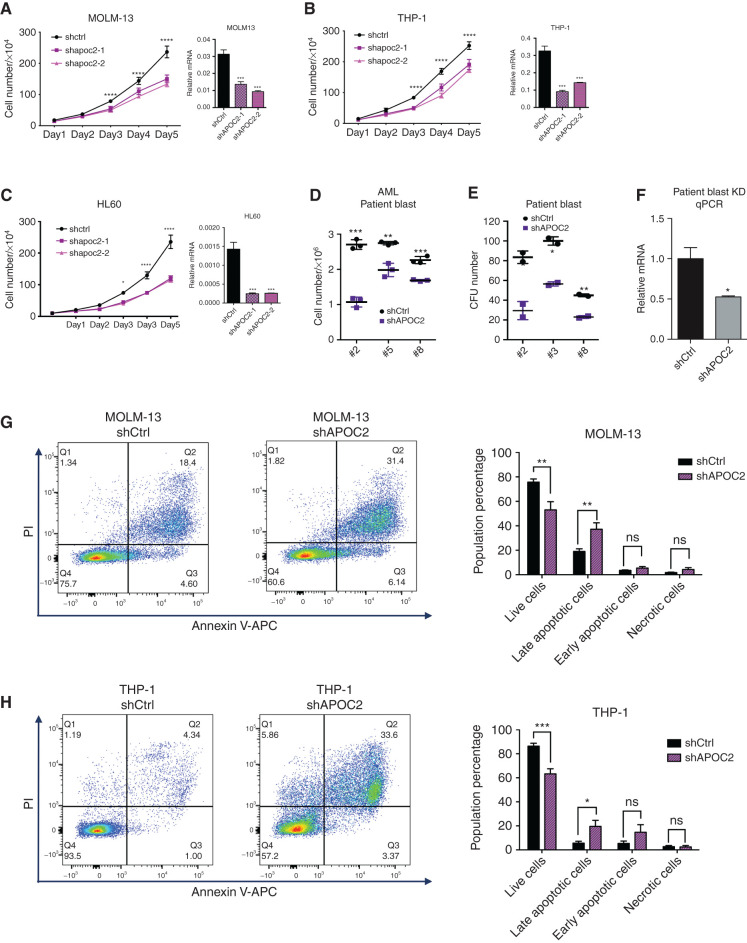
Next, we assessed the effect of APOC2 knockdown on leukemia cells using shRNA lentiviral plasmids. APOC2-shRNA–infected AML cell lines (MOLM-13, KG-1, and THP-1)exhibited significant reductions in proliferation compared with their respective controls, which were transduced with scramble sequence (Supplementary Fig. S4A–S4E, MOLM-13, 58%, P = 0.0036; THP-1, 33%, P = 0.014). Because the viability of the shAPOC2 cells was significantly reduced over time, we were unable to generate stable cells with APOC2 knockdown after puromycin selection. We therefore engineered a tetracycline-on (tet-on) lentiviral plasmid for the inducible expression of shRNA in which the APOC2 gene was targeted by two different shRNAs. Consistently, in three cell lines (MOLM-13, THP-1, and HL60), both shRNAs against APOC2 inhibited cell proliferation compared with the scramble control group, which further validates that the observed effect is unlikely to be due to off-target knockdowns (Fig. 2A–C, MOLM-13, 37.7%, P = 0.017; THP-1, 34%, P = 0.017; HL60, 49%, P = 0.00016). The knockdown of APOC2 mRNA levels was confirmed by qPCR. The knockdown of APOC2 also inhibited cell proliferation in AML primary blasts (Fig. 2D, means: #2: 2.7 × 106 vs. 1.07 × 106, P = 0.0001; #3: 2.74 × 106 vs. 1.98 × 106, P = 0.0026; #8: 2.26 × 106 vs. 1.69 × 106, P = 0.0007). CFC assays that were performed in primary blasts obtained from three different patients with AML showed that the knockdown of APOC2 significantly decreased the number of colonies (Fig. 2E, means: #2: 83.5 vs. 29.5, P = 0.02; #3: 100.0 vs. 56.5, P = 0.006; #8: 45.0 vs. 23.0, P = 0.004). The efficiency of the APOC2 knockdown was measured by qPCR (Fig. 2F). Moreover, Annexin V and PI staining analysis by flow cytometry showed that the APOC2 knockdown in MOLM-13 and THP-1 cells increased the population of late apoptotic cells on post viral transduction day 9 and day 4, respectively, compared with the respective controls (Fig. 2G, MOLM-13: shCtrl vs. shAPOC2, live cells, 75.71% vs. 53.06%, P = 0.008; late apoptotic cells, 19% vs. 37.24%, P = 0.007; Fig. 2H, THP-1: shCtrl vs. shAPOC2, live cells, 86.35% vs. 63.3%, P = 0.003; late apoptotic cells, 5.59% vs. 19.55%, P = 0.0387).
Figure 2.
APOC2 knockdown inhibits AML cell survival and induces apoptosis in vitro. Cell proliferation assays in MOLM-13 cells (A), THP-1 cells (B), and HL60 cells (C) infected with two different tet-on APOC2-shRNAs or control shRNA. The knockdown efficiency of APOC2 was measured by qPCR (compared by two-way ANOVA). D, Cell viability assay results for the knockdown of APOC2 in primary blasts from patients with AML. E, Colony formation assay results for the knockdown of APOC2 in AML primary blasts. F, Measurement of APOC2 knockdown in patient blasts by qPCR. G and H, Annexin V and PI staining levels were measured by flow cytometry to assess cell apoptosis in MOLM-13 and THP-1 cells infected with APOC2 shRNA or control shRNA. The quantification of early and late apoptotic cell population changes is presented in the panel on the right. The differences between the groups were analyzed using unpaired t tests (****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant).
APOC2 Cooperates with CD36 to Promote Leukemia Growth
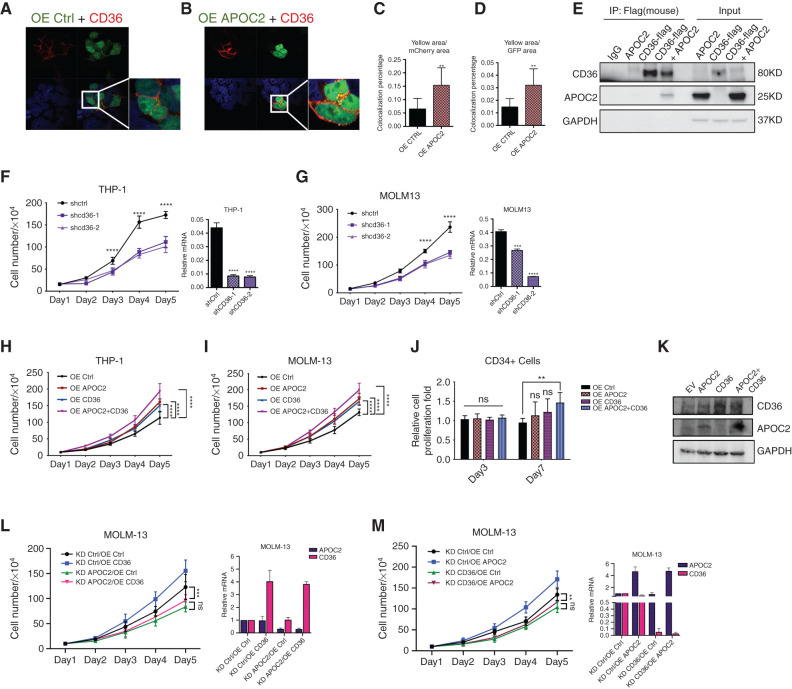
Fibrillar APOC2 was previously found to both interact with CD36 and trigger downstream pathways in atherosclerosis (11). To test whether the action of APOC2 is similar in AML, we first performed immunofluorescence microscopy to identify colocalizations of APOC2 and CD36 in HEK 293T cells. Both the confocal pictures and quantitative data showed mCherry-CD36 colocalized with APOC2-GFP but not empty-GFP (Fig. 3A–D, yellow area/mCherry area, 2.4-fold, P = 0.0053; yellow area/GFP area, 2.2-fold, P = 0.005), indicating that APOC2 and CD36 are colocalized. The physical interaction between APOC2 and CD36 was further confirmed by coimmunoprecipitation, showing that APOC2 was pulled down by flag-tagged CD36 (Fig. 3E). Then, we examined the effect of CD36 gain or loss of function in AML cells by utilizing the tetracycline-on lentiviral system for inducible expression of CD36 shRNAs (shCD36) and lentiviral system for CD36 overexpression (OE CD36). Similar to the effect of APOC2 knockdown, CD36 knockdown inhibited cell proliferation in both THP-1 and MOLM-13 cells (Fig. 3F and G; THP-1, 37.4%, P = 0.0096; MOLM-13, 40.8%, P = 0.0175).
Figure 3.
APOC2 interacts and cooperates with CD36 to promote leukemia growth. A and B, Confocal microscopy images showing the colocalization of CD36 and APOC2. 293T cells were cotransfected with mCherry-CD36 and empty-GFP or mCherry-CD36 and APOC2-GFP plasmids and imaged 48 hours later by confocal microscopy. C and D, Quantitative analyses of colocalization area conducted by ImageJ. Pictures were taken from ten different fields for each slide (duplicate slides were used for each condition) and combined results of two different experiments were reported. E, Immunoprecipitation (IP) assay demonstrating the interaction between flag-CD36 and APOC2. 293T cells were cotransfected with flag-CD36 and APOC2 plasmids; flag antibody was used to pull-down CD36; APOC2 antibody was used to detect APOC2 in IP lysates as well as input in Western blot. F and G, Effects of tetracycline-controlled CD36 knockdown on cell proliferation in THP-1 and MOLM-13 cells. H and I, Effects of ectopic expression of APOC2, CD36, and both on cell proliferation in THP-1 and MOLM-13 cells. J, Viability of CD34+ cells ectopically expressing empty vector, APOC2, CD36, and both on days 3 and 7. K, The expression level of APOC2 and CD36 was detected by Western blot. L and M, Effects of knocking down APOC2 and CD36 on the proliferation of OE APOC2 and CD36 stable cells, respectively. mRNA levels of APOC2 and CD36 changes were confirmed by qPCR. The difference between groups was analyzed by unpaired t test (****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant).
To determine whether CD36 cooperates with APOC2 to enhance cell proliferation, we conducted ectopic expression for either APOC2 or CD36, alone or combined (OE APOC2+CD36) in THP-1 and MOLM-13 cells. We found that OE APOC2+CD36 cells have significantly greater increase in proliferation compared with OE APOC2 or OE CD36 cells (Fig. 3H and I). We also transduced CD34+ cord blood cells with empty vector, APOC2, CD36 or the combined lentiviral particles. Assessment of cell count showed significant increase in the number of viable cell in CD34+ cells transduced with both APOC2 and CD36 compared with OE Ctrl cells and cells transduced with either APOC2 or CD36 lentiviral particles alone at day 7 post transduction (Fig. 3J, 30% increase, P = 0.0012). OE APOC2 or OE CD36 resulted in only slight increase in cell proliferation. Overexpression was confirmed by Western blot analysis (Fig. 3K).
To assess whether CD36 expression is specific to the leukemia stem cell fraction, we used the GSE30377 dataset to compare CD36 mRNA levels in AML cells from 23 patients that were sorted into the following populations: CD34+CD38−, CD34+CD38+, CD34−CD38−, and CD34−CD38+. CD36 mRNA levels were not significantly different among the groups (Supplementary Fig. S5A). Because high APOC2 level was associated with poor overall survival, we asked whether high CD36 level is also associated with clinical outcome. When patients were dichotomized on the basis of CD36 median expression, high CD36 expression patients had shorter, but not statistically significant, overall survival (P = 0.114); however, when patients in TCGA dataset were dichotomized on the basis of their APOC2 and/or CD36 mRNA expression Z-score into high (Z-score ≥2) and low (Z-Score <2), patients with high APOC2 and/or CD36 levels had significantly shorter OS than patients with low APOC2 and low CD36 expression (Supplementary Fig. S5B, median OS 9.2 vs. 21.5 months, P = 0.017). To confirm the functional association between APOC2 and CD36, shAPOC2 or shCD36 cells were transduced with CD36 or APOC2 overexpression viral particles, respectively. Doxycycline induced knockdown of APOC2 or CD36 abolished the enhanced proliferation induced by ectopic expression of the partner protein CD36 and APOC2, respectively. Overexpression and knockdown of APOC2 and CD36 were confirmed by qPCR (Fig. 3L and M, MOLM-13 KD APOC2/OE Ctrl vs. KD APOC2/OE CD36, ns; MOLM-13 KD CD36/OE Ctrl vs. KD CD36/OE APOC2, ns). This suggests that both APOC2 and CD36 are required for the enhanced proliferation and the presence of either protein is not sufficient to rescue the effect of depleting the other.
Targeting CD36 Function Induces Apoptosis and Abrogates APOC2 Proleukemic Effects
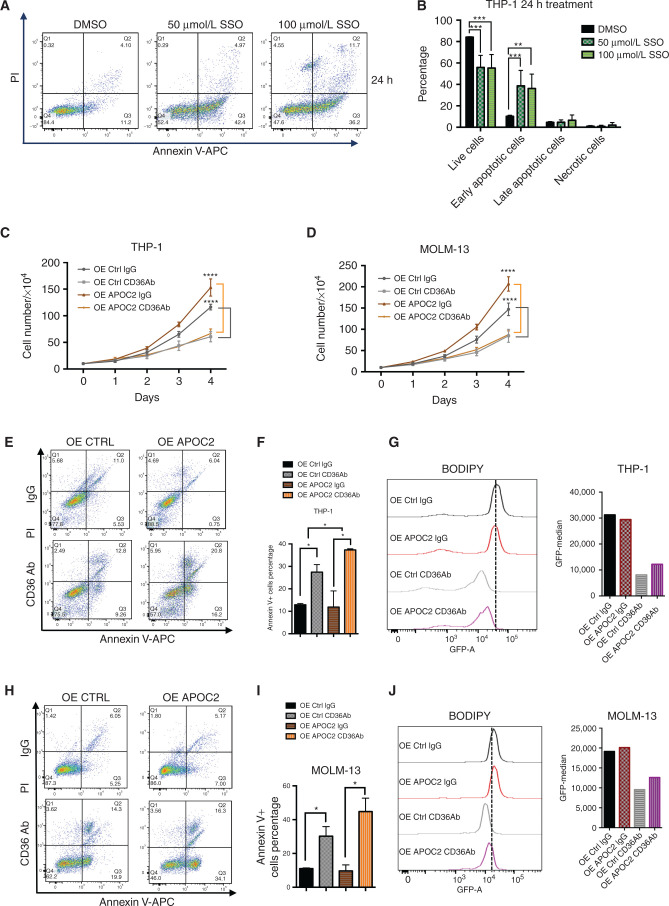
CD36 is a multifunctional receptor. One main function of CD36 is fatty acids transportation. We therefore treated AML cells with sulfo-N-succinimidyl oleate sodium (SSO), the irreversible inhibitor of CD36 for fatty acid translocation (12), to assess whether blocking fatty acids transportation causes a similar effect to that of CD36 knockdown. SSO at 50 μmol/L and 100 μmol/L concentrations induced apoptosis 24 hours post-treatment, and the proportion of living cells decreased significantly in treated cells (Fig. 4A and B, THP-1 cells: early apoptosis: 10.2% vs. 38.6% for DMSO vs. 50 μmol/L SSO, respectively, P = 0.027; 10.2% vs. 36.2% for DMSO vs. 100 μmol/L SSO, respectively, P = 0.029; Supplementary Fig. S6A, MOLM-13 cells: 6.6% vs. 18% for DMSO vs. 50 μmol/L SSO, respectively, P = 0.05; 6.6% vs. 17.8% for DMSO vs. 100 μmol/L SSO, respectively, P = 0.023). This apoptotic effect was transient, however, and disappeared 48 hours posttreatment (Supplementary Fig. S6B–S6D).
Figure 4.
Blocking CD36 induces apoptosis in AML cells and abrogates APOC2 proleukemic effects. A, Results of the Annexin V and PI staining apoptosis assays in THP-1 cells 24 hours after treatment with either 50 μmol/L or 100 μmol/L of SSO. B, Quantification of apoptotic populations in THP-1 cells 24 hours after SSO treatment. C, The effects of CD36-blocking antibody on cell proliferation in THP-1 OE Ctrl and OE APOC2 cells. D, The effects of CD36-blocking antibody on cell proliferation in MOLM-13 OE Ctrl and OE APOC2 cells. E, The effects of CD36 blocking antibody on apoptosis 24 hours posttreatment in THP-1 OE Ctrl and OE APOC2 cells. F, The quantification of Annexin V+ cell populations in THP-1 OE Ctrl and OE APOC2 cells treated with IgG or CD36Ab. G, A comparison of BODIPY staining between THP-1 OE Ctrl and OE APOC2 cells treated with IgG or CD36Ab. H, The effects of CD36 blocking antibody on apoptosis 24 hours posttreatment in MOLM-13 OE Ctrl and OE APOC2 cells. I, The quantification of Annexin V+ cell populations in MOLM-13 OE Ctrl and OE APOC2 cells treated with IgG or CD36Ab. J, A comparison of BODIPY staining between MOLM-13 OE Ctrl and OE APOC2 cells treated with IgG or CD36Ab. The differences between the groups were analyzed using unpaired t tests (****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05).
THP-1 and MOLM-13 cells both express relatively high levels of CD36. We therefore treated THP-1 and MOLM-13 cells ectopically expressing APOC2 or their respective controls with CD36-blocking antibody (2 μg/mL). Both groups of cells exhibited a significant decrease in cell proliferation four days post incubation with the CD36-blocking antibody (Fig. 4C, THP-1: OE Ctrl, 47.96%, P < 0.0001; OE APOC2, 56.54%, P < 0.0001; Fig. 4D, MOLM-13: OE Ctrl, 42.35%, P = 0.0011; OE APOC2, 57.68%, P < 0.0001). These data suggest that treatment with the CD36-blocking antibody abrogated the enhanced proliferation effects observed with APOC2 overexpression. Furthermore, CD36-blocking antibody induced significant apoptosis in both control cells and in cells overexpressing APOC2 (Fig. 4E and F, IgG vs. CD36Ab: THP-1, Annexin V+ cells, OE Ctrl: 12.93% vs. 27.44%, P = 0.0265; OE APOC2: 11.89% vs. 37.35%, P = 0.0379). Cells in the OE APOC2 groups exhibited a higher percentage of Annexin V than cells in the OE Ctrl groups when treated with CD36 antibody (P = 0.0318 for THP-1). Cells treated with the CD36 blocking antibody also exhibited a reduction in BODIPY staining, indicating less cellular uptake of fatty acids (Fig. 4G). Similar results were found for the MOLM-13 cells (Fig. 4H–J, IgG vs. CD36Ab: Annexin V+ cells, OE Ctrl: 11.01% vs. 30.21%, P = 0.041; OE APOC2: 9.67% vs. 44.77%, P = 0.0294).
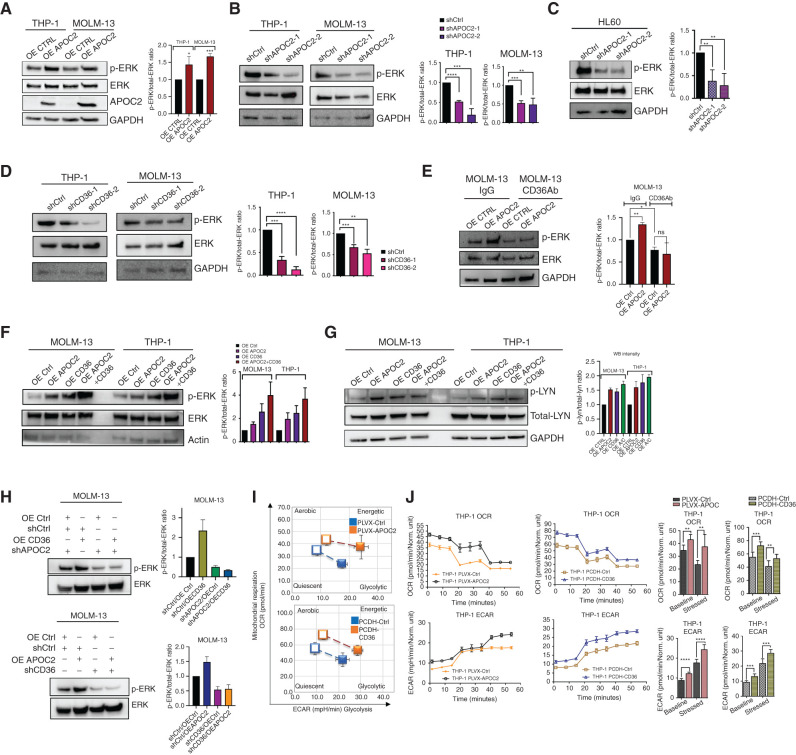
APOC2–CD36 Activates the ERK Signaling Pathway and Enhances Metabolic Activity of Leukemic Cells
To address whether APOC2 triggers signaling pathways downstream of CD36, we assessed the effect of APOC2 gain-of-function and loss-of-function on MAPK (ERK) phosphorylation. We found that OE APOC2 cells, both THP-1 and MOLM-13, exhibited higher levels of phospho-ERK (p-ERK), compared with their respective control cells, while total-ERK did not change (Fig. 5A, p-ERK/total-ERK ratio change, THP-1 OE APOC2, 1.43 fold increase, P = 0.0343; MOLM13 OE APOC2, 1.67 fold increase, P = 0.0001). Conversely, p-ERK levels decreased when endogenous expression of APOC2 was knocked down by two different APOC2 shRNAs (Fig. 5B, THP-1 p-ERK/total-ERK ratio change, shAPOC2-1, 47.56% decrease, P < 0.0001; shAPOC2-2, 81.50% decrease, P = 0.0008; MOLM13 p-ERK/total-ERK ratio change, shAPOC2-1, 47.65% decrease, P = 0.0002; shAPOC2-2, 51.39% decrease, P = 0.0063). Similarly, in HL60, the knockdown of APOC2 resulted in a significant decrease in p-ERK protein levels (Fig. 5C, p-ERK/total-ERK ratio change, shAPOC2-1, 62.25% decrease, P = 0.0025; p-ERK/total-ERK ratio change, shAPOC2-2, 72.00% decrease, P = 0.0017). A significant decrease in p-ERK protein levels also was observed when CD36 was depleted by two different CD36 shRNAs (Fig. 5D, THP-1: p-ERK/total-ERK ratio change, shCD36-1, 66.27% decrease, P = 0.0001; shCD36-2, 87.32% decrease, P < 0.0001; MOLM13: p-ERK/total-ERK ratio change, shCD36-1, 32.56% decrease, P = 0.0007; shCD36-2, 46.90% decrease, P = 0.001). Consistently, treatment with the CD36 blocking antibody caused a decrease in p-ERK in OE Ctrl compared with IgG treatment (p-ERK/total-ERK ratio change 22.58%, P = 0.036) and abrogated the enhanced level of p-ERK in OE APOC2 cells (Fig. 5E). Furthermore, the simultaneous overexpression of APOC2 and CD36 triggered greater increase of ERK phosphorylation in both THP-1 and MOLM-13 cells (Fig. 5F, MOLM13: p-ERK/total-ERK ratio change, OE APOC2, 1.5-fold increase; OE CD36, 1.6-fold increase; OE both, 3.0-fold increase, P = 0.0032; THP-1, p-ERK/total-ERK ratio change, OE APOC2, 2.0-fold increase; OE CD36, 2.5-fold increase; OE both, 3.7-fold increase, P = 0.0485). Overexpression of APOC2, CD36, or both activates the CD36 downstream target LYN by increasing the p-LYN levels (Fig. 5G, MOLM13: p-LYN/total-LYN ratio change, OE APOC2, 1.5-fold increase; OE CD36, 1.4-fold increase; OE both, 1.7-fold increase, P = 0.0032; THP-1, p-LYN/total-LYN ratio change, OE APOC2, 1.6-fold increase; OE CD36, 1.8-fold increase; OE both, 2.0-fold increase, P = 0.0197). Knockdown of APOC2 or CD36 abolished activation of ERK induced by OE CD36 or OE APOC2, respectively, and caused a reduction in p-ERK level compared with control (Fig. 5H, p-ERK/total-ERK ratio change, shCtrl/OE CD36, 2.6-fold increase; shAPOC2/OE Ctrl, 49.38% decrease; shAPOC2/OE CD36, 66.22% decrease, P = 0.0061; shCtrl/OE APOC2, 1.5-fold increase, shCD36/OE Ctrl, 45.63% decrease; shCD36/OE APOC2, 43.29% decrease, P = 0.0052).
Figure 5.
APOC2–CD36 activates the ERK signaling pathway and enhances metabolic activity of leukemic cells. A, The levels of p-ERK and total-ERK proteins detected by Western blot in THP-1 and MOLM-13 cells ectopically expressing APOC2 compared with empty vector control cells. GAPDH was used as an internal control (quantification of three independent repeats). B, The levels of p-ERK and total-ERK proteins detected by Western blot in THP-1 and MOLM-13 APOC2 knockdown and scramble control cells. GAPDH was used as an internal control (quantification of three independent repeats). C, Thelevels of p-ERK and total-ERK proteins detected by Western blot in HL60 APOC2 knockdown and scramble control cells. Right, the p-ERK/total-ERK ratio in the control and knockdown APOC2 cells (quantification of four independent repeats). D, The levels of p-ERK and total-ERK proteins detected by Western blot in THP-1 and MOLM-13 CD36 knockdown and scramble control cells. GAPDH was used as an internal control (quantification of three independent repeats). E, The levels of p-ERK and total-ERK proteins detected by Western blot in MOLM-13 OE Ctrl and OE APOC2 cells treated with IgG or CD36Ab. GAPDH was used as an internal control (quantification of two independent repeats). F, The levels of p-ERK and total-ERK proteins detected by Western blot in THP-1 and MOLM-13 OE Ctrl, OE APOC2, OE CD36, and OE APOC2+CD36 cells. Actin was used as an internal control (quantification of two independent repeats). G, The levels of p-LYN and total-LYN proteins detected by Western blot in MOLM-13 and THP-1 OE Ctrl, OE APOC2, OE CD36, and OE APOC2+CD36 cells. GAPDH was used as an internal control (quantification of two independent repeats). H, The levels of p-ERK and total-ERK proteins detected by Western blot in CD36 OE MOLM-13 cells transduced with shAPOC2 tet-on viral particles and in APOC2 OE cells transduced with shCD36 tet-on viral particles (quantification of two independent repeats). I, Cell energy phenotype of PLVX-Ctrl, PLVX-APOC2 and PCDH- Ctrl, and PCDH- CD36 THP-1 cells. Baseline phenotype is indicated by an open marker. Stressed phenotype is indicated by a filled marker. J and K, OCR (to quantify mitochondrial respiration) and ECAR (indicator of glycolysis) of PLVX-Ctrl, PLVX-APOC2 and PCDH- Ctrl and PCDH- CD36 THP-1 cells determined by Seahorse XF cell analysis. The differences between the groups were analyzed using unpaired t tests and one-way ANOVA (****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant).
Given the roles of APOC2 and CD36 in lipid metabolism and transport, we speculated that modulating their expression in AML cells would impact their bioenergetic profile. Live-cell metabolic measurements performed by Seahorse XF showed that OE APOC2 or OE CD36 THP-1 and MOLM-13 cells exhibit higher energetic metabolic phenotype compared with OE Ctrl cells (Fig. 5I; Supplementary Fig. S7A). An increase in mitochondrial respiration rates assessed by the oxygen consumption rates (OCR) was observed in OE APOC2 and OE CD36 at their basal conditions (Fig. 5J; Supplementary Fig. S7B). Similarly, extracellular acidification rates (ECAR) were also increased in OE APOC2 and OE CD36 cells compared with OE Ctrl cells under stressed condition (Fig. 5K; Supplementary Fig. S7C).
Conditional Knockdown of APOC2 and CD36 Decrease Leukemia Burden in Murine Leukemia Models
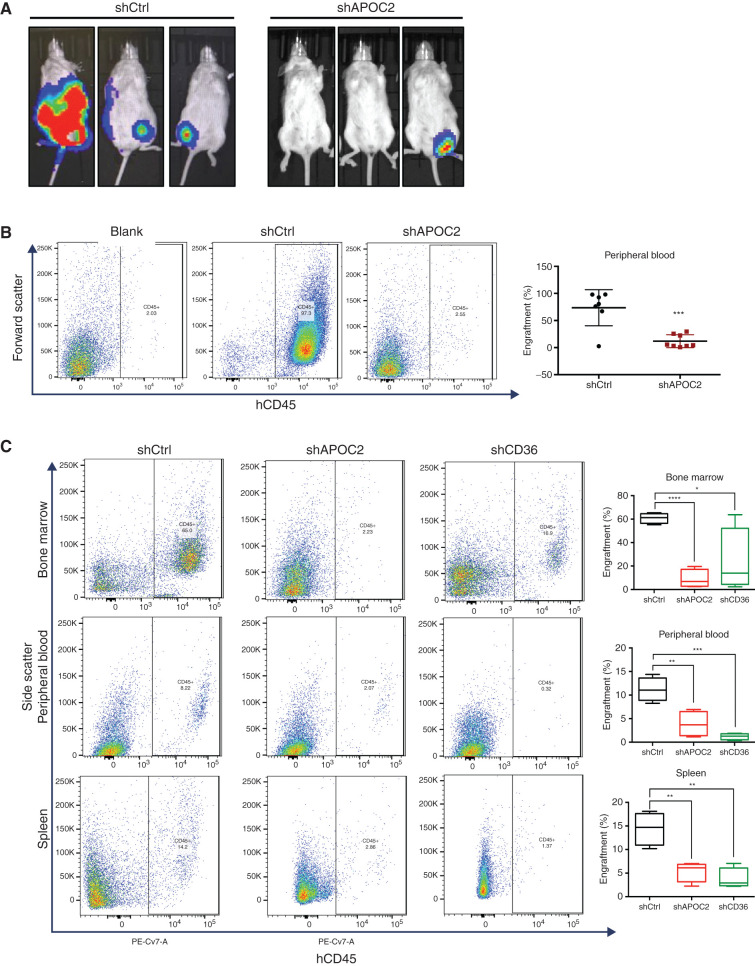
To examine the effect of targeting APOC2 in murine models of AML, luciferase-positive HL60 cells (HL60luci+) with either tet-on-inducible APOC2 shRNAs or tet-on-inducible scramble sequences were intravenously injected into NOD-scid/Il2rg−/− (NSG) mice to generate human AML xenografts (n = 7 in shCtrl group, n = 8 in shAPOC2 group). Three days later, mice were treated once every other day with doxycycline via oral gavage. Mice were monitored and imaged on week four and five posttransplant. Mice in the shCtrl group exhibited a stronger bioluminescent signal compared with mice in the shAPOC2 group (Fig. 6A). Mice were sacrificed on day 36, when signs of disease burden appeared. We found a lower percentage of leukemic cell engraftments in the shAPOC2 group compared with the shCtrl group. The shAPOC2 group exhibited significantly less leukemic burden in peripheral blood (PB) compared with the shCtrl group (Fig. 6B; Supplementary Fig. S8A and S8B, hCD45+ cells: 73.6% vs. 11.8%, P = 0.0003). Minimal engraftment of HL60 was observed in the spleens and BM of mice in both groups (Supplementary Fig. S9A and S9B). Six out of seven mice in the shCtrl group exhibited more than three secondary tumors, located on the back, lower abdomen, and legs. In contrast, only one out of eight mice in the shAPOC2 group exhibited secondary tumors (Supplementary Fig. S9C).
Figure 6.
Conditional knockdown of APOC2 or CD36 reduced leukemia engraftment in AML NSG murine models. A, Bioluminescent imaging of NSG mice intravenously injected with HL60 shCtrl or shAPOC2 luciferase cells. B, Representative flow cytometry showing hCD45-positive engraftment in the PB of blank, HL60 shCtrl, and HL60 shAPOC2 luciferase cells. Quantification of engraftment of HL60 cells in the PB. C, Representative flow cytometry showing hCD45-positive engraftment in the BM, PB, and spleen of MOLM-13 shCtrl, MOLM-13 shAPOC2, and MOLM-13 shCD36 luciferase cells of NSG mice. Quantification of engraftment of MOLM-13 cells in the BM, PB, and spleen. The differences between the groups were analyzed using unpaired t tests (****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05).
We also validated the effect of targeting APOC2 and CD36 in the MOLM-13 murine model, in which MOLM-13 cells stably expressing tet-on-inducible APOC2 shRNA, CD36 shRNA, or scramble sequences were intravenously injected into NSG mice to generate human AML xenografts (n = 4 per group) and treated with doxycycline grain–based rodent diet continuously. Mice were sacrificed on day 25. Mice from the shAPOC2 and shCD36 groups exhibited significantly less leukemia engraftment in the BM, PB, and spleen compared with the shCtrl group (Fig. 6C, % BM engraftment means, 60.8 vs. 8.9 vs. 23.5 for shCtrl vs. shAPOC2 vs. shCD36, P < 0.0001 and P = 0.0368; % PB engraftment means, 11.2 vs. 3.9 vs. 1.2, P = 0.0073 and P = 0.0003; % spleen engraftment means, 14.4 vs. 5.4 vs. 3.8, P = 0.0044 and P = 0.0021), indicating that the knockdowns of APOC2 and CD36 reduced the leukemia burden in vivo (Supplementary Fig. S10–12). There was no significant difference in the weights of the livers and spleens between the groups (Supplementary Fig. S13A–S13C).
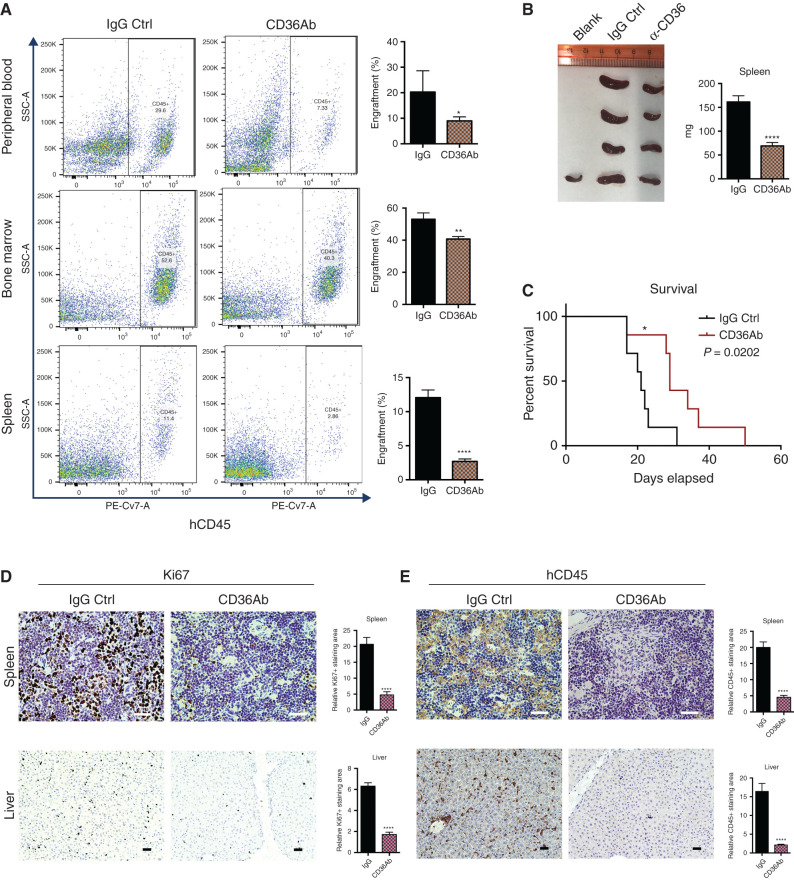
Anti-CD36 Antibody Treatment Reduces Leukemia Progression and Increases Overall Survival of AML Murine Model
APOC2 cooperates with CD36 to activate ERK signaling pathway leading to enhanced leukemia growth. In order to test the role of CD36 in vivo and verify the potential of targeting the APOC2–CD36 signaling pathway for AML treatment, CD36 blocking antibody was used to treat MOLM-13 engrafted NSG mice. Seven days after injection of MOLM-13 cells into NSG mice, mice were treated with CD36 blocking antibody or IgG control once every three days for five doses (n = 4 per group). Mice were sacrificed on day 21. CD36 antibody treatment group had less leukemia burden in BM, PB, and spleen (Fig. 7A, % PB engraftment means, 20.27 vs. 8.98 for IgG vs. CD36 antibody, P = 0.373; % BM engraftment means, 53.08 vs. 40.70 for IgG vs. CD36 antibody, P = 0.0011; % spleen engraftment means, 12.05 vs. 2.70 for IgG vs. CD36 antibody, P < 0.0001). The spleens in CD36 antibody treatment group were smaller and weighed less than the ones in IgG group (Fig. 7B, spleen weight means, 161.5 mg vs. 69.15 mg, P < 0.0001). No significant difference in liver weight was observed between the treatment groups Supplementary Fig. S13D) In addition, to determine whether blocking CD36 potentially delays leukemia progression, engrafted NSG mice were treated with IgG and CD36 blocking antibody the same as above for survival analysis (n = 7 per group). With CD36 antibody treatment, mice engrafted with MOLM-13 cells survived significantly longer than the IgG group (Fig. 7C, P = 0.0202). Consistently, Ki67 and hCD45 staining of spleens and livers presented a lower percentage of infiltrated blasts compared with tissues obtained from the IgG-treated group (Fig. 7D and E, spleen Ki67 staining, 4.35-fold, P < 0.0001; liver Ki67 staining, 3.67-fold, P < 0.0001; spleen CD45 staining, 4.38-fold, P < 0.0001; liver CD45 staining, 7.64-fold, P < 0.0001).
Figure 7.
Treatment of CD36 blocking antibody delays leukemia engraftment in NSG murine model. A, NSG mice were engrafted with MOLM-13 cells for 7 days and then treated with CD36 antibody (CD36Ab) or IgG control. Representative flow cytometry results showing the engraftment of BM, PB, and spleen, quantification of engraftment in BM, PB, and spleen. B, Spleens collected from mice engrafted with MOLM-13 and treated with IgG and CD36 antibody and comparison of spleen weight between IgG group and CD36 antibody group. C, Survival data from mice engrafted with MOLM-13 and treated with IgG and CD36 antibody. D, Histology staining of Ki67 antibody on spleen and liver sections from mice engrafted with MOLM-13 and treated with IgG and CD36 antibody. Quantification analysis and representative staining of tissue sections obtained from the different organs. E, Histology staining of human CD45 antibody on spleen and liver sections from mice engrafted with MOLM-13 and treated with IgG and CD36 antibody. Quantification analysis and representative staining of tissue sections obtained from the different organs. The differences between the groups were analyzed using unpaired t tests (****, P < 0.0001; **, P < 0.01; *, P < 0.05).
Discussion
The link between lipid metabolism and cancer is well established (13, 14, 15), yet the role of APOC2 in cancer has not been reported previously. In this study, we identified APOC2 as a gene that is hypomethylated and upregulated in AML. In normal tissues, APOC2 is expressed primarily in the liver and secreted into the plasma. APOC2 is also expressed at lower levels by other tissues, including the intestine, macrophages, and adipose tissue (6). Characterization of APOC2 mRNA levels in AML showed that overexpression of APOC2 is significantly associated with MLL rearrangements, likely due to an epigenetic mechanism regulating its expression. MLL rearrangements are associated with poor clinical outcomes in AML, which is consistent with the observed association between high APOC2 expression and shorter overall survival in patients with AML. In macrophages, the APOC2 gene is transcriptionally regulated by the liver X receptor (LXR) and STAT1 (6), two proteins known to be involved in cancer progression (16, 17). A recent study has implicated APOC2 and its carrier LPL in hematopoietic stem progenitor cell (HSPC) maintenance via docosahexaenoic acid (DHA) supply; knockout of either LPL or its obligatory cofactor APOC2 results in reduced HSPC expansion and defective hematopoiesis in zebrafish (18).
Using both gain- and loss-of-function approaches in several cellular and murine models, we demonstrated that APOC2 upregulation enhances leukemia growth, and conversely, that knockdown of APOC2 inhibits cell survival and triggers apoptosis. APOC2 overexpression activates the CD36-ERK pathways, which mechanistically explain the observed phenotype. While little is known about the role of APOC2 in cancer, the role of its partner CD36 is more established (19). CD36 is a transmembrane glycoprotein that is present on the surface of various cell types, including adipocytes, myocytes, hepatocytes, and mononuclear phagocytes. As a receptor, CD36 has been reported to recognize different ligands, including phospholipids, lipoproteins, long-chain fatty acids, and amyloid fibrils (20). In response to ligands, CD36 recruits and activates its downstream non-receptor tyrosine kinases, MAPK kinases (21). CD36 has been shown to confer chemotherapy resistance and metastasis initiation potential in cancer cells (22, 23). In chronic myeloid leukemia (CML), the fatty acids transporter CD36 is associated with leukemia stem cells that evade chemotherapy by residing in the niche of adipose tissue (23, 24). Furthermore, in chronic lymphoblastic leukemia (CLL) cells, STAT3 was shown to activate CD36 by binding to its promoter, facilitating fatty acid uptake (25). In addition, AraC-resistant cells exhibit increased fatty-acid oxidation, upregulated CD36 expression, and high oxidative phosphorylation (OXPHOS); targeting mitochondrial metabolism via the CD36-FAO-OXPHOS axis enhanced the antileukemic effects of AraC (26). Our study establishes the link between a novel target in AML, APOC2, and the fatty acids transporter, CD36, and establishes ERK as a potential signaling pathway resulting from the interaction of APOC2 and CD36. Our study shows that targeting the APOC2–CD36–ERK pathway with either CD36 antibodies or SSO leads to antileukemia effects. The use of these treatments at high concentrations, and their short half-life, limit their potential activity and suggests a need for more effective drugs to modulate the APOC2–CD36 axis.
In summary, our clinical association analysis, combined with preclinical functional and mechanistic studies, has uncovered a novel role of APOC2 upregulation in AML. APOC2 is highly expressed in AML and is associated with poor clinical outcomes. Furthermore, APOC2 promotes leukemia growth via the CD36–ERK signaling pathway. Our study indicates that the APOC2–CD36 signaling axis may be an actionable therapeutic target in AML.
Methods
Study Approval
The use of human materials was approved by the Institutional Review Board of the University of Southern California (USC, Los Angeles, CA) in accordance with the Helsinki Declaration. All animal protocols were approved by the Institution for Animal Care and Use Committee (IACUC) of USC.
Patient Datasets
Molecular data and clinical outcomes were available in 173 patients with AML from TCGA from cBioPortal (27, 28). We dichotomized patients in TCGA data set into high (Z ≥ 1) and low (Z < 1) mRNA expression groups based on their APOC2 mRNA expression Z-score (RNA Seq V2 RSEM). All the patients were diagnosed and have received treatment, according to the National Comprehensive Cancer Network (NCCN) guidelines, between November 2001 and March 2010. Additional patient data from the GSE7186 (29), GSE13159 (30), GSE1159 (31), GSE13164 (30), GSE17855 (13), GSE63409 (10), and GSE30377 (32) datasets were downloaded from the Gene Expression Omnibus (GEO) database.
Patient Samples
Blood samples were obtained from AML patients at the time of diagnosis from the Norris Comprehensive Cancer Center at USC. All samples were collected after obtaining written informed consent. The use of human materials was approved by the Institutional Review Boards of USC, in accordance with the Helsinki Declaration.
AML Murine Xenograft Models
NSG mice were purchased from The Jackson Laboratory and kept in pathogen-free conditions. To generate AML engraftment, HL-60 and MOLM-13 knockdown APOC2 (shAPOC2) cells, as well as control cells (shCtrl; 2 × 106 per mouse), were injected intravenously (through the tail vein) into 4 to 6-week-old male and female NSG mice. For shRNA expression, mice received doxycycline via oral gavage or Bio-Serv Doxycycline Grain-Based Rodent Diet (Thermo Fisher Scientific, 14-727-450). Engraftments were monitored by bioluminescence imaging once per week. Once mice reached the endpoint, when signs of disease burdens appeared, the blood and BM were collected for flow cytometry analysis. Leukemia progression was monitored by bioluminescence imaging and flow cytometry.
To conduct CD36Ab treatment in vivo, MOLM-13 cells (2 × 106 per mouse) were first engrafted in mice. Starting from day 7, mice were injected intravenously with 100 μL of PBS containing either 5 μg of monoclonal CD36Ab FA6.152 or 5 μg of mouse control IgG every 3 days for 5 doses. Experiments were ended on day 21.
Cell Culture and Transfection
AML cell lines THP-1, MV4-11, and KG-1 were obtained from ATCC. MOLM-13, U937, and KG-1A cells were kindly provided by Dr. Wendy Stock's Laboratory. All AML cell lines were authenticated at the University of Arizona Cell Authentication Core and tested for mycoplasma contamination in our lab using PCR-based method (SouthernBiotech). All cell lines were Mycoplasma free. AML cell lines were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and 1% antibiotics (Invitrogen). Primary samples were cultured in RPMI1640 supplemented with 20% FBS and StemSpan CC100, which contains a combination of both early- and late-acting recombinant human (rh) cytokines (Flt3L, SCF, IL3, and IL6). HEK293T cells were cultured in DMEM (Invitrogen) supplemented with 20% FBS and 1% antibiotics. Transfection was performed with Calcium Phosphate Transfection Kits (Clontech), according to the manufacturer's instruction.
To conduct the CD36Ab treatment assay, THP-1 and MOLM-13 cells were treated with either 20 μg/mL of a monoclonal CD36Ab FA6.152 (Abcam) or a corresponding mouse monoclonal IgG (Invitrogen) for the first 30 minutes. Cells were then released in RPMI supplemented with 10% FBS in a final concentration of 2 μg/mL CD36Ab (22).
To conduct the SSO treatment assay, SSO (SML2148; Sigma) was freshly dissolved in DMSO at a concentration of 100 mmol/L and then immediately applied to the cells at final concentrations of 50 μmol/L and 100 μmol/L.
Statistical Analysis
All experiments were independently repeated at least three times except when otherwise mentioned in the figure legend. Data were represented as the mean ± standard error of the mean (SEM). Statistical significance was calculated using the Student t test, Mann–Whitney test, one-way ANOVA, two-way ANOVA in GraphPad Prism 6.0 (GraphPad Software, Inc.). The statistical test was indicated in the figure legend for each analysis.
Survival analysis was conducted in STATA 12.0 SE using the Cox proportional hazards model to assess the association between APOC2 expression and overall survival in patients with AML, after adjusting for other factors, such as sex, cytogenetic status, and genetic mutations. The associations between the APOC2 expression group and the molecular and mutational status were analyzed using both Student t test and Fisher exact test. A P value less than 0.05 was considered statistically significant for all tests. P value was adjusted according for multiple testing in the methylation analysis.
Additional materials and methods are included in Supplementary Data.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Authors' Contributions
T. Zhang: Conceptualization, formal analysis, validation, investigation, methodology, writing–original draft, writing–review, and editing. J. Yang: Validation and methodology. V. Vaikari: Validation and methodology. J. Beckford: Formal analysis and methodology. S. Wu: Formal analysis. M. Akhtari: Resources. H. Alachkar: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, project administration, writing–review, and editing.
Supplementary Material
Acknowledgments
We thank the Bioinformatics Core at Norris Medical Library, University of Southern California. We acknowledge the funding support for H. Alachkar from the University of Southern California, School of Pharmacy Seed Fund, The Norris Cancer Center pilot fund, STOP Cancer pilot funding, and The Ming Hsieh Institute foundation grants (for H. Alachkar). H. Alachkar was also supported by grant UL1TR001855 from the National Center for Advancing Translational Science (NCATS) of the NIH. We would also like to acknowledge the statistician in our department, Mimi Lou, MS (applied statistics), who was consulted on the interpretation of statistical analysis.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Blood Cancer Discov 2020;1:198–213
References
- 1. De Kouchkovsky I, Abdul-Hay M. Acute myeloid leukemia: a comprehensive review and 2016 update. Blood Cancer J 2016;6:e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Longo DL, Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med 2015;373:1136–52. [DOI] [PubMed] [Google Scholar]
- 3. Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 2010;115:453–74. [DOI] [PubMed] [Google Scholar]
- 4. Nimer SD, MacGrogan D, Jhanwar S, Alvarez S. Chromosome 19 abnormalities are commonly seen in AML, M7. Blood 2002;100:3838. [DOI] [PubMed] [Google Scholar]
- 5. Kei AA, Filippatos TD, Tsimihodimos V, Elisaf MS. A review of the role of apolipoprotein C-II in lipoprotein metabolism and cardiovascular disease. Metabolism 2012;61:906–21. [DOI] [PubMed] [Google Scholar]
- 6. Wolska A, Dunbar RL, Freeman LA, Ueda M, Amar MJ, Sviridov DO, et al. Apolipoprotein C-II: New findings related to genetics, biochemistry, and role in triglyceride metabolism. Atherosclerosis 2017;267:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mak PA, Laffitte BA, Desrumaux C, Joseph SB, Curtiss LK, Mangelsdorf DJ, et al. Regulated expression of the apolipoprotein E/C-I/C-IV/C-II gene cluster in murine and human macrophages. A critical role for nuclear liver X receptors alpha and beta. J Biol Chem 2002;277:31900–8. [DOI] [PubMed] [Google Scholar]
- 8. Musliner TA, Herbert PN, Church EC. Activation of lipoprotein lipase by native and acylated peptides of apolipoprotein C-II. Biochim Biophys Acta 1979;573:501–9. [DOI] [PubMed] [Google Scholar]
- 9. Beil FU, Fojo SS, Brewer HB, Jr, Greten H, Beisiegel U. Apolipoprotein C-II deficiency syndrome due to apo C-IIHamburg: clinical and biochemical features and HphI restriction enzyme polymorphism. Eur J Clin Invest 1992;22:88–95. [DOI] [PubMed] [Google Scholar]
- 10. Jung N, Dai B, Gentles AJ, Majeti R, Feinberg AP. An LSC epigenetic signature is largely mutation independent and implicates the HOXA cluster in AML pathogenesis. Nat Commun 2015;6:8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Medeiros LA, Khan T, El Khoury JB, Pham CL, Hatters DM, Howlett GJ, et al. Fibrillar amyloid protein present in atheroma activates CD36 signal transduction. J Biol Chem 2004;279:10643–8. [DOI] [PubMed] [Google Scholar]
- 12. Coort SL, Willems J, Coumans WA, van der Vusse GJ, Bonen A, Glatz JF, et al. Sulfo-N-succinimidyl esters of long chain fatty acids specifically inhibit fatty acid translocase (FAT/CD36)-mediated cellular fatty acid uptake. Mol Cell Biochem 2002;239:213–9. [PubMed] [Google Scholar]
- 13. Balgobind BV, Van den Heuvel-Eibrink MM, De Menezes RX, Reinhardt D, Hollink IH, Arentsen-Peters ST, et al. Evaluation of gene expression signatures predictive of cytogenetic and molecular subtypes of pediatric acute myeloid leukemia. Haematologica 2011;96:221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Slattery M, Sweeney C, Murtaugh M, Ma K, Potter J, Levin T, et al. Associations between apoE genotype and colon and rectal cancer. Carcinogenesis 2005;26:1422–9. [DOI] [PubMed] [Google Scholar]
- 15. Su WP, Chen YT, Lai WW, Lin CC, Yan JJ, Su WC. Apolipoprotein E expression promotes lung adenocarcinoma proliferation and migration and as a potential survival marker in lung cancer. Lung Cancer 2011;71:28–33. [DOI] [PubMed] [Google Scholar]
- 16. Lin CY, Gustafsson JA. Targeting liver X receptors in cancer therapeutics. Nat Rev Cancer 2015;15:216–24. [DOI] [PubMed] [Google Scholar]
- 17. Kovacic B, Stoiber D, Moriggl R, Weisz E, Ott RG, Kreibich R, et al. STAT1 acts as a tumor promoter for leukemia development. Cancer Cell 2006;10:77–87. [DOI] [PubMed] [Google Scholar]
- 18. Liu C, Han T, Stachura DL, Wang H, Vaisman BL, Kim J, et al. Lipoprotein lipase regulates hematopoietic stem progenitor cell maintenance through DHA supply. Nat Commun 2018;9:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liang Y, Han H, Liu L, Duan Y, Yang X, Ma C, et al. CD36 plays a critical role in proliferation, migration and tamoxifen-inhibited growth of ER-positive breast cancer cells. Oncogenesis 2018;7:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal 2009;2:re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rahaman SO, Lennon DJ, Febbraio M, Podrez EA, Hazen SL, Silverstein RL. A CD36-dependent signaling cascade is necessary for macrophage foam cell formation. Cell Metab 2006;4:211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pascual G, Avgustinova A, Mejetta S, Martin M, Castellanos A, Attolini CS, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 2017;541:41–5. [DOI] [PubMed] [Google Scholar]
- 23. Ye H, Adane B, Khan N, Sullivan T, Minhajuddin M, Gasparetto M, et al. Leukemic stem cells evade chemotherapy by metabolic adaptation to an adipose tissue niche. Cell Stem Cell 2016;19:23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Landberg N, von Palffy S, Askmyr M, Lilljebjorn H, Sanden C, Rissler M, et al. CD36 defines primitive chronic myeloid leukemia cells less responsive to imatinib but vulnerable to antibody-based therapeutic targeting. Haematologica 2018;103:447–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rozovski U, Harris DM, Li P, Liu Z, Jain P, Ferrajoli A, et al. STAT3-activated CD36 facilitates fatty acid uptake in chronic lymphocytic leukemia cells. Oncotarget 2018;9:21268–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov 2017;7:716–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Andersson A, Ritz C, Lindgren D, Edén P, Lassen C, Heldrup J, et al. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia 2007;21:1198. [DOI] [PubMed] [Google Scholar]
- 30. Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Bene MC, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol 2010;28:2529–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 2004;350:1617–28. [DOI] [PubMed] [Google Scholar]
- 32. Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med 2011;17:1086–93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.