Abstract
Purpose
Ovarian cancer is the leading cause of death in gynecologic malignancies. Growing evidences demonstrate that a complicated relationship exists between the gut microbiota and cancer treatment. However, there are few studies explored the alterations of gut microbiota in ovarian cancer patients following anti-cancer treatments. Therefore, we aim to analyze the changes of the gut microbiota in ovarian cancer patients treated with radical surgery and chemotherapy.
Patients and Methods
The microbial genes were examined from a total of 75 fecal samples from 18 ovarian cancer patients, including 10 preoperative fecal samples (Group B), 4 postoperative fecal samples (Group M0), as well as 61 fecal samples after first to fifth cycles of chemotherapy, using 16S rRNA sequencing.
Results
Our results showed that fecal samples collected in postoperative (Group M0) exhibited significant decreases in abundance of Bacteroidetes and Firmicutes, while a significant increase in abundance of Proteobacteria compared with preoperative (Group B) fecal samples. LEfSe analysis identified that Bilophila and Faecalibacterium are the key genera in Group B, while Klebsiella and Enterococcus are the key genus in Group M0. Compared with before chemotherapy, the abundance of Bacteroidetes and Firmicutes increased, and the abundance of Proteobacteria decreased after chemotherapy. In addition, anaerobic bacteria, such as Bacteroides, Collinsella and Blautia, exhibited significant increases after chemotherapy. Moreover, we observed that certain bacterial genera were significantly correlated with clinicopathological characteristics of ovarian cancer patients.
Conclusion
Our study suggested that radical surgery and chemotherapy altered the composition of gut microbiota in ovarian cancer patients. Therapeutic strategies targeting the gut microbiota may be beneficial for the clinical treatment of ovarian cancer.
Keywords: ovarian cancer, intestinal microbiota, radical surgery, chemotherapy, 16S rRNA sequencing
Introduction
Ovarian cancer (OV) remains the most common cause of death in gynecologic malignancies worldwide, causing up to 295,414 cancer cases and 184,799 cancer deaths in the world in 2018.1 Due to lack of effective screening method and early detection, most ovarian cancers are diagnosed at an advanced stage and the 5-year survival rate is lamentable, only about 40% after diagnosis. Ovarian cancer is a highly heterogeneous cancer that varies in histology, molecular biology and numerous other characteristics. Epithelial ovarian cancer accounts for 90% of ovarian cancers and can be divided into two types, among which, type II tumors always present in high grade, advanced stage and are highly aggressive.2 At present, aggressive cytoreductive surgery followed by platinum-based chemotherapy are the gold standard for the treatment of ovarian cancer, whereas patients treated with chemotherapy may develop intestinal micro-ecological disorders that induce intestinal mucosal inflammation.3 Such side effects of chemotherapy gravely worsen the patients’ quality of life and even reduce the treatment efficiency.4 Thus, it is extremely urgent to analyze the changes of microbial composition before and after ovarian cancer treatment, and ameliorate treatment plan in order to improve the quality of life of ovarian cancer patients.
In the past two decades, the primary role of platinum-based chemotherapy in the first-line management of advanced ovarian cancer has not changed, including regimens of cisplatin plus paclitaxel, combined carboplatin therapy plus either paclitaxel or docetaxel.5 However, the majority of chemotherapeutic drugs currently available have cytotoxic effects, and cause gastrointestinal mucositis, including ulcers, nausea, vomiting, bloating, diarrhea and constipation, challenging the efficiency of chemotherapeutic treatment of ovarian cancer. Previous studies reported that cisplatin have antibiotic effects on both Gram-negative and Gram-positive bacterial strains, including some Bacillus and E. coli.6 Besides, cisplatin may also damage the intestinal mucosa through binding to DNA and forming crosslinks that impair DNA replications.7 Campbell et al revealed that intestinal microbiota is associated with other common adverse side effects of cisplatin, like ototoxicity and weight loss.8 Zhao et al demonstrated that altered intestinal microbiota, particularly the decreased Firmicutes, would probably be the cause of cisplatin-associated side effects, such as body weight loss and cardiac dysfunction. And Lactobacillus supplementation potentially prevents cisplatin-induced cardiotoxicity possibly via inflammation inhibition.9 Another study discovered that the abundance of Ruminococcus gnavus significantly decreased in mice treated with cisplatin and restored with oral gavage of fecal pellet.3 Meanwhile, intestinal microbiota may also influence the anti-cancer activity of cisplatin. Gram-positive bacteria was found to play a crucial role in the process of tumor-related inflammatory cells producing reactive oxygen species (ROS), which is essential for platinum-induced DNA damage. Several studies in animals had confirmed that anti-Grampositive antibiotics treatment the reduced anti-cancer efficacy of platinum in mice.10
Recent studies have shown that the intestinal microbiota may be disrupted by the use of antibiotics or bowel preparations in the process of surgery.11 Ohigashi et al reported that the abundance of obligate anaerobes was decreased after surgery in colorectal cancer patients, whereas the abundance of Enterobacteriaceae, Enterococcus, Staphylococcus and Pseudomonas was observed to be increased.12 Meanwhile, gut microbiota may potentially impact cancer outcomes after surgery.13 However, current studies regarding the effects of surgery and chemotherapy on the gut microbiota mainly use animal models, and no study has been found exploring the effects of surgery and chemotherapy in gut microbiota of ovarian cancer patients. We therefore performed 16S rRNA sequencing in order to explore the alterations of the intestinal microbiota in ovarian cancer patients treated with radical surgery and chemotherapy.
Patients and Methods
Patients and Fecal Samples Collection
Patients diagnosed with ovarian cancer were recruited between December 2015 and September 2016 in Sir Run Run Shaw Hospital, China. Details of the FIGO 2009 stage, tumor grade, histological type, ascites, lymph node metastasis, chemotherapeutic scheme, gastrointestinal reaction and recurrence were taken from the clinical records (Supplementary Table S1). All participants had received radical surgery performed by the same surgeons, followed by the standardized chemotherapeutic scheme TC (day 1, carboplatin, AUC 6–7.5, intravenous injection; day 1, paclitaxel, 175 mg/m2 over 3 hours, intravenous injection; q-21 days × 6 cycles) or TP (day 1, cisplatin, 75 mg/m2, intravenous injection; day 1, paclitaxel, 135 mg/m2 over 24 hours, intravenous injection; q-21 days × 6 cycles). A total of 75 fecal samples were obtained from 18 ovarian cancer patients (Table 1), including 10 preoperative fecal samples (denoted as Group B), 4 samples collected four weeks after surgery and before the first cycle of chemotherapy (denoted as Group M0), and 61 fecal samples obtained after the first to fifth cycles of chemotherapy (denoted as groups M1-5), containing 18, 17, 13, 7, and 6 samples, respectively. Fecal samples were collected in a sterile tube and immediately stored at −80°C until microbial analysis. The ethics committee of Sir Run Run Shaw Hospital reviewed and approved the study protocols, and all patients provided their informed consent before they participated in this study.
Table 1.
Clinical Characteristics of Ovarian Cancer Patients (Mean ± SD) in Each Group
| Group | Preoperative Group | Postoperative Group | Chemotherapy Group | p value |
|---|---|---|---|---|
| No. of patients | 10 | 4 | 18 | |
| Age (years) | 56.2±5.5 | 60.5±2.1 | 56.3±6.8 | >0.05 |
| BMI (kg·m−2) | 23.1±6.0 | 21.4±2.5 | 23.2±4.7 | >0.05 |
Abbreviations: BMI, body mass index; SD, standard deviation.
Genomic DNA Extraction, PCR Amplification and 16S rRNA Sequencing
Genomic DNA was extracted using the E.Z.N.A.® Stool DNA Kit (Omega Bio-Tek, Norcross, Georgia, USA) according to the manufacturer’s protocol. Subsequently, the DNA concentration and purity were evaluated by a NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, MA, USA), and then the DNA quality was monitored on 2% agarose gel electrophoresis. The bacterial genomic DNA was amplified with the bacterial universal primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) specific for the V4-V5 hypervariable regions of the 16S rRNA gene. PCR was performed in a 20 μL mixture, containing 10 μL 2x Premix Taq, 0.5 μL FastPfu polymerase, 1.5 μL of each primer (forward and reverse primer), 20 ng template DNA and the remaining volume of ddH2O. The cycling parameters were as follows: 98°C of 30 s, 30 cycles of 15 s at 98°C, 15 s at 58°C, and 15 s at 72°C, and a final extension at 72°C for 1 min. The PCR products were analyzed by 2% agarose gel and then quantified on the Qubit® 2.0 Fluorometer (Life Technologies, USA). Pooled amplicons underwent paired-end sequencing (2× 250 nt) on an Illumina MiSeq platform (Illumina, San Diego, CA, USA) according to the standard instructions.
Bioinformatic Analysis
Raw sequencing files were demultiplexed, quality filtered and merged by QIIME.14 Sequences with ≥97% similarity were clustered into the same operational taxonomic units (OTUs). Alpha diversity (Observed species, Shannon, Simpson, Chao1, ACE, Good’s coverage) and Beta diversity were calculated with QIIME software and displayed with R (version 3.5.1). Linear discriminant analysis effect size (LEfSe) method was used to discover a metagenomic biomarker.15 The heat map of the relative abundance of 61 bacterial genera was drawn on the basis of OTUs using R (version 3.5.1). Spearman correlation coefficients were analyzed by using SPSS software (version 24.0) and the networks were carried out by using Cytoscape (version 3.6.1).16
Statistical Analysis
All statistical analyses were implemented by SPSS software (version 24.0; IBM, Chicago, IL, USA) and GraphPad Prism software (version 7.0.4; La Jolla, CA, USA). The data of age and body mass index (BMI) were expressed as mean ± standard deviation (SD) and compared by one-way ANOVA analysis and Tukey’s post hoc test. The non-parametric Mann–Whitney U-test (for unpaired two groups) or Kruskal–Wallis omnibus test (for more than two groups) was used to test the differences between groups. A two-tailed p value <0.05 was considered statistically significant.
Results
Analysis of the Alpha and Beta Diversity of Microbial Community in Fecal Samples
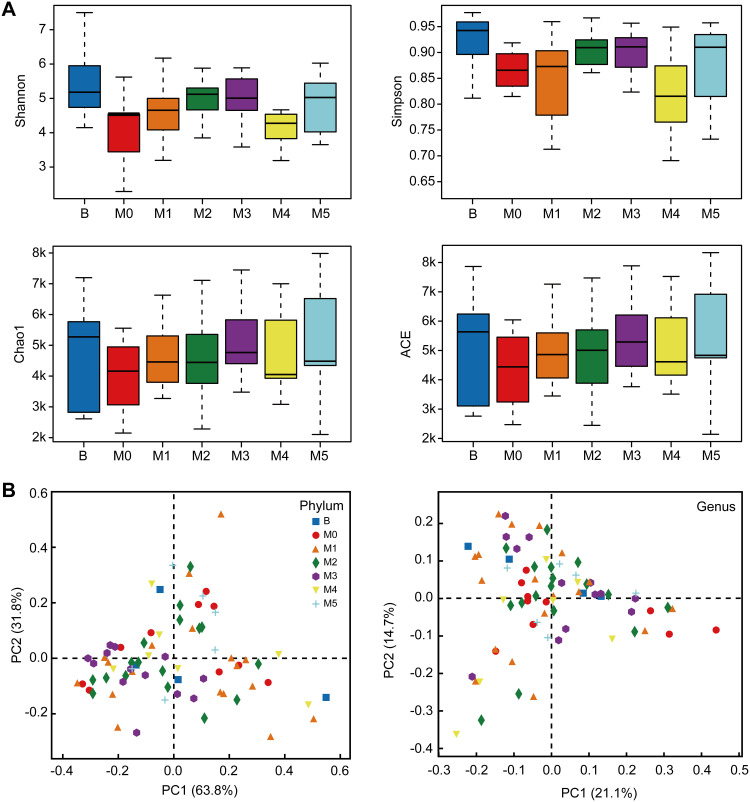
To evaluate the bacterial community diversity and richness, we compared the Shannon, Simpson, Chao1 and ACE index in preoperative, postoperative and after the first to fifth round of postoperative chemotherapy groups. There were significant differences in Shannon (p = 0.025) and Simpson (p = 0.023) index value in overall seven groups, while no significant difference was identified between preoperative and postoperative groups, or among postoperative and chemotherapy groups (Figure 1A). Principal co-ordinate analysis (PCoA) suggested that a separation existed (PC1: 63.8%, PC2: 31.8%) at the phylum level and (PC1: 21.1%, PC2: 14.7%) at the genus level among these seven groups (Figure 1B, Supplementary Figure S1).
Figure 1.
Alpha and beta diversity of bacterial populations in preoperative Group B, pre-chemotherapy Group M0 and groups M1–5 during stages of chemotherapy. (A) Boxplots of observed OTUs (≥97% identity level), Shannon index, Simpson index, Chao1 estimator richness and ACE values of the fecal samples in seven groups. (B) PCoA plots obtained from sequencing the microbial in fecal samples.
Comparison of Gut Microbiota Before and After Surgery in Ovarian Cancer Patients
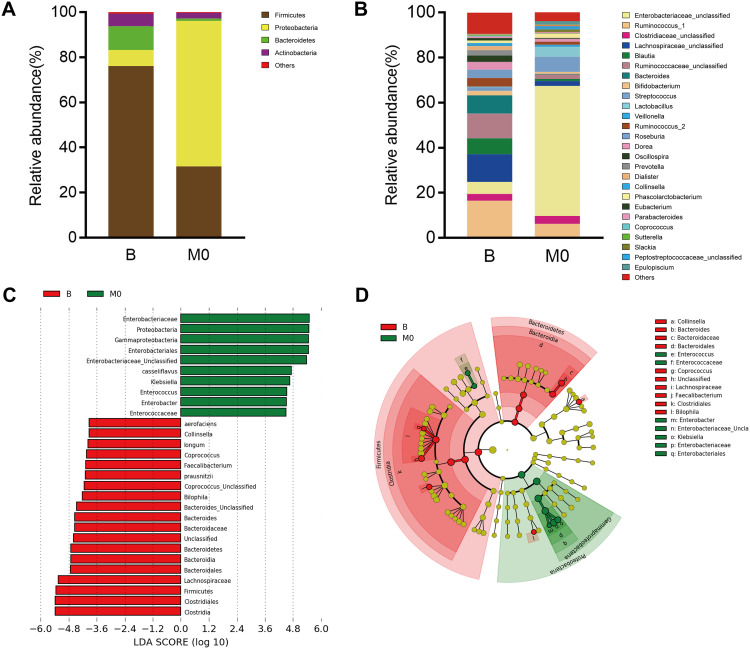
We compared the total fecal bacteria between preoperative (Group B) and postoperative (Group M0) groups. At the phylum level, the abundance of Proteobacteria increased markedly after radical surgery (Group B vs M0: 7.13% vs 64.50%, p = 0.002), while the abundance of Bacteroidetes (Group B vs M0: 10.59% vs 0.99%, p = 0.002) and Firmicutes (Group B vs M0: 76.05% vs 31.64%, p = 0.002) decreased significantly. Meanwhile, the abundance of Actinobacterial decreased after radical surgery but insignificantly (Group B vs M0: 5.48% vs 2.59%) (Figure 2A). At the genus level, the number of Bacteroides (Group B vs M0: 6.89% vs 0.17%, p < 0.01), Faecalibacterium (Group B vs M0: 2.22% vs 0.06%, p < 0.05), Bilophila (Group B vs M0: 0.31% vs 0.00%, p < 0.05), Collinsella (Group B vs M0: 1.28% vs 0%, p < 0.05) and Coprococcus (Group B vs M0: 0.39% vs 0.02%, p < 0.05) were significantly lower in Group M0 than those in Group B, while Enterobacter (Group B vs M0: 0% vs 0.14%, p < 0.05), Klebsiella (Group B vs M0: 0.22% vs 6.40%, p < 0.05) and Enterococcus (Group B vs M0: 0.03% vs 0.49%, p < 0.05) were significantly higher in Group M0 than those in Group B (Figure 2B). Besides, the abundance Lachnospiraceae_Ruminococcus (Group B vs M0: 17.40% vs 4.51%), Blautia (Group B vs M0: 6.03% vs 0.93%), Roseburia (Group B vs M0: 3.20% vs 0.01%), Prevotella (Group B vs M0: 2.13% vs 0.16%) and Collinsella (Group B vs M0: 1.28% vs 0.00%) decreased after surgery but without significant difference. To determine the intestinal microbial changes associated with radical surgery at the taxonomical level, we used LEfSe analysis to analyze the microbial clade differences (Figure 2C and D). The dominant phyla identified in Group B and Group M0 were Firmicutes and Proteobacteria, respectively. The key genus in Group B was Bilophila, followed by Faecalibacterium. The key genus in Group M0 was Klebsiella, followed by Enterococcus.
Figure 2.
Comparison of intestinal microbiota between preoperative (Group B) and postoperative (Group M0) ovarian cancer patients. Relative abundance of the dominant phyla (A) and genera (B) in these two groups. (C) Histogram of linear discriminant analysis (LDA) scores for differentially abundant genera. (D) Cladogram of LDA coupled with effective size measurement showing differentially abundant genera.
Comparison of Gut Microbiota Before and After Chemotherapy in Ovarian Cancer Patients
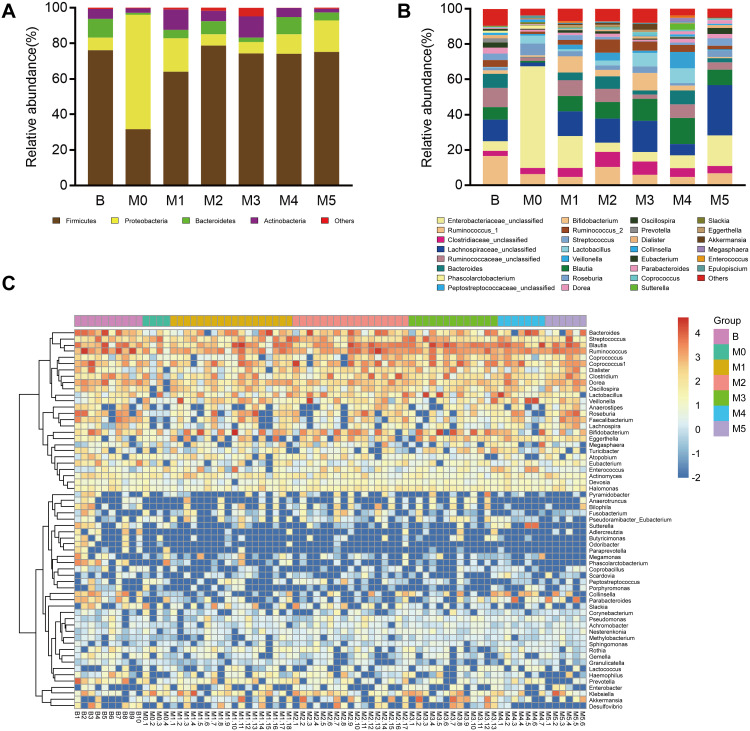
The distribution and composition of gut microbial were significantly different among Group M0 and groups M1-M5. At the phylum level, Group M0 was mainly enriched by Proteobacteria, whereas M1-5 was primarily characterized by Firmicutes. Compared with Group M0, Proteobacteria showed a trend of decreasing after chemotherapy (M0, 64.50%; M1, 18.77%; M2, 6.41%; M3, 6.43%; M4, 10.97%; M5, 17.76%), while Firmicutes (M0, 31.64%; M1, 64.05%; M2, 78.70%; M3, 74.28%; M4, 74.11%; M5, 75.14%) and Bacteroidetes (M0, 0.99%; M1, 4.72%; M2, 7.32%; M3, 2.52%; M4, 9.69%; M5, 4.44%) were more abundant than before chemotherapy (Figure 3A). At the genus level, the composition of the intestinal microbial altered markedly after multiple cycles of chemotherapy. On the one hand, chemotherapeutic drugs dramatically reduced some originally dominant bacteria, such as Enterobacteriaceae_unclassified (M0, 57.96%; M1, 18.41%; M2, 6.76%; M3, 6.02%; M4, 6.38%; M5, 14.49%), Klebsiella (M0, 6.40%; M1, 2.26%; M2, 0.79%; M3, 0.99%; M4, 0.35%; M5, 3.20%), and Enterobacter (M0, 0.14%; M1, 0.08%; M2, 0.02%; M3, 0.01%; M4, 0.00%; M5, 0.04%). On the other hand, chemotherapeutic drugs caused a remarkable increase in conditionally pathogenic bacteria, like Bacteroides (M0, 0.17%; M1, 4.08%; M2, 5.61%; M3, 2.07%; M4, 8.62%; M5, 6.35%), Bilophila (M0, 0.00%; M1, 0.09%; M2, 0.19%; M3, 0.09%; M4, 0.00%; M5, 0.43%), Collinsella (M0, 0.00%; M1, 0.10%; M2, 0.31%; M3, 0.53%; M4, 1.20%; M5, 0.69%), Faecalibacterium (M0, 0.06%; M1, 0.55%; M2, 0.53%; M3, 0.12%; M4, 0.42%; M5, 2.20%), and Coprococcus (M0, 0.02%; M1, 0.30%; M2, 0.87%; M3, 1.46%; M4, 0.76%; M5, 0.66%) (Figure 3B).
Figure 3.
Comparison of intestinal microbiota among preoperative Group B, postoperative Group M0 and groups M1–5 during stages of chemotherapy. Relative abundance of the dominant phyla (A) and genera (B) in the seven groups. (C) Heat map analysis of the bacterial distribution based on hierarchical clustering of the 75 fecal samples in preoperative Group B, postoperative Group M0 and groups M1–5.
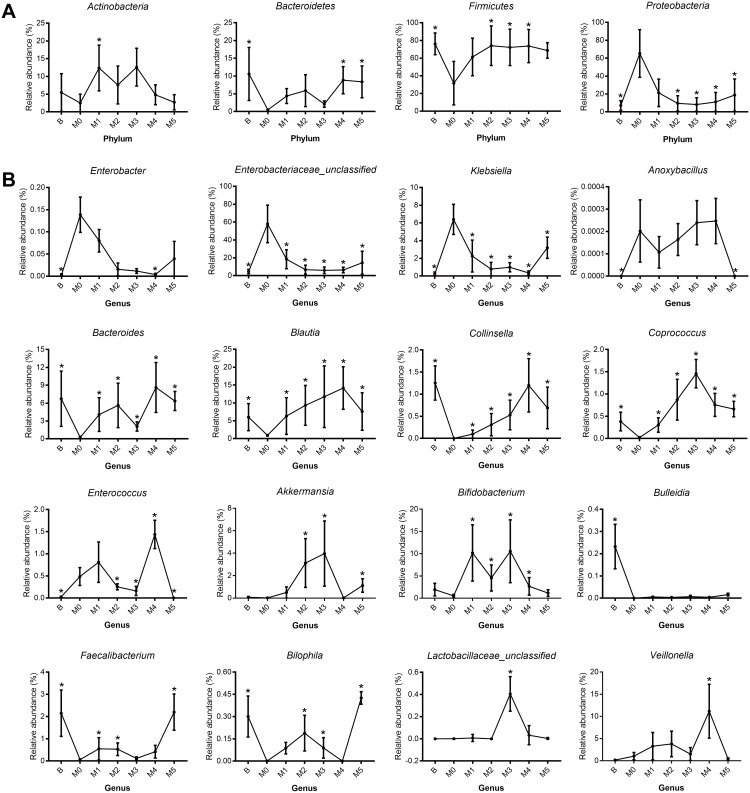
We next exhibited the alterations of representative microbial phyla and genera during the process of cancer treatment by the relative abundance trend charts (Figure 4). The abundance of some bacteria including Enterobacter, Enterobacteriaceae_unclassified, Klebsiella, Bilophila, Enterococcus, Coprococcus, Veillonella, Bifidobacterium, Akkermansia and Lactobacillaceae_unclassified fluctuated greatly in the first two cycles of chemotherapy but gradually returned to the original levels subsequently. Moreover, genus Bacteroides, Blautia and Collinsella tended to increase while genus Enterobacter, Enterobacteriaceae_unclassified and Klebsiella tended to decrease following multiple cycles of chemotherapy, though without significant differences. The results indicated that the abundance of some bacterial genera may restore the original level after multiple cycles of chemotherapy, while that of other bacterial genera may deviate, indicating chemotherapy may have differential influence on the gut microbiota (Figure 3C).
Figure 4.
Real-time quantitative polymerase chain reaction (PCR) for (A) four major phyla and (B) twelve genera, included the preoperative Group B, pre-chemotherapy Group M0, and groups M1–5 during stages of chemotherapy. (A) Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria. (B) Enterobacter, Enterobacteriaceae_unclassified, Klebsiella, Anoxybacillus, Bacteroides, Blautia, Collinsella, Coprococcus, Enterococcus, Akkermansia, Bifidobacterium, Bulleidia, Faecalibacterium, Bilophila, Lactobacillaceae _unclassified and Veillonella. *p < 0.05.
Co-Occurrence Networks of Bacterial Genera in Different Groups
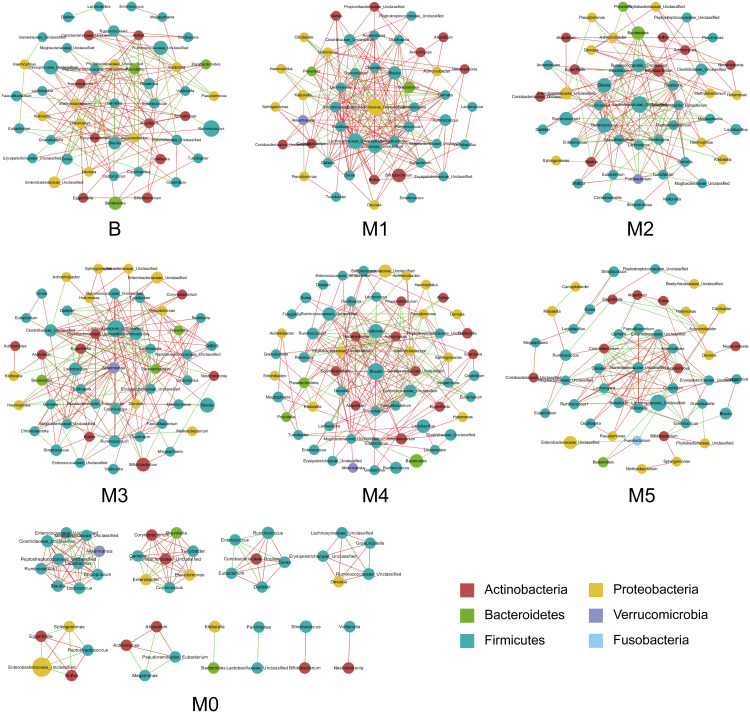
The co-occurrence networks were generated according to the abundance and correlations of bacterial genera in the seven groups, based on Spearman correlation coefficient that R > 0.6, P < 0.05 (Figure 5). The results suggested that the majority of genera in these groups were from the phylum Firmicutes, which exhibited a decreased tendency in postoperative fecal samples and exhibited more abundance in the multiple cycles of chemotherapy. Moreover, the most correlated bacterial genera in each group were shown as follows: Corynebacterium and Klebsiella (r=0.95) in Group B; Erysipelotrichaceae_Unclassified and Ruminococcus (r = 0.91) in Group M1; Clostridiaceae_Unclassified and SMB53 (r = 0.89) in Group M2; Lactococcus and Prevotella (r = 0.93) in Group M3; Prevotella and Rothia (r = 0.99) in Group M4; as well as Gemella and Mogibacteriaceae_Unclassified (r = 0.99) in Group M5.
Figure 5.
Co-occurrence network of genera in preoperative Group B, pre-chemotherapy Group M0 and groups M1–5 during stages of chemotherapy. Each node represents a genus and the size of which is proportional to the relative abundance; each node is colored based on their taxonomic level. The color of the nodes represents their taxonomic assignment. Spearman correlation coefficient (red or green) was used to indicate connection of paired nodes.
Abbreviations: OV, ovarian cancer; HGSC, high-grade serous carcinoma; LEfSe, linear discriminant analysis effect size; PCoA, principal co-ordinate analysis; OUT, operational taxonomic unit.
Correlations of Gut Microbiota and Clinical Characteristics
We analyzed the correlations between the gut microbiota and clinicopathological characteristics, including lymph nodes metastasis, gastrointestinal reactions and recurrence (Supplementary Figure S2). We observed that Bifidobacterium (p < 0.001), Akkermansia (p < 0.05), Desulfovibrio (p < 0.05), Enterococcus (p < 0.05) and Dorea (p < 0.05) were significantly associated with lymph nodes metastasis, while Ruminococcus (p < 0.05), Ruminococcaceae_unclassified (p < 0.01), Desulfovibrio (p < 0.05), Clostridiaceae_unclassified (p < 0.001) and Lactobacillus (p < 0.05) were significantly correlated with gastrointestinal reactions. Moreover, Bifidobacterium (p < 0.05), Megamonas (p < 0.001) and Pseudomonas (p < 0.01) were significantly higher in patients with shorter survival period, and Klebsiella (p < 0.001) and Fusobacterium (p < 0.01) were lower in patients with longer survival period.
Discussion
Accumulating evidence indicates that microbiota plays a pivotal role in the occurrence and development of cancer, and microbiota dysbiosis is associated with cancer therapies.17 A recent study observed reduced richness and diversity of microbiota in ovarian cancer tissues compared to normal fallopian tube tissues, suggesting that microbial composition alterations may be correlated with the initiation and development of ovarian cancer.18 Another study suggested that pelvic radiotherapy remarkably changed the microbiota community and severely reduced the species-level taxa of gut microbiota in gynecological cancer patients.19 However, the effects of surgery and chemotherapy on gut microbiota in ovarian cancer patients were still unclear. Here, we reported for the first time the alterations of intestinal microbiota composition in ovarian cancer patients receiving radical surgery and chemotherapy via using 16S rRNA sequencing.
In the present study, we found that the composition and diversity of intestinal microbiota in postoperative ovarian cancer patients significantly differed from that in preoperative ovarian cancer patients. At the phylum level, the relative abundance of Proteobacteria was strikingly higher in postoperative fecal samples than that in preoperative samples. A bloom of Proteobacteria in gut was often correlated with microbiota dysbiosis and was a potential biomarker for intestinal inflammation.20 Chen et al had also confirmed that Proteobacteria increased significantly after radical surgery in colorectal cancer patients.21 Besides, the abundance of Bacteroidetes and Firmicutes was found decreased significantly after radical surgery in this study. Recent studies suggested that patients with a higher abundance of Firmicutes and lower abundance of Bacteroidetes had a high risk of colitis on immunotherapy.22 At the genus level, the Gram-negative bacteria, such as Enterobacter and Klebsiella, belonging to Proteobacteria phylum, were drastically increased. Moreover, the abundance of short-chain fatty acids (SCFAs) producing bacteria, such as Bacteroidetes, Faecalibacterium, Blautia, Roseburia and Prevotella decreased after surgery. SCFAs, such as acetate, propionate, butyrate and lactate have been proven to have profound anti-inflammatory, anti-tumor and immune-regulatory functions.23,24 It is noteworthy that Enterobacteriaceae, a kind of aerobic bacteria that could reflect intestinal inflammation, increased after surgery.25 Furthermore, we noticed that fecal samples collected after surgery were marked with an increase in Enterobacteriaceae family, which may be closely associated with the inflammatory states.26
Platinum-taxane-based chemotherapy plays a primary role in the first-line management of ovarian cancer. Evidence is growing that chemotherapeutic drugs have significant effects on the intestinal microorganisms, leading to adverse side effects and reduced efficiency. Meanwhile, the intestinal microbiota can modulate the host response to chemotherapeutic drugs.27 Thus, our study next analyzes the impacts of chemotherapy on gut microbiota. We found that the abundance of anaerobic bacteria, such as Bacteroides, Collinsella and Blautia, exhibited an increasing tendency after multiple cycles of chemotherapy. Bacteroides fragilis are opportunistic human pathogens, including enterotoxigenic B. fragilis (ETBF) and nontoxigenic B. fragilis (NTBF). ETBF was reported to mediate pathogenesis of colorectal cancer.28 A study in mice found that cepharanthine hydrochloride and cisplatin combined chemotherapy reduced the abundance of Bacteroides, indicating that cepharanthine hydrochloride can regulate the intestinal dysbiosis caused by cisplatin chemotherapy.29 Sheng et al reported that colorectal cancer patients exhibited a high abundance of Collinsella compared with healthy controls.30 Beside, Collinsella has been proven to increase intestinal permeability and enhance the pro-inflammatory effects, leading to the loss of intestinal epithelial integrity.31 Several studies have confirmed the gut commensal Blautia as a potential beneficial player that higher abundance of Blautia was correlated with reduced lethal graft versus host disease (GVHD) and improved overall survival.32 Noriho et al revealed that the abundance of Blautia in gut was associated positively with prognosis of hepatocellular carcinoma patients treated chemotherapy. Their study also demonstrated that overuse of antianaerobic drug was an independent factor of poor prognosis for hepatocellular carcinoma patients.33
Moreover, we also observed that the abundance of Veillonella, Lachnospiraceae_unclassified, Roseburia, Akkermansia and Bifidobacterium increased at the first to third cycles of chemotherapy and decreased at the subsequence cycles of chemotherapy. Veillonella has been reported to co-occur with Streptococcus in various microbial ecosystems related to humans and was believed to have metabolic interactions.34 A decreased abundance of Roseburia can predict the occurrence of inflammatory bowel disease (IBD) and they might play roles in the prevention and treatment of other diseases associated with inflammation by producing butyrate.35 Akkermansia was a potential pain-causing bacterium that can promote barrier integrity.36 A study in mice revealed that Paclitaxel could alter the brain function via decreasing the abundance of Akkermansia muciniphila, leading to neuropathic pain.37 Bifidobacterium have been noted to be key players in the intestinal microbial balance of the healthy state38 and related to the antitumor effects. A study found that Bifidobacterium-treated Taconic Farms (TAC) mice exhibited markedly improved tumor control compared with non-Bifidobacterium treated mice via modulating the anti-tumor immunity.39
Cong et al had previously revealed that Fusobacterium and Klebsiella were significantly associated with lymphatic invasion in colorectal cancer patients.40 In this study, we found certain bacterial genera that may be correlated with lymph nodes metastasis, gastrointestinal reactions and overall survival. However, the number of patients in our study is relatively small to validate the results. Yet, our study provided preliminary understanding on the relationship between gut microbiota and clinical prognosis.
Several limitations in this study should be acknowledged. First, the sample size is relatively small to validate the effects of surgery and chemotherapy on intestinal microbiota, as well as to explore the correlation between gut microbiota and clinicopathological characteristics. Thus, fecal samples of the same patient preoperative, postoperative and after every cycle of chemotherapy are necessary to collect. Second, 16S rRNA sequencing did not allow us to study the bacteria at the species level. Furthermore, we could improve it by more accurate techniques and measure metabolites of gut microbiota. Third, this study includes TC or TP chemotherapy schemes and failed to assess the effects of a specific chemotherapeutic scheme on intestinal microbiota. A well-designed chemotherapy scheme and more clinical samples are needed in the future.
Conclusion
Our study evaluated the changes of intestinal microbiota in ovarian cancer patients undergoing radical surgery and chemotherapy. The compositions of intestinal microbial in postoperative ovarian cancer patients were notably different from that in preoperative ovarian cancer patients, showing a dramatic increase of Proteobacteria, and decrease of Bacteroidetes and Firmicutes. Moreover, chemotherapy can also change the composition and diversity of gut microbiota. These results indicated that anti-cancer treatment had a non-negligible impact on the intestinal microbiota of ovarian cancer patients and gut microbiota may potentially correlate with clinical prognosis of ovarian cancer patients. Using probiotics or antibiotics during chemotherapy could potentially enhance the therapeutic efficiency in ovarian cancer patients.
Funding Statement
This study was supported by National Key Research and Development Program of China (2018YFC1004800).
Ethics Statement
IRB of Sir Run Run Shaw Hospital approved this study (20200331-43), and all study subjects provided informed consent. All procedures were in accordance with the Declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas. All authors took part in drafting, revising or critically reviewing the article. All authors gave final approval of the version to be published. All authors have agreed on the journal to which the article has been submitted and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Kurman RJ, Shih M. The dualistic model of ovarian carcinogenesis: revisited, revised, and expanded. Am J Pathol. 2016;186(4):733–747. doi: 10.1016/j.ajpath.2015.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perales-Puchalt A, Perez-Sanz J, Payne KK, et al. Frontline Science: microbiota reconstitution restores intestinal integrity after cisplatin therapy. J Leukoc Biol. 2018;103(5):799–805. doi: 10.1002/JLB.5HI1117-446RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jordan KR, Loman BR, Bailey MT, et al. Gut microbiota-immune-brain interactions in chemotherapy-associated behavioral comorbidities. Cancer. 2018;124(20):3990–3999. doi: 10.1002/cncr.31584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA Cancer J Clin. 2019;69(4):280–304. doi: 10.3322/caac.21559 [DOI] [PubMed] [Google Scholar]
- 6.Joyce K, Saxena S, Williams A, et al. Antimicrobial spectrum of the antitumor agent, cisplatin. J Antibiot (Tokyo). 2010;63(8):530–532. doi: 10.1038/ja.2010.64 [DOI] [PubMed] [Google Scholar]
- 7.Gori S, Inno A, Belluomini L, et al. Gut microbiota and cancer: how gut microbiota modulates activity, efficacy and toxicity of antitumoral therapy. Crit Rev Oncol Hematol. 2019;143:139–147. doi: 10.1016/j.critrevonc.2019.09.003 [DOI] [PubMed] [Google Scholar]
- 8.Campbell KC, Rybak LP, Meech RP, et al. D-methionine provides excellent protection from cisplatin ototoxicity in the rat. Hear Res. 1996;102(1–2):90–98. doi: 10.1016/S0378-5955(96)00152-9 [DOI] [PubMed] [Google Scholar]
- 9.Zhao L, Xing C, Sun W, et al. Lactobacillus supplementation prevents cisplatin-induced cardiotoxicity possibly by inflammation inhibition. Cancer Chemother Pharmacol. 2018;82:999–1008. doi: 10.1007/s00280-018-3691-8 [DOI] [PubMed] [Google Scholar]
- 10.Pflug N, Kluth S, Vehreschild JJ, et al. Efficacy of antineoplastic treatment is associated with the use of antibiotics that modulate intestinal microbiota. Oncoimmunology. 2016;5(6):e1150399. doi: 10.1080/2162402X.2016.1150399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaines S, Shao C, Hyman N, et al. Gut microbiome influences on anastomotic leak and recurrence rates following colorectal cancer surgery. Br J Surg. 2018;105(2):e131–e141. doi: 10.1002/bjs.10760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohigashi S, Sudo K, Kobayashi D, et al. Significant changes in the intestinal environment after surgery in patients with colorectal cancer. J Gastrointest Surg. 2013;17(9):1657–1664. doi: 10.1007/s11605-013-2270-x [DOI] [PubMed] [Google Scholar]
- 13.Lauka L, Reitano E, Carra MC, et al. Role of the intestinal microbiome in colorectal cancer surgery outcomes. World J Surg Oncol. 2019;17(1):204. doi: 10.1186/s12957-019-1754-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otasek D, Morris JH, Boucas J, et al. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019;20(1):185. doi: 10.1186/s13059-019-1758-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–563.e516. doi: 10.1016/j.cell.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou B, Sun C, Huang J, et al. The biodiversity composition of microbiome in ovarian carcinoma patients. Sci Rep. 2019;9(1):1691. doi: 10.1038/s41598-018-38031-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nam YD, H J K, J G S, et al. Impact of pelvic radiotherapy on gut microbiota of gynecological cancer patients revealed by massive pyrosequencing. PLoS One. 2013;8(12):e82659. doi: 10.1371/journal.pone.0082659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin NR, Whon TW, Bae J. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33(9):496–503. doi: 10.1016/j.tibtech.2015.06.011 [DOI] [PubMed] [Google Scholar]
- 21.Kong C, Gao R, Yan X, et al. Alterations in intestinal microbiota of colorectal cancer patients receiving radical surgery combined with adjuvant CapeOx therapy. Sci China Life Sci. 2019;62(9):1178–1193. doi: 10.1007/s11427-018-9456-x [DOI] [PubMed] [Google Scholar]
- 22.Frankel AE, Coughlin LA, Kim J, et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia. 2017;19(10):848–855. doi: 10.1016/j.neo.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan J, McKenzie C, Potamitis M, et al. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. [DOI] [PubMed] [Google Scholar]
- 24.Routy B, Gopalakrishnan V, Daillere R, et al. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 2018;15(6):382–396. doi: 10.1038/s41571-018-0006-2 [DOI] [PubMed] [Google Scholar]
- 25.Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2(2):119–129. doi: 10.1016/j.chom.2007.06.010 [DOI] [PubMed] [Google Scholar]
- 26.Deng X, Li Z, Li G, et al. Comparison of microbiota in patients treated by surgery or chemotherapy by 16S rRNA sequencing reveals potential biomarkers for colorectal cancer therapy. Front Microbiol. 2018;9:1607. doi: 10.3389/fmicb.2018.01607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexander JL, Wilson ID, Teare J, et al. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. 2017;14(6):356–365. doi: 10.1038/nrgastro.2017.20 [DOI] [PubMed] [Google Scholar]
- 28.Ou J, Carbonero F, Zoetendal EG, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr. 2013;98(1):111–120. doi: 10.3945/ajcn.112.056689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou P, Li Z, Xu D, et al. Cepharanthine hydrochloride improves cisplatin chemotherapy and enhances immunity by regulating intestinal microbes in mice. Front Cell Infect Microbiol. 2019;9:225. doi: 10.3389/fcimb.2019.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheng Q, Du H, Cheng X, et al. Characteristics of fecal gut microbiota in patients with colorectal cancer at different stages and different sites. Oncol Lett. 2019;18:4834–4844. doi: 10.3892/ol.2019.10841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J, Wright K, J M D, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8(1):43. doi: 10.1186/s13073-016-0299-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.R R J, Taur Y, S M D, et al. Intestinal blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant. 2015;21(8):1373–1383. doi: 10.1016/j.bbmt.2015.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iida N, Mizukoshi E, Yamashita T, et al. Overuse of antianaerobic drug is associated with poor postchemotherapy prognosis of patients with hepatocellular carcinoma. Int J Cancer. 2019;145(10):2701–2711. doi: 10.1002/ijc.32339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egland PG, Palmer RJ, Kolenbrander PE. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc Natl Acad Sci USA. 2004;101(48):16917–16922. doi: 10.1073/pnas.0407457101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imhann F, Vich Vila A, Bonder MJ, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2018;67(1):108–119. doi: 10.1136/gutjnl-2016-312135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cruz-Aguliar RM, Wantia N, Clavel T, et al. An open-labeled study on fecal microbiota transfer in irritable bowel syndrome patients reveals improvement in abdominal pain associated with the relative abundance of Akkermansia Muciniphila. Digestion. 2019;100(2):127–138. doi: 10.1159/000494252 [DOI] [PubMed] [Google Scholar]
- 37.Ramakrishna C, Corleto J, Ruegger PM, et al. Dominant role of the gut microbiota in chemotherapy induced neuropathic pain. Sci Rep. 2019;9(1):20324. doi: 10.1038/s41598-019-56832-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Florez AB, Sierra M, Ruas-Madiedo P, et al. Susceptibility of lactic acid bacteria, bifidobacteria and other bacteria of intestinal origin to chemotherapeutic agents. Int J Antimicrob Agents. 2016;48(5):547–550. doi: 10.1016/j.ijantimicag.2016.07.011 [DOI] [PubMed] [Google Scholar]
- 39.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cong J, Zhu H, Liu D, et al. A pilot study: changes of gut microbiota in post-surgery colorectal cancer patients. Front Microbiol. 2018;9:2777. doi: 10.3389/fmicb.2018.02777 [DOI] [PMC free article] [PubMed] [Google Scholar]