Brain atlases reveal neuroanatomical and activity signatures associated with behavioral evolution in cavefish.
Abstract
Environmental perturbation can drive behavioral evolution and associated changes in brain structure and function. The Mexican fish species, Astyanax mexicanus, includes eyed river-dwelling surface populations and multiple independently evolved populations of blind cavefish. We used whole-brain imaging and neuronal mapping of 684 larval fish to generate neuroanatomical atlases of surface fish and three different cave populations. Analyses of brain region volume and neural circuits associated with cavefish behavior identified evolutionary convergence in hindbrain and hypothalamic expansion, and changes in neurotransmitter systems, including increased numbers of catecholamine and hypocretin/orexin neurons. To define evolutionary changes in brain function, we performed whole-brain activity mapping associated with behavior. Hunting behavior evoked activity in sensory processing centers, while sleep-associated activity differed in the rostral zone of the hypothalamus and tegmentum. These atlases represent a comparative brain-wide study of intraspecies variation in vertebrates and provide a resource for studying the neural basis of behavioral evolution.
INTRODUCTION
Brain function and behavior are influenced by evolutionary history and ecological environment (1). Robust differences in gross anatomy, neural connectivity, and gene expression have been associated with behavioral evolution in closely related species (2, 3). Most studies using comparative anatomy have focused on a small number of brain regions, limiting insight into brain-wide changes in brain structure and function. The recent generation of whole-brain atlases and connectomes has provided an increased understanding of how neural circuits function. These resources have largely been limited to a select number of genetically accessible research organisms and have not been applied to a diverse set of models commonly used to study trait evolution. The generation of whole-brain atlases in closely related species, or even independent populations of the same species, has potential to provide insight into the principles governing the evolution of brain structure and neural circuit connectivity associated with behavioral diversity.
The Mexican tetra Astyanax mexicanus comprises eyed surface fish that inhabit rivers throughout Mexico and at least 29 populations that are largely hydrologically separated in limestone caves in the San Luis Potosi region of northeast Mexico (4). Within the past 1 million years, multiple colonizations of caves by eyed surface ancestors have yielded independent cave populations that are geographically and hydrologically isolated from one another (5). While trait evolution in cavefish has been studied for over a century, our understanding of neuroanatomical evolution is limited to anatomical changes in a few brain regions, including reduced size of the optic tectum and hypothalamic expansion in cavefish populations; moreover, differences in brain activity have not been examined (6, 7). Cavefish have evolved numerous behavioral changes, including sleep loss, reduced social behaviors, widespread changes in sensory processing, and alterations in foraging behavior (8). Despite the long-standing focus on characterizing differences in behavior and morphology between cave populations, unexpectedly little is known about the brain anatomy and neural circuits associated with these behavioral changes in cave populations.
Many behaviors differ between surface and cavefish populations, suggesting widespread differences in brain structure and function (8). Sleep and foraging are two homeostatically regulated behaviors that are essential for many aspects of biological function (9). These behaviors interact at the genetic and neural circuit levels, and loss of sleep is associated with metabolism-related disorders (10). Although little is known about the genetic and evolutionary basis underlying the integration of sleep and foraging behaviors, it is hypothesized that the need for sleep is reduced in animals with greater foraging demands (11, 12). We have previously identified the convergent evolution of multiple cavefish populations with regard to sleep loss, with cavefish displaying as much as an 80% reduction in sleep duration compared with surface fish counterparts (13). In addition, hunting behaviors differ markedly between surface fish and cavefish. These differences include changes in the sensory modalities used to identify and capture prey in larval and adult fish, and hyperphagia in multiple adult cavefish populations (14–16). Identifying the neural changes associated with the evolution of sleep and foraging behaviors may provide insight into fundamental principles governing the evolution of neural circuits and brain function.
Recently, image registration has been applied to generate high-resolution reference brains that label neural circuits (17, 18). In the zebrafish, multiple brain atlases have been developed that map neural circuits and putative connectivity between behaviorally relevant neurons (17, 19, 20). These resources have provided unparalleled insight into brain function, but this approach has not yet been applied to study how evolution shapes brain development and function.
Like zebrafish, larval A. mexicanus are transparent and amenable to whole-brain imaging in intact fish (21, 22). In this study, we combined morphometric analysis, imaging of neural circuits, and whole-brain activity mapping to generate standard brains for river-dwelling surface fish and three independently evolved populations of cavefish. Using these reference brains, we quantified volume of defined neuroanatomical regions and specific neuronal populations that contribute to sleep and feeding. We also mapped patterns of neuronal activity to standard brains to generate a brain-wide map of activity differences in waking, feeding, and sleeping fish using phosphorylated extracellular signal–regulated kinase (pERK) labeling (20). Together, these studies are the first comparative brain-wide analyses identifying differences in brain anatomy and function between populations with highly divergent behaviors.
RESULTS
Behavioral and neuroanatomical evolution in larval A. mexicanus
To compare brain anatomy across independently evolved populations of A. mexicanus, we performed whole-brain confocal imaging in surface fish and fish from Tinaja, Molino, and Pachón cavefish populations (Fig. 1A). We first sought to determine whether differences in sleep and foraging were present across all three cavefish populations at 6 days postfertilization (dpf) when the fish are transparent, and their brains are accessible to intact whole-brain imaging. In agreement with previous findings at different developmental stages (13, 23), sleep was reduced across all three cave populations relative to surface fish (Fig. 1B). The reduction in sleep is due to decreases in both total sleep bout number and average bout duration (and fig. S1). We similarly sought to examine whether the increased strike angle associated with prey capture that we previously identified in Pachón cavefish is present in other cavefish populations (14). Strike angle was increased in Pachón and Tinaja, while Molino did not substantially differ relative to surface fish (Fig. 1C). Therefore, multiple independently evolved populations of cavefish have convergently evolved differences in sleep and feeding behaviors that manifest as early as at 6 dpf.
Fig. 1. Behavioral and neuroanatomical evolution in larval A. mexicanus.
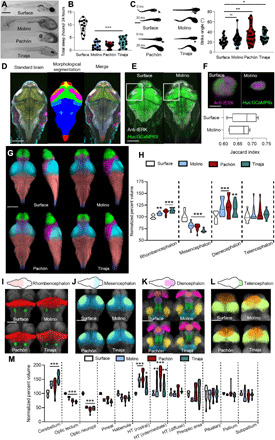
(A) Image of 6–dpf (days postfertilization) fish. Scale bar, 500 μm. (B) Total sleep over 24 hours in 6-dpf A. mexicanus [one-way analysis of variance (ANOVA), F = 51.53, P < 0.0001; Dunnett’s to surface: Molino, P < 0.001; Pachòn, P < 0.001; and Tinaja, P < 0.001). (C) Feeding angle orienting to prey (0 ms) and then immediately after strike (20 ms) (Kruskal-Wallis test = 13.39, P = 0.003; Dunnett’s to surface: Molino, P > 0.56; Pachón, P < 0.01; and Tinaja, P < 0.03). ns, not significant. (D) Standard brain with labels registered to the template brain (left). Segmentations applied to the template brain (middle). Merge of standard brain with anatomical segmentations (right). (E) Registrations of Huc:GCaMP6s reveals overlap of tERK with transgenic label. (F) Projection of anatomical overlap between Huc:GCaMP (green) and tERK (magenta) of optic neuropil. (G) Segmentation of developmental regions of brains using tERK staining: telencephalon (green), diencephalon (magenta), mesencephalon (cyan), rhombencephalon (red), and spine (blue). Scale bar, 300 μm. (H) Developmental regions relative to whole-brain size. Two-way ANOVA: genotype, F = 13.91, P < 0.001; brain region, F = 3134, P < 0.001; interaction, F = 0.65, P = 0.58. Dunnett’s: rhombencephalon, Molino, P < 0.01; Pachòn, P < 0.001; and Tinaja, P < 0.001. Mesencephalon, Molino, P < 0.001; Pachòn, P < 0.001; and Tinaja, P < 0.001. Diencephalon, Molino, P < 0.001; Pachòn, P < 0.001; and Tinaja, P < 0.01. Telencephalon, Molino, P = 0.74; Pachòn, P = 0.68; Tinaja, P > 0.89. (I) Volumetric projections of nuclei within the rhombencephalon: cerebellum (red) and locus coeruleus (green). Scale bar, 200 μm. (J) Nuclei within the mesencephalon. Optic tectum (blue) and optic neuropil (light green). Scale bar, 200 μm. (K) Nuclei in the diencephalon: pineal gland (light red), habenula (pink), preoptic hypothalamus (purple), rostral zone of the hypothalamus (green), intermediate zone of the hypothalamus (blue), diffuse nucleus of the hypothalamus (yellow), and pituitary complex (dark blue). Scale bar, 100 μm. (L) Nuclei within telencephalon: subpallium (orange) and pallium (light green). Scale bar, 100 μm. (M) Quantifications of (F) to (I) normalized to surface fish. *P ≤ 0.05, **P < 0.01, and ***P < 0.005 for indicated comparisons in all statistical tests.
To characterize differences in brain anatomy between A. mexicanus populations, we quantified the size of individual brain regions within each A. mexicanus population. Immunostaining for total ERK (tERK) has been established in zebrafish as a method for labeling broad anatomical regions within the central nervous system (20). At 6 dpf, we immunolabeled larvae for tERK and obtained whole-mount confocal images of the entire brain. tERK was expressed throughout the brain in all A. mexicanus populations tested (fig. S2A), confirming that tERK immunostaining also serves as a broad neuroanatomical marker in A. mexicanus. Total brain volume did not differ between surface fish and fish from the three cave populations (fig. S2B).
To assist in localizing and quantifying the volume of regions and nuclei within the brain, we generated a common reference brain for each population using tERK staining to perform image registration. Each reference brain contained six to seven labels per population, including anti-tERK, anti-pERK, anti-hypocretin/orexin (HCRT), anti–tyrosine hydroxylase (TH), anti–agouti-related protein (AgRP), anti–α-melanocyte–stimulating hormone (α-MSH), and transgenically labeled Huc:GCaMP6s, which resulted in an anatomical atlas containing 18 regions of the brain (Fig. 1D and fig. S3A). To confirm accuracy of image registrations and neural segmentations, we implemented the Jaccard image similarity analysis, which measures the volume of intersection between a registered brain and the template. We tested whole-brain registration quality of numerous tERK image stacks for each population that we tested. This technique yielded a Jaccard index of 0.67 for surface, 0.71 for Molino, 0.69 for Pachón, and 0.70 for Tinaja, indicating that the registration algorithm was accurate and that there were no significant differences in the quality of registration among the populations (fig. S3B and movie S3).
Transgenic technology has recently been applied to A. mexicanus, allowing tissue labeling of specific tissues with genetic precision (21, 22). To broadly label the brain, we generated transgenic A. mexicanus surface and Molino cavefish harboring Huc:GCaMP6s, which has previously been reported to label most neurons in zebrafish (24). We found similarly broad expression in surface fish and Molino cavefish, in which most neurons across the brain are labeled (Fig. 1E). To verify that our registrations and segmentations of small nuclei were accurate, we segmented the optic neuropil of five tERK-labeled fish and five Huc:GCaMP6s for both surface and each cavefish population, which resulted in similar volumes with a Jaccard index of 0.68 for surface fish and 0.66 for cavefish (Fig. 1F). These findings confirm the accuracy of image registration and confirm broad expression of the tERK marker.
To further validate registration accuracy, we measured positioning of TH neurons within the locus coeruleus, which revealed robust alignment of TH+ neurons across populations of A. mexicanus, providing an average three-dimensional positioning error of 5.6 μm in surface, 6.4 μm in Molino, 5.8 μm in Pachón, and 6.3 μm in Tinaja, comparable to published values in the zebrafish brain atlas (fig. S3, C and E) (20). Together, these data demonstrate the ability to perform accurate image registration, allowing for the generation of a standard brain with multiple labels, which guided segmentation of brains into distinct neural regions and nuclei in four different populations of A. mexicanus.
To determine whether identifiable brain regions were different in size between populations, select regions were identified using the standard brains we developed as a reference map, along with a brain atlas made for adult A. mexicanus (25). The volume of each region was then normalized to the volume of the entire brain. Volumetric quantification revealed convergence on changes in the major brain subdivisions that are established during neurodevelopment across all three cave populations relative to surface fish. The rhombencephalon and the diencephalon were expanded, and the mesencephalon was reduced in fish from all three cave populations relative to surface fish (Fig. 1, G and H, fig. S4, and table S1). These observations suggest that broad changes in brain structure are shared across independently evolved cavefish populations.
To determine whether there are changes in the size of brain regions that may be associated with evolved behavioral differences, we quantified 13 additional brain regions, including the tectum, cerebellum, pallium, and four regions of the hypothalamus (Fig. 1, I to M, table S1, and movies S1 and S2), in accordance with previously described nomenclature (26). Consistent with previous reports, the optic tectum and neuropil were reduced, and the total hypothalamus volume was enlarged in all three cavefish populations (6, 25, 27). The increase in hypothalamus volume was because of an enlargement of the rostral and intermediate zones of the hypothalamus, with no differences between surface fish and cavefish populations in the volume of the diffuse nucleus of the hypothalamus (Fig. 1M). Together, these data suggest that cavefish from different populations have repeatedly evolved many of the same neuroanatomical changes in behaviorally relevant brain regions. Several other nuclei were consistently smaller in cavefish, including the pineal gland and habenulae, but did not meet statistical significance, raising the possibility that differences in neural structure are broadly present between surface fish and cavefish (Fig. 1M).
Evolved changes in modulatory neurons associated with sleep and feeding
The circuitry underlying sleep/wake regulation is highly conserved across vertebrate species (28, 29). Studies in zebrafish have identified a central wake-promoting role for the catecholamines dopamine and norepinephrine, and hypothalamic neurons expressing HCRT that consolidate wakefulness (30, 31). We previously reported that functional differences in β-adrenergic and HCRT signaling contribute to sleep loss in Pachón cavefish (32, 33), but the role of these signaling pathways in sleep regulation and the neuroanatomy of catecholamine and HCRT neurons have not been characterized in other populations of cavefish.
We first characterized the neuroanatomy of catecholamine neurons in each of the brains used in generating the standard reference atlas (Fig. 2A). The number of TH+ neurons in the locus coeruleus, a highly conserved wake-promoting region, did not differ between any of the A. mexicanus populations we examined (Fig. 2B). TH+ neurons were more abundant in the telencephalon of both the Pachón and Tinaja populations, but the number of neurons in the telencephalon did not differ between Molino and surface fish (Fig. 2C). Furthermore, the number of TH+ cells in the pretectal area of the brain was significantly reduced in all cavefish populations (Fig. 2D), and hypothalamic TH+ neurons were more abundant in all three populations of cavefish than in surface fish (Fig. 2E). Last, in the medial octavolateralis nucleus, a primary integration site of lateral line afferents, TH+ cell number was significantly greater in Molino and Pachón than in surface fish, but did not differ significantly between Tinaja and surface fish (Fig. 2F and fig. S5). Together, these findings reveal that there are evolved differences in the numbers of TH+ cells in multiple cavefish populations and that differences in the number of neurons expressing TH can be independently regulated among distinct brain regions.
Fig. 2. Whole-brain imaging of circuits associated with sleep and feeding.
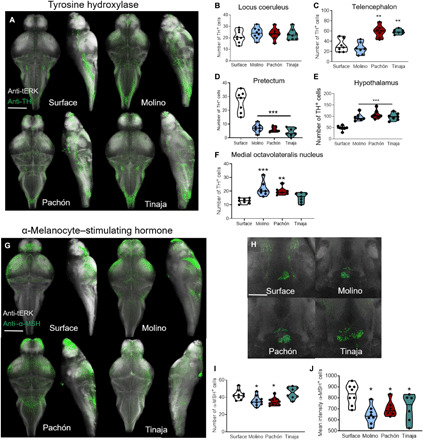
(A) Whole-brain volumetric reconstructions of confocal imaging with anti-tERK (white) and anti-TH (green) for surface, Molino, Pachón, and Tinaja cavefish, with dorsal (left) and sagittal (right) views. Scale bar, 250 μm. (B to F) Numbers of cells expressing TH in distinct regions of the brain (B). TH+ cell quantification in the locus coeruleus (one-way ANOVA, F = 0.509, P > 0.68; Molino, P > 0.64; Pachón, P > 0.54; and Tinaja, P > 0.83). (C) TH+ cell number in the telencephalon (one-way ANOVA, F = 18.87; P < 0.001; Molino, P > 0.66; Pachón, P < 0.001; and Tinaja, P < 0.001). (D) Quantification of pretectal TH+ neuron cells (one-way ANOVA, F = 35.19, P < 0.001; Molino, P < 0.001; Pachón, P < 0.001; and Tinaja, P < 0.001). (E) Number of TH+ cells in the hypothalamus (one-way ANOVA, F = 20.16, P < 0.001; Molino, P < 0.001; Pachón, P < 0.001; and Tinaja, P < 0.001). (F) TH+ cell number in the medial octavolateralis nucleus (one-way ANOVA, F = 9.532, P < 0.001; Molino, P < 0.001; Pachón, P < 0.002; and Tinaja, P > 0.44). (G) Whole-brain volumetric reconstructions of confocal imaging with anti-tERK (white) and anti–α-MSH (green) for four populations of A. mexicanus. Scale bar, 300 μm. (H) Single-plane view of the α-MSH (green) cell cluster in the pituitary complex in surface fish and Molino, Pachón, and Tinaja cavefish. Scale bar, 50 μm. (I) Total number of cells expressing α-MSH in the pituitary complex. (J) Mean fluorescence intensity of α-MSH+ individual cells from (I). All comparisons were carried with n > 8, and all post hoc tests compared cavefish to surface fish. N > 8 for all measurements. *P ≤ 0.05, **P < 0.01, and ***P < 0.005 for indicated comparisons in all statistical tests.
In zebrafish, hcrt-expressing neurons localize to the rostral zone and preoptic area of the hypothalamus, and these neurons localized to similar regions across all A. mexicanus populations (34, 35).
In all three cave populations, HCRT neurons were more abundant in both of these brain regions (fig. S6, A to C), and the HCRT signal per cell was significantly elevated (fig. S6D). A descending pathway along the midline that connects the midbrain to the spine stained strongly for HCRT in all cavefish populations, but not in surface fish (fig. S6E). In surface fish, and all three cavefish populations, HCRT-immunoreactive fibers localized to the locus coeruleus, as well as the lateral and intermediate zones of the hypothalamus, with ascending projections into the telencephalon (fig. S6E).
To determine whether cavefish evolved differences in neuropeptides that regulate feeding behavior, we examined the neuroanatomy of several conserved neuropeptides that regulate appetite. Genetic variants in the melanocortin receptor MC4R have been implicated in the regulation of feeding in diverse species, including A. mexicanus (15). The neuropeptide α-MSH antagonizes MC4R to inhibit feeding (36), and we identified an antibody that selectively labels α-MSH neurons based on its known expression pattern in zebrafish (37). Immunostaining for α-MSH in surface fish predominantly labels neurons in the pituitary complex with projections that ramify throughout the hypothalamus (Fig. 2G). The number of α-MSH+ neurons was significantly reduced in both Molino and Pachón cavefish but did not differ between Tinaja and surface fish (Fig. 2H). We identified differences in signal from α-MSH projections in a number of brain regions, including higher immunoreactivity in the cerebellum of surface fish than in all cavefish populations (fig. S7). In addition, the intensity of the α-MSH signal from ascending projections that run laterally along the rostral zone of the hypothalamus into the forebrain was reduced in Pachón and Molino cavefish relative to surface fish (fig. S7), and the intensity of labeling within the tectum was reduced in all three cavefish populations. Together, the number of α-MSH neurons is reduced in the multiple cavefish populations, but not Tinaja, revealing differences in the evolution of feeding circuits among cavefish populations.
The neuropeptide AgRP opposes α-MSH signaling and functions as an inverse agonist of MC4R (36). We found that AGRP localizes to the hypothalamus in surface fish and in all three populations of cavefish (fig. S8A), consistent with its expression in zebrafish (38), and AgRP+ cells were more abundant in all three populations of cavefish than in surface fish (fig. S8B); in addition, the fluorescence intensity per cell was significantly higher in all populations of cavefish compared with surface (fig. S8C), indicating an increased number of AGRP+ cells and an increase in neuropeptide synthesis in cave populations. The projections of AgRP+ neurons shared many similarities with those of α-MSH neurons: AgRP+ fibers ran laterally in the medial hypothalamus cell bundle, with ascending fibers in the telencephalon forebrain bundle apparent in all populations (fig. S8D). We identified several significant differences in projections between populations, including tracts that connected the diffuse nucleus of the hypothalamus to the hindbrain in all three cavefish populations but were absent in surface fish (fig. S8D). Together, these findings are consistent with the adaptation of cavefish to a limited food environment and suggest changes in feeding circuitry that may underlie differences in prey-seeking behavior.
Brain atlas reveals altered landscape of neural activity
The robust differences in behavior and neuroanatomy raise the possibility that brain activity differs between A. mexicanus populations. In zebrafish and mice, pERK accurately reflects neuronal activity with temporal resolution on the order of minutes, with strong induction within 2 min and total abrogation by 30 min (20, 39, 40). To establish baseline differences in neural activity among A. mexicanus populations, we performed whole-mount immunostaining for pERK and tERK. A. mexicanus do not require food in their first week of life, and we collected nonfed 6-dpf fish between zeitgeber time 4 (ZT4) 4 and ZT6 (Fig. 3A). To localize pERK signal to distinct regions of the brain, we performed registrations to the standard brains we developed, which allowed us to extract 18 anatomical regions across the entire brain and measure changes in neural activity across these regions.
Fig. 3. Whole-brain pERK neural activity imaging reveals altered landscape of brain activity.

(A) Average pERK activity maps overlaid onto standard brains with segmentations in major brain subdivisions in the indicated population of A. mexicanus. Scale bar, 200 μm. (B) Scree plot for principal components analysis (PCA). Together, principal component 1 (PC1) and PC2 explain 44.78% of the variation in the PCA; further components do not meet the Kaiser criterion for further analysis. (C) Loading plot for PCA analysis showing the correlations between regions and the amount each region contributes to the PCs. Vectors that form small angles show correlated neural activity. (D) PCA of whole-brain neural activity in the brain of free-swimming fish. PC1 (one-way ANOVA, F = 8.019, P < 0.001; Dunnett’s post hoc: Molino, P < 0.003; Pachón, P < 0.001; and Tinaja, P > 0.95). PC2 (one-way ANOVA, F = 8.786, P = 0.0001; Dunnett’s post hoc: Molino, P < 0.001; Pachón, P < 0.05; and Tinaja, P = 0.001). Percentages indicate the amount of variance in neural activity explained by each PC. (E) Maximum-intensity projections of mean pERK signal in the rostral zone of the hypothalamus. Scale bar, 100 μm. (F) Quantification of pERK signal in rostral zone of the hypothalamus (one-way ANOVA, F = 4.69, P < 0.01; Molino, P < 0.01; Pachón, P < 0.04; and Tinaja, P ≥ 0.95). (G) Maximum-intensity projections of pERK signal in habenula. Scale bar, 100 um. (H) Quantification of pERK activity in the habenula (one-way ANOVA, F = 4.16, P = 0.012; Molino, P = 0.99; Pachón, P = 0.018; and Tinaja, P < 0.02). (I) Maximum-intensity projections of pERK signal in the pallium. Scale bar, 100 μm. (J) Quantification of pallial neural activity (one-way ANOVA, F = 6.18, P = 0.001; Molino, P < 0.001; Pachón, P < 0.03; and Tinaja, P < 0.04). N > 10 for all pERK activity mapping. *P ≤ 0.05, **P < 0.01, and ***P < 0.005 for indicated comparisons in all statistical tests.
We applied unbiased statistical methodology to identify differences in neural activity across the entire brain using principal components analysis (PCA) on pERK/tERK imaging data across multiple individuals of four populations of A. mexicanus. PCA effectivity reduces dimensionality of data and allowed us to capture the essence of variation across the 18 regions of the brain we segmented within a few components, similar to methods used in reducing data dimensionality in numerous species for neural imaging. This dimensionality reduction can be measured with scree plot analysis, which quantifies the amount of variation captured from each principal component (PC). Eigenvalues measure the covariance of the data and allow for ranking of each PC from highest to lowest. Using these analyses, we found that the first two PCs contained 44.78% total variance of the data, with small contributions in each additional PC, allowing us to focus the rest of our analysis within these first two PCs (Fig. 3B). PCA loading plot analysis revealed the contribution and correlation of brain activity in each brain region on each PC. Rhombencephalon, diencephalon, and hypothalamic regions were highly correlated and strongest along PC1, while pallium, subpallium, habenula, and optic tectum were strongest in PC2 (Fig. 3C). The PCA revealed distinctive clustering patterns of neural activity between individual A. mexicanus populations. For example, Molino cavefish formed a cluster that was distinct from surface fish in both the first and second PCs (PC1 and PC2), whereas Pachón formed a unique cluster shifted to the right from surface fish in PC1 (Fig. 3B). Tinaja cavefish formed a cluster below surface fish in PC2. PCA variable analysis revealed the candidate regions most strongly associated with altered neural activity in each PC, including the rostral zone of the hypothalamus in PC1 (fig. S9, A and B) and pallium and habenula in PC2 (fig. S9C). Collectively, the PCA of pERK/tERK imaging from A. mexicanus revealed distinct evolved patterns of neural activity within PC space, suggesting that cave-adapted fish have unique neural activity profiles compared with evolutionarily older surface fish.
To further characterize differences in neural activity among populations, we directly compared the levels of pERK activity in specific regions identified by PCA. As the rostral zone of the hypothalamus is thought to be homologous to the lateral hypothalamus in mammals, which serves as a critical regulator of both sleep and feeding behavior (41, 42), we speculated that activity within this region might differ between cavefish populations. We observed a significant increase in pERK activity in the rostral zone of the hypothalamus in Molino and Pachón cavefish populations relative to surface fish, but no differences between surface fish and Tinaja (Fig. 3, C and D). Furthermore, neural activity in the habenula, a region involved in stress response (43, 44), was significantly reduced relative to surface fish in Pachón and Tinaja, but unaltered in Molino (Fig. 3, E and F). Last, activity within the pallium, an area analogous to the mammalian amygdala and hippocampus that has been associated with emotion, motivation, and recently sleep regulation in zebrafish (45, 46), was significantly reduced in all populations of cavefish relative to surface fish (Fig. 3, G and H). We also quantified activity in 12 additional brain regions (fig. S10 and table S3). Together, this analysis reveals brain region–specific changes in neural activity associated with behaviors that have diverged between surface fish and cavefish.
Neural activity associated with hunting behavior
To determine how brain activity differs during a multimodal sensory behavior, we quantified the effects of prey capture on neural activity. We compared brain-wide pERK levels in fish-fed brine shrimp (Artemia) for 10 min with fish that had not been fed and were freely moving before euthanasia (Fig. 4A). We applied PCA to whole-brain activity patterns to determine whether hunting would create unique activity signatures in each population. Pachón and Tinaja cavefish formed distinct clusters in PC1 relative to surface fish, suggesting that they have evolved distinct neural activity patterns associated with hunting. By contrast, the evolutionarily younger Molino cavefish did not significantly differ from surface fish in either PC1 or PC2 (Fig. 4B). Loading plots from the PCA analysis revealed that brain regions clustered tightly in either PC1, which was made up of areas with differing responses across populations. Alternatively, PC2 contained nuclei that exhibited similar neural activity among both surface and cave populations (fig. S12, A and B). The diffuse nucleus of the hypothalamus was identified as the most significant variable in PC2, suggesting that the hypothalamus integrates multimodal sensory inputs, which are activated by hunting in all populations (fig. S12C).
Fig. 4. pERK neural activity during feeding reveals sensory transformation in cavefish and convergence on hypothalamic circuitry.
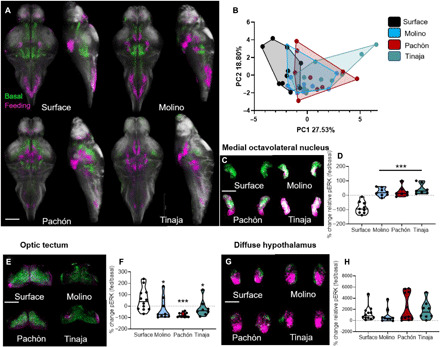
(A) Average whole-brain pERK activity patterns registered to standard brains (white) in nonfeeding fish (green) and fish undergoing a 10-min feeding assay (magenta). Scale bar, 200 μm. (B) PCA of whole-brain activity (reflected by pERK signal) in fish undergoing 10-min feeding assay PC1 explained 27.53% of the variability of the brain activity (one-way ANOVA, F = 6.652, P = 0.001; Molino, P > 0.40; Pachón, P < 0.03; and Tinaja, P < 0.001). There were no statistical differences between populations along PC2, which explained 18.80% of the variability. (C) Maximum-intensity projection of pERK neural activity in medial octavolateralis nucleus (MON) activity for nonfed (green) and fed (magenta) fish. Scale bar, 50 μm. (D) Quantitation of the change in pERK activity in the MON during feeding (one-way ANOVA, F = 22.14, P < 0.001; Molino, Pachón, and Tinaja, P < 0.001). (E) Maximum-intensity projection of pERK activity in the optic tectum of nonfed (green) and feeding (magenta) fish. Scale bar, 200 μm. (F) Quantification of change in pERK activity in the optic tectum during feeding (one-way ANOVA, F = 6.13, P = 0.002; Molino, P > 0.02; Pachón, P < 0.001; and Tinaja, P > 0.04). (G) Maximum-intensity projection of pERK activity in the diffuse nucleus of the hypothalamus of nonfed (green) and feeding (magenta) fish. Scale bar, 100 μm. (H) Quantification of change in pERK activity in the MON during feeding (one-way ANOVA, F = 1.43, P = 0.24; Molino, P > 0.91; Pachón, P > 0.33; and Tinaja, P > 0.8). N > 10 for all feeding pERK neural activity.
All cavefish populations exhibited a significant increase in medial octavolateralis nucleus activity following hunting behavior, whereas surface fish exhibited a reduction, suggesting opposing polarities of hunting-induced medial octavolateralis nucleus activity between surface fish and all three cavefish populations (Fig. 4, C and D). Surface fish, like zebrafish, use visual cues to orient relative to prey. pERK levels (i.e., neural activity) in the optic tectum were significantly higher in surface fish than in all three cavefish, suggesting that the tectum is not an input for hunting-associated behavior in cave populations (Fig. 4, E and F). Last, feeding on brine shrimp induced a robust increase in neural activity in the diffuse nucleus of the hypothalamus across all populations (Fig. 4, G and H), suggesting that this nucleus integrates multiple sensory modalities during hunting. Together, these findings highlight the evolution of brain-wide changes during hunting behavior across multiple cave-adapted populations of A. mexicanus.
Evolution of sleep-associated neural activity
Mapping brain activity during sleep in cavefish is difficult because individuals from these populations sleep for limited periods. However, the small size and relatively permeable blood-brain barrier of A. mexicanus allow for measuring the effects of drugs on sleep regulation, similar to approaches for imaging neural correlates of sleep previously used in zebrafish (33, 45, 47). To compare brain activity in sleeping surface fish and cavefish, we pharmacologically induced sleep in all populations of cavefish. Previously, we showed that moderate concentrations of the β-adrenergic antagonist propranolol and HCRT receptor inhibitor N-ethyl-2-[(6-methoxy-pyridin-3-yl)-(toluene-2-sulphonyl)-amino]-N-pyridin-3-ylmethyl-acetamide (EMPA) restore sleep to Pachón cavefish without affecting sleep in surface fish, suggesting enhanced sensitivity to inhibitors of β-adrenergic and HCRT signaling (32, 33). Both target receptors have been implicated in human sleep, suggesting the effects of these drugs may be conserved from fish to mammals (48, 49).
Treatment with β-adrenergic antagonist propranolol and the HCRT receptor inhibitor EMPA restored sleep in all three cavefish populations, suggesting that conserved signaling pathways contribute to sleep loss in independently evolved cavefish populations (Fig. 5A). Both drugs increased sleep bout length and bout number without affecting waking activity, suggesting that the elevation of sleep is not due to lethargy (fig. S13). To determine whether the two drugs induce similar or distinct changes in neural activity, we compared neural activity between awake dimethyl sulfoxide (DMSO)–treated fish and pharmacologically induced sleeping fish treated with EMPA or propranolol (Fig. 5B). Both EMPA- and propranolol-treated sleeping surface fish and cavefish exhibited an overall reduction in neural activity compared with awake fish, similar to what was recently reported for drug-treated sleeping zebrafish and naturally occurring nighttime sleep (45, 50). In all populations, the PCA results significantly differed between asleep/drug-treated and awake/DMSO-treated fish, suggesting that pERK is a robust marker for detecting neural activity differences associated with treatment groups (fig. S14A).
Fig. 5. Whole-brain activity imaging of sleep-like state reveals heterogeneous neural signatures.
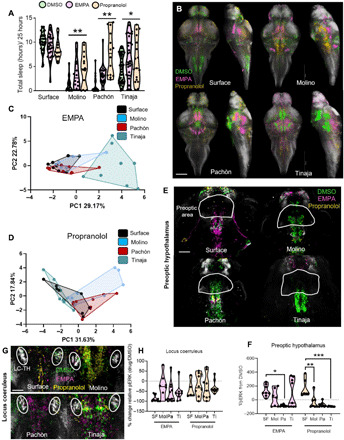
(A) A 24-hour sleep quantification with control dimethyl sulfoxide (DMSO; green), EMPA (magenta), and propranolol (yellow) treatments (two-way ANOVA, genotype, F = 13.18, P < 0.01; drug treatment, F = 34.50, P < 0.001; genotype-phenotype, F = 2.13, P = 0.234; Dunn’s analysis: surface EMPA, P = 0.894; surface propranolol, P < 0.42; Molino EMPA, P < 0.02; Molino propranolol, P < 0.01; Pachòn EMPA, P = 0.011; Pachòn propranolol, P < 0.001; Tinaja EMPA, P < 0.001; and Tinaja propranolol, P = 0.034). (B) Whole-brain activity in DMSO (green), EMPA (magenta), or propranolol (yellow). Scale bar, 200 μm. (C) PCA of neural activity in fish treated with EMPA. PC1 explains 29.17% of the PCA variance (one-way ANOVA, F = 16.75, P < 0.001; Molino, P > 0.56; Pachón, P > 0.96; and Tinaja, P < 0.001). PC2 explains 22.78% of the variance (one-way ANOVA, F = 0.849, P > 0.47; Molino, P > 0.84; Pachòn, P > 0.74; and Tinaja, P > 0.90). (D) PCA of neural activity in sleeping fish treated with propranolol PC1 explains 31.94% of the variation (one-way ANOVA, F = 7.475, P < 0.001; Molino, P < 0.02; Pachón, P < 0.05; and Tinaja, P > 0.61). PC2 explains 18.60% of the neural activity variation (one-way ANOVA, F = 2.03, P = 0.131; Molino, P = 0.435; Pachòn, P = 0.579; and Tinaja, P = 0.921). (E) Average neural activity in preoptic area of the hypothalamus in awake DMSO (green) and sleeping EMPA (magenta) and sleeping propranolol (yellow). Scale bar, 100 μm. (F) Quantification of the change in pERK neural activity in the preoptic area of the hypothalamus in sleeping versus waking fish (two-way ANOVA, F = 6.959, P < 0.001; for EMPA treatment: Molino, P > 0.34; Pachón, P < 0.001; and Tinaja, P > 0.95; propranolol treatment: Molino, P < 0.02; Pachón, P < 0.001; and Tinaja, P < 0.001). (G) Hindbrain area containing TH+ locus coeruleus neurons (white circles) with average neural activity of awake DMSO (green) and sleeping EMPA-treated (magenta) or EMPA-propranolol–treated fish (yellow). Scale bar, 50 μm (H) Quantification of the change in pERK signal (two-way ANOVA, drug treatment, F = 1.71, P = 0.124; EMPA: Molino, P = 0.1; Pachòn, P = 0.743; and Tinaja, P = 0.727; for propranolol treatment: Molino, P > 0.97; Pachón, P > 0.99; and Tinaja, P > 0.99). *P ≤ 0.05, **P < 0.01, and ***P < 0.005 for indicated comparisons in all statistical tests.
Next, we sought to determine whether drug-treated fish in a sleep-like state converged upon shared or independent patterns of neural activity in each population. PCA analysis of EMPA-treated sleeping fish revealed that surface and Molino cavefish clustered tightly together, whereas Pachón cavefish formed a separate cluster in PC2 and Tinaja formed a separate cluster to the right in PC1 (Fig. 5C). Variable analysis derived from PCA revealed that the main regions driving the changes along PC1 in Tinaja were in the telencephalon, including the pallium and subpallium, while the most significant variables for Pachón in PC2 were in the diencephalon, including several known sleep centers of the brain, such as the rostral zone and preoptic area of the hypothalamus (fig. S14, B to D). Propranolol treatment also resulted in unique neural activity profiles across populations of A. mexicanus, with Molino and Pachón forming clusters of sleep-associated activity distinct from both surface fish and Tinaja cavefish in PC1 (Fig. 5D). PCA variable analysis revealed several highly associated brain regions for both propranolol and EMPA sleep conditions. These included regions that have been implicated in zebrafish or mammalian sleep regulation, including the rostral zone and preoptic areas of the hypothalamus, and the locus coeruleus, indicating that shared regions of neural activity may be associated with sleep in A. mexicanus (fig. S14, B to G).
We next quantified the changes in pERK activity in sleeping/drug-treated fish relative to waking DMSO-treated fish in different brain regions. In mammals, preoptic regions of the hypothalamus promote sleep (51). Activity in the preoptic hypothalamus was robustly elevated in sleeping surface fish but was reduced or unchanged in all sleeping cavefish populations, revealing the presence of differentially evolved sleep signatures between surface and cave forms (Fig. 5, E and F). In mammals and zebrafish, the locus coeruleus promotes wakefulness and receives inputs from wake-promoting HCRT neurons (30, 52). We observed a significant reduction in neural activity in locus coeruleus TH+ neurons in surface and all cave populations treated with either drug (Fig. 5, G and H). In sleeping fish, pERK activity was robustly elevated in a large area of the tegmentum, a sleep-promoting area in both mammals and zebrafish (43, 53). In all populations, treatment with EMPA and propranolol increased tegmentum activity during sleep relative to DMSO-treated fish (fig. S15). In addition, activity was reduced during sleep across surface and all three cavefish populations in the rostral zone of the hypothalamus, a region containing HCRT neurons (fig. S15). Activity during sleep in numerous other regions, including the pallium, subpallium, intermediate zone of the hypothalamus, and cerebellum, differed among cavefish populations (fig. S15). Together, these results demonstrate unique activity signatures associated with sleep-like states across different A. mexicanus populations.
DISCUSSION
In this study, we used whole-brain imaging in fixed samples of independently evolved populations of A. mexicanus, an evolutionary model in which the cave and surface forms exhibit significant differences in complex behaviors, including sleep and hunting. Our systematic approach has revealed evolved alterations in neuronal organization at several levels, including morphology, circuitry, and neural activity. This work provides a basis for investigating the mechanisms by which evolution has altered brain morphology, and how these morphological changes are related to changes in behavior. In addition, our atlas will facilitate an unbiased examination of the relationship between the function and anatomy of different brain regions and their relationship to the ecologies of each of the four populations studied.
Hunting behavior induced broad changes in brain activity across all four A. mexicanus populations, and likely activates brain regions associated with sensory processing, satiety, and motivation. While we identified differences in strike angle in two of the three populations of cavefish studied, it is possible that differences in brain activity or hunting-associated neurons are related to other aspects of feeding, such as overall consumption, or vibration attraction behavior that emerge later in development. Alterations in feeding-associated circuitry identified here occurring early in development suggest that evolutionary changes are unique among different cavefish populations, and future studies will directly address the interaction of evolutionarily altered circuits with behavioral differences. The evolution of sensory systems is particularly prominent in cavefish, including intrapopulation differences in vision, mechanosensation, taste, and smell, in the regulation of behavior (8). The identification of differences in anatomy and activity of numerous brain regions associated with the processing of sensory information, including the optic tectum and the medial octavolateralis nucleus, which receives information from the lateral line. Both of these regions were differentially active between surface and cavefish during feeding, suggesting that the two forms rely in different sensory modalities or that these modalities are differentially processed.
In zebrafish, hypnotic pharmacological agents have been shown to induce heterogeneous effects both behaviorally, to sleep architecture, and neuronally, to cellular activity patterns during sleep (45, 47, 50). We examined the effects of several sleep-promoting drugs on brain activity in multiple A. mexicanus populations. Pharmacological manipulations can act on numerous populations of neurons and have secondary effects. For example, propranolol, a drug that potently induces sleep in cavefish, is used to treat both anxiety and heart disorders (33, 54, 55). It is therefore possible that the neural signatures shown here are representing a wide range of physiological responses in regions such as the habenulae and pallium, which underlie stress regulation or other responses in addition to their measurable effect on sleep (table S5). Future experiments harnessing genetic technology for in vivo neural recording with drug treatments will further reveal how neural activity patterns have evolved across populations of A. mexicanus. To date, sleep in A. mexicanus larvae and adults has been defined largely based on canonical methodology from zebrafish, which uses behavioral criteria such as quiescence and arousal threshold to define sleep (13, 23). Recently, neural correlates of non–REM (rapid eye movement) and REM sleep have been found in zebrafish using fluorescence-based polysomnography (45). These studies localized synchronous activity associated with sleep to the dorsal pallium, raising the possibility that this region is analogous to the mammalian cortex (45). Although the pERK imaging method used in this study does not have the temporal resolution to detect these events, we identified differences in neural activity within the dorsal pallium between A. mexicanus populations. The application of in vivo imaging, using the GCaMP6s transgenic lines described here, will allow for greater temporal resolution of the differences in neural activity identified in this study, providing an opportunity to define sleep based on neural synchrony, similarly to methods commonly used in mammals.
Whole-brain morphometric brain atlases enable localization of neuroanatomical regions associated with different behaviors (17, 19, 20). To date, these atlases have generated a number of species, allowing brains from different individuals or signals from whole-brain Ca2+ imaging to be mapped onto a single standard brain (17, 19, 56). The generation of these brain atlases in A. mexicanus represents the first use of whole-brain morphometrics to compare brain anatomy between different populations. This approach could be applied in other research organisms, including zebrafish and fruit flies, to identify differences in neuroanatomy between independent strains. The generation of brain atlases for individual populations may provide insights into the neural mechanisms underlying these behavioral differences.
The development of a functional brain atlas in A. mexicanus will facilitate future efforts to better understand how evolution of the brain has led to behavioral divergence. A. mexicanus are a powerful system for the study of behavioral evolution, allowing for comparisons between surface and cavefish, as well as between independently evolved cavefish populations. While our understanding of the divergence time between populations remains incomplete, a recent analysis using whole-genome mapping suggests ~167 to 200,000 years (57). Using individuals of the same species with a span of behavioral alterations will allow for direct interrogation of genotype-phenotype interactions; previously, these interactions have been difficult to parse by comparative approaches because of the relatively large phylogenetic divergences in other systems and the lack of functional tools. Isolated A. mexicanus populations represent diverse members of the same species, which is genetically amenable to transgenesis and mutagenesis techniques including the Tol2 transposase system and CRISPR-Cas9 engineering (22, 58, 59). The brain atlas could be used as an anatomical marker to align whole-brain GCaMP imaging at a cellular resolution. The development of a functional brain atlas in A. mexicanus will facilitate future efforts to better understand how evolution of the brain has led to behavioral divergence. Here, our analyses in A. mexicanus are reliant on manual segmentation of brain regions, and our quantification consisted of 18 brain regions. In zebrafish, automated analysis, aided in part by increased resolution afforded by transgenic lines, has allowed for segmentation into hundreds of different brain structures (19, 20, 26). The application of this technology, in combination with the use of genetically expressed anatomical marker, such as pan-neuronally expressed GCaMP, has potential to compare the evolution of over 200 brain regions between populations.
Together these studies identify large-scale differences between surface fish and cavefish populations of A. mexicanus, as well as between different populations of cavefish. This represents the first whole-brain anatomical brain atlas comparing intraspecies differences in brain structure and function. This resource has potential to provide information about the fundamental principles guiding the relationship between the evolution of brain function and behavior, as well as the contributions of naturally occurring variation in brain function that underlies behavioral differences between individuals.
MATERIALS AND METHODS
Fish care
Animal husbandry was carried out, as previously described (22), and all protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of Florida Atlantic University. Adult breeding fish were housed in the university core fish facilities at a water temperature of 21° ± 1°C. Lights were maintained on a 14:10 light-dark cycle throughout all experiments. Daylight intensity was between 25 and 40 lux for both rearing and behavioral experiments. After nighttime breeding, larval fish were raised in an incubator at 23°C until 6 dpf to ensure consistent development. Fish were not fed until 6 dpf, and unless noted, fish were not in the fed state when euthanized for imaging.
Sleep behavior
Sleep behavior was assayed, as previously described in (32). Briefly, 6-dpf fish were individually housed in 24-well tissue culture plates (catalog no. 662-102, CellStar) and acclimated for 18 to 24 hours before the beginning of the experiment at ZT0. Fish were recorded at 15 frames per second (fps) using a universal serial bus (USB) webcam equipped with a zoom lens and an infrared-pass filter. Videos were saved as .avi files using the VirtualDub software and then processed using the EthoVision XT (v12) behavioral profiling software. Raw locomotor data were exported as Unicode text and then processed by custom-written code to calculate sleep parameters.
Quantification of prey capture
At 6 dpf, larval fish were individually placed into circular wells with a diameter of 16 mm and a depth of 3 mm. After an acclimation period of 2 min, approximately 30 brine shrimp (Artemia) of the first instar stage were added to the well, and prey capture behavior was recorded from above at 100 fps for a period of 2 min. Recordings were acquired using a USB 3.0 camera (Grasshopper3, FLIR Systems) fitted with a zoom lens (75-mm DG Series Fixed Focal Length Lens, Edmund Optics Worldwide) and recorded with FlyCapture2 software (v2.11.3.163, FLIR Systems). To quantify prey capture dynamics, the angle of prey capture (strike angle) was measured for all successful feeding events in the 2-min recording interval using the native “Angle” tool in ImageJ [National Institutes of Health (NIH), v.1.51]. All measurements were made in the frame before initiation of movement toward the prey. Strike angle was defined as the angle between the line segment extending down the fish’s midline and terminating parallel with the pectoral fins, and the line segment extending from this point to the center of the prey. Measurements of each strike were averaged to calculate the mean strike angle for that individual, and any recording with fewer than three feeding events was excluded from analysis.
Quantification of brain activity during feeding behavior
Fish at 6 dpf were individually placed in 24-well plates (catalog no. 662-102, CellStar). After the fish were left undisturbed for 1 hour, approximately 30 brine shrimp were added to each well, and the fish were allowed to feed for 10 min. Fish were then immediately fixed in a 4% paraformaldehyde (PFA) solution and immunostained. Before immunostaining, feeding was visually confirmed on the basis of the presence of brine shrimp in the gut of the fish.
Pharmacology
All drug treatments were approved by the Florida Atlantic University IACUC committee (protocols A15-32 and A16-04). For behavioral recording experiments, all fish were placed into individual wells of a 24-well plate and allowed to acclimate overnight before the beginning of the experiment. At ZT0, fish were treated with either solvent control, 0.1% DMSO, or freshly prepared propranolol (Sigma-Aldrich) or EMPA (Tocris Biosciences). Both drugs were dissolved in 100% DMSO and then diluted to final concentrations of 0.1% DMSO and 30 μM propranolol or 100 μM EMPA. Behavior was then recorded for 24 hours across light-dark phases. For imaging experiments, all fish were treated with solvent, 0.1% DMSO, or freshly prepared propranolol or EMPA dissolved in DMSO. Fish were monitored from ZT2 to ZT4 for bouts of inactivity associated with sleep (>60 s). Fish were euthanized for imaging if they displayed a bout of inactivity of >120 s. Control DMSO fish were euthanized for imaging at any time after undergoing a swim bout during the 2-hour assay. Data are presented as DMSO (waking) and propranolol or EMPA (sleep like) to characterize whole-brain activity under these unique conditions.
Immunohistochemistry
Briefly, 6-dpf fish were strained through a plastic mesh sieve and then dropped into ice-cold 4% PFA to kill them quickly before pERK activity resulting from euthanasia could be detected. The fish were fixed overnight, rinsed (here and below, rinses were in 0.3% PBT (phosphate-buffered saline with Tween 20), performed three times for 15 min each), treated with 150 mM tris-HCl (pH 9.0) for 15 min at 70°C, rinsed, incubated for 30 min on ice in 0.05% trypsin-EDTA, rinsed, placed in 3% H2O2 with 1% KOH for 15 min at room temperature to bleach pigmentation, and rinsed a final time. Fish were then placed in 0.3% PBT containing 2% DMSO, 1% BSA, and primary antibody at the indicated dilution: mouse anti-tERK (1:500), rabbit anti-pERK (1:500), rabbit anti-HCRT (1:500), rabbit anti-TH (1:500), sheep anti–α-MSH (1:5000), or rabbit anti-Agrp (1:400). Secondary antibodies were as follows: Alexa Fluor 488–conjugated anti-sheep immunoglobulin G (IgG) H + L, Alexa Fluor 488–conjugated anti-rabbit IgG H + L, and Alexa Fluor 561–conjugated anti-mouse IgG2a. See table S2 for a complete list of concentrations and product numbers. Special care was taken when fish were being imaged for pERK, which is a fast indicator of neuronal activity; larval fish were euthanized as quickly and consistently as possible and then processed essentially as described in (20).
Molecular cloning and transgenesis
The A. mexicanus Tg(Huc:GCaMP6s) was generated using the previously published zebrafish Tg(elav3:H2B-GCaMP6s) (24). Briefly, the zebrafish elavl3 promoter with the subcloned HSB, which restricts expression to the nucleus, was cloned to a Tol2 vector. The transgene plasmid and transposase RNA were injected into one to four cell-stage embryos at 25 ng/μl to a 1-nl volume, as previously described (22). Parental (F0) founders were bred, and transgenic lines were isolated by high expression of fluorescence in the F1 and F2 generations. All transgenic animals were raised under standard conditions.
Image acquisition and analysis
All images were procured on a Nikon A1 upright confocal microscope equipped with a motorized piezo x-y-z stage and controlled by the Nikon Elements software. Fish were mounted dorsal side up in 2% low–melting temperature agarose (Sigma-Aldrich, A9414) on a microscope slide (Fisher, 12-518-101) in a glass-bottomed chamber. Individual fish were held in 100 to 150 μl of agarose. A tiling function was used to image the entire brain, and images were stitched together with both images having a 15% overlap on the x-y plane. All images were acquired at 2-μm steps. Cell counts and intensity were quantified using the Nikon Elements software (4.5). Individual regions of interest (ROIs) were drawn over each detected cell. Mean intensity was calculated by subtracting background intensity, extracting the entire stack signal into Excel, and then restricting the quantified signal to ROIs matching the cells. To segregate neuronal populations within anatomical regions, brains were registered and overlaid with the label field. Cells within specific nuclei were then quantified within that region.
Morphometric analysis
All morphometric image analysis was performed using the FEI Amira software. Confocal stacks were imported into Fiji/ImageJ (1.52) and then imported to Amira (6.2.1). A mask was applied to include only neural tissue in the field of view. Brain regions were then manually segmented using the “lasso tool” with automatic edge detection. A developmental map was created that included the main divisions of the brain, including spine, rhombencephalon, mesencephalon, diencephalon, and telencephalon. These large divisions were segmented with tERK antibody, which defines physical divisions between regions. This result was saved as a label field for both the template brains and for each animal that was segmented. A second anatomical map of smaller nuclei was generated, including the cerebellum, optic tectum, optic neuropil, habenula, pineal gland, rostral zone of the hypothalamus, diffuse nucleus of the hypothalamus, intermediate zone of the hypothalamus, preoptic nucleus, pallium, subpallium, and pituitary complex. For the template brain of each population, six different cell markers (anti-tERK, anti-TH, anti-HCRT, anti–α-MSH, anti-AgRP, and anti-pERK) were used to guide segmentation of regions by expression pattern. The template maps for each population were then used to guide all other segmentations. The material statistics, containing all raw data regarding sizes and locations of regions, were then exported, and percentage of brain volume was calculated for each region. Data were analyzed in MATLAB (2019b) or GraphPad (v8) and visualized by ranking regions by size and graphing in a MATLAB ribbon plot. Statistical differences in region size were determined by performing one-way analysis of variance (ANOVA) with a Dunnett’s post hoc test to detect changes in region volume across populations.
Image registration
A template brain for each population of A. mexicanus was imaged using tERK immunological stain to label all neural tissue. The voxel size for the template was 0.61 × 0.61 × 2 μm3 (x × y × z). The template and transformation brains were loaded into Amira, and the “Register images” module was loaded. The transformation model included 12 degrees of freedom to account for ridged, isotropic, anisotropic, and shearing transformations. The outside threshold was set to 0.8. A correlation metric was used as the model for the transformations. A histogram filter was applied between 100 and 4095 to remove dark background pixels from the transformation calculation, which significantly reduced registration time. The coarsest resampling rate was set to either 16 × 14 × 6 or 14 × 16 × 6 depending on the reference brain used, and “ignore finest resolution” was unchecked to increase registration accuracy. Population averages of each antibody stain were generated by loading registered stacks into Amira and selecting the “average volumes” module. A single image stack was then made to represent the average expression pattern of 3 to 10 individuals for each antibody stain from each population.
pERK activity mapping
All analyses were performed in Fiji/ImageJ, Amira, and MATLAB. After registration, the pERK channel was divided by the tERK channel. Histogram restoration was performed to restore the original span of pixels acquired on the confocal. A Gaussian filter was then applied to smooth pixels, with pixels saturation set to 70% of the maximum pixel intensity and cut at 5% of the peak lowest pixel intensity to delete background noise, similar to previous analysis in zebrafish (20). This range of pixels generated a cell mask of pERK-positive cells and removed background from quantitative analysis (fig. S3). Individual image stacks were quantified by extracting all voxels and determining the mean signal of voxels per anatomical region denoted within the template brains.
Principal components analysis
Using PCA, we can consider each individual as a coordinate in a space whose axes are linearly independent combinations of regional brain activity ranked according to total interindividual variance of their activity as characterized by pERK expression. We transformed the 18-dimensional activity space into just two dimensions where each PC (dimension) comprises a combination of brain regions grouped by their alignment (correlation) with each other and ranked according to the magnitude of their variance. PCA was performed in MATLAB 2019a and XLStat. All pERK voxels were isolated by brain regions, including developmental regions, as well as all smaller regions, resulting in 20 different components for the PCA. The first two components accounted for between 47.3 and 58% of the total variability across all brains. Statistical differences between populations by PCA were detected using a one-way ANOVA with post hoc analysis.
Generation of brain atlas
The standard brain for each population was generated by registering all brains with tERK to a tERK+ template brain, with a separate template for each population. Each brain represents a unique transformation to align to the template. Thus, individual brains were registered, and then the transformation matrix generated by the tERK channel was applied to the second channel, which imaged the protein of interest. Briefly, the steps in Amira were as follows. Population averages for each protein marker were calculated from between 3 and 18 fish. To generate the average, the “average volumes” module was loaded, and then the transformed stacks were loaded, with the resultant single image representing the average of all images processed. This was performed for each population for HCRT, TH, AgRP, α-MSH, and pERK for baseline conditions and feeding or drug treatments. Each of these average stacks was saved to represent the average expression pattern for that protein for each population. If any expression outside the brain was present in a stack, then it was excluded by generating a mask to delete it from view.
Supplementary Material
Acknowledgments
We are grateful for support from J. Boerner (FAU) for image analysis and H. Burgess (NIH) for the helpful discussion. In addition, A. Loppato and P. Lewis (FAU) provided critical animal care support. Funding: This work was supported by NIH grants R21NS105071 (to A.C.K. and E.R.D.) and R01GM127872 (to A.C.K.), NSF awards DEB 174231 (to A.C.K. and J.E.K.), IOS 165674 (to A.C.K.), and IOS 1923372 (to A.C.K. and J.E.K.), and BSF award SP#2018-190 (to A.C.K. and L.A.). Author contributions: Conceptualization: J.B.J., J.E.K., L.A., E.R.D., E.L., and A.C.K. Data acquisition and analysis: J.B.J., E.L., A.Y., A.P., and Y.F. All authors contributed to the writing of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/38/eaba3126/DC1
REFERENCES AND NOTES
- 1.G. F. Striedter, A history of ideas in evolutionary neuroscience, in Evolution of Nervous Systems (2007), vol. 1, pp. 1–15. [Google Scholar]
- 2.O’Connell L., Hofmann H. A., Evolution of a vertebrate social decision-making network. Science 336, 1154–1157 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Seeholzer L. F., Seppo M., Stern D. L., Ruta V., Evolution of a central neural circuit underlies Drosophila mate preferences. Nature 559, 564–569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keene A. C., Joiner W. J., Neurodegeneration: Paying it off with sleep. Curr. Biol. 25, R234–R236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gross J. B., The complex origin of Astyanax cavefish. BMC Evol. Biol. 12, 105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soares D., Yamamoto Y., Strickler A. G., Jeffery W. R., The lens has a specific influence on optic nerve and tectum development in the blind cavefish Astyanax. Dev. Neurosci. 26, 308–317 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Menuet A., Alunni A., Joly J.-S., Jeffery W. R., Rétaux S., Expanded expression of Sonic Hedgehog in Astyanax cavefish: Multiple consequences on forebrain development and evolution. Development 134, 845–855 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Yoshizawa M., Behaviors of cavefish offer insight into developmental evolution. Mol. Reprod. Dev. 82, 268–280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth T. C. II, Rattenborg N. C., Pravosudov V. V., The ecological relevance of sleep: The trade-off between sleep, memory and energy conservation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 945–959 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arble D. M., Bass J., Behn C. D., Butler M. P., Challet E., Czeisler C., Depner C. M., Elmquist J., Franken P., Grandner M. A., Hanlon E. C., Keene A. C., Joyner M. J., Karatsoreos I., Kern P. A., Klein S., Morris C. J., Pack A. I., Panda S., Ptacek L. J., Punjabi N. M., Sassone-Corsi P., Scheer F. A., Saxena R., Seaquest E. R., Thimgan M. S., Van Cauter E., Wright K. P., Impact of sleep and circadian disruption on energy balance and diabetes : A summary of workshop discussions. Sleep 38, 1849–1860 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capellini I., Nunn C. L., McNamara P., Preston B. T., Barton R. A., Energetic constraints, not predation, influence the evolution of sleep patterning in mammals. Funct. Ecol. 22, 847–853 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siegel J. M., Clues to the functions of mammalian sleep. Nature 437, 1264–1271 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duboué E. R., Keene A. C., Borowsky R. L., Evolutionary convergence on sleep loss in cavefish populations. Curr. Biol. 21, 671–676 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Lloyd E., Olive C., Stahl B. A., Jaggard J. B., Amaral P., Duboué E. R., Keene A. C., Evolutionary shift towards lateral line dependent prey capture behavior in the blind Mexican cavefish. Dev. Biol. 441, 328–337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aspiras A. C., Rohner N., Martineau B., Borowsky R. L., Tabin C. J., Melanocortin 4 receptor mutations contribute to the adaptation of cavefish to nutrient-poor conditions. Proc. Natl. Acad. Sci. U.S.A. 112, 9688–9673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshizawa M., Goricki S., Soares D., Jeffery W. R., Evolution of a behavioral shift mediated by superficial neuromasts helps cavefish find food in darkness. Curr. Biol. 20, 1631–1636 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marquart G. D., Tabor K. M., Brown M., Strykowski J. L., Varshney G. K., La Fave M. C., Mueller T., Burgess S. M., Higashijima S.-I., Burgess H. A., A 3D searchable database of transgenic zebrafish gal4 and cre lines for functional neuroanatomy studies. Front. Neural Circuits 9, 78 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuan L., Li Y., Lau C., Feng D., Bernard A., Sunkin S. M., Zeng H., Dang C., Hawrylycz M., Ng L., Neuroinformatics of the Allen Mouse Brain Connectivity Atlas. Methods 73, 4–17 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Kunst M., Laurell E., Mokayes N., Kramer A., Kubo F., Fernandes A. M., Förster D., Maschio M. D., Baier H., A cellular-resolution atlas of the larval zebrafish brain. Neuron 103, 21–38.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Randlett O., Wee C. L., Naumann E. A., Nnaemeka O., Schoppik D., Fitzgerald J. E., Portugues R., Lacoste A. M. B., Riegler C., Engert F., Schier A. F., Whole-brain activity mapping onto a zebrafish brain atlas. Nat. Methods 12, 1039–1046 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stahl B. A., Peuß R., Dole B. M., Kenzior A., Jaggard J. B., Gaudenz K., Krishnan J., McGaugh S. E., Duboue E. R., Keene A. C., Rohner N., Stable transgenesis in Astyanax mexicanus using the Tol2 transposase system. Dev. Dyn. 248, 679–687 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stahl B. A., Jaggard J. B., Chin J. S. R., Kowalko J. E., Keene A. C., Duboué E. R., Manipulation of gene function in Mexican cavefish. J. Vis. Exp. 10.3791/59093, (2019). [DOI] [PubMed] [Google Scholar]
- 23.Yoshizawa M., Robinson B. G., Duboué E. R., Masek P., Jaggard J. B., O’Quin K. E., Borowsky R. L., Jeffery W. R., Keene A. C., Distinct genetic architecture underlies the emergence of sleep loss and prey-seeking behavior in the Mexican cavefish. BMC Biol. 13, 15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freeman J., Vladimirov N., Kawashima T., Mu Y., Sofroniew N. J., Bennett D. V., Rosen J., Yang C.-T., Looger L. L., Ahrens M. B., Mapping brain activity at scale with cluster computing. Nat. Methods 11, 941–950 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Loomis C., Peuß R., Jaggard J. B., Wang Y., McKinney S. A., Raftopoulos S. C., Raftopoulos A., Whu D., Green M., McGaugh S. E., Rohner N., Keene A. C., Duboue E. R., An adult brain atlas reveals broad neuroanatomical changes in independently evolved populations of mexican cavefish. Front. Neuroanat. 13, 88 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta T., Marquart G. D., Horstick E. J., Tabor K. M., Pajevic S., Burgess H. A., Morphometric analysis and neuroanatomical mapping of the zebrafish brain. Methods 150, 49–62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alié A., Devos L., Torres-Paz J., Prunier L., Boulet F., Blin M., Elipot Y., Retaux S., Developmental evolution of the forebrain in cavefish, from natural variations in neuropeptides to behavior. eLife 7, e32808 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joiner W. J., Unraveling the evolutionary determinants of sleep. Curr. Biol. 26, R1073–R1087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A. C. Keene, L. Appelbaum, Sleep in Fish Models, in Handbook of Behavioral Neuroscience, H. C. Dringenberg, Ed. (Elsevier Academic Press, 2019). [Google Scholar]
- 30.Singh C., Oikonomou G., Prober D. A., Norepinephrine is required to promote wakefulness and for hypocretin-induced arousal in zebrafish. eLife 4, e07000 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elbaz I., Levitas-Djerbi T., Appelbaum L., The hypocretin/orexin neuronal networks in zebrafish. Curr. Top. Behav. Neurosci. 33, 75–92 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Jaggard J. B., Stahl B. A., Lloyd E., Prober D. A., Duboue E. R., Keene A. C., Hypocretin underlies the evolution of sleep loss in the Mexican cavefish. eLife 7, e32637 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duboué E. R., Borowsky R. L., Keene A. C., β-adrenergic signaling regulates evolutionarily derived sleep loss in the Mexican cavefish. Brain Behav. Evol. 80, 233–243 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Kaslin J., Nystedt J. M., Ostergård M., Peitsaro N., Panula P., The orexin/hypocretin system in zebrafish is connected to the aminergic and cholinergic systems. J. Neurosci. 24, 2678–2689 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Appelbaum L., Wang G. X., Maro G. S., Mori R., Tovin A., Marin W., Yokogawa T., Kawakami K., Smith S. J., Gothilf Y., Mignot E., Mourrain P., Sleep-wake regulation and hypocretin-melatonin interaction in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 106, 21942–21947 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adan R. A. H., Tiesjema B., Hillebrand J. J. G., la Fleur S. E., Kas M. J. H., de Krom M., The MC4 receptor and control of appetite. Br. J. Pharmacol. 149, 815–827 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forlano P. M., Cone R. D., Conserved neurochemical pathways involved in hypothalamic control of energy homeostasis. J. Comp. Neurol. 505, 235–248 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Shainer I., Michel M., Marquart G. D., Bhandiwad A. A., Zmora N., Livne Z. B.-M., Zohar Y., Hazak A., Mazon Y., Förster D., Hollander-Cohen L., Cone R. D., Burgess H. A., Gothilf Y., Agouti-related protein 2 is a new player in the teleost stress response system. Curr. Biol. 29, 2009–2019.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji R.-R., Baba H., Brenner G. J., Woolf C. J., Nociceptive-specific activation of ERK in spinal neurons contributes to pain hypersensitivity. Nat. Neurosci. 2, 1114–1119 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Dai Y., Iwata K., Fukuoka T., Kondo E., Tokunaga A., Yamanaka H., Tachibana T., Liu Y., Noguchi K., Phosphorylation of extracellular signal-regulated kinase in primary afferent neurons by noxious stimuli and its involvement in peripheral sensitization. J. Neurosci. 22, 7737–7745 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arrigoni E., Chee M. J. S., Fuller P. M., To eat or to sleep: That is a lateral hypothalamic question. Neuropharmacology 154, 34–49 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Herrera C. G., Ponomarenko A., Korotkova T., Burdakov D., Adamantidis A., Sleep & metabolism: The multitasking ability of lateral hypothalamic inhibitory circuitries. Front. Neuroendocrinol. 44, 27–34 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Andalman A. S., Burns V. M., Lovett-Barron M., Broxton M., Poole B., Yang S. J., Grosenick L., Lerner T. N., Chen R., Benster T., Mourrain P., Levoy M., Rajan K., Deisseroth K., Neuronal dynamics regulating brain and behavioral state transitions. Cell 177, 970–985.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Facchin L., Duboué E. R., Halpern M. E., Disruption of epithalamic left-right asymmetry increases anxiety in Zebrafish. J. Neurosci. 35, 15847–15859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leung L. C., Wang G. X., Madelaine R., Skariah G., Kawakami K., Deisseroth K., Urban A. E., Mourrain P., Neural signatures of sleep in zebrafish. Nature 571, 198–204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Trotha J. W., Vernier P., Bally-Cuif L., Emotions and motivated behavior converge on an amygdala-like structure in the zebrafish. Eur. J. Neurosci. 40, 3302–3315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rihel J., Prober D. A., Arvanites A., Lam K., Zimmerman S., Jang S., Haggarty S. J., Kokel D., Rubin L. L., Peterson R. T., Schier A. F., Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 327, 348–351 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi G., Xing L., Wu D., Bhattacharyya B. J., Jones C. R., McMahon T., Chong S. Y. C., Chen J. A., Coppola G., Geschwind D., Krystal A., Ptáček L. J., Fu Y.-H., A rare mutation of β1-adrenergic receptor affects sleep/wake behaviors. Neuron 103, 1044–1055.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson M. D., Comings D. E., Abu-Ghazalah R., Jereseh Y., Lin L., Wade J., Sakurai T., Tokita S., Yoshida T., Tanaka H., Yanagisawa M., Burnham W. M., Moldofsky H., Variants of the orexin2/hcrt2 receptor gene identified in patients with excessive daytime sleepiness and patients with Tourette’s syndrome comorbidity. Am. J. Med. Genet. B Neuropsychiatr. Genet. 129B, 69–75 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Zada D., Bronshtein I., Lerer-Goldshtein T., Garini Y., Appelbaum L., Sleep increases chromosome dynamics to enable reduction of accumulating DNA damage in single neurons. Nat. Commun. 10, 895 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saper C. B., Scammell T. E., Lu J., Hypothalamic regulation of sleep and circadian rhythms. Nature 437, 1257–1263 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Carter M. E., Brill J., Bonnavion P., Huguenard J. R., Huerta R., de Lecea L., Mechanism for Hypocretin-mediated sleep-to-wake transitions. Proc. Natl. Acad. Sci. U.S.A. 109, E2635–E2644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fuller P. M., Saper C. B., Lu J., The pontine REM switch: Past and present. J. Physiol. 584, 735–741 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srinivasan A. V., Propranolol: A 50-year historical perspective. Ann. Indian Acad. Neurol. 22, 21–26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steenen S. A., van Wijk A. J., van der Heijden G. J. M. G., van Westrhenen R., de Lange J., de Jongh A., Propranolol for the treatment of anxiety disorders: Systematic review and meta-analysis. J. Psychopharmacol. 30, 128–139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ng L., Bernard A., Lau C., Overly C. C., Dong H.-W., Kuan C., Pathak S., Sunkin S. M., Dang C., Bohland J. W., Bokil H., Mitra P. P., Puelles L., Hohmann J., Anderson D. J., Lein E. S., Jones A. R., Hawrylycz M., An anatomic gene expression atlas of the adult mouse brain. Nat. Neurosci. 12, 356–362 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Herman A., Brandvain Y., Weagley J., Jeffery W. R., Keene A. C., Kono T. J. Y., Bilandžija H., Borowsky R., Espinasa L., O’Quin K., Ornelas-García C. P., Yoshizawa M., Carlson B., Maldonado E., Gross J. B., Cartwright R. A., Rohner N., Warren W. C., McGaugh S. E., The role of gene flow in rapid and repeated evolution of cave-related traits in Mexican tetra, Astyanax mexicanus. Mol. Ecol. 22, 4397–4416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elipot Y., Legendre L., Père S., Sohm F., Rétaux S., Astyanax transgenesis and husbandry: How cavefish enters the laboratory. Zebrafish 11, 291–299 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Klaassen H., Wang Y., Adamski K., Rohner N., Kowalko J. E., CRISPR mutagenesis confirms the role of oca2 in melanin pigmentation in Astyanax mexicanus. Dev. Biol. 441, 313–318 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/38/eaba3126/DC1


