Within mycobacteria, polysaccharide chain length determines periplasm size and modulates antibiotic susceptibility.
Abstract
Bacteria control the length of their polysaccharides, which can control cell viability, physiology, virulence, and immune evasion. Polysaccharide chain length affects immunomodulation, but its impact on bacterial physiology and antibiotic susceptibility was unclear. We probed the consequences of truncating the mycobacterial galactan, an essential linear polysaccharide of about 30 residues. Galactan covalently bridges cell envelope layers, with the outermost cell wall linkage point occurring at residue 12. Reducing galactan chain length by approximately half compromises fitness, alters cell morphology, and increases the potency of hydrophobic antibiotics. Systematic variation of the galactan chain length revealed that it determines periplasm size. Thus, glycan chain length can directly affect cellular physiology and antibiotic activity, and mycobacterial glycans, not proteins, regulate periplasm size.
INTRODUCTION
Bacteria generate cell surface polysaccharides of controlled size. Biosynthesis of these glycans occurs through processes that regulate chain length (1), indicating that polysaccharide length is linked to function. The one known function for saccharide chain length is to influence the potency of immunomodulatory glycans. For instance, in Salmonella enterica, O-antigens of >100 repeat units serve as physical barriers that facilitate bacterial evasion of complement-mediated killing (2). Alternatively, in mycobacteria, the cell surface glycoconjugate lipoarabinomannan (LAM) affects host immunomodulation (3). When arabinan branching is disrupted, the mannan core of LAM is more efficiently recognized by macrophages as foreign; consequently, the host mounts a proinflammatory response (4). Beyond immunomodulatory glycans, however, the effects of polysaccharide chain length on bacterial physiology and survival are not apparent.
An essential polysaccharide whose length is controlled is the mycobacterial galactan. The galactan’s only known function is to covalently link the peptidoglycan (PG) and arabinan (5). The mycobacterial galactan anchors to the PG through the galactan reducing end, and genetic perturbation studies indicate that the 8th, 10th, and 12th β, d-galactofuranose (Galf) residues of the polysaccharide act as branch points for the arabinan (Fig. 1A) (6). The galactan consists of approximately 30 Galf residues such that it extends far beyond the 12 residues needed for branching (6–9). The “extra” length of this glycan appears to be disconnected from immune regulation because the galactan is positioned within the mycobacterial periplasm and not on the cell surface (10, 11). To assess the importance of galactan length, we tested the consequences of its truncation.
Fig. 1. The galactan and the mycobacterial cell envelope.
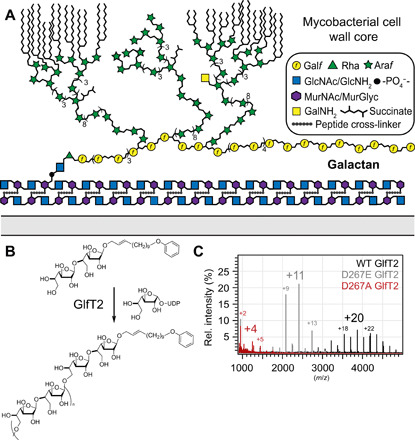
(A) Schematic depiction of the envelope of mycobacteria highlighting the mAGP (mycolyl-arabinogalactan-peptidoglycan). The glycans are depicted with the standard nomenclature. (B) Galactofuranosyl transferase 2 (GlfT2) extends the galactan to its final length. Polymerase activity can be probed in vitro using purified protein, synthetic acceptor, and sugar donor. (C) Overlayed matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) spectra produced by GlfT2 metal ion binding variants. Number of Galf residues added to acceptor noted above major product peaks.
To engineer galactan chain length, we focused on galactofuranosyl transferase 2 (GlfT2), the biosynthetic polymerase responsible for galactan elongation (Fig. 1B) (12). This GT2-family glycosyltransferase acts on a Galf-primed precursor in the cytoplasm and appears to control galactan length. Recombinant Mycobacterium tuberculosis GlfT2 can process a synthetic acceptor to yield glycans of physiologically relevant chain lengths (12). The number of saccharide residues in the glycan product was altered by introducing single-point substitutions into the tri-aspartate (DDD) metal ion binding motif involved in binding of the sugar donor uridine diphosphate (UDP)–Galf) (13, 14). We hypothesized that replacing wild-type (WT) GlfT2 in Mycobacterium smegmatis with a variant could afford cells with a truncated galactan, and that these mutant cells would reveal the contributions of galactan chain length to bacterial physiology.
RESULTS
To generate mutants with shorter galactan, we characterized M. smeg GlfT2 (gene ID: Msmeg_6403). Chain elongation experiments with recombinant WT M. smeg GlfT2 afforded polysaccharides of similar length as those generated by M. tuberculosis GlfT2 (Fig. 1, B and C). We generated D267E and D267A variants of M. smeg GlfT2, as these variants have substitutions at the metal ion binding motif; therefore, they should bind UDP-Galf less well and afford shorter galactan chains (fig. S1) (14). In a chain elongation assay, D267A GlfT2 was only able to transfer a few Galf residues to acceptor, but D267E GlfT2 afforded products about half the length of those obtained using WT GlfT2 (Fig. 1C). We therefore introduced glfT2 point mutants to an M. smeg strain lacking endogenous glfT2. Using a modified approach to mycobacterial recombineering (15), we deleted glfT2 at its endogenous site while providing complementation through exogenous expression of Flag-glfT2 from an integration proficient plasmid (16). Expression of D267A Flag-glfT2 did not complement, but the D267E Flag-glfT2 construct did (fig. S2).
To determine whether the galactan length had been altered in cells, we analyzed the monosaccharide composition of WT, Flag-glfT2 complemented (WTcomp), and D267E Flag-glfT2 complemented (D267Ecomp) cells (fig. S3) (17). Because each galactan chain contains one rhamnose (Rha) residue, the ratio of galactose to rhamnose (Gal:Rha) is a surrogate for average galactan chain length. Applying this analysis revealed that D267Ecomp cells generate galactan chains approximately twofold shorter than WT cells (Table 1 and fig. S3). The arabinosylation of the D267Ecomp strain was within 5% of the WTcomp strain, implying that the galactan chain in the mutant undergoes arabinan formation. Next, we determined whether any other saccharide units or lipids in the D267Ecomp cell envelope had changed. No change in the identity of covalently associated lipids or the relative amount of galactan appended to the cell wall was detected. Any changes in composition of free lipids and lipoarabinomannan (LAM) were minimal (fig. S3). Thus, the major perturbation of D267Ecomp cells is a truncated galactan.
Table 1. Monosaccharide composition analysis of cell envelope isolated from WT, WTcomp, and D267Ecomp strains.
Ratios are the average of three biological replicates
| Ara | Gal | Rha | |
| WT | 64 | 34 | 1 |
| WTcomp | 59 | 33 | 1 |
| D267Ecomp | 56 | 16 | 1 |
A growth defect of D267Ecomp cells was readily apparent, as their doubling time was approximately 1.5-fold slower than that of WT (Fig. 2A). Extracellular glycans can influence a cell’s ability to withstand osmotic pressure (18, 19); therefore, we tested whether hyperosmotic stress would further exacerbate this defect. We evaluated osmolyte tolerance for WT, WTcomp, and D267Ecomp cells and found that the mutant strain was much less resistant to hyperosmotic stress (fig. S4). When exposed to high salt concentration (~500 mM sodium chloride), the growth rate was severely restricted.
Fig. 2. Impact of galactan truncation on fitness.
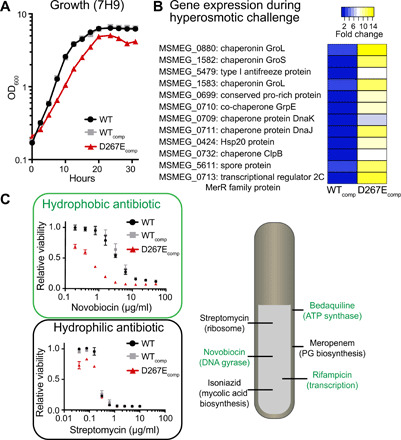
(A) Growth rate monitored in standard 7H9 medium. (B) RNA-seq data highlighting genes substantially up-regulated in D267Ecomp cells during hyperosmotic challenge. (C) Changes in the minimum inhibitory concentrations (MICs) of hydrophobic drugs (green), like novobiocin, versus hydrophilic drugs (black), like streptomycin. Antibiotic targets are listed.
We profiled cells cultured in hyperosmotic medium containing 250 mM sodium chloride (about 15-fold higher than standard conditions) using RNA sequencing (RNA-seq). Data from the surviving population indicate that the genes associated with nutrient acquisition (Fig. 2B and table S1) and proteostasis are up-regulated. Increasing the levels of proteins that mediate proteostasis can mitigate osmotic stress (20). The mutant cells strongly up-regulate these pathways, thereby underscoring their high sensitivity.
M. smeg mutants with defects in cell wall biosynthesis are hypersensitive to select antibiotics (7, 21). We, therefore, assessed the viability of the D267Ecomp strain after antibiotic exposure (Fig. 2C). We reasoned that loss of proper glycan chain length could result in either an alteration of cell envelope permeability to facilitate antibiotic uptake or an increase in the susceptibility of the relevant biosynthetic pathway to inhibition. Consequently, we tested a panel of antibiotics with diverse hydrophobicities and mechanisms of action. D267Ecomp cells were more susceptible than WT to numerous antibiotics (fig. S5); however, those antibiotics did not share a related mechanism of action. For example, no potency increases were observed for antibiotics targeting the cell envelope. In addition, hydrophilic antibiotics, such as streptomycin or meropenem, exhibited no change in activity. These findings align with results from our lipid analysis experiments indicating that the mutant has an intact mycolic acid–rich outer membrane, as this layer is likely the major barrier for hydrophilic antibiotics. In contrast, hydrophobic antibiotics were more effective on the mutant. Specifically, hydrophobic agents with unrelated mechanisms of action, such as rifampicin and novobiocin, were more potent (fig. S5).
Deletion of nonessential genes involved in cell wall polysaccharide biosynthesis can alter bacterial cell shape and cell envelope architecture (19, 22). Consequently, we evaluated cell shape of D267Ecomp cells using microscopy. Using a membrane stain to illuminate the cell outline, we found that relative cell length had decreased (fig. S6). To better observe any defects in cell growth and division, we next conducted time-lapse microscopy in a microfluidics chamber (movies S1 to S6). M. smeg cells are normally long, thin rods; however, defects in cell envelope biosynthesis can result in “blebbing,” a phenotype characterized by the appearance of circular protrusions (Fig. 3A). Analysis of time-lapse microscopy revealed that D267Ecomp cells undergo blebbing, and numerous instances of cell rupture were detected during division. These phenotypes also have been observed with mutants of PG biosynthesis, in which atomic force microscopy indicated a substantial decrease in cell wall rigidity (23). These data underscore the underappreciated contributions the galactan makes to cell structure and rigidity.
Fig. 3. Galactan chain length affects cell shape and periplasm size.
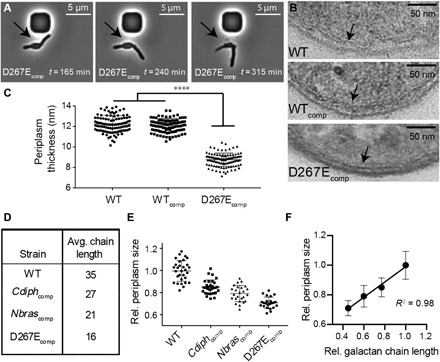
(A) Time-lapse microscopy images of M. smegmatis glfT2 mutants. Galactan truncation led to blebbing morphology (arrows) and, in multiple instances, cell rupture during division. (B) TEM of M. smegmatis glfT2 mutants highlighting the periplasm (arrows). (C) Galactan truncation decreases periplasm size in M. smegmatis (P < 0.0001). (D) Introduction of glfT2 orthologs affords access to strains with altered galactan chain length. (E) TEM images were used to quantify periplasm thickness across ortholog complemented strains. (F) Periplasm size and galactan chain length correlate linearly.
The effect of the galactan on cell length and rigidity prompted us to examine the architecture of the mutant cell envelope. We used transmission electron microscopy (TEM) to visualize WT, WTcomp, and D267Ecomp cells. The morphology of multiple D267Ecomp cells was indicative of blebbing, consistent with our previous observations (fig. S6). In the mutant, the periplasm, the region where the galactan resides, had contracted (Fig. 3B and fig. S6). On average, the thickness of the periplasm of D267Ecomp cells was decreased by about 30% (Fig. 3C). To better understand the relationship between galactan chain length and cell shape, we generated a set of strains whose galactan length varied systematically.
Orthologs of GlfT2 have been identified across the Corynebacterineae suborder, and previous investigations indicate that orthologous GlfT2s have distinct chain length control (24). We introduced the glfT2 genes from either Nocardia brasiliensis (Nbras) or Corynebacterium diphtheriae (Cdiph) into our glfT2-deficient M. smeg strain. These orthologs complemented growth, and in both Cdiphcomp and Nbrascomp strains, the galactan chain length was shorter than that of WT (Fig. 3D and fig. S7). The frequency of cell blebbing increases as galactan size decreases (fig. S6). We therefore imaged these cells using TEM. The results showed that periplasm thickness and galactan chain length were correlated (Fig. 3, E and F).
To validate the relationship between galactan chain length and periplasm size, we examined these strains using cryo-TEM. Because chemical fixation, dehydration, or embedding of samples was not required, the cells can be imaged in a near-native state. We visualized WT, WTcomp, Cdiphcomp, Nbrascomp, and D267Ecomp strains using cryo-TEM and measured average periplasm size for each strain (Fig. 4A). The data from this analysis recapitulated the trend seen using conventional TEM (Fig. 4, B and C). Among these strains, galactan chain length correlated directly with periplasm size, with an average contribution of 3.2 Å per Galf residue.
Fig. 4. Native-state imaging of glfT2 engineered M. smeg.
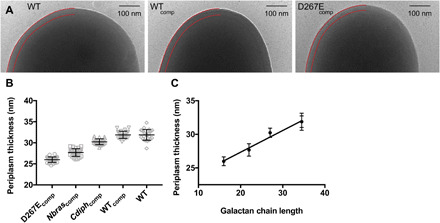
(A) Cryo-TEM micrographs of WT, WTcomp, and D267Ecomp strains. Boundaries of periplasm are highlighted with red lines (left). (B) Quantification of periplasm size. (C) Cryo-TEM data analysis correlates with galactan chain length and periplasm size.
DISCUSSION
Many polysaccharides, including the galactan, are assembled in the cytoplasm and then transported into the extracellular space; therefore, export and chain length control are tightly coupled (25, 26). Our ability to isolate a mycobacterial strain with a truncated galactan indicates that there is flexibility in chain length required for transport. This flexibility would endow mycobacteria the freedom to modulate chain length in response to environmental stimuli without creating a need to modify other cellular machinery. M. smeg strains with truncated arabinan increase the length of their galactan, perhaps as a compensatory mechanism (7).
Our finding that galactan truncation results in hypersensitivity to hydrophobic antibiotics underscores an important role for bacterial polysaccharides. In M. smeg, deletion mutants lacking arabinofuranosyl transferases are similarly sensitized to hydrophobic antibiotics (7). Still, the role of the glycan in these arabinan-deficient strains was difficult to address as these strains lack the ability to generate some glycan linkages altogether and harbor outer membrane defects. Antibiotic hypersensitivity of D267Ecomp cells indicates that changes in glycan chain length alone increase antibiotic permeability. For both Gram-positive and Gram-negative bacteria, cell surface polysaccharides appear to form a barrier against hydrophobic antibiotics (27, 28). This role for glycans is apparent in the combinations of tuberculosis frontline antibiotics that act synergistically. Ethambutol, an inhibitor of arabinan biosynthesis, synergizes with the hydrophobic drug rifampicin, whereas isoniazid, an inhibitor of mycolic acid biosynthesis, does not (29). Together, these data demonstrate the role cell envelope polysaccharides play in determining permeability and illustrate chain length as an important determinant of polysaccharide biological function.
We characterized features of the cell envelope of the D267Ecomp strain and found that galactan chain length is the major alteration. Some minor changes were also observed. An unidentified lipid species appeared to increase, and we surmise that up-regulation of this lipid helps compensate for galactan truncation. In addition, LAM produced by the D267Ecomp strain migrates as a higher molecular weight species during SDS–polyacrylamide gel electrophoresis (PAGE) analysis. This finding is intriguing because LAM appeared different, yet the immediate LAM precursor lipomannan was unchanged. This result implies that between these strains, changes occurred to either LAM arabinosylation or LAM tailoring (30). Both of these processes occur within the periplasm, suggesting that LAM biosynthesis is affected by reduction of periplasm size. Emerging work, particularly the identification of a galactan hydrolase (31), also indicates that a dynamic remodeling of the cell envelope happens within the periplasm. Thus, exploring important biological periplasmic processes, including LAM biosynthesis as well as PG assembly and remodeling, could provide insight into cell envelope assembly.
The importance of the periplasm in Gram-negative bacteria is well established; this compartment has roles in processes that include nutrient acquisition, protein secretion, and cell wall assembly (32). Still, the consensus that mycobacteria have a periplasm has only emerged in the past decade (10, 11) and comparatively little is known regarding its function. Gram-negative and mycobacterial periplasm biology highlights important similarities. In Gram-negative bacteria, the periplasm size is dictated by the protein Lpp such that one end of the protein is covalently linked to the PG, while the other contains a lipid anchor to the outer membrane (33). In contrast, in mycobacteria, a glycan functions as the bridge between the PG and the outer membrane, and our data indicate that the galactan dictates periplasm size. Changing the dimensions of the periplasm has consequences. For example, in S. enterica, increasing or decreasing periplasm size impairs motility and changes cell morphology (33). For mycobacteria, decreasing periplasm size appears to disrupt normal nutrient uptake, as D267Ecomp cells subjected to hyperosmotic challenge strongly up-regulate nutrient acquisition machinery. Last, in Escherichia coli, the Rcs system detects periplasm stress, in part by the protein RcsF (34). In M. smeg, there are two proteins with sequence similarity to RcsF (table S1 and fig. S8). The transcript for one (Msmeg_0699) is among the most highly up-regulated in D267Ecomp cells during normal growth and hyperosmotic challenge. This finding suggests that periplasm size is a cellular attribute that mycobacteria maintain and monitor.
Polysaccharides decorate the surface of all cells. The length of the glycan chains is controlled, but how chain length affects bacterial physiology has been unclear. Before this study, the only known role of chain length had been to influence immunomodulatory properties (1). We found that polysaccharide chain length affects bacterial physiology and numerous cellular attributes, including cell permeability, morphology, mechanical integrity, resistance to environmental stress, periplasm size, and antibiotic susceptibility. Our findings highlight how bacteria can modulate polysaccharide length to respond to environmental stresses. The observation that strains of bacteria can modulate galactan chain length (7) highlights the flexibility endowed by the template-independent synthesis used to create glycans. This flexibility could be an advantage in responding to dynamic changes in the environment. Last, our data suggest that agents that block galactan biosynthesis could enhance susceptibility to known hydrophobic antibiotics.
MATERIALS AND METHODS
Bacterial strains and culture conditions
M. smegmatis mc2155 and derived mutants were grown with shaking at 37°C in Middlebrook 7H9 broth (BD, Franklin Lake, NJ) supplemented with 0.2% (w/v) glucose, 0.2% (v/v) glycerol, 0.5% (w/v) bovine serum albumin (United States Biological, Salem, MA), catalase (4 mg/liter) (Sigma-Aldrich), 15 mM sodium chloride, and 0.05% (v/v) Tween 80. Cells were maintained on agar plates using 7H10 agar (BD, Franklin Lake, NJ) supplemented with 0.2% (v/v) glucose, 0.2% (v/v) glycerol, 0.5% (w/v) bovine serum albumin (United States Biological, Salem, MA), catalase (4 mg/liter) (Sigma Aldrich), and 15 mM sodium chloride. When necessary, antibiotic selections were performed using kanamycin (20 μg/ml), hygromycin B (50 μg/ml), and gentamicin (2.5 μg/ml).
Molecular biology methods and cloning
All cloning was performed using DH5α (NEB) grown in LB supplemented with kanamycin (50 μg/ml) or ampicillin (100 μg/ml) when appropriate. Plasmids and oligonucleotide primers used in this study are described in table S2. For protein expression and purification, M. smegmatis glfT2 was cloned into pET28a. Site-directed mutation constructs were amplified by polymerase chain reaction (PCR), phosphorylated by T4 Polynucleotide Kinase (NEB), ligated by Fermentas T4 DNA Ligase (Thermo Fisher Scientific), digested with Dpn-1 (NEB), and transformed into DH5α. Correct constructs were identified by Sanger sequencing. For expression of constructs within M. smegmatis, glfT2 was cloned into pAMJ7, an integration-proficient vector. pAMJ7 was generated by replacement of the kanamycin resistance cassette in pTTP1B with a gentamicin resistance cassette. The same primers used to develop point variants for pET vector plasmids were used to develop point variants of integration-proficient vectors.
Recombineering was used to replace glfT2 on the M. smegmatis chromosome with a hygromycin B resistance cassette. Briefly, a modified recombineering plasmid (pAMJ23) was created by introduction of glfT2 into pJV48. The resultant plasmid was electroporated into WT M. smeg. The allelic exchange substrate (AES) was generated by creation of a plasmid (pAMJ36) containing a hygromycin resistance cassette flanked by 500 base pairs (bp) on each side containing homology to the 5′ and 3′ ends of glfT2, respectively. The glfT2 deletion fragment was liberated from pAMJ36 by restriction digest with Afe I and Sbf I and purified by agarose gel extraction. M. smegmatis transformed with pAMJ23 were grown to mid-log in 7H9 supplemented with glycerol and Tween 80 only. Once at mid-log, induction of recombineering proteins was initiated by addition of acetamide to a final concentration of 0.2% (w/v). After a 3-hour induction, electrocompetent cells were made from this culture, and these cells were then electroporated with 200 ng of AES. Transformed cells were recovered in complete 7H9 for 4 hours at 37°C and were then plated onto 7H10 plates supplemented with hygromycin (50 μg/ml). Colonies that grew were screened by PCR using primers described in table S2. PCR product that matched the predicted size for a glfT2 deletion strain (pAMJ36 M. smegmatis) was sequenced by Sanger sequencing. Legitimacy of this strain was validated by next-generation sequencing.
Integration-proficient vectors harboring glfT2 mutations were electroporated into pAMJ36 M. smegmatis. The original recombineering plasmid, pAMJ23, was then removed by plating these cells on 7H10 plates supplemented with ADC (albumin-dextrose-catalase supplement) and 10% sucrose (w/v). Because pAMJ23 contains sacB, a negative selectable marker lethal in the presence of sucrose, only those cells that expel pAMJ23 will grow. However, cells lacking a functional copy of glfT2 will lose viability following expulsion of pAMJ23. To determine viability following exposure to sucrose, colony-forming units (CFU) were determined. In some cases, suppressor mutations can silence sacB lethality, preventing efficient removal of pAMJ23 from the cells. In such cases, cells retain kanamycin resistance. Consequently, experiments that found high bacterial viability combined with kanamycin resistance following sucrose exposure were removed from analysis, as they were likely reflective of sacB suppressor mutations, rather than efficient clearance of pAMJ23 from the cell. Expression of Flag-glfT2 constructs was validated using Western blotting with a FLAG-M2-HRP (horseradish peroxidase) conjugate antibody (diluted 1:1000, Sigma-Aldrich).
Protein expression and purification
A homology model for M. smeg GlfT2 was generated using SWISS-MODEL (35). Protein was expressed in DE3 Tuner cells. Starter cultures were used to inoculate 1-liter cultures of LB supplemented with kanamycin to 50 μg/ml. These cultures were grown to mid-log at 37°C, and once reaching OD600 (optical density at 600 nm) ~ 0.6, they were chilled on ice. Protein production was induced by addition of IPTG (isopropyl-β-D-thiogalactopyranoside) to 0.1 mM. Cultures were then incubated overnight at 16°C. Cells were then harvested by spinning at 4000 relative centrifugal force (RCF) for 10 min. Cell pellets were frozen at −80°C until purification.
Cell pellets were thawed on ice and resuspended in buffer A [50 mM sodium phosphate, 400 mM NaCl, 20 mM imidazole, 15% glycerol (pH 7.6)] supplemented with lysozyme (1 mg/ml), 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.1% Triton X-100, and 1250 U benzonase (Millipore Sigma). Cell lysis was completed by mechanical disruption at 22 kpsi (Constant Systems Cell Disruptor). The lysate was clarified by centrifugation at 20,000 RCF, and the supernatant was filtered using a 0.22-μm PES EasyFlow syringe-drive filter (Millipore Sigma). GlfT2 constructs with N-terminal hexahistidine tags were purified using IMAC (immobilized metal affinity chromatography). The filtered supernatant was loaded onto an ÄKTA fast protein liquid chromatography system and applied to a 1-ml HisTrap HP column (GE Healthcare). A linear, 5%/min gradient of buffer B [50 mM sodium phosphate, 400 mM NaCl, 400 mM imidazole, 15% glycerol (pH 7.6)] was used to elute GlfT2 constructs. Fractions containing GlfT2 constructs were identified by SDS-PAGE, pooled, vitrified, and stored at −80°C.
GlfT2 in vitro assays
GlfT2 acceptor elongation experiments were set up similarly to previous reports (12). Kinetics parameters were determined using UDP-Glo (Promega). Enzyme acceptor binding was saturated by addition of acceptor to 200 μM, and donor sugar (UDP-Galf) concentration varied from 15 to 1500 μM with UDP-Galf. Kinetic reactions were initiated by the addition of GlfT1 to 50 nM and quenched with UDP-Glo reagent after 40 min at room temperature. Reaction volumes were 10 μl and were performed in quadruplicate. This time point was chosen to avoid any artifacts owing to quenching the reaction during kinetic lag phase (36).
Cell length determination
Cells were grown in supplemented 7H9 medium and incubated with FM-464 (5 μg/ml) to stain the cell membrane. Cells were placed on a 1.0% agarose pad. Images were collected using a Nikon Eclipse Ti-E epifluorescence microscope using a 100× objective. Length determination was determined using NIS-Elements software. Cell boundaries were determined using thresholding for bright objects, and of the cells selected, only single cells were measured. Statistical significance was determined by unpaired t test (P < 0.0001).
Transmission electron microscopy
Cells were grown to mid-log in supplemented 7H9 medium. Fixation, dehydration, embedding, sectioning, and microscopy were performed similar to as previously described (37). Periplasm size was determined using ImageJ. Distance between the inner and outer membranes was measured in at least 10 locations per cell, and the average of those individual measurements was taken as the periplasm size for that cell. For initial imaging using WT, WTcomp, and D267Ecomp strains, 100 cells were analyzed for each strain. Statistical significance was determined by unpaired t test (P <0.0001). Subsequent imaging using WT, Cdiphcomp, Nbrascomp, and D267Ecomp strains analyzed 30, 30, 25, and 23 cells, respectively.
Cryo-TEM
Cells were cultured to mid-log in complete 7H9 and concentrated 50-fold by pelleting by centrifugation and resuspending in a 50-fold smaller volume. Concentrated cells were plunge-frozen in liquid ethane using an automated Leica EM GP system (Leica Microsystems) using R2/2 200 mesh grids (Quantifoil) and imaged using a Talos 120 kV cryo-transmission electron microscope. For quantification, the distance between the inner and outer membranes was measured an average of 10 individual locations per cell. The mean of those individual measurements was taken as the periplasm size for that cell. Twenty-five cells were analyzed for each strain.
Time-lapse microscopy
Cells were grown to mid-log and diluted to OD600 of 0.1. Cells were loaded into a CellASIC (B04A) plate with constant fully supplemented 7H9 flow in a 37°C chamber. Images were taken every 15 min on an inverted Nikon TI-E microscope using a 60× objective. Cells were imaged using phase contrast. For analyzing frequency of blebbing, cells were loaded into the B04A plate, and images from the initial time point were used to score for blebbing. Undistorted rod-shaped cells were counted as phenotypically normal, and cells with circular protrusions were counted as undergoing blebbing. Scoring was performed in technical quadruplicate.
Minimum inhibitory concentration determination
Antibiotic sensitivity was determined using an alamarBlue assay similar to previous reports (38). Briefly, cells were cultured to late-log phase using supplemented 7H9 medium and diluted to OD600 = 0.05. An antibiotic of interest was then added and diluted serially twofold in a black 96-well plate (Corning). The final volume of cultures was 100 μl. Cells were incubated with shaking for 24 hours at 37°C. Cell viability was determined by adding 6 μl of alamarBlue (Thermo Fisher Scientific) in each well. The plate was then incubated at 37°C for 1 hour, and fluorescence (emission, 590 nm) was measured.
SDS sensitivity
Minimum inhibitory concentration (MIC) for SDS sensitivity was conducted similar to antibiotics. Viability of cells following exposure to SDS was determined by culturing strains to mid-log, supplementing with SDS to 0.05% (w/v), and incubating for 4 hours at room temperature. CFU were then determined for each strain, with and without SDS added. Concentration of SDS was chosen based on previous hypersusceptibility phenotypes.
RNA sequencing
Cells were grown to mid-log in either supplemented 7H9 or supplemented 7H9 with 250 mM NaCl. Cells were harvested by centrifugation at 3000 RCF for 10 min, and cell pellets were resuspended in TRIzol. Cell suspensions were frozen at −80°C. RNA extraction, library preparation, and sequencing were performed as previously described (39). Raw data analysis was performed similar to previous work (40). Statistical significance was determined by unpaired t test.
Mycolyl-arabinogalactan-peptidoglycan isolation
Cell envelope material was extracted similar to previous protocols. Isolation of mycolyl-arabinogalactan-peptidoglycan (mAGP) was conducted similar to previous reports (8, 41). Briefly, cell pellets were thawed on ice and resuspended in 50 ml of phosphate-buffered saline (PBS) supplemented with 2% Triton X-100. Cells were lysed at 22 kpsi with a benchtop cell disruptor from Constant Systems (Kennesaw, GA), and lysate was clarified by centrifugation (20,000 RCF for 1 hour). The supernatant was discarded, and insoluble pellet was extracted three times with 2% SDS in PBS for 1 hour at 95°C. Following the third extraction, mAGP material was washed with milli-Q water, 80% acetone in water, and 100% acetone. Residual acetone was allowed to evaporate overnight.
Cell envelope composition analysis
Galactan length determination
Composition analysis was performed using 300 to 400 μg of sample. A 10-μg aliquot of inositol was added to the samples, which were then dried. Samples were hydrolyzed in 2 M trifluoroacetic acid (TFA) for 2 hours in a sealed tube at 121°C, reduced with NaBD4, and acetylated using acetic anhydride/TFA. Resulting alditol acetates were analyzed on an Agilent 7890A gas chromatograph (GC) interfaced to a 5975C MSD, electron impact ionization mode. Separation was performed on a 30-m Supelco SP-2331 bonded phase fused silica capillary column. Alongside the samples, a standard containing 1 μg of rhamnose, 15 μg of arabinose, 10 μg of inositol, and 10 μg of galactose was run. The reported Ara:Gal:Rha ratios were an average of either three biological replicates (WT and D267Ecomp) or two biological replicates (WTcomp, Cdiphcomp, and Nbrascomp).
Quantification of MurNAc residues
Composition analysis was performed by combined GC/mass spectrometry (GC/MS) of heptafluorobutyric acid (HFBA) derivatives of monosaccharide methyl glycosides produced from acidic methanolysis. Briefly, 700 to 900 μg of mAGP were heated with 6 M HCl for 17 hours at 105°C. After heating, the sample was dried thoroughly by a stream of nitrogen and repeated additions of methanol. Methanolysis was carried out by addition of methanolic HCl for 15 min at 80°C. After cooling and removal of the solvent with nitrogen, the sample was treated with 200 μl of acetonitrile and 50 μl of the HFBA reagent. After reacting for 30 min at 105°C, the sample was dried and dissolved in acetonitrile. GC/MS analysis of the HFBA methyl glycosides was performed on an Agilent 7890A GC interfaced to a 5975C MSD, using a Supelco DB-5 fused silica capillary column (30 m × 0.25 mm). For quantitation, 2-aminoadipic acid was used as an internal standard. To create a comparison of Rha/MurNAc residues, relative abundance of these monosaccharides was calculated as a percentage of total mAGP mass.
Biolog determination of osmosensitivity
Cells were cultured in 7H9 medium to mid-log phase. They were diluted to OD600 = 0.1, diluted approximately 15-fold further into 7H9 broth supplemented with Dye G (Biolog). This mixture was then added to PM09 plates, with each well containing 100 μl of inoculated culture. Three plates were used per strain as technical replicates. Plates were incubated at 30°C for 72 hours. Data were analyzed subtracting background signal measured from uninoculated wells that acted as negative controls.
Lipid isolation and analysis
Cells were cultured (50 ml of complete 7H9) to mid-log and harvested, and cell pellets were frozen at −80°C until further use. Lipid extraction began with resuspension of each cell pellet in 3 ml of 2:1 methanol: chloroform. The initial extraction took place overnight at room temperature with agitation. Delipidated cells were pelleted by centrifugation at 300 RCF, and the supernatant was carefully removed. Delipidation continued with addition of 3 ml of 1:2 methanol:chloroform. Extraction proceeded with agitation at 55°C for 2 hours. Delipidated cells were pelleted by centrifugation at 300 RCF, and the supernatant was carefully removed. This extraction was then repeated a second time, for a total of three extractions. Both the delipidated cells and supernatant were then evaporated overnight under nitrogen. After evaporation, the material from the supernatant fraction was subjected to a Folch wash. It was resuspended in 4:2:1 chloroform:methanol:water, with water being supplemented with 0.9% NaCl (w/v). Following extraction, the two phases were separated and evaporated under nitrogen flow. The aqueous layer was carried forward for LM (lipomannan)/LAM (lipoarabinomannan) analysis, whereas the organic layer contained freely extractable lipids. These lipids were dissolved in 2:1 chloroform:methanol and were analyzed by thin-layer chromatography (TLC) using 20:4:0.5 chloroform:methanol:water. The plate was developed by staining in 10% cupric sulfate and charring. Assignments of lipids were performed using Madacki et al. (42) as a reference.
Delipidated cells from the first series of extractions were then extracted for LM/LAM. Extractions used 3 ml of 50% ethanol at 85°C, and three extractions were performed in total. The first lasted for 3 hours, the second lasted for 2 hours, and the third lasted for 1 hour. Between washes, insoluble material was pelleted by centrifugation at 300 RCF. The 9 ml of 50% ethanol was combined with the aqueous layer of the Folch wash. These combined fractions were then evaporated under nitrogen. The dried material was redissolved in 2 ml of water and extracted three times with 2 ml of PBS-saturated phenol at 80°C. The samples were then dialyzed, separated via SDS-PAGE, and stained with the Pro-Q Emerald stain (Thermo Fisher Scientific). Assignment of bands was determined using Jankute et al. (43) as a reference.
Last, mycolic acids covalently appended to the mAGP were analyzed. Insoluble delipidated cells were resuspended in 1 ml of 15% (w/v) tetrabutylammonium hydroxide. To release free mycolic acids, these suspensions were heated at 98°C overnight. Permethylation was performed by addition of dichloromethane (DCM) (1.5 ml), milli-Q filtered H2O (1.0 ml), and iodomethane (150 μl). This reaction proceeded for 4 hours at room temperature, yielding mycolic acid methyl esters (MAMEs). After two washes with water (2 ml each), the organic phase was evaporated under nitrogen. Diethyl ether (2.0 mL) was added to each sample to extract MAMEs and after evaporation under nitrogen, MAMEs were resuspended in DCM (100 μl) and analyzed via TLC using 95:5 hexanes:ethyl acetate as the mobile phase. Plate was developed by staining in 10% cupric sulfate and charring. Assignments of lipids were performed using Madacki et al. (42) as a reference.
Supplementary Material
Acknowledgments
We thank E. Grevstad, N. Watson, C. Baranowski, and C. Dulberger for technical assistance with microscopy. We thank the Broad Institute MOC for technical assistance with RNA-seq and data analysis. We thank D. Wesener for helpful discussions. MALDI-TOF MS data were collected at UW-Madison MS Facility (NCRR 1S10RR024601-01). Recombineering vectors were provided by G. Hatfull. Funding: This project was supported by the NIAID (R01-AI126592) and the NIH Common Fund (U01 GM125288). Glycosyl composition and linkage analysis were performed at the Complex Carbohydrate Research Center and were supported by the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, U.S. Department of Energy grant (DE-SC0015662) to P.A. A.M.J. thanks T32 GM08349 at UW-Madison for support. H.L.H. was supported by the NIGMS F31 GM 108408 and T32 GM008505. E.U. was supported by the profile area “Antibiotics” from the Faculty of Science, Leiden University. Author contributions: A.M.J. and L.L.K. conceptualized and designed the study. A.M.J., L.M.K., H.L.H., P.W.S., S.P., E.U., and I.B. performed experiments. All authors participated in data analysis. A.M.J. and L.L.K. wrote the manuscript with editing input from L.M.K., H.L.H., P.W.S., A.B., S.P., E.U., I.B., and P.A. Competing interests: The authors declare that they have no competing interests. Data and materials availability: RNA sequencing data are available on the NCBI Sequencing Read Archive (project ID: PRJNA631850). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/38/eaba4015/DC1
REFERENCES AND NOTES
- 1.Greenfield L. K., Whitfield C., Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr. Res. 356, 12–24 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Murray G. L., Attridge S. R., Morona R., Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J. Bacteriol. 188, 2735–2739 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pitarque S., Larrouy-Maumus G., Payré B., Jackson M., Puzo G., Nigou J., The immunomodulatory lipoglycans, lipoarabinomannan and lipomannan, are exposed at the mycobacterial cell surface. Tuberculosis 88, 560–565 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birch H. L., Alderwick L. J., Appelmelk B. J., Maaskant J., Bhatt A., Singh A., Nigou J., Eggeling L., Geurtsen J., Besra G. S., A truncated lipoglycan from mycobacteria with altered immunological properties. Proc. Natl. Acad. Sci. U.S.A. 107, 2634–2639 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan F., Jackson M., Ma Y., McNeil M., Cell wall core galactofuran synthesis is essential for growth of mycobacteria. J. Bacteriol. 183, 3991–3998 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alderwick L. J., Radmacher E., Seidel M., Gande R., Hitchen P. G., Morris H. R., Dell A., Sahm H., Eggeling L., Besra G. S., Deletion of Cg-emb in corynebacterianeae leads to a novel truncated cell wall arabinogalactan, whereas inactivation of Cg-ubiA results in an arabinan-deficient mutant with a cell wall galactan core. J. Biol. Chem. 280, 32362–32371 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Escuyer V. E., Lety M.-A., Torrelles J. B., Khoo K.-H., Tang J.-B., Rithner C. D., Frehel C., McNeil M. R., Brennan P. J., Chatterjee D., The role of the embA and embB gene products in the biosynthesis of the terminal hexaarabinofuranosyl motif of Mycobacterium smegmatis arabinogalactan. J. Biol. Chem. 276, 48854–48862 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Besra G. S., Khoo K.-H., McNeil M. R., Dell A., Morris H. R., Brennan P. J., A new interpretation of the structure of the mycolyl-arabinogalactan complex of Mycobacterium tuberculosis as revealed through characterization of oligoglycosylalditol fragments by fast-atom bombardment mass spectrometry and 1H nuclear magnetic resonance spectroscopy. Biochemistry 34, 4257–4266 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Jankute M., Cox J. A., Harrison J., Besra G. S., Assembly of the mycobacterial cell wall. Annu. Rev. Microbiol. 69, 405–423 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Zuber B., Chami M., Houssin C., Dubochet J., Griffiths G., Daffé M., Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 190, 5672–5680 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann C., Leis A., Niederweis M., Plitzko J. M., Engelhardt H., Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. U.S.A. 105, 3963–3967 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.May J. F., Splain R. A., Brotschi C., Kiessling L. L., A tethering mechanism for length control in a processive carbohydrate polymerization. Proc. Natl. Acad. Sci. U.S.A. 106, 11851–11856 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.May J. F., Levengood M. R., Splain R. A., Brown C. D., Kiessling L. L., A processive carbohydrate polymerase that mediates bifunctional catalysis using a single active site. Biochemistry 51, 1148–1159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheatley R. W., Zheng R. B., Richards M. R., Lowary T. L., Ng K. K., Tetrameric structure of the GlfT2 galactofuranosyltransferase reveals a scaffold for the assembly of mycobacterial Arabinogalactan. J. Biol. Chem. 287, 28132–28143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Kessel J. C., Hatfull G. F., Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Pham T. T., Jacobs-Sera D., Pedulla M. L., Hendrix R. W., Hatfull G. F., Comparative genomic analysis of mycobacteriophage Tweety: Evolutionary insights and construction of compatible site-specific integration vectors for mycobacteria. Microbiology 153, 2711–2723 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peña M. J., Tuomivaara S. T., Urbanowicz B. R., O’Neill M. A., York W. S., Methods for structural characterization of the products of cellulose- and xyloglucan-hydrolyzing enzymes. Methods Enzymol. 510, 121–139 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Auer G. K., Lee T. K., Rajendram M., Cesar S., Miguel A., Huang K. C., Weibel D. B., Mechanical genomics identifies diverse modulators of bacterial cell stiffness. Cell Syst. 2, 402–411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown S., Santa Maria J. P. Jr., Walker S., Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 67, 313–336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feder M. E., Hofmann G. E., Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 61, 243–282 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Kieser K. J., Baranowski C., Chao M. C., Long J. E., Sassetti C. M., Waldor M. K., Sacchettini J. C., Ioerger T. R., Rubin E. J., Peptidoglycan synthesis in Mycobacterium tuberculosis is organized into networks with varying drug susceptibility. Proc. Natl. Acad. Sci. U.S.A. 112, 13087–13092 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhavsar A. P., Beveridge T. J., Brown E. D., Precise deletion of tagD and controlled depletion of its product, glycerol 3-phosphate cytidylyltransferase, leads to irregular morphology and lysis of Bacillus subtilis grown at physiological temperature. J. Bacteriol. 183, 6688–6693 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baranowski C., Welsh M. A., Sham L.-T., Eskandarian H. A., Lim H. C., Kieser K. J., Wagner J. C., McKinney J. D., Fantner G. E., Ioerger T. R., Walker S., Bernhardt T. G., Rubin E. J., Rego E. H., Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. eLife 7, e37516 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wesener D. A., Levengood M. R., Kiessling L. L., Comparing galactan biosynthesis in Mycobacterium tuberculosis and Corynebacterium diphtheriae. J. Biol. Chem. 292, 2944–2955 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagelueken G., Huang H., Clarke B. R., Lebl T., Whitfield C., Naismith J. H., Structure of WbdD: A bifunctional kinase and methyltransferase that regulates the chain length of the O antigen in Escherichia coli O9a. Mol. Microbiol. 86, 730–742 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams D. M., Ovchinnikova O. G., Koizumi A., Mainprize I. L., Kimber M. S., Lowary T. L., Whitfield C., Single polysaccharide assembly protein that integrates polymerization, termination, and chain-length quality control. Proc. Natl. Acad. Sci. U.S.A. 114, E1215–E1223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geisinger E., Isberg R. R., Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLOS Pathog. 11, e1004691 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohler T., Weidenmaier C., Peschel A., Wall teichoic acid protects Staphylococcus aureus against antimicrobial fatty acids from human skin. J. Bacteriol. 191, 4482–4484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffner S. E., Svenson S. B., Källenius G., Synergistic effects of antimycobacterial drug combinations on Mycobacterium avium complex determined radiometrically in liquid medium. Eur. J. Clin. Microbiol. 6, 530–535 (1987). [DOI] [PubMed] [Google Scholar]
- 30.Palčekova Z., Angala S. K., Belardinelli J. M., Eskandarian H. A., Joe M., Brunton R., Rithner C., Jones V., Nigou J., Lowary T. L., Gilleron M., McNeil M. M., Jackson M., Disruption of the SucT acyltransferase in Mycobacterium smegmatis abrogates succinylation of cell envelope polysaccharides. J. Biol. Chem. 294, 10325–10335 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen L., Viljoen A., Villaume S., Joe M., Halloum I., Chêne L., Méry A., Fabre E., Takegawa K., Lowary T. L., Vincent S. P., Kremer L., Guérardel Y., Mariller C., The endogenous galactofuranosidase GlfH1 hydrolyzes mycobacterial arabinogalactan. J. Biol. Chem. 295, 5110–5123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikaido H., Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen E. J., Ferreira J. L., Ladinsky M. S., Beeby M., Hughes K. T., Nanoscale-length control of the flagellar driveshaft requires hitting the tethered outer membrane. Science 356, 197–200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asmar A. T., Ferreira J. L., Cohen E. J., Cho S.-H., Beeby M., Hughes K. T., Collet J.-F., Communication across the bacterial cell envelope depends on the size of the periplasm. PLOS Biol. 15, e2004303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F. T., de Beer T. A. P., Rempfer C., Bordoli L., Lepore R., Schwede T., SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levengood M. R., Splain R. A., Kiessling L. L., Monitoring processivity and length control of a carbohydrate polymerase. J. Am. Chem. Soc. 133, 12758–12766 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian J., Sinskey A. J., Stubbe J., Kinetic studies of polyhydroxybutyrate granule formation in Wautersia eutropha H16 by transmission electron microscopy. J. Bacteriol. 187, 3814–3824 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kincaid V. A., London N., Wangkanont K., Wesener D. A., Marcus S. A., Héroux A., Nedyalkova L., Talaat A. M., Forest K. T., Shoichet B. K., Kiessling L. L., Virtual screening for UDP-galactopyranose mutase ligands identifies a new class of antimycobacterial agents. ACS Chem. Biol. 10, 2209–2218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shishkin A. A., Giannoukos G., Kucukural A., Ciulla D., Busby M., Surka C., Chen J., Bhattacharyya R. P., Rudy R. F., Patel M. M., Novod N., Hung D. T., Gnirke A., Garber M., Guttman M., Livny J., Simultaneous generation of many RNA-seq libraries in a single reaction. Nat. Methods 12, 323–325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandlik A., Livny J., Robins W. P., Ritchie J. M., Mekalanos J. J., Waldor M. K., RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10, 165–174 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daffe M., Brennan P. J., McNeil M., Predominant structural features of the cell wall arabinogalactan of Mycobacterium tuberculosis as revealed through characterization of oligoglycosyl alditol fragments by gas chromatography/mass spectrometry and by 1H and 13C NMR analyses. J. Biol. Chem. 265, 6734–6743 (1990). [PubMed] [Google Scholar]
- 42.Madacki J., Laval F., Grzegorzewicz A., Lemassu A., Záhorszká M., Arand M., Neil M. M., Daffé M., Jackson M., Lanéelle M.-A., Korduláková J., Impact of the epoxide hydrolase EphD on the metabolism of mycolic acids in mycobacteria. J. Biol. Chem. 293, 5172–5184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jankute M., Alderwick L. J., Noack S., Veerapen N., Nigou J., Besra G. S., Disruption of mycobacterial AftB results in complete loss of terminal β(1 → 2) arabinofuranose residues of lipoarabinomannan. ACS Chem. Biol. 12, 183–190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/38/eaba4015/DC1


