ABSTRACT
Helicobacter pylori is a fastidious Gram-negative bacterium that infects over half of the world's population, causing chronic gastritis and is a risk factor for stomach cancer. In developing and rural regions where prevalence rate exceeds 60%, persistence and waterborne transmission are often linked to poor sanitation conditions. Here we demonstrate that H. pylori not only survives but also replicates within acidified free-living amoebal phagosomes. Bacterial counts of the clinical isolate H. pylori G27 increased over 50-fold after three days in co-culture with amoebae. In contrast, a H. pylori mutant deficient in a cagPAI gene (cagE) showed little growth within amoebae, demonstrating the likely importance of a type IV secretion system in H. pylori for amoebal infection. We also demonstrate that H. pylori can be packaged by amoebae and released in extracellular vesicles. Furthermore, and for the first time, we successfully demonstrate the ability of two free-living amoebae to revert and recover viable but non-cultivable coccoid (VBNC)-H. pylori to a culturable state. Our studies provide evidence to support the hypothesis that amoebae and perhaps other free-living protozoa contribute to the replication and persistence of human-pathogenic H. pylori by providing a protected intracellular microenvironment for this pathogen to persist in natural aquatic environments and engineered water systems, thereby H. pylori potentially uses amoeba as a carrier and a vector of transmission.
Keywords: Helicobacter pylori, free-living amphizoic amoebae, intracellular multiplication, extracellular vesicles, phagosomal pH, bacterial recovery
Free-living amoebae were shown to play a role in the survival and recovery of H. pylori in aquatic environments, demonstrating a new mechanism for their distribution and persistence in these habitats.
INTRODUCTION
Recently, the World Health Organization (WHO) published a catalogue of 12 families of antimicrobial resistant bacteria that pose the greatest threat to human health, one of which was Helicobacter pylori (Willyard 2017). Helicobacter pylori is an antibiotic resistant, gastric carcinogenic pathogen, described in 1984 as the principal cause of peptic ulcer disease (Blaser 1997; Dunn, Cohen and Blaser 1997). This fastidious Gram-negative bacterium colonizes the gastric mucosa of over half of the world's population, yet its microbial ecology outside of the human host is poorly understood (Aziz, Khalifa and Sharaf 2015). Over the last decade or so, however, various studies implicate or support the theory that free-living amoebae (FLA) could be aquatic environmental hosts for H. pylori (Winiecka-Krusnell et al. 2002; Smith and Ashbolt 2012; Ashbolt 2015; Moreno-Mesonero et al. 2016, 2017).
FLA are one of the most ubiquitous organisms found in aquatic environments, being primary predators controlling bacterial populations (Rodriguez-Zaragoza 1994; Samba-Louaka et al. 2019). However, they are known natural environmental reservoirs for a range of amoeba-resisting bacterial pathogens (ARB) and a vector of human infection for, water-associated infections (Thomas et al. 2010; Thomas and Ashbolt 2011; Dey, Hoffman and Glomski 2012). Interestingly, the intracellular growth of free-living bacteria within FLA has been shown to increase bacterial resistance to antibiotics (Maurin, Bryskier and Raoult 2002) and biocides (King et al. 1988) and upregulate virulence (Molmeret et al. 2005). In addition to FLA being present in water systems, they have been identified within the gastrointestinal track of humans from the stomach to faeces (Thamprasert, Khunamornpong and Morakote 1993; Bradbury 2014), they are considered as an amphizoic organisms and live as parasites within host tissue (ability to live endozoically) (Page 1967; Jadin 1973; Martinez and Visvesvara 1997).
The significance of FLA in human intestinal tract infection goes back to 1912 when Chatton and Lalung-Bonnaire isolated the FLA Vahlkampfia sp. from a human intestine and proposed the idea that the amoeba could be the pathogen and/or an important vector of infection (Chatton and Lalung-Bonnaire 1912). Almost a century later, Steele and colleagues reported the implication of the FLA Willaertia magna in the gastric infection of a dog (Steele et al. 1997).
Since H. pylori and FLA co-occur in human intestinal tract, we considered the possibility that FLA might serve as natural hosts and be vectors for H. pylori transmission. We tested this hypothesis by studying the interaction between H. pylori and two different amoebae species that are also facultative parasites of humans and animals. The selected FLA are, W. magna belonging to the Vahlkampfiidae family and Vermamoeba vermiformis, belonging to Vermamoebidae family and one of the most common free-living protists in human environments (de Moura et al. 1985; Bradbury 2014; Delafont et al. 2018). Moreover, many reports have implicated the survival of H. pylori in aquatic environments within FLA, hence this pathogen is considered to be an ARB (Giao et al. 2011; Santiago, Moreno and Ferrus 2015; Moreno-Mesonero et al. 2016, 2017). However, no direct demonstration of H. pylori replication within amoebae hots had been provided.
It has been concluded that during harsh environmental conditions H. pylori adopts a dormant stage and remains infectious, which represent a potential public health concern (Nilsson et al. 2002), transforming from a spiral shape to the coccoid form that includes viable but non-culturable (VBNC) cells (Sarem and Corti 2016) and subsequently becomes highly tolerant to antibiotics (Bates, Adams and Oliver 2003). However, one of the most important challenges has been determining the mechanism of the coccoid form reactivation (Loke et al. 2016). Interestingly, it has been speculated that VBNC enteric bacteria can be recovered by co-culture with eukaryotic cells (Senoh et al. 2012). Similar results were obtained with VBNC Legionella pneumophila when co-cultured with FLA (Garcia et al. 2007). To investigate this further, we analyzed the morphological changes of H. pylori in co-culture with FLA using an imaging flow cytometric approach, that we already have developed (Dey et al. 2019). In addition, we investigated growth and survival strategies of H. pylori in co-culture with FLA by culture, molecular and imaging techniques.
MATERIALS AND METHODS
H. pylori strains and growth conditions
A total of four different H. pylori strains were used in this study: H. pylori G27; two derivative mutant strains (∆cagE and ∆cagM) obtained from Dr Monika Keelan, Faculty of Medicine and Dentistry, University of Alberta; and Green Fluorescent Protein (GFP) expressing H. pylori from Dr John Kao, University of Michigan, USA. Helicobacter pylori were grown on Brain Heart Infusion (BHI) agar plates (BactoTM, Le Pont de Claix) supplemented with 15 mg/mL of Amphotericin B and Vancomycin to suppress the growth of other bacteria and fungi (Jiang and Doyle 2000), the plates incubated under microaerophilic conditions (anaerobic jars filled with 5% CO2, 5% H2 and balance N2) at 37°C and the bacteria cultivated for 3–4 days to maintain a stationary growth phase for bacterial optimal growth (Worku et al. 1999).
Amoebae growth conditions
The amoebae used in this study were W. magna (ATCC 50035) isolated from bovine faeces and Vermamoeba vermiformis (ATCC 50237) isolated from a hospital cooling tower drain. Amoebae were grown in sterile tissue culture flasks (Thermo Scientific, Edmonton, Canada 130192 or 130193) in SCGYEM (Serum-Casein-Glucose-Yeast-Extract-Medium: ATCC medium 1021) at 25°C in a 5% CO2 incubator. The trophozoites were maintained in exponential growth phase by sub-culturing every 3–4 days in fresh SCGYEM, and then harvested by tapping the flasks and washing three times with sterile distilled water by centrifugation at 2000 g for 10 min to remove carried-over nutrients in the supernatants. Experiments were carried out in sterile 15 mL screw-cap tubes, using 3 mL of medium. FLA were used at a final cell concentration of 105/mL determined by a haemocytometer counting (Hausser Scientific, Horsham, USA HSR3110) and distributed in autoclaved lake water (autoclaved for 20 min at 121°C) from the Edmonton area (Alberta, Canada).
Co-culture of H. pylori with FLA
Bacterial suspensions (H. pylori G27, ∆cagE and ∆cagM) in sterile distilled water (OD550 1.0, ∼109/mL; Kavermann et al. 2003) were used to inoculate amoeba cultures at a multiplicity of infection (MOI) of 100. Tubes (FALCON, Fischer Scientific, Edmonton, Canada 3033) containing 3 mL of autoclaved lake water were seeded with 105/mL trophozoites of the different amoebae, low speed centrifugation at 500 g for 5 min was used to initiate physical interaction between bacteria and amoebae and then the co-cultures were incubated under microaerophilic conditions at 25 and 37°C and performed in triplicate (n = 4–6). After 2 h incubation, the co-cultures were treated with 100 μg/mL of sterile gentamicin (Sigma-Aldrich, Ontario, Canada G1397) for 1 h in order to kill extracellular H. pylori and subsequently quantify intracellular bacteria. After centrifugation (10 min at 2500 g) the medium was removed and the amoebae pellet was washed twice in sterile Phosphate Buffered Saline (PBS) medium to eliminate residual gentamicin and the possible remaining extracellular bacteria. The co-cultures were resuspended in sterile lake water, which was unable to support bacterial outgrowth in the absence of amoebae and to establish conditions that mimic aquatic environments.
The concentrations of the amoebae and H. pylori were determined at 24 h intervals from days 0 to 4 (d0–d4). The growth of amoebae was determined by counting resuspended cells per unit volume with a haemocytometer after staining with trypan blue (Gibco, Edmonton, Canada fischer scientific 15 250–061) for amoebae exposed and unexposed to H. pylori. Simultaneously, the bacterial concentrations (CFU/mL and genomic copies/mL) with or without FLA from aliquots of the same experimental samples were estimated with culture and qPCR methods after lysing the FLA by passing the sample through a sterile 20-gauge syringe needles (BD, Franklin lakes, USA 304827) 5–6 times.
The total number of H. pylori (with or without FLA) was established by performing serial 10-fold dilutions of the co-culture medium containing lysed amoeba and bacteria with sterile distilled water that were subsequently spread in triplicate on BHI agar plates supplemented with Amphotericin B and Vancomycin and incubated at 37°C under microaerophilic conditions.
Cytotoxicity of H. pylori
The cytotoxicity of H. pylori towards FLA was studied qualitatively by fluorescent microscopy EVOS FL (ThermoFisher Scientific, Edmonton, Canada) using the GFP-H. pylori, amoebae cells quantified with a haemocytometer (Hausser Scientific HSR3110) and trypan blue (Gibco 15 250–061) staining as previously described (Dey et al. 2009). The influence of bacteria on amoebal monolayer formation in 6-well plates containing 105 amoebae/well infected with GFP-H. pylori at a MOI of 100 were photographed after 72 h of co-culture.
Transwell assay
To examine whether H. pylori can grow extracellularly and damage the host cells by direct contact with amoebae, a transwell plate assay was used and H. pylori separated from amoebae (V. vermiformis or W. magna) by a 0.4 µm membrane. The transwell plates consisted of 6 wells (Corning, New york, USA), each well containing two chambers and separated by a 0.4 µm polycarbonate membrane. FLA (V. vermiformis or W. magna) were placed in the apical compartment and H. pylori G27 strain placed in the basal compartment (amoebae free) in sterile lake water and with the same concentration (FLA/bacteria) as described in co-culture section. After 24 h of incubation, H. pylori cells were plated on BHI agar plates supplemented with Amphotericin B and Vancomycin, and incubated at 37°C under microaerophilic conditions for Colony Forming Unit (CFU) determination.
Quantitative PCR (qPCR) analysis
A total of 1 mL of the co-culture samples were taken at different post-infection times and lysed mechanically by pumping several times through a 20-gauge syringe needle (BD 304827) prior to the DNA extraction process. A total of 100 µL from the original volume (1 mL) were taken for DNA extraction using DNeasy blood and tissue extraction kit (Qiagen, Toronto, Canada). Abundance of total H. pylori was quantified in triplicate using a qPCR assay, designed and optimized in this study to be specific to the 23S rRNA gene of H. pylori, but no other Helicobacter, Arcobacter or Campylobacter species (see Supplementary data SD 1). The limit of detection (LOD) is <10 copies/qPCR rxn with the following sequences:
HpyP3: 5′- 6FAM-AAGATATATGAGAATTGTATCCGCC—NFQMGB -3′
HpyF3: 5′- GCGGTATGAAGTGAGCATGCA -3′
HpyR3: 5′- GACCATCGCGTAGGAAACCTT -3′
qPCR amplifications were performed using an ABI 7500 Fast cycler (ThermoFisher®, Waltham, USA) in a 20 μL volume containing 1x IDT PrimeTime qPCR Mastermix (ThermoFisher®), 450 nM of each primer, 200 nM of probe and 5 μL of DNA template. The amplification conditions were as follows: 95°C for 2 min, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s.
pH measurement
For the intracellular pH measurement, H. pylori G27 was used and the infected amoebae (W. magna) were incubated with 10 μM of pHrodo Green or Red intracellular pH indicators for 30 min at 37°C according to the manufacturer's protocol (Life technologies, Edmonton, Canada P35373). Standard curves of cytosolic pH were created using pHrodo Green or Red with intracellular pH calibration buffer kit (Life technologies P35379) of pH 4.5. After incubation, the samples treated with pHrodo Red were washed and re-suspended in PBS prior to processing through the Attune NxT acoustic focusing flow cytometer with (blue/red/violet/yellow) lasers (Thermo Fisher Scientific) and the pH Rodo data was collected on the YL2 detector. The fluorescence images of the cells treated with pHrodo Green were osecbtained using an EVOS FL fluorescent cell imaging system (Thermo Fisher Scientific), that capture the green fluorescence channel combined with transmitted light channel. Images were acquired at 100x magnification.
Induction of viable but nonculturable state in H. pylori cells
Entry into the VBNC state was induced by incubation of GFP-H. pylori suspensions in sterile lake water (5 mL), covered in the dark at 4°C. To assess the culturability of the bacteria, samples were taken at intervals and inoculated on BHI agar plates with amphotericin and vancomycin and incubated at 37°C under microaerophilic conditions. The GFP-H. pylori entered into the VBNC state after 5 months incubation and was confirmed by the absence of growth on BHI, in accordance with previous studies which reported similar prolonged incubation times to induce VBNC cells (Asakura et al. 2007; Su et al. 2015), and demonstrated by absence of any colonies. Apparent VBNC cells and spiral GFP-H. pylori were co-cultured with amoebae and compared to controls (VBNC and spiral cells alone), with a MOI of 100 and incubated at 25 ⁰C for up to 72 h. H. pylori cells (VBNC and spiral) were plated on BHI agar plates supplemented with Amphotericin B and Vancomycin, and incubated at 37°C under microaerophilic conditions for CFU determination. To aid in resolving ‘viable’ and damaged cells, 1.5 µL/mL of propidium iodide (Thermo Fisher Scientific P3566) was added to the samples after incubation time (VBNC cells and reverted cells), followed by mixing and 5–10 min incubation in the dark.
Morphological characteristics of assumed VBNC (coccoid) cells and reverted (spiral) cells were analyzed by imaging flow cytometer (ImageStreamx® MarkII, Millipore Sigma, Seattle, USA).
Imaging Flow Cytometry (IFC) analysis
Intracellular locations of GFP-H. pylori in W. magna coculture was performed using ImageStream® cytometry as previously described (Dey et al. 2019). Briefly, FLA were infected for 2 h with viable spiral GFP-H. pylori at a MOI of 100, washed and re-suspended in PBS prior to processing through the ImageStream®X Mark II (Millipore Sigma). Cells were acquired at 60x magnification. Analysis was performed using the IDEAS software (Amnis, Seattle, USA) and cells were identified on the basis of bright field morphology and size. For the recovering analysis and after 30 min of coinfection with FLA, GFP-H. pylori were gated based on small size and low side scatter (to remove any internal control SpeedBeads® from the gate). They were then further gated based on GFP signal, degree of focus and all doublets/ clumps were removed from the analysis. Spiral and coccoid bacteria were then gated based on Aspect Ratio Intensity (Ch01) and Major Axis Intensity (Ch01) to discriminate between the two bacterial cell morphologies (spiral vs VBNC cell forms). The bacteria gating strategy as previously described (Dey et al. 2019) and briefly provided in (Supplementary data SD. 2).
MICROSCOPY
Fluorescence microscopy
Co-cultures of GFP-H. pylori and FLA in 12-well tissue culture plates (Fisher Scientific 130185) overlaid with microscopy cover slips (Fisher Scientific 12–546-1) were incubated at 37°C with 5% CO2. After 2 h of infection, the sterile lake water medium was removed and cells fixed with 4% paraformaldehyde for 5 min at room temperature and then washed with PBS three times. Images were taken with an EVOS FL fluorescent cell imaging system (ThermoFisher Scientific).
Scanning electron microscopy (SEM)
The overnight co-culture samples (H. pylori G27-W. magna) were fixed for 15–60 min with glutaraldehyde (Electron Microscopy Sciences, Hatfield, USA glutaraldehyde 16537–10) and followed by three 5-min rinse steps in 1x Dulbecco's Phosphate Buffered Saline (DPBS). Samples were then dehydrated using increasing concentrations of ethanol (Fisher BioReagents BP82031GAL), 5 min each in 10, 30, 50, 70, 80, 90 and 100% (ethanol in distilled water). The samples were further dried for 10 min in hexamethyldisilazane (Sigma-Aldrich H4875). Finally, the membranes were stuck to SEM stubs with adhesive carbon strips and sputter coated with gold using a Leica EM ACE600 sputter coater. The final samples were imaged using a Sigma VP HD Field-emission SEM (Zeiss, Pleasanton, CA) at 10 000x magnification through the University of Alberta Microscopy Facility.
Transmission electron microscopy (TEM)
Axenic cultures of amoebae were infected for 6 h with H. pylori G27 at a MOI of 100 on Thermanox cover slips (Thermo Fisher 174985). After decanting the medium, amoebae were fixed at room temperature with 2.5% glutaraldehyde and 0.1 M sodium cacodylate buffer (Electron Microscopy Sciences 15960). Samples were submitted for processing at the imaging core at University of Alberta, Faculty of Medicine and Dentistry.
Statistical analysis
Data were analyzed with a two-tailed Student's t-test (Microsoft Excel 2007), with an alpha value of 0.05, under the assumption that data were normally distributed and had equal variances (see details in figure legends).
RESULTS AND DISCUSSION
Intracellular multiplication of H. pylori and their effect on FLA viability
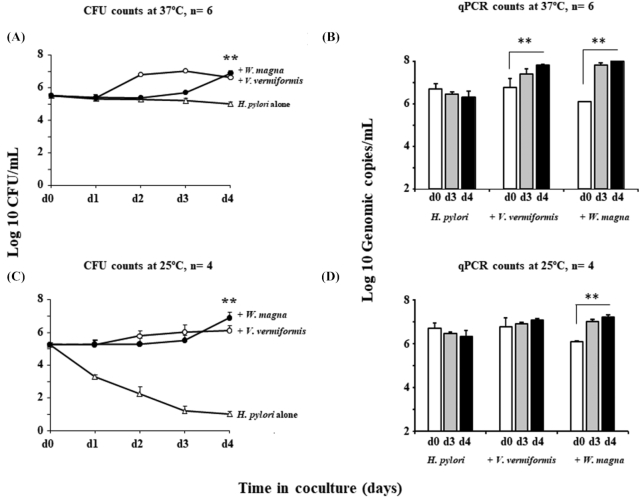
To determine the number of H. pylori in amoebae, estimates were made by CFUs and qPCR targeting its 23S rRNA gene. Analysis showed the bacterial numbers of H. pylori from both FLA species significantly increased over 50-fold by day 4 at 37C (Fig. 1A and B), suggesting that FLA contribute to the amplification of H. pylori and promote their multiplication.
Figure 1.
Intracellular growth of H. pylori in co-culture with FLA. (A) and (C): Growth Kinetics of H. pylori G27 with V. vermiformis (open circles) and W. magna (closed circles) or without amoebae (open triangles) by CFU counts at 37°C and 25°C respectively for up to 4 days. (B) and (D): Growth Kinetics of H. pylori G27 with or without amoebae by qPCR at 37°C and 25°C respectively for up to 4 days, white bars represent the number of H. pylori on day 0 of experiments; grey and black bars represent, respectively, day 3 and day 4 of co-culture. Data are the mean ± SEM, n = 4–6 performed in triplicate. Statistical differences by Student's t-test comparing the indicated strains to the control in the absence of amoeba after 4 days (**: P < 0.001).
Similar qPCR analysis results were also obtained at 25 ⁰C, but with somewhat slower kinetics. However, as seen in Fig. 1C, viable counts did not increase (P< 0.001), and viable H. pylori without amoeba hosts also decreased by over 4 log10 after 3 days, suggesting that H. pylori cells may entered the VBNC state period after time; This suggested that lower temperature may affect the growth of H. pylori and increase the likelihood of VBNC cell formation. The reported optimum temperature range for growth of H. pylori is from 30 to 37°C (Jiang and Doyle 1998), which is consistent with our observation that the growth of H. pylori alone declined in lake water and viable cells only persisted for 2 days at 25°C. In contrast, at the same temperature but in the presence of amoebae H. pylori continue to grow and survived for up to 4 days.
Therefore, it was of interest to further view, by electron microscopy, the formation of numerous replicative phagosomes within V. vermiformis and W. magna filled with H. pylori (Fig. 2A and B).
Figure 2.
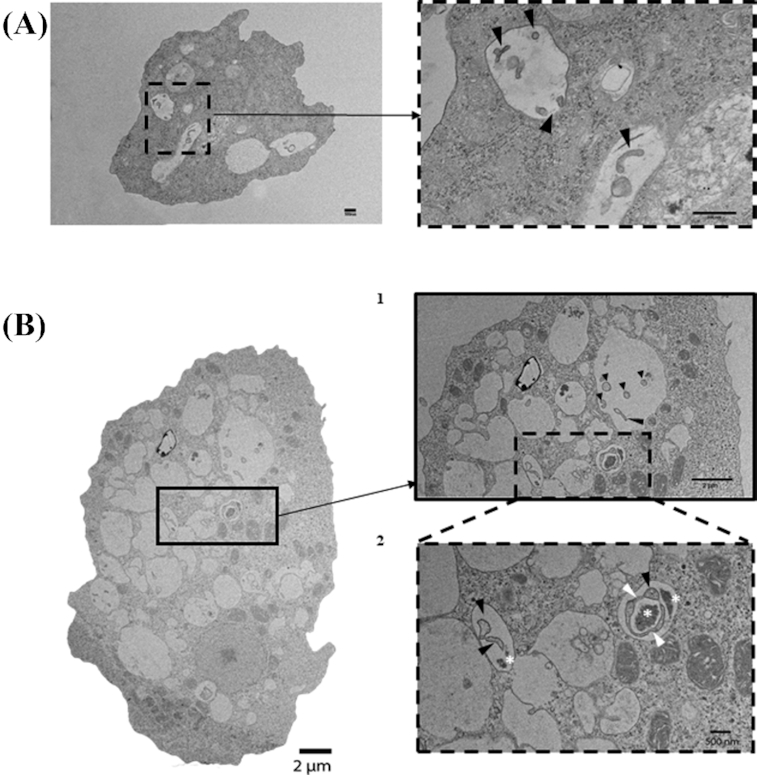
Transmission Electron Microscopy (TEM) of spiral shaped rod morphology of H. pylori contained within (A)V. vermiformis and (B1 and B2) W. magna phagosomes after 6 h of co-culture at 37°C (black arrowheads). Note, B2 demonstrates H. pylori in the process of septation (white arrowheads) as well as the bacterial remnants (white stars).
Since the cag Pathogenicity Island (cagPAI) that encodes a type IV secretion system (T4SS), is considered to be involved in bacterial internalization and is a major putative virulence factor in H. pylori (Tegtmeyer, Wessler and Backert 2011; Boonyanugomol et al. 2013; Tegtmeyer et al. 2017), we tested whether two different genes belonging to the cagPAI family, cagE and cagM were required for bacterial multiplication in presence of FLA.
As seen in Fig. 3 and based on qPCR analysis, H. pylori mutant deficient in cagPAI gene cagE (∆ cagE) failed to multiply in amoebae and growth kinetics dropped by over two log10 by day 3 post infection compared to ∆ cagM mutant (Fig. 3 A and B); suggesting that deficient cagE gene cells within FLA are eventually digested but not with deficient cagM cells, showing the importance of cagE gene in H. pylori infection and multiplication within amoebae.
Figure 3.
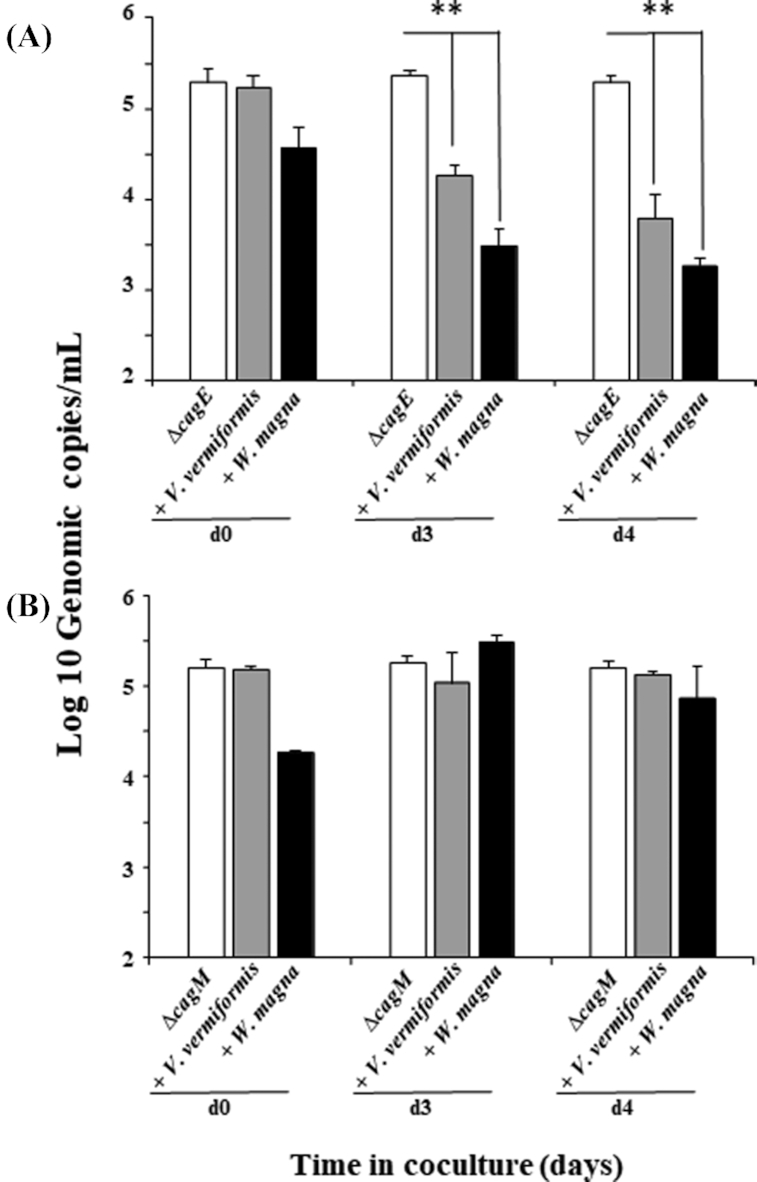
qPCR analysis of H. pylori mutant deficient in cagPAI genes growth in co-culture with FLA. (A) Growth Kinetics of H. pylori mutant ∆cagE with V. vermiformis (grey bars) and W. magna (black bars) or without amoebae (open bars) respectively at 37°C for up to 4 days. (B) Growth Kinetics of H. pylori mutant ∆cagM with V. vermiformis (grey bars) and W. magna (black bars) or without amoebae (open bars) respectively at 37°C for up to 4 days. Data are the mean ± SEM, n = 3 performed in triplicate. Statistical differences by Student's t-test comparing the indicated strains to the control in the absence of amoeba after 4 days (**: P < 0.001).
Similar results were also obtained at 25 ⁰C, indicating that the observed phenomenon was not affected by a lower temperature (Supplementary data SD 3).
Interestingly, several studies indicate that a functional crosstalk exists between the two virulence factors, vacuolating toxin VacA and the effector protein CagA (encoded by the cag pathogenicity island together with a T4SS); this relationship could be a strategy to improve the bacterium's fitness within the hostile gastric environment (Gangwer et al. 2010; Boquet and Ricci 2012; Djekic and Muller 2016; Ricci 2016). Indeed, our results demonstrates that CagE-deficient mutants did exhibit an intracellular proliferation defect. It has been demonstrated that CagE and CagM are required for CagA translocation through Cag-T4SS and are also needed for the secretion of interleukin-8 (IL-8) by the host gastric epithelial cells (Fischer et al. 2001; Shariq et al. 2015; Bats et al. 2018). Unexpectedly, the CagM-deficient gene did not affect the growth of the bacteria; implying that CagE gene may be essential for H. pylori survival within amoebae and additional studies will be required to understand the genes involved in H. pylori entering and replicating within amoebae.
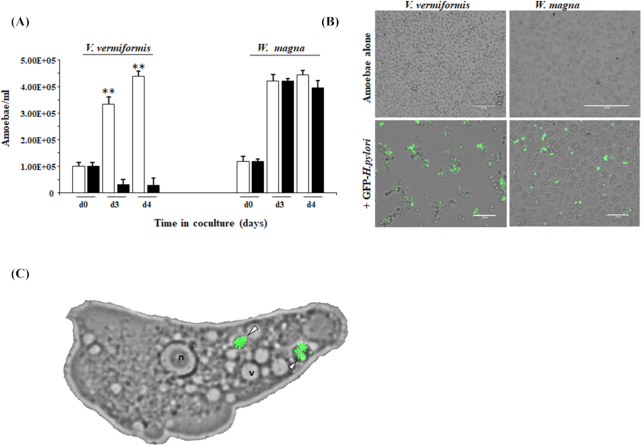
Furthermore, we screened the impact of H. pylori infection on the growth of amoebae. Within 3 days the number of viable V. vermiformis trophozoites in co-culture with GFP-H. pylori had dropped significantly, the growth was reduced by 94% compared to those of uninfected amoebae or those cocultured with W. magna (Fig. 4A). The effect of H. pylori towards V. vermiformis is further illustrated in Fig. 4B, showing that H. pylori prevented the formation of a V. vermiformis cell monolayer and affected their appearance (encystment) after 3 days of infection in sterile lake water, while W. magna was apparently unaffected.
Figure 4.
Effect of H. pylori on the amoebic viability. (A) Effect of H. pylori infection on the growth of amoebae: The different amoebae were cultured at 37°C for 4 days either with (black bars) or without (white bars) H. pylori at MOI of 100 as described in methods. The data are expressed as the number of amoebae/mL of medium and are the average ± SEM of four independent experiments performed in triplicate. Statistical differences (Student t-test) between the growth of amoebae cultured either with or without H. pylori are indicated (**P < 0.001). (B) Effect of H. pylori on monolayer formation by amoebae: Representative fluorescent images of the two amoebae cultured in 6-well plates either with or without H. pylori after 72 h. (C) Micrograph (60x) of W. magna infected with GFP–H. pylori after 6 h of co-culture. The white arrowhead points to replicative phagosomes containing H. pylori. nucleus (n), vacuole (v).
Helicobacter pylori has long been viewed as an extracellular bacterium, attached to gastric epithelial cells and adapting different mechanisms to avoid phagocytosis and evade immune responses (Allen 2000, 2007). Based on our transwell assay results, no bacterial increase was observed in amoeba-free compartments (Supplementary data SD 4), suggesting that H. pylori replicates intracellularly within the two amoebae tested. On closer observation using fluorescent microscopy showed W. magna phagosomes filled with GFP-H. pylori (Fig. 4C). Interestingly, our study demonstrates that intracellular replication of H. pylori had a cytotoxic effect on the V. vermiformis host by disrupting the culture-flask monolayer and damaging the host cells. However, our results also demonstrate that H. pylori is able to replicate and escape from within W. magna without killing the host cell.
Release of packaged H. pylori by W. magna
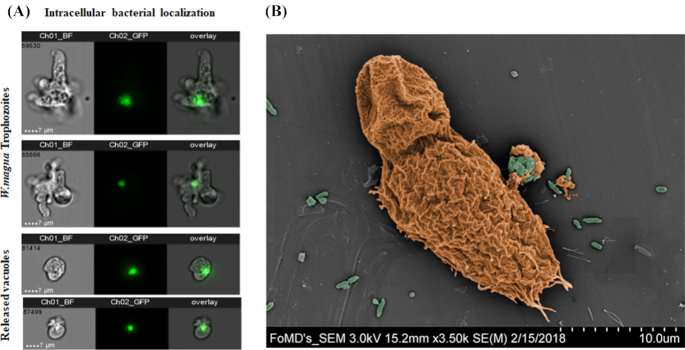
From the results depicted in Fig. 4 (A and B) showing unaffected W. magna cells exposed to H. pylori and the successful growth of the bacterium, it is reasonable to hypothesize that H. pylori is able to replicate and escape from within W. magna without killing the host cell. To this end, we applied an imaging flow cytometry approach to study infected amoebae and localize phagocytized GFP- H. pylori cells. Fig. 5A shows live images of GFP-H. pylori internalized by W. magna and located in amoebal-released vesicles. Using scanning microscopy, we can see W. magna in the process of releasing a vesicle filled with bacteria (Fig. 5B). Tracked by fluorescence microscopy, different stages of H. pylori contained within W. magna phagosomes were observed (Supplementary data SD. 5). Overall these observations demonstrate that H. pylori indeed most likely replicates within W. magna and exits via released vesicles.
Figure 5.
Packaging of H. pylori by amoebae. (A) Internalized GFP-H. pylori in live W. magna and located in released vacuoles using ImageStream flow cytometry. (B) Colorized scanning electron micrograph of H. pylori packaged in excreted vesicles by W. magna. H. pylori (highlighted in green) was cocultured with W. magna (in orange) for 24 h, at an MOI of 1:100.
Interestingly, this phenomenon was first observed with L. pneumophila as a survival strategy that enable them to avoid digestion and to be packaged in the egested pellets from FLA and ciliates (Rowbotham 1980; Berk et al. 1998, 2008; Bouyer et al. 2007). More importantly, the released vesicles can serve as natural reservoirs for many opportunistic bacteria pathogens, including respiratory strains when expelled within aerosolized amoebal vesicles (Berk et al. 1998; Shaheen and Ashbolt 2018). Hence the growth outcome of H. pylori is likely dependent upon the species of host amoebae and its growth environment (Allen 2000; Dey et al. 2009).
H. pylori adaptation within acidified phagosomes
Considering that H. pylori is a neutralophile known to colonize the acidic human stomach by using a variety of acid-adaptive mechanisms (Wen et al. 2003; Scott et al. 2007), we next examined and tracked the intracellular pH of amoebae after infection. Previous studies showed that GFP fluorescence decreases significantly under acidic conditions (Campbell and Choy 2001; Roberts et al. 2016), hence we used an intracellular pH indicator (green or red at pH 4–5) and flow cytometry to illustrate phagosome acidification.
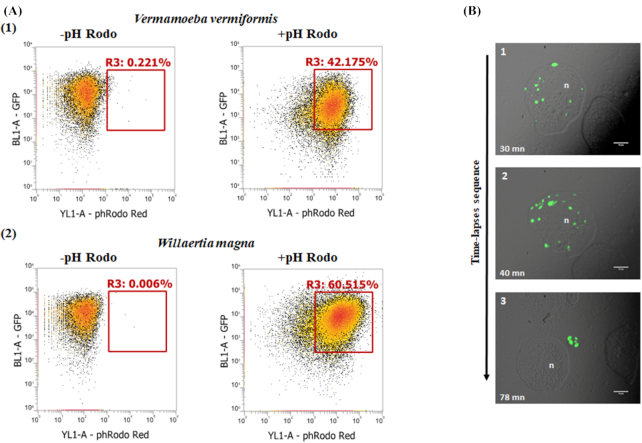
Figure 6 (A1 and A2) shows the co-culture samples of GFP H. pylori-amoebae incubated with pHrodo Red becoming increasingly fluorescent over time. There was a clear distinction between positive and negative pHrodo, indicating the presence of GFP-H. pylori in acidified amoeba phagosomes. In a separate experiment, we tracked the intracellular pH of W. magna after infection with a non-GFP H. pylori using a green intracellular pH indicator (pHrodo green). As seen in Fig. 6B1 and after 30 min of incubation, the formation of acidified phagosomes followed ingestion by W. magna. Our observations suggest that H. pylori adapt the acidified phagosomal compartment and resist digestion by free-living amoebae.
Figure 6.
Amoebal intracellular pH analysis. (A) Amoebae infected with GFP-H. pylori for 2 h at 37⁰C, analysis of intracellular pH dynamics evaluated by flow cytometry. The fluorescence percentage represents the occurrence of acidic phagosomes with or without treatment with red fluorescent intracellular pH indicator (pHrodo Red); (1) V. vermiformis (2) W. magna. (B) Green fluorescent intracellular pH indicator in W. magna infected with non GFP-H. pylori and treated with pHrodo Green. Time-lapse fluorescent micrographs representing: Panel (1) and (2) acidic phagosomes formation; and (3) release of phagosome containing H. pylori from trophozoite after 78 min; n, nucleus; (100x objective).
Culturability recovering of VBNC H. pylori cells
Helicobacter pylori is well known to exist in two morphologies in the culturable and nonculturable states; spiral and coccoid forms (Nilius et al. 1993; Benaissa et al. 1996; Willen et al. 2000; Young, Allaker and Hardie 2001; Saito et al. 2003), the coccoid form has also been described for a VBNC form of H. pylori (Nilius et al. 1993; Vijayakumari et al. 1995; Mizoguchi, Fujioka and Nasu 1999). Morphological change to the coccoid form appears to be a survival strategy that allows VBNC cells to persist under harsh environmental conditions (Mizunoe et al. 2000; Oliver 2010; Zeng et al. 2013; Zhao et al. 2013).
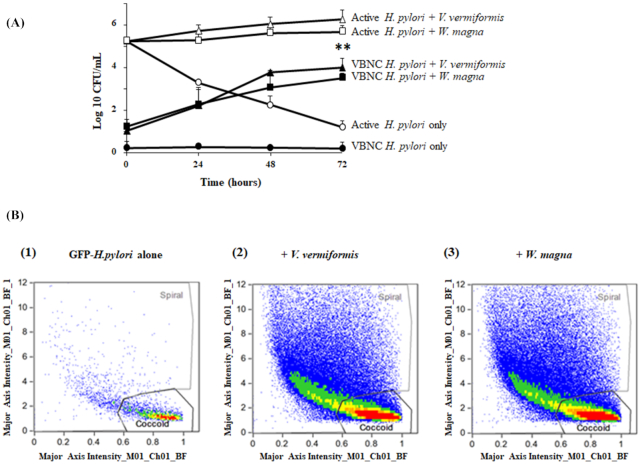
As we have previously observed (Fig. 1C), H. pylori lost almost all culturability after 3 days at 25⁰C in the absence of amoeba host but the cells persisted and were detectable by qPCR. This discrepancy between the two methods could be explained by the fact that qPCR is able to detect viable, dead and VBNC cells, while the culture method detects only the living culturable bacteria. Interestingly, it has been previously reported that amoebae are able to reactivate VBNC bacteria into a viable pathogenic state (Steinert et al. 1997; Garcia et al. 2007; Dey et al. 2019). We thus investigated VBNC-H. pylori reactivation in co-culture with amoebae using culture and imaging flow cytometry. As shown in Fig. 7A, VBNC cells cocultured with V. vermiformis and W. magna trophozoites appeared to revert to a culturable state, and that coculture with amoebae led to over 3-log10 increase (P < 0.001) of culturable H. pylori after 48 h compared to VBNC H. pylori alone.
Figure 7.
VBNC H. pylori recovery analysis. (A) Growth kinetics of active or VBNC H. pylori with V. vermiformis (open and closed triangles) and W. magna (open and closed squares) or without amoebae (open and closed circles respectively) by CFU counts at 25°C for up to 72 h. Data are the mean ± SEM, n = 3 (biological repeats) performed in triplicate. Statistical differences by Student's t-test comparing the indicated strains to the control in the absence of amoeba after 72 h (**P < 0.001). (B) ImageStream flow cytometry analysis of the VBNC GFP-H. pylori reactivation induced by adding amoebae. Percentage of coccoid and spiral shape bacteria after 30 mn incubation: without amoebae (1), with V. vermiformis (2) and with W. magna (3).
Morphological changes between spiral and VBNC H. pylori were detected by using Imaging flow cytometry (Supplementary data SD 6). After 2 h of incubation time, the VBNC H. pylori-amoebae co-culture was treated with gentamicin and then cultured for another 30 min, at which time the bacterial population analyzed by IFC. In Fig. 7B–D and based on bacterial morphology and size, the co-cultures with V. vermiformis and W. magna resulted in a significant increase in spiral-shape GFP-H. pylori (50%) when compared with GFP-H. pylori alone (26%). Taken together, the recovery from VBNC cells by co-incubation with amoebae was associated with a morphological change from coccoid to spiral-shape cells as described by IFC. Hence we demonstrate for the first time the ability of free-living amoebae to revert VBNC-H. pylori cells into a viable spiral form. Thus, FLA may also be a promising tool for the enrichment of environmentally-stressed H. pylori, particularly from water samples with low concentrations, as used for other amoeba-resisting bacterial pathogen recovery from waters (Corsaro et al. 2010).
Our study demonstrates that H. pylori is likely to replicate and survive in FLA under conditions that simulate aquatic environments in which H. pylori has been previously detected by PCR (Linke et al. 2010; Moreno and Ferrus 2012; El-Sharouny, El-Shazli and Olama 2015). Prospectively, FLA could be a promising cell model for the identification of the genes encoding the proteins involved in the adaptation of helicobacters to withstand digestion within acidic phagosomes. We also report evidence that FLA may act as a biological mediator of activation of dormant, VBNC H. pylori. Our findings also highlight and support the hypothesis put forth by Chatton over 100 years ago that amoebae could serve as vectors (Chatton and Lalung-Bonnaire 1912), and in this case sustain water-associated H. pylori transmission. Further studies are necessary in order to examine the transmissibility of VBNC and spiral H. pylori contained within FLA (trophozoites and/or vesicles), which would help inform future risk assessments of H. pylori in water.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Monika Keelan (University of Alberta) and John Kao (University of Michigan) for supplying the different Helicobacter strains used in this study. We are also very grateful for the technical support provided by Sieren Wang, Candis Scott and Homun Yee. Flow cytometry was performed at the University of Alberta, Faculty of Medicine and Dentistry Flow Cytometry Facility, which receives financial support from the Faculty of Medicine and Dentistry and Canadian Foundation for Innovation (CFI) awards to contributing investigators. Aja Rieger is an ISAC Shared Resource Lab Emerging Leader (2017–2021).
Contributor Information
Rafik Dey, School of Public Health, University of Alberta,11405-87 Avenue, Edmonton, Alberta T6G 1C9, Canada; Deparment of Medical Microbiology and Immunology, University of Alberta, Edmonton, Alberta T6G 2E1, Canada.
Aja Rieger, Deparment of Medical Microbiology and Immunology, University of Alberta, Edmonton, Alberta T6G 2E1, Canada.
Graham Banting, School of Public Health, University of Alberta,11405-87 Avenue, Edmonton, Alberta T6G 1C9, Canada.
Nicholas J Ashbolt, School of Public Health, University of Alberta,11405-87 Avenue, Edmonton, Alberta T6G 1C9, Canada; Deparment of Medical Microbiology and Immunology, University of Alberta, Edmonton, Alberta T6G 2E1, Canada; Provincial Laboratory for Public Health (ProvLab), Alberta Health Services, Edmonton, Canada; School of Environmental, Sciense and Engineering, Southern Cross University, Lismore NSW, Australia.
FUNDING
These studies were supported by Alberta Innovates (grant number 201300490), Alberta, Canada.
Conflicts of interest
None declared.
REFERENCES
- Allen LA. Modulating phagocyte activation: the pros and cons of Helicobacter pylori virulence factors. J Exp Med. 2000;191:1451–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen LA. Phagocytosis and persistence of Helicobacter pylori. Cell Microbiol. 2007;9:817–28. [DOI] [PubMed] [Google Scholar]
- Asakura H, Ishiwa A, Arakawa E et al. Gene expression profile of Vibrio cholerae in the cold stress-induced viable but non-culturable state. Environ Microbiol. 2007;9:869–79. [DOI] [PubMed] [Google Scholar]
- Ashbolt NJ. Microbial contamination of drinking water and human health from community water systems. Curr Environ Health Rep. 2015;2:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Khalifa MM, Sharaf RR. Contaminated water as a source of Helicobacter pylori infection:A review. J Adv Res. 2015;6:539–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TC, Adams B, Oliver JD. Survival of VBNC Helicobacter pylori cocci following antibiotic treatment. International Conference on Helicobacter pylori, Aarhus, Denmark, 2003. [Google Scholar]
- Bats SH, Berge C, Coombs N et al. Biochemical characterization of the Helicobacter pylori Cag Type 4 Secretion System protein CagN and its interaction partner CagM. Int J Med Microbiol. 2018;308:425–37. [DOI] [PubMed] [Google Scholar]
- Benaissa M, Babin P, Quellard N et al. Changes in Helicobacter pylori ultrastructure and antigens during conversion from the bacillary to the coccoid form. Infect Immun. 1996;64:2331–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk SG, Faulkner G, Garduno E et al. Packaging of live Legionella pneumophila into pellets expelled by Tetrahymena spp. does not require bacterial replication and depends on a Dot/Icm-mediated survival mechanism. Appl Environ Microbiol. 2008;74:2187–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk SG, Ting RS, Turner GW et al. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl Environ Microbiol. 1998;64:279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ. Ecology of Helicobacter pylori in the human stomach. J Clin Invest. 1997;100:759–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyanugomol W, Chomvarin C, Hahnvajanawong C et al. Helicobacter pylori cag pathogenicity island (cagPAI) involved in bacterial internalization and IL-8 induced responses via NOD1- and MyD88-dependent mechanisms in human biliary epithelial cells. PLoS One. 2013;8:e77358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boquet P, Ricci V. Intoxication strategy of Helicobacter pylori VacA toxin. Trends Microbiol. 2012;20:165–74. [DOI] [PubMed] [Google Scholar]
- Bouyer S, Imbert C, Rodier MH et al. Long-term survival of Legionella pneumophila associated with Acanthamoeba castellanii vesicles. Environ Microbiol. 2007;9:1341–4. [DOI] [PubMed] [Google Scholar]
- Bradbury RS. Free-living amoebae recovered from human stool samples in Strongyloides agar culture. J Clin Microbiol. 2014;52:699–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell TN, Choy FYM. The Effect of pH on green fluorescent protein: a Brief Review. Molecular Biology Today. 2001;2:1–4. [Google Scholar]
- Chatton E, Lalung-Bonnaire P. Amibe limax (Vahlkampfia n. gen.) dans l'intestin humaine. Son importance pour l'interprétation des amibes de culture. Bull Soc path exot. 1912;5:135–43. [Google Scholar]
- Corsaro D, Pages GS, Catalan V et al. Biodiversity of amoebae and amoeba-associated bacteria in water treatment plants. Int J Hyg Environ Health. 2010;213:158–66. [DOI] [PubMed] [Google Scholar]
- Delafont V, Rodier MH, Maisonneuve E et al. Vermamoeba vermiformis: a Free-Living Amoeba of Interest. Microb Ecol. 2018;76:991–1001. [DOI] [PubMed] [Google Scholar]
- de Moura H, Salazar HC, Fernandes O et al. [Free-living amoeba in the human intestine. Evidences of parasitism]. Rev Inst Med Trop Sao Paulo. 1985;27:150–6. [DOI] [PubMed] [Google Scholar]
- Dey R, Bodennec J, Mameri MO et al. Free-living freshwater amoebae differ in their susceptibility to the pathogenic bacterium Legionella pneumophila. FEMS Microbiol Lett. 2009;290:10–17. [DOI] [PubMed] [Google Scholar]
- Dey R, Hoffman PS, Glomski IJ. Germination and amplification of anthrax spores by soil-dwelling amoebas. Appl Environ Microbiol. 2012;78:8075–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey R, Rieger AM, Stephens C et al. Interactions of Pseudomonas aeruginosa with Acanthamoeba polyphaga Observed by Imaging Flow Cytometry. Cytometry A. 2019;95:555–64. [DOI] [PubMed] [Google Scholar]
- Djekic A, Muller A. The immunomodulator VacA promotes immune tolerance and persistent Helicobacter pylori infection through its activities on T-cells and antigen-presenting cells. Toxins (Basel). 2016;8:pii: E187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sharouny E, El-Shazli H, Olama Z. Detection of Helicobacter pylori DNA in Some Egyptian water systems and its incidence of transmission to individuals. Iran J Public Health. 2015;44:203–10. [PMC free article] [PubMed] [Google Scholar]
- Fischer W, Puls J, Buhrdorf R et al. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol. 2001;42:1337–48. [DOI] [PubMed] [Google Scholar]
- Gangwer KA, Shaffer CL, Suerbaum S et al. Molecular evolution of the Helicobacter pylori vacuolating toxin gene vacA. J Bacteriol. 2010;192:6126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MT, Jones S, Pelaz C et al. Acanthamoeba polyphaga resuscitates viable non-culturable Legionella pneumophila after disinfection. Environ Microbiol. 2007;9:1267–77. [DOI] [PubMed] [Google Scholar]
- Giao MS, Azevedo NF, Wilks SA et al. Interaction of Legionella pneumophila and Helicobacter pylori with bacterial species isolated from drinking water biofilms. BMC Microbiol. 2011;11:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadin JB. [Hypotheses on the adaptation of amoebas of the limax group to man and animals]. Ann Parasitol Hum Comp. 1973;48:199–204. [PubMed] [Google Scholar]
- Jiang X, Doyle MP. Effect of environmental and substrate factors on survival and growth of Helicobacter pylori. J Food Prot. 1998;61:929–33. [DOI] [PubMed] [Google Scholar]
- Jiang X, Doyle MP. Growth supplements for Helicobacter pylori. J Clin Microbiol. 2000;38:1984–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavermann H, Burns BP, Angermuller K et al. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J Exp Med. 2003;197:813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CH, Shotts EB Jr., Wooley RE et al. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl Environ Microbiol. 1988;54:3023–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke S, Lenz J, Gemein S et al. Detection of Helicobacter pylori in biofilms by real-time PCR. Int J Hyg Environ Health. 2010;213:176–82. [DOI] [PubMed] [Google Scholar]
- Loke MF, Ng CG, Vilashni Y et al. Understanding the dimorphic lifestyles of human gastric pathogen Helicobacter pylori using the SWATH-based proteomics approach. Sci Rep. 2016;6:26784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez AJ, Visvesvara GS. Free-living, amphizoic and opportunistic amebas. Brain Pathol. 1997;7:583–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurin M, Bryskier A, Raoult D. Antibiotic susceptibilities of Parachlamydia acanthamoeba in amoebae. Antimicrob Agents Chemother. 2002;46:3065–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi H, Fujioka T, Nasu M. Evidence for viability of coccoid forms of Helicobacter pylori. J Gastroenterol. 1999;34:32–36. [PubMed] [Google Scholar]
- Mizunoe Y, Wai SN, Ishikawa T et al. Resuscitation of viable but nonculturable cells of Vibrio parahaemolyticus induced at low temperature under starvation. FEMS Microbiol Lett. 2000;186:115–20. [DOI] [PubMed] [Google Scholar]
- Molmeret M, Horn M, Wagner M et al. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol. 2005;71:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mesonero L, Moreno Y, Alonso JL et al. Detection of viable Helicobacter pylori inside free-living amoebae in wastewater and drinking water samples from Eastern Spain. Environ Microbiol. 2017;19:4103–12. [DOI] [PubMed] [Google Scholar]
- Moreno-Mesonero L, Moreno Y, Alonso JL et al. DVC-FISH and PMA-qPCR techniques to assess the survival of Helicobacter pylori inside Acanthamoeba castellanii. Res Microbiol. 2016;167:29–34. [DOI] [PubMed] [Google Scholar]
- Moreno Y, Ferrus MA. Specific detection of cultivable Helicobacter pylori cells from wastewater treatment plants. Helicobacter. 2012;17:327–32. [DOI] [PubMed] [Google Scholar]
- Nilius M, Strohle A, Bode G et al. Coccoid like forms (CLF) of Helicobacter pylori. Enzyme activity and antigenicity. Zentralbl Bakteriol. 1993;280:259–72. [DOI] [PubMed] [Google Scholar]
- Nilsson HO, Blom J, Abu-Al-Soud W et al. Effect of cold starvation, acid stress, and nutrients on metabolic activity of Helicobacter pylori. Appl Environ Microbiol. 2002;68:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JD. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev. 2010;34:415–25. [DOI] [PubMed] [Google Scholar]
- Page FC. Taxonomic criteria for limax amoebae, with descriptions of 3 new species of Hartmannella and 3 of Vahlkampfia. J Protozool. 1967;14:499–521. [DOI] [PubMed] [Google Scholar]
- Ricci V. Relationship between VacA toxin and host cell autophagy in Helicobacter pylori infection of the human stomach: a few answers, many questions. Toxins (Basel). 2016;8:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TM, Rudolf F, Meyer A et al. Identification and Characterisation of a pH-stable GFP. Sci Rep. 2016;6:28166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Zaragoza S. Ecology of free-living amoebae. Crit Rev Microbiol. 1994;20:225–41. [DOI] [PubMed] [Google Scholar]
- Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol. 1980;33:1179–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito N, Konishi K, Sato F et al. Plural transformation-processes from spiral to coccoid Helicobacter pylori and its viability. J Infect. 2003;46:49–55. [DOI] [PubMed] [Google Scholar]
- Samba-Louaka A, Delafont V, Rodier MH et al. Free-living amoebae and squatters in the wild: ecological and molecular features. FEMS Microbiol Rev. 2019;43:415–34. [DOI] [PubMed] [Google Scholar]
- Santiago P, Moreno Y, Ferrus MA. Identification of viable Helicobacter pylori in drinking water supplies by cultural and molecular techniques. Helicobacter. 2015;20:252–9. [DOI] [PubMed] [Google Scholar]
- Sarem M, Corti R. Role of Helicobacter pylori coccoid forms in infection and recrudescence. Gastroenterol Hepatol. 2016;39:28–35. [DOI] [PubMed] [Google Scholar]
- Scott DR, Marcus EA, Wen Y et al. Gene expression in vivo shows that Helicobacter pylori colonizes an acidic niche on the gastric surface. Proc Natl Acad Sci USA. 2007;104:7235–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoh M, Ghosh-Banerjee J, Ramamurthy T et al. Conversion of viable but nonculturable enteric bacteria to culturable by co-culture with eukaryotic cells. Microbiol Immunol. 2012;56:342–5. [DOI] [PubMed] [Google Scholar]
- Shaheen M, Ashbolt NJ. Free-Living Amoebae supporting intracellular growth may produce vesicle-bound respirable doses of Legionella within drinking water systems. Exposure and Health. 2018;10:201–9. [Google Scholar]
- Shariq M, Kumar N, Kumari R et al. Biochemical Analysis of CagE: A VirB4 Homologue of Helicobacter pylori Cag-T4SS. PLoS One. 2015;10:e0142606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD, Ashbolt NJ. The fate of Helicobacter pylori phagocytized by Acanthamoeba polyphaga demonstrated by fluorescent in situ hybridization and quantitative polymerization chain reaction tests. Curr Microbiol. 2012;65:805–12. [DOI] [PubMed] [Google Scholar]
- Steele KE, Visvesvera GS, Bradley GA et al. Amebiasis in a dog with gastric ulcers and adenocarcinoma. J Vet Diagn Invest. 1997;9:91–93. [DOI] [PubMed] [Google Scholar]
- Steinert M, Emody L, Amann R et al. Resuscitation of viable but nonculturable Legionella pneumophila Philadelphia JR32 by Acanthamoeba castellanii. Appl Environ Microbiol. 1997;63:2047–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Sun F, Wang Y et al. Identification, characterization and molecular analysis of the viable but nonculturable Rhodococcus biphenylivorans. Sci Rep. 2015;5:18590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer N, Wessler S, Backert S. Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 2011;278:1190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer N, Wessler S, Necchi V et al. Helicobacter pylori employs a unique basolateral type IV secretion mechanism for CagA delivery. Cell Host Microbe. 2017;22:552–60. e555. [DOI] [PubMed] [Google Scholar]
- Thamprasert K, Khunamornpong S, Morakote N. Acanthamoeba infection of peptic ulcer. Ann Trop Med Parasitol. 1993;87:403–5. [DOI] [PubMed] [Google Scholar]
- Thomas JM, Ashbolt NJ. Do free-living amoebae in treated drinking water systems present an emerging health risk? Environ Sci Technol. 2011;45:860–9. [DOI] [PubMed] [Google Scholar]
- Thomas V, McDonnell G, Denyer SP et al. Free-living amoebae and their intracellular pathogenic microorganisms: risks for water quality. FEMS Microbiol Rev. 2010;34:231–59. [DOI] [PubMed] [Google Scholar]
- Vijayakumari S, Khin MM, Jiang B et al. The pathogenic role of the coccoid form of Helicobacter pylori. Cytobios. 1995;82:251–60. [PubMed] [Google Scholar]
- Wen Y, Marcus EA, Matrubutham U et al. Acid-adaptive genes of Helicobacter pylori. Infect Immun. 2003;71:5921–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willen R, Carlen B, Wang X et al. Morphologic conversion of Helicobacter pylori from spiral to coccoid form. Scanning (SEM) and transmission electron microscopy (TEM) suggest viability. Ups J Med Sci. 2000;105:31–40. [DOI] [PubMed] [Google Scholar]
- Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nature. 2017;543:15. [DOI] [PubMed] [Google Scholar]
- Winiecka-Krusnell J, Wreiber K, von Euler A et al. Free-living amoebae promote growth and survival of Helicobacter pylori. Scand J Infect Dis. 2002;34:253–6. [DOI] [PubMed] [Google Scholar]
- Worku ML, Sidebotham RL, Walker MM et al. The relationship between Helicobacter pylori motility, morphology and phase of growth: implications for gastric colonization and pathology. Microbiology. 1999;145:2803–11. [DOI] [PubMed] [Google Scholar]
- Young KA, Allaker RP, Hardie JM. Morphological analysis of Helicobacter pylori from gastric biopsies and dental plaque by scanning electron microscopy. Oral Microbiol Immunol. 2001;16:178–81. [DOI] [PubMed] [Google Scholar]
- Zeng B, Zhao G, Cao X et al. Formation and resuscitation of viable but nonculturable Salmonella typhi. Biomed Res Int. 2013;2013:907170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Bi X, Hao Y et al. Induction of viable but nonculturable Escherichia coli O157:H7 by high pressure CO2 and its characteristics. PLoS One. 2013;8:e62388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.