Paleogenetics reveals that imported domestic horses replaced native wild ones in Transcaucasia and Anatolia before 2000 BCE.
Abstract
Despite the important roles that horses have played in human history, particularly in the spread of languages and cultures, and correspondingly intensive research on this topic, the origin of domestic horses remains elusive. Several domestication centers have been hypothesized, but most of these have been invalidated through recent paleogenetic studies. Anatolia is a region with an extended history of horse exploitation that has been considered a candidate for the origins of domestic horses but has never been subject to detailed investigation. Our paleogenetic study of pre- and protohistoric horses in Anatolia and the Caucasus, based on a diachronic sample from the early Neolithic to the Iron Age (~8000 to ~1000 BCE) that encompasses the presumed transition from wild to domestic horses (4000 to 3000 BCE), shows the rapid and large-scale introduction of domestic horses at the end of the third millennium BCE. Thus, our results argue strongly against autochthonous independent domestication of horses in Anatolia.
INTRODUCTION
The domestication of the horse ca. 5500 years ago represents one of the most important technological innovations in the ancient world (1, 2). With the harnessing of horsepower, political, economic, and social relationships throughout the ancient world were transformed as horses revolutionized transportation and affected patterns of trade, warfare, and migration [e.g., (1–4)]. Archaeological, organic residue, and genetic analyses suggest that the domestic horse originated in the Central Asian steppes, then spread into eastern Europe and later into southwest Asia (SWA) [e.g., (2, 5–11)]. In particular, data from the Botai hunter-gatherer culture in Kazakhstan suggest that, by the mid to late fourth millennium BCE, horses were bitted, milked, selected for the TRPM1 coat color locus, and kept in enclosures and were therefore under intensive management (10, 12–14). A recent study of ancient horse genomes, however, challenged the view that modern domestic horses derived from Central Asia, because “Botai-like” horses were shown to be the ancestors of northeast Asian Przewalski’s horses but not the main source of ancient or modern domestic horses (14, 15). Another recent study ruled out a second potential horse domestication center, the Iberian Peninsula, showing that Iberian wild horses went extinct without leaving notable traces in the genomes of modern horses (16). There are, however, two more areas that have been proposed as domestication centers for modern horses: the Pontic-Caspian steppe (17) and Anatolia (8, 18). Although the former has been long hypothesized as the likely source of domestic horses (12, 13, 19), the latter region has been poorly explored regarding its role in horse domestication processes (20–22), despite its long history of wild horse exploitation and its reputation for breeding valuable horses in Classical Antiquity (23).
The origin of the domestic horse in Anatolia, and more generally in SWA, continues to represent a complex archaeological puzzle. A combination of textual, iconographic, and archaeozoological data suggest that, by the mid to late third millennium BCE, domestic horses were introduced from neighboring mountain regions into Mesopotamia (modern Iraq and northeast Syria), where they were often referred to in cuneiform texts as ANŠE-KURRA (“donkey of the mountain”) (24–28). Initially kept only in small numbers, horses rose to prominence across SWA within a few centuries in association with the spread of chariots, a technological innovation of the second millennium BCE (1, 29). Because horse domestication in the Eurasian steppes, a region historically known for its “horse cultures,” likely began in the fourth or perhaps even fifth millennium BCE (17, 30–32), it has long been argued that southwest Asian horses are the descendants of these early domesticates, which arrived in the region via poorly understood participation in Pontic-Caspian-Transcaucasian interaction spheres or population movements (33, 34).
Another hypothesis argues that Anatolia played a central role in the transmission of domestic horses into Syro-Mesopotamia (33, 35, 36), and an Anatolian contribution to early domestic populations has been suggested (37–40). Archaeological data indicate the widespread presence of wild horses (Equus ferus) and also of so-called hydruntines [a subspecies of the Asiatic wild ass named Equus hemionus hydruntinus (41)] in early and middle Holocene Anatolia that were regularly exploited (20, 22, 40, 42). The continuity of human-horse interactions from the ninth millennium through the second millennium BCE, when domesticates are known from archaeological contexts, led to the hypothesis that Anatolian wild horses may have been a potential source population for domestic horses. The lack of reliable osteomorphological criteria for differentiating the skeletal remains of wild and domestic horses, however, has hampered attempts to address the hypothesis of a local domestication. Therefore, the cultural processes and mechanisms triggering the widespread appearance of domesticates in the late third millennium are still elusive. In this study, we take advantage of the abundance of archaeological horse remains from the central Anatolian plateau (20, 42–46) to provide the first rigorous test of the hypothesis of Anatolian horse domestication applying paleogenetics.
Complete present-day mitochondrial genomes have revealed 18 major haplogroups (A to R), the radiation times of which date mostly to the Neolithic and later periods (47). In contrast, studies of the mitochondrial hypervariable region in ancient horses have shown that domestic horses exhibit a much higher amount of genetic variation in mitochondrial lineages compared to cattle, sheep, and pigs (48–51). Furthermore, most mitochondrial lineages observed in domestic horses already existed before domestication (52). These analyses of ancient horses did not yield a clear phylogeographic structure that would allow the spatiotemporal origin(s) of horse domestication to be identified. These findings interpreted as suggesting that the mobility of wild horses in northern Eurasia allowed constant population reshuffling and repeated recruitment of wild local mares, precluding the establishment of a phylogeographic structure (52, 53).
In contrast, extant domestic horses exhibit remarkably little variation in the male Y chromosome line, with only one haplotype so far identified in modern domesticates, which led to the early claim of a single domestication event for horses (54, 55). Paleogenomic analyses of ancient specimens, however, observed additional male lineages in prehistoric populations before domestication and revealed that genetically diverse male founders were involved in early domestication (15, 56). This diversity was subsequently reduced, likely as a result of more directed human selection likely starting in the Iron Age and continuing during Roman times (57), and again during the Islamic conquest and the Byzantine-Sassanid war after 7th to 9th c. CE (16).
Paleogenetic evidence from genetic loci associated with coat color in horses argues for a diversification of coat color starting in the Bronze Age and is considered associated with an early stage of the domestication process (11, 58). Because the appearance of new coat colors is common in domestic taxa compared to their wild counterparts, they provide a useful marker for identifying domestic horses in archaeological assemblages.
Up to now, the origins of domestic horses in Anatolia have remained elusive; but careful recovery of horse remains from well-stratified archaeological contexts in Anatolia and in the neighboring Caucasus, together with progress in paleogenetic approaches, now makes it possible to specifically address the processes responsible for the origins of domestic horses in this part of western Asia. For this project, we combined morphological classification of equid remains, which can be hampered by a lack of diagnostic anatomical and/or biometrical criteria (41, 59), with paleogenetic analysis of mitochondrial, Y chromosome DNA, and autosomal DNA markers related to coat color to trace the spatiotemporal dynamics of the emergence of domestic horses in Anatolia. We analyzed over 100 equid remains from 14 prehistoric sites in central Anatolia and the Caucasus covering most of the Holocene (9000 BCE to 1000 CE) to gain insights into the origins of domestic horses in Anatolia, a pivotal issue in near Eastern history.
RESULTS
On the basis of our extensive experience with ancient animal remains from SWA including Anatolia, we expected DNA to be highly degraded in most samples that we had collected, of which only a few were petrous bones. DNA in osseous remains from SWA is notoriously poorly preserved, petrous bones being an exception, although not all of them contain preserved endogenous DNA. For this reason, we decided to rely on a highly optimized metabarcoding approach combining the sensitivity of polymerase chain reaction (PCR) and the efficiency of next-generation sequencing (NGS) specifically tailored to highly degraded ancient DNA (60). This approach has been shown to recover DNA molecules that escape shotgun sequencing (15) or sequence capture (61) and, if primers are optimized in silico and in vitro, is highly locus specific (60). Used in combination with methods minimizing contamination (62), as well as statistically sufficient replications (60) (see also Materials and Methods and the Supplementary Materials), it at least equals or can occasionally be superior to DNA capture methods, which are plagued by biases [e.g., (63)]. We have used this approach for the study of various species, including horses (15, 60, 64, 65).
Here, we analyzed 111 equid remains from eight sites in central Anatolia and six sites in the Caucasus dating from the Early Neolithic to the Iron Age (ca. 9000 to 500 BCE; see table S1), with a few samples dating to later, historic periods. This approach had been developed and optimized previously to produce reliable data from highly degraded samples (60, 64, 65), and it has been used successfully in situations where shotgun sequencing was not effective enough to genotype a large proportion of phenotype-associated single-nucleotide polymorphisms (SNPs) (15). We targeted the mitochondrial hypervariable region and 18 specific SNP regions diagnostic for the 18 major mitochondrial haplogroups considered diagnostic in earlier studies [(47) and table S2]. These SNPs are sufficiently diagnostic to recapitulate the essential features of the mitogenome phylogeny (Fig. 1). Moreover, we analyzed six regions of the Y chromosome, four anonymous Y-linked fragments, and two fragments of the amelogenin gene to evaluate male inheritance [(54, 56, 57) and table S3]. Last, we chose a set of eight diagnostic SNPs in seven genes associated with the coat color in horses, including basic colors (bay, black, chestnut, and gray), diluted phenotypes (silver and cream), spotted or painted phenotypes (overo, tobiano, and sabino), and leopard spotting (table S4) (11).
Fig. 1. Comparative maximum likelihood phylogenetic analyses of horse mitogenome using either the complete mitogenome sequences (left side) or the concatenated mitogenome fragments used for genotyping ancient remains (right side).
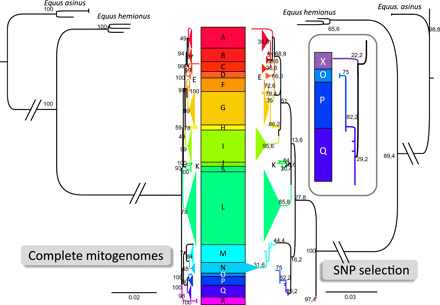
The nomenclature of the horse mitochondrial haplogroups from A to R is as defined by Achilli et al. (47). Haplogroup S corresponds to an additional haplogroup obtained when adding Przewalski’s horse sequences not belonging to the F haplogroup (98). The scale bar represents the number of nucleotide substitution per site as indicated. The branches separating horse, hemione, and donkey sequences are not drawn to scale as indicated by the intersecting parallels. A magnified view of the O-P-Q subtree of the concatenated fragments is represented in the box on the right side. The magnified view additionally reveals the position of the X sequence found in two ancient Anatolian remains. The numbers by the nodes indicate their corresponding bootstrap values.
Genotyping versus osteological determination
From the 111 analyzed equid remains, 77 (70%) yielded ancient DNA results and could be genotyped in independent triplicate PCR experiments (table S1). We obtained 14 different caballine (Equus caballus) mitochondrial haplogroups previously defined in present-day horses (47) and a previously unidentified haplogroup, here termed X, that belongs to the subtree of the O, P, and Q haplotypes (Fig. 1 and table S2). Moreover, from 10 specimens, we obtained haplotypes characteristic of donkeys (Equus asinus). Last, seven specimens yielded haplotypes belonging to E. hemionus clade H1, which Bennett et al. (41) assigned to E. hemionus hydruntinus and are therefore referred to as “hydruntine” below.
Genotyping and osteological determination agreed in 48 of the 57 remains that were assigned osteologically, with various degrees of certainty, to one of the equid species (84%) (table S1). In particular, 38 of the 40 remains assigned osteologically to wild or domestic horses showed the corresponding mitochondrial DNA (mtDNA) (95%). Agreement was also obtained for 3 of 6 hydruntine and 7 of 11 donkey remains. Of the 20 remains that could not be assigned osteologically to one of the aforementioned species, we identified 16 horses, 2 donkeys, and 2 hydruntines through mtDNA typing. Genotyping and osteological determination disagreed in only four osteologically unambiguously assigned cases: Two bone specimens determined osteologically as hydruntines yielded horse mtDNA, whereas two others classified osteologically as horses carried a donkey and a hydruntine mtDNA. Last, one unassigned equid was determined genetically as a hybrid, more precisely a mule, because it carried horse mitochondrial and donkey Y chromosomal DNA (table S1: specimen CD6189).
Diachronic pattern of maternal lineages
The 12 Anatolian horse remains predating ~4500 BCE carried either the mitochondrial haplogroup P or a previously unidentified mitotype, termed here X, not previously identified in modern or ancient horses (Figs. 1 and 2 and tables S1 and S2). Neither the P nor the X haplogroup has been documented so far beyond Anatolia in contemporaneous or older samples (48, 50, 66–69). This strongly suggests that these two haplogroups represent the unique signature of a local wild horses native to the Anatolia plateau. After 2200 BCE, this pattern changed profoundly, with 13 new mitochondrial haplogroups appearing in faunal assemblages from the Bronze and Iron Ages (Fig. 2 and table S1). Among the Bronze and Iron Age specimens, the pre-Bronze Age haplogroup P represents only 6% of the obtained haplotype spectrum (2 of the 33 remains), with both specimens dating to the earliest phase of this period (c. 2000 BCE). Moreover, in post-3300 BCE specimens, haplotype X is no longer detected. The novel haplotypes detected in our archaeological sample correspond mainly to haplogroups Q (11 remains), G (5 remains), and N (5 remains), while haplogroups A, B, D, E, H, I, L, and Q account for the remaining 20 specimens (table S1). These results indicate a nearly complete population turnover from the late third millennium BCE onward and correspond well with iconographic and textual evidence for the appearance and dispersal of horse management in Anatolia and Mesopotamia (26, 28).
Fig. 2. Mitochondrial and coat color diversity before (top) and after (bottom) 2000 BCE.
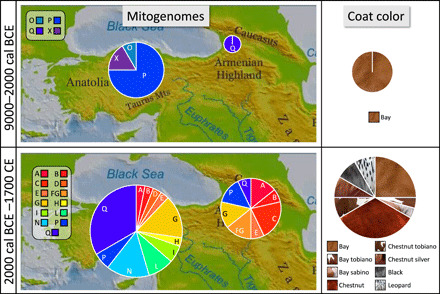
(Left) Evolution of mitochondrial haplotype diversity of horses in Anatolia and the southern Caucasus. (Right) Evolution of coat color genetic diversity in these two geographic regions in the same time ranges. The area of the circles is proportional to the number of individuals present in each category.
In the Caucasus, the earliest specimen that yielded a genetic result dates to the third millennium BCE and corresponds to haplogroup Q. Of the remaining 13 specimens, all excavated from archaeological contexts dating to the second millennium BCE, and 11 represent a diverse array of haplogroups including A, B, C, E, FG, G, and Q. Together, this change in haplotypes in both Anatolia and the Caucasus is statistically highly significant (P = 5.7 × 10 −6, Fisher’s test). The remaining two samples were identified as haplogroup P, presumably representing a continuation of the native Anatolian matriline into the Late Bronze Age.
Paternal lineages and hybrids
Paternal lineages were genotyped through six different loci on the Y chromosome (table S3). Y chromosomal DNA data are less numerous, and none were obtained from remains older than the Bronze Age, which must be due to poor DNA preservation. In 19 specimens dating to the Bronze Age or subsequent periods, however, the Y chromosome haplotype could be determined. Of these, 12 belong to E. caballus and 6 to E. asinus, and one specimen that was identified more generally as asinine (tables S1 and S3).
We could attribute the horse Y chromosomal sequences to two of a total of four horse haplotypes that have been described previously (57): Five remains were carriers of haplotype Y-HT-1, which is the major haplogroup present in modern horses, while four carried the extinct haplotype Y-HT-3 and three could not be determined due to SNPs that did not yield sufficient sequence coverage.
One specimen originating from Çadır Höyük yielded Y chromosomal SNPs corresponding to a jackass, whereas the mtDNA corresponded to a horse (tables S1 and S3), thus reflecting the presence of a hybrid (mule) dating to the Iron Age. The mitotype of this individual was L, a mitotype not encountered in SWA before the Bronze Age.
Coat color
We genotyped SNPs associated with coat color variations (11, 58). As discussed above, retrieval of nuclear DNA data in addition to mtDNA requires better ancient DNA preservation. Therefore, as for the Y chromosome, nuclear SNPs could not be genotyped in a reliable manner in samples predating the Early Bronze Age (tables S1 and S4). Together, we obtained SNPs from 43 specimens, allowing us to infer the coat color for 33 individuals, including 25 horses, 6 donkeys, 1 hydruntine, and 1 mule. In our dataset, we identified the mutant allele for all but two of the eight interrogated SNPs. In particular, only the mutant alleles for overo and cream were missing, while the other six genetic variants were present. Consequently, a large part of the diversity of mutations affecting the coat color already observed in ancient northern Eurasia (11) proved present in Bronze Age horses in SWA (table S4). Our results allowed us to attribute a coat color to 25 horses from the Bronze Age and later periods. We identified seven horses with a wild-type bay-colored coat; one with a bay sabino; eight with a chestnut-colored coat; two each with the colors chestnut tobiano, chestnut silver, leopard, and black; one with a bay tobiano coat color; and one specimen whose DNA preservation was not good enough to discriminate between chestnut and bay (tables S1 and S4). This diversification in the coat color distribution is statistically significant (P = 1.25 × 10−3, Fisher’s test).
As expected, the six donkeys and the hydruntine did not harbor any of the mutant SNPs that humans selected for in domestic horses. The sample that was identified as a mule carried one mutant allele in both the ASIP and MC1R genes, most likely originating from its horse mother, which are associated with a bay tobiano coat in horses (tables S1 and S4). In specimen AC8811 from Early Bronze Age Acemhöyük, a chestnut coat color is combined with mitotype P, representing the local Anatolian wild horse matriline. This combination indicates that local Anatolian mares were incorporated into domestic herds in the Early Bronze Age.
DISCUSSION
Wild and domesticated horses in Anatolia
Our genotyping of 60 ancient horses was designed to elucidate the long-standing question of a local domestication of horses in Anatolia. Our results allow us to conclude that domestic horses were introduced into the Caucasus and Anatolia by at least 2000 BCE, presumably from the Eurasian steppes. This conclusion is based on the fact that, in Anatolia, local horse populations before ca. 4500 cal BCE carried only two mitochondrial haplogroups, P and X, the latter being a previously unrecorded haplotype that belongs to the O-P-Q subtree (Fig. 1). So far, these haplogroups have not been encountered elsewhere in Eurasia in contemporaneous or earlier contexts. Furthermore, haplotype X likely had a limited temporal occurrence in Holocene Anatolia possibly disappearing after 5500 BCE. The foregoing supports our conclusion that these two haplogroups reflect the local mitochondrial signature of wild horses hunted in Anatolia in the early and middle Holocene (Fig. 2). We propose that the P and X haplogroups evolved independently in Anatolia during the late Pleistocene and early Holocene with little or no gene flow from neighboring wild horse populations due to geographic barriers separating Anatolia from northern Eurasia, namely, the Bosporus as well as the Caucasus and the Zagros mountain ranges. Thus, our study provides the first evidence showing that Anatolia was home to a genetically distinct population of wild horses, which, based on archaeozoological findings, were widely exploited during the Neolithic and Chalcolithic periods (20, 22, 42). Equid remains from early Neolithic Aşıklı Höyük, Neolithic/Chalcolithic Köşk Höyük, and Late Chalcolithic/Early Bronze Age Çadır Höyük are representative of these wild Anatolian horses.
Around 2000 BCE, we observe a statistically significant decline in the frequency of this local wild horse mitochondrial signature as the P haplogroup becomes rare and the X haplotype disappears completely. Presumably, the low frequency of haplotype X in pre-Bronze Age horse assemblages (2 of 11 in our dataset) offers an explanation as to why it did not survive in Anatolia into historic times.
Parallel to this, the diversity of maternal lineages in archaeological horse remains from the Caucasus and Anatolia increased markedly from 2 to 14 (Fig. 2), all of which identified previously in present-day horses (47) as well as in Eneolithic, Chalcolithic, and Early Bronze Age horses from southeast Europe and Kazakhstan (50). In these studies, mitochondrial haplogroups display no phylogeographic structure in Eurasia, which is consistent with the absence of significant physical barriers across the vast Eurasian steppes. This suggests that horse populations in the Eurasian steppes were panmictic, likely explaining the high diversity of present-day domestic horse populations. It would also account for the rapid diversification observed in our dataset upon introduction in the Caucasus and Anatolia. This sudden appearance of allochthonous lineages coincides with the emergence of iconographic and epigraphic evidence for horses and horse riding at the end of the third millennium BCE and argues for substantial imports of domestic forms and hence against an independent local domestication process (26–28).
The results we obtained from genotyping of alleles associated with variations in coat color reinforce this conclusion, because Bronze Age horses in Anatolia and the Caucasus show mutations corresponding to coat color variants thought to have been selected during domestication in the Eurasian steppes, such as chestnut, black, and silver (11). Mutations associated with coat color dilutions or spotting appeared later in our dataset (after 1200 BCE). Thus, we conclude from our genetic data that the domestic horses introduced to Anatolia in the Bronze Age carried mutations found earlier in northern Eurasia (11) and therefore derived predominantly from stocks imported from this vast region. Although the ultimate geographic origins of this allochthonous population cannot be defined with the data at hand, the Eurasian steppes north of the Black Sea seems a most plausible candidate.
We found that the local Anatolian P haplotype persisted in Bronze Age domestic horses in Anatolia and the Caucasus at low frequency (8% in our dataset) as well as in present-day horses, suggesting that wild Anatolian mares were incorporated into domestic herds probably very soon after the introduction of domestic horses in the region and before their local extinction in the wild. This possibility is in agreement with our observation that at least two of the four Bronze Age horses carry the P haplogroup. Because these individuals also carry a mutant coat color allele (AC8811 and TS2; table S1), it is most likely that they represent domestic horses whose maternal ancestors were recruited from local wild Anatolian populations, which follows a pattern observed elsewhere, whereby local mares were proposed to be recruited into domestic herds resulting in high mitogenomic diversity (48, 50). Our results suggest a rapid shift from hunting to herding following the introduction of domestic horses in the Early Bronze Age, which likely correlates with a terminal decline in the wild horse population in Anatolia and the Caucasus indicated by the low frequencies of their remains in faunal assemblages (70, 71).
The diachronic pattern in Anatolia and the Caucasus of the two Y chromosomal haplotypes, Y-HT-1 and Y-HT-3, hints at the possibility of population dynamics similar to that documented for northern Eurasia. In this latter region, Y-HT-1, constituting the predominant Y haplotype in present-day horses, markedly increased after the onset of domestication, reaching fixation in the gene pool of the domestic horse by the Middle Ages, while Y-HT-3 declined over time until its disappearance (57). In our Anatolian dataset, both haplotypes occurred at comparable frequencies around 2000 BCE; but here too, Y-HT-3 declined over time, with the most recent horse that carried Y-HT-3 originating from the Caucasus at around 1300 BCE. This reduction of Y chromosome diversity is presumably the result of strong selection of stallions (16).
Because northern Eurasia, and in particular the Pontic-Caspian steppe, is currently the most likely origin for the domestic horses brought into Anatolia, there are two possible introductory routes, one via southeastern Europe and one via the Caucasus. The route across the Bosporus has been postulated on the basis of the earliest zooarchaeological evidence for domestic horses in the southern Balkans at the Early Bronze Age site of Kanligeçit around 2600 to 2300 BCE (8). The coat colors of 10 horses from this site were genotyped and revealed a highly biased distribution of coat color mutations with 6 of the 10 homozygous black horses (a/a), 4 of which also show the leopard spotting (LP), plus 2 bay-colored horses with leopard spotting; no chestnut mutation was detected (72). This pattern strongly contrasts with our results in Anatolia and the Caucasus, where chestnut (e/e) is the earliest coat color variant, while black and leopard mutants remain very rare (only 2 of 25). These differences argue against the introduction of a domestic horse population similar to that found at Kanligeçit. Moreover, there is no archaeological evidence for horse management in western Anatolia in the third millennium BCE, providing little additional support for the hypothesis of an early introductory route across the Bosporus (73, 74).
In contrast, our identification of several allochthonous mitochondrial lineages and coat color mutations appearing broadly contemporaneously in the southern Caucasus and in central Anatolia argues in favor of a dispersal route via the Caucasus. The abundance of horse bones and images of horses in Maikop culture settlements and burials of c. 3300 BCE in the northern Caucasus led to the suggestion that horseback riding began in the Maikop period (75). In addition, recent studies of ancient human genomes showed continuous gene flow between Copper Age steppes and Caucasus peoples (3, 4, 76, 77), and later, during the Bronze Age, between Mesopotamia, Anatolia, the southern and northern Caucasus, and the steppes (78). This exchange between human groups appears to intensify during a century-long period of cooling and desertification, known as the 4.2-ka (thousand year) event [e.g., (79) but see (80)], which may have affected subsistence strategies and social networks in the steppe zone [e.g., (78, 81)]. On present evidence, this climatic event seems to be broadly contemporaneous with the arrival of nonlocal horse mitochondrial haplogroups and coat colors in the Caucasus and Anatolia and linked to the expansion of horse husbandry and possibly Indo-European languages [e.g., (14, 75) but see (82)]. Although the cultural processes initiating the dispersal of horse husbandry south of the Caucasus are currently difficult to address, it may relate to human population movements into the Caucasus and subsequently into Anatolia beginning in the late third millennium BCE.
Other equids
The present study identified a mule, offspring of a female horse and a male donkey, from an Early Iron Age context (dated to 1100 to 800 BCE) at central Anatolian Çadır Höyük. Mules have been identified on the basis of osteological criteria and, more recently, on genomic data in the European Iron Age and Roman periods (16, 83–85). Although several specimens have been tentatively identified as mules from Bronze and Iron Age contexts in SWA (86–88), this is the most ancient genomic evidence for a mule in SWA. Because wild donkeys are not native to Anatolia, the jackass involved must have been a domestic animal (29, 71). The parent mare of the mule from Çadır Höyük was also a domestic animal, as it carried an allochthonous mitochondrial haplogroup and contributed two coat color mutant alleles to its offspring. Although not a surprise, the presence of a mule in the Anatolian Iron Age reflects a new role for domestic horses that emerged as they moved into SWA where domestic donkeys had been used since the fourth millennium BCE (89) and where a tradition of equid (donkey × hemione) hybridization emerged in the third millennium BC (28). This situation reflects a true integration of horses into southwest Asian equid economies and is an early example of intentional livestock engineering. Being the most expensive species mentioned on a price list for livestock dating to the Hittite period (~1600 to ~1178 BCE (90), mules were obviously highly valued.
In addition to horses, our results provide unambiguous evidence that Neolithic, Chalcolithic, and Bronze Age people in Anatolia also hunted hydruntines, the hemione subspecies E. hemionus hydruntinus, which once inhabited large parts of Anatolia (41). This is not a trivial result, because osteological identification of the hydruntine is not obvious (41, 59), an observation confirmed in our study illustrating disagreement between genotyping and osteological determination in six cases. At the same time, it also confirmed the results of an earlier genetic study from our group illustrating that no other subspecies of hemione populated the Anatolian plateau (41). Hence, the most recent evidence in our dataset for an Anatolian hydruntine dates to ~2200 BCE, which is in agreement with results reported earlier (41). The combined data thus suggest that hydruntines went extinct in Anatolia during the Late Bronze Age more or less at the same time as local wild horses perhaps in response to the same combination of factors including increased aridity associated with the 4.2-ka event, competition for pasture resources with growing numbers of livestock, and hunting pressure perhaps related to practices of elites hunting (28).
CONCLUSIONS
Our study of ancient equid remains from Anatolia and the southern Caucasus covering ~9000 years of the Holocene analyzed the dynamics over time of mitochondrial lineages and tested the hypothesis that Anatolia was a center of horse domestication. We were able to identify mitotypes characteristic of local Anatolian wild horses, which were regularly exploited in the early and middle Holocene. However, we identified a pattern of genetic change that does not reflect a gradual process involving the local population but rather a sudden appearance ~2000 BCE of nonlocal lineages that are still present in domestic horses. We also show that these imported horses exhibited coat colors that are absent in local wild horses before domestication. Moreover, continuation of Anatolian maternal lineage P into the Bronze Age implies some limited incorporation of local mares into domestic herds. These patterns of change indicate that domestic horses were introduced into Anatolia perhaps via the Caucasus region during the Bronze Age and provide a date for the beginning of the exploitation of domestic horses in Anatolia and Transcaucasia. They also argue against local independent domestication of the horse in this region. Our results strongly suggest that Anatolia was not a primary source for domestic horse lineages, but, as observed in other regions, local matrilines were incorporated into herds of imported domestic horses, which were also hybridized with local donkeys to create mules. The ultimate geographic origins of the imported domestic herds remain to be determined, but eliminating Anatolia as a source of domestication directs further attention to the adjacent regions of the Black Sea.
MATERIAL AND METHODS
Archaeological bones
The provenance of the analyzed horse specimens is described in section S1 and table S1.
Genetic analysis
Sample preparation, DNA extraction, DNA purification, and the following pre-PCR procedures were carried out in the high-containment facility of the Jacques Monod Institute in Paris physically separated from areas where modern samples are analyzed and dedicated exclusively to ancient DNA analysis using the strict procedures for contamination prevention previously described (62, 91). Bone specimens were cleaned and ground in a freezer mill, and DNA-extracted and purified as previously described (see section S2.1) (61, 92). Genetic analyses were performed using aMPlex Torrent, a multiplex PCR assay coupled to NGS optimized to genotype reliably highly damaged ancient DNA at high throughput (60, 64, 65). The approach relies on careful design of primers that were optimized in silico and in vitro and on the systematic replication in triplicates of each PCR [(60) and Supplementary Materials]. Moreover, we used it in combination with rigorous methods of elimination of DNA molecules contaminating commercially purchased reagents (62) and of previous PCR amplification products with uracil-DNA glycosylase (UDG) treatment (93), which also removes cytosine deamination products (94). This approach proved extremely useful to retrieve selected markers more efficiently and at much lower costs than with shotgun sequencing (15). Compared to DNA capture approaches, it provides a higher yield, because capture approaches tend to leave gaps in the genomic regions that are targeted, which then have to be filled via PCR approaches [(61) and Supplementary Materials].
To amplify the mitochondrial genome, primers were designed to cover the complete mitochondrial genome with minimal primer dimer propensity from a multiple alignment of all the equid mitogenome sequences present in GenBank in 2014 using the software Oligo 7 as previously described (60). The primers were then tested for efficiency and dimer formation using quantitative real-time PCR optimized as described before (60) and combined in three different multiplex reactions (see sections S2.2.1, S3.1, and S3.4.1). We proceeded in the same way to analyze the other genetic markers. To analyze the coat color, we chose a set of eight SNPs in seven genes for detecting basic colors (bay, black, chestnut, and gray), diluted phenotypes (silver and cream), spotted or painted phenotypes (overo, tobiano, and sabino), and leopard spotting, as described previously (see sections S2.2.2, S3.2, and S3.4.2) (11). For the analysis of the Y chromosome, we selected four anonymous Y-linked fragments (Y2B17, Y3B1, Y3B12, and Y3B19) and two fragments of the amelogenin gene (AME2 and AME3), as previously described (see sections S2.2.3, S3.3, and S3.4.3) (54). To protect against cross-contamination between samples, we used the UDG-coupled PCR system (see sections S2.2.1 to S2.2.3). For each sample, we then performed the six multiplex PCRs with different primer combinations (mtDNA, coat color, and Y chromosome) (see section S3.4). Each extract was amplified in triplicate, and the triplicates were pooled so that, at the end, there were three pools for each extract. The PCR products were pooled in a 96-well plate, and DNA libraries were prepared in an automate Tecan Freedom EVO 100 ligating sample-specific Ion Torrent barcoded adaptors (see section S2.3). After amplification, the size distribution and concentration of the library were assessed on the Agilent 2100 Bioanalyzer. Emulsion PCR and Ion Sphere Particle enrichment were conducted with the Ion OneTouch System (Life Technologies) using the Ion OneTouch 200 Template kit v2 DL (see section S2.3). The DNA library was sequenced on the Ion Torrent Personal Genome Machine (PGM) Sequencer using the Ion PGM 200 Sequencing Kit and Ion 314 semiconductor sequencing chips (Life Technologies) (see section S2.3). Consensus sequences were established typically from several tens to several hundreds of sequences, and only replicated sequences were considered here.
Phylogenetic analysis of the mtDNA
A maximal likelihood phylogenetic tree was constructed from aligned complete Equus mitogenomes using RAxML (95) and a general time reversible nucleotide substitution model, with gamma categories and an estimated proportion of invariant sites. Following reduction of the mitogenomes to the fragment amplified by PCR after primer removal, a phylogenetic tree was constructed using PhyML (96) with four gamma categories and an estimated proportion of invariant sites and a Tamura-Nei substitution model (97).
Statistical analysis
The probability (two-tailed P values) that the changes of mtDNA haplotype and coat color diversity in Anatolia and the Caucasus after ~2000 BCE could have been observed simply by chance was estimated with the Fisher’s exact test using as nominal variables time (before and after 2000 BCE) and genotype (OPQX versus other haplogroups for mitochondrial haplotypes, bay versus other colors for coat colors).
Supplementary Material
Acknowledgments
We thank the directors of excavations H. Haupmann† (Norşun Tepe) and M. Özbaşaran (Aşıklı Höyük) for access to samples as well as the Department of Archaeology at Bitlis Eren University, the Çorum Müzesi, A. Schachner, and T. Emre Şerifoğlu for their support. Permission to export samples was granted by the Turkish Ministry of Culture and Tourism and the Aksaray Museum. We also thank R. Badalyan and A. T. Smith for permission to analyze the equids from the Project ArAGATS excavations. Funding: The study was supported by the NSF (BCS-1311551). The paleogenomic facility of the Institut Jacques Monod obtained support from the University Paris Diderot within the program “Actions de recherches structurantes.” The sequencing facility of the Institut Jacques Monod, Paris, is supported by grants from the University Paris Diderot, the Fondation pour la Recherche Médicale (DGE20111123014), and the Région Île-de-France (11015901). Moreover, we acknowledge support from the French National Research Center CNRS, the National Geographic Society, the American Research Institute in Turkey, the University of North Carolina at Chapel Hill, the Baylor University, and the Deutsche Forschungsgemeinschaft (DFG) to J.P. (PE 424/10,1-3). S.E.A. was supported by a Fulbright IIE Grant and the University of Chicago. Author contributions: B.S.A. and E.-M.G. initiated and coordinated the project. E.-M.G. and T.G. designed research and supervised experiments. S.G. performed experiments. E.-M.G., T.G., and S.G. analyzed data. S.E.A., H.B., H.C., N.M., and H.-P.U. provided samples and archaeological contexts. E.-M.G., T.G., and B.S.A. wrote the paper with input from J.P. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/38/eabb0030/DC1
REFERENCES AND NOTES
- 1.R. Drews, Early Riders: The Beginnings of Mounted Warfare in Asia and Europe (Routledge, 2004). [Google Scholar]
- 2.M. A. Levine, in Late Prehistoric Exploitation of the Eurasian Steppe, M. Levine, Y. Rassamakin, A. M. Kislenko, N. S. Tatarintseva, Eds. (Cambridge McDonald Institute, 2005), pp. 5–58. [Google Scholar]
- 3.Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., Brandt G., Nordenfelt S., Harney E., Stewardson K., Fu Q., Mittnik A., Bánffy E., Economou C., Francken M., Friederich S., Pena R. G., Hallgren F., Khartanovich V., Khokhlov A., Kunst M., Kuznetsov P., Meller H., Mochalov O., Moiseyev V., Nicklisch N., Pichler S. L., Risch R., Rojo Guerra M. A., Roth C., Szécsényi-Nagy A., Wahl J., Meyer M., Krause J., Brown D., Anthony D., Cooper A., Alt K. W., Reich D., Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allentoft M. E., Sikora M., Sjögren K. G., Rasmussen S., Rasmussen M., Stenderup J., Damgaard P. B., Schroeder H., Ahlström T., Vinner L., Malaspinas A. S., Margaryan A., Higham T., Chivall D., Lynnerup N., Harvig L., Baron J., Casa P. D., Dąbrowski P., Duffy P. R., Ebel A. V., Epimakhov A., Frei K., Furmanek M., Gralak T., Gromov A., Gronkiewicz S., Grupe G., Hajdu T., Jarysz R., Khartanovich V., Khokhlov A., Kiss V., Kolář J., Kriiska A., Lasak I., Longhi C., McGlynn G., Merkevicius A., Merkyte I., Metspalu M., Mkrtchyan R., Moiseyev V., Paja L., Pálfi G., Pokutta D., Pospieszny Ł., Price T. D., Saag L., Sablin M., Shishlina N., Smrčka V., Soenov V. I., Szeverényi V., Tóth G., Trifanova S. V., Varul L., Vicze M., Yepiskoposyan L., Zhitenev V., Orlando L., Sicheritz-Pontén T., Brunak S., Nielsen R., Kristiansen K., Willerslev E., Population genomics of Bronze Age Eurasia. Nature 522, 167–172 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Bökönyi S., The earliest waves of domestic horses in East Europe. J. Indo Eur. Stud. 6, 17–76 (1978). [Google Scholar]
- 6.N. Benecke, Der Mensch und seine Haustiere (Konrad Theiss Verlag GmbH & Co, 1994). [Google Scholar]
- 7.Uerpmann H.-P., Die Domestikation des Pferdes im Chalkolithikum West- und Mitteleuropas. Madrid. Mitt. 31, 109–153 (2001). [Google Scholar]
- 8.N. Benecke, On the beginning of horse husbandry in the southern Balkan Peninsula-the horse bones from Kırklareli -Kanligeçi̇t (Turkish Thrace), in Equids in Time and Space: Papers in Honour of Véra Eisenmann, M. Mashkour, Ed. (Oxbow Books, Oxford, 2006).
- 9.H. J. Greenfield, in Horses and Humans: The Evolution of Human-Equine Relationships, S. L. Olsen, S. Grant, A. Choyke, L. Bartosiewicz, Eds. (BAR International Series 1560, 2006), pp. 221–244. [Google Scholar]
- 10.Outram A. K., Stear N. A., Bendrey R., Olsen S., Kasparov A., Zaibert V., Thorpe N., Evershed R. P., The earliest horse harnessing and milking. Science 323, 1332–1335 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Ludwig A., Pruvost M., Reissmann M., Benecke N., Brockmann G. A., Castanos P., Cieslak M., Lippold S., Llorente L., Malaspinas A. S., Slatkin M., Hofreiter M., Coat color variation at the beginning of horse domestication. Science 324, 485 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anthony D. W., Brown D., Bit wear, horseback riding, and the Botai site in Kazakstan. J. Archaeol. Sci. 25, 331–347 (1998). [Google Scholar]
- 13.S. J. Olsen, in Prehistoric Steppe Adaptation and the Horse, M. Levine, C. Renfrew, K. Boyle, Eds. (McDonald Institute Monographs, 2003), pp. 83–102. [Google Scholar]
- 14.Gaunitz C., Fages A., Hanghøj K., Albrechtsen A., Khan N., Schubert M., Seguin-Orlando A., Owens I. J., Felkel S., Bignon-Lau O., de Barros Damgaard P., Mittnik A., Mohaseb A. F., Davoudi H., Alquraishi S., Alfarhan A. H., al-Rasheid K. A. S., Crubézy E., Benecke N., Olsen S., Brown D., Anthony D., Massy K., Pitulko V., Kasparov A., Brem G., Hofreiter M., Mukhtarova G., Baimukhanov N., Lõugas L., Onar V., Stockhammer P. W., Krause J., Boldgiv B., Undrakhbold S., Erdenebaatar D., Lepetz S., Mashkour M., Ludwig A., Wallner B., Merz V., Merz I., Zaibert V., Willerslev E., Librado P., Outram A. K., Orlando L., Ancient genomes revisit the ancestry of domestic and Przewalski’s horses. Science 360, 111–114 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Librado P., Gamba C., Gaunitz C., der Sarkissian C., Pruvost M., Albrechtsen A., Fages A., Khan N., Schubert M., Jagannathan V., Serres-Armero A., Kuderna L. F. K., Povolotskaya I. S., Seguin-Orlando A., Lepetz S., Neuditschko M., Thèves C., Alquraishi S., Alfarhan A. H., al-Rasheid K., Rieder S., Samashev Z., Francfort H. P., Benecke N., Hofreiter M., Ludwig A., Keyser C., Marques-Bonet T., Ludes B., Crubézy E., Leeb T., Willerslev E., Orlando L., Ancient genomic changes associated with domestication of the horse. Science 356, 442–445 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Fages A., Hanghøj K., Khan N., Gaunitz C., Seguin-Orlando A., Leonardi M., McCrory Constantz C., Gamba C., al-Rasheid K. A. S., Albizuri S., Alfarhan A. H., Allentoft M., Alquraishi S., Anthony D., Baimukhanov N., Barrett J. H., Bayarsaikhan J., Benecke N., Bernáldez-Sánchez E., Berrocal-Rangel L., Biglari F., Boessenkool S., Boldgiv B., Brem G., Brown D., Burger J., Crubézy E., Daugnora L., Davoudi H., de Barros Damgaard P., de los Ángeles de Chorro y de Villa-Ceballos M., Deschler-Erb S., Detry C., Dill N., do Mar Oom M., Dohr A., Ellingvåg S., Erdenebaatar D., Fathi H., Felkel S., Fernández-Rodríguez C., García-Viñas E., Germonpré M., Granado J. D., Hallsson J. H., Hemmer H., Hofreiter M., Kasparov A., Khasanov M., Khazaeli R., Kosintsev P., Kristiansen K., Kubatbek T., Kuderna L., Kuznetsov P., Laleh H., Leonard J. A., Lhuillier J., Liesau von Lettow-Vorbeck C., Logvin A., Lõugas L., Ludwig A., Luis C., Arruda A. M., Marques-Bonet T., Matoso Silva R., Merz V., Mijiddorj E., Miller B. K., Monchalov O., Mohaseb F. A., Morales A., Nieto-Espinet A., Nistelberger H., Onar V., Pálsdóttir A. H., Pitulko V., Pitskhelauri K., Pruvost M., Rajic Sikanjic P., Rapan Papeša A., Roslyakova N., Sardari A., Sauer E., Schafberg R., Scheu A., Schibler J., Schlumbaum A., Serrand N., Serres-Armero A., Shapiro B., Sheikhi Seno S., Shevnina I., Shidrang S., Southon J., Star B., Sykes N., Taheri K., Taylor W., Teegen W. R., Trbojević Vukičević T., Trixl S., Tumen D., Undrakhbold S., Usmanova E., Vahdati A., Valenzuela-Lamas S., Viegas C., Wallner B., Weinstock J., Zaibert V., Clavel B., Lepetz S., Mashkour M., Helgason A., Stefánsson K., Barrey E., Willerslev E., Outram A. K., Librado P., Orlando L., Tracking five millennia of horse management with extensive ancient genome time series. Cell 177, 1419–1435.e31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D. Anthony, The Horse, the Wheel, and Language: How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World (Princeton Univ. Press, 2007). [Google Scholar]
- 18.Arbuckle B. S., Animals and inequality in Chalcolithic central Anatolia. J. Anthropol. Archaeol. 31, 302–313 (2012). [Google Scholar]
- 19.N. S. Shaler, Domesticated Animals. Their Relation to Man and to His Advancement in Civilization (Charles Scribner’s Sons, 1895). [Google Scholar]
- 20.L. Martin, N. Russell, in Horses and Humans: The Evolution of Human-Equine Relationships, S. J. Olsen, Ed. (British Archaeological Reports International Series 1560, 2006), pp. 115–126. [Google Scholar]
- 21.A. Ünal, Eski Anadolu’da At Hititçe Kikkuli at eğitimi metinleri ve “Tavlaya çekmek”le ilgili teknik bir ayrıntı. Çorum Kültür Sanat: Bilim, Kültür, Sanat, Tarih, ve Turiz, Dergisi II, 40–66 (2013–2014).
- 22.B. S. Arbuckle, A. Öztan, in Archaeozoology of the Near East XII, C. Çakırlar, J. Chahoud, R. Berthon, S. E. Pilaar, Eds. (Barkhuis Publishing & University of Groningen, 2018), pp. 42–59. [Google Scholar]
- 23.J. M. C. Toynbee, M. R. Alföldi, D. Misselbeck, Tierwelt der Antike - Bestiarium Romanum (Verlag Philipp von Zabern, 1983). [Google Scholar]
- 24.Boessneck J., von den Driesch A., Pferde im 4/3. Jahrtausend v. Chr. in Ostanatolien. Säugetierkd. Mitt. 24, 81–87 (1976). [Google Scholar]
- 25.M. A. Littauer, J. H. Crouwel, Wheeled Vehicles and Ridden Animals in the Ancient Near East (Brill, 1979). [Google Scholar]
- 26.Owen D. I., The first equestrian: An Ur III glyptic scene. Acta Sumerologica 13, 259–273 (1991). [Google Scholar]
- 27.J. Oates, in Prehistoric Steppe Adaptation and the Horse, M. Levine, C. Renfrew, K. Boyle, Eds. (McDonald Institute Monograph, 2003), pp. 115–125. [Google Scholar]
- 28.J. Zarins, The Domestication of Equidae in Third-Millennium BCE Mesopotamia. Cornell University Studies in Assyriology and Sumerology (Cornell University, 2014), vol. 24. [Google Scholar]
- 29.J. Clutton-Brock, Horse Power. A History of the Horse and the Donkey in Human Societies (Harvard Univ. Press, 1992). [Google Scholar]
- 30.S. Bökönyi, in Die Indogermanen und Das Pferd, B. Hänsel, S. Zimmer, Eds. (Akten Internationalen Interdisziplinären Kolloquiums Freie Universität Bernfried Schlerath, 1994), pp. 115–122. [Google Scholar]
- 31.F. E. Zeuner, A History of Domesticated Animals (Hutchinson, 1963). [Google Scholar]
- 32.H.-P. Uerpmann, Domestication of the Horse: When, Where, and Why, L. Bodson, Ed., Colloque d’histoire des connaissances zoologiques (Le cheval et les autres équidés: Aspects de l’histoire de leur insertion dans les activités humains) (University of Liege, 1995). [Google Scholar]
- 33.A. Sherratt, in Prehistoric Steppe Adaptation and the Horse, M. Levine, C. Renfrew, K. Boyle, Eds. (McDonald Institute Monographs, 2003), pp. 233–252. [Google Scholar]
- 34.D. W. Anthony, The Horse, the Wheel, and Language: How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World (Princeton Univ. Press, 2007). [Google Scholar]
- 35.C. Becker, in Die Indogermanen und Das Pferd, B. Hänsel, S. Zimmer, Eds. (Akten Internationalen Interdisziplinären Kolloquiums Freie Universität, Bernfried Schlerath, 1994), pp. 145–177. [Google Scholar]
- 36.H.-P. Uerpmann, The Ancient Distribution of Ungulate Mammals in the Middle East. Beihefte zum Tübinger Atlas des Vorderen Orients Reihe A (Naturwissenschaften) Nr. 27, Wiesbaden, Dr. Ludwig Reichert, (1987).
- 37.S. Bökönyi, in Equids in the Ancient World Volume II, R. H. Meadow, H.-P. Uerpmann, Eds. (Dr. Ludwig Reichert Verlag, 1991), vol. Beihefte zum Tübinger Atlas des Vorderen Orients Reihe A (Naturwisseschaften) 19/2, pp. 123–131. [Google Scholar]
- 38.N. Benecke, in Archaeozoology of the Near East III. Proceedings of the Third International Symposium on the Archaeozoology of Southwestern Asia and Adjacent Areas, H. Buitenhuis, L. Bartosiewicz, A. Choyke, Eds. (ARC Publication 18, 1998), pp. 172–179. [Google Scholar]
- 39.E. Vila, in Equids in Time and Space: Papers in Honour of Vera Eisenmann, M. Mashkour, Ed. (Oxbow Books, 2006), pp. 102–123. [Google Scholar]
- 40.Arbuckle B. S., Zooarchaeology at Acemhöyük 2013. Anadolu/Anatolia 39, 55–68 (2013). [Google Scholar]
- 41.Bennett E. A., Champlot S., Peters J., Arbuckle B. S., Guimaraes S., Pruvost M., Bar-David S., Davis S. J. M., Gautier M., Kaczensky P., Kuehn R., Mashkour M., Morales-Muñiz A., Pucher E., Tournepiche J. F., Uerpmann H. P., Bălăşescu A., Germonpré M., Gündem C. Y., Hemami M.-R., Moullé P.-E., Ötzan A., Uerpmann M., Walzer C., Grange T., Geigl E. M., Taming the late Quaternary phylogeography of the Eurasiatic wild ass through ancient and modern DNA. PLOS ONE 12, e0174216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.H. Buitenhuis, J. Peters, N. Pöllath, M. C. Stiner, N. D. Munro, Ö. Saritas, The Faunal Remains from Levels 3 and 2 of Aşıklı Höyük: Evidence for Emerging Management Practices, in The Early Settlement at Aşıklı Höyük; Essays in Honor of Ufuk Esin, M. Özbașaran, G. Duru, M. C. Stiner, Eds. (Phoibos Verlag Wien, 2018), pp. 282–323. [Google Scholar]
- 43.S. Payne, in Equids in the Ancient World, R. H. Meadow, H. P. Uerpmann, Eds. (Dr. Ludwig Reichert Verlag, 1991), vol. II, pp. 132–165. [Google Scholar]
- 44.H.-P. Uerpmann, in The Salvage Excavations at Orman Fidanlıgı: A Chalcolithic Site in Inland Northwestern Anatolia, T. Efe, Ed. (TASK Vakfı Yayınları, 2001), pp. 187–210. [Google Scholar]
- 45.Arbuckle B. S., Zooarchaeology at Kösk Höyük. Kazı Sonuçları Toplantısı 27, 124–136 (2008). [Google Scholar]
- 46.Arbuckle B. S., Chalcolithic caprines, Dark Age dairy and Byzantine beef. Anatolica 35, 179–224 (2009). [Google Scholar]
- 47.Achilli A., Olivieri A., Soares P., Lancioni H., Kashani B. H., Perego U. A., Nergadze S. G., Carossa V., Santagostino M., Capomaccio S., Felicetti M., al-Achkar W., Penedo M. C. T., Verini-Supplizi A., Houshmand M., Woodward S. R., Semino O., Silvestrelli M., Giulotto E., Pereira L., Bandelt H. J., Torroni A., Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc. Natl. Acad. Sci. U.S.A. 109, 2449–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vila C., Leonard J. A., Gotherstrom A., Marklund S., Sandberg K., Liden K., Wayne R. K., Ellegren H., Widespread origins of domestic horse lineages. Science 291, 474–477 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Jansen T., Forster P., Levine M. A., Oelke H., Hurles M., Renfrew C., Weber J., Olek K., Mitochondrial DNA and the origins of the domestic horse. Proc. Natl. Acad. Sci. U.S.A. 99, 10905–10910 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cieslak M., Pruvost M., Benecke N., Hofreiter M., Morales A., Reissmann M., Ludwig A., Origin and history of mitochondrial DNA lineages in domestic horses. PLOS ONE 5, e15311 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lira J., Linderholm A., Olaria C., Durling M. B., M. T. P. Gilbert, Ellegren H., Willerslev E., Lidén K., Arsuaga J. L., Götherström A., Ancient DNA reveals traces of Iberian Neolithic and Bronze Age lineages in modern Iberian horses. Mol. Ecol. 19, 64–78 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Lippold S., Matzke N. J., Reissmann M., Hofreiter M., Whole mitochondrial genome sequencing of domestic horses reveals incorporation of extensive wild horse diversity during domestication. BMC Evol. Biol. 11, 328 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warmuth V., Eriksson A., Bower M. A., Barker G., Barrett E., Hanks B. K., Li S., Lomitashvili D., Ochir-Goryaeva M., Sizonov G. V., Soyonov V., Manica A., Reconstructing the origin and spread of horse domestication in the Eurasian steppe. Proc. Natl. Acad. Sci. U.S.A. 109, 8202–8206 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindgren G., Backström N., Swinburne J., Hellborg L., Einarsson A., Sandberg K., Cothran G., Vilà C., Binns M., Ellegren H., Limited number of patrilines in horse domestication. Nat. Genet. 36, 335–336 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Kavar T., Dovç P., Domestication of the horse: Genetic relationships between domestic and wild horses. Livest. Sci. 116, 1–14 (2008). [Google Scholar]
- 56.Lippold S., Knapp M., Kuznetsova T., Leonard J. A., Benecke N., Ludwig A., Rasmussen M., Cooper A., Weinstock J., Willerslev E., Shapiro B., Hofreiter M., Discovery of lost diversity of paternal horse lineages using ancient DNA. Nat. Commun. 2, 450 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Wutke S., Sandoval-Castellanos E., Benecke N., Döhle H. J., Friederich S., Gonzalez J., Hofreiter M., Lõugas L., Magnell O., Malaspinas A. S., Morales-Muñiz A., Orlando L., Reissmann M., Trinks A., Ludwig A., Decline of genetic diversity in ancient domestic stallions in Europe. Sci. Adv. 4, eaap9691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pruvost M., Bellone R., Benecke N., Sandoval-Castellanos E., Cieslak M., Kuznetsova T., Morales-Muñiz A., O’Connor T., Reissmann M., Hofreiter M., Ludwig A., Genotypes of predomestic horses match phenotypes painted in Paleolithic works of cave art. Proc. Natl. Acad. Sci. U.S.A. 108, 18626–18630 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geigl E. M., Grange T., Eurasian wild asses in time and space: Morphological versus genetic diversity. Ann. Anat. 194, 88–102 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Guimaraes S., Pruvost M., Daligault J., Stoetzel E., Bennett E. A., Côté N. M. L., Nicolas V., Lalis A., Denys C., Geigl E. M., Grange T., A cost-effective high-throughput metabarcoding approach powerful enough to genotype ~44 000 year-old rodent remains from Northern Africa. Mol. Ecol. Resour. 17, 405–417 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Massilani D., Guimaraes S., Brugal J.-P., Bennett E. A., Tokarska M., Arbogast R.-M., Baryshnikov G., Boeskorov G., Castel J.-C., Davydov S., Madelaine S., Putelat O., Spasskaya N. N., Uerpmann H.-P., Grange T., Geigl E.-M., Past climate changes, population dynamics and the origin of Bison in Europe. BMC Biol. 14, 93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Champlot S., Berthelot C., Pruvost M., Bennett E. A., Grange T., Geigl E. M., An efficient multistrategy DNA decontamination procedure of PCR reagents for hypersensitive PCR applications. PLOS ONE 5, e13042, e13042 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Albrechtsen A., Nielsen F. C., Nielsen R., Ascertainment biases in SNP chips affect measures of population divergence. Mol. Biol. Evol. 27, 2534–2547 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coté N. M., Daligault J., Pruvost M., Bennett E. A., Gorgé O., Guimaraes S., Capelli N., Bailly M. L., Geigl E.-M., Grange T., A new high-throughput approach to genotype ancient human gastrointestinal parasites. PLOS ONE 11, e0146230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ottoni C., van Neer W., de Cupere B., Daligault J., Guimaraes S., Peters J., Spassov N., Prendergast M. E., Boivin N., Morales-Muñiz A., Bălăşescu A., Becker C., Benecke N., Boroneant A., Buitenhuis H., Chahoud J., Crowther A., Llorente L., Manaseryan N., Monchot H., Onar V., Osypińska M., Putelat O., Quintana Morales E. M., Studer J., Wierer U., Decorte R., Grange T., Geigl E.-M., The palaeogenetics of cat dispersal in the ancient world. Nat. Ecol. Evol. 1, 0139 (2017). [Google Scholar]
- 66.Weinstock J., Willerslev E., Sher A., Tong W., Ho S. Y. W., Rubenstein D., Storer J., Burns J., Martin L., Bravi C., Prieto A., Froese D., Scott E., Xulong L., Cooper A., Evolution, systematics, and phylogeography of Pleistocene horses in the new world: A molecular perspective. PLOS Biol. 3, e241 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lorenzen E. D., Nogués-Bravo D., Orlando L., Weinstock J., Binladen J., Marske K. A., Ugan A., Borregaard M. K., Gilbert M. T., Nielsen R., Ho S. Y., Goebel T., Graf K. E., Byers D., Stenderup J. T., Rasmussen M., Campos P. F., Leonard J. A., Koepfli K. P., Froese D., Zazula G., Stafford TW Jr, Aaris-Sørensen K., Batra P., Haywood A. M., Singarayer J. S., Valdes P. J., Boeskorov G., Burns J. A., Davydov S. P., Haile J., Jenkins D. L., Kosintsev P., Kuznetsova T., Lai X., Martin L. D., McDonald H., Mol D., Meldgaard M., Munch K., Stephan E., Sablin M., Sommer R. S., Sipko T., Scott E., Suchard M. A., Tikhonov A., Willerslev R., Wayne R. K., Cooper A., Hofreiter M., Sher A., Shapiro B., Rahbek C., Willerslev E., Species-specific responses of Late Quaternary megafauna to climate and humans. Nature 479, 359–364 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Orlando L., Ginolhac A., Zhang G., Froese D., Albrechtsen A., Stiller M., Schubert M., Cappellini E., Petersen B., Moltke I., Johnson P. L. F., Fumagalli M., Vilstrup J. T., Raghavan M., Korneliussen T., Malaspinas A. S., Vogt J., Szklarczyk D., Kelstrup C. D., Vinther J., Dolocan A., Stenderup J., Velazquez A. M. V., Cahill J., Rasmussen M., Wang X., Min J., Zazula G. D., Seguin-Orlando A., Mortensen C., Magnussen K., Thompson J. F., Weinstock J., Gregersen K., Røed K. H., Eisenmann V., Rubin C. J., Miller D. C., Antczak D. F., Bertelsen M. F., Brunak S., al-Rasheid K. A. S., Ryder O., Andersson L., Mundy J., Krogh A., Gilbert M. T. P., Kjær K., Sicheritz-Ponten T., Jensen L. J., Olsen J. V., Hofreiter M., Nielsen R., Shapiro B., Wang J., Willerslev E., Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499, 74–78 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Elsner J., Hofreiter M., Schibler J., Schlumbaum A., Ancient mtDNA diversity reveals specific population development of wild horses in Switzerland after the Last Glacial Maximum. PLOS ONE 12, e0177458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boessneck J., Driesch A. v. d., Tierknochen und Molluskenfunde aus Munbāqa. Mitteilungen der Deutschen Orient-Gesellschaft 118, 147–160 (1986). [Google Scholar]
- 71.H. P. Uerpmann, The Ancient Distribution of Ungulate Mammals in the Middle East, Beihefte zum Tübinger Atlas des Vorderen Orients. Reihe A (Naturwissenschaften) Nr. 27 (Dr. Ludwig Reichert Verlag, 1987). [Google Scholar]
- 72.Ludwig A., Reissmann M., Benecke N., Bellone R., Sandoval-Castellanos E., Cieslak M., Fortes G. G., Morales-Muñiz A., Hofreiter M., Pruvost M., Twenty-five thousand years of fluctuating selection on leopard complex spotting and congenital night blindness in horses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20130386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.H. P. Uerpmann, in Troia and the Troad: Scientific Approaches, G. A. Wagner, E. Pernicka, H. P. Uerpmann, Eds. (Springer, 2013). [Google Scholar]
- 74.C. Çakırlar, L. Atici, Patterns of animal exploitation in western Turkey: From Paleolithic molluscs to Byzantine elephants, in The Oxford Handbook of Zooarchaeology, U. Albarella, H. Russ, K. Vickers, S. Viner-Daniels, Eds., (Oxford Univ. Press, 2017). [Google Scholar]
- 75.Lazaridis I., Nadel D., Rollefson G., Merrett D. C., Rohland N., Mallick S., Fernandes D., Novak M., Gamarra B., Sirak K., Connell S., Stewardson K., Harney E., Fu Q., Gonzalez-Fortes G., Jones E. R., Roodenberg S. A., Lengyel G., Bocquentin F., Gasparian B., Monge J. M., Gregg M., Eshed V., Mizrahi A. S., Meiklejohn C., Gerritsen F., Bejenaru L., Blüher M., Campbell A., Cavalleri G., Comas D., Froguel P., Gilbert E., Kerr S. M., Kovacs P., Krause J., McGettigan D., Merrigan M., Merriwether D. A., O’Reilly S., Richards M. B., Semino O., Shamoon-Pour M., Stefanescu G., Stumvoll M., Tönjes A., Torroni A., Wilson J. F., Yengo L., Hovhannisyan N. A., Patterson N., Pinhasi R., Reich D., Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Barros Damgaard P., Martiniano R., Kamm J., Moreno-Mayar J. V., Kroonen G., Peyrot M., Barjamovic G., Rasmussen S., Zacho C., Baimukhanov N., Zaibert V., Merz V., Biddanda A., Merz I., Loman V., Evdokimov V., Usmanova E., Hemphill B., Seguin-Orlando A., Yediay F. E., Ullah I., Sjögren K. G., Iversen K. H., Choin J., de la Fuente C., Ilardo M., Schroeder H., Moiseyev V., Gromov A., Polyakov A., Omura S., Senyurt S. Y., Ahmad H., McKenzie C., Margaryan A., Hameed A., Samad A., Gul N., Khokhar M. H., Goriunova O. I., Bazaliiskii V. I., Novembre J., Weber A. W., Orlando L., Allentoft M. E., Nielsen R., Kristiansen K., Sikora M., Outram A. K., Durbin R., Willerslev E., The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science 360, eaar7711 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang C. C., Reinhold S., Kalmykov A., Wissgott A., Brandt G., Jeong C., Cheronet O., Ferry M., Harney E., Keating D., Mallick S., Rohland N., Stewardson K., Kantorovich A. R., Maslov V. E., Petrenko V. G., Erlikh V. R., Atabiev B. C., Magomedov R. G., Kohl P. L., Alt K. W., Pichler S. L., Gerling C., Meller H., Vardanyan B., Yeganyan L., Rezepkin A. D., Mariaschk D., Berezina N., Gresky J., Fuchs K., Knipper C., Schiffels S., Balanovska E., Balanovsky O., Mathieson I., Higham T., Berezin Y. B., Buzhilova A., Trifonov V., Pinhasi R., Belinskij A. B., Reich D., Hansen S., Krause J., Haak W., Ancient human genome-wide data from a 3000-year interval in the Caucasus corresponds with eco-geographic regions. Nat. Commun. 10, 590 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Szczęsny T. J., Was the 4.2 ka event an anthropogenic disaster? Open J. Ecol. 6, 613–631 (2016). [Google Scholar]
- 79.Voosen P., New geological age comes under fire. Science 361, 537–538 (2018). [DOI] [PubMed] [Google Scholar]
- 80.N. Shishlina, in Counterpoint: Essays in Archaeology and Heritage Studies in Honour of Professor Kristian Kristiansen, S. Bergerbrant, S. Sabatini, Eds. (Archaeopress, 2013), pp. 53–60. [Google Scholar]
- 81.D. W. Anthony, The Horse, the Wheel and Language. How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World (Princeton Univ. Press, 2007), pp. 847. [Google Scholar]
- 82.J.-P. Démoule, Mais où sont passés les Indo-Européens ? Le mythe d’origine de l’Occident (Editions du Seuil, 2014), pp. 823. [Google Scholar]
- 83.Armitage P. L., Chapman H., Roman mules. Lond. Archaeol. 3, 339–346 (1979). [Google Scholar]
- 84.J. Peters, Römische Tierhaltung und Tierzucht. Eine Synthese aus archäozoologischer Untersuchung und schriftlich-bildlicher Überlieferung. Passauer Universitätsschriften zur Archäologie (Rahden/Westf.: Leidorf., 1998), vol. 5.
- 85.C. Johnstone, A biometric study of equids in the Roman world, PhD thesis, Department of Archaeology, University of York (2004). [Google Scholar]
- 86.J. Peters, N. Pöllath, A. von den Driesch, in Madaba Plains Project: The 1994 Season at Tall Al-Umayri and Subsequent Studies, L. Herr, Ed. (Andrews Univ. Press, 2002), vol. Madaba Plains Project series 5, pp. 305–347. [Google Scholar]
- 87.C. Becker, in Archaeozoology of the Near East VIII, E. Vila, L. Gourichon, A. Choyke, H. Buitenhuis, Eds. (Maison de l’Orient et de la Mediterranee, 2008), pp. 561–580. [Google Scholar]
- 88.F. A. Mohaseb, M. Mashkour, in Elamica: On History and Culture of Elam and Its Neighboring Regions, B. Mofidi-Nasrabadi, Ed. (Verlag Franzbecker, 2012), vol. 2, pp. 33–54. [Google Scholar]
- 89.Boessneck J., von den Driesch A., Steger U., Tierknochenfunde der Ausgrabungen des Deutschen Archäologischen Instituts Baghdad in Uruk-Warka, Iraq. Baghdader Mitt. 15, 149–189 (1984). [Google Scholar]
- 90.H. Otten, in Kulturgeschichte des alten Orients: Mesopotamien, Hethiterreich, Syrien-Palästina, Urartu, H. Schmökel, Ed. (Kröner, 1961), pp. 311–446. [Google Scholar]
- 91.Bennett E. A., Massilani D., Daligault J., Geigl E.-M., Grange T., Library construction for ancient genomics: Single strand or double strand? Biotechniques 56, 289–290 (2014). [DOI] [PubMed] [Google Scholar]
- 92.Gorgé O., Bennett E. A., Massilani D., Daligault J., Pruvost M., Geigl E. M., Grange T., Analysis of ancient DNA in microbial ecology. Methods Mol. Biol. 1399, 289–315 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Pruvost M., Grange T., Geigl E.-M., Minimizing DNA contamination by using UNG-coupled quantitative real-time PCR on degraded DNA samples: Application to ancient DNA studies. Biotechniques 38, 569–575 (2005). [DOI] [PubMed] [Google Scholar]
- 94.Pruvost M., Schwarz R., Bessa Correia V., Champlot S., Grange T., Geigl E.-M., DNA diagenesis and palaeogenetic analysis: Critical assessment and methodological progress. Palaeogeogr. Palaeoclimatol. Palaeoecol. 266, 211–219 (2008). [Google Scholar]
- 95.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O., New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 97.A. J. Drummond, B. Ashton, S. Buxton, M. Cheung, A. Cooper, C. Duran, M. Field, J. Heled, M. Kearse, S. Markowitz, R. Moir, S. Stones-Havas, S. Sturrock, T. Thierer, A. Wilson, Geneious v5.4 (2011); http://www.geneious.com/. [DOI] [PMC free article] [PubMed]
- 98.Der Sarkissian C., Ermini L., Schubert M., Yang M. A., Librado P., Fumagalli M., Jónsson H., Bar-Gal G. K., Albrechtsen A., Vieira F. G., Petersen B., Ginolhac A., Seguin-Orlando A., Magnussen K., Fages A., Gamba C., Lorente-Galdos B., Polani S., Steiner C., Neuditschko M., Jagannathan V., Feh C., Greenblatt C. L., Ludwig A., Abramson N. I., Zimmermann W., Schafberg R., Tikhonov A., Sicheritz-Ponten T., Willerslev E., Marques-Bonet T., Ryder O. A., Cue M. M., Rieder S., Leeb T., Slatkin M., Orlando L., Evolutionary genomics and conservation of the endangered Przewalski’s horse. Curr. Biol. 25, 2577–2583 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/38/eabb0030/DC1


