SUMMARY
Megasporogenesis is essential for female fertility, and requires the accomplishment of meiosis and the formation of functional megaspores. The inaccessibility and low abundance of female meiocytes make it particularly difficult to elucidate the molecular basis underlying megasporogenesis. We used high-throughput tag-sequencing analysis to identify genes expressed in female meiocytes (FMs) by comparing gene expression profiles from wild-type ovules undergoing megasporogenesis with those from the spl mutant ovules, which lack megasporogenesis. A total of 862 genes were identified as FMs, with levels that are consistently reduced in spl ovules in two biological replicates. Fluorescence-assisted cell sorting followed by RNA-seq analysis of DMC1:GFP-labeled female meiocytes confirmed that 90% of the FMs are indeed detected in the female meiocyte protoplast profiling. We performed reverse genetic analysis of 120 candidate genes and identified four FM genes with a function in female meiosis progression in Arabidopsis. We further revealed that KLU, a putative cytochrome P450 monooxygenase, is involved in chromosome pairing during female meiosis, most likely by affecting the normal expression pattern of DMC1 in ovules during female meiosis. Our studies provide valuable information for functional genomic analyses of plant germline development as well as insights into meiosis.
Keywords: female meiocyte, megasporogenesis, meiosis, transcriptome, Arabidopsis
INTRODUCTION
As in other angiosperms, Arabidopsis has two morphologically distinct gametophytes that are embedded within different sexual organs of a flower. The female meiocyte (also called the megasporocyte or megaspore mother cell, MMC) differentiate from an archesporial cell arising in the subepidermal cell layer at the tip of each ovule primordium. Female meiosis produces a tetrad of four haploid megaspores. Three of the megaspores degenerate and the persistent functional megaspore undergoes three sequential rounds of mitotic division, forming the female gametophyte: the seven-celled embryo sac (Yadegari and Drews, 2004). Male meiocytes (also called the microsporocytes or pollen mother cells, PMCs) differentiate from progenitor cells in the anther primordium and undergo meiosis to form a tetrad of four haploid microspores. Each microspore undergoes asymmetric mitosis to produce a vegetative cell and a generative cell. The generative cell undergoes one more round of mitosis to from two sperm cells, and the resulting male gametophyte, the pollen grain, is composed of a three-celled male germ unit (McCormick, 1993). Therefore, both male and female gametophyte development occur over two phases: the first phase, microsporogenesis or megasporogenesis, which starts from reproductive organ differentiation, and ends after meiosis by haploid spore formation; and the second phase, microgametogenesis or megagametogenesis, which consists of several rounds of haploid cell mitosis, leading to the formation of mature gametes.
Complex gene regulatory networks are expected to be required for the highly coordinated processes of cell division, differentiation, expansion, and degeneration that occur in both female and male gametophyte development. Transcriptome analyses of male and female gametophytes at different developmental stages revealed the distinct sets of genes that are expressed in these two types of gametophytes, and provided a framework for the elucidation of transcriptional networks that are linked to cellular identity and gene function during gametogenesis (Hennig et al., 2004; Honys and Twell, 2004; Pagnussat et al., 2005; Pina et al., 2005; Yu et al., 2005; Johnston et al., 2007; Steffen et al., 2007; Wuest et al., 2010; Sanchez-Leon et al., 2012). Sporogenesis occurs early in development within tissues of specialized floral organs. Microsporogenesis takes place in the anther at stages 1–7 of anther development (Sanders et al., 1999), involving the stages from the initiation of stamen primordium to meiosis (Ma, 2005). Comparative transcriptomic studies using wild type (WT) and microsporogenesis-arrested mutant anthers identified groups of male meiocyte-enriched genes (Alves-Ferreira et al., 2007; Wijeratne et al., 2007). Recent genome-wide gene expression analyses of isolated microsporocytes uncovered novel regulators that are involved in the control of key developmental processes during microsporogenesis (Chen et al., 2010; Libeau et al., 2011; Yang et al., 2011). In contrast, little is known about genes that are implicated in the control and elaboration of the early steps of female gamete production, especially during megasporogenesis. The inaccessibility, small size and low abundance of female meiocytes have hampered the elucidation of the molecular basis of early female reproductive development for decades. Molecular mechanisms and gene regulatory networks controlling megasporogenesis remain elusive.
Here, we identified genes expressed in female meiocyte (called FMs) through gene profiling by comparing WT ovules undergoing megasporogenesis with spl ovules, which lack megasporogenesis, using high-throughput tag-sequencing (Tag-seq). The expression of FMs in female meiocyte was confirmed by RNA-seq analysis of the fluorescence-assisted cell-sorted female meiocytes. We functionally validated the identified candidate genes by reverse-genetic analysis and identified four genes involved in megasporogenesis. We further revealed that a cytochrome P450, KLU, regulates chromosome pairing during female meiosis, and that this is likely to occur by modulating the expression of DMC1 in a cell type-specific manner in Arabidopsis ovules. Our studies provide valuable information to gain a deeper understanding of the regulatory networks underlying megasporogenesis.
RESULTS
Identification of genes expressed in female meiocytes (FMs)
We performed differential gene expression profiling to identify genes with higher expression levels in WT ovules with placenta (simplified as ovule) undergoing megasporogenesis, compared with those from homozygous spl mutants, using high-throughput Tag-seq analysis. spl is a recessive sporophytic mutation that completely abolishes both male and female sporogenesis (Schiefthaler et al., 1999; Yang et al., 1999b). At stages 10–11, WT flower ovules develop into stages 2I–2IV (Figure S1a–d). During these stages, the archesporial cell elongates, differentiates into the female meiocyte and then undergoes meiosis (Smyth et al., 1990; Schneitz et al., 1995). In homozygous spl mutants, female meiocyte development is impaired in the archesporial cell, which fails to undergo female meiocyte differentiation and subsequent meiosis (Figure S1e–h). Two biological replicates of ovules at stages 2II–2IV from WT and spl buds (Smyth et al., 1990; Schneitz et al., 1995; Christensen et al., 1997) were collected for RNA extraction. Typically, ~15 μg of total RNA was obtained from the ovule tissue of 220 pistils, and ~6 μg of total RNA for each sample was used for the 3′-tag digital gene expression library construction and Illumina sequencing analysis. The high-throughput sequencing data were reproducible, and Pearson correlation coefficients in the two WT and spl replicates were 0.94 and 0.80, respectively (Figure S2).
To identify female meiocyte-expressed genes (FMs), we performed statistical analysis of the sequence data to screen for genes exhibiting reduced expression in spl ovules relative to WT ovules (Audic and Claverie, 1997). We first used stringent values for the false discovery rate (FDR) < 0.001 and for log2 WT/SPL ≥ 1 as the threshold to estimate the FMs in both replicates. A total of 79 genes satisfied this criterion (Table S1). To avoid missing potentially important but moderately expressed genes that had been excluded by the stringent cut-off because of the variation in expression between replicates, we used less stringent criteria to define the FMs: (i) in both replicates, expression levels are reduced in spl ovules compared with WT ovules;(ii) at least in one replicate, expression levels are twofold greater in WT ovules compared with spl mutant ovules, and the FDR value is lower than 0.001. With these criteria, 862 genes were identified as FMs (Table S2). This less stringent cut-off is the result of balancing the number of false negatives and true FMs. We compared our data set with previously published data and found that out of the 862 FMs, 487 genes (Table S3) were detected in the laser-captured MMC microarray profiling (Schmidt et al., 2011), and 835 genes (Table S3) were present in the male meiocyte RNA-seq profiling (Chen et al., 2010; Yang et al., 2011). These data suggest that a subset of candidate genes involved in megasporogenesis was revealed by the Tag-seq-based comparative gene expression profiling analysis, and they indicate that most of the FMs are also expressed in male meiocytes.
Functional classification of the FMs
We functionally classified the 862 identified FMs on the basis of their biological or biochemical function using the gene ontology (GO) annotation of the Arabidopsis genome provided by the Arabidopsis Information Resource (http://www.arabidopsis.org). We found that nine GO terms, including those related to biogenesis, metabolism, structure and transporter activity, were greatly over-represented in female meiocytes compared with the Arabidopsis genome (P < 0.05 regarded as significantl P = 0.05465 as nearly significant; Table 1). This finding suggested that female meiocytes are synthetically and metabolically very active, and that material transport and gene expression are highly promoted to support the progression of megasporogenesis. The complete genes list within each over-represented GO term is provided in Table S4.
Table 1.
Gene ontology analysis of female meiocyte-expressed genes (FMs)
| GO term | GO categorya | Significant | Expected | P |
|---|---|---|---|---|
| Structural molecule activity | MF | 36 | 15.2 | 0.0004 |
| Protein transmembrane transporter activity | MF | 8 | 1.6 | 0.0547 |
| Cellular component biogenesis at cellular level | BP | 22 | 8.0 | 0.0076 |
| Ribonucleoprotein complex biogenesis | BP | 17 | 5.5 | 0.0142 |
| Macromolecule metabolic process | BP | 249 | 204 | 0.0168 |
| Non-membrane-bounded organelle | CC | 63 | 31.6 | 1.3e−05 |
| Ribosome | CC | 36 | 15.5 | 0.0003 |
| Intracellular organelle part | CC | 131 | 91.0 | 0.0007 |
| Nucleus | CC | 48 | 24.4 | 0.0009 |
GO category classifications: BP, biological process; CC, cellular component; MF, molecular function. P < 0.05, significant; P = 0.0547, near significant.
We next searched for Pfam-defined protein domains that are over-represented in female meiocytes. A total of 624 Pfam domains were found in the 862 FMs, indicating that the transcriptome of female meiocytes are functionally complex (Table S5). Nine Pfam domains were significantly over-represented (a family-wise error rate ≤0.05) among the FM genes (Table 2), including energy transfer (mitochondrial carrier), biosynthesis (cosyl transferase), DNA binding (WRC and HMG-box), calmodulin binding and protein–protein interaction (QLQ and WD40). These findings suggest the involvement of diverse molecular regulation mechanisms during megasporogenesis, and the large number of FMs identified in this study may provide a framework for the functional analysis of the genes implicated in megasporogenesis.
Table 2.
The nine most enriched Pfam domains in Arabidopsis female meiocytes
| Pfam_domain | Observed | Expected | P |
|---|---|---|---|
| Mito_carr | 19 | 5.3 | 1.86e−6 |
| Glycos_transf_4 | 3 | 0.1 | 3.10e−5 |
| WRC | 5 | 0.4 | 3.17e−5 |
| QLQ | 4 | 0.3 | 0.0001 |
| CaM_binding | 4 | 0.3 | 0.0002 |
| HMG box | 5 | 0.6 | 0.0002 |
| Ribosomal_S13 | 3 | 0.2 | 0.0003 |
| WD40 | 17 | 6.7 | 0.0005 |
Validation of comparative gene expression profiling of the FMs
To verify differentially expressed genes obtained from the Tag-seq analysis, we randomly selected 11 downregulated genes in spl ovules from the 862 identified FMs, and two unchanged genes and one upregulated gene in spl ovules, for quantitative RT-PCR (qRT–PCR) analysis. The relative expression levels for the 14 genes in WT and spl ovules were consistent with the Tag-seq data (Figure 1a; Table S2). We also performed three-dimensional whole-mount in situ hybridization analyses for two genes (AT1G15460 and AT2G33750) that were downregulated in spl ovules compared with WT ovules. As predicted, both genes were expressed in WT female meiocytes (Figure 1b,c). To investigate the expression pattern of the FMs in the whole plant, we also analyzed the expression levels of six spl downregulated genes in different tissues by qRT-PCR and found that all of the six FM genes are not only expressed in meiosis ovules, but also in mature ovules, anthers and vegetative tissues (Figure 1d).
Figure 1.
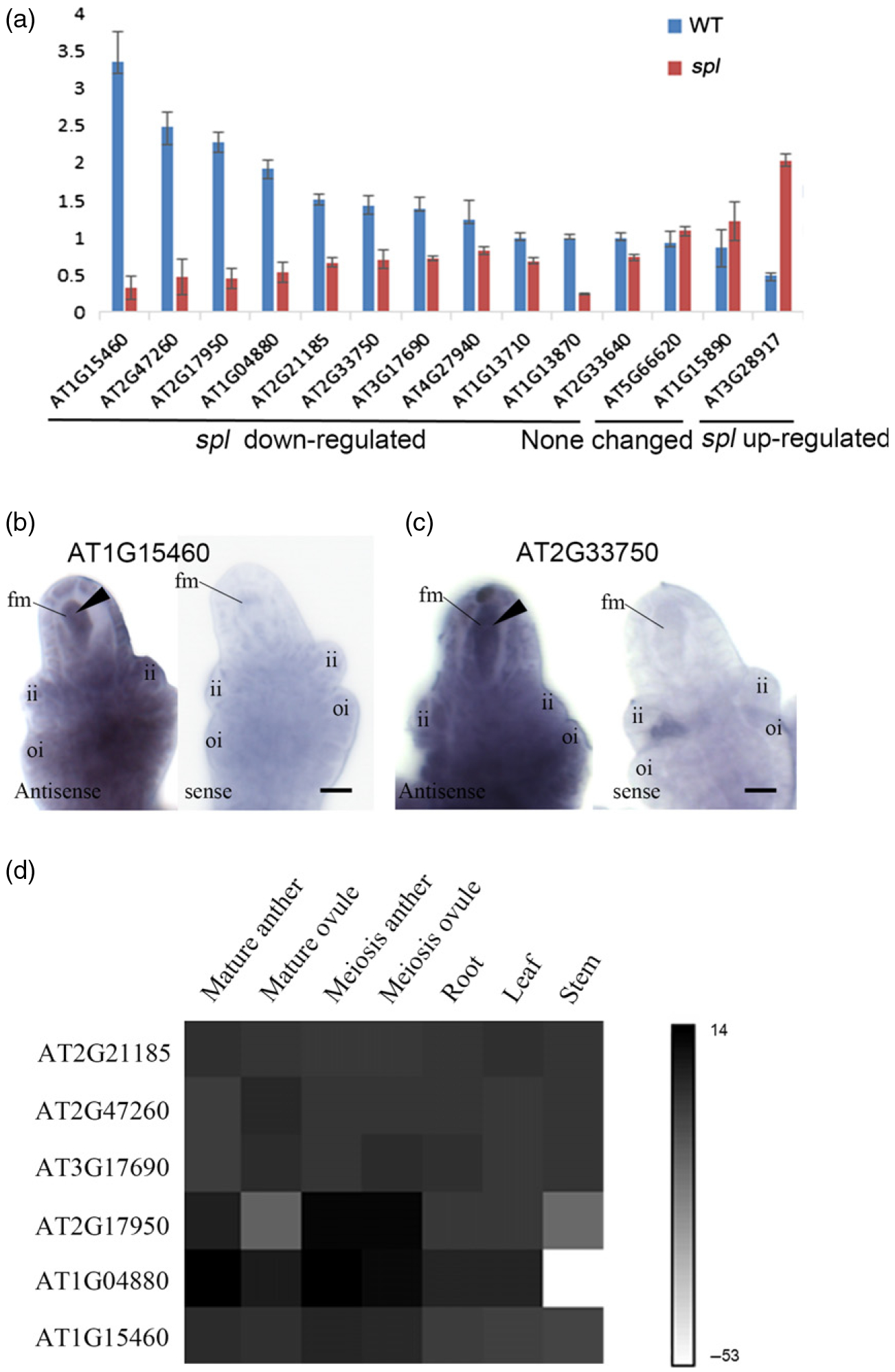
Validation of sequencing results by real-time PCR and in situ hybridization.
(a) Transcript levels of 14 genes in wild-type (WT) and spl ovules. Transcript levels were measured by qRT-PCR and normalized to HK2. Each value represents the mean of two biological replicates and two technical replicates. Error bars represent standard deviation.
(b) Whole-mount in situ hybridization of At1g15460.
(c) Whole-mount in situ hybridization of At2 g33750. Arrowheads point to the enriched expression pattern in the female meioctye (fm). Scale bars: 10 μm. Abbreviations: ii, inner integument; oi, outer integument.
(d) Heat map of the relative expression levels of six FM genes in different tissues measured by qRT-PCR. Colors are scaled by log2-transformed mean expression values from two biological replicates and two technical replicates. Black denotes high expression and white denotes low expression.
DMC1 encodes a RecA homolog and is expressed specifically in female meiocyte in ovule tissues (Figure 2a; Klimyuk and Jones, 1997; Pittman et al., 1998; Couteau et al., 1999). To further evaluate whether FMs are indeed expressed in female meiocytes, we performed fluorescence-assisted cell sorting to isolate DMC1:GFP-labeled female meiocytes (Figure 2b,c; Qin et al., 2014). Following RNA extraction and amplification, transcript-profiling studies based on RNA-seq analysis were performed. Of the 862 FMs identified, 775 (90%) were detected at the levels of at least one read per kilo-base of mRNA length per million mapped reads (RPKM ≥ 1) in the female meiocyte RNA-seq database in two biological replicates (Table S6). Taken together, the results of qRT-PCR and in situ RNA hybridization are consistent with the Tag-seq data, indicating high reliability of the comparative transcriptome analysis in this study.
Figure 2.

Fluorescence-assisted cell sorting of DMC1:GFP-labeled female meiocytes.
(a) In ovules at stage 2III/IV, DMC1:GFP is detected exclusively in the female meiocyte (fm).
(b) Protoplasts released from ovules after enzyme digestion.
(c) Sorted female meiocytes with DMC1:GFP signal. Scale bars: 20 μm.
Reverse genetic analysis of the FMs
To assess the function of the FMs during megasporogenesis, we performed reverse genetic analysis on a selected set of FMs using T-DNA insertional mutants. Considering the difficulty in completing reverse genetic analysis for all 862 genes, we subjected our data set to several filtering criteria to narrow the scope of our initial functional studies. First, we filtered out the genes that were not consistently detected in several independent analyses, and narrowed our sample down to the 448 genes (Table S7) that are present in all four data sets: (i) spl downregulated genes that are most likely expressed in female meiocytes (Table S2);(ii) genes that are expressed in the sorted female meiocytes identified in this study (Table S6); (iii) previously reported laser-dissected MMC-expressed genes (Schmidt et al., 2011); and (iv) ovule primordial-expressed genes (Matias-Hernandez et al., 2010). Although more than 400 genes were efficiently excluded by this filter, it should be noted that genes that are not represented on the ATH1 array, not preserved after paraffin embedding and laser dissection, and not expressed in megasprocyte protoplasma were also excluded by this filter. Further investigations would be necessary to determine whether these genes are required for megasporogenesis. To identify the genes that are most likely to be involved in megasporogenesis, we next filtered out genes that are enriched in the developing female gametophyte (Yu et al., 2005), generated from the functional megaspore produced after megasporogenesis in mature embryo sacs (Johnston et al., 2007; Steffen et al., 2007) or in female gametophyte cells (Wuest et al., 2010). This further reduced the number of candidate genes to 414 (Table S7). We were able to obtain T-DNA insertional mutants for 120 genes (Table S7) of these 414 genes from Joseph Ecker’s SALK confirmed T-DNA collection (CS27941, CS27942 and CS27943; ABRC, http://abrc.osu.edu). The 120 mutant lines were named fm mutants (Table S7). We then screened for reduced fertility phenotypes that could result from mutations affecting megasporogenesis. For every T-DNA line, 18 individual plants were planted and three immature siliques per plant were dissected to identify consistent sterility defects in the sibling adult plants. We discarded two mutant lines with aborted brown seeds (Table S7), likely to have resulted from embryo lethality. We also discarded five mutant lines that only displayed one or two plants with reduced fertility (Table S7), suggesting that the T-DNA insertion and the reduced seed-set phenotype are not linked. Finally, four mutants with consistently reduced fertility phenotypes were obtained (Table S7). We then further confirmed the correlation between the insertion site and the reduced fertility phenotype for the four mutant lines. RT-PCR on flower-bud cDNA from homozygote mutant plants indicated that the expressions of the corresponding four female meiocyte-expressed genes, FM5, FM12, FM13 and FM43, in the screened mutant lines are indeed knocked out (Figure S3; Table 3).
Table 3.
Reduced fertility mutants corresponding to female meiocyte-expressed genes (FMs)
| Mutant name | Insertional line | Gene ID | log2 Ratio (WT/spl) | Gene description |
|---|---|---|---|---|
| fm5 | SALK_057612C | At1 g04880 | 2.4 | HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain |
| fm12 | SALK_024697C | At1 g13710 | 2.0 | CYP78A5, KLU | cytochrome P450, family 78, subfamily A, polypeptide 5 |
| fm13 | SALK_140551C | At1 g13870 | 2.2 | DRL1, AtKTI12 | calmodulin binding; purine nucleotide binding |
| fm43 | SALK_016521C | At2 g33640 | 2.7 | DHHC-type zinc finger family |
To establish whether the observed fertility deficiency resulted from male or female defects, we performed reciprocal crosses for these mutant lines. Our data, summarized in Table 4, indicate that reduced fertility in three out of the four mutants (fm5, fm12 and fm13) was observed only when a homozygous mutant was used as a female parent, suggesting that defects result from the female. Self-pollinated heterozygous fm/+ mutants exhibited full fertility, indicating that these mutant female gametes (fm5, fm12 and fm13) are fully functional (Table 4). Reciprocal crosses between fm43 and WT plants showed that both male and female reproductive tissues were completely infertile (Table 4). In contrast to the three mutants described above, heterozygous fm43/+ pistils pollinated with WT pollen result in a low seed set phenotype (Table 4). In the progeny of self-pollinated fm43/+ heterozygous plants, the segregation ratio of +/+ : fm43/+ : fm43/ fm43 (22 : 42 : 5) is distorted from the expected 1 : 2 : 1 ration according to Mendelian law, suggesting that fm43 mutation causes gametophytic defects. We further tested the transmission efficiency of fm43 through male and female by determining the segregation ratio of the F1 progeny of the reciprocal crossed siliques between heterozygous fm43/+ and WT (Table 4) through PCR-based genotyping. No significant differences from the expected 1 : 1 segregation of fm43/+ : +/+ (175 : 192) were detected when fm43/+ was used as the male donor (χ2 test; P > 0.05). Whereas distorted segregation of fm43/+ : +/+ (36 : 78) was detected when fm43/+ was used as the female donor (χ2 test; P < 0.01). These data suggested that the function of the female gametophyte is disrupted more severely than that of the male gametophyte in the fm43/+ mutant. Taken together, the mutation in fm5, fm12 and fm13 are female sporophytic, whereas the mutation in fm43 is gametophytic.
Table 4.
Reduced seed-set phenotypes result from female sporophytic tissue in fm5, fm12 and fm13, and male and female defects in fm43
| Female parent | Male parent | Pollination | Developed seeds (%) | Undeveloped ovules (%) | Siliques counted (n) |
|---|---|---|---|---|---|
| +/+ | +/+ | Auto self | 57.4 ± 3.1 (99.1) | 0.6 ± 0.7 (0.9) | 20 |
| +/+ | +/+ | Manual self | 49.8 ± 3.9 (93.1) | 3.7 ± 1.6 (6.9) | 10 |
| fm5/+ | fm5/+ | Auto self | 57.2 ± 2.9 (98.6) | 0.8 ± 0.8 (1.4) | 10 |
| fm5/fm5 | fm5/fm5 | Auto self | 45.6 ± 2.6 (87.2) | 6.7 ± 2.1 (12.8) | 10 |
| +/+ | fm5/fm5 | Manual | 52.9 ± 3.3 (91.5) | 4.9 ± 2.5 (8.5) | 12 |
| fm5/fm5 | +/+ | Manual | 36.7 ± 4.2 (74.0) | 12.9 ± 2.7 (26.0) | 10 |
| fm12/+ | fm12/+ | Auto self | 57.6 ± 3.6 (98.3) | 1.0 ± 0.8 (1.7) | 10 |
| fm12/fm12 | fm12/fm12 | Auto self | 28.6 ± 2.9 (62.6) | 17.1 ± 5.5 (37.4) | 11 |
| +/+ | fm12/fm12 | Manual | 52.0 ± 3.8 (91.4) | 4.9 ± 2.9 (8.6) | 13 |
| fm12/fm12 | +/+ | Manual | 15.1 ± 5.3 (45.9) | 32.8 ± 4.5 (68.5) | 16 |
| fm13/+ | fm13/+ | Auto self | 61.7 ± 3.4(98.1) | 1.2 ± 0.9 (1.9) | 10 |
| fm13/fm13 | fm13/fm13 | Auto self | 32.1 ± 5.6(55.1) | 26.8 ± 7.5 (45.5) | 14 |
| +/+ | fm13/fm13 | Manual | 50.2 ± 5.1 (93.4) | 3.54 ± 1.7 (6.6) | 13 |
| fm13/fm13 | +/+ | Manual | 31.1 ± 6.3 (60.7) | 20.1 ± 8.5 (39.3) | 10 |
| fm43/+ | fm43/+ | Auto self | 29.1 ± 3.2 (53.1) | 25.7 ± 2.8 (46.9) | 10 |
| +/+ | fm43/+ | Manual | 47.4 ± 3.1 (92.2) | 4.0 ± 1.0 (7.8) | 16 |
| fm43/+ | +/+ | Manual | 24.7 ± 2.8 (45.4) | 29.8 ± 2.9 (54.6) | 13 |
| fm43/fm43 | fm43/fm43 | Auto self | 0 (0) | 45.9 ± 2.8 (100) | 10 |
| +/+ | fm43/fm43 | Manual | 0 (0) | 54.1 ± 2.9 (100) | 11 |
| fm43/fm43 | +/+ | Manual | 0 (0) | 46.3 ± 2.7 (100) | 9 |
Parent genotypes were confirmed by PCR and progeny segregation.
Megasporogenesis is affected in fm mutants
To further investigate the defects of female gametophyte development in the four identified fm mutants, we examined female gametophyte structure using both confocal laser scanning microscopy (CLSM) and differential interference contrast (DIC) microscopy. In mature female gametophyte development stage 7 (FG7; Christensen et al., 1997), WT female gametophyte consists of a central cell, an egg cell and two synergids (Figures 3a and S4a). In contrast,13.6% of fm5, 39.8% of fm12, 45.7% of fm13 and 45.4% of fm43/+ mutant ovules exhibited an absence of the female gametophyte (Figures 3a,c and S4a), which consequently causes a failure in penetration by the pollen tube and subsequent abortion.
Figure 3.
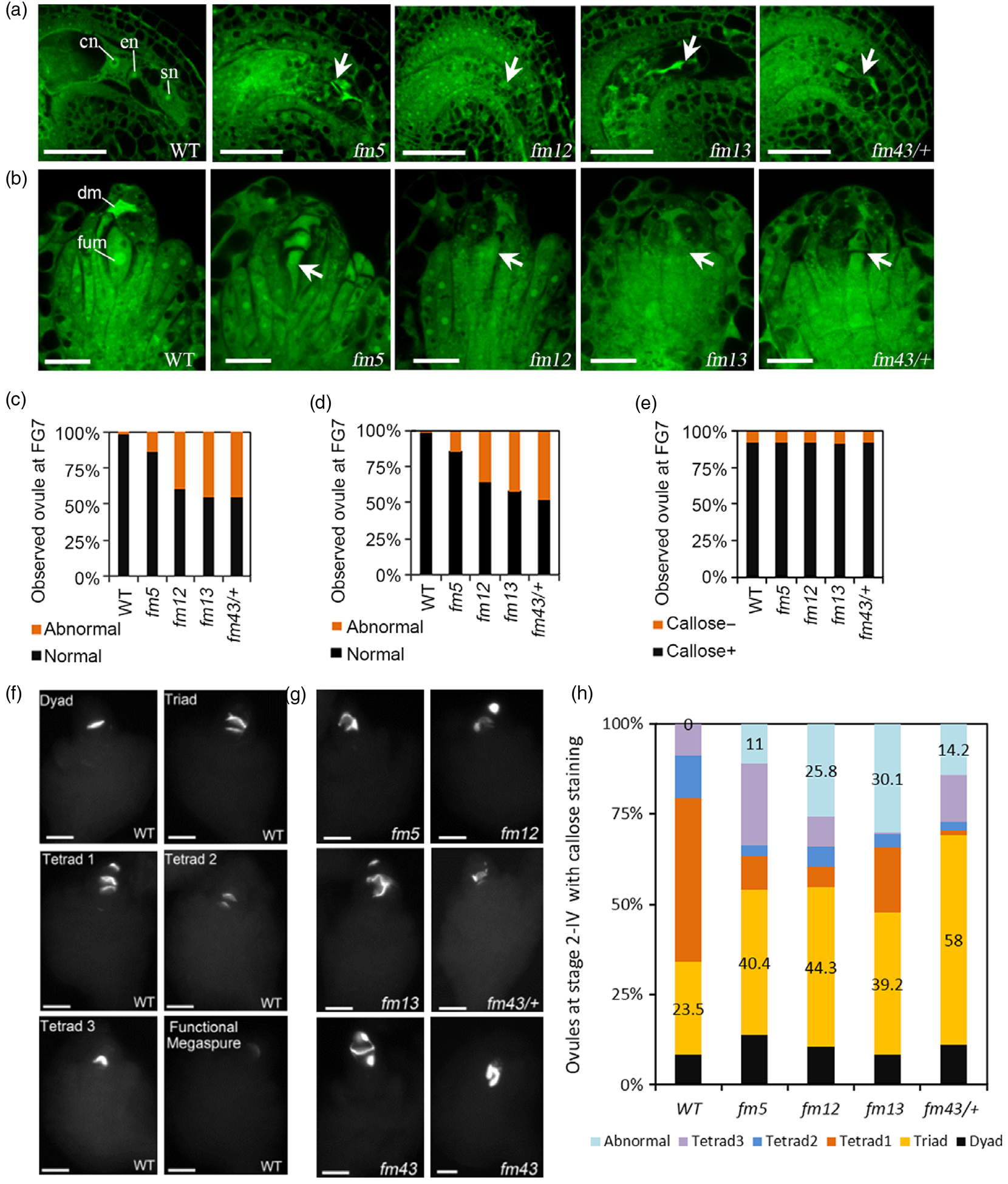
Female gametophyte development and callose deposition during megasporogenesis are defective in the fm5, fm12, fm13 and fm43/+ mutant ovules.
(a) and (b) CLSM analysis of female gametophyte in wild-type (WT) and fm5, fm12, fm13 and fm43/+ mutant ovules at stage FG7 and FG1, respectively. Arrows indicate the absence of a female gametophyte in (a), or an aborted functional megaspore in (b). Abbreviations: cn, central cell nucleus; dm, degenerating megaspore cells; en, egg cell nucleus; fum, functional megaspore; sn, synergid nucleus.
(c, d) Quantification of normal and aberrant female gametophyte at FG7 and FG1, respectively, in more than 100 ovules for each sample.
(e) Quantification of callose staining-positive (+) and callose staining-negative (–) ovules at stage 2IV, in which female meiocyte undergoes meiosis, in more than 100 ovules for each sample.
(f) Callose-stained wall deposition in WT ovules at stage 2IV.
(g) Abnormal callose staining is observed in fm5, fm12, fm13, fm43/+ and fm43 ovules during meiosis. Callose persists in fm43 ovule at later stages of development (bottom right).
(h) Quantitative profile of callose deposition at various stages of meiosis in more than 100 ovules for each sample. Scale bars: (a) 30 μm; (b) 10 μm; (f, g) 20 μm.
To determine when the defects of female gametophyte development manifest, we examined the female gametophyte structure of the WT and fm mutants at the early stages of female gametophyte development. In WT ovules at female gametophyte development stage 1 (FG1; Christensen et al., 1997), the female gametophyte is uninucleate. The functional megaspore produced after meiosis is in a teardrop shape and is accompanied with degenerated megaspores in the distal end (Figures 3b and S4b). A subset of fm5 (14.4%), fm12 (35.3%), fm13 (42.1%) and fm43/+ (47.9%) mutants contained aborted functional megaspores (Figures 3b,d and S4b), suggesting that defects in these mutants manifest before the functional megaspore is formed, probably during female meiosis. Microspore tetrad appeared normal in the anthers of these four mutants (Figure S5), implying that male meiosis is not affected in these mutants, whereas the fm43/+ plant produced 28.4% (119/419) of aborted pollen (Figure S5), indicating that pollen development after meiosis is affected by fm43 mutation. The comparable male transmission efficiency of fm43/+ to wild type, however, as shown above, suggests that the fm43 mutant pollen tubes have even higher competence to target female gametophyte than wild-type pollen tubes, probably with higher growth speed, as the fewer fm43 male gametes fertilized female gametes as efficiently as wild-type male gametes.
Cytological analysis of female meiosis was conducted to determine whether the fm mutations affected megasporogenesis. We first investigated whether female meiocytes in the fm ovules were competent to enter meiosis by monitoring the presence of callose, a convenient cytological marker for meiosis as it is deposited at the cell plate of cells undergoing meiosis prior to cytokinesis. Comparable percentages of callose-positive staining were scored in WT, fm5, fm12, fm13 and fm43/+ ovules at stage 2IV (Figure 3e; Christensen et al., 1997), suggesting that female meiocytes in these four fm mutants entered meiosis similarly to WT. To determine whether subsequent steps in meiosis were affected in the fm mutant ovules, we examined callose deposition during the progression of meiosis. In WT ovule (Figure 3f), the callose signal first appears at the cell plate that separates the two daughter cells of female meiocyte in a dyad, followed by accumulation at the cell plates that separate the cells of a triad and a tetrad (called tetrad 1). After meiosis, the callose signal disappeared from the cell plate separating the four megaspores forming tetrad 2 and tetrad 3. When a functional megaspore was formed, the callose signal almost disappeared (Figure 3f). In the fm mutants (Figure 3g), depositions of callose during megasporogenesis were markedly different from those observed in the WT: fm5 (11.0%), fm12 (25.8%), fm13 (30.1%) and fm43/+ (14.2%) ovules exhibited abnormal callose staining (Figure 3g), suggesting that cell plate formation during megasporogenesis is disturbed in the four fm mutants. In addition to the aberrant callose-staining signal during the meiosis of fm43 ovules, we observed that callose fluorescence persists in fm43 ovules, even at later stages of development (Figure 3g, bottom right), beyond the time when it would normally be found in the WT. Moreover, upon analysis of more than 100 ovules at stage 2IV, increased percentages of the fm5 (40.4%), fm12 (44.3%), fm13 (39.2%) and fm43/+ (58.0%) mutant ovules analyzed were at the triad stage compared with the WT (23.5%; Figure 3h, Table S8), implying that the progression of meiosis is perturbed in these fm mutant ovules.
KLU is required for chromosome pairing and organization at female meiosis I
Considering the difficulty in characterizing meiotic defects in all four fm mutants, we selected fm12 for fluorescence in situ hybridization (FISH) using a 180-bp centromeric repeat-specific probe (Zhang et al., 2012), as FISH helps in distinguishing megasporocytes from the somatic cells (Armstrong et al., 2001), enabling a more reliable analysis of meiotic progression. The gene mutated in fm12 encodes the cytochrome P450 monooxygenase CYP78A5 named KLU, the promoter activity of which was detected at the base of the nucellus in the region initiating the inner integument flanking the female meiocytes in a previous study (Adamski et al., 2009). We hypothesize that klu may provide a good tool to study cell–cell communication between the female meiocyte and the surrounding somatic cells during megasporogenesis.
In leptotene, WT spreads (Figure 4a,e) showed between eight and 10 unpaired centromeres. In the pachytene stage of prophase I, full synapsis between homologous chromosomes resulted in five pairs of centromeres (Figure 4b,f). Centromere pairing persisted even as chromosomes condensed during the diakinesis stage of prophase I (Figure 4c,g). Beginning at late diakinesis, the homologous centromeres were separated such that 10 signals were typically observed from metaphase I (Figure 4d,h). In klu, FISH analysis identified two types of female meiocytes at prophase I: some were similar to those in the WT (Figure 4i–p), but the rest contained defective centromere morphology (Figure 4q–t). We observed that klu female meiocytes at pachytene exhibited six centromeres, containing two unpaired centromeres (Figure 4q,r, white arrows). Diakinesis in some of the klu female meiocytes also contained a mixture of paired and unpaired centromeres (Figure 4s,t, white arrows). Out of the 59 observed klu female meiocytes, 16 (27.1%) exhibited abnormal centromere behaviors. In contrast to the disturbed female meiosis in klu female meiocytes, we failed to identify differences between WT and klu male meiosis by centromere FISH analysis on more than 100 microsporocytes (Figure S6). Taken together, these results indicated that chromosome pairing and organization are perturbed in klu female meiosis, whereas male meiosis during microsporogenesis does not appear to be affected in klu plants. To further prove that the klu mutation is responsible for these meiotic defects, we complemented the klu reduced-fertility phenotype by transforming full-length genomic fragments of KLU driven by its own promoter into mutant plants. Ten transgenic lines were obtained and the seed set of the klu KLU:KLU was restored to 93.2–100% in independent lines, compared with 62.6% in klu plants, indicating a role for KLU in megasporogenesis through the control of female meiosis.
Figure 4.
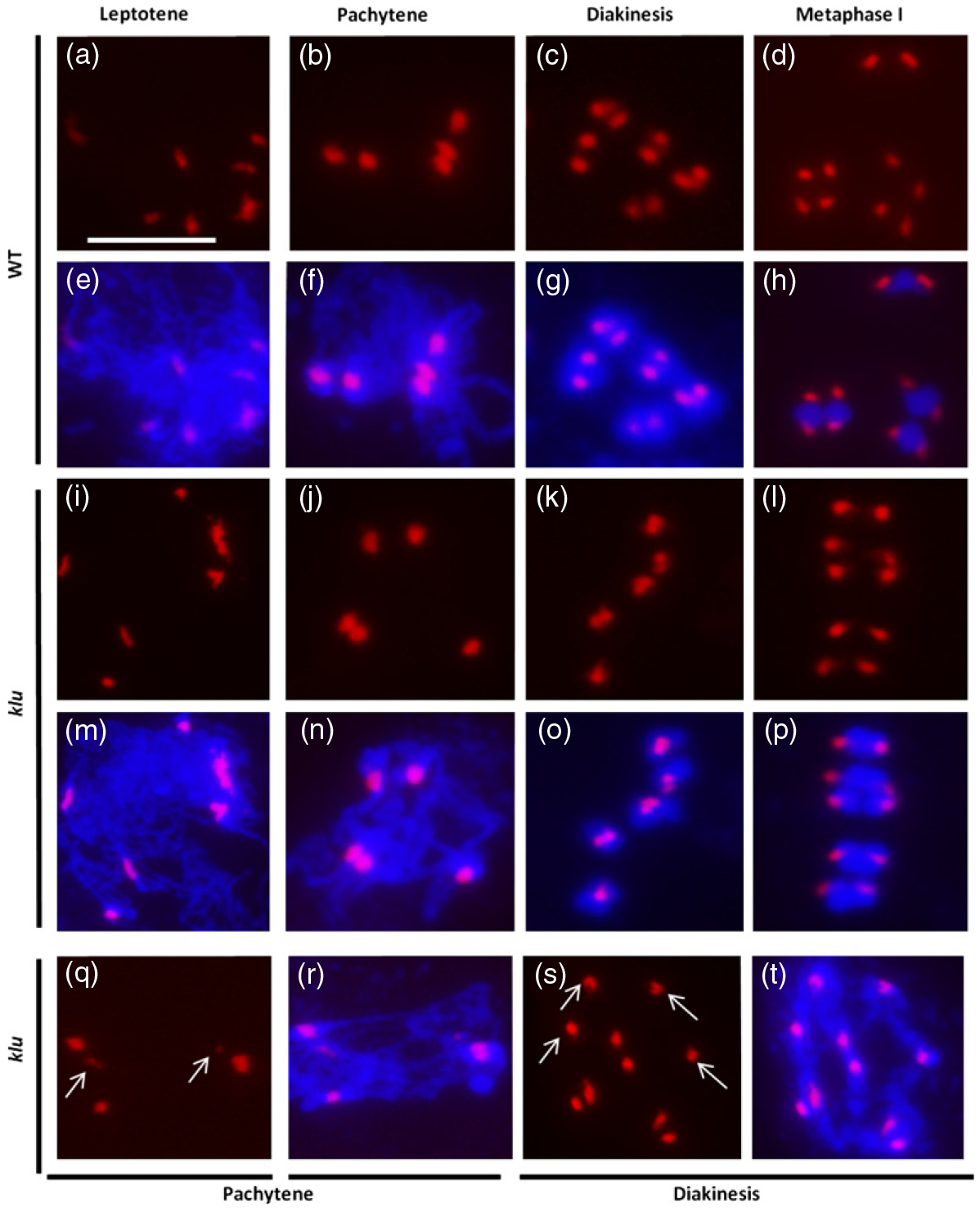
Defective centromere pairing during meiosis I in klu female meiocytes.
(a–d, i–l, q, s) Epifluorescence microscopy images of female meiotic spreads subjected to FISH analysis using a centromere-specific probe (red signal).
(e–h, m–p, r, t) Merged epifluorescence microscopy images of centromere (red) and 4′,6-diamidino-2-phenylindole (DAPI)-stained chromosomes (blue) in female meiotic spreads. (a, e), (b, f), (c, g) and (d, h) Wild-type female meiocyte at leptotene, pachytene, diakinesis and metaphase I, respectively.
(i, m), (j, n), (k, o) and (l, p) Apparently normal centromere signal in klu female meiocytes at leptotene, pachytene, diakinesis and metaphase I, respectively.
(q, r) klu female meiocyte at pachytene with unparied centromeres (white arrows).
(s, t) klu female meiocyte at diakinesis with a mixture of univalents (white arrows) and bivalents. Scale bars: 10 μm.
KLU is required for normal DMC1 expression pattern in ovules undergoing megasporogenesis
DMC1 is a highly conserved gene in eukaryotes and encodes a RecA (Recombinase A) protein that promotes interhomolog recombination during meiosis (Bishop et al., 1992). In Arabidopsis, DMC1 is required for meiotic recombination in both male and female meiocytes (Klimyuk and Jones, 1997; Couteau et al., 1999). In ovule primordium, DMC1 is preferentially expressed in the female meiocyte (Klimyuk and Jones, 1997; Siddiqi et al., 2000) and facilitates centromere pairing (Da Ines et al., 2012) and formation, and/or stabilization of bivalents (Couteau et al., 1999), aspects of meiosis that are also disrupted in klu megasporocytes (Figure 4). We introduced DMC1:GFP into klu plants by crossing. Conspicuous discrepancies in the expression patterns of DMC1:GFP in WT and in the klu ovules were examined. In WT ovules, the DMC1:GFP signal was observed exclusively in the female meiocytes in ovules at stage 2III/IV (Figure 5a,c), as previously reported (Siddiqi et al., 2000; Qin et al., 2014); however, only 64.9% (n = 114) of klu ovules exhibited DMC1:GFP activity in female meiocytes (Figure 5b,d). The remaining35.1% of klu ovules showed an absence of DMC1:GFP signal in the female meiocytes (Figure 5e, white arrow-head). Interestingly, an increased DMC1:GFP signal was observed in the somatic tissues of all klu ovules (Figure 5b,d,e). Unlike in WT (Figure 5c), the most extensive DMC1:GFP signal in the klu ovule was detected in the inner integument primordia (Figure 5b,d,e, white arrows), where KLU:YFP (Figure 5f) and KLU mRNA were preferentially expressed (Figure 5g,h). These results indicate that klu mutation results in the loss of DMC1 expression in a substantial number of klu female meiocytes and causes ectopic expression of DMC1 in the surrounding somatic cells of all klu ovules.
Figure 5.

Expression of DMC1 during megasporogenesis is altered in klu mutants.
(a–e) CLSM images of DMC1:GFP expression in wild-type (WT; a, c) and klu (b, d, e) ovules at stage 2III/IV. The highest levels of DMC1:GFP ectopic expression in klu ovules are detected in the inner integument primordium (c, d, e, white arrows). The white arrowhead in (e) indicates no expression of DMC1:GFP in the klu female meiocyte.
(f) CLSM images of KLU:YFP expression in WT ovules at stage 2III/IV.
(c–f) FM4–64 staining (red) showing the outline of the ovule.
(g, h) Whole-mount in situ hybridization of KLU mRNA in WT ovules at stage 2III/IV. Arrow in (g) points to enriched expression of KLU in inner integument primordim. Abbreviations: fm, female meiocyte; ii, inner integument; oi, outer integument.
Scale bars: (a, b) 50 μm; (c–h) 10 μm.
DISCUSSION
A Tag-seq-based comparative genetic subtraction approach identifies a large number of FM genes
In this study, we explored the comparative transcriptome-based discovery of genes involved in megasporogenesis using the spl mutant lacking megasporogenesis, and high-throughput Tag-seq. This approach identified 862 female meiocyte expressed genes. The reliability of our transcriptomic data was supported by the detection of 90% of these FM genes by fluorescence-assisted cell-sorted female meiocyte protoplasts, and by the results of real-time qRT-PCR and RNA in situ analysis.
Our sequencing profiles detected nearly all of the 9115 female meiocyte-expressed genes recently revealed by transcriptomic analysis using laser-captured MMCs (Schmidt et al., 2011). In addition, our sequencing-based strategies uncovered 375 FM genes that had not been identified from previous studies based on microarray analysis (Schmidt et al., 2011). These new FMs include 111 genes that are not represented in the ATH1 array and many genes expressed at low levels that may also play an important role during megasporogenesis, but that are undetectable by microarray analysis. These findings expand our understanding of plant germline formation and indicate that much remains to be discovered about this important biological process.
The previously characterized homeodomain transcription factor WUS, which acts downstream of SPL/NZZ in the ovule, is essential for female meiocyte formation because of its induction of the expression of two small peptides, WIH1 and WIH2. In addition to its function in the establishment of a stem cell niche in the shoot and floral meristems (Laux et al., 1996; Mayer et al., 1998), WUS is also expressed at the distal region of ovules and plays a central role in integument formation and megagametogenesis (Gross-Hardt et al., 2002). Consistently, we successfully identified WUS through comparative transcriptome analysis in this study. Several known meiotic genes, including the chromatin condensation gene SMC2 (Siddiqui et al., 2003), the cohesion protein encoding gene SYN3 (Yuan et al., 2012), homologous chromosome recombination regulating genes MSH3/7 (Culligan and Hays, 2000; Higgins et al., 2004) and RFC1 (Liu et al., 2012; Wang et al., 2012), and the cell cycle regulator OSD1 (d’Erfurth et al., 2009), were also found in our FMs gene set. This analysis implies that female meiocyte-expressed genes may play a crucial role in megasporogenesis and also supports the need to functionally characterize novel FMs.
Are mechanisms regulating male and female meiosis identical?
As meiosis is a fundamental process, it is reasonable to expect that the molecular mechanisms underlying this process must be shared during microsporogenesis and megasporogenesis. Indeed, many mutations that disrupt male meiosis in Arabidopsis also affect female meiosis (Bai et al., 1999; Couteau et al., 1999; Li et al., 2005; Stacey et al., 2006; Sebastian et al., 2009). Consistently, comparison of our FM data sets with the male meiocyte profiling (Chen et al., 2010; Yang et al., 2011) reveals that the majority of FM genes (96.9%) are also expressed in male meiocytes. This analysis suggests that male and female meiocytes share highly similar gene profiles, and that male and female meiosis are likely to be governed by many conserved molecular signaling pathways, even though most of the data on meiosis in plants originates from studies on the male meiocyte, because of the inaccessibility of the female counterpart. Interestingly, some genes known to be involved in male meiosis do not seem to have a role in female meiosis: these include MMD1 (Yang et al., 2003) and ASK1 (Yang et al., 1999a). These genes have similar expression values in both the male and female meiocytes: MMD1 has an RPKM value of 14.6 in the female meiocyte and 4.7 in the male meiocyte; ASK1 shows an RPKM value of 69.9 in the female meiocyte and 79.2 in the male meiocyte (Chen et al., 2010). It is likely that there are some homologs or other genes that can substitute the function of these genes in female meiocytes. These data imply that specific regulating mechanisms exist in male and female meiosis. Dramatic differences in the distribution and the rate of crossovers along chromosomes in male and female meiosis further support this notion (Giraut et al., 2011).
Reports on genes specifically involved in female meiosis have been very limited. swi1–1, an allele of switch1/dyad (Siddiqi et al., 2000; Mercier et al., 2001), causes failure to complete meiosis in the female meiocyte, where meiosis arrests at the end of meiosis I, whereas the mutation has no effects on male meiosis. arp6–1 and arp6–2, two mutant alleles of a subunit in chromatin-remodeling complex SWR1, exhibit meiosis defects specifically in female meiocyte (Qin et al., 2014), whereas the arp6–3 allele disrupts meiosis during both microsporogenesis and megasporogenesis (Rosa et al., 2013). The distinct mechanisms that are needed for female meiosis remain largely unknown. A forward-genetic screen identified 80 meiotic mutants from a collection of 55 000 T-DNA insertion lines (0.15%; De Muyt et al., 2009); however, it is unknown whether female meiosis is also affected in those mutants. In this study, we identified four mutations that specifically affect female meiosis, but not male meiosis, from a population of 120 T-DNA insertions in female meiocyte-expressed genes (3.3%), which represents a ~22-fold enrichment in the identification of functionally significant genes over the reported forward-genetic screen. Most importantly, many meiotic mutants identified by the reported forward-genetic screen turned out to be known meiotic genes (De Muyt et al., 2009). In contrast, the four genes implicated in female meiosis identified in this study have not previously been identified. These examples further illustrate that although male and female meiocytes express a great number of common genes and share many conserved regulatory mechanisms, the fine molecular details underlying megasporogenesis and microsprogenesis have diverged.
KLU controls female-specific meiosis in megasporogenesis
KLU was previously shown to be involved in the generation of a unique mobile growth stimulator to promote the growth of leaves and floral organs, to prolong the plastochron and to stimulate cell proliferation during seed development in a non-cell autonomous manner (Anastasiou et al., 2007; Wang et al., 2008; Adamski et al., 2009); however, it remains unknown why the expression of KLU in outer integuments using INNER NO OUTER (INO) promoter is sufficient to rescue the small seed size phenotype of the klu mutant, but fails to complement the reduced seed set phenotype, which was also shown to be determined by maternal control (Adamski et al., 2009). We revealed that the reduced female fertility in klu results from defective chromosome pairing and organization during female meiosis. KLU preferentially expressed in the inner integument, and klu mutant ovules show megasporogensis defects, suggesting that developmental coordination between the female meiocyte and the surrounding somatic cells is crucial for female gametophyte development. DMC1 was shown to be essential for chromosome pairing and univalent formation (Couteau et al., 1999; Sanchez-Moran et al., 2007). We further determined that KLU is required for normal DMC1 expression in ovule during megasporogenesis. A similarly altered DMC1 expression pattern during megasporogenesis is also apparent in arp6 mutant ovules (Qin et al., 2014). It will be interesting to investigate whether KLU coordinates with the chromatin remodeling complex SWR1 to regulate DMC1 expression.
EXPERIMENTAL PROCEDURES
Plants, materials and growth conditions
Seeds were spread on Murashige and Skoog plates after surface sterilization using chlorine gas for 2 h. Eight-day-old seedlings were transferred to soil with a 16-h light/8-h dark cycle at 22°C and 70% humidity.
Tissue collection and RNA isolation
spl homozygous plants were screened from progeny of heterozygous spl/+ (CS877147) obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu) and genotyped with SPL primers (Table S9). Flower buds at stages 10–11 were harvested from WT (Columbia ecotypes) and spl plants (Smyth et al., 1990; Christensen et al., 1997). Ovules at stages 2II–2IV with placenta were excised under a dissection microscope with a 27.5-gauge needle, from the ovaries that were held horizontally on double-sided tape. Excised ovules were immediately collected and stored in Qiagen RNAlater RNA stabilization reagent (http://www.qiagen.com) at 4°C. Total RNA was extracted from the ovule tissues using the Qiagen RNAeasy Plant Mini kit. Yield and RNA purity were determined using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, http://www.thermoscientific.com). RNA integrity was detected using an Agilent 2100 Bioanalyzer (http://www.genomics.agilent.com).
Sequencing and data processing
The 3′-tag digital gene expression libraries were constructed by BGI (formerly the Beijing Genomics Institute, http://www.genomics.cn/index) using the Digital Gene Expression Tag Profiling Kit (Illumina Inc., http://www.illumina.com) according to the manufacturer’s instructions. RNA (6 lg in total) from each sample was used for Illumina Sequencing, performed as previously described (Wu et al., 2010). Raw sequences were transformed into Clean Tags by filtering off empty reads, low-quality tags (containing ambiguous bases), adaptor-only tags and tags that occurred only once (probably as a result from sequencing error). The remaining high-quality sequences were mapped to the Arabidopsis thaliana genome (TAIR10, ftp://ftp.arabidopsis.org/Genes/TAIR10_gen-ome_release). For conservative and precise annotation, sequences with perfect homology or only 1-nt mismatch were further analysed. Clean tags aligned to multiple transcripts were excluded from our analysis. The remaining clean tags were designed as unambiguous clean tags. The number of unambiguous clean tags for each gene was calculated and normalized to the number of transcripts per million clean tags (TPM; Morrissy et al., 2009). When multiple types of tags were mapped to the different positions of the same gene, the gene expression levels were represented by the sum of all hit numbers.
Gene ongtology enrichment analysis for female meiocyte-expressed genes
Arabidopsis gene-to-GO annotations were downloaded from the GO site (revision 1.1.2512). To look for significantly enriched GO terms in female meiocyte-expressed genes compared with the genome background, the hypergeometric test was applied to map female meiocyte-expressed genes to the terms of the GO database (Horan et al., 2008). GO terms with P < 0.05 were considered significantly over-represented in female meiocytes, and are reported in Table 1. The complete results of the GO-term enrichment analysis are provided in Table S4.
Pfam domain enrichment analysis of female meiocyte-expressed genes
The protein domain annotations in the Pfam database were obtained from http://pfam.sanger.ac.uk (Finn et al., 2010). Arabidopsis protein sequences were searched against protein family models in the Pfam-A database, and a putative domain was accepted if the E-value was below 1e−7. Fisher’s exact test was used to assess the statistical significance of Pfam domain enrichment in the female meiocyte-expressed genes compared with the whole genome background.
Validation of sequencing data by real-time RT-PCR and in situ hybridization
RNA extracted from two independent biological samples was reverse transcribed and used to perform real-time qRT-PCR with SYBR Green I detection on an STEP-ONE system (Applied Biosystems, http://www.appliedbiosystems.com), as previously described (Qin et al., 2009). The sequences of all used primers for real-time RT-PCR and in situ hybridization probes are presented in Table S9. For in situ hybridization, DNA fragments of 309-bp AT1G15460, 291-bp AT2G33750 and 275-bp KLU were used for RNA probe synthesis. Whole-mount in situ hybridization experiments were carried out as previously described (Hejatko et al., 2006) on InsituPro VSi working station (Intavis AG, http://www.intavis.com).
Fluorescence-assisted cell sorting of DMC1:GFP labeled female meiocytes
Ovules from stage 10–11 flower buds of DMC1:GFP transgenic lines were dissected, transferred immediately to a Petri dish containing the digestion enzyme solution [43 g L−1 MS powder,30.5 g L−1 glucose, 30.5 g L−1 manitol, 0.65 g L−1 2-(N-morpholino)ethanesulphonic acid (MES), 1% cellulase R-10 and 0.2% macerozyme R-10] and incubated at 28°C for 2 h on a bench-top orbital shaker set at about 1 g as previously described, with minor modifications (Qin and Zhao, 2006). After rinsing three times, female meiocytes showing a strong GFP fluorescent signal were released from ovules and collected with a micropipette under an OLYMPUS IX2 fluorescence microscope (Olympus, http://www.olympus-global.com). RNA was extracted from ~150 female meiocytes with the PicoPure RNA isolation kit (Arcturus/Molecular Devices, http://www.moleculardevices.com) and amplified using a TargetAmp two-round amino-allyl antisense RNA (aRNA) amplification kit (Epicentre Bio-technologies, http://www.epibio.com) with SuperScript III and SuperScript II reverse transcriptases (Invitrogen, now Life Technologies, http://www.lifetechnologies.com). RNA samples from two independent biological replicates were assessed by an Agilent 2100 Bioanalyzer and sequenced using an Illumina sequencing platform.
Reverse genetic analysis
Mutant lines from ABRC with consistent reduced fertility were obtained and confirmed the insertion loci in a PCR reaction, as previously described (Qin et al., 2009), with the primers listed in Table S9. CLSM and DIC observation of female gametophyte, aniline blue staining of callose during megasprogenesis, meiotic chromosome spreading and centromere FISH analysis are conducted as previously described (Qin et al., 2014). The 180-bp repetitive sequence of centromere was previously described (Zhang et al., 2012). Observation of expression of DMC1:GFP in ovules and FM4–64 staining of ovule outline was conducted according to the published procedure (Lin et al., 2012).
Complementation of klu
The KLU full-length genomic fragment was amplified by PCR (primer sequence in Table S9), sequenced, cloned in the pGWB404 vector, and introduced in Agrobacterium tumefaciens. klu plants were transformed by floral dip (Clough and Bent, 1998). After screening on kanamycin plates, positive transformants were chosen for subsequent analysis.
Supplementary Material
Figure S2. Reproducibility analysis of two parallel experiments.
Figure S3. RT-PCR analysis of the transcripts of the corresponding FM genes in the four fm mutants.
Figure S1. Comparison of the spl mutant ovule with the WT ovule from stage 2I to stage 2IV.
Figure S4. Female gametophyte development is defective in the fm5, fm12, fm13 and fm43/+ mutant ovules.
Figure S5. Microspore tetrads and mature pollens from the fm5, fm12, fm13 and fm43/+ mutants.
Figure S6. Centromere behavior in male meiosis is apparently normal in the klu mutant.
Table S1. Raw gene expression data for the 79 FMs identified with stringent cut-off.
Table S2. Raw gene expression data for the 862 FMs identified with a less stringent cut-off.
Table S3. List of the FMs detected in other published analyses.
Table S4. Information on the FMs located within each over-represented GO term.
Table S5. Pfam domains matched to the FMs.
Table S6. The 775 FMs present in the female meiocyte protoplast profiling.
Table S7. List of genes and mutants for reverse genetic analysis.
Table S8. Callose deposition during female meiosis is defective in the four fm mutants.
Table S9. Primers used in this study.
ACKNOWLEDGEMENTS
We are grateful to: Dr Ramin Yadegari, University of Arizona, USA, for providing the DMC1:GFP seeds; and Dr Michael Lenhard, University of Potsdam, Germany, for providing the KLU:YFP seeds. We thank Dr Shingo Nagawa for assistance with in situ hybridization analysis, and Drs Ravishankar Palanivelu and Irene Lavagi for critical comments on the article. This work was supported by the Ministry of Science and Technology of China (2011CB944603), National Natural Science Foundation of China (31170290) and by funds from the Center for Genomics and Biotechnology, Fujian Agriculture and Forestry University. The authors have no conflicts of interest to declare.
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Adamski NM, Anastasiou E, Eriksson S, O’Neill CM and Lenhard M (2009) Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl Acad. Sci. USA, 106, 20115–20120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Ferreira M, Wellmer F, Banhara A, Kumar V, Riechmann JL and Meyerowitz EM (2007) Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol. 145, 747–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C and Lenhard M (2007) Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell, 13, 843–856. [DOI] [PubMed] [Google Scholar]
- Armstrong SJ, Franklin FC and Jones GH (2001) Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J. Cell Sci 114, 4207–4217. [DOI] [PubMed] [Google Scholar]
- Audic S and Claverie JM (1997) The significance of digital gene expression profiles. Genome Res. 7, 986–995. [DOI] [PubMed] [Google Scholar]
- Bai X, Peirson BN, Dong F, Xue C and Makaroff CA (1999) Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell, 11, 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L and Kleckner N (1992) DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell, 69, 439–456. [DOI] [PubMed] [Google Scholar]
- Chen C, Farmer AD, Langley RJ, Mudge J, Crow JA, May GD, Huntley J, Smith AG and Retzel EF (2010) Meiosis-specific gene discovery in plants: RNA-Seq applied to isolated Arabidopsis male meiocytes. BMC Plant Biol. 10, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CA, King EJ, Jordan JR and Drews GN (1997) Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod 10, 49–64. [Google Scholar]
- Clough SJ and Bent AF (1998) Floral dip: a simplified method for Agro-bacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D and Doutriaux MP (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell, 11, 1623–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM and Hays JB (2000) Arabidopsis MutS homologs-AtMSH2, AtMSH3, AtMSH6, and a novel AtMSH7-form three distinct protein heterodimers with different specificities for mismatched DNA. Plant Cell, 12, 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Ines O, Abe K, Goubely C, Gallego ME and White CI (2012) Differing requirements for RAD51 and DMC1 in meiotic pairing of centromeres and chromosome arms in Arabidopsis thaliana. PLoS Genet. 8, e1002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Pereira L, Vezon D et al. (2009) A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet. 5, e1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M and Mercier R (2009) Turning meiosis into mitosis. PLoS Biol. 7, e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J et al. (2010) The Pfam protein families database. Nucleic Acids Res. 38, D211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraut L, Falque M, Drouaud J, Pereira L, Martin OC and Mezard C (2011) Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 7, e1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt R, Lenhard M and Laux T (2002) WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 16, 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejatko J, Blilou I, Brewer PB, Friml J, Scheres B and Benkova E (2006) In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat. Protoc 1, 1939–1946. [DOI] [PubMed] [Google Scholar]
- Hennig L, Gruissem W, Grossniklaus U and Kohler C (2004) Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol. 135, 1765–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FC and Jones GH (2004) The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 18, 2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D and Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5, R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan K, Jang C, Bailey-Serres J, Mittler R, Shelton C, Harper JF, Zhu JK, Cushman JC, Gollery M and Girke T (2008) Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol. 147, 41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AJ, Meier P, Gheyselinck J, Wuest SE, Federer M, Schlagenhauf E, Becker JD and Grossniklaus U (2007) Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biol. 8, R204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimyuk VI and Jones JD (1997) AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J. 11, 1–14. [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J and Jurgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development, 122, 87–96. [DOI] [PubMed] [Google Scholar]
- Li W, Yang X, Lin Z, Timofejeva L, Xiao R, Makaroff CA and Ma H (2005) The AtRAD51C gene is required for normal meiotic chromosome synapsis and double-stranded break repair in Arabidopsis. Plant Physiol. 138, 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libeau P, Durandet M, Granier F, Marquis C, Berthome R, Renou JP, Taconnat-Soubirou L and Horlow C (2011) Gene expression profiling of Arabidopsis meiocytes. Plant Biol . (Stuttg), 13, 784–793. [DOI] [PubMed] [Google Scholar]
- Lin D, Nagawa S, Chen J et al. (2012) A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr. Biol 22, 1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Deng Y, Li G and Zhao J (2012) Replication factor C1 (RFC1) is required for double-strand break repair during meiotic homologous recombination in Arabidopsis. Plant J. 73, 154–165. [DOI] [PubMed] [Google Scholar]
- Ma H (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol 56, 393–434. [DOI] [PubMed] [Google Scholar]
- Matias-Hernandez L, Battaglia R, Galbiati F, Rubes M, Eichenberger C, Grossniklaus U, Kater MM and Colombo L (2010) VERDANDI is a direct target of the MADS domain ovule identity complex and affects embryo sac differentiation in Arabidopsis. Plant Cell, 22, 1702–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G and Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell, 95, 805–815. [DOI] [PubMed] [Google Scholar]
- McCormick S (1993) Male gametophyte development. Plant Cell, 5, 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Vezon D, Bullier E, Motamayor JC, Sellier A, Lefevre F, Pelletier G and Horlow C (2001) SWITCH1 (SWI1): a novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev. 15, 1859–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissy AS, Morin RD, Delaney A, Zeng T, McDonald H, Jones S, Zhao Y, Hirst M and Marra MA (2009) Next-generation tag sequencing for cancer gene expression profiling. Genome Res. 19, 1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D and Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development, 132, 603–614. [DOI] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijo JA and Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 138, 744–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA and Schimenti JC (1998) Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell, 1, 697–705. [DOI] [PubMed] [Google Scholar]
- Qin Y and Zhao J (2006) Localization of arabinogalactan proteins in egg cells, zygotes, and two-celled proembryos and effects of beta-D-glucosyl Yariv reagent on egg cell fertilization and zygote division in Nicotiana tabacum L. J. Exp. Bot 57, 2061–2074. [DOI] [PubMed] [Google Scholar]
- Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA and Palanivelu R (2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet. 5, e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Zhao L, Skaggs M et al. (2014) ACTIN-RELATED PROTEIN6 regulates female meiosis by modulating meiotic gene expression in Arabidopsis. Plant Cell, 26, 1612–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M, Von Harder M, Cigliano RA, Schlogelhofer P and Mittelsten Scheid O (2013) The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell, 25, 1990–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Leon N, Arteaga-Vazquez M, Alvarez-Mejia C et al. (2012) Transcriptional analysis of the Arabidopsis ovule by massively parallel signature sequencing. J. Exp. Bot 63, 3829–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Moran E, Santos JL, Jones GH and Franklin FC (2007) ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 21, 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P, Bui A, Weterings K, McIntire K, Hsu Y, Lee P, Truong M, Beals T and Goldberg R (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11, 297–322. [Google Scholar]
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E and Schneitz K (1999) Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 96, 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Wuest SE, Vijverberg K, Baroux C, Kleen D and Grossniklaus U (2011) Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development. PLoS Biol. 9, e1001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz K, Hulskamp M and Pruitt RE (1995) Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole--mount tissue. Plant J. 7, 731–749. [Google Scholar]
- Sebastian J, Ravi M, Andreuzza S, Panoli AP, Marimuthu MP and Siddiqi I (2009) The plant adherin AtSCC2 is required for embryogenesis and sister-chromatid cohesion during meiosis in Arabidopsis. Plant J. 59, 1–13. [DOI] [PubMed] [Google Scholar]
- Siddiqi I, Ganesh G, Grossniklaus U and Subbiah V (2000) The dyad gene is required for progression through female meiosis in Arabidopsis. Development, 127, 197–207. [DOI] [PubMed] [Google Scholar]
- Siddiqui NU, Stronghill PE, Dengler RE, Hasenkampf CA and Riggs CD (2003) Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development, 130, 3283–3295. [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL and Meyerowitz EM (1990) Early flower development in Arab[notdef]ıdopsis. Plant Cell, 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey NJ, Kuromori T, Azumi Y, Roberts G, Breuer C, Wada T, Maxwell A, Roberts K and Sugimoto-Shirasu K (2006) Arabidopsis SPO11–2 functions with SPO11–1 in meiotic recombination. Plant J. 48, 206–216. [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Macfarlane J and Drews GN (2007) Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51, 281–292. [DOI] [PubMed] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E and Weigel D (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell, 20, 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cheng Z, Huang J, Shi Q, Hong Y, Copenhaver GP, Gong Z and Ma H (2012) The DNA replication factor RFC1 is required for interference-sensitive meiotic crossovers in Arabidopsis thaliana. PLoS Genet. 8, e1003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne AJ, Zhang W, Sun Y, Liu W, Albert R, Zheng Z, Oppenheimer DG, Zhao D and Ma H (2007) Differential gene expression in Arabidopsis wild-type and mutant anthers: insights into anther cell differentiation and regulatory networks. Plant J. 52, 14–29. [DOI] [PubMed] [Google Scholar]
- Wu J, Zhang Y, Zhang H, Huang H, Folta KM and Lu J (2010) Whole genome wide expression profiles of Vitis amurensis grape responding to downy mildew by using Solexa sequencing technology. BMC Plant Biol. 10, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenfuhrer J, von Mering C and Grossniklaus U (2010) Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol 20, 506–512. [DOI] [PubMed] [Google Scholar]
- Yadegari R and Drews GN (2004) Female gametophyte development. Plant Cell, 16(Suppl), S133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Hu Y, Lodhi M, McCombie WR and Ma H (1999a) The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc. Natl Acad. Sci. USA, 96, 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J and Sundaresan V (1999b) The SPOROCYTE-LESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13, 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Makaroff CA and Ma H (2003) The Arabidopsis MALE MEIO-CYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell, 15, 1281–1295. [PMC free article] [PubMed] [Google Scholar]
- Yang H, Lu P, Wang Y and Ma H (2011) The transcriptome landscape of Arabidopsis male meiocytes from high-throughput sequencing: the complexity and evolution of the meiotic process. Plant J. 65, 503–516. [DOI] [PubMed] [Google Scholar]
- Yu HJ, Hogan P and Sundaresan V (2005) Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol. 139, 1853–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Yang X, Ellis JL, Fisher NM and Makaroff CA (2012) The Arabidopsis SYN3 cohesin protein is important for early meiotic events. Plant J. 71, 147–160. [DOI] [PubMed] [Google Scholar]
- Zhang C, Song Y, Cheng ZH, Wang YX, Zhu J, Ma H, Xu L and Yang ZN (2012) The Arabidopsis thaliana DSB formation (AtDFO) gene is required for meiotic double-strand break formation. Plant J. 72, 271–281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S2. Reproducibility analysis of two parallel experiments.
Figure S3. RT-PCR analysis of the transcripts of the corresponding FM genes in the four fm mutants.
Figure S1. Comparison of the spl mutant ovule with the WT ovule from stage 2I to stage 2IV.
Figure S4. Female gametophyte development is defective in the fm5, fm12, fm13 and fm43/+ mutant ovules.
Figure S5. Microspore tetrads and mature pollens from the fm5, fm12, fm13 and fm43/+ mutants.
Figure S6. Centromere behavior in male meiosis is apparently normal in the klu mutant.
Table S1. Raw gene expression data for the 79 FMs identified with stringent cut-off.
Table S2. Raw gene expression data for the 862 FMs identified with a less stringent cut-off.
Table S3. List of the FMs detected in other published analyses.
Table S4. Information on the FMs located within each over-represented GO term.
Table S5. Pfam domains matched to the FMs.
Table S6. The 775 FMs present in the female meiocyte protoplast profiling.
Table S7. List of genes and mutants for reverse genetic analysis.
Table S8. Callose deposition during female meiosis is defective in the four fm mutants.
Table S9. Primers used in this study.


