Abstract
Background
RmtF, as 16S rRNA methyltransferase, leads to high-level resistance to aminoglycoside and is now barely reported.
Methods and Results
Three rmtF-positive Klebsiella pneumoniae isolates, belonging to the pandemic clone sequence type 15, were isolated from children and coproduced blaOXA-232 and blaCTX-M-15. The rmtF gene was located on an IncFIB transformable plasmid of 128,536-bp and blaOXA-232 was on a 6141-bp ColKP3 plasmid, respectively.
Conclusion
Plasmids with rmtF found worldwide, shared relatively low similarity, and merely matched partly in their multidrug resistance region. Notably, clinical isolates coproducing rmtF and blaOXA-232 are gradually increasing in China.
Keywords: Klebsiella pneumoniae, RmtF, blaOXA-232, ST15, aminoglycoside
Introduction
Carbapenemase-producing Klebsiella pneumoniae have become a great challenge for antimicrobial chemotherapy, while aminoglycosides can lower the mortality rate effectively in combination therapy of them.1 16S rRNA methyltransferase (16S-RMTase), which induces high-level resistance to aminoglycosides, is now commonly encountered in Enterobacterales worldwide. In China, 16S-RMTases have been found in Enterobacterales from both humans and animals with similar isolation rates, among them rmtB was the most common followed by armA.2 RmtF was a 16S-RMTase firstly identified in France in 2012,3 and thereafter only happened individually in countries like the United States, India and some European countries.4–6
Methods and Results
Recently, three Klebsiella pneumoniae strains K60, K65 and K77 producing RmtF coupled with OXA-232 were isolated from neonates in Shanghai, China. Ethics committee approval was obtained from the institutional review board of Huashan hospital for these isolates, and verbal informed consent from patients’ parents was also accepted and approved by Huashan Hospital.
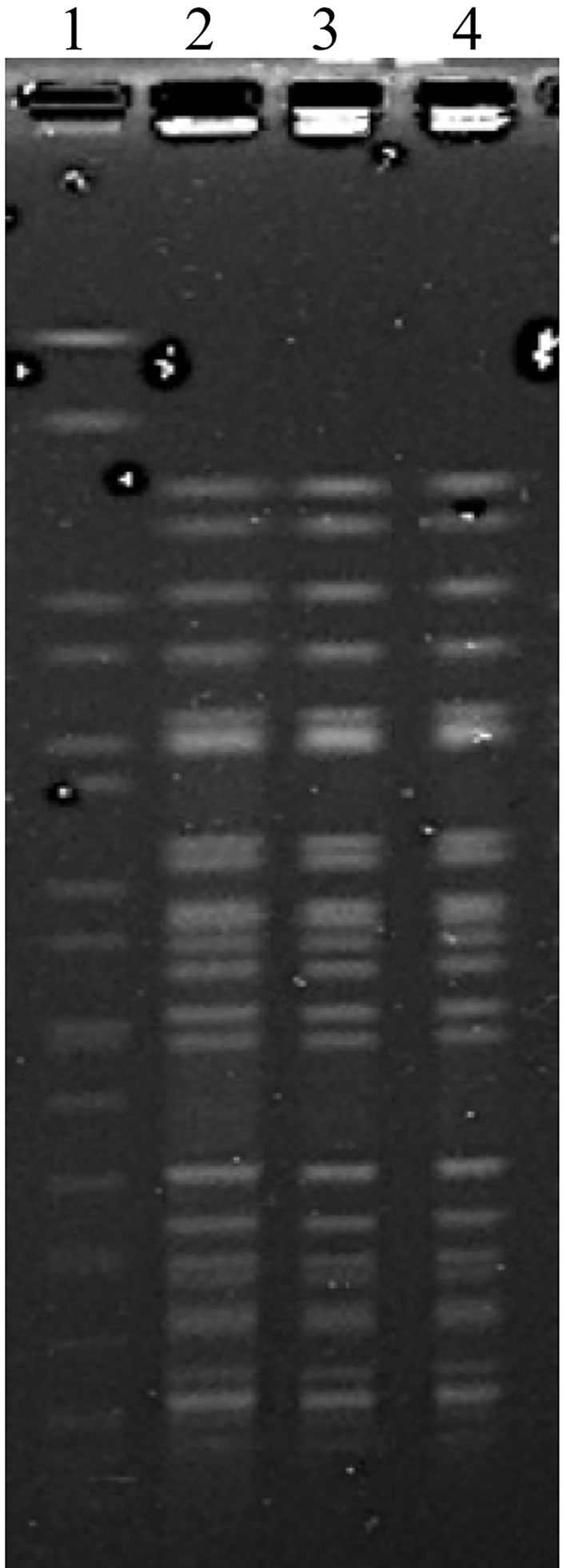
As determined by the reference Clinical and Laboratory Standards Institute (CLSI) broth microdilution method,7 all three K. pneumoniae isolates were highly resistant to aminoglycosides and most antimicrobial agents tested, except carbapenems, to which the resistance produced in low degree (Table 1). The consistency of the pulsed-field gel electrophoresis (PFGE) image of strain K60, K65 and K77 indicated that they were identical (Figure 1).
Table 1.
Minimal Inhibitory Concentrations (MICs) of K. Pneumoniae K60, K65, K77 and Their rmtF-Positive Escherichia Coli Transconjugants
| Antimicrobials | MIC (μg/mL) for: | ||||||
|---|---|---|---|---|---|---|---|
| Recipient | Donors | Transconjugants | |||||
| J53AziR | K60 | K65 | K77 | J60 | J65 | J77 | |
| Amikacin | ≤1 | >128 | >128 | >128 | >128 | >128 | >128 |
| Ertapenem | ≤0.25 | 8 | 8 | 16 | ≤0.25 | ≤0.25 | ≤0.25 |
| Imipenem | 0.25 | 1 | 1 | 1 | 0.25 | 0.25 | 0.25 |
| Meropenem | ≤0.03 | 4 | 4 | 4 | ≤0.03 | ≤0.03 | ≤0.03 |
| Cefmetazole | 2 | 16 | 16 | 32 | 2 | 2 | 2 |
| Cefazolin | 4 | >32 | >32 | >32 | >32 | >32 | >32 |
| Cefuroxime | 16 | >64 | >64 | >64 | >64 | >64 | >64 |
| Ceftriaxone | ≤0.25 | >32 | >32 | >32 | >32 | >32 | >32 |
| Ceftazidime | 0.5 | >32 | >32 | >32 | 32 | 32 | 32 |
| Cefepime | ≤0.06 | >128 | >128 | >128 | 32 | 32 | 32 |
| Ceftazidime/avibactam | 0.25 | 0.5 | 0.5 | 0.5 | 0.25 | 0.25 | 0.5 |
| Cefoperazone/sulbactam | ≤1 | >128 | >128 | >128 | 32 | 16 | 16 |
| Tigecycline | 0.25 | 2 | 2 | 2 | 0.25 | 0.25 | 0.25 |
| Polymyxin B | 0.25 | 0.5 | 0.5 | 0.5 | 0.5 | 0.25 | 0.25 |
| Aztreonam | ≤1 | >128 | >128 | >128 | 64 | 64 | 64 |
| Ciprofloxacin | ≤0.06 | >8 | >8 | >8 | 0.25 | 0.25 | 0.5 |
| Levofloxacin | ≤0.125 | >16 | >16 | >16 | 0.5 | 0.5 | 0.5 |
| Trimethoprim/sulfamethoxazole | ≤0.25 | >32 | >32 | >32 | >32 | >32 | >32 |
Figure 1.
PFGE of K. pneumoniae K60, K65 and K77. Lanes 1, marker Salmonella braenderup H9812; line 2 to 4, PFGE image of K. pneumoniae K60, K65 and K77.
Multiplex PCRs were performed to detect Ambler class A, B and D β-lactamase-encoding genes and 16S rRNA methyltransferase-encoding genes6,8,9 and followed by DNA sequencing. RmtF and blaOXA-232 genes were positive for all three K. pneumoniae isolates. A cloning experiment was then performed. E. coli DH5α was transformed with these plasmids, which yielded the vector pBad33 with a 1.5-kb insert containing rmtF,3 and then their resistance to aminoglycosides transferred from sensitive to highly resistant (Table 1).
Mating-out assays were performed to establish the transferability of the rmtF using the azide-resistant E. coli J53 as recipient strain (selected with gentamicin at 25 mg/L and sodium azide at 150mg/L). Transconjugants were highly resistant to aminoglycosides, cephalosporins and trimethoprim/sulfamethoxazole, while the plasmid containing blaOXA-232 was not obtained.
Genomic DNA of K. pneumoniae K60, K65 and K77 were subjected to whole-genome sequencing (WGS) through Illumina (Illumina, San Diego, CA, USA) short-read sequencing and Nanopore (Oxford, UK) long-read sequencing. Both Genome and plasmids of these strains showed substantial homology, only with the difference of single-nucleotide polymorphism (SNP) level. The genome of these strains was ca. 5335-kb in length and was mapped to CP008929.1(Nepal), CP015990.1(China), CP022127.1(United States), with the proportion over 98% of each, suggesting the possibility of widespread. MLST showed that it belonged to the pandemic clone sequence type 15 (ST15) (https://cge.cbs.dtu.dk/services/MLST/), which is one of the dominant global type, associated with a range of beta-lactamases, including OXA,10 NDM11 and CTX-M.12
Each strain contained three plasmids carried resistance genes. P1 was a 136,315-bp IncFII plasmid harboring aph (3”)-Ib, aph (6)-Id, qnrB1, blaCTX-M-15, blaTEM-1, dfrA14 and sul2, causing resistance to aminoglycosides, quinolones, beta-lactams and trimethoprim/sulfamethoxazole. P2 was a 128,536-bp IncFIB plasmid harboring aac (6ʹ)-Ib, rmtF, arr-2 and cat, causing resistance to aminoglycosides, rifampicin, chloramphenicol. The blaOXA-232 was carried on P3, a 6141-bp ColKP3 nonconjugative plasmid identical to that identified previously,13 and was now widely reported in the world.14,15 In China, such plasmid with blaOXA-232 first emerged in 2017,16 and further appeared in the clonal dissemination of ST15 carbapenem-resistant K. pneumoniae among elderly patients.17 Notably, in this study, blaCTX-M-15 and rmtF transmitted by plasmid P1 and P2 in the conjugation experiment, while blaOXA-232 on P3 could not cotransfer with them. This result was completely consistent with previous reports, but in which the rmtF and blaCTX-M-15 genes were located on the same 160 kb plasmid.6 Among them, plasmid P2 with rmtF was barely reported in China.
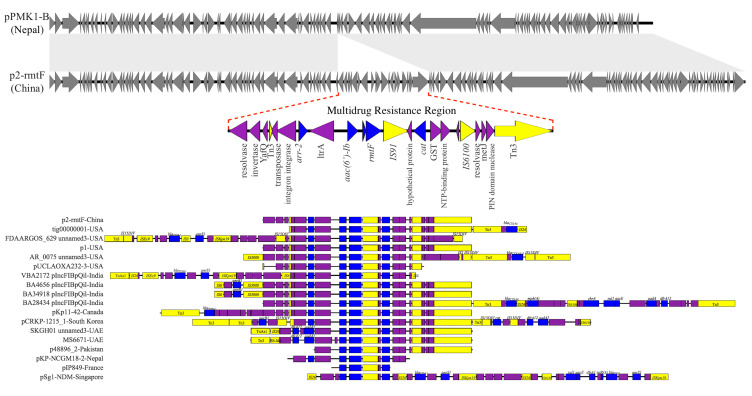
P2 was perfectly mapped to pPMK1-B (GenBank accession no. CP008931.1),14 except a multidrug resistance region (MRR) of 16,839-bp carrying all the resistance genes of P2 (Figure 2). The MRR was flanked by genes of Tn3 family transposases on both sides, and also contained IS91 and IS6100. Mobile elements like these can cluster and be combined with resistance genes, bringing about multiple resistance transfer of plasmids.18 Genes encoding YafQ family toxin proteins were also found in the MRR, such toxin-antitoxin proteins were frequently located on plasmids where they serve to promote plasmid’s stability and maintenance in the bacterial host.
Figure 2.
Major structural features of P2, pPMK1-B and the MRR of other plasmids with rmtF gene. Plasmids sequenced analyzed were compared as follows: P2 was compared with pPMK1-B, the MRR of P2 was compared with the MRR of other plasmids carrying rmtF gene: tig00000001-USA (121,057-bp, NZ_CP021758.1), FDAARGOS_629 unnamed3-USA (138,560-bp, CP044045.1), p1-USA (142,764-bp, CP033947.1), AR_0075 unnamed3-USA (93,870-bp, CP032188.1), pUCLAOXA232-3-USA (83,730-bp, CP012569.1), VBA2172 pIncFIBpQil-India (127,300-bp, CP036321.1), BA4656 pIncFIBpQil-India (164,210-bp, CP035907.1), BA34918 pIncFIBpQil-India (164,195-bp, CP036193.1), BA28434 pIncFIBpQil-India (130,775-bp, CP036328.1), pKp11-42-Canada (146,695-bp, KF295829.1), pCRKP-1215_1-South Korea (130,922-bp, CP024839.1), SKGH01 unnamed 3-United Arab Emirates (84,941-bp, NZ_CP015503.1), MS6671-United Arab Emirates (84,940-bp, LN824138.1), p48896_2-Pakistan (114,815-bp, CP024431.1), pKP-NCGM18-2-Nepal (9812-bp, AB824739.1), pIP849-France (4710-bp, JQ808129.1), pSg1-NDM-Singapore (90,103-bp, NZ_CP011839.1). Grey symbols indicate identical plasmid regions of P2 and pPMK1-B, while gray shading indicates >99% identity of them, and the red dotted line indicates their different component. Resistance genes are indicated by blue symbols. Transposon-related genes the class 1 integrase gene, and insertion sequences are indicated by yellow symbols. Other genes are indicated by violet symbols. Labels common to these MRRs appear only in the P2 diagram.
In all, 17 full sequences of plasmids with rmtF were found in GenBank data, which were reported worldwide, and then they were compared to P2 using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Except for pSg1-NDM (GenBank accession no. CP008931.1), an IncR plasmid found in Singapore, the rest of these plasmids were IncF plasmids with only one MRR. As one of the most frequent plasmid types, plasmids of the IncF group have a primary role in the antimicrobial resistance of Enterobacterales and show rapid evolution.19 However, these plasmids shared relatively low similarity, merely matched imperfectly in their MRR, while all of these MRR were jointed to mobile elements like Tn3 family transposases (Figure 2).
Conclusion
Worryingly, the co-occurrence of 16S rRNA methyltransferases and carbapenemases has been increasingly reported among K. pneumoniae in recent years.20 Enterobacterales isolates producing rmtF used to be extremely rare in China, but in recent years relevant reports have emerged and always accompanied with coproduction of OXA-232.21,22 In countries like India and the UK, the detection rate of rmtF increased rapidly, suggesting the possibility of its speedy spread. Plasmids with rmtF gene often featured resistance genes acquiring through mobile elements and plasmid addiction modules made up of toxin-antitoxin proteins, which led to the stable persistence of clinical isolates and ultimately resulted in multidrug resistance to almost all of the clinically available antibiotics. In this study, all three clinical isolates were susceptible only to cefepime/zidebactam, ceftazidime/avibactam, tigecycline, polymyxin B and imipenem. This made clinical choices extremely limited if economic cost and medicine availability were taken into consideration. This represented the worry toward the distribution and transmission of clinical Isolates with analogous plasmids as a global public threat. To screen these resistance genes and comprehend their transmissibility in time, plasmid analysis may be a useful supplementary method for medical institutions. Based on correlative results, measures like contact isolation and environmental cleaning can also be performed to avoid nosocomial outbreak.
Funding Statement
This work was supported by the National Natural Science Foundation of China (grant no. 81871690, 81902101) and the National Mega-project for Innovative Drugs (2019ZX09721001-006-004). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
- 1.Daikos GL, Sophia T, Tzouvelekis LS, et al. Carbapenemase-producing Klebsiella pneumoniae bloodstream infections: lowering mortality by antibiotic combination schemes and the role of carbapenems. Antimicrob Agents Chemother. 2014;58(4):2322–2328. doi: 10.1128/AAC.02166-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu F, Dan Y, Pan J. High prevalence of plasmid-mediated 16S rRNA methylase gene rmtB among Escherichia coli clinical isolates from a Chinese teaching hospital. BMC Infect Dis. 2010;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galimand M, Courvalin P, Lambert T. RmtF, a new member of the aminoglycoside resistance 16S rRNA N7 G1405 methyltransferase family. Antimicrob Agents Chemother. 2012;56(7):3960–3962. doi: 10.1128/AAC.00660-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hidalgo L, Hopkins KL, Gutierrez B, et al. Association of the novel aminoglycoside resistance determinant RmtF with NDM carbapenemase in enterobacteriaceae isolated in India and the UK. J Antimicrob Chemother. 2013;68(7):1543–1550. doi: 10.1093/jac/dkt078 [DOI] [PubMed] [Google Scholar]
- 5.Lee CS, Vasoo S, Hu F, Patel R, Doi Y. Klebsiella pneumoniae ST147 coproducing NDM-7 carbapenemase and RmtF 16S rRNA methyltransferase in Minnesota. J Clin Microbiol. 2014;52(11):4109–4110. doi: 10.1128/JCM.01404-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mancini S, Poirel L, Tritten ML, Lienhard R, Bassi C, Nordmann P. Emergence of an MDR Klebsiella pneumoniae ST231 producing OXA-232 and RmtF in Switzerland. J Antimicrob Chemother. 2017. doi: 10.1093/jac/dkx428 [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 29th Ed. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2019. [Google Scholar]
- 8.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi: 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 9.Berçot B, Poirel L, Nordmann P. Updated multiplex polymerase chain reaction for detection of 16S rRNA methylases: high prevalence among NDM-1 producers. Diagn Microbiol Infect Dis. 2011;71(4):442–445. doi: 10.1016/j.diagmicrobio.2011.08.016 [DOI] [PubMed] [Google Scholar]
- 10.Pitout JDD, Peirano G, Kock MM, Strydom K-A, Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev. 2019;33(1):e00102–00119. doi: 10.1128/CMR.00102-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peirano G, Ahmed-Bentley J, Fuller J, Rubin JE, Pitout JDD. Travel-related carbapenemase-producing gram-negative bacteria in Alberta, Canada: the first 3 years. J Clin Microbiol. 2014;52(5):1575–1581. doi: 10.1128/JCM.00162-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SY, Ko KS. Diverse plasmids harboring blaCTX-M-15 in Klebsiella pneumoniae ST11 isolates from several Asian countries. Microb Drug Resist. 2019;25(2):227–232. doi: 10.1089/mdr.2018.0020 [DOI] [PubMed] [Google Scholar]
- 13.Potron A, Rondinaud E, Poirel L, et al. Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D β-lactamase from enterobacteriaceae. Int J Antimicrob Agents. 2013;41(4):325–329. doi: 10.1016/j.ijantimicag.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 14.Stoesser N, Giess A, Batty EM, et al. Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae isolates from neonatal infections in a Nepali hospital characterizes the extent of community- versus hospital-associated transmission in an endemic setting. Antimicrob Agents Chemother. 2014;58(12):7347–7357. doi: 10.1128/aac.03900-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nahid F, Zahra R, Sandegren L, Chang Y-F. A blaOXA-181-harbouring multi-resistant ST147 Klebsiella pneumoniae isolate from Pakistan that represent an intermediate stage towards pan-drug resistance. PLoS One. 2017;12(12):e0189438. doi: 10.1371/journal.pone.0189438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin D, Dong D, Li K, et al. Clonal dissemination of OXA-232 carbapenemase-producing Klebsiella pneumoniae in neonates. Antimicrob Agents Chemother. 2017;61(8). doi: 10.1128/AAC.00385-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shu L, Dong N, Lu J, et al. Emergence of OXA-232 carbapenemase-producing Klebsiella pneumoniae that carries a pLVPK-like virulence plasmid among elderly patients in China. Antimicrob Agents Chemother. 2019;63(3). doi: 10.1128/aac.02246-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Partridge SR. Analysis of antibiotic resistance regions in gram-negative bacteria. FEMS Microbiol Rev. 2011. 35(5):1574–6976. [DOI] [PubMed] [Google Scholar]
- 19.Johnson TJ, Nolan LK. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol Rev. 2010;74(3):477–478. doi: 10.1128/MMBR.00002-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor E, Sriskandan S, Woodford N, Hopkins KL. High prevalence of 16S rRNA methyltransferases among carbapenemase-producing Enterobacteriaceae in the UK and Ireland. Int J Antimicrob Agents. 2018;52(2):278–282. doi: 10.1016/j.ijantimicag.2018.03.016 [DOI] [PubMed] [Google Scholar]
- 21.Abdul Momin MHF, Liakopoulos A, Phee LM, Wareham DW. Emergence and nosocomial spread of carbapenem-resistant OXA-232-producing Klebsiella pneumoniae in Brunei Darussalam. J Glob Antimicrob Resist. 2017. S2213716517300516. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Ma W, Qin Q, et al. Nosocomial spread of OXA-232-producing Klebsiella pneumoniae ST15 in a teaching hospital, Shanghai, China. BMC Microbiol. 2019;19(1):235. doi: 10.1186/s12866-019-1609-1 [DOI] [PMC free article] [PubMed] [Google Scholar]