BNAbs of VH1-69 germline origin from a Chinese patient recognize the V3 glycan supersite via a disulfide-locked long HCDR3 loop.
Abstract
An oligomannose patch around the V3 base of HIV-1 envelope glycoprotein (Env) is recognized by multiple classes of broadly neutralizing antibodies (bNAbs). Here, we investigated the bNAb response to the V3 glycan supersite in an HIV-1–infected Chinese donor by Env-specific single B cell sorting, structural and functional studies, and longitudinal analysis of antibody and virus repertoires. Monoclonal antibodies 438-B11 and 438-D5 were isolated that potently neutralize HIV-1 with moderate breadth, are encoded by the VH1-69 germline gene, and have a disulfide-linked long HCDR3 loop. Crystal structures of Env-bound and unbound antibodies revealed heavy chain–mediated recognition of the glycan supersite with a unique angle of approach and a critical role of the intra-HCDR3 disulfide. The mechanism of viral escape was examined via single-genome amplification/sequencing and glycan mutations around the N332 supersite. Our findings further emphasize the V3 glycan supersite as a prominent target for Env-based vaccine design.
INTRODUCTION
During HIV-1 infection, a small fraction of individuals mount effective antibody responses against coevolving viruses, which eventually become broadly neutralizing antibodies (bNAbs) capable of countering diverse HIV-1 isolates with exceptional potency (1–3). Most of the known HIV-1 bNAbs have unusual characteristics such as selective germline gene usage, high degree of somatic hypermutation (SHM), and long heavy-chain complementarity-determining region 3 (HCDR3) loops. Precursors of bNAbs may emerge in the early stage of infection and later achieve neutralization breadth and potency through an extensive maturation process (4–7). Such bNAb precursors often display low levels of autologous neutralization but can initiate the process of recognizing conserved epitopes on the envelope glycoprotein (Env) in response to a constantly evolving viral population (5, 6, 8). Multiple bNAb epitopes on HIV-1 Env have been identified, such as the CD4 binding site (CD4bs), glycan-rich epitopes at the V2 apex and V3 base, gp120-gp41 interface, fusion peptide, and membrane-proximal external region (MPER) (9), in addition to the silent face (10, 11).
HIV-1 cohort studies found ~15% of individuals with broad serum neutralizing activity and nearly half of these broad neutralizers showing bNAb responses to the oligomannose-rich patch centered at Asn332 (12, 13), suggesting that the immune system can effectively recognize this glycan-rich V3 epitope on the Env spike. Panels of human antibodies to this site have been isolated that exhibit moderate [PCDN lineage (6)] to high potency [PGT121 (13), PGT128 (14, 15), 10–1074 (16), and BG18 (2)], as well as substantial breadth (28 to 68%) (17). These bNAbs penetrate the glycan shield of the Env spike usually with their light-chain CDR1/2 (LCDR1/2), HCDR2, and long HCDR3 loops and interact with both oligomannose glycans and peptide components of the N332/V3 supersite. Furthermore, structural studies (18–21) have revealed various angles of approach by which these bNAbs access the CCR5 coreceptor binding motif, GDIR (22, 23). Env trimer–based immunogens RC1 (24) and N332-GT (25) were designed to elicit human bNAb precursor antibodies targeting the N332/V3-glycan patch. It has been reported that a common mechanism used by HIV-1 to escape the N332/V3-specific bNAbs is through mutations of Asn332 (6), Asn295, and Asn301 (26) that eliminate N-linked glycans; a glycan shift from position 332 to 334 (4, 27); or an unusual elongation of the V1 loop with additional glycans and disulfide bonds (28). However, some HIV-1–infected individuals can develop broad serum neutralization against the N332/V3 supersite by switching the glycan from position 334 to 332 (27). It is, therefore, imperative to study more elite donors to understand how bNAbs recognize the shifting glycans at or around the V3 base and how the coevolving viruses counter it. One of the key questions is whether any other features could be used by bNAbs to enhance the glycan epitope recognition and, hence, HIV-1 neutralization, in addition to diverse angles of approach and long CDR loops.
In this study, we examined an N332/V3-directed bNAb response in an HIV-1 subtype B–infected Chinese donor, CBJC438. This individual showed sustained plasma neutralizing activity over a period of 8 years from 2005 to 2012. Using a heptad repeat 1 (HR1)–stabilized BG505 trimer (29) as bait, we isolated two monoclonal antibodies (mAbs) (438-B11 and 438-D5) from the 2008 sample that exhibited exceptional neutralizing potency with moderate breadth. To understand how these two related bNAbs recognize and neutralize HIV-1, we determined their structures in the unbound form and in complex with an uncleaved prefusion-optimized (UFO) BG505 Env trimer. Crystal structures revealed how these bNAbs approach the N332/V3 supersite on the trimer with a disulfide-locked HCDR3 loop and a unique angle of approach relying on heavy chain–dependent recognition. Longitudinal repertoire analysis uncovered a diverging bNAb lineage that expanded and peaked in 2008 but diminished to only a few clones in 2012 due to viral escape in and around the N332 supersite. These results depict a dynamic host-virus coevolution process in which a VH1-69–originated bNAb lineage emerged in the donor B cell repertoire by targeting the N332/V3-glycan patch on the HIV-1 Env. The viruses responded by shifting the glycan from position 332 to 334 and minimizing the involvement of other glycans, such as N301, in epitope recognition by bNAbs, thereby enabling the viruses to successfully circumvent this bNAb lineage.
RESULTS
BNAbs 438-B11 and 438-D5 are isolated from a Chinese donor, CBJC438
A Chinese donor, CBJC438, was diagnosed in the early 1990s with an HIV-1 subtype B infection through a contaminated plasma donation. A natural history study cohort of more than 400 HIV-1–infected plasma donors with a CD4 count of more than 350 cells/μl was established in 2005 and followed every 6 months to investigate the HIV-1 disease progression correlates, including innate immunity, cytotoxic T lymphocyte responses, and neutralizing antibodies. Patients were followed until their CD4 count was below 200 (before 2008) or 350 cells/μl (after 2012), reaching the criteria of World Health Organization’s antiviral guideline for developing countries. As a member of this cohort and after 15 years of infection without antiviral treatment, CBJC438 displayed a CD4+ T cell count ranging from 345 to 827 cells/μl with a viral load from 18,700 to 225,000 copies/ml during 2005 to 2012 (Fig. 1A). Serum neutralization was tested against a global panel of 12 pseudoviruses (30) at all time points studied (2005 to 2012). The neutralizing activity of the donor’s plasma from different time points showed 100% breadth against the global virus panel (Fig. 1B and table S1A). The average potency was between 50 and 352 geometric mean titers. The 2008 samples were used for antibody isolation by single cell–based sorting to identify Env-specific mAbs. An HR1-redesigned BG505 trimer (29) was used as the sorting bait. The same trimer was used previously (31) as the panning antigen to analyze the single-chain antibody fragment libraries derived from donor 17, the source of PGT121-class bNAbs (13). Here, this BG505 Env bait was used to sort immunoglobulin G–positive (IgG+) B cells from CBJC438 peripheral blood mononuclear cells (PBMCs) with a set of selected cell markers. Native trimer-specific single B cells with phenotype CD19+CD20+CD14−CD3−CD8−IgM−IgG+BG505+ were gated and sorted into a 96-well polymerase chain reaction (PCR) plate (Fig. 1C). The frequency of BG505+ B cells was 0.024% (63 of 264,025) in live CD19+CD20+ cells. Four memory B cells were sorted, amplified with matching heavy- and light-chain genes, and cloned into the full IgG1 expression vectors. MAbs were expressed transiently in human embryonic kidney–293F (HEK293F) cells. Of the four mAbs, 438-B11 and 438-D5 bound the BG505 bait with high affinity in enzyme-linked immunosorbent assay (ELISA), whereas 438-D6 and 438-F11 showed negligible binding (fig. S1A). Consistently, both 438-B11 and 438-D5 exhibited potent neutralizing activity [50% inhibitory concentration (IC50) = 0.18 μg/ml] against the global panel with a breadth of 67% (table S1B), whereas 438-D6 and 438-F11 failed to neutralize any isolate on this panel. Therefore, 438-D6 and 438-F11 were most likely nonspecific mAbs as a result of high background in the B cell sorting experiment. The neutralizing activity of 438-B11 was further characterized against a panel of 129 isolates (32), with two N332-specific bNAbs (PGT121 and PGT128) included for comparison. On this large panel, PGT121 and PGT128 neutralized 62.8 and 68.2% of the isolates with IC50 values of 0.028 and 0.013 μg/ml, respectively, whereas 438-B11 was able to neutralize 41.9% of the isolates with an IC50 of 0.027 μg/ml (Fig. 1D and table S1C). Therefore, in terms of potency, 438-B11 is on par with the two extensively studied bNAbs targeting the N332/V3 supersite but shows a 20.9 to 26.3% reduction in breadth. Sequence analysis showed that 438-B11 and 438-D5 belong to the same clonal family, having a heavy chain of IGHV1-69*01 allelic origin with ~25% SHM and a 21–amino acid HCDR3 loop (Kabat numbering) and a light chain of IGKV3-20*01 allelic origin with ~13% SHM and a 9–amino acid LCDR3 loop (Fig. 1E and table S1D). 438-B11 and 438-D5 differ by one residue (Arg12 versus Lys12) in framework region 1 (FR1) of the heavy chain and one residue (Phe83 versus Leu83) in FR3 of the light chain (Fig. 1E). In addition, 438-B11 and 438-D5 both contain cysteines at positions 98 and 100a in HCDR3, suggesting the presence of a disulfide bond. Notably, disulfide-linked HCDR3 loops have been found in several somatic variants (VRC03/6/8) (33) and the inferred unmutated common ancestor (34) of the VRC01 lineage, the CAP256-VRC26 lineage (35), and bovine bNAbs against the CD4bs (36). Therefore, 438-B11 and 438-D5 represent another class of human HIV-1 bNAbs containing an extra disulfide bond in HCDR3.
Fig. 1. Isolation of neutralizing antibodies 438-B11 and 438-D5 from donor CBJC438.
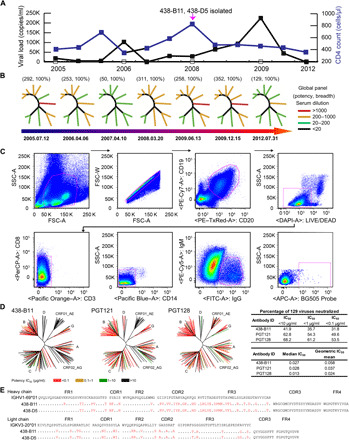
(A) The number of CD4+ T cells (blue) and viral loads (represented on y axes) were measured from 2005 to 2012 and represented in the line graph. The mAbs were isolated in 2008. (B) The neutralizing activity of donor CBJC438 serum from seven time points from 2005 to 2012 was tested against pseudoviruses on a global panel. (C) Antigen-specific single B cells were isolated by flow cytometry. Around 10 million PBMCs from CBJC438 were incubated with the sorting probe and cell markers. The gated cells were sorted into a 96-well PCR plate. (D) The neutralizing breadth and potency of 438-B11 were tested on a 129-virus panel. The N332-specific bNAbs PGT121 and PGT128 were used as controls. (E) Sequence analysis of mAbs 438-B11 and 438-D5 with alignment to their respective germline genes. FR and CDR are based on Kabat nomenclature. The symbol “.” denotes conserved amino acids.
Crystallography reveals the epitope and angle of approach for bNAb 438-B11
Before structural analysis, ELISA was performed to probe the 438-B11 and 438-D5 epitopes on the HIV-1 Env (fig. S1A). Both bNAbs showed high affinity for the BG505 UFO.664 trimer (29) and for the gp120-ferritin nanoparticle but low to no affinity for the V1V2-ferritin nanoparticle (37). These results suggest that the 438-B11/D5 epitopes are located in gp120 but are not part of the V1V2 region and the gp120-gp41 interface. We then determined the crystal structure of antigen-binding fragments (Fabs) of 438-B11 and another bNAb, 35O22 (38) (to aid in crystallization), in complex with the BG505 UFO.664 trimer (29) at 3.80 Å resolution in a hexagonal (H32) crystal lattice. The complex structure showed that three 438-B11 Fabs were bound to one BG505 UFO.664 trimer (Fig. 2A) by recognizing the N332/V3-glycan supersite involving both Asn301 and Asn332 (Fig. 2B, upper inset). The heavy chain of 438-B11 interacts with almost all glycan moieties of the oligomannose at Asn301 and Asn332 and protects them from the partial deglycosylation of BG505 UFO.664 by endoglycosidase H carried out before crystallization. Electron density was visible for GlcNAc2Man8 at Asn301 and GlcNAc2Man9 at Asn332. Both Asn301 and Asn332 glycans were predominantly buried by the heavy chain of 438-B11 (Fig. 2B, glycan schematics), except for the D2 arm of Asn332 and the terminal glycan moieties of Asn301 D1 and D2 arms. Similar to PGT128 [Protein Data Bank (PDB) ID: 3TYG], HCDR2 of 438-B11 interacts with the predominantly apolar face of the Asn301 glycan. Its hydrophobic tip (Val53, 33 Å2; Val54, 8 Å2) was slightly buried inside the Asn301 glycan pocket (fig. S1B). The 21–amino acid–long HCDR3 loop enables 438-B11 to access the protein surface around the base of the Asn332 glycan on gp120. Notably, the extended, U-shaped HCDR3 loop differentiates 438-B11 from other bNAbs directed to the N332/V3 supersite such as PGT121, PGT128, 10-1074, and BG18 (16, 21, 39, 40), whose HCDR3 loops adopt a protruding, two-stranded β-hairpin conformation (fig. S1C). The CCR5 coreceptor binding motif, GDIR, is mainly recognized by CDR1, CDR3, FR1, and FR3 of the 438-B11 heavy chain (Fig. 2C), whereas both heavy- and light-chain CDRs are required by other bNAbs [PGT121 (LCDR1/2/3 and HCDR3), 10-1074 (LCDR1/2/3 and HCDR3), and BG18 (LCDR2 and HCDR3)] except for PGT128 (HCDR3). The tip of 438-B11 HCDR3 (Tyr100c) is deeply buried [buried surface area (BSA), 124 Å2] inside the Asn332 glycan pocket similar to that observed for PGT121, 10-1074, and BG18 (Fig. 2D and fig. S1C).
Fig. 2. Structural epitope mapping of 438-B11 on HIV-1 Env.
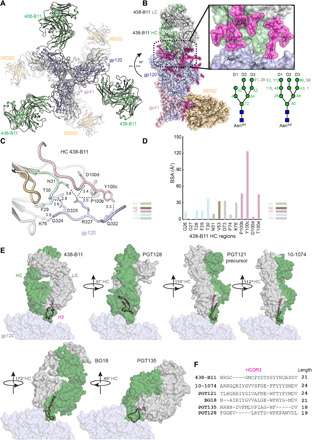
(A) Crystal structure at 3.8 Å of the BG505 UFO.664 (gp120, blue; gp41, pink) Env trimer in complex with Fabs 438-B11 (green) at the N332/V3 supersite and 35O22 (yellow) at the gp120/gp41 interface. (B) Side view of one protomer of the BG505 UFO.664 trimer in complex with Fab 438-B11 (HC, green; and LC, gray) and Fab 35O22 (yellow) Fabs. The cartoon representation of the structure is overlaid on the transparent molecular surface. The right inset shows the interaction of the 438-B11 Fab with Asn301 and Asn332 glycans (pink). The glycans are shown in sticks and overlaid on the transparent molecular surface. Below the inset is a schematic of Asn301 and Asn332 glycans observed in the structure with GlcNAc moieties as dark blue squares and mannose residues as green circles. The surface area (Å2) on each glycan moiety that is buried by 438-B11 (HC, green; LC, gray) is shown. (C) Recognition of the GDIR motif (blue) at the base of V3 by Fab 438-B11. The heavy chain (HC) 438-B11 is represented in cartoon and sticks with the CDRs and FRs (H1, green; H2, brown; H3, pink; FR1, cyan; and FR3, gray) that interact with gp120 (blue). (D) Buried surface area (Å2) of residues on the CDR loops and FRs of 438-B11 when bound to gp120 on Env. (E) Comparison of the mode of binding of N332/V3-class bNAbs to the surface of gp120 (blue) of HIV-1 Env. A distinct orientation of 438-B11 (HC, green; LC, gray) on HIV-Env is observed compared to other bNAbs of this class. HCDR3 loops (pink) are shown in cartoon representation. (F) Alignment of the amino acid sequences of HCDR3 of various N332/V3-class bNAbs.
To define the angle of approach for 438-B11, we compared the rotation of the heavy chains between 438-B11 and bNAbs PGT128 (PDB ID: 5C7K), PGT121 precursor (PDB ID: 5CEZ), 10-1074 (PDB ID: 6OKP), BG18 (PDB ID: 6CH7), and PGT135 (PDB ID: 4JM2) (Fig. 2E). We observed differences between 438-B11 and other bNAbs in terms of HCDR3 orientation and conformation using the Env trimer as a reference in three-dimensional (3D) space (Fig. 2E and fig. S2A). Their orientations differ substantially and are rotated around the long axis of the Fab by approximately 90° to 120° (PGT128, PGT121 precursor, and 10-1074) to 180° (BG18), where the relative disposition of the heavy and light chains on the gp120 surface is reversed. PGT135 is also much more tilted compared to the other bNAbs (Fig. 2E and fig. S2B). To engage both the N332/V3 glycan patch and the GDIR motif, a protruding two-stranded β-hairpin HCDR3 and light-chain CDRs are vital for the PGT121 precursor, 10-1074, and BG18. As a result, both their heavy- and light-chain CDRs are involved in targeting this epitope, as reflected by their contributions to the buried surface area on the protein surface of gp120 (fig. S2C). In contrast, bNAbs (438-B11, PGT128, and PGT135), which mainly use their heavy chains to interface with both the glycan patch and the GDIR motif, require a larger buried surface area of their HCDRs on the protein surface of gp120 (Fig. 2 and fig. S2C). Notably, the heavy chain accounts for 83% of total buried surface of 438-B11, similar to that of PGT128 (85%) (fig. S2C). Of note, the hydrophobic residue at the tip of HCDR3 in all bNAbs of the N332/V3 class (PhePGT121precursor, Phe10-1074, LeuBG18, and Tyr438-B11) interacts with a “neutral face” in proximity to the Asn332 glycan on the Env, except for Tyr100B at the tip of PGT128 HCDR3, which is positioned closer to the GDIR motif (fig. S2D). Thus, the distinct angle of approach and the HCDR-mediated interactions facilitate 438-B11 recognition of the N332/V3-glycan supersite on the HIV-1 Env.
BG505 UFO.664 trimer has a similar architecture to BG505 SOSIP and NFL trimers
Previously, we determined the structure of a cleaved, HR1-redesigned BG505 gp140 trimer (29). Here, we obtained a 3.8-Å-resolution structure for the BG505 UFO.664 trimer, which contained both a redesigned HR1 bend and a long 10–amino acid (G4S)2 linker at the furin cleavage site. This atomic structure thus provided an opportunity for comparison of all three native-like trimer platforms (41, 42): the cleaved SOSIP.664 trimer, the uncleaved native flexible linker (NFL) trimer, and the UFO trimer based on the same BG505 backbone (fig. S3A). Upon superimposition, little difference was observed for the protomer conformation or the overall trimer packing, with Cα root mean square deviation (RMSD) ranging from 0.5 to 0.7 Å, suggesting that all three designs have preserved the native-like Env structure. While UFO shares the same gp120-gp41 disulfide bond (Ala501Cys-Ala605Cys) (43) with SOSIP and the long cleavage-site linker with NFL (44), shortening of the 21–amino acid HR1 N-terminal region (HR1N) to an 8–amino acid loop and its computational optimization (fig. S3B) substantially improved trimer yield, purity, and stability, supporting the notion that HR1N is the main determinant of HIV-1 Env metastability (29, 45). The nearly identical trimer structures in three complexes, SOSIP/PGT121 precursor (18), NFL/PGT122 (46), and UFO/438-B11, suggest that 438-B11 would not affect the Env conformation upon binding similar to the PGT121 class of bNAbs.
Germline reversion shows differential effects of heavy and light chains on function
VH1-69 is a common germline gene for human bNAbs targeting the conserved stalk region of influenza virus hemagglutinin and the neutralizing face on glycoprotein E2 of hepatitis C virus (HCV) (47, 48). Hydrophobic residues in HCDR2 at positions 53 (mostly Ile for influenza antibodies) and 54 (Phe for both influenza and HCV) are crucial for the recognition of conserved viral epitopes by these VH1-69 bNAbs (47). HIV-1 antibodies of the VH1-69 origin recognize the MPER in gp41 [4E10 (49–51)], the receptor binding site [8ANC195 (52)], the coreceptor binding site [17b (53)], and the CD4bs [VRC13 (54)]. All of these HIV-1 antibodies contain hydrophobic residues at positions 53 and 54 except for VRC13 (Ser53 and Thr54). In 438-B11 and 438-D5, Ile53 and Phe54 within the VH1-69 germline gene are substituted to smaller hydrophobic residues (Val) by somatic mutation, suggesting a similar involvement of HCDR2 in epitope recognition.
To assess the functional role of 438-B11 heavy and light chains in Env recognition, we created three 438-B11 variants by pairing germline (gl) and mature (m) antibody chains, namely, mHC/glKC, glHC/mKC, and glHC/glKC. The glHC and glKC constructs were created by reverting the variable (V) and joining (J) genes to their putative germline genes while retaining the CDR3 segments, as found in the mature antibody (fig. S4A). These chimeric antibodies would also inform on how much the CDR3 region, versus the rest of the variable domain, contributes to the antibody function. A double-cysteine mutant (Cys98Ala and Cys100aAla), termed ΔSS, was also included to examine the functional role of the HCDR3 disulfide bond. In a biolayer interferometry (BLI) assay, each 438-B11 variant was tested against five UFO or UFO-BG trimers of diverse subtypes (fig. S4B) (45). Overall, a strain-specific pattern was observed, with clade A BG505 UFO and clade B H078.14 UFO-BG trimers recognized by 438-B11 and variants more effectively than other trimers, consistent with the moderate neutralizing breadth of 438-B11/D5 on the large virus panel (Fig. 1D). Among the tested 438-B11 variants, mHC/glKC showed some degree of reduction in binding to BG505 UFO compared to wild-type (WT) 438-B11, whereas no trimer binding was observed for glHC/gKC and glHC/mKC, confirming that the heavy chain and HCDR3-mediated interactions are essential to the 438-B11 recognition of the N332/V3 supersite on the Env trimer. Unexpectedly, the 438-B11ΔSS mutant showed comparable binding to BG505 UFO and a substantially reduced, but still detectable, binding to H078.14 UFO-BG, suggesting that the intra-HCDR3 disulfide bond exerts a differential effect on epitope recognition depending on the Env. We also examined the effect of the maturation of individual antibody chains on the neutralizing activity of 438-B11 using the global panel (fig. S4C). Consistently, light-chain reversion appeared to have less effect on antibody function than heavy-chain reversion, with 438-B11 mHC/glKC showing 17% reduction in breadth and 2.5-fold reduction in potency compared to WT 438-B11 or 438-B11 mHC/mKC. Of note, an S27N mutation from the germline V gene of κ light chain, IGKV3-20*1, produces an N-linked glycan in 438-B11 LCDR1. In our structural analysis, no contacts were observed between this glycan and the Env trimer, suggesting that this glycan may not be directly involved in Env recognition.
Structural comparison of bound and unbound 438-D5 and 438-B11
To investigate whether 438-D5 and 438-B11 undergo conformational changes during Env recognition, we determined crystal structures of unbound 438-B11 and 438-D5 Fabs at resolutions of 2.05 and 2.70 Å, respectively (Fig. 3, A and B). Of note, the unbound 438-B11 was stabilized using protein G to facilitate crystallization and structure determination (Fig. 3B). Overall, the two bNAbs showed highly similar structures with Cα RMSD of 0.5 Å (Fig. 3C). The amino acid differences between 438-B11 and 438-D5 in heavy-chain FR1 (Fig. 3C, upper inset) and light-chain FR3 (Fig. 3C, lower inset) had, therefore, little effect on the backbone structure. While the variable regions (Fv) of 438-B11 and 438-D5 share a high degree of structural similarity (≤0.2 Å), some flexibility was observed for HCDR3 (Fig. 3D). One of the distinctive features of the 438-B11 and 438-D5 paratopes is the disulfide bond between Cys98 and Cys100a in the long 21–amino acid HCDR3 (Fig. 3D), which forms an extended U-shaped loop (fig. S5A). Only a small deviation in the tip of HCDR3 was observed for the unbound 438-B11 and 438-D5 upon superimposition (5.5 Å between Tyr100c Cα atoms), reflecting different crystal packing interactions (Fig. 3D, inset). A feature shared by all bNAbs of the N332 class is a long HCDR3 loop, which forms either a two-stranded β-hairpin [PGT121 (PDB ID: 4FQ1) and 10-1074 (PDB ID: 4FQ2)] or an extended loop [PGT128 (PDB ID: 3TV3) and BG18 (PDB ID: 5UD9)] to penetrate the oligomannose patch around Asn332 and to recognize the GDIR motif. In comparison with PGT121 and 10-1074, which have 24–amino acid HCDR3 loops, 438-B11 has a 3–amino acid shorter HCDR3 loop that is tilted toward the variable region of immunoglobulin heavy chain (VH) domain for GDIR recognition (Fig. 3E). This tilting in 438-B11 closes a potential gap between HCDR3 and HCDR1/2, whereas a cleft is observed in this region for PGT121 and 10-1074 (16). While 438-B11 relies only on the heavy-chain recognition of the protein component of the Env surface, PGT121 and 10-1074 recruit their light chains to bind the GDIR motif. In terms of HCDR3 tilting toward the VH domain, resemblance can be seen with PGT128 (19, 40) and BG18 (2). Some variations in HCDR3 length (438-B11, 21 amino acids; PGT128, 19 amino acids; and BG18, 21 amino acids) and HCDR2 length (438-B11, 17 amino acids; PGT128, 22 amino acids; and BG18, 9 amino acids) are also observed (Fig. 3E). The longer HCDR2s in 438-B11 and PGT128 facilitate antibody engagement with the N332/V3 glycan patch, as indicated by their increased BSA [438-B11, 35% (385 of 1094 Å2); PGT128, 51% (635 of 1240 Å2)] (fig. S2C). All LCDRs of BG18 (BSA, 262 Å2) are involved in Env recognition, whereas notably fewer but important light-chain interactions are observed for PGT128 and 438-B11. Nevertheless, the long HCDR2 in PGT128 and its noncanonical disulfide bond with HCDR1 are critical for recognizing glycans at Asn301 and Asn332. For BG18, HCDR1/3 and LCDR2 interact with the glycan at Asn332, while LCDR3 interacts with the D2 arm of the N392 glycan. For 438-B11, HCDR3 and LCDR1/3 are responsible for the recognition of the Asn332 glycan, with HCDR2 and LCDR3 interacting with the glycan at Asn301. Together, 438-B11 and 438-D5 predominantly use their HCDR loops to achieve high potency and appreciable breadth with modest levels of SHM (table S1D).
Fig. 3. Structural analysis of unbound 438-B11 and 438-D5.
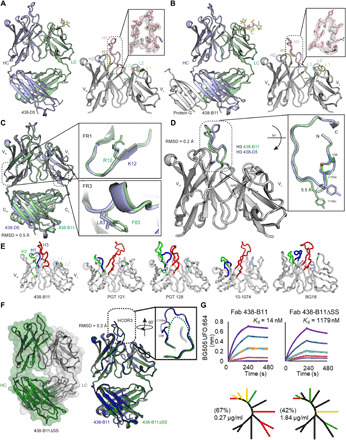
(A) Crystal structure of 438-D5 Fab at 2.0 Å is shown in cartoon. The variable domain of the 438-D5 Fab is shown with HCDR loops labeled and colored for H1 (orange), H2 (blue), and H3 (pink) and LCDR loops for L1 (yellow), L2 (cyan), and L3 (green). The right inset shows the 2Fo − Fc composite omit map (contoured at 1.0σ) of HCDR3. VL, variable region of immunoglobulin light chain. (B) Crystal structure of 438-B11 Fab in complex with protein G (gray) at 2.7 Å shown in cartoon. The right inset shows the 2Fo − Fc composite omit map (contoured at 1.0 s) of HCDR3. (C) Superposition of 438-B11 (green) and 438-D5 (blue) Fab crystal structures with single-residue differences in heavy-chain FR1 and light-chain (LC) FR3 (see insets). (D) Superposition of variable domains of protein G-bound 438-B11 (green) and 438-D5 (blue) illustrates some differences at the HCDR3 apex that may be due to differences in crystal packing (see insets). (E) The variable domains from the various N332/V3-class bNAbs are shown with HCDR loops labeled and colored for H1 (blue), H2 (green), and H3 (red) and LCDR loops for L1 (light blue), L2 (light green), and L3 (light pink). (F) Crystal structure of 438-B11ΔSS Fab at 2.1 Å shown in cartoon and overlaid with transparent molecular surface representation of heavy chain (HC; green) and light chain (LC; gray). The right panel shows the superposition of unbound 438-B11 (blue) and 438-B11ΔSS (green). The inset shows the superposition of HCDR3. The disordered HCDR3 tip is shown as a dashed line. (G) (Top) Binding kinetics of 438-B11 and 438-B11ΔSS Fabs to soluble BG505 UFO.664. (Bottom) Neutralization breadth (%) and potency (μg/ml) of 438-B11 and 438-B11ΔSS on a global panel.
Previous studies have demonstrated the impact of somatic mutations in antibody elbow and FRs on interdomain flexibility (55–57), antigen affinity, potency, and breadth (58, 59). In particular, a Phe83Val mutation was reported in the κ light chain of human antibodies that could increase both antigen affinity and stability (56). To examine this possibility, we introduced the F83V mutation in the mature 438-B11 light chain and determined the thermostability of Fab 438-B11KC-F83V by differential scanning calorimetry (DSC). The melting temperature (Tm) of 438-B11KC-F83V was 3°C greater than that of 438-B11 (73.5° versus 70.5°C) (fig. S5B). In BLI assays, a slight increase in trimer affinity was observed for 438-B11 KC-F83V (Kd = 12.7 nM) with respect to the WT 438-B11 (Kd = 14.0 nM), with slightly lower binding signals (fig. S5C). In TZM-bl assays, the Phe83Val mutant showed almost identical neutralization profiles against the 12-virus global panel (fig. S5D). Our analysis thus revealed that the unbound 438-B11 and 438-D5, both containing a disulfide bond in their HCDR3 loops, are highly similar in structure.
The disulfide bond in 438-B11/D5 HCDR3 is important for binding and neutralization
To examine the role of the HCDR3 disulfide bond, the structure of the unbound disulfide mutant, 438-B11ΔSS (Cys98Ala/Cys100cAla), was determined at 2.10 Å (Fig. 3F). 438-B11ΔSS HCDR3 could not be modeled because of the weak electron density for this region, indicating increased flexibility of the HCDR3 loop without the disulfide bond (Fig. 3F, inset). Disulfide bonds have been found to stabilize ultra-long HCDR3 loops in human HIV-1 bNAbs such as CAP256-VRC26 (35) and the VRC01 lineage from donor 45 (33), as well as in bovine bNAbs (33, 36, 60). Little structural difference was observed (RMSD, 0.2 Å) upon superposition of unbound 438-B11 and 438-B11ΔSS crystal structures except for HCDR3 (Fig. 3F). Although 438-B11 and 438-B11ΔSS IgGs showed little difference in trimer binding profiles in BLI when trimers were used in solution as the analyte (fig. S4B), an 85-fold reduction in binding affinity (mainly caused by a faster off-rate) was observed because of the disulfide mutation when Fabs were used as analytes and trimers were immobilized on the BLI biosensors (Fig. 3G, top). Consistently, a notable reduction in the neutralizing potency was observed for 438-B11ΔSS on the global panel (Fig. 3G, bottom). To determine whether and how the disulfide mutation affects the 438-B11–Env interactions, we determined the crystal structure of the BG505 UFO.664 trimer in complex with 438-B11ΔSS and 35O22 Fabs at low resolution (6.50 Å) (fig. S6A and table S3E). Despite the low resolution, the crystal structure shows no difference in the angle of approach for the 438-B11 mutant. However, a small upward displacement (~1 Å) was observed for the 438-B11 mutant upon superimposition of 438-B11– and 438-B11ΔSS–bound Envs (fig. S6B, left inset). This movement may explain the reduced trimer binding observed for the 438-B11 mutant (Fig. 3G, top). The loss of potency observed for 438-B11ΔSS (Fig. 3G, bottom) is likely caused by the disruption of critical hydrogen bonds (Fig. 2C) between HCDR3 and the GDIR. Our analysis reveals that the disulfide bond is essential to the conformational rigidity of HCDR3 and Env recognition via a “lock-and-key” model, consistent with the high potency but moderate breadth of 438-B11 and 438-D5.
Longitudinal analysis of CBJC438 B cell repertoires over 8 years
Previously, we used next-generation sequencing (NGS) and unbiased repertoire capture (61) to probe the memory B cell repertoire of an HIV-1–infected Chinese patient, DRVI01 (who developed VRC01-like bNAbs), across three time points (3). Here, we followed a similar strategy to generate B cell repertoire profiles for CBJC438 at time points 2005, 2008, and 2012. For each time point, the total RNA obtained from 10 million to 20 million PBMCs was used to prepare antibody H, κ, and λ chain libraries using 5′–rapid amplification of cDNA ends (RACE) PCR and reverse primers (61). Antibody NGS was performed on the Ion GeneStudio S5 system (62) that generated more than 14 million raw reads (table S1E). After data processing with the Antibodyomics 2.0 pipeline (31), 3.73 to 4.15 million antibody chains were obtained for the three time points studied. In terms of germline gene usage, we observed a substantial change in gene frequency over time (fig. S7A). The 2005 HC repertoire was highly skewed toward three germline genes, IGHV1-2 (11.2%), IGHV3-21 (27.2%), and IGHV3-7 (15.0%), while IGHV1-69, the germline family of 438-B11/D5 HC, only accounted for 5.0% of the total population. Notably, IGHV1-69 was predominantly used at the 2008 time point, representing 11.4% of the HC repertoire, and then decreased to 6.2% at 2012. The high frequency of IGHV1-69 suggests that the 438-B11/D5 lineage might have undergone a significant expansion by 2008, which coincided with the lowest viral load around the same time (Fig. 1A). Consistently, the KC repertoire exhibited a similar pattern to the HC repertoire, with 2005 having a significantly different germline gene distribution than in 2008 and 2012. IGKV3-20, the germline family of 438-B11/D5 KC, increased in the repertoire over time from 6.7% at 2005 to 11.1% at 2012. The LC repertoire showed relatively steady distributions across three time points, with the frequency of IGLV2-14 notably increasing from 9.1% at 2005 to 16.0% at 2012. In terms of germline divergence or degree of SHM (fig. S7B), the repertoire appeared to contain more germline-like antibodies at 2008 and 2012 than at 2005. The frequency of germline-like HCs peaked at 2008, while κ and λ light chains showed an elevated frequency of germline-like sequences at 2012, suggesting that HC may take precedence in antibody maturation as it is often more essential to antigen interactions. IGHV1-69 and IGKV3-20 exhibited SHM patterns consistent with those observed for their respective repertoires, indicating a repertoire-level preference for antibody maturation. In terms of HCDR3 loop length (fig. S7C), we observed a rapid change in distribution between 2005 and the latter two time points, with an average length of 13.2, 13.5, and 12.5 amino acids at the repertoire level and 13.4, 14.2, and 14.1 amino acids in the IGHV1-69 family for 2005, 2008, and 2012, respectively. The greater frequency of 19– to 21–amino acid HCDR3 loops at 2008 compared to 2005 and 2012 is consistent with the finding that 438-B11 and 438-D5 were isolated from 2008 and had 21–amino acid HCDR3 loops. In summary, antibody NGS demonstrated a dynamic B cell repertoire in CBJC438 similar to DRVI01 (3), even though bNAbs elicited in these two HIV-1–infected donors were targeting two distinct epitopes (glycan supersite versus CD4bs). However, it is possible that bNAbs of other epitope specificities were developed in these two patients since the neutralizing mAbs could not fully recapitulate the serum neutralization breadth (Fig. 1B) (3).
Longitudinal tracing reveals rapid 438-B11/D5 lineage development in the donor repertoire
As 438-B11 and 438-D5 were isolated from 2008, the repertoire data obtained from 2005, 2008, and 2012 offered an opportunity to investigate the development of this N332-directed bNAb lineage over an 8-year time period (Fig. 4A). The HC and KC repertoires with respect to 438-438-B11 are visualized using the standard identity/divergence two-dimensional (2D) plots (3, 63–65). Here, the 438-B11/D5 lineage was defined by a combined metrics of V/J gene usage and HCDR3 identity (for HCs only), in addition to sequence identity and germline divergence. D gene was not used because of the ambiguity in gene assignment. For time point 2005 (Fig. 4A, left), we observed a distinct island on the 2D plot of HCs with a germline divergence of 18.6 to 31.5% (on average 20.7%) and identity to 438-B11 from 75.1 to 83.3% (on average 80.6%). Most of the HCs from this island showed the same HCDR3 loop length (21 amino acids) as 438-B11 despite an HCDR3 identity of 85% or lower, suggesting that they may share the same B cell precursor as 438-B11. On the 2D plot of IGHV1-69 HCs, 1602 have HCDR3 loops of 20 to 25 amino acids, with 796 of these HCs overlapping with this sequence island and using the same J gene (JH3*01) as 438-B11/D5, suggesting that ~50% of the long HCDR3 antibodies in this germline family at 2005 were related to 438-B11/D5. While no separate island was observed on the 2D plot of total KC repertoire, a group of 743 KCs of the IGKV3-20 origin showed a 438-B11 KCDR3 identity of 85% or greater and could be recognized on the 2D plot of the IGKV3-20 family. For 2008 (Fig. 4A, middle), the presence of two 438-B11/D5–related sublineages in the donor repertoire was evident. Two distinct sequence islands were found on the 2D plot of HCs: The top island, which is closely related to 438-B11 and 438-D5, shows a germline divergence of 24.2 to 34.8% (on average 26.7%) and 438-B11 identity of 86.9 to 99% (on average 96.7%), while the bottom island, which resembles the island on the 2005 2D plot, shows a divergence of 23.3 to 38.3% (on average 26.8%) and 438-B11 identity of 75.1 to 83.6% (on average 81.0%). Further analysis of the IGHV1-69 family revealed a large group of HCs (9036) with long HCDR3 loops (20 to 25 amino acids), of which 1038 contain 21–amino acid HCDR3 loops with a 438-B11 HCDR3 identity of 90% or greater and overlap with the top island on the 2D plot. In addition to HCDR3 similarity, all HCs from these two islands shared the same germline genes, IGHV1-69 and IGHJ3*01, with 438-B11/D5 HCs, confirming that they belong to the same lineage. The 2008 KC repertoire, as well as the IGKV3-20–derived KCs, demonstrated a pattern consistent with their HC counterparts. For example, KCs with a 438-B11 KCDR3 identity of 85% or greater can be found in two visually separate populations, one occupying the top island, which is closely related to 438-B11/D5, while the other forms an extension from the main sequence population, which corresponds to a sublineage more remotely related to 438-B11/D5. However, the dramatic lineage expansion and diversification observed for 2008 was not sustained. As a result, the 438-B11/D5 lineage could not be detected in the 2012 repertoire using the current NGS protocol (Fig. 4A, right). Only two 438-B11/D5–related HCs, with an SHM of 26 to 27% but notably different 438-B11 identities (97.4 versus 80.3%, corresponding to two sublineages), were identified from 1516 IGHV1-69–derived HCs with 20 to 25–amino acid HCDR3 loops. Similarly, only six KCs with a 438-B11 KCDR3 identity of more than 80% were found in the 2012 KC repertoire, but none was 438-B11/D5–like.
Fig. 4. Longitudinal analysis of the 438-B11/D5 lineage development.
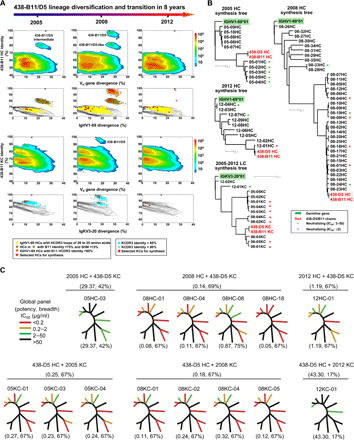
(A) Identity-divergence analysis of the unbiased heavy-chain (H) and light-chain (κ) repertoires of donor CBJC438 at time points 2005, 2008, and 2012. For the whole-repertoire analysis (rows 1 and 3), sequences are plotted as a function of sequence identity to 438-B11 and sequence divergence from putative germline genes. Color coding denotes sequence density. For analysis of the 438-B11–encoding germline gene families (rows 2 and 4), sequences are plotted as a function of sequence identity to 438-B11 and sequence divergence from IgHV1-69 and IgKV3-20 for the heavy and light chains, respectively. The germline gene family distribution is presented as black contour lines. 438-B11 and sequences identified on the basis of various criteria (see legend below figure) are shown on the black contours with the number of sequences labeled accordingly. (B) Dendrograms of functionally tested heavy and light chains are rooted by their respective germline allelic genes, IgHV1-69 and IgKV3-20, respectively. Heavy chains from 2005, 2008, and 2012 are presented in three separate dendrograms, while light chains from all three time points are shown in one dendrogram. (C) Neutralization breadth (%) and potency (μg/ml) of NGS-derived heavy and light chains paired with their respective 438-D5 partner chains on a global panel. The color-coding scheme is based on their potency.
In our previous study, longitudinal lineage analysis revealed how a VRC01-class antibody, DRVIA7, emerged in a Chinese patient (3). Here, we observed a rather different lineage pattern for 438-B11 and 438-D5, which target the N332 supersite. While 438-B11 and 438-D5 were isolated from a 2008 donor sample, they were not the direct descendants of antibody lineages found in the analysis of 2005 donor repertoire; instead, they belonged to a branch, or sublineage, that diverged from the main lineage after 2005. Unexpectedly, while at 2008, the 438-B11/D5 sublineage and main lineage shared the same level of SHM (~27%), the difference in their nucleotide sequences can be as significant as 20% or greater, suggesting that these diverged rapidly from their common ancestor within only 3 to 4 years of the process of antibody maturation. Despite the high mutational rate, the disulfide bond within HCDR3 remained conserved in both sublineages, confirming the functional importance of this relatively rare structural feature (Fig. 2). The expansion of the N332-directed 438-B11/D5 lineage and the presence of more germline-like HCs at 2008 (fig. S7B) suggest an actively evolving B cell repertoire in response to chronic HIV-1 infection. The fact that the 438-B11/D5–like antibodies became almost undetectable in the 2012 donor repertoire suggested a drastic shift in B cell repertoire response, in which the decline of the 438-B11/D5 lineage must be accompanied by the rise of other antibody lineages that were better suited to cope with the escaping viruses. This is evident from the large population of near-germline light chains observed for the 2012 repertoire (fig. S7B). Limited by sample availability, we cannot trace this antibody lineage before 2005 and can only speculate that, on the basis of the high degree of SHM (~21%) at 2005, this lineage must have existed in the donor repertoire for an extended period.
Functional characterization of antibodies identified from the NGS-derived repertoire
To investigate the functional variation within the rapidly evolving 438-B11/D5 lineage, we devised a bioinformatics approach to select HC and KC sequences from the NGS-derived repertoire at each time point to cover both sublineages (fig. S8 and table S2). Of note, the most representative clusters within the main population of IGHV1-69 and IGKV3-20 sequences were also selected for synthesis (fig. S8). While some of these sequences may not be evolutionarily related, dendrograms were generated using phylogenetic tools to cluster these HCs and KCs. Three dendrograms rooted by IGHV1-69*01 were constructed for 10 HCs from 2005, 35 HCs from 2008, and 9 HCs from 2012, with 438-B11 and 438-D5 HCs included as anchors (Fig. 4B, top). A single dendrogram rooted by IGKV3-20*01 was constructed for a total of 12 KCs (5 from 2005, 5 from 2008, and 2 from 2012) with 438-B11 and 438-D5 KCs included as anchors (Fig. 4B, bottom). After pairing HCs and KCs with the respective partner chains from WT 438-D5, the reconstituted antibodies were tested for neutralization against a global panel of 12 primary isolates and two tier 1 isolates, SF162 and MW965 (table S3, A and B). Neutralizers with the most notable activity against the global panel appeared to arise from both sublineages identified in the 2D analysis, with more KC variants showing WT-like neutralization than HC variants (Fig. 4, B and C). In the 2005 HC dendrogram, all five HC variants derived from the low 438-B11 identity sublineage could neutralize primary isolates and tier 1 isolates, albeit weakly. In the 2008 HC dendrogram, multiple HC variants showed WT-like potency, including one (08-04HC) with a low 438-B11 identity of 81.5% (Fig. 4, B and C). Notably, an HC (08-33HC) with a low SHM of 9.1% and 438-B11 identity of 70% neutralized four primary isolates on the global panel and tier 1 SF162 weakly, suggesting that other IGHV1-69–origin NAb lineages might be present in the 2008 repertoire. In the 2012 HC dendrogram, two germline-like HCs, 12-04HC (with a disulfide bond in HCDR3 at a similar position) and 12-07HC (with a similar HCDR loop to 438-B11 but without the disulfide bond), neutralized clade A p398F1 on the global panel and tier 1 SF162 weakly with an IC50 value of 16.1 to 47.4 μg/ml. In contrast, multiple KC variants from different time points exhibited WT-like neutralization potency, suggesting that the antibody function of this bNAb lineage is more sensitive to HC mutation. Together, antibody synthesis not only validated the function of both 438-B11/D5 sublineages but also confirmed HC-mediated Env recognition, consistent with the structural analysis (Fig. 2).
SGA and mutational analyses reveal viral diversity and potential escape mechanism
Following our previously reported strategy (3), we used single genome amplification (SGA) (66) to investigate viral diversity and virus-antibody coevolution in donor CBJC438. A total of 53 full-length Envs were amplified and sequenced from plasma viral RNA at years 2006 (16), 2008 (19), and 2009 (18). Of note, the 2012 sample failed to produce any usable Env sequences for this analysis. All of the Env sequences were used to construct a neighbor-joining (NJ) phylogenetic tree. Overall, we observed rapid diversification of HIV-1 Env sequences from 2006 to 2009, as indicated by large genetic distances (Fig. 5A, left). This simple phylogenetic analysis also revealed the emergence and evolution of multiple viral subpopulations at each time point, as represented by distinct branching patterns (Fig. 5A, right).
Fig. 5. Virus escape through glycan shift revealed by SGA and mutations.
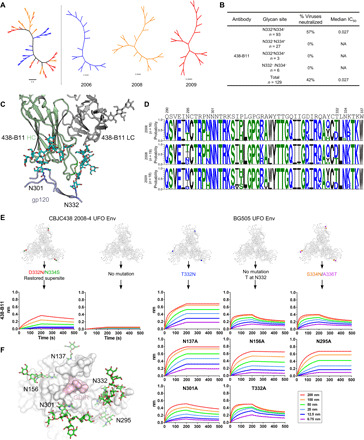
(A) NJ phylogenetic tree of 53 HIV-1 Env sequences isolated from donor CBJC438 by SGA. (B) Percent neutralization breadth and median IC50 in μg/ml were determined at an IC50 cutoff of 10 μg/ml for 438-B11. Viruses were separated into those containing an N-linked glycan at N332 but not N334 (N332+N334−), at N334 but not N332 (N332−N334+), at both N332 and N334 (N332+N334+), and at neither N332 nor N334 (N332−N334−). NA, not applicable. (C) Interaction of 438-B11 with the N332 and N301 glycans. (D) Sequence logo of the V3 region from three time points. (E) Binding affinity of 438-B11 antibodies to glycan mutants of CBJC438 UFO Env trimer and BG505 UFO Env trimer. Sensorgrams were obtained from an Octet RED96 instrument using a trimer titration series of six concentrations (200 to 6.25 nM by twofold dilution). For the CBJC438 Env, the D332N/N334S mutation restored the N332 supersite, whereas for the BG505 Env, the T332N mutation restored the N332 supersite. For BG505, a panel of glycan mutants was used to determine which glycan(s) surrounding the N332 supersite may be involved in epitope recognition. (F) Glycans surrounding the N332 supersite on the surface of BG505 Env trimer. A top view of the BG505 UFO.664 trimer surface with the GDIR motif colored pink. Glycans N137, N156, N295, N301, and N332 are represented with stick models and labeled.
We then analyzed the 438-B11 neutralization data derived from a large panel of 129 viruses to understand how viruses within CBJC438 escaped this potent N332-restricted NAb lineage (Fig. 5B). As expected, all 36 viruses on the panel that have glycans at N334 or lack the N332 glycan were resistant to 438-B11. More than half (57%) of the remaining 93 viruses contain an N-linked glycan at N332 and could be neutralized by 438-B11. Together, the results clearly indicated the indispensable role of N332 in the 438-B11/D5 recognition of HIV-1 Env but also suggest potential involvement of other glycans and amino acids (Fig. 5B). Our crystal structure of BG505 UFO.664 in complex with Fab 438-B11 also supports the notion that glycans at N332 and N301 are crucial for epitope recognition by 438-B11 (Fig. 5C). We then determined the frequency of N-X-S/T sequon at both N332 and N334 for potential N-glycosylation site by aligning the SGA-derived Env sequences obtained from three time points (Fig. 5D). Unexpectedly, an N-X-T sequon shift from N332 to N334 was observed at all three time points. To validate this observation, we created a UFO.664 trimer on the basis of a full-length Env sequence (Env08-4) selected from the 2008 subset. A double mutation, D332N/N334S, was introduced to mimic the Env of an autologous viral ancestor that might be sensitive to the N332-directed bNAbs. This inferred ancestral Env bound 438-B11 at a modest level compared to previous antigenic profiling for 10 Envs of five HIV-1 subtypes (45), whereas the WT Env trimer, which had presumably escaped, was not recognized by 438-B11 (Fig. 5E, two leftmost panels). While this result appeared to indicate that N332 is the key determinant for 438-B11 recognition and neutralization of HIV-1, the rapid 438-B11/D5 lineage expansion in the 2008 repertoire suggested that other Env elements might be involved. To explore this possibility, we first tested three glycan variants of the BG505 UFO.664 Env trimer, namely, T332N (N332-senstive Env), WT (Env without the N332 glycan), and a double mutant (S334N/A336T to mimic the N332-to-N334 glycan shift). As expected, the BG505.T332N Env bound 438-B11 strongly with a fast on-rate and a plateaued dissociation curve in BLI. Unexpectedly, WT Env (without N332) and the N332-to-N334 glycan shift Env could still bind 438-B11 but at a notably reduced level, with a fast on-rate and a dissociation curve in BLI (Fig. 5E, three rightmost panels), suggesting that glycans other than N332 may be involved in Env recognition by 438-B11. To this end, we tested another five BG505 Env mutants including N137A, N156A, N295A, N301A, and T332A (as a control). While N137A, N156A, and N295A bound to 438-B11 with WT-equivalent profiles, N301A showed a reduction in binding signal and visible dissociation curves (Fig. 5E, bottom right). Together, our analyses suggest that viruses within CBJC438 likely escaped the 438-B11/D5 lineage by mutating N332 to N334 and by impeding the 438-B11/D5 recognition of other glycans in the N332 V3 supersite such as N301 (Fig. 5F), which led to the marginalization of this lineage in the donor repertoire. However, patient samples before 2005 would be needed to capture these critical events to validate our hypothesis. Nonetheless, such an escape mechanism with cooperative viral mutations may be common to HIV-1–infected patients who develop a serum NAb response to the N332/V3-glycan supersite.
DISCUSSION
The HIV-1 Env is covered by N-linked glycans, many of which are of the oligomannose type (67, 68). With long HCDR3 loops and diverse angles of approach, the N332 class of bNAbs recognizes the conserved “GDIR” motif at the base of the V3 loop (22). However, this motif is shielded by a glycan patch centered around the position Asn332 and, thus, was termed the N332 glycan supersite (69). Here, we investigated the antibody-virus coevolution in a Chinese donor (CBJC438), who was infected by subtype-B HIV-1 and developed potent bNAb responses to the N332/V3 supersite. Using single B cell sorting, we isolated two N332 bNAbs from CBJC438 and characterized their structure and function before antibody NGS, which revealed a dynamic B cell response. However, analyses of both antibody and virus repertoires suggested that the viruses within this donor escaped the 438-B11/D5 lineage even before the first time point (2005). In addition, since the neutralization breadth of 438-B11/D5 could not fully recapitulate that of the patient sera, other bNAb lineages might be present in the patient repertoire that were not identified in B cell sorting using a clade A Env probe. Nonetheless, our study shows that the N332/V3-glycan supersite can be recognized by B cell receptors of a common germline gene, VH1-69, which may acquire unusual features such as a long, disulfide-locked HCDR3 loop to potently neutralize HIV-1.
The high-resolution crystal structure of 438-B11 bound to the BG505 UFO trimer reveals how its 21–amino acid–long HCDR3 loop penetrates a glycan patch involving N332, N301, N386, N392, N137, and N156. 438-B11 engages N301 and N332 glycans in Env binding, similar to PGT128 (40) and bNAbs of the PCDN lineage (6). The only other N332 bNAb from a clade B donor, BG18, requires the N332 glycan but not the N301 glycan (2). Nonetheless, 438-B11 binds this supersite with a unique angle of approach and relies mainly on its heavy chain to mediate Env interaction, similar to PGT124 and PGT128. However, the intra-HCDR3 disulfide bond has not been seen in other N332-directed bNAbs. This disulfide bond stabilizes the HCDR3 of 438-B11 in a U-shaped conformation and plays a critical role in Env binding and HIV-1 neutralization. Such intra-HCDR3 disulfide bonds have been found in somatic variants and the inferred precursor of VRC01 (33, 34). In mature VRC01 and NIH45-46 bNAbs, Cys98 in HCDR3 forms a disulfide bond with a cysteine in HCDR1 (70, 71), suggesting an increased structural rigidity of HCDR3 and the paratope. While intra-HCDR3 disulfide bonds have been found for a panel of human NAbs against viral pathogens (table S3C), 438-B11 and 438-D5 represent only the third class of such human HIV-1 bNAbs. In addition to antibody analysis, the first high-resolution structure of BG505 UFO.664 trimer has also enabled a detailed comparison of native-like Env trimers across design platforms.
Unbiased longitudinal NGS analysis (3, 61, 72) revealed a dynamic B cell repertoire from 2005 to 2012, first with a shift in germline gene usage and then with increased germline antibodies at the later time points likely to cope with rapidly evolving viruses that had escaped the previous generation of bNAbs (fig. S7B). This repertoire dynamics coincided with the 438-B11/D5 lineage development, with a notable lineage expansion at 2008 followed by a sharp decline at 2012 (Fig. 4A), as indicated by the frequency of 438-B11/D5–like HCs within the IGHV1-69 family (2005, 1.57%; 2008, 6.30%; and 2012, 0.002%). 438-B11 and 438-D5 appeared to be from a sublineage that was not present or had a low frequency in the 2005 repertoire. The rapid maturation (~6%) of the 438-B11/D5 lineage from 2005 to 2008 is reminiscent of the VRC01-like DRVIA7 lineage (3), suggesting a common pattern in chronic HIV-1 infections. However, while the use of 5′-RACE PCR and a single reverse 3′ primer in antibody NGS can eliminate primer bias and other issues associated with multiplex PCR, sequencing errors still occur and may lead to artificially increased lineage diversity (61, 72). To this end, NGS-derived antibodies were synthesized to validate their function. The SGA analysis of Envs isolated from CBJC438 suggested that the N332-to-N334 glycan shift most likely occurred before 2005, which explained the continuous decline of the 438-B11/D5 lineage. A similar viral escape mechanism has been reported for subtype C infections (27). Further analysis of 438-B11 binding to diverse Env mutants revealed that glycans other than Asn332 may be involved in Env recognition, consistent with the 438-B11/D5 lineage expansion at 2008 despite the glycan shift from N332 to N334 in all SGA-identified Envs.
In summary, we have identified two novel bNAbs that target the N332/V3-glycan supersite with high potency, modest breadth, and a markedly different disposition. The detailed analyses of this bNAb lineage provided valuable information for vaccine design targeting this glycan supersite. While the VH1-69 germline gene is readily available in the human B cell repertoire, the generation of a disulfide-linked long HCDR3 loop may pose the most significant challenge for eliciting a 438-B11/D5–like bNAb response. Nonetheless, future investigations may be directed to the evaluation of a VH1-69 germline–targeting vaccine strategy (73) in a bNAb knock-in mouse model (74). Such vaccines could be native-like trimers, N332-focused immunogens [e.g., epitope scaffolds (75)], or a combination of both in a prime-boost strategy (76) to enhance the bNAb responses.
MATERIALS AND METHODS
Chinese donor CBJC438 samples
The sera and PBMCs described in this study were collected from a Chinese patient (CBJC438) in an HIV-1–infected former plasma donor study cohort, which was established by the Chinese National Natural Science Foundation and the National Major Project on Infectious Disease. This study has been reviewed and approved by the Institutional Review Board of the National Center for AIDS/STD Control and Prevention, Chinese Center for Disease Control and Prevention. A detailed analysis of serum neutralization has been reported for the samples collected from this study cohort (77).
Cell lines
HEK293F cells and ExpiCHO cells (Thermo Fisher Scientific) were used to produce HIV-1 Env proteins, Fabs, and IgGs. HEK293T/17 cells (American Type Culture Collection) were used to produce HIV-1 pseudoviruses. TZM-bl cells (National Institutes of Health AIDS Reagent Program; www.aidsreagent.org/) were used to test the neutralizing activity of sera and antibodies.
HIV-1 neutralizing assay
Neutralizing activity of the plasma and the mAbs was measured using a TZM-bl/pseudovirus neutralizing assay as previously described (78, 79). Briefly, plasma samples or mAbs were serially diluted in 10% Dulbecco’s modified Eagle’s medium (DMEM), and 100 μl per well was added into 96-well flat bottom plate. Pseudovirus of 200 TCID50 (median tissue culture infectious dose) in 50-μl volume was added to each well of the plate. The plate was then incubated at 37°C, 5% CO2 for 1 hour. TZM-bl cells were added at 1 × 104 per well (in 100-μl volume) in 10% DMEM growth medium containing DEAE-dextran (7.5 μg/ml). Cell controls (TZM-bl cell only) and virus controls (TZM-bl and pseudovirus) were included in the assay. The plates were then incubated at 37°C, 5% CO2 for 48 hours. After 150 μl of the supernatant was removed, 100 μl of Bright-Glo luciferase reagent (Promega, Madison, WI) was added to each well. Last, after lysis, the luminescence was measured for the cell lysate using a PerkinElmer VICTOR3 luminometer or a BioTek microplate reader. The 50% inhibitory dose (ID50) or IC50 was calculated as the plasma dilution or mAb concentration that resulted in a 50% reduction in relative luminescence units, with the cell control subtracted from both the sample and the virus control. All neutralizing assays were conducted following the standard operating procedure according to the Good Clinical Laboratory Practice compliance.
Identification of antigen-specific mAbs by single B cell sorting
Single B cell sorting was followed as previously described (3, 71). Briefly, PBMCs from donor CBJC438 were incubated with an antibody cocktail consisting of CD19-PE-Cy7, CD8-PerCP, IgM-PE-Cy5, IgG-FITC (all from BD Biosciences), CD20-ECD (Beckman), CD3–Pacific Orange (Invitrogen), CD14–eFluor 450 (eBioscience), BG505-APC (purified in laboratory), and LIVE/DEAD Fixable Dead Cell Stain Kit (Invitrogen) to exclude dead cells. The PBMCs were first stained with 50 μl of phosphate-buffered saline (PBS) with LIVE/DEAD at 4°C in the dark for 30 min and then stained with 50 μl of PBS with an antibody cocktail for 1 hour at 4°C in the dark. The stained cells were washed and resuspended in PBS and then passed through a 70-μm cell mesh (BD Biosciences). Antigen-specific single B cells were gated as CD19+CD20+CD8−CD3−CD14−IgM−IgG+BG505+ and sorted into a 96-well PCR plate containing 20 μl of lysis buffer: 5 μl of 5× first-strand buffer, 0.5 μl of RNaseOUT, 1.25 μl of 0.1 M dithiothreitol (all from Invitrogen), and 0.0625 μl of IGEPAL (Sigma-Aldrich) per well. The sorted plate was snap-frozen on dry ice and stored at −80°C before reverse transcription (RT) reaction.
Single B cell PCR, cloning, and expression of mAbs
The variable heavy- and light-chain genes were amplified and cloned into expression vectors to produce full IgG1 antibodies as previously described (3, 63, 80, 81). Briefly, RT-PCR was used to acquire the cDNA of each clone: 3 μl of random hexamers (150 ng/μl) (Promega), 2 μl of deoxynucleotide triphosphate (dNTP) mix (10 mM each) (Invitrogen), and 1 μl of SuperScript III (Invitrogen), following the program: 42°C for 10 min, 25°C for 10 min, 50°C for 60 min, and 94°C for 5 min. Nested PCR was used to amplify antibody variable genes: 2.5 μl of 10× buffer, 0.5 μl of 10 mM dNTP mix, 0.5 μl of 25 mM MgCl2, 0.5 μl of 25 μM 5′ primers and 0.5 μl of 25 μM 3′ primers (Liao et al., 2009; Tiller et al., 2008), 0.2 μl of HotStarTaq Plus (Qiagen), 17.8 μl of H2O, and 2.5 μl of cDNA, following the program: 94°C for 5 min, 50 cycles of 94°C for 30 s, 52°C for 45 s, and 72°C for 1 min, and 1 cycle of 72°C for 10 min. Next, 1.75 μl of first-round PCR product was mixed with 2.5 μl of 10× CL buffer, 0.5 μl of 10 mM dNTP mix, 5 μl of 5× Q-solution, 0.5 μl of 25 μM 5′ primers and 0.5 μl of 25 μM 3′ primers (Liao et al., 2009; Tiller et al., 2008), 0.2 μl of HotStarTaq Plus (Qiagen), and 14.3 μl of H2O for second-round PCR, following the program: 94°C for 5 min, 50 cycles of 94°C for 30 s, 52°C for 45 s, and 72°C for 1 min, and 1 cycle of 72°C for 10 min. The paired heavy- and light-chain PCR products were sequenced with 5′ and 3′ primers of second-round PCR (Beijing Tianyi Huiyuan Life Science and Technology Inc.). Synthesized specific 5′ and 3′ primers with restriction digest sites (heavy chain, 5′–Age I/3′–Sal I; κ chain, 5′–Age I/3′–Bsi WI; λ chain, 5′–Age I/3′–Xho I) were used to reamplify second-PCR products: 10 μl of 5× PrimeSTAR buffer (Mg2+ Plus), 4 μl of 2.5 mM dNTP mix, 1 μl of each 5′- and 3′-specific primer (25 μM), 0.5 μl of PrimeSTAR HS DNA Polymerase (TAKARA), 1.5 μl of first-PCR product, and 32 μl of H2O, following the program: 98°C for 15 s, 30 cycles of 98°C for 10 s, 52°C for 15 s, and 72°C for 45 s, and 1 cycle of 72°C for 7 min. The specific PCR products were cloned into full-length IgG1 expression vectors. MAbs were expressed by transient transfection of 293 F cells (Life Technologies) with equal amount of paired heavy- and light-chain plasmids and purified from culture supernatants after 5 days using protein A columns (National Engineering Research Center for Biotechnology, Beijing) according to the manufacturer’s protocol.
Germline gene usage of mAbs
The program IMGT/V-QUEST (www.imgt.org/IMGT_vquest/vquest) was applied to analyze gene germline. The CDR3 length was determined on the basis of the Kabat numbering. The SHM frequency was calculated from the mutated nucleotides.
ELISA binding of mAbs to the BG505 sorting probe
The BG505 sorting probe was diluted in PBS at a final concentration of 2 μg/ml and added to 96-well ELISA plates (100 μl per well). The plates were coated at 4°C overnight, washed five times with PBS-T (PBS containing 0.05% Tween 20), and blocked at room temperature for 1 hour with blocking buffer (PBS containing 5% skim milk and 2% BSA). MAbs were serially fivefold diluted from 10 μg/ml in blocking buffer and added into wells (100 μl per well) and incubated at 37°C for 1 hour. The plates were washed five times with PBS-T, and 100 μl per well of horseradish peroxidase–conjugated goat anti-human IgG antibodies (ZSGB-BIO, Beijing) (diluted 1:5000 in blocking buffer) was added and incubated at 37°C for 1 hour. The wells were washed five times with PBS-T, and 100 μl of trimethylboron substrate (Kinghawk, Beijing) was added and incubated at room temperature for 20 min, and the reaction was stopped by the addition of 2 M H2SO4 to each well. The readout was measured at a wavelength of 450 nm.
Synthesis and production of antibody variants selected from repertoire sequencing
Briefly, the selected heavy- and light-chain genes from the deep sequence analysis were synthesized (Beijing Tianyi Huiyuan Life Science and Technology Inc.) and cloned into the full-length IgG1 expression vectors containing the constant regions. Antibody production was performed using a previously described protocol (3, 81). The heavy chains selected from donor CBJC438 were paired with the 438-D5 light chain, and the selected light chains were paired with the 438-D5 heavy chain. MAbs were expressed by cotransfection of 293 F cells with equal amount of paired heavy- and light-chain plasmids and purified from culture supernatants using protein A columns after 5 days (National Engineering Research Center for Biotechnology, Beijing) according to the manufacturer’s instructions.
Expression, purification of HIV-1 Env, bNAbs, and complex formation
Clade A isolate BG505 UFO.664 WT HIV-1 Env was expressed in FreeStyle HEK293S cells. Secreted protein was harvested from media and purified with a 2G12-coupled affinity matrix followed by size exclusion chromatography (SEC) on a Superdex 200 column (GE Healthcare). All Fabs of bNAbs and their variants [438-B11, 438-D5, 438-B11ΔSS, 438-B11KC-F83V, and 35O22 (38)] were produced by transient transfection of FreeStyle HEK293F cells (Invitrogen) and purified first using either a κ or λ capture select column (GE Healthcare), which was followed by ion-exchange chromatography (GE Healthcare) and SEC on a Superdex 75 column. Of note, the 438-B11ΔSS mutant (with the Cys98Ala and Cys100aAla substitutions in 438-B11) was generated using a PCR-based mutagenesis approach. Domain III of protein G with a C-terminal His6-tag was cloned into the pet22b vector and expressed in BL21 Star (DE3) (Thermo Fisher Scientific) cells. Cells were harvested at 4000g for 20 min and lysed three times at 15,000 psi in PBS using an Avestin Emulsiflex C3 cell disruptor, followed by centrifugation at 10,000 rpm in a JLA10.5 rotor for 1 hour. The supernatant was filtered and purified using affinity (Ni-NTA) and SEC on a Superdex 75 column (GE Healthcare). The unbound Fabs 438-B11, 438-D5, and 438-B11ΔSS proteins were subjected to crystallization trials with and without protein G to minimize the flexibility of the constant domain of the Fabs with respect to the variable domain.
Crystallization and data collection
Two BG505 UFO.664 antibody complexes were formed by mixing Env with 438-B11 or 438-B11ΔSS and 35O22 Fabs in a molar ratio of 1:3.5:3.5 (UFO:438-B11 or 438-B11ΔSS:35O22) at room temperature for 30 min to form a complex. These complexes were then partially deglycosylated using endoglycosidase H digestion (New England Biolabs) (82) at 37°C for 1 hour and then purified on a Superdex 200 column. Both trimer complexes were SEC-purified in 50 mM tris-HCl and 150 mM NaCl (pH 7.4) and concentrated to ∼10 mg/ml before sending to crystallization trials. Samples were set up at both 4° and 20°C using our Rigaku CrystalMation robotic system (83). High-quality crystals of Fabs 438-B11 and 35O22 bound to BG505 UFO.664 were obtained in 0.2 M NaCl, 36% PEG-400 (polyethylene glycol 400), and 0.1 M potassium phosphate–sodium phosphate (pH 5.7) at 20°C. However, crystals of Fabs 438-B11ΔSS and 35O22 bound to BG505 UFO.664 were obtained in 12% 1-propanol, 0.1 M MES (pH 6.5), 20% PEG monomethyl ether 5000, and 25% glycerol at 4°C. The unbound SEC-purified Fabs 438-D5 (~6 mg/ml), 438-B11ΔSS (~25 mg/ml), and 438-B11 (~6 mg/ml) in 50 mM tris-HCl and 150 mM NaCl (pH 7.4) were concentrated. PG was mixed with unbound concentrated Fab 438-B11 in a 1:1 molar ratio at 37°C for 30 min before subjected to crystallization trials. The unbound Fab 438-D5 crystallized in 16% 2-propanol, 5% glycerol, 17% PEG-4000, and 0.095 M sodium citrate–citric acid (pH 6.2) at 20°C. The unbound Fab 438-B11ΔSS crystallized in 0.1 M sodium acetate (pH 4.16), 0.2 M lithium sulfate, and 40% PEG-400 at 4°C. The PG bound to the constant domain of Fab 438-B11 crystallized in 0.085 M sodium acetate (pH 4.0), 0.17 M ammonium acetate, 5% glycerol, 27.88% PEG-4000, and 15% glycerol. Data were collected at Advanced Photon Source 23-IDD and 23-IDB and Stanford Synchrotron Radiation Lightsource beamline 12-2.
Structure determination and refinement
Two BG505 UFO.664 complexes with Fabs 438-B11 or 438-B11ΔSS and 35O22 and unbound Fabs 438-D5, 438-B11+PG, and 438-B11ΔSS crystals were diffracted to 3.8-, 6.5-, 2.0-, 2.7-, and 2.1-Å resolutions and were processed (indexed, integrated, and scaled) with HKL2000 (84) in H32, H3, P212121, P21, and P21 space groups, respectively. The 438-D5 Fab structure was determined by molecular replacement (MR) using the program Phaser (85) with VH1-69 germline antibody CR9114 (PDB ID: 5WL2) as the initial MR model. The BG505 UFO.664 complex with Fabs 438-B11 and 35O22 was determined by using PDB 5CEZ (18) for Env BG505 UFO.664 gp140, PDB 4TOY (38) for 35O22, and the Fab 438-D5 structure for Fab 438-B11 as MR search models. For the Fab 438-B11ΔSS–bound UFO.664 trimer complex, the WT 438-B11–bound UFO.664 complex crystal structure was used as the MR search model. Given the limited resolution of the datasets, grouped B-factor refinement for each residue and positional coordinate refinement were enforced using a reference model set of restraints. The Fab 438-D5 structure was also used as the initial MR model for unbound Fabs 438-B11+PG and 438-B11ΔSS and PDB 1UWX (86) for PG. Model building was carried out with Coot, and refinement was carried out with Phenix (87, 88). The UFO.664-bound Fab 438-B11 complex crystal structure was refined to an Rcryst/Rfree of 29.9%/31.7% and an overall completeness of 99.6% (table S3D). The Fab 438-B11ΔSS–bound UFO.664 trimer crystal structure was refined to an Rcryst/Rfree of 30.7%/35.5% and an overall completeness of 97.5% (table S3E). The unbound 438-D5 crystal structure was refined to an Rcryst/Rfree of 20.0%/24.5% and a completeness of 99.8% (table S3D). The unbound 438-B11+PG crystal structure was refined to an Rcryst/Rfree of 22.7%/26.0% and a completeness of 96.6% (table S3D). The unbound 438-B11ΔSS crystal structure was refined to an Rcryst/Rfree of 22.9%/26.8% and a completeness of 99.9% (table S3D). Structure quality was determined by MolProbity (89). For the Fabs, residues were numbered according to the Kabat numbering scheme (90), and Env was numbered according to the HXB2 system (91). Structure validation was performed using the wwPDB Validation Service (validate.wwpdb.org), pdb-care (92), and Privateer (93). Data collection and refinement statistics are shown in table S3 (D and E).
Biolayer interferometry
BLI assays were performed using an Octet RED96 instrument (FortéBio) with agitation set to 1000 rpm in FortéBio 1× kinetic buffer. The final volume for all the solutions was 200 μl per well. Assays were performed at 30°C in solid black 96-well plates (Geiger Bio-One). To determine Kd values for antibody-Env interactions, soluble BG505 UFO.664 (containing an Avi tag at the C terminus) was loaded onto streptavidin biosensors at a concentration between ~10 and 50 μg/ml in kinetics buffer [1× PBS (pH 7.4), 0.01% BSA (bovine serum albumin), and 0.002% Tween 20]. The binding kinetics for the association of the Fabs were measured in a dilution series between 800 and 50 nM for 438-B11 and 438-B11KC-F83V, and between 10,000 and 312 nM for 438-B11ΔSS. Octet data were processed by FortéBio’s data acquisition software v.8.1 and fit with a 1:1 global fitting model to calculate the Kd values. To estimate antibody binding to trimers derived from different subtypes or with mutations at the N332 supersite, 5 μg ml−1 of IgG antibody in 1× kinetics buffer was loaded onto the surface of anti-human Fc Capture Biosensors (AHC) for 300 s. A 60-s biosensor baseline step was applied before the analysis of the association of the antibody on the biosensor to the antigen in solution for 200 s. A twofold concentration gradient of untagged soluble trimer, starting at 200 nM, was used in a dilution series of six. The dissociation of the interaction was followed for 300 s. Correction of baseline drift was performed by subtracting the mean value of shifts recorded for a sensor loaded with IgG antibody but not incubated with trimer and for a sensor without IgG antibody but incubated with trimer. Octet data were also processed by FortéBio’s data acquisition software v.8.1 and fitted with a 2:1 model to achieve the optimal fitting results. Because of the avidity effect caused by the presence of two Fabs within each IgG, the Kd values cannot be determined and the sensorgrams serve as a qualitative assessment of antibody-Env interaction.
Differential scanning calorimetry
The thermal stability of unbound Fabs 438-B11, 438-B11ΔSS, and 438-B11KC-F83V in TBS buffer from 20° to 120°C was measured using a MicroCal VP-Capillary calorimeter (Malvern) at a scanning rate of 90°C/hour. Data were analyzed using the VP-Capillary DSC automated data analysis software. The resulting data were fit to a non–two-state model.
Antibody sequencing library preparation using 5′-RACE PCR
To ensure the unbiased analysis of the donor B cell repertoires (3, 61, 72), an improved version of the 5′-RACE PCR protocol for sample preparation has been reported in our recent study (31). Here, total RNA was extracted from 1 million to 5 million PBMCs into 30 μl of water with RNeasy Mini Kits (Qiagen, Valencia, CA). For unbiased repertoire analysis, 5′-RACE was performed with SMARTer RACE cDNA Amplification Kit (Clontech, Mountain View, CA). The cDNA was purified and eluted in 20 μl of elution buffer (NucleoSpin PCR Clean-up Kit, Clontech). The immunoglobulin PCRs were set up with Platinum Taq High-Fidelity DNA Polymerase (Life Technologies, Carlsbad, CA) in a total volume of 50 μl, with 5 μl of cDNA as template, 1 μl of 5′-RACE primer or gene-specific forward primers, and 1 μl of 10 μM reverse primer. To facilitate NGS on the Ion GeneStudio S5 system, the forward 5′-RACE primer contained a P1 adaptor, while the reverse primer contained an A adaptor and an Ion Xpress barcode (Life Technologies) to differentiate libraries from different time points. A total of 25 cycles of PCRs were performed, and the 5′-RACE PCR products at ~600 base pairs were gel-purified (Qiagen, Valencia, CA).
NGS and antibodyomics analysis
Antibody NGS has been adapted to the Ion GeneStudio S5 system (31, 62). Briefly, the antibody heavy-chain and light-chain (κ and λ) libraries were quantitated using a Qubit 2.0 Fluorometer with the Qubit dsDNA HS Assay Kit. Equal amounts of heavy (H), κ, and λ chain libraries (1:1:1) from various time points were mixed and loaded onto an Ion 530 chip to increase the sequencing depth and to eliminate run-to-run variation. Template preparation and Ion 530 chip loading were performed on the Ion Chef system using the Ion 530 ExT Kit, followed by S5 sequencing with the default settings. Raw sequencing data were processed without 3′-end trimming in base calling to extend the read length. The human Antibodyomics pipeline version 1.0 (3, 61, 63) has been modified to improve computational efficiency and data accuracy (31, 62). This new pipeline was used to process and annotate the CBJC438 antibody NGS data for repertoire profiling and lineage tracing. The distributions of germline genes, germline divergence or degree of SHM, and CDR3 loop length were derived from antibody NGS data as general repertoire profiles. The divergence/identity 2D plots were constructed to visualize 438-B11 lineages in the context of CBJC438 antibody repertoire. A stepwise screening approach was devised to identify sequences from germline gene families of 438-B11 and 438-D5 (IGHV1-69 and IGKV3-20) for antibody synthesis and functional characterization. Dendrograms were generated in phylogenetic analysis using the software PHYLIP as previously described (3).
SGA and sequencing
HIV-1 RNA was isolated from plasma using the QIAamp viral RNA minikit (Qiagen, Valencia, CA) and reverse-transcribed to cDNA using the SuperScript III reverse transcriptase (Invitrogen Life Technologies, Grand Island, NY). The HIV-1 envelope SGA was performed as previously described (3, 66). PCR products were gel-purified by using a QIAquick gel extraction kit (Qiagen, Valencia, CA) and then sequenced on an ABI 3770 sequencer (Applied Biosciences). The full-length envelope sequences were assembled and edited using Sequencher 4.7 (Gene Codes, Ann Arbor, MI). Sequences were aligned using Gene Cutter (www.hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html) and CLUSTALW together with selected subtypes and then additionally edited by hand when needed. Phylogenetic analysis of envelope sequences was performed as previously described (3).
Supplementary Material
Acknowledgments
We thank Y. Hua, H. Tien, R. Stanfield, X. Dai, and M. Elsliger for excellent technical help. Diffraction data were collected at Advanced Photon Source (APS) beamline 23-IDD and Stanford Synchrotron Radiation Lightsource (SSRL) beamline 12-2. Use of the APS was supported by the DOE, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357. Use of the SSRL was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-76SF00515. Funding: This work was supported by the National Major Project (2012ZX10001008 and 2018ZX10731101-001-019) (Y.S.), the National Postdoctoral Program for Innovative Talents (BX201600078) (B.J.), the State Key Laboratory Innovative Project (2012SKLID103) (Y.S.), the HIV Vaccine Research and Design (HIVRAD) program (P01 AI110657) (I.A.W.), the International AIDS Vaccine Initiative Neutralizing Antibody Center and CAVD, the Scripps Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID UM1 AI100663) (I.A.W.), the Scripps Consortium for HIV/AIDS Vaccine Development (CHAVD 1UM1 AI144462) (I.A.W.), the HIV Vaccine Research and Design (HIVRAD) program (P01 AI124337) (J.Z.), and NIH Grants R01 AI129698 (J.Z.), R01 AI140844 (J.Z.), and R01 AI43563 (M.B.Z.). Author contributions: Project design was performed by S.K., B.J., J.Z., I.A.W., and Y.S. Cohort maintenance and sample collection were carried out by D.L., Y.F., and B. Su. Serum and antibody neutralization was performed by L.R., K.H., L.Z., and J.H. The 129-virus panel was provided by L.Z., E.L., D.S., and M.B.Z. Neutralization against the 129-virus panel was performed by B. Shapero and X.L. Sorting probe design and production were carried out by L.H. and J.Z. Single B cell sorting and cloning were done by B.J., D.L., and S.W. Antibody isolation, synthesis, expression, and characterization were performed by B.J. Antibody expression and purification were carried out by B.J., Y.H., and X.G. BG505 UFO.664 gp140 construct design was performed by L.H. and J.Z. Antibody and Env expression and purification were carried out by S.K., A.S., X.L., B. Shapero, C.S., C.J.M., C.N., and L.H. X-ray crystallography was performed by S.K. BLI was done by S.K., X.L., L.H., and J.Z. DSC and data analysis were carried out by S.K. and I.A.W. NGS, antibody selection, and synthesis were performed by L.H. and J.Z. NGS antibody neutralization was done by L.H., X.L., and L.R. SGA and sequencing were performed by Z.Z. and Y.F. Manuscript was written by S.K., B.J., L.H., J.Z., I.A.W., and Y.S. All authors were asked to comment on the manuscript. The TSRI manuscript number is 29923. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data and code to understand and assess the conclusions of this research are available in the main text, the Supplementary Materials, and the PDB (accession codes 6UTK, 6UUH, 6UUM, 6UUL, and 6V6W). Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/38/eabb1328/DC1
REFERENCES AND NOTES
- 1.Burton D. R., Mascola J. R., Antibody responses to envelope glycoproteins in HIV-1 infection. Nat. Immunol. 16, 571–576 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freund N. T., Wang H., Scharf L., Nogueira L., Horwitz J. A., Bar-On Y., Golijanin J., Sievers S. A., Sok D., Cai H., Lorenzi J. C. C., Halper-Stromberg A., Toth I., Piechocka-Trocha A., Gristick H. B., van Gils M. J., Sanders R. W., Wang L.-X., Seaman M. S., Burton D. R., Gazumyan A., Walker B. D., West A. P. Jr., Bjorkman P. J., Nussenzweig M. C., Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller. Sci. Transl. Med. 9, eaa12144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kong L., Ju B., Chen Y., He L., Ren L., Liu J., Hong K., Su B., Wang Z., Ozorowski G., Ji X., Hua Y., Chen Y., Deller M. C., Hao Y., Feng Y., Garces F., Wilson R., Dai K., O’Dell S., Kee K. M., Mascola J. R., Ward A. B., Wyatt R. T., Li Y., Wilson I. A., Zhu J., Shao Y., Key gp120 glycans pose roadblocks to the rapid development of VRC01-class antibodies in an HIV-1-infected Chinese donor. Immunity 44, 939–950 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonsignori M., Zhou T., Sheng Z., Chen L., Gao F., Joyce M. G., Ozorowski G., Chuang G.-Y., Schramm C. A., Wiehe K., Alam S. M., Bradley T., Gladden M. A., Hwang K.-K., Iyengar S., Kumar A., Lu X., Luo K., Mangiapani M. C., Parks R. J., Song H., Acharya P., Bailer R. T., Cao A., Druz A., Georgiev I. S., Kwon Y. D., Louder M. K., Zhang B., Zheng A., Hill B. J., Kong R.; Cinque Soto NISC Comparative Sequencing Program, Mullikin J. C., Douek D. C., Montefiori D. C., Moody M. A., Shaw G. M., Hahn B. H., Kelsoe G., Hraber P. T., Korber B. T., Boyd S. D., Fire A. Z., Kepler T. B., Shapiro L., Ward A. B., Mascola J. R., Liao H.-X., Kwong P. D., Haynes B. F., Maturation pathway from germline to broad HIV-1 neutralizer of a CD4-mimic antibody. Cell 165, 449–463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao H.-X., Lynch R., Zhou T., Gao F., Alam S. M., Boyd S. D., Fire A. Z., Roskin K. M., Schramm C. A., Zhang Z., Zhu J., Shapiro L.; NISC Comparative Sequencing Program, Mullikin J. C., Gnanakaran S., Hraber P., Wiehe K., Kelsoe G., Yang G., Xia S.-M., Montefiori D. C., Parks R., Lloyd K. E., Scearce R. M., Soderberg K. A., Cohen M., Kamanga G., Louder M. K., Tran L. M., Chen Y., Cai F., Chen S., Moquin S., Du X., Joyce M. G., Srivatsan S., Zhang B., Zheng A., Shaw G. M., Hahn B. H., Kepler T. B., Korber B. T. M., Kwong P. D., Mascola J. R., Haynes B. F., Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacLeod D. T., Choi N. M., Briney B., Garces F., Ver L. S., Landais E., Murrell B., Wrin T., Kilembe W., Liang C.-H., Ramos A., Bian C. B., Wickramasinghe L., Kong L., Eren K., Wu C.-Y., Wong C.-H.; The IAVI Protocol C Investigators & The IAVI African HIV Research Network, Kosakovsky Pond S. L., Wilson I. A., Burton D. R., Poignard P., Early antibody lineage diversification and independent limb maturation lead to broad HIV-1 neutralization targeting the Env high-mannose patch. Immunity 44, 1215–1226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wibmer C. K., Bhiman J. N., Gray E. S., Tumba N., Abdool Karim S. S., Williamson C., Morris L., Moore P. L., Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLOS Pathog. 9, e1003738 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sather D. N., Carbonetti S., Malherbe D. C., Pissani F., Stuart A. B., Hessell A. J., Gray M. D., Mikell I., Kalams S. A., Haigwood N. L., Stamatatos L., Emergence of broadly neutralizing antibodies and viral coevolution in two subjects during the early stages of infection with human immunodeficiency virus type 1. J. Virol. 88, 12968–12981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwong P. D., Mascola J. R., HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity 48, 855–871 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Schoofs T., Barnes C. O., Suh-Toma N., Golijanin J., Schommers P., Gruell H., West A. P. Jr., Bach F., Lee Y. E., Nogueira L., Georgiev I. S., Bailer R. T., Czartoski J., Mascola J. R., Seaman M. S., McElrath M. J., Doria-Rose N. A., Klein F., Nussenzweig M. C., Broad and potent neutralizing antibodies recognize the silent face of the HIV envelope. Immunity 50, 1513–1529.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou T., Zheng A., Baxa U., Chuang G.-Y., Georgiev I. S., Kong R., O’Dell S., Shahzad-ul-Hussan S., Shen C.-H., Tsybovsky Y., Bailer R. T., Gift S. K., Louder M. K., Kee K. M., Rawi R., Stevenson C. H., Stewart-Jones G. B. E., Taft J. D., Waltari E., Yang Y., Zhang B., Shivatare S. S., Shivatare V. S., Lee C.-C. D., Wu C.-Y.; NISC Comparative Sequencing Program, Mullikin J. C., Bewley C. A., Burton D. R., Polonis V. R., Shapiro L., Wong C.-H., Mascola J. R., Kwong P. D., Wu X., A neutralizing antibody recognizing primarily N-linked glycan targets the silent face of the HIV envelope. Immunity 48, 500–513.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landais E., Huang X., Havenar-Daughton C., Murrell B., Price M. A., Wickramasinghe L., Ramos A., Bian C. B., Simek M., Allen S., Karita E., Kilembe W., Lakhi S., Inambao M., Kamali A., Sanders E. J., Anzala O., Edward V., Bekker L.-G., Tang J., Gilmour J., Kosakovsky-Pond S. L., Phung P., Wrin T., Crotty S., Godzik A., Poignard P., Broadly neutralizing antibody responses in a large longitudinal sub-saharan HIV primary infection cohort. PLOS Pathog. 12, e1005369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker L. M., Huber M., Doores K. J., Falkowska E., Pejchal R., Julien J.-P., Wang S.-K., Ramos A., Chan-Hui P.-Y., Moyle M., Mitcham J. L., Hammond P. W., Olsen O. A., Phung P., Fling S., Wong C.-H., Phogat S., Wrin T., Simek M. D.; Protocol G. Principal Investigators, Koff W. C., Wilson I. A., Burton D. R., Poignard P., Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker L. M., Phogat S. K., Chan-Hui P.-Y., Wagner D., Phung P., Goss J. L., Wrin T., Simek M. D., Fling S., Mitcham J. L., Lehrman J. K., Priddy F. H., Olsen O. A., Frey S. M., Hammond P. W.; Protocol G Principal Investigators, Kaminsky S., Zamb T., Moyle M., Koff W. C., Poignard P., Burton D. R., Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker L. M., Simek M. D., Priddy F., Gach J. S., Wagner D., Zwick M. B., Phogat S. K., Poignard P., Burton D. R., A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLOS Pathog. 6, e1001028 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mouquet H., Scharf L., Euler Z., Liu Y., Eden C., Scheid J. F., Halper-Stromberg A., Gnanapragasam P. N. P., Spencer D. I. R., Seaman M. S., Schuitemaker H., Feizi T., Nussenzweig M. C., Bjorkman P. J., Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. U.S.A. 109, E3268–E3277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sok D., Burton D. R., Recent progress in broadly neutralizing antibodies to HIV. Nat. Immunol. 19, 1179–1188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garces F., Lee J. H., de Val N., de la Pena A. T., Kong L., Puchades C., Hua Y., Stanfield R. L., Burton D. R., Moore J. P., Sanders R. W., Ward A. B., Wilson I. A., Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity 43, 1053–1063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong L., de la Peña A. T., Deller M. C., Garces F., Sliepen K., Hua Y., Stanfield R. L., Sanders R. W., Wilson I. A., Complete epitopes for vaccine design derived from a crystal structure of the broadly neutralizing antibodies PGT128 and 8ANC195 in complex with an HIV-1 Env trimer. Acta Crystallogr. D Biol. Crystallogr. 71, 2099–2108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonich C. A., Williams K. L., Verkerke H. P., Williams J. A., Nduati R., Lee K. K., Overbaugh J., HIV-1 neutralizing antibodies with limited hypermutation from an infant. Cell 166, 77–87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnes C. O., Gristick H. B., Freund N. T., Escolano A., Lyubimov A. Y., Hartweger H., West A. P. Jr., Cohen A. E., Nussenzweig M. C., Bjorkman P. J., Structural characterization of a highly-potent V3-glycan broadly neutralizing antibody bound to natively-glycosylated HIV-1 envelope. Nat. Commun. 9, 1251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sok D., Pauthner M., Briney B., Lee J. H., Saye-Francisco K. L., Hsueh J., Ramos A., Le K. M., Jones M., Jardine J. G., Bastidas R., Sarkar A., Liang C.-H., Shivatare S. S., Wu C.-Y., Schief W. R., Wong C.-H., Wilson I. A., Ward A. B., Zhu J., Poignard P., Burton D. R., A prominent site of antibody vulnerability on HIV envelope incorporates a motif associated with CCR5 binding and its camouflaging glycans. Immunity 45, 31–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaik M. M., Peng H., Lu J., Rits-Volloch S., Xu C., Liao M., Chen B., Structural basis of coreceptor recognition by HIV-1 envelope spike. Nature 565, 318–323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Escolano A., Gristick H. B., Abernathy M. E., Merkenschlager J., Gautam R., Oliveira T. Y., Pai J., West A. P. Jr., Barnes C. O., Cohen A. A., Wang H., Golijanin J., Yost D., Keeffe J. R., Wang Z., Zhao P., Yao K.-H., Bauer J., Nogueira L., Gao H., Voll A. V., Montefiori D. C., Seaman M. S., Gazumyan A., Silva M., Mc Guire A. T., Stamatatos L., Irvine D. J., Wells L., Martin M. A., Bjorkman P. J., Nussenzweig M. C., Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques. Nature 570, 468–473 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steichen J. M., Lin Y.-C., Havenar-Daughton C., Pecetta S., Ozorowski G., Willis J. R., Toy L., Sok D., Liguori A., Kratochvil S., Torres J. L., Kalyuzhniy O., Melzi E., Kulp D. W., Raemisch S., Hu X., Bernard S. M., Georgeson E., Phelps N., Adachi Y., Kubitz M., Landais E., Umotoy J., Robinson A., Briney B., Wilson I. A., Burton D. R., Ward A. B., Crotty S., Batista F. D., Schief W. R., A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Science 366, eaax4380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krumm S. A., Mohammed H., Le K. M., Crispin M., Wrin T., Poignard P., Burton D. R., Doores K. J., Mechanisms of escape from the PGT128 family of anti-HIV broadly neutralizing antibodies. Retrovirology 13, 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore P. L., Gray E. S., Wibmer C. K., Bhiman J. N., Nonyane M., Sheward D. J., Hermanus T., Bajimaya S., Tumba N. L., Abrahams M.-R., Lambson B. E., Ranchobe N., Ping L., Ngandu N., Karim Q. A., Karim S. S. A., Swanstrom R. I., Seaman M. S., Williamson C., Morris L., Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat. Med. 18, 1688–1692 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Kerkhof T. L. G. M., de Taeye S. W., Boeser-Nunnink B. D., Burton D. R., Kootstra N. A., Schuitemaker H., Sanders R. W., van Gils M. J., HIV-1 escapes from N332-directed antibody neutralization in an elite neutralizer by envelope glycoprotein elongation and introduction of unusual disulfide bonds. Retrovirology 13, 48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong L., He L., de Val N., Vora N., Morris C. D., Azadnia P., Sok D., Zhou B., Burton D. R., Ward A. B., Wilson I. A., Zhu J., Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat. Commun. 7, 12040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.deCamp A., Hraber P., Bailer R. T., Seaman M. S., Ochsenbauer C., Kappes J., Gottardo R., Edlefsen P., Self S., Tang H., Greene K., Gao H., Daniell X., Sarzotti-Kelsoe M., Gorny M. K., Zolla-Pazner S., La Branche C. C., Mascola J. R., Korber B. T., Montefiori D. C., Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 88, 2489–2507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He L., Lin X., de Val N., Saye-Francisco K. L., Mann C. J., Augst R., Morris C. D., Azadnia P., Zhou B., Sok D., Ozorowski G., Ward A. B., Burton D. R., Zhu J., Hidden lineage complexity of glycan-dependent HIV-1 broadly neutralizing antibodies uncovered by digital panning and native-like gp140 trimer. Front. Immunol. 8, 1025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seaman M. S., Janes H., Hawkins N., Grandpre L. E., Devoy C., Giri A., Coffey R. T., Harris L., Wood B., Daniels M. G., Bhattacharya T., Lapedes A., Polonis V. R., McCutchan F. E., Gilbert P. B., Self S. G., Korber B. T., Montefiori D. C., Mascola J. R., Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84, 1439–1452 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X., Zhang Z., Schramm C. A., Joyce M. G., Kwon Y. D., Zhou T., Sheng Z., Zhang B., O’Dell S., Kee K. M., Georgiev I. S., Chuang G.-Y., Longo N. S., Lynch R. M., Saunders K. O., Soto C., Srivatsan S., Yang Y., Bailer R. T., Louder M. K.; NISC Comparative Sequencing Program, Mullikin J. C., Connors M., Kwong P. D., Mascola J. R., Shapiro L., Maturation and diversity of the VRC01-antibody lineage over 15 years of chronic HIV-1 infection. Cell 161, 470–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonsignori M., Scott E., Wiehe K., Easterhoff D., Alam S. M., Hwang K.-K., Cooper M., Xia S.-M., Zhang R., Montefiori D. C., Henderson R., Nie X., Kelsoe G., Moody M. A., Chen X., Joyce M. G., Kwong P. D., Connors M., Mascola J. R., McGuire A. T., Stamatatos L., Medina-Ramírez M., Sanders R. W., Saunders K. O., Kepler T. B., Haynes B. F., Inference of the HIV-1 VRC01 antibody lineage unmutated common ancestor reveals alternative pathways to overcome a key glycan barrier. Immunity 49, 1162–1174.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doria-Rose N. A., Schramm C. A., Gorman J., Moore P. L., Bhiman J. N., De Kosky B. J., Ernandes M. J., Georgiev I. S., Kim H. J., Pancera M., Staupe R. P., Altae-Tran H. R., Bailer R. T., Crooks E. T., Cupo A., Druz A., Garrett N. J., Hoi K. H., Kong R., Louder M. K., Longo N. S., Kee K. M., Nonyane M., O’Dell S., Roark R. S., Rudicell R. S., Schmidt S. D., Sheward D. J., Soto C., Wibmer C. K., Yang Y., Zhang Z.; NISC Comparative Sequencing, Mullikin J. C., Binley J. M., Sanders R. W., Wilson I. A., Moore J. P., Ward A. B., Georgiou G., Williamson C., Abdool Karim S. S., Morris L., Kwong P. D., Shapiro L., Mascola J. R., Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509, 55–62 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sok D., Le K. M., Vadnais M., Saye-Francisco K. L., Jardine J. G., Torres J. L., Berndsen Z. T., Kong L., Stanfield R., Ruiz J., Ramos A., Liang C.-H., Chen P. L., Criscitiello M. F., Mwangi W., Wilson I. A., Ward A. B., Smider V. V., Burton D. R., Rapid elicitation of broadly neutralizing antibodies to HIV by immunization in cows. Nature 548, 108–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He L., de Val N., Morris C. D., Vora N., Thinnes T. C., Kong L., Azadnia P., Sok D., Zhou B., Burton D. R., Wilson I. A., Nemazee D., Ward A. B., Zhu J., Presenting native-like trimeric HIV-1 antigens with self-assembling nanoparticles. Nat. Commun. 7, 12041 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang J., Kang B. H., Pancera M., Lee J. H., Tong T., Feng Y., Imamichi H., Georgiev I. S., Chuang G.-Y., Druz A., Doria-Rose N. A., Laub L., Sliepen K., van Gils M. J., de la Peña A. T., Derking R., Klasse P.-J., Migueles S. A., Bailer R. T., Alam M., Pugach P., Haynes B. F., Wyatt R. T., Sanders R. W., Binley J. M., Ward A. B., Mascola J. R., Kwong P. D., Connors M., Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515, 138–142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Julien J.-P., Sok D., Khayat R., Lee J. H., Doores K. J., Walker L. M., Ramos A., Diwanji D. C., Pejchal R., Cupo A., Katpally U., Depetris R. S., Stanfield R. L., Bride R. M., Marozsan A. J., Paulson J. C., Sanders R. W., Moore J. P., Burton D. R., Poignard P., Ward A. B., Wilson I. A., Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLOS Pathog. 9, e1003342 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pejchal R., Doores K. J., Walker L. M., Khayat R., Huang P.-S., Wang S.-K., Stanfield R. L., Julien J.-P., Ramos A., Crispin M., Depetris R., Katpally U., Marozsan A., Cupo A., Maloveste S., Liu Y., Bride R. M., Ito Y., Sanders R. W., Ogohara C., Paulson J. C., Feizi T., Scanlan C. N., Wong C.-H., Moore J. P., Olson W. C., Ward A. B., Poignard P., Schief W. R., Burton D. R., Wilson I. A., A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334, 1097–1103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanders R. W., Moore J. P., Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol. Rev. 275, 161–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward A. B., Wilson I. A., The HIV-1 envelope glycoprotein structure: Nailing down a moving target. Immunol. Rev. 275, 21–32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Binley J. M., Sanders R. W., Clas B., Schuelke N., Master A., Guo Y., Kajumo F., Anselma D. J., Maddon P. J., Olson W. C., Moore J. P., A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74, 627–643 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma S. K., de Val N., Bale S., Guenaga J., Tran K., Feng Y., Dubrovskaya V., Ward A. B., Wyatt R. T., Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 11, 539–550 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He L., Kumar S., Allen J. D., Huang D., Lin X., Mann C. J., Saye-Francisco K. L., Copps J., Sarkar A., Blizard G. S., Ozorowski G., Sok D., Crispin M., Ward A. B., Nemazee D., Burton D. R., Wilson I. A., Zhu J., HIV-1 vaccine design through minimizing envelope metastability. Sci. Adv. 4, eaau6769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarkar A., Bale S., Behrens A.-J., Kumar S., Sharma S. K., de Val N., Pallesen J., Irimia A., Diwanji D. C., Stanfield R. L., Ward A. B., Crispin M., Wyatt R. T., Wilson I. A., Structure of a cleavage-independent HIV Env recapitulates the glycoprotein architecture of the native cleaved trimer. Nat. Commun. 9, 1956 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen F., Tzarum N., Wilson I. A., Law M., VH1-69 antiviral broadly neutralizing antibodies: Genetics, structures, and relevance to rational vaccine design. Curr. Opin. Virol. 34, 149–159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lang S., Xie J., Zhu X., Wu N. C., Lerner R. A., Wilson I. A., Antibody 27F3 broadly targets influenza A group 1 and 2 hemagglutinins through a further variation in V(H)1-69 antibody orientation on the HA. Cell Rep. 20, 2935–2943 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irimia A., Sarkar A., Stanfield R. L., Wilson I. A., Crystallographic identification of lipid as an integral component of the epitope of HIV broadly neutralizing antibody 4E10. Immunity 44, 21–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cardoso R. M. F., Zwick M. B., Stanfield R. L., Kunert R., Binley J. M., Katinger H., Burton D. R., Wilson I. A., Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22, 163–173 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Zhang L., Irimia A., He L., Landais E., Rantalainen K., Leaman D. P., Vollbrecht T., Stano A., Sands D. I., Kim A. S.; IAVI Protocol G Investigators, Poignard P., Burton D. R., Murrell B., Ward A. B., Zhu J., Wilson I. A., Zwick M. B., An MPER antibody neutralizes HIV-1 using germline features shared among donors. Nat. Commun. 10, 5389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheid J. F., Mouquet H., Ueberheide B., Diskin R., Klein F., Oliveira T. Y. K., Pietzsch J., Fenyo D., Abadir A., Velinzon K., Hurley A., Myung S., Boulad F., Poignard P., Burton D. R., Pereyra F., Ho D. D., Walker B. D., Seaman M. S., Bjorkman P. J., Chait B. T., Nussenzweig M. C., Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang C.-c., Venturi M., Majeed S., Moore M. J., Phogat S., Zhang M.-Y., Dimitrov D. S., Hendrickson W. A., Robinson J., Sodroski J., Wyatt R., Choe H., Farzan M., Kwong P. D., Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc. Natl. Acad. Sci. U.S.A. 101, 2706–2711 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou T., Lynch R. M., Chen L., Acharya P., Wu X., Doria-Rose N. A., Joyce M. G., Lingwood D., Soto C., Bailer R. T., Ernandes M. J., Kong R., Longo N. S., Louder M. K., Kee K. M., O’Dell S., Schmidt S. D., Tran L., Yang Z., Druz A., Luongo T. S., Moquin S., Srivatsan S., Yang Y., Zhang B., Zheng A., Pancera M., Kirys T., Georgiev I. S., Gindin T., Peng H.-P., Yang A.-S.; NISC Comparative Sequencing Program, Mullikin J. C., Gray M. D., Stamatatos L., Burton D. R., Koff W. C., Cohen M. S., Haynes B. F., Casazza J. P., Connors M., Corti D., Lanzavecchia A., Sattentau Q. J., Weiss R. A., West A. P. Jr., Bjorkman P. J., Scheid J. F., Nussenzweig M. C., Shapiro L., Mascola J. R., Kwong P. D., Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell 161, 1280–1292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stanfield R. L., Takimoto-Kamimura M., Rini J. M., Profy A. T., Wilson I. A., Major antigen-induced domain rearrangements in an antibody. Structure 1, 83–93 (1993). [DOI] [PubMed] [Google Scholar]
- 56.Koenig P., Lee C. V., Walters B. T., Janakiraman V., Stinson J., Patapoff T. W., Fuh G., Mutational landscape of antibody variable domains reveals a switch modulating the interdomain conformational dynamics and antigen binding. Proc. Natl. Acad. Sci. U.S.A. 114, E486–E495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henderson R., Watts B. E., Ergin H. N., Anasti K., Parks R., Xia S.-M., Trama A., Liao H.-X., Saunders K. O., Bonsignori M., Wiehe K., Haynes B. F., Alam S. M., Selection of immunoglobulin elbow region mutations impacts interdomain conformational flexibility in HIV-1 broadly neutralizing antibodies. Nat. Commun. 10, 654 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klein F., Diskin R., Scheid J. F., Gaebler C., Mouquet H., Georgiev I. S., Pancera M., Zhou T., Incesu R.-B., Fu B. Z., Gnanapragasam P. N. P., Oliveira T. Y., Seaman M. S., Kwong P. D., Bjorkman P. J., Nussenzweig M. C., Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153, 126–138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ovchinnikov V., Louveau J. E., Barton J. P., Karplus M., Chakraborty A. K., Role of framework mutations and antibody flexibility in the evolution of broadly neutralizing antibodies. eLife 7, e33038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stanfield R. L., Wilson I. A., Smider V. V., Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies. Sci. Immunol. 1, aaf7962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He L., Sok D., Azadnia P., Hsueh J., Landais E., Simek M., Koff W. C., Poignard P., Burton D. R., Zhu J., Toward a more accurate view of human B-cell repertoire by next-generation sequencing, unbiased repertoire capture and single-molecule barcoding. Sci. Rep. 4, 6778 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tzarum N., Giang E., Kong L., He L., Prentoe J., Augestad E., Hua Y., Castillo S., Lauer G. M., Bukh J., Zhu J., Wilson I. A., Law M., Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies. Sci. Adv. 5, eaav1882 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu X., Zhou T., Zhu J., Zhang B., Georgiev I., Wang C., Chen X., Longo N. S., Louder M., Kee K. M., O’Dell S., Perfetto S., Schmidt S. D., Shi W., Wu L., Yang Y., Yang Z.-Y., Yang Z., Zhang Z., Bonsignori M., Crump J. A., Kapiga S. H., Sam N. E., Haynes B. F., Simek M., Burton D. R., Koff W. C., Doria-Rose N. A., Connors M.; NISC Comparative Sequencing Program, Mullikin J. C., Nabel G. J., Roederer M., Shapiro L., Kwong P. D., Mascola J. R., Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou T., Zhu J., Wu X., Moquin S., Zhang B., Acharya P., Georgiev I. S., Altae-Tran H. R., Chuang G.-Y., Joyce M. G., Kwon Y. D., Longo N. S., Louder M. K., Luongo T., Kee K. M., Schramm C. A., Skinner J., Yang Y., Yang Z., Zhang Z., Zheng A., Bonsignori M., Haynes B. F., Scheid J. F., Nussenzweig M. C., Simek M., Burton D. R., Koff W. C.; NISC Comparative Sequencing Program, Mullikin J. C., Connors M., Shapiro L., Nabel G. J., Mascola J. R., Kwong P. D., Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245–258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu J., Wu X., Zhang B., Kee K. M., O’Dell S., Soto C., Zhou T., Casazza J. P.; NISC Comparative Sequencing Program, Mullikin J. C., Kwong P. D., Mascola J. R., Shapiro L., De novo identification of VRC01 class HIV-1-neutralizing antibodies by next-generation sequencing of B-cell transcripts. Proc. Natl. Acad. Sci. U.S.A. 110, E4088–E4097 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salazar-Gonzalez J. F., Bailes E., Pham K. T., Salazar M. G., Guffey M. B., Keele B. F., Derdeyn C. A., Farmer P., Hunter E., Allen S., Manigart O., Mulenga J., Anderson J. A., Swanstrom R., Haynes B. F., Athreya G. S., Korber B. T. M., Sharp P. M., Shaw G. M., Hahn B. H., Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82, 3952–3970 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Struwe W. B., Chertova E., Allen J. D., Seabright G. E., Watanabe Y., Harvey D. J., Medina-Ramirez M., Roser J. D., Smith R., Westcott D., Keele B. F., Bess J. W. Jr., Sanders R. W., Lifson J. D., Moore J. P., Crispin M., Site-specific glycosylation of virion-derived HIV-1 Env Is mimicked by a soluble trimeric immunogen. Cell Rep. 24, 1958–1966.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crispin M., Ward A. B., Wilson I. A., Structure and immune recognition of the HIV glycan shield. Annu. Rev. Biophys. 47, 499–523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kong L., Lee J. H., Doores K. J., Murin C. D., Julien J.-P., Bride R. M., Liu Y., Marozsan A., Cupo A., Klasse P.-J., Hoffenberg S., Caulfield M., King C. R., Hua Y., Le K. M., Khayat R., Deller M. C., Clayton T., Tien H., Feizi T., Sanders R. W., Paulson J. C., Moore J. P., Stanfield R. L., Burton D. R., Ward A. B., Wilson I. A., Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat. Struct. Mol. Biol. 20, 796–803 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diskin R., Scheid J. F., Marcovecchio P. M., West A. P. Jr., Klein F., Gao H., Gnanapragasam P. N. P., Abadir A., Seaman M. S., Nussenzweig M. C., Bjorkman P. J., Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334, 1289–1293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu X., Yang Z.-Y., Li Y., Hogerkorp C.-M., Schief W. R., Seaman M. S., Zhou T., Schmidt S. D., Wu L., Xu L., Longo N. S., Kee K. M., O’Dell S., Louder M. K., Wycuff D. L., Feng Y., Nason M., Doria-Rose N., Connors M., Kwong P. D., Roederer M., Wyatt R. T., Nabel G. J., Mascola J. R., Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dai K., He L., Khan S. N., O’Dell S., Kee K. M., Tran K., Li Y., Sundling C., Morris C. D., Mascola J. R., Hedestam G. B. K., Wyatt R. T., Zhu J., Rhesus macaque B-cell responses to an HIV-1 trimer vaccine revealed by unbiased longitudinal repertoire analysis. MBio 6, e01375-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Steichen J. M., Kulp D. W., Tokatlian T., Escolano A., Dosenovic P., Stanfield R. L., McCoy L. E., Ozorowski G., Hu X., Kalyuzhniy O., Briney B., Schiffner T., Garces F., Freund N. T., Gitlin A. D., Menis S., Georgeson E., Kubitz M., Adachi Y., Jones M., Mutafyan A. A., Yun D. S., Mayer C. T., Ward A. B., Burton D. R., Wilson I. A., Irvine D. J., Nussenzweig M. C., Schief W. R., HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity 45, 483–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Escolano A., Steichen J. M., Dosenovic P., Kulp D. W., Golijanin J., Sok D., Freund N. T., Gitlin A. D., Oliveira T., Araki T., Lowe S., Chen S. T., Heinemann J., Yao K.-H., Georgeson E., Saye-Francisco K. L., Gazumyan A., Adachi Y., Kubitz M., Burton D. R., Schief W. R., Nussenzweig M. C., Sequential immunization elicits broadly neutralizing anti-HIV-1 antibodies in Ig knockin mice. Cell 166, 1445–1458.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morris C. D., Azadnia P., de Val N., Vora N., Honda A., Giang E., Saye-Francisco K., Cheng Y., Lin X., Mann C. J., Tang J., Sok D., Burton D. R., Law M., Ward A. B., He L., Zhu J., Differential antibody responses to conserved HIV-1 neutralizing epitopes in the context of multivalent scaffolds and native-like gp140 trimers. MBio 8, e00036-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu K., Acharya P., Kong R., Cheng C., Chuang G.-Y., Liu K., Louder M. K., O’Dell S., Rawi R., Sastry M., Shen C.-H., Zhang B., Zhou T., Asokan M., Bailer R. T., Chambers M., Chen X., Choi C. W., Dandey V. P., Doria-Rose N. A., Druz A., Eng E. T., Farney S. K., Foulds K. E., Geng H., Georgiev I. S., Gorman J., Hill K. R., Jafari A. J., Kwon Y. D., Lai Y.-T., Lemmin T., Kee K. M., Ohr T. Y., Ou L., Peng D., Rowshan A. P., Sheng Z., Todd J.-P., Tsybovsky Y., Viox E. G., Wang Y., Wei H., Yang Y., Zhou A. F., Chen R., Yang L., Scorpio D. G., McDermott A. B., Shapiro L., Carragher B., Potter C. S., Mascola J. R., Kwong P. D., Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat. Med. 24, 857–867 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hu X., Hong K., Zhao C., Zheng Y., Ma L., Ruan Y., Gao H., Greene K., Sarzotti-Kelsoe M., Montefiori D. C., Shao Y., Profiles of neutralizing antibody response in chronically human immunodeficiency virus type 1 clade B'-infected former plasma donors from China naive to antiretroviral therapy. J. Gen. Virol. 93, 2267–2278 (2012). [DOI] [PubMed] [Google Scholar]
- 78.Montefiori D. C., Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. Chapter 12, Unit 12.11 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Li M., Gao F., Mascola J. R., Stamatatos L., Polonis V. R., Koutsoukos M., Voss G., Goepfert P., Gilbert P., Greene K. M., Bilska M., Kothe D. L., Salazar-Gonzalez J. F., Wei X., Decker J. M., Hahn B. H., Montefiori D. C., Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79, 10108–10125 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liao H.-X., Levesque M. C., Nagel A., Dixon A., Zhang R., Walter E., Parks R., Whitesides J., Marshall D. J., Hwang K.-K., Yang Y., Chen X., Gao F., Munshaw S., Kepler T. B., Denny T., Anthony Moody M., Haynes B. F., High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J. Virol. Methods 158, 171–179 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M. C., Wardemann H., Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol Methods 329, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Julien J.-P., Cupo A., Sok D., Stanfield R. L., Lyumkis D., Deller M. C., Klasse P.-J., Burton D. R., Sanders R. W., Moore J. P., Ward A. B., Wilson I. A., Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Elsliger M.-A., Deacon A. M., Godzik A., Lesley S. A., Wooley J., Wüthrich K., Wilson I. A., The JCSG high-throughput structural biology pipeline. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1137–1142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 85.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tzitzilonis C., Prince S. M., Collins R. F., Achtman M., Feavers I. M., Maiden M. C. J., Derrick J. P., Structural variation and immune recognition of the P1.2 subtype meningococcal antigen. Proteins 62, 947–955 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 88.Adams P. D., Gopal K., Grosse-Kunstleve R. W., Hung L.-W., Ioerger T. R., McCoy A. J., Moriarty N. W., Pai R. K., Read R. J., Romo T. D., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C., Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 11, 53–55 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Davis I. W., Murray L. W., Richardson J. S., Richardson D. C., MOLPROBITY: Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martin A. C., Accessing the Kabat antibody sequence database by computer. Proteins 25, 130–133 (1996). [DOI] [PubMed] [Google Scholar]
- 91.Ratner L., Fisher A., Jagodzinski L. L., Mitsuya H., Liou R. S., Gallo R. C., Wong-Staal F., Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res. Hum. Retrovir. 3, 57–69 (1987). [DOI] [PubMed] [Google Scholar]
- 92.Lutteke T., von der Lieth C. W., pdb-care (PDB carbohydrate residue check): A program to support annotation of complex carbohydrate structures in PDB files. BMC Bioinformatics 5, 69 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Agirre J., Strategies for carbohydrate model building, refinement and validation. Acta Crystallogr. D Struct. Biol. 73, 171–186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/38/eabb1328/DC1


