MIR15A/MIR16-1 deletion promotes multiple myeloma (MM) initiation and progression in mice, identifying it as a key tumor suppressor gene associated with human chromosome 13 loss in MM.
Abstract
The most common genetic abnormality in multiple myeloma is the deletion of chromosome 13, seen in almost half of newly diagnosed patients. Unlike chronic lymphocytic leukemia, where a recurrent minimally deleted region including MIR15A/MIR16-1 has been mapped, the deletions in multiple myeloma predominantly involve the entire chromosome and no specific driver gene has been identified. Additional candidate loci include RB1 and DIS3, but while biallelic deletion of RB1 is associated with disease progression, DIS3 is a common essential gene and complete inactivation is not observed. The Vk*MYC transgenic mouse model of multiple myeloma spontaneously acquires del(14), syntenic to human chromosome 13, and Rb1 complete inactivation, but not Dis3 mutations. Taking advantage of this model, we explored the role in multiple myeloma initiation and progression of two candidate loci on chromosome 13: RB1 and MIR15A/MIR16-1. Monoallelic deletion of Mir15a/Mir16-1, but not Rb1, was sufficient to accelerate the development of monoclonal gammopathy in wild-type mice and the progression of multiple myeloma in Vk*MYC mice, resulting in increased expression of Mir15a/Mir16-1 target genes and plasma cell proliferation, which was similarly observed in patients with multiple myeloma.
Significance:
In the absence of a defined, minimally deleted region the significance of del(13) in multiple myeloma has remained controversial. Here we show that haploinsufficiency of Mir15a/Mir16-1, but not Rb1, upregulates the cell cycle–regulatory network, inducing monoclonal gammopathy in mice and promoting multiple myeloma progression in both mice and men.
See related commentary by Walker, p. 16 .
This article is highlighted in the In This Issue feature, p. 5
Introduction
Multiple myeloma is a tumor of monoclonal antibody (mAb) producing plasma cells (PC) in the bone marrow (BM). It is preceded by a common premalignant condition called monoclonal gammopathy of undetermined significance (MGUS) that shares with multiple myeloma the presence of recurrent chromosomal translocations to the immunoglobulin loci, resulting in dysregulated expression of D-type Cyclins (CCND1–3), NSD2/MMSET and FGFR3, the MAF family of transcription factors (MAF, MAFA and MAFB), or hyperdiploidy of odd numbered chromosomes, both considered primary, tumor-initiating events (1).
We have shown that overexpression of MYC and MYC target genes distinguishes MGUS from patients with multiple myeloma (2–4). Rearrangements of the MYC locus occur in approximately 40% of patients with multiple myeloma and result in MYC-dysregulated expression, driven by a variety of juxtaposed enhancers and super-enhancers active in PCs (5). By forcing sporadic MYC activation in germinal center B cells of C57BL/6 mice, we generated a clinically predictive and biologically faithful transgenic mouse model of multiple myeloma, Vk*MYC, characterized by a progressive monoclonal PC expansion in the BM associated with the presence of abundant monoclonal immunoglobulins detectable on serum protein electrophoresis as an M-spike, as well as anemia, bone disease, and renal impairment (2, 6–8). Notably, no multiple myeloma develops in Vk*MYC mice when backcrossed from a C57BL/6 strain, prone to develop MGUS, to a Balb/c strain that does not, indicating that MYC dysregulation is not an MGUS-initiating event but rather drives the progression of MGUS to multiple myeloma (5). Additional genetic events are involved in multiple myeloma progression and the most common include activating mutations of genes in the RAS and MAPK or NFκB pathways, or inactivation of tumor suppressor genes like DIS3, FAM46C, RB1, CDKN2C, or TP53 (9).
Monosomy 13, or del(13q), occurs in approximately 40% of patients with multiple myeloma, but is found at much higher frequency in patients with t(4;14) translocations (80%–94%) or MAF family translocations (75%–80%), both considered high-risk features (10, 11). The prognostic significance of monosomy 13 remains controversial, with earlier studies indicating a lack of evidence for independent association with increased risk (12) and a more recent study showing shorter overall survival (OS) in patients with monosomy 13, even after adjusting for age, sex, International Staging System (ISS) stage, first-line therapy, and high-risk features (11).
In chronic lymphocytic leukemia (CLL) and its precursor condition, monoclonal B-cell lymphocytosis, the chromosome 13 deletion is found in 58% and 39% of cases, respectively, with a minimally deleted region (MDR) mapped to 13q14 (13). In contrast, no MDR has been identified in multiple myeloma, and most of the tumors with chromosome 13 deletion do not have mutations of RB1 or DIS3 (the two genes on chromosome 13 recurrently mutated in multiple myeloma). This suggests that haploinsufficiency of one or more tumor suppressor genes drives the loss of chromosome 13 in this disease (14).
The MIR15A/MIR16-1 cluster is located within the CLL 13q14 MDR and it has been shown to directly regulate the expression of several cell cycle–controlling genes (CCND1–3, CDK4 and 6, CHK1, MCM5, and CDC25A). Consistently, loss of MIR15A/MIR16-1 drives B-cell expansion in vitro and in vivo by promoting G0–G1–S transition in both murine and human B cells and PCs (15–17). Furthermore, Mir-15a/16-1null mice develop lymphoproliferative pathologies, although with low penetrance and indolent disease course. A minority of mice also develop tumors with plasmacytic features, suggesting a role for MIR15A/MIR16-1 deletion in PC tumorigenesis (15). Here, we take advantage of available mouse strains and examine in vivo the impact of two candidate loci on chromosome 13, RB1 and MIR15A/MIR16-1, and their respective contributions to gene expression and tumor progression in Vk*MYC mice and in a large cohorts of MGUS and multiple myeloma patients observed at Mayo Clinic or enrolled in the CoMMpass clinical trial (http://research.themmrf.org).
Results
Frequent Monosomy 14 and Focal Rb1 Deletionin Vk*MYC Mice
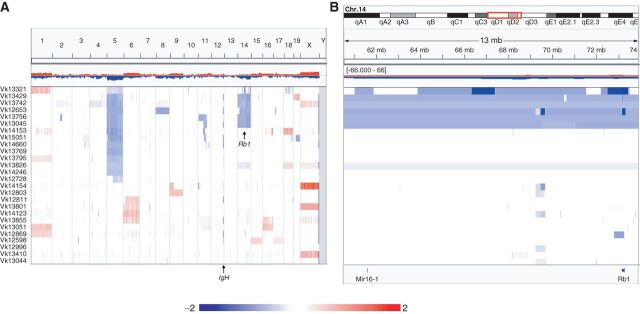
To investigate the genomic alterations that cooperate with MYC in driving multiple myeloma progression, we performed array-based comparative genomic hybridization (aCGH) analysis on 26 independent CD138-selected multiple myeloma tumors from 17 aged Vk*MYC mice (de novo tumors) and 9 Vk*MYC tumor–bearing mice (transplants) using the mouse Agilent 244K array (Fig. 1A). Visual identification of biallelic deletions within the IgH locus, physiologically occurring during B-cell development (18), confirmed the flow cytometric estimation of purity (>85%) for all but one sample (Supplementary Fig. S1A). Overall, the genetic complexity of Vk*MYC tumors is low. Like human multiple myeloma, a common abnormality identified in Vk*MYC multiple myeloma was loss of a sex chromosome: 7 of 12 male and 9 of 14 female mice had loss of Y and X, respectively. Although we found no evidence of hyperdiploidy, present in approximately 50% of human patients with multiple myeloma, we detected frequent copy number gains of chromosomes 1, 6, 16, and 18 in three, three, two, and two tumors, respectively. We also noted monosomy of chromosomes 5 and 14 in 13 and six tumors, respectively. In particular, the mouse chromosome 14 has a large MDR syntenic to the human chromosome 13, which includes Dis3, Rb1, and the Mir15a/Mir16-1 cluster. Of the six cases with monosomy 14, two had focal Rb1 biallelic deletions. In addition, one tumor without monosomy 14 presented with focal biallelic Rb1 and monoallelic Mir15a/Mir16-1deletion and another tumor with monoallelic Mir15a/Mir16-1 loss (Fig. 1B). Additional recurrent focal biallelic deletions identified in Vk*MYC tumors include Kdm6a/Utx, found in seven tumors (Supplementary Fig. S1B), and Cdkn2a, found in two tumors (Supplementary Fig. S1C). The summary of recurrent chromosome copy-number abnormalities (CNA) and biallelic deletions detected in Vk*MYC tumors is reported in Table 1. While a complete analysis of genomic aberrations in Vk*MYC multiple myeloma is ongoing, a focused examination of candidate genes on mouse chromosome 14 did not identify any single-nucleotide variants (SNV) of Rb1, Dis3, or Mir15a/Mir16-1 in 29 independent tumors.
Figure 1.
Recurrent monosomy 14, syntenic to human chromosome 13, and Rb1 focal deletions in multiple myeloma tumors from Vk*MYC mice. Graphic representation in IGV of copy number abnormalities detected by aCGH in 26 independent CD138+ multiple myeloma tumors from Vk*MYC mice shown at whole genome level (A) or zoomed on the Mir15a/16-1 and Rb1 loci on chromosome 14 (B). Red and blue indicate copy number gain and loss, respectively, and color scale (log2) is indicated. Each line represents an individual tumor sample.
Table 1.
Tabulation of recurrent CNAs observed in Vk*MYC tumors
| Sample | Origin | Sex | Trisomy | Monosomy | Sex chr. loss | Biallelic deletions |
|---|---|---|---|---|---|---|
| Vk12598 | Transplant | M | 15,17 | Y | Kdm6a | |
| Vk12653 | Transplant | M | 5,8,14 | Y | Cdkn2a, Kdm6a, Rb1 | |
| Vk12728 | De novo | F | ||||
| Vk12803 | De novo | F | 9 | X | ||
| Vk12811 | De novo | F | X | Kdm6a | ||
| V12869 | Transplant | M | 1 | Y | Rb1 (1N) | |
| V12996 | De novo | M | ||||
| Vk13044 | Transplant | F | X | |||
| Vk13045 | Transplant | F | 5,14 | X | Kdm6a | |
| Vk13051 | Transplant | F | 1,16 | X | ||
| Vk13321 | Transplant | M | 5 | Rb1, Mir15a/Mir16-1 (1N) | ||
| VK13410 | De novo | F | ||||
| Vk13429 | De novo | M | 19 | 5,14 | Y | Rb1 |
| Vk13742 | De novo | F | 5,14 | |||
| Vk13756 | De novo | M | 5,14 | Y | Kdm6a | |
| V13769 | De novo | M | 5 | |||
| Vk13795 | De novo | M | 1 | 5 | ||
| V13801 | De novo | F | 6 | |||
| V13826 | De novo | F | 18 | 5,14 | X | |
| Vk13855 | De novo | F | 6,16 | X | ||
| Vk14123 | Transplant | F | 6 | X | ||
| V14153 | De novo | M | 18 | 5 | Y | Kdm6a |
| V14154 | Transplant | F | Cdkn2a | |||
| V14246 | De novo | M | 5 | |||
| Vk14660 | De novo | F | 5 | X | Kdm6a | |
| Vk15051 | De novo | M | 5 | Y |
Analysis of Human Multiple Myeloma Identifies a Similar Frequency of DIS3 Mutation at Diagnosis and Relapse, but Increased Frequency of RB1 Inactivation with Disease Progression
Analysis of patients with multiple myeloma enrolled in the CoMMpass study identified nonsynonymous (NS) SNV of DIS3 in 10% of both newly diagnosed multiple myeloma (NDMM) and relapsed multiple myeloma (RMM) cases. These were almost exclusively missense mutations, with no nonsense or frameshift mutations, no copy-neutral loss of heterozygosity, or biallelic deletions. A third of the mutations occurred in three codons (D479, D488, and R780), almost never associated with del(13) (Fig. 2A). In contrast, the remaining two-thirds of DIS3 mutations were distributed across the various exons, almost always associated with del(13). Furthermore, the Dependency Map (http://depmap.org) identifies DIS3 as a common essential gene across almost all cell lines examined, indicating that complete inactivation is not tolerated. Altogether, this analysis highlights a complicated relationship between different DIS3 mutations and del(13). In contrast, NS-SNV of RB1 were predominantly truncating (10/19 NS-SNV are stop-gain or frameshift) and biallelic inactivation of RB1 [NS-SNV with loss of heterozygosity (LOH) or biallelic deletion] was identified in 31 of 887 (3.5%) of NDMM and 18 of 121 (15%) of RMM, where biallelic deletion predominates (12%; Fig. 2B). CoMMpass analysis also identified copy-number–dependent expression of RB1 (median log2 TPM levels for 0, 1, 2, and 3 or more copies of 1.83, 4.62, 5.41, and 6.26, respectively, with high significance according to Jonckheere–Terpstrata nonparametric trend test; Fig. 2C) with infrequent biallelic inactivation associated with poor prognosis (Fig. 2D), matching results from FoundationOne panel data (19).
Figure 2.
Rb1 haploinsufficiency does not contribute to multiple myeloma initiation and progression. A, Copy-number abnormalities and NS-SNV are shown for 91 NDMM with DIS3 mutations. Top, the complete 28kb DIS3 genomic locus; bottom, codons R780, D488, and D479. The patients are ordered vertically by degree of copy loss, with light blue indicating one copy loss, and white no copy loss; 101 missense SNVs are colored red, 2 splice-donor site SNVs purple, 2 splice-acceptor site SNVs light green, and 3 start-loss SNVs dark green. B, RB1 CNA and NS-SNV are shown for 35 NDMM (orange bar) and 20 RMM (green bar), the color scale accentuates biallelic deletion in dark blue [log2(CN/2) ≤ 1.5], one copy loss in white [−1.5<log2(CN/2) ≤ 0.5], and no copy loss in pink and red [log2(CN/2) > −0.5]. RB1 is biallelically inactivated in 31 of 924 NDMM (10 NS-SNV associated with LOH, and 21 with biallelic deletion), and in 18 of 121 RMM (3 NS-SNV associated with LOH, and 15 with biallelic deletion); 8 stop-gain SNV are colored red, 2 frameshift SNV light green, 4 missense SNV dark green, 4 splice-donor SNV pink, and 1 splice acceptor SNV brown. C, Visualization of copynumber–dependent expression of RB1 (log2 TPM) in patients with newly diagnosed multiple myeloma enrolled in the CoMMpass clinical study. Vertical lines indicate median RB1 expression for a given copy number level. D, PFS of patients with newly diagnosed multiple myeloma enrolled in the CoMMpass clinical study sorted by RB1 copy number (overall log-rank P < 0.001). E, OS in weeks of mice of the indicated genotype (n indicates number of mice analyzed for each group). F, Incidence of M-spike for the same mice as in C, detected by serum protein electrophoresis performed at the indicated week. n indicates number of sera analyzed for each group. For E and F, P values for log-rank (Mantel–Cox) test are shown with *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, nonsignificant.
Rb1 Haploinsufficiency Does Not Contribute to Myeloma Progression
The prevalence of monosomy 14 in Vk*MYC mice, coupled with Rb1 biallelic deletions (Fig. 1B), prompted us to investigate the contribution of Rb1 loss to multiple myeloma progression in Vk*MYC mice. Because the Vk*MYC mice carry LoxP sites flanking the transgenic 3′ kappa enhancer responsible for driving MYC expression in PCs (Supplementary Fig. S2), any genetic cross with mice expressing CRE recombinase in B cells will cause floxing of the transgene and suppression of MYC transcription. We therefore crossed Vk*MYC mice with constitutive Rb1het mice to generate multiple myeloma tumors lacking one copy of Rb1. Constitutive Rb1 biallelic loss is embryonic lethal, and Rb1het mice have been shown to develop pituitary tumors in which the remaining copy of Rb1 is lost (20). Consistently, we observed a shorter overall survival (OS) in Rb1het mice (median OS, 57 weeks) compared with wild-type (WT) mice (120 weeks). Loss of one Rb1 copy also significantly reduced the OS of Vk*MYC mice (83 weeks for Vk*MYC vs. 67 weeks for Vk*MYCxRb1het mice; Fig. 2E). Similar to Rb1het mice, the early mortality noted in Vk*MYCxRb1het mice was attributable to the development of pituitary tumors and not to the acceleration of multiple myeloma progression. We previously reported that multiple myeloma cells in Vk*MYC mice secrete a large amount of monoclonal immunoglobulin detectable by serum protein electrophoresis (SPEP) as M-spikes, which are a clinically useful marker for tumor burden (2). We therefore compared the incidence of M-spike development between Vk*MYC and Vk*MYCxRb1het mice by serial SPEP analysis conducted over time in cohorts of mice across genotypes. Loss of one copy of Rb1 did not accelerate the development of monoclonal gammopathy in Vk*MYC mice. Similarly, no differences were noted in the incidence of M-spike between WT and Rbhet mice (Fig. 2F). In an attempt to isolate the multiple myeloma phenotype and propagate multiple myeloma cells, we transplanted BM mononuclear cells isolated from moribund Vk*MYCxRb1het mice into WT recipient mice and monitored them by SPEP. No M-spike was detected up to 1 year after transplantation, indicating that tumor engraftment did not occur (Supplementary Fig. S3A). This was likely due to the low tumor burden of the donor mice (average BM PC content was 5.79%), which were euthanized before aggressive multiple myeloma had developed. In a retrospective analysis of Vk*MYC tumor transplantation, we observed that successful multiple myeloma engraftment into recipient mice is more likely to occur when donor organs (either spleen, BM, or lymph nodes) have higher PC content (mean PC is 13.1% for nonengrafted and 28.2% for engrafted tumors; Supplementary Fig. S3A). Significantly, the odds of engraftment for donor tumor PC content >10% were over seven times greater than for cases with donor tumor PC content <10% [OR 7.79 (1.69–35.92); Fisher P < 0.01]. On the other hand, the age of the donor mouse did not affect the ability of the tumor to engraft, although Vk*MYCxRb1het donors were significantly younger than the Vk*MYC donors in our analysis (Supplementary Fig. S3B). We therefore conclude that loss of one Rb1 copy does not contribute to multiple myeloma initiation or progression. However, the presence of focal Rb1 biallelic deletions observed in multiple myeloma tumors from Vk*MYC mice, as well as in human multiple myeloma, clearly indicates that complete Rb1 inactivation is selected by multiple myeloma cells and likely plays a role in tumor progression.
Monoallelic Loss of Mir15a/Mir16-1 Accelerates Monoclonal Gammopathy in Wild-Type C57BL/6 Mice
Having excluded RB1 as a key gene driving chromosome 13 loss in multiple myeloma, we investigated the contribution of MIR15A/MIR16-1 haploinsufficiency to multiple myeloma initiation and progression. The generation of mice carrying germline or conditional deletion of either the CLL chromosome 13 MDR, containing the Mir15a/Mir16-1 cluster and the DLeu2 gene (hereinafter referred to as MDR mice) or the Mir15a/Mir16-1 cluster alone (referred to as MIR mice) has been described, where MIR and MDR mice develop B-cell–autonomous clonal lymphoproliferative conditions with shorter OS for MDRnull mice only (15). We aged a cohort of littermate WT, MDRhet, and MDRnull mice and analyzed them by SPEP, performed every 10 weeks, for evidence of monoclonal gammopathy. C57BL/6 WT mice, known to spontaneously develop a gammopathy with age (21), had a median time to develop an M-spike (time to spike, TTS) of 96 weeks (Fig. 3A). Loss of one copy of Mir15a/Mir16-1 cluster in MDRhet mice significantly accelerated M-spike development, reducing the TTS to 57 weeks, while complete Mir15a/Mir16-1loss in MDRnull mice further reduced the TTS to 48 weeks. Notably, the intensity of the observed M-spikes remained stable over time in all the cohorts analyzed, with a gamma/albumin ratio ≦ 0.1 indicating that losing one copy of Mir15a/Mir16-1 promotes clonal PC accumulation in WT mice, but it is not sufficient to drive its malignant transformation to multiple myeloma.
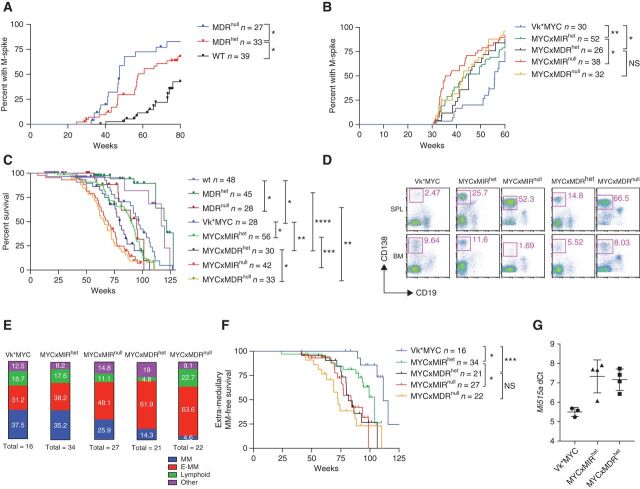
Figure 3.
Loss of one copy of the Mir15a/Mir16-1 cluster accelerates M-spike development and promotes multiple myeloma progression and extra-medullary dissemination. Incidence of M-spikes over time (weeks) in cohorts of WT, MDRhet, or MDRnull littermate mice (A) or in Vk*MYC mice alone or crossed with mice lacking one or two copies of the Mir15a/Mir16-1 cluster only (MYCxMIR) or both the dLeu2 gene and the Mir15a/Mir16-1 cluster (MYCxMDR; B). n indicates number of sera for each group analyzed by serum protein electrophoresis at the indicated week. C, Kaplan–Meier overall survival plot in weeks for the same cohort of mice as in B. n indicates number of mice in each group. D, Representative flow cytometric analysis performed on spleen and BM cells collected from moribund mice of the indicated genotype. Numbers indicate the percentage of plasma cells (CD19−CD138+) within the pink gates. E, Incidence of multiple myeloma (MM), extra-medullary multiple myeloma (E-MM), lymphoproliferative disease (Lymphoid), and other conditions (Other) detected by flow cytometric analysis performed at necropsy on moribund mice of the indicated genotype. Numbers within each box indicate the percentage of each condition, and the total number of necropsied mice is shown. F, Extra-medullary multiple myeloma–free survival (in weeks) for the same cohorts of mice as in C. n indicates number of mice in each group. G, miR-15a expression evaluated by qPCR (TaqMan assay) in multiple myeloma tumors harvested from Vk*MYC, MYCxMIRhet, or MYCxMDRhet mice. ΔCt (threshold cycle) values were normalized to U6 RNA. Each dot represents the average value from an individual tumor assayed in triplicate. Lines indicate mean with SD. P values for log-rank (Mantel–Cox) test are shown, with *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Monoallelic Loss of Mir15a/Mir16-1 Accelerates Myeloma Progression in Vk*MYC Mice
Having observed a role for Mir15a/Mir16-1 haploinsufficiency in PC expansion, we crossed both MIR and MDR constitutive mice with Vk*MYC mice and monitored them over time for multiple myeloma development and progression. Vk*MYC mice, as reported previously, developed M-spikes beginning at 30 weeks of age, with 50% of the mice having a detectable M-spike by 56 weeks (2). Biallelic loss of the miR15-a/16-1 cluster significantly accelerated the TTS, with 50% of Vk*MYCxMIRnull and Vk*MYCxMDRnull mice developing M-spikes at 36 and 43 weeks, respectively. An acceleration in M-spike development was evident even in mice with one copy of Mir15a/Mir16-1, with 50% of Vk*MYCxMIRhet and Vk*MYCxMDRhet mice developing M-spikes at 44 and 45 weeks, respectively (Fig. 3B).
Consistent with the increase in tumor burden, the copy number level of Mir15a/Mir16-1 also had a significant effect on the OS of the mice. As previously reported, loss of one copy of the MDR region did not reduce the OS of WT mice, while biallelic loss did (117 vs. 95 weeks; ref. 15). The 97-week median survival observed for the Vk*MYC mice was reduced to 90 and 68 weeks by the loss of one or two copies of the Mir15a/Mir16-1 cluster, respectively, or to 80 and 66 weeks by the loss of one or two copies of the MDR region (which includes DLeu2; Fig. 3C). Flow cytometric and histologic analysis performed at necropsy identified the presence of extra-medullary (E-MM) PC expansion, defined by the presence of more than 8% PC in extra-medullary sites (most commonly spleen and mesenteric lymph nodes), in Vk*MYC mice with monoallelic or biallelic Mir15a/Mir16-1 deletion, with increased frequency in the latter (Fig. 3D and E). Overall, loss of Mir15a/Mir16-1 did not alter the nature of MYC driven PC dyscrasia and expansion of monoclonal CD19− CD138+ PCs occurring with an approximated incidence of 70% across all the genotypes analyzed. However, loss of Mir15a/Mir16-1 accelerated its manifestation and promoted extra-medullary disease dissemination in a dose-dependent manner. Consistently, the E-multiple myeloma–free survival was significantly shorter in mice lacking one or two copies of the Mir15a/Mir16-1 cluster compared with Vk*MYC mice (Fig. 3F).
Next, we wondered whether multiple myeloma tumors that arose in Vk*MYCxMIRhet and Vk*MYCxMDRhet mice had retained the second copy of the Mir15a/Mir16-1 cluster or had acquired Mir15a/Mir16-1 complete inactivation. We employed a TaqMan qPCR assay and measured Mir15a expression in total RNA isolated from CD138+ multiple myeloma cells. ΔCt values normalized against U6 snRNA indicated a maintained expression of Mir15a in the heterozygous tumors with decreased expression levels compared with Vk*MYC tumors with two copies of Mir15a (Fig. 3G). As expected, no Mir15a expression was detected in tumors from Vk*MYCxMIRnull and Vk*MYCxMDRnull mice. We concluded that Mir15a/Mir16-1 haploinsufficiency accelerates multiple myeloma development in Vk*MYC mice.
It was previously noted that mice carrying constitutive or B-cell–specific loss of Mir15a/Mir16-1 cluster had an identical B-cell-autonomous–driven phenotype (15). Because we were not able to evaluate conditional Mir15a/Mir16-1 deletion in Vk*MYC mice, we wondered whether reduced Mir15a/Mir16-1 expression in the multiple myeloma microenvironment could contribute to the phenotype we observed. Specifically, it has been reported that multiple myeloma cell lines cocultured with BM mesenchymal cells derived from MIRnull, but not WT mice, display higher proliferation rates (22) and that Mir16 levels regulate the polarization state of tumor-promoting M2 macrophages (23, 24). We therefore compared engraftment rate in WT, MDRhet, and MDRnull littermate recipient mice transplanted with Vk12598 multiple myeloma tumors lacking del(14), and found no differences in the time of appearance of M-spikes and their progression over time among the three cohorts analyzed (Supplementary Fig. S4A), although MDRnull recipients had shorter OS (43.5 days compared with 53 days for MDRhet and 55 days for WT recipients; Supplementary Fig. S4B). We therefore concluded that although complete loss of Mir15a/Mir16-1 in the tumor microenvironment may favor myelomagenesis, Mir15a/Mir16-1haploinsufficiency has a clear tumor cell–autonomous role in promoting multiple myeloma progression. Overall, these data indicate that Mir15a/Mir16-1 regulates multiple myeloma initiation and progression in a copy number–dependent way, with a more aggressive tumor course in mice lacking both copies of the Mir15a/Mir16-1 cluster.
Loss of the Mir15a/Mir16-1 Cluster Induces Myeloma Cell Proliferation in Vk*MYC Mice
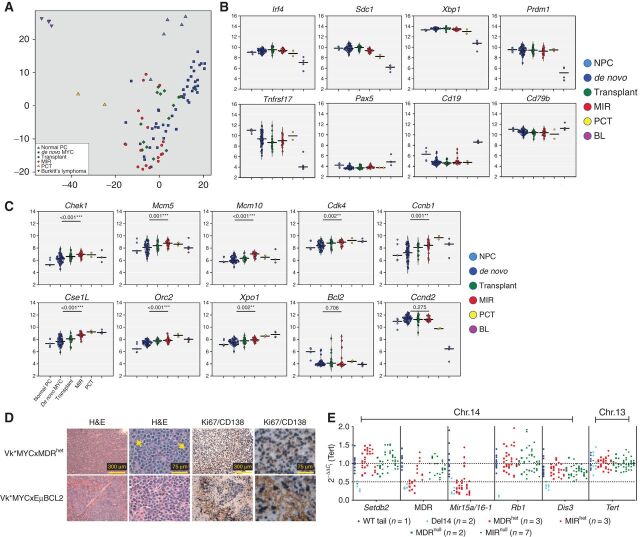
Next, we sought to characterize the effects of Mir15a/Mir16-1 deletion at the molecular level in multiple myeloma and compared gene expression profile (Affymetrix) in CD138+ PCs harvested from 45 independent Vk*MYC mice with multiple myeloma (de novo), 16 Vk*MYC tumor–bearing mice (transplants), 18 Vk*MYCxMIRnull and Vk*MYCxMDRnull de novo mice (MIR), and 5 age-matched WT controls (normal PC from BM and spleen). As a reference, we also included in our analysis two Balb/c plasmacytoma (PCT) cell lines (XRPC24 and LS136) and four Burkitt's lymphoma tumors that arose spontaneously in Vk*MYC mice in our analysis (2). Principal component analysis of differentially expressed genes clearly clustered the samples based on their cell of origin (B cell vs. PC), with the Burkitt's lymphoma samples more spatially separated and normal PCs (NPC) and PCT samples distinct from Vk*MYC–derived multiple myeloma samples, which clustered closer together (Fig. 4A). Consistently, the expression of PC (Irf4, Sdc1, Xbp1, Prdm1, Tnfrsf17) versus B-cell (Pax5, Cd19, Cd79b) lineage–specific genes underscored the terminally differentiated status of Vk*MYC and Vk*MYCxMIR multiple myeloma tumors (Fig. 4B). Analysis of differentially expressed genes in multiple myeloma cells from de novo mice lacking one or two copies of the Mir15a/Mir16-1 cluster, Vk*MYCxMIR, and Vk*MYCxMDR, compared with Vk*MYC mice with two copies of Mir15a/Mir16-1, identified cell-cycle genes previously reported to be direct Mir15a/Mir16-1 targets in CLL and multiple myeloma (Fig. 4C; Supplementary Table S1; refs. 15, 17, 25). Genes upregulated in Vk*MYCxMIR and Vk*MYCxMDR mice include Chek1, Chek2, Mcm5, Mcm10, Cdk4, and Cse1l/Xpo2, along with Mcm2 and Mcm7, Ccnb1, Ccna2, Birc5, and Cdkn2a. In addition, the origin recognition complex genes Orc5 and Orc2, which are essential for initiation of DNA replication and recruitment of MCM proteins that form the DNA helicase complex, and the melanoma-associated antigen, Magea4, shown to abrogate p53 induced cell-cycle arrest (26), were also upregulated in MIR and MDR mice tumors. Consistently, gene set enrichment analysis identified significant enrichment in additional genes in the cell-cycle and DNA replication pathways (Supplementary Fig. S5A and S5B). Unexpectedly, pathway analysis also identified upregulation of genes involved in RNA transport and protein translation initiation, including the Ras-related nuclear protein, Ran; the exportins Xpo1/Crm1 and Xpot, which shuttle RNAs and ribosomal subunits from the nucleus to the cytoplasm, hydrolize GTP, and return to the nucleus (27, 28); the mRNA export factor Rae1 and several genes encoding for the nuclear pore complex, previously implicated in tumorigenesis (29); the protein arginine methyltransferases Prmt5 and two members of the SMN complex Ddx20/Gemin3; Gemin5 required for correct assembly of the spliceosome (30); and several subunits of eukaryotic initiation factors eIF3, which control translation of cell proliferation mRNAs (31), for example, eIF5, eIF1, and eIF2b (Supplementary Fig. S6). Bcl2, Vegfa, Tab3, and Cabin1, previously reported to be direct miR15-a/16-1 targets in multiple myeloma or CLL, were not differentially expressed in MYCxMIR tumors (17, 32–34). However, Bcl2 was not generally expressed in Vk*MYC tumors, with appreciable expression in 3 of 45 de novo Vk*MYC compared with 3 of 16 transplant and 3 of 18 Vk*MYCxMIR and Vk*MYCxMDR tumors, where the expression levels were 2- to 4-fold higher. In addition, we did not observe significant upregulation of Ccnd1, -2, or -3 in Vk*MYCxMIR and Vk*MYCxMDR tumors, above the high levels of Ccnd2 found in all Vk*MYC tumors (mean log2 RMA-normalized Ccnd2 levels = 11.402; Fig. 4C). Surprisingly, we observed high levels of Ccnd2 also in five NPCs harvested from aged C57BL/6 WT mice (= 10.783), which were not seen in four Burkitt's lymphoma tumors that spontaneously developed in Vk*MYC mice (= 6.180; ref. 2).
Figure 4.
Loss of one copy of the Mir15a/Mir16-1 cluster promotes multiple myeloma cell proliferation. Visualization of the two most variable principle components from the top 5,000 most variable genes (A) and differential expression of plasma cells and B-cell lineage–specific genes (B) or cell-cycle genes (C), in NPCs from aged WT mice, de novo, or transplant Vk*MYC mice alone or crossed with mice lacking one or two copies of the Mir15a/Mir16-1 cluster only or of both the dLeu2 gene and the Mir15a/Mir16-1 cluster, all grouped together (MIR), Balb/c plasmacytoma lines (PCT) or Vk*MYC derived Burkitt's lymphoma tumors (BL). P values comparing de novo Vk*MYC to MIR mice are indicated. D, Assessment of proliferation by ki67 staining (blue) in formalinc-fixed, paraffin-embedded tumor tissues from representative Vk*MYCxMDRhet or Vk*MYCxEμBCL2 mice. Magnification bars are shown. E, qPCR analysis performed on genomic DNA from WT, Vk*MYC, Vk*MYCxMIR, or Vk*MYCxMDR mice. ΔCt values for the indicated genes, displayed in their genomic order on chromosome 14, were normalized to Tert as a diploid control. Each dot represents the calculated 2−ΔΔCt gene levels from an individual tumor assayed in triplicate. The number of independent tumors analyzed for each genotype is indicated. The dotted horizontal lines mark haploid and diploid gene levels.
Overall, the pattern of gene expression found in NPC, multiple myeloma samples harvested from de novo Vk*MYC, or from transplant Vk*MYC mice demonstrated an increasingly more proliferative disease, at least in part, supported by Cdkn2a biallelic inactivation found in transplant, but not de novo Vk*MYC tumors (Table 1), and consistent with the previously reported more aggressive and drug-resistant nature of the transplanted model (6). Interestingly, de novo Vk*MYCxMIR and Vk*MYCxMDR tumors displayed even higher levels of proliferation markers. Histologic examination of spleen sections collected from Vk*MYCxMIR and Vk*MYCxMDR mice with multiple myeloma and stained with hematoxylin and eosin revealed the presence of characteristic mitotic figures, absent in control spleen sections from age-matched Vk*MYC mice or even Vk*MYC crossed with Eμ-BCL2 mice (Vk*MYCxEμ-BCL2), chosen as a control for extra-medullary multiple myeloma (2). Furthermore, double staining with antibodies against CD138, which identifies PCs, and Ki67, a marker of cell proliferation, identified cycling multiple myeloma cells in the spleen of Vk*MYCxMIR or Vk*MYCxMDR mice, but not Vk*MYCxEμ-BCL2 mice (Fig. 4D).
Vk*MYC Tumors with Mono or Biallelic Loss of Mir15a/Mir16-1 Cluster Do Not Acquire Monosomy 14
Subsequently, we considered whether multiple myeloma tumors harvested from Vk*MYCxMIR and Vk*MYCxMDR mice developed del(14) with the same frequency as Vk*MYC mice. We designed and validated qPCR primer sets against genes distributed along chromosome 14 (Setdb2, Rb1, Dis3, as well as Mir15a/Mir16-1 and MDR) and included Tert as a normalizer diploid locus. The qPCR analysis correctly identified monosomies of chromosome 14 tested in two multiple myeloma tumors with known del(14) detected by aCGH, but not in WT genomic DNA (Fig. 4E). The assay also confirmed monosomic Mir15a/Mir16-1 and MDR loci in multiple myeloma tumors from Vk*MYCxMIRhet (n = 3) and Vk*MYCxMDRhet (n = 3) mice. However, no other regions of chromosome 14 loss were identified in any of the multiple myeloma tumors from Vk*MYCxMIR (n = 10) or Vk*MYCxMDR (n = 5) mice. Similarly, no NS-SNV were identified in targeted sequence analysis of Rb1, Dis3, or Mir15a/Mir16-1.
Deletion of MIR15A/MIR16-1 Is Present in MGUS Patients
Having identified a role for reduced Mir15a/Mir16-1 expression in monosomy 14 associated with increased multiple myeloma cell proliferation and disease progression in mice, we sought to investigate the frequency of monosomy 13 in the major molecular subtypes of MGUS and multiple myeloma. In an analysis of patients treated or observed at Mayo Clinic, patients were first classified into six distinct molecular groups based on the presence of a recurrent IgH translocation [CCND: t(11;14) and t(6;14); MAF: t(14;16) and t(14;20); MMSET: t(4;14)], or hyperdiploidy, divided into two main types based on the presence of trisomy 11 [unsupervised hierarchical clustering of copy number abnormalities in multiple myeloma by Walker and colleagues (9) identified cluster 1 (HRD11–) vs. cluster 2 (HRD11+)], with the remaining patients without hyperdiploidy or recurrent IgH translocations classified as nonhyperdiploid (nHRD2). FISH was performed on 382 patients with MGUS and 1,899 with multiple myeloma (11, 35). Similar to previous analyses (10, 11), there was a marked difference in the frequency of del(13), identified by loss of an RB1 probe, based on the molecular groups, with the lowest frequency (19%) in HRD11+ and highest (80%) in the MMSET and MAF groups (Table 2). Within each molecular group, the frequency of del(13) was similar in MGUS versus multiple myeloma, with the exception of the CCND (20% vs. 37%) and MAF (38% vs. 76%) groups where del(13) is clearly associated with progression from MGUS to multiple myeloma. We concluded that del(13) occurs at a very early stage in the pathogenesis of multiple myeloma and loss of one copy of MIR15a/MIR16-1 is likely to contribute to the immortalization of the MGUS clone, as we noted in mice with one copy of Mir15a/Mir16-1, which displayed an increased incidence in monoclonal gammopathy (Fig. 3A).
Table 2.
Frequency of del(13q) by FISH analysis in molecular subtypes of MGUS and multiple myeloma
| Total | % Total | % excluding insuff PC | % ≥1 FISH abnl | Del13/13q Present | % del13 | Fisher test | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic classification | MGUS | MM | MGUS | MM | MGUS | MM | MGUS | MM | MGUS | MM | MGUS | MM | P |
| Insufficient PC | 107 | 190 | 28% | 10% | |||||||||
| No FISH abnormality | 72 | 118 | 19% | 6% | 26% | 7% | |||||||
| CCND: t(11;14) or t(6;14) | 64 | 341 | 17% | 18% | 23% | 20% | 32% | 21% | 13 | 125 | 20% | 37% | 0.014 |
| MMSET: t(4;14) | 8 | 155 | 2% | 8% | 3% | 9% | 4% | 10% | 6 | 127 | 75% | 82% | 0.640 |
| MAF: t(14;16) or t(14;20) | 16 | 75 | 4% | 4% | 6% | 4% | 8% | 5% | 6 | 57 | 38% | 76% | 0.005 |
| HRD11+ | 43 | 444 | 11% | 23% | 16% | 26% | 21% | 28% | 7 | 86 | 16% | 19% | 0.839 |
| HRD11− | 13 | 217 | 3% | 11% | 5% | 13% | 6% | 14% | 9 | 107 | 69% | 49% | 0.253 |
| nHRD2 | 59 | 359 | 15% | 19% | 21% | 21% | 29% | 23% | 36 | 219 | 61% | 61% | 1.000 |
| Total | 382 | 1899 | 382 | 1899 | 275 | 1709 | 203 | 1591 | 77 | 721 | 38% | 45% | 0.051 |
Note: Patients assigned to mutually exclusive groups, starting from the top, after excluding those with insufficient PCs or with no FISH abnormalities. Seventeen patients had a report of hyperdiploidy alone without mention of specific chromosomes and were excluded from the analysis. HRD11+: 11+ and at one of 3+,7+,9+,15+. nHRD2, the remaining patients (no recurrent translocation or hyperdiploidy).
Abbreviation: MM, multiple myeloma.
Deletion of MIR15A/MIR16-1 Is Associated with Increased Proliferation in Patients with Multiple Myeloma
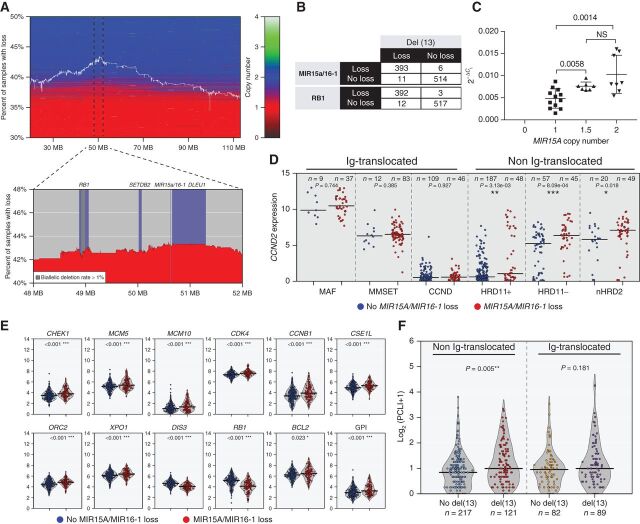
The analysis of copy number changes in the IA14 CoMMpass dataset found evidence of del(13), defined by a mono or biallelic loss of more than 10% of the length of chromosome 13, in 44% of 924 NDMM, with a newly identified peak region of deletion occurring around MIR15A/MIR16-1 (Fig. 5A). While most of the samples with loss of MIR15A/MIR16-1 or RB1 also have del(13), 6/399 and 3/395 samples, respectively, showed specific copy number loss of MIR15A/MIR16-1 or RB1 without del(13) (Fig. 5B). Having identified copy number dependent expression of MIR15A in a panel of multiple myeloma cell lines (Fig. 5C), we decided to compare gene expression in patients with multiple myeloma with and without MIR15A/MIR16-1 loss. The most striking difference we noted was the increased expression of CCND2 associated with MIR15A/MIR16-1 loss (Supplementary Fig. S7A). However, we reasoned that this observation could simply be a reflection of the fact that del(13) multiple myeloma cases are enriched by those belonging to the MAF or MMSET groups (78% of MAF, 88% of MMSET), which have a unique transcriptional profile characterized by increased CCND2 expression (36). Furthermore, other multiple myeloma cases carry IgH chromosome translocations directly dysregulating D-type cyclins, known MIR15A/MIR16-1 target genes. To isolate the transcriptional effects associated to MIR15A/MIR16-1 loss, we therefore restricted our analysis to the remaining 58% CoMMpass cases, altogether defined as non-Ig translocated and comprising HRD11+, HRD11−, and nHRD2 groups. Even in this controlled analysis, we observed significantly higher CCND2 expression associated to MIR15A/MIR16-1 loss in the non-Ig–translocated group (Fig. 5D). When we looked at the expression of other MIR15A/MIR16-1 target genes in the same group of patients, we found that MIR15A/MIR16-1 loss was strikingly associated with increased expression of the exact same proliferation and nuclear transport markers we identified in Vk*MYCxMIR and Vk*MYCxMDR mice with copy number loss of Mir15a/Mir16-1 (Figs. 4C and 5E). Overall, the upregulation of cell-cycle genes resulted in a marked increase in the gene proliferation index (GPI), as defined previously (ref. 37; Fig. 5E). In contrast, with the exception of CDK4, no difference in expression of MIR15A/MIR16-1 target genes was observed in NDMM cases with Ig translocations (Supplementary Figs. S5D and S7B). Finally, in an independent cohort of 612 multiple myeloma patients treated at the Mayo Clinic with available FISH and PC labeling index (PCLI) data collected one month before or within 6 months from initial diagnosis, we identified a significant association between presence of del(13) and increased PCLI for the non-Ig-translocated group (Fig. 5F). We therefore concluded that aberrations to chromosome 13 provide significant positive contribution to proliferation in patients with multiple myeloma lacking Ig translocation.
Figure 5.
MIR15A/MIR16-1 monosomy is associated with increased proliferation in non-Ig–translocated NDMM. A, Investigation of CoMMpass NDMM copy number levels of chromosome 13 show region surrounding MIR15A/MIR16-1 to be the most frequently lost. The white line indicates the percentage of all cases with a loss at specific locations along chromosome 13 where horizontal shading reproduces segmentation data for each patient sorted vertically according to the percentage of total loss of chromosome 13. B, Distribution of NDMM cases with or without copy number loss of MIR15A/16-1 or RB1 relative to presence or absence of del(13). C, Relative expression of MIR15a (normalized to U6 snRNA) in multiple myeloma cell lines relative to MIR15A copy number. Each dot represents the average MIR15A value calculated by TaqMan assay using the 2−ΔCt method from triplicate assays for each cell line tested. P values for nonparametric t test are shown. D, Log2-transformed TPM data for NDMM reveal increased CCND2 expression in non-Ig-translocated cases with loss of MIR15A/MIR16-1. E, Increased expression of proliferation targets (CHEK1, MCM5, MCM10, CDK4, and CCNB1), DNA replication (ORC2), nuclear transport (XPO1 and CSE1L), and antiapoptotic (BCL2) genes for NDMM non-Ig–translocated cases with loss of MIR15A/MIR16-1, resulting in overall increase in GPI. In addition, expression of markers on chromosome 13 (DIS3, RB1) shows copy number dependency. The number of patients within each group and Wilcoxon P values for each comparison are shown for D and E. F, Plasma cell labeling index calculated by flow cytometry in clonal PCs collected at the time of diagnosis in the indicated subgroups, divided into Ig-translocated and not translocated, with or without del(13) according to FISH analysis. The number of patients within each group and Wilcoxon P values for each comparison are shown.
Discussion
Genomics analysis of multiple myeloma tumors harvested from Vk*MYC mice showed similarity with human multiple myeloma. Like human multiple myeloma, Vk*MYC multiple myeloma tumors express canonical markers of PC terminal differentiation, including Sdc1, Irf4, Xbp1, Prdm1, and Tnfrsf17 (Bcma), and lack expression of B-cell markers like Cd19 and Pax5. As we have identified universal CCND dysregulation in human multiple myeloma tumors (36), we found that Vk*MYC multiple myeloma also display high levels of Ccnd2 expression. Among other genomic changes reported in human multiple myeloma, albeit at low frequency, we identified inactivation of Rb1 in three, Cdkn2a (p16/ARF) in two and of Kdm6a/Utx in seven of 26 tumors. While deletion of RB1 and CDKN2A are rare in multiple myeloma cell lines, which more frequently inactivate CDKN2C/p18, inactivation of KDM6A is quite common, at 30% to 40% (keatslab.org). Altogether, these data show that although MYC is the primary driver of Vk*MYC multiple myeloma, which is also dysregulated in approximately 40% of patients with multiple myeloma, the molecular pathogenesis of Vk*MYC multiple myeloma is heterogeneous, with some shared features with human multiple myeloma, including del(14), syntenic to human chromosome 13. More detailed genomic studies are in progress to evaluate the significance of other CNAs identified in Vk*MYC tumors, including that of monosomy 5, seen in all of the cases with monosomy 14, which has been also identified in the 5T model of multiple myeloma, with a MDR including Fgfr3 and Nsd2/Mmset syntenic to human chromosome 4 (38).
Del(13) is a commonly reported abnormality in multiple myeloma; however, without a well-defined MDR its biological consequences have remained undefined. Its clinical significance has also been controversial. Del(13) was the first chromosomal abnormality to be associated with poor prognosis in multiple myeloma or with the transition from MGUS to multiple myeloma (39, 40). However, it was later found that in the absence of other confounding high risk multiple myeloma features like del(17), t(4;14), or t(14;16) translocations, del(13) by itself was not an independent prognostic factor (41). The most frequently mutated gene on chromosome 13 is DIS3, seen in 10% of NDMM and RMM. A recurrent mutation of codon R780K has been shown to significantly alter DIS3 exoribonucleolytic activity, while other mutations (V504G, I845I) have little, if any effect (42). Interestingly, we found that recurrent mutations of three codons (R780, R488, and R479) are almost never associated with LOH, which, together with the absence of biallelic deletion of DIS3, suggests a selection for partial, but not complete inactivation of DIS3. Functional dissection in vivo of DIS3 mutations in multiple myeloma will depend on the development of mouse models that reflect this complexity. Our work aimed therefore to dissect in vivo, in a biologically faithful mouse model of multiple myeloma, the contribution of available alleles to multiple myeloma pathogenesis. First, we confirmed the Knudson hypothesis and showed that Rb1 haploinsufficiency does not promote the development of gammopathy nor contribute to multiple myeloma initiation and progression. On the other hand, biallelic Rb1 inactivation is clearly selected by tumor cells during multiple myeloma progression in patients, in Vk*MYC mice as well as in the 5T murine model of multiple myeloma (38), and RB1 biallelic inactivation confers very poor prognosis to patients with multiple myeloma. In contrast, loss of one copy of the Mir15a/Mir16-1 cluster is sufficient to promote the development of monoclonal gammopathy in WT mice and drives multiple myeloma progression and extra-medullary disease dissemination in Vk*MYC mice with high penetrance, suggesting that multiple myeloma cells have acquired the ability to expand independently on a permissive BM niche. Interestingly, the presence of the MYC transgene is dominant over the loss of Mir15a/Mir16-1 and clearly dictates the phenotype of the resulting tumors, which are universally multiple myeloma and not CLL. Importantly, while monosomy of chromosome 14, syntenic to human chromosome 13, spontaneously occurs in a fraction of multiple myeloma tumors from Vk*MYC mice, it never develops in Vk*MYCxMIR or Vk*MYCxMDR tumors lacking one or both copies of the Mir15a/Mir16-1 cluster, suggesting that loss of MIR15A/MIR16-1 expression drives the selection for chromosome 13 loss in human multiple myeloma.
Direct Mir15a/Mir16-1 target genes have been previously identified in various model systems, and converge on regulators of cell-cycle progression. Consistently, in a comparative analysis of murine and human multiple myeloma, we show that loss of MIR15A/MIR16-1 is associated with increased multiple myeloma cell proliferation and more aggressive disease course. To our knowledge, this is the first time that a specific gene signature associated to MIR15A/MIR16-1 loss is identified in murine or human tumors. The clinical significance of such increased proliferation, noted in two independent datasets and with two different methodologies, remains to be systematically determined, as well as the specific effects of MIR15A/MIR16-1 expression on multiple myeloma survival. In previous studies, the levels of exosomal MIR-16 were found to be a significant risk factor for progression free survival, but not OS, in a univariate analysis adjusted for ISS stage and cytogenetic abnormalities, while high levels of circulating miR-16 in the serum of patients with NDMM were independent prognosticators of OS (43, 44). Furthermore, the presence of del(13) was found to be independently associated with shorter overall and progression-free survival (11) and in a new model aimed to predict risk of progression of patients with smoldering multiple myeloma, presented by the International Myeloma Working Group, t(4;14), t(14;16), 1q gain, or del(13) were identified in a multivariate Cox regression analysis to predict outcome (45).
As we defined a role for Mir15a/Mir16-1 in the development of monoclonal gammopathy in mice, we sought to assess the contribution of del(13) in MGUS. The frequency of del(13) between MGUS and multiple myeloma was similar in most genetic subgroups, indicating that del(13) is an early event in multiple myeloma pathogenesis. Consistently, PC levels of MIR15 and MIR16 were not found to vary between MGUS and multiple myeloma (46). However, it was clearly underrepresented in the CCND and MAF subgroups of patients, suggesting that these MGUS patients with del(13) are more likely to rapidly progress to multiple myeloma.
Overall, we identify a role for del(13) in multiple myeloma pathogenesis in all but the MMSET subgroups, where the very low number of patients lacking del(13) reduces the power of the analysis. We also wonder whether loss of MIR15A/MIR16-1 in the MMSET subgroup represents a prerequisite initiating event leading to the expansion of the pool of germinal center B cells amenable to acquire t(4;14) translocation. It is also possible that loss of expression of other genes on chromosome 13 drives the selection for del(13) in these tumors, which are associated with higher frequency of mutations in DIS3 (9).
In summary, we show that one of the most common genomic abnormalities in multiple myeloma, del(13), is driven in large part by loss of one copy of the MIR15A/MIR16-1 cluster, resulting in reduced miR-15a/16-1 levels and increased expression of cell-cycle–regulatory genes, which drive disease progression. Therefore, while the contribution to multiple myeloma progression of other genes on chromosome 13 (e.g., DIS3) is likely, this study establishes a prominent role for the loss of MIR15A/MIR16-1 in multiple myeloma pathogenesis.
Methods
Mice
All in vivo experiments were performed under the Mayo Foundation Institutional Animal Care and Use Committee approval. The generation, characterization, and maintenance of Vk*MYC mice (GenBank accession no. MN894515), Vk*MYCxEμBCL2 mice, and of the Vk12598 transplantable line have been reported elsewhere (2, 6, 8). The B6.129S2-Rb1tm1Tyj/J mice that constitutively lack one Rb1 allele (20), here referred to as Rbhet mice, were purchased from The Jackson Laboratory (stock 02102). Mice that constitutively carry a deletion of the dLeu2 gene and Mir15a/Mir16-1 cluster, here referred to as MDR mice, or the Mir15a/Mir16-1 cluster only, MIR mice, were kindly donated by Riccardo Dalla-Favera, Institute for Cancer Genetics, Columbia University, New York, NY (15). All mice were maintained in C57BL/6J strain and crossed with each other or with Vk*MYC mice. SPEP was periodically performed on sex-matched mice as described previously (8). Mice were necropsied when moribund to assess the presence of phenotypic abnormalities by flow cytometry and IHC (6, 8). Kaplan–Meier survival curves were generated using Prism software (GraphPad Software Inc.) and compared by log-rank test.
aCGH of Murine Multiple Myeloma Tumors
High-resolution aCGH was performed on Puregene (Qiagen) purified DNA obtained from CD138 selected (8) multiple myeloma cells from 27 mice using the SurePrint G3 Mouse CGH 244K Microarray kit (Agilent Technologies). Digestion, labeling, and hybridization steps were done according to the manufacturer's protocols with some modifications. Briefly, 1.2 μg of DNA from CD138 selected Vk*MYC multiple myeloma tumors or pooled spleens of young transgenic mice were separately digested with bovine DNase I (Ambion) for 12 minutes at room temperature. Next, random primers and exo-Klenow Fragment were used to differentially label tumor (Cy5) and reference (Cy3) genomic DNA samples (Agilent Technologies). Labeled genomic reactions were cleaned up with Microcon YM-30 Columns (Millipore) and hybridized at 65°C for 40 hours. Microarrays were scanned in a DNA Microarray Scanner (Agilent Technologies). Feature extraction was performed with Feature Extraction Software, version 9.5 (Agilent Technologies). Log2 ratio data were imported and analyzed using DNA Analytics Version 4.0.85 software (Agilent Technologies). Copy-number abnormalities were calculated using aberration detection module-1 algorithm with a threshold of 7.5. A 2 probe, 0.25-log2 filters were used in the aberration detection, obtaining an average genomic resolution of 17 kb. Visualization of the data aligned to mm10 was performed using IGV software (Broad Institute). The data have been submitted with GEO submission #GSE110954.
Gene Expression Profiling of Murine Multiple Myeloma Samples
RNA from CD138-selected multiple myeloma samples was extracted from TRIzol and further purified on Purelink Micro-Mini Columns (Invitrogen), with an On-Column DNAse Digestion Step. Gene expression profiling was performed on the Affymetrix mouse 430 2.0 array following the manufacturer's suggested protocol. Datasets have been deposited to GEO under submission no. GSE111921, and further analysis was conducted on a subset of samples restricted to normal PC, Vk*MYC, Vk*MYCxMIR, or Vk*MYCxMDR tumors. Gene-level expression was derived from CEL files using custom CDF from brainarray, version 23, and RMA normalization (47). Gene-set enrichment analysis was performed using the GAGE (generally applicable gene set enrichment for pathway analysis) R package (48).
Quantization of miR-15a Expression in Murine Multiple Myeloma Tumors
Ten nanograms of TRIzol-extracted RNA from CD138-selected multiple myeloma samples were retro-transcribed to cDNA using the TaqMan microRNA Reverse Transcription kit with RT stem-loop primers specific to mature miR-15a (assay 0389) or U6 snRNA (assay 01973, both Applied Biosystems), according to the manufacturer's instructions. The obtained cDNA was diluted 1:3 and 3 μL were amplified in a TaqMan PCR reaction using a Rotor-Gene 3000 instrument (Corbett Research). Relative miR-15a expression was normalized to U6 snRNA and quantified using the ΔCt method (miR-15a Ct – U6 Ct). Each sample was tested in triplicate.
Assessment of Chromosome 14 Ploidy by qPCR in Murine Multiple Myeloma Tumors
The efficiencies of SYBR Green PCR reactions for each target gene were calculated on serially diluted WT DNAs, and the accepted Ct range for each reaction was determined using a Rotor-Gene 3000 instrument (Corbett Research). Then, 3 ng of genomic tumor DNA were amplified in triplicate using the validated primer set. The ploidy of chr14 genes was calculated by the 2−ΔΔCt method against Tert2, chosen as normal diploid control because it is located in a genomic locus not affected by copy number changes. Primer sequences and PCR efficiency are listed in Supplementary Table S2.
Copy-number–dependent Quantization of MIR15A Expression in Human Myeloma Cell Lines
Human myeloma cell lines have been described previously (49) and were maintained in RPMI1640 supplemented with 5% FBS and glutamine without antibiotics. None of these lines are listed on the International Cell Line Authentication Committee database of commonly misidentified cell lines and are routinely fingerprinted by assessment of copy-number polymorphisms by PCR and tested for Mycoplasma contamination using the MycoAlert kit (Promega). The copy number for MIR15A in HMCL was extracted from the segmented copy number data described previously (49) using IGV, Gitools Heatmap, Export Gene Matrix TDM (50). RNA was extracted from five million logarithmically growing cells using the MirVana miRNA isolation kit (Invitrogen) following manufacturer's instructions. Relative MIR15A expression was normalized to the average of U6 snRNA and RNU48 snoRNA (assay 01006) expression and quantified using the 2−ΔCt method. Each sample was tested in triplicate.
Genomic Analysis of Human Multiple Myeloma Tumors
Clinical variables and genetic data, including whole-exome sequencing (WES), biallelic frequency segmented WES for estimation of LOH, RNA-Seq (Salmon TPM), WGS-segmented files (copy number estimates), and long-insert WGS were downloaded from MMRF CoMMpass (IA14 release data from http://research.themmrf.org). From the available samples, we focus on 924 baseline BM samples with available whole-exome or whole-genome copy number data and RNA-Seq Salmon TPM expression data. For analysis of samples at disease progression, WES on 146 samples from 121 patients from IA15 release was used.
Segmentation Analysis of Human Chromosome 13 in Multiple Myeloma Tumors
Del(13) determinations are based on the total length of chromosome 13′s q arm with a copy-number level, according to whole-exome segmentation file, below a given threshold, where samples with del(13) have 10% or more of the span of 13 below 1.30. This del(13) determination is positive for 98.5% of cases with loss of MIR15A/MIR16-1, 99% of cases with loss of RB1, and 100% of cases with loss of DIS3. To graphically search for a minimally deleted region of 13, copy-number levels from segmentation files were queried at 25k base-pair increments across all of 13, with even greater refinement for the 14q.2 band and the section of 13 surrounding MIR15A/MIR16-1 (Fig. 5A).
Disclosure of Potential Conflicts of Interest
S.K. Kumar is a consultant/advisory board member at Celgene, Takeda, AbbVie, Roche, GlaxoSmithKline, and Amgen. No potential conflicts of interest were disclosed by the other authors.
Authors' Contributions
M. Chesi: Conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review, and editing. C.K. Stein: Conceptualization, data curation, formal analysis, writing–original draft, writing–review, and editing. V.M. Garbitt: Investigation. M.E. Sharik: Investigation. Y.W. Asmann: Data curation, formal analysis, investigation, and methodology. M. Bergsagel: Methodology. D.L. Riggs: Writing–review and editing. S.J. Welsh: Writing–review and editing. E.W. Meermeier: Writing–review and editing. S.K. Kumar: Resources, data curation, and formal analysis. E. Braggio: Data curation, formal analysis, investigation, methodology, writing–review, and editing. P.L. Bergsagel: Data curation, formal analysis, supervision, writing–review, and editing.
Supplementary Material
Visualization in IgV of aCGH data
Diagram of Vk*MYC vector
Engraftment comparison of Vk*MYC donor mice
Engraftment of Vk12598 tumor in WT, MDRhet or MDRnull recipient mice
GSEA and KEGG cell cycle pathway analysis
KEGG RNA transport pathway
GEP of MIR target genes in NDMM
Gene differentially expressed in Vk*MYC versus Vk*MYCxMIR mice
Primer sequences and efficiency
Acknowledgments
We are grateful to Riccardo Dalla-Favera for providing the MIR and MDR mice. We thank W. Mike Kuehl for critical interpretation of the data. This work has been supported by the following NIH/NCI grants: CA234181 (to M. Chesi, C.K. Stein, V.M. Garbitt, M.E. Sharik, E.W. Meermeier, and M. Bergsagel), CA186781 (to M. Chesi, C.K. Stein, D.L. Riggs, and P.L. Bergsagel), CA190045 (to M. Chesi, V.M. Garbitt, and M.E. Sharik), and CA224018 (to C.K. Stein, D.L. Riggs, and P.L. Bergsagel).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online https://bloodcancerdiscov.aacrjournals.org/.
References
- 1. Kuehl WM, Bergsagel PL. Molecular pathogenesis of multiple myeloma and its premalignant precursor. J Clin Invest 2012;122:3456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chesi M, Robbiani DF, Sebag M, Chng WJ, Affer M, Tiedemann R, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell 2008;13:167–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chng WJ, Huang GF, Chung TH, Ng SB, Gonzalez-Paz N, Troska-Price T, et al. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia 2011;25:1026–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Misund K, Keane N, Stein CK, Asmann YW, Day G, Welsh S, et al. MYC dysregulation in the progression of multiple myeloma. Leukemia 2020;34:322–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Affer M, Chesi M, Chen WD, Keats JJ, Demchenko YN, Tamizhmani K, et al. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia 2014;28:1725–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chesi M, Matthews GM, Garbitt VM, Palmer SE, Shortt J, Lefebure M, et al. Drug response in a genetically engineered mouse model of multiple myeloma is predictive of clinical efficacy. Blood 2012;120:376–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calcinotto A, Ponzoni M, Ria R, Grioni M, Cattaneo E, Villa I, et al. Modifications of the mouse bone marrow microenvironment favor angiogenesis and correlate with disease progression from asymptomatic to symptomatic multiple myeloma. Oncoimmunology 2015;4:e1008850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chesi M, Mirza NN, Garbitt VM, Sharik ME, Dueck AC, Asmann YW, et al. IAP antagonists induce anti-tumor immunity in multiple myeloma. Nat Med 2016;22:1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walker BA, Mavrommatis K, Wardell CP, Ashby TC, Bauer M, Davies FE, et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 2018;132:587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiecchio L, Dagrada GP, Ibrahim AH, Dachs Cabanas E, Protheroe RK, Stockley DM, et al. Timing of acquisition of deletion 13 in plasma cell dyscrasias is dependent on genetic context. Haematologica 2009;94:1708–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Binder M, Rajkumar SV, Ketterling RP, Greipp PT, Dispenzieri A, Lacy MQ, et al. Prognostic implications of abnormalities of chromosome 13 and the presence of multiple cytogenetic high-risk abnormalities in newly diagnosed multiple myeloma. Blood Cancer J 2017;7:e600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood 2007;109:3489–95. [DOI] [PubMed] [Google Scholar]
- 13. Rawstron AC, Bennett FL, O'Connor SJ, Kwok M, Fenton JA, Plummer M, et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med 2008;359:575–83. [DOI] [PubMed] [Google Scholar]
- 14. Fonseca R, Oken MM, Harrington D, Bailey RJ, Van Wier SA, Henderson KJ, et al. Deletions of chromosome 13 in multiple myeloma identified by interphase FISH usually denote large deletions of the q arm or monosomy. Leukemia 2001;15:981–6. [DOI] [PubMed] [Google Scholar]
- 15. Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, et al. The DLEU2/miR-15a/16–1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 2010;17:28–40. [DOI] [PubMed] [Google Scholar]
- 16. Lindner SE, Lohmuller M, Kotkamp B, Schuler F, Knust Z, Villunger A, et al. The miR-15 family reinforces the transition from proliferation to differentiation in pre-B cells. EMBO Rep 2017;18:1604–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roccaro AM, Sacco A, Thompson B, Leleu X, Azab AK, Azab F, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood 2009;113:6669–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bergsagel PL, Kuehl WM. Degree of focal immunoglobulin heavy chain locus deletion as a measure of B-cell tumor purity. Leukemia 2013;27:2067–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chavan S, He J, Tytarenko R, Deshpande S, Patel P, Bailey M, et al. Bi-allelic inactivation is more prevalent at relapse in multiple myeloma, identifying RB1 as an independent prognostic marker. Blood Cancer J 2017;7:e535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature 1992;359:295–300. [DOI] [PubMed] [Google Scholar]
- 21. Radl J, Hollander CF. Homogeneous immunoglobulins in sera of mice during aging. J Immunol 1974;112:2271–3. [PubMed] [Google Scholar]
- 22. Roccaro AM, Sacco A, Maiso P, Azab AK, Tai YT, Reagan M, et al. BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J Clin Invest 2013;123:1542–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jia X, Li X, Shen Y, Miao J, Liu H, Li G, et al. MiR-16 regulates mouse peritoneal macrophage polarization and affects T-cell activation. J Cell Mol Med 2016;20:1898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khalife J, Ghose J, Martella M, Viola D, Rocci A, Troadec E, et al. MiR-16 regulates crosstalk in NF-kB tolerogenic inflammatory signaling between myeloma cells and bone marrow macrophages. JCI Insight 2019;4(21):e129615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, et al. MiR-15a and miR-16–1 cluster functions in human leukemia. Proc Natl Acad Sci U S A 2008;105:5166–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bhan S, Chuang A, Negi SS, Glazer CA, Califano JA. MAGEA4 induces growth in normal oral keratinocytes by inhibiting growth arrest and apoptosis. Oncol Rep 2012;28:1498–502. [DOI] [PubMed] [Google Scholar]
- 27. Sun Q, Chen X, Zhou Q, Burstein E, Yang S, Jia D. Inhibiting cancer cell hallmark features through nuclear export inhibition. Signal Transduct Target Ther 2016;1:16010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schmidt J, Braggio E, Kortuem KM, Egan JB, Zhu YX, Xin CS, et al. Genome-wide studies in multiple myeloma identify XPO1/CRM1 as a critical target validated using the selective nuclear export inhibitor KPT-276. Leukemia 2013;27:2357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rodriguez-Bravo V, Pippa R, Song WM, Carceles-Cordon M, Dominguez-Andres A, Fujiwara N, et al. Nuclear pores promote lethal prostate cancer by increasing POM121-driven E2F1, MYC, and AR nuclear import. Cell 2018;174:1200– 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett 2008;582:1997–2003. [DOI] [PubMed] [Google Scholar]
- 31. Lee AS, Kranzusch PJ, Cate JH. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 2015;522:111–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A 2005;102:13944–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun CY, She XM, Qin Y, Chu ZB, Chen L, Ai LS, et al. miR-15a and miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis 2013;34:426–35. [DOI] [PubMed] [Google Scholar]
- 34. Zhang L, Zhou L, Shi M, Kuang Y, Fang L. Downregulation of miRNA-15a and miRNA-16 promote tumor proliferation in multiple myeloma by increasing CABIN1 expression. Oncol Lett 2018;15:1287–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lakshman A, Paul S, Rajkumar SV, Ketterling RP, Greipp PT, Dispenzieri A, et al. Prognostic significance of interphase FISH in monoclonal gammopathy of undetermined significance. Leukemia 2018;32:1811–5. [DOI] [PubMed] [Google Scholar]
- 36. Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J Jr. Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood 2005;106:296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hose D, Reme T, Hielscher T, Moreaux J, Messner T, Seckinger A, et al. Proliferation is a central independent prognostic factor and target for personalized and risk-adapted treatment in multiple myeloma. Haematologica 2011;96:87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maes K, Boeckx B, Vlummens P, De Veirman K, Menu E, Vanderkerken K, et al. The genetic landscape of 5T models for multiple myeloma. Sci Rep 2018;8:15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Avet-Loiseau H, Li JY, Morineau N, Facon T, Brigaudeau C, Harousseau JL, et al. Monosomy 13 is associated with the transition of monoclonal gammopathy of undetermined significance to multiple myeloma. Intergroupe Francophone du Myelome. Blood 1999;94:2583–9. [PubMed] [Google Scholar]
- 40. Tricot G, Barlogie B, Jagannath S, Bracy D, Mattox S, Vesole DH, et al. Poor prognosis in multiple myeloma is associated only with partial or complete deletions of chromosome 13 or abnormalities involving 11q and not with other karyotype abnormalities. Blood 1995;86:4250–6. [PubMed] [Google Scholar]
- 41. Gutierrez NC, Castellanos MV, Martin ML, Mateos MV, Hernandez JM, Fernandez M, et al. Prognostic and biological implications of genetic abnormalities in multiple myeloma undergoing autologous stem cell transplantation: t(4;14) is the most relevant adverse prognostic factor, whereas RB deletion as a unique abnormality is not associated with adverse prognosis. Leukemia 2007;21:143–50. [DOI] [PubMed] [Google Scholar]
- 42. Tomecki R, Drazkowska K, Kucinski I, Stodus K, Szczesny RJ, Gruchota J, et al. Multiple myeloma-associated hDIS3 mutations cause perturbations in cellular RNA metabolism and suggest hDIS3 PIN domain as a potential drug target. Nucleic Acids Res 2014;42:1270–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Manier S, Liu CJ, Avet-Loiseau H, Park J, Shi J, Campigotto F, et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood 2017;129:2429–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rocci A, Hofmeister CC, Geyer S, Stiff A, Gambella M, Cascione L, et al. Circulating miRNA markers show promise as new prognosticators for multiple myeloma. Leukemia 2014;28:1922–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miguel JS, Mateos MV, Gonzalez V, Dimopoulos MA, Kastritis E, Hajek R, et al. Updated risk stratification model for smoldering multiple myeloma (SMM) incorporating the revised IMWG diagnostic criteria. J Clin Oncol 2019;37(15_suppl):8000. [Google Scholar]
- 46. Seckinger A, Meissner T, Moreaux J, Benes V, Hillengass J, Castoldi M, et al. miRNAs in multiple myeloma–a survival relevant complex regulator of gene expression. Oncotarget 2015;6:39165–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 2005;33:e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 2009;10:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell 2007;12:131–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Visualization in IgV of aCGH data
Diagram of Vk*MYC vector
Engraftment comparison of Vk*MYC donor mice
Engraftment of Vk12598 tumor in WT, MDRhet or MDRnull recipient mice
GSEA and KEGG cell cycle pathway analysis
KEGG RNA transport pathway
GEP of MIR target genes in NDMM
Gene differentially expressed in Vk*MYC versus Vk*MYCxMIR mice
Primer sequences and efficiency



![Figure 2. Rb1 haploinsufficiency does not contribute to multiple myeloma initiation and progression. A, Copy-number abnormalities and NS-SNV are shown for 91 NDMM with DIS3 mutations. Top, the complete 28kb DIS3 genomic locus; bottom, codons R780, D488, and D479. The patients are ordered vertically by degree of copy loss, with light blue indicating one copy loss, and white no copy loss; 101 missense SNVs are colored red, 2 splice-donor site SNVs purple, 2 splice-acceptor site SNVs light green, and 3 start-loss SNVs dark green. B, RB1 CNA and NS-SNV are shown for 35 NDMM (orange bar) and 20 RMM (green bar), the color scale accentuates biallelic deletion in dark blue [log2(CN/2) ≤ 1.5], one copy loss in white [−1.5<log2(CN/2) ≤ 0.5], and no copy loss in pink and red [log2(CN/2) > −0.5]. RB1 is biallelically inactivated in 31 of 924 NDMM (10 NS-SNV associated with LOH, and 21 with biallelic deletion), and in 18 of 121 RMM (3 NS-SNV associated with LOH, and 15 with biallelic deletion); 8 stop-gain SNV are colored red, 2 frameshift SNV light green, 4 missense SNV dark green, 4 splice-donor SNV pink, and 1 splice acceptor SNV brown. C, Visualization of copynumber–dependent expression of RB1 (log2 TPM) in patients with newly diagnosed multiple myeloma enrolled in the CoMMpass clinical study. Vertical lines indicate median RB1 expression for a given copy number level. D, PFS of patients with newly diagnosed multiple myeloma enrolled in the CoMMpass clinical study sorted by RB1 copy number (overall log-rank P < 0.001). E, OS in weeks of mice of the indicated genotype (n indicates number of mice analyzed for each group). F, Incidence of M-spike for the same mice as in C, detected by serum protein electrophoresis performed at the indicated week. n indicates number of sera analyzed for each group. For E and F, P values for log-rank (Mantel–Cox) test are shown with *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, nonsignificant.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c445/8500718/ea4f96ba52f2/68fig2.jpg)