Abstract
The physiological processes of cells and organisms are regulated by various biological macromolecules, including long-noncoding RNAs (lncRNAs), which cannot be translated into protein and are different from small-noncoding RNAs on their length. In animals, lncRNAs are involved in development, metabolism, reproduction, aging and other life events by cis or trans effects. For many functional lncRNAs, there is growing evidence that they play different roles on cellular level and organismal level. On the other hand, many annotated lncRNAs are not essential and could be transcription noises. In this minireview, we investigate the physiological function of lncRNAs in cells and focus on their functions and functional mechanisms on the organismal level. The studies on lncRNAs using different classic animal models such as worms and flies are summarized and discussed in this article.
Keywords: Long noncoding RNA, LincRNA, CircRNA, CRISPR, Phenotype, Physiological function
1. Introduction
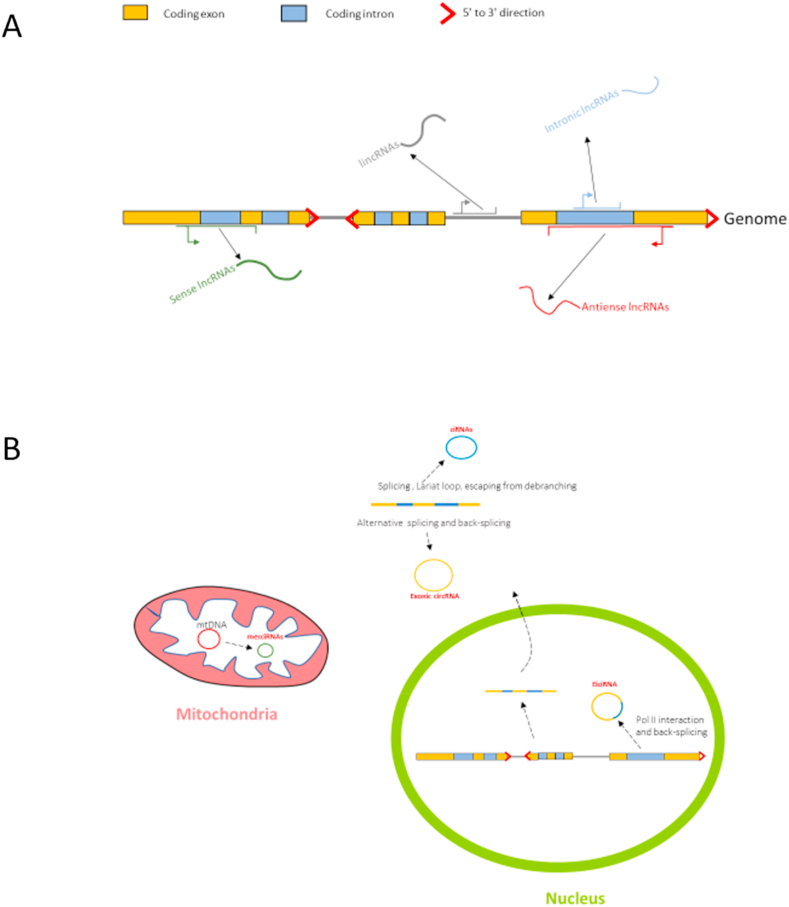
Recent studies illustrate varieties of noncoding RNAs which lack ORFs and cannot translate. Noncoding RNAs can be classified according to their location, function, size or secondary structure [1,2]. The noncoding RNAs with the length of hundreds (200) nt or more are long(large) noncoding RNAs (LncRNAs) [3,4]. LncRNAs can be linear or circular, distributed in the nucleus, cytoplasm and mitochondria [[5], [6], [7], [8]](Fig. 1). According to the distribution of lncRNAs in the genome, relative positions with nearby coding genes and transcription directions, they can be divided into four groups: long intervening/intergenic noncoding RNAs(lincRNAs), intronic lncRNAs, sense lncRNAs and antisense lncRNAs [3]. According to their functions, lncRNAs can be roughly divided into: functional lncRNAs, whose transcripts can regulate genes expression in cis or in trans; lncRNAs play their roles during transcription, but their transcripts have no functions; no functions lncRNAs which might be transcription noises [[9], [10], [11]].
Fig. 1.
Types of long-noncoding RNA(linear and circular). (A) 4 groups of lncRNA according to the distribution of lncRNAs in the genome, relative positions with nearby coding genes and transcription directions. (B) 4 groups of circRNA in cytoplasm, nucleus and mitochondria.
In cells, lncRNAs which located in the nucleus or cytoplasm interact with DNAs, proteins or other RNAs. They involve in the cell's proliferation, differentiation and apoptosis [12]. In animals, lncRNAs play their functional roles in development, reproduction, aging and disease [[12], [13], [14]]. Interestingly, in some cases, the key roles of lncRNAs in cells do not match their importance at the whole-organism scale, for example, knockout almost all lincRNAs one by one in nematodes did not exhibit critical phenotypes [15]. In mammals, a widely studied long noncoding RNA, Hotair regulates epidermal cell differentiation and interacts with epigenetic factors such as PRC2 to participate in tumor metastasis [16,17]. Although Hotair plays a key role in cultural cells, one study suggested that Hotair were dispensable for whole animals [18]. Hotair knockout mice did not exhibit the expecting phenotypes suggesting they are not necessary for mouse development and embryonic survival [18]. The same also occurred in the study of Malat1 [[19], [20], [21], [22]].With the advances of technology, especially CPRISPR gene editing and Next-generation sequencing, a growing body of lncRNA researches are conducted at the whole-organism scale. In this review, we summarize the physiological functions and mechanisms of lncRNAs in cells and animal models.
2. Cellular physiological functions of LncRNAs
2.1. lincRNAs
Long intervening/intergenic noncoding RNAs(lincRNAs) are lncRNAs located between coding genes and have no overlap with any annotated protein-coding sequences. LincRNAs exercise physiological functions in cells such as carcinogenesis, infection and inflammation. For example, NEAT (Nuclear Enriched Abundant Transcript) RNAs including Neat1 and Malat1(also named as Neat2), are classic examples of lincRNAs in mammalian cells and are involved in carcinogenesis [[23], [24], [25], [26], [27]]. They are conserved across various mammals and locate in the nuclear and participate in many biological processes such as paraspeckle formation, cell cycle regulation, alternative splicing and cancer cells migration [28,29]. Another extensively studied lincRNA Hotair which located in HOXC gene cluster, is a highly expressed gene in metastatic breast cancers [30,31]. Hotair interacts with Polycomb-group proteins and reprogram chromatin state in trans [32,33]. LincRNA-p21 activated by tumor suppressor P53, is another important transcriptional repressor that binds to the hnRNP family protein hnRNP-K after DNA damage and participates in maintaining of the P53 induced genome stability [34]. Not only lincRNAs could be taken as regulators and biomarkers in tumor cells, but also functional molecules for cellular physiology. For instance, lincRNAs with elevated expression patterns in iPS and ES cells suggest their functions in establishment and maintenance of pluripotency [35,36]. In pluripotent stem cells, lincRNA-RoR modulates reprogramming as the direct targets of key transcription factors [35]. In other studies, lincRNAs played their roles in immunomodulation such as lincRNA-EPS. It acts as an important regulator of immune response genes in immune cells in trans [37].
2.2. Circular RNAs
Not only linear but also circular long noncoding RNAs modulate cellular physiological processes [[38], [39], [40]]. Circular RNAs are generated by back-splicing from precursor mRNAs and displayed special expression pattern in tissues and developmental stages [[41], [42], [43], [44]]. In general, circular RNAs are roughly divided into four categories: Exonic circRNA, circular intronic RNAs (ciRNAs), exon–intron circRNAs (EiciRNAs) and mitochondria-encoded circRNA(mecciRNAs) [7,8,45,46]. Circular RNAs can function as microRNA sponges and regulate genes expression in trans [47,48]. For example, CDR1as(ciRS-7) can bind miR-7 and miR-671 to regulate the expression of their target genes and functions in cellular proliferation and apoptosis [[49], [50], [51]]. CDR1as knockout mice displayed abnormal brain function due to the defect of synaptic neurotransmission [52]. CDR1as is also a regulator of insulin secretion and oncogene [53,54]. On the other hand, Circular RNAs can act as protein sponge, decoy or scaffold [55,56]. For example, Cia-cGAS acts as nuclear cGAS sponge to block its enzymatic activity in hematopoietic stem cells to protect their homeostasis [55]. In another research, circ-Foxo3 constructs circ-Foxo3-p21-CDK2 complex to block cell cycle progression by suppressing CDK2 [56]. Circular RNAs are also able to function in cis. Take EiciRNAs as an example, which are circularized with introns “retained” between their exons [7]. EiciRNAs such as circEIF3J and circPAIP2 can hold U1 snRNP by specific RNA-RNA interaction, then the complexes further interact with the Pol II at the promoters of parental genes to enhance their expression level and arise a positive feedback in genes expression [7]. In addition to the nuclear genome, the circular RNAs encoded by mitochondrial genome which are termed as mecciRNAs also have important roles [8].The mecciRNAs promote mitochondrial importation of nuclear-encoded proteins, by interacting with TOM40 and PNPASE serving as molecular chaperones [8]. Dynamic expression of mecciND1 under stress regulated cellular physiology by increasing the RPA70 and RPA32 protein levels in mitochondria [8].
2.3. Other LncRNAs
In addition to lincRNAs, there are other forms of lncRNAs which modulate cellular physiological functions, such as antisense lncRNAs and long intronic noncoding RNAs [[57], [58], [59], [60], [61]]. For example, nuclear-enriched AS Uchl RNA in dopaminergic neurons upregulates UCHL1 protein levels via the SINEB2 repeat element [57]. The inhibition of mTORC1 by rapamycin increased UCHL1 protein levels by AS Uchl implied a mechanism of antisense lncRNAs in the control of cellular stress signaling pathways and their roles in neurodegenerative diseases [57]. In other researches, long intronic transcripts take their roles as precursors of small RNAs, cofactors of alternative promoters and regulators of alternative pre-mRNA splicing [58]. Take SAF as an example, a 1500 nt intronic lncRNA transcribed from the opposite strand of FAS gene intron 1 regulates the alternative splicing of FAS in cis to protect cells from membrane-mediated apoptosis [59]. In addition, overlapped transcripts are also involved in cellular events such as 5S-OT(5S rRNA overlapped transcript) [60]. 5S-OT modulates 5S rRNA transcription in mice and humans by cis effect, and it is intriguing that human 5S-OT regulates alternative splicing of numerous genes by U2AF65 and Alu pairing in trans [60]. In a classic human macrophage differentiation model, knockdown of 5S-OT decreased THP-1 cells differentiation efficiency [60]. LncRNAs also participate in cell division, for example, 171 nt α-satellite RNAs which are transcribed from centromeric repeats are managed by a RNAi pathway and function in chromosome segregation [61].
3. The physiological functions of LncRNA in animals
3.1. Caenorhabditis elegans
As simplest one of animal models, Caenorhabditis elegans has many advantages in genetics and molecular biology researches [[62], [63], [64], [65]]. Using C. elegans, researchers found rncs-1, an 800 nt lincRNA(long intervening noncoding RNAs) is up-regulated after starvation which is expressed in intestine and hypodermis, and inhibits Dicer cleavage in vitro and in vivo [66].Overexpression of rncs-1 led to an increased frequency of males during starvation indicated its functional roles of lncRNAs in response to stress [66]. LncRNAs can regulate development and sexual maturation. Lep-5, a 600 nt cytoplasmic lincRNA, regulates developmental timing as a scaffold to bring LEP-2 into its target gene LIN-28, and takes part in tail tip morphogenesis of males regulating sexual maturation cell-autonomously in nervous system [67,68]. Lep-5 is conserved across Caenorhabditis uncovering evidence for its function in evolution [67,68].
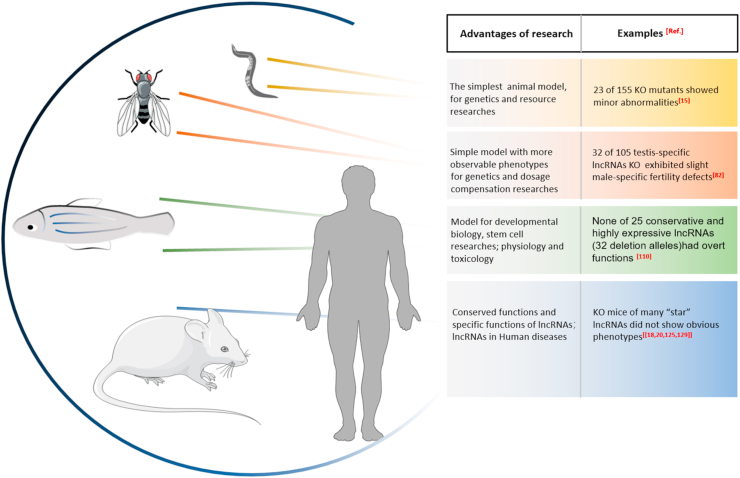
As model organism which can be handled easily, C. elegans is used in resource research [69]. Using available RNA-seq and other techniques, 170 lincRNAs and 60 ancRNAs(antisense lncRNAs) were identified in C. elegans [70]. LincRNAs of C. elegans are expressed in a stage-specific manner, and many of them are dauer stage-specific or sperm-specific molecules [15,70]. To investigate their spatiotemporal expression, transgenic reporter strains and RNA-seq were generated showing that the expression patterns of lincRNAs are more specific and heterogeneous than transcription factors [71]. LincRNAs of C. elegans can be detected in different developmental stages and tissues including intestine cells, hypodermal cells, muscles and neurons [71]. Using CPRISPR knockout strains, the functions of lincRNAs in C. elegans were systematically evaluated by our group, several representative phenotypes were tested in these lincRNAs KO animals, and the global features such as their exon numbers, conservation, and length were described [15]. 23 of 155 KO mutants showed minor abnormalities in locomotion, defecation, pharyngeal pumping, egg retention, development and offspring numbers. Mechanistically, some of these lincRNAs played cis roles to regulate the expression neighboring genes, some of them could function as ceRNAs against microRNAs in trans. By bioinformatics analysis from ChIP-seq datasets (modENCODE), the 23 phenotypic lincRNAs are regulated by more transcription factors than the others indicating that lincRNAs are targets of TFs in neurons to control their function directly [15,72,73].
3.2. Drosophila melanogaster
Like C. elegans, Drosophila melanogaster is also a kind of classic and simple animal model but have more observable phenotypes in genetics [[74], [75], [76], [77]]. Drosophila is taken as a research platform to investigate the in vivo functions of noncoding RNAs for decades [[78], [79], [80], [81]]. Transcription of many Drosophila lncRNAs occurs during embryogenesis and display spatiotemporally expression [78]. As a resource study, Wen et al. identified 128 testis-specific lncRNAs in which 105 of them were knocked out by CRISPR. Among the KO mutants, only 33 (31%) exhibited male-specific fertility defects most of them (32) just have partially decreased male fertility [82].
One of the important functions of lncRNAs is to regulate the chromatin state. LncRNAs involving in the X chromosome dosage compensation were elucidated in the studies of Drosophila [83]. RoX1 and roX2 genes produce male-specific lncRNAs that co-localize with the MSL (Male-Specific Lethal) protein complex. They form a stable association with the protein complex and activate the expression of X-linked genes in males to equalize genes expression between two sexes [83]. The ChIRP (Chromatin Isolation by RNA Purification) -seq analysis displayed the Drosophila roX genes binding sites on X chromosome directly [84]. Interestingly, both roX1 and roX2 are non-essential. Deletion roX1 or roX2 in both sexes had no significant phenotypes [83]. Males of roX double mutants were disrupted in development. Males carrying roX - chromosomes were lethal and only 5% of them were survival. Although the double mutant showed the male-specific lethal phenotypes, the females of them were not affected, either roX1 or roX2 cDNAs could rescue the male-specific phenotypes of the double mutants [83]. At molecular level, roX1 and roX2 intact with some important proteins (MSL1-3, MOF, MLE) and form MSL complex to regulate epigenetic modification such as histone acetylation [85].
Evidence in Drosophila shows that lncRNAs participate in the cellar response to stress. For example, one of the heat shock proteins hsr-omega encoding a nuclear lncRNA, participates in the reorganization of nucleoplasmic omega speckles after heat shock [86]. It functions as a hub and accumulate hnRNPs. Hsr-omega nullisomic mutants resulted in embryonic lethality [[86], [87], [88]]. Recent studies also demonstrated the regulation of lncRNAs in neurogenesis and their molecular mechanism in flies [89,90]. Neurogenic lncRNAs are expressed specifically during early stages of nervous system development and mark specific subsets of neurogenic cell types including neurons and glia [89]. Another study indicated that lncRNAs controlled by Hox genes participated in the formation of anteroposterior (AP) axis of Drosophila. A 92k nt lncRNA encoded by the intergenic region isolating Abd-A and Abd-B was identified [90]. This CNS-specific lincRNA(iab8ncRNA) suppresses the expression of Abd-A genes by two redundant mechanisms: the first way is mediated by mir-iab-8, a microRNA encoded by the intronic sequence within iab8ncRNA; on the other hand, the transcriptional interference by iab8ncRNA on Abd-A promoter is involved in the regulation [90].In addition, lncRNAs act not only in the formation and function of nervous system but also in the behavior of Drosophila [81,91,92].For instance, the cytoplasmic yellow-achaete intergenic RNA (yar) which is conserved in Drosophila regulates the sleep behavior, the phenotypic rescue by a yar transgene suggests that it functions in trans [81].
3.3. Zebrafish
Compared to simple models such as nematodes and flies, Danio rerio(zebrafish) belongs to vertebrates and is closer to mammals. Zebrafish is one of the most classic model vertebrates [[93], [94], [95]]. It has many features such as ease of feeding and embryo transparency that make it an excellent model for research of developmental biology, stem cell research, physiology and toxicology [[96], [97], [98], [99]].LncRNAs of zebrafish were identified using RNA deep sequencing approaches in three independent studies resent years [100]. Ulitsky et al. annotated 567 lincRNAs, by using RNA-seq, ploy(A) mapping and chromatin marks. Among them, only 29 had putative mammalian orthologs, but most of them displayed tissue-specific expression [101]. Using MO (morpholino antisense oligos) knockdown protocol, two conserved lincRNAs exhibited functional roles and the MO resulted in embryonic defects: Linc-oip5 was required for the normal size of head, eyes and tail; linc-birc6 was required for brain and eyes development [101]. Pauli et al. performed RNA-seq experiments at 8 developmental time points of zebrafish and identified lncRNAs expressed during embryogenesis [102]. LncRNAs of zebrafish were expressed at lower levels but in narrower time windows compared with coding genes in early embryos and showed tissue-specific and subcellularly restricted expression patterns [102]. By RNA-seq of 5 different tissues from adult zebrafish, Kaushik et al. annotated 442 predicted lncRNAs with 419 were newly annotated [103]. 77 lncRNAs were tissue-specific and the adult brain enriched the most tissue-specific lncRNAs [103].
Some evidence suggests that not only lincRNAs regulate the development of zebrafish, but also antisense lncRNAs [102,104,105]. In zebrafish, an antisense lncRNA, tie-1AS which is expressed spatiotemporally can bind tie-1 mRNA selectively to form tie-1: tie-1AS hybrid to regulate tie-1 transcript levels [104]. Overexpression of tie-1AS led to defects in the formation of contact junctions in endothelial cells and abnormal vascular development. In addition, tie-1AS is conserved in humans and mice [104,106]. As vertebrates, zebrafish is used for a model to reveal the conserved functional lncRNAs in humans and their roles in diseases [[106], [107], [108]]. The roles of lncRNAs in the regulation of sexual reproduction and behavior were analyzed by Yuan et al. [109]. In the brain of zebrafish, there were numerous gender-specific lncRNAs like humans with 12 new lncRNAs were annotated [109]. Even though several lncRNAs may be critical and essential in fish, a more recently resource study using CRISPR KO mutants indicated that the majority of individual lncRNAs in zebrafish had no key roles, and the phenotypes of the KO mutants such as embryogenesis, viability and fertility had no overt abnormalities [110].
3.4. Mammals
The atlas of biological functions of several “star” lncRNAs is drawing both in vivo and in vitro, using both cultured cells and mammalian models such as mice and rats [[111], [112], [113], [114]]. Mus musculus(mouse) were usually utilized as a mammalian model in genetics and molecular biology for decades [115]. Mice and humans share more than 90% conserved regions in the genomes, but in the transcription level, lncRNAs are expressed at a lower level and less conservation in sequences comparing with coding genes. However, there are thousands of conserved orthologous lncRNAs [116]. The X-chromosome dosage in mammals is controlled by a long noncoding RNA, Xist [117,118]. It is similar to roX genes in Drosophila, but Xist effects in an opposite way: roX1 and roX2 activate X-linked genes in males, however Xist inactivate X-linked genes in females in embryonic development [[119], [120], [121]]. Xist RNA can coat and accumulate on one X chromosome (where it is expressed), recruit a series of epigenetic regulators then transcriptional silencing rapidly ensues [122,123]. Mutations of Xist in mice result in females embryonic lethal inheriting paternal allele but males without any phenotypes [124,125]. Another lncRNA which was discovered for several decades is H19, a 2.5 kb untranslated transcript from the distal region of chromosome 7 in mice [126]. It is expressed at a very high level in embryonic tissues including endoderm and mesoderm, and its expression level maintains during several days after birth then disappears in adult [117,118]. H19 is an imprinted and exclusive maternal origin allele gene [[127], [128], [129]]. The deletion of H19 in mice led to no obvious phenotypes except slightly increased growth in homozygous mutants [124,129]. For other widely studied lncRNAs, for example, Neat1 and Malat1 are globally expressed and have cellular functions, the mutant mice do not exhibit overt abnormalities except for the defects of paraspeckles [130,131]. In another study, the KO mice of Hotair were fertile and viable with slight skeletal abnormalities [132].
For the whole organism, there are several lncRNAs which are essential in mammals. In one study, the function of 18 mammalian lincRNAs candidates were evaluated by mice mutants [22]. 3(Fendrr, Peril, and Mdgt)of these were critical and the mutants displayed embryonic and postnatal lethal phenotypes. Fendrr and mdgt might have functions in multiple organs, and Peril might have functions in ESCs (embryonic stem cells) [22]. However, most of lncRNAs are not essential for their loss-of-function mutants are viable and fertile [22,124]. But on the other hand, they in turn participate in the regulation of many physiological and pathological processes [133,134]. LncRNAs are involved in pathogen infection [135,136]. For example, Peng et al. reported that the mice infected by SARS-CoV showed significant different expression of lncRNAs which were similar to influenza virus infection [135]. LncRNAs are also involved in the cellular responses to bacterial infection such as Sros1 which could sensitize mice to L. monocytogenes [136]. LncRNA also play their roles in cancers [137]. Malat-1 is named for its function in metastasis of lung cancer cells, deletion of malat-1 in mice impaired tumor cells metastasis. Malat-1 could also be taken as a predictive marker clinically [137]. LncRNAs are involved in metabolism [138]. For instance, lipid metabolism regulated by lncRNAs associates with obesity and hepatic steatosis, Muret et al. summarized 60 lncRNAs in mice and humans involved in lipid metabolism and their functions in diseases [139].Additionally, there is evidence that lncRNAs can be regulator in neuroregeneration suggesting their roles in neurodegenerative diseases [140]. Perry et al. reported the lncRNAs expressed during neuroregeneration in dorsal root ganglia of mice and found two key lncRNAs, Silc1 and Norris1 [141]. Silc1 regulated transcription factor Sox11 in cis, Silc1 KO mice displayed delayed regeneration following injury [141].
4. Summary
For cellular physiology, lncRNAs function in proliferation, differentiation, stress, aging, and apoptosis by epigenetic, transcriptional, and post-transcriptional regulation. They can be various forms and play trans and cis roles in cells [5,10]. The functions and functional mechanism of lncRNAs in physiology of animals are revealed using classic animal models [62,74,93]. Much evidence exhibits the differences of their effects between cellular levels and whole organism levels. Even though lncRNAs show their important roles in many biological processes, depletion of them impact fewer phenotypes than expected [124]. The resource research using C. elegans, Drosophila and zebrafish suggest that lncRNAs are not essential for whole animal in most cases [15,82,110]. Interestingly, the KO mice of many “star” lncRNAs do not show obvious phenotypes [18,20,125,129] (Fig. 2). However, lncRNAs may play their functional roles under particular physiological and pathological conditions, making them potential key molecule in organisms.
Fig. 2.
LncRNAs Researches in animal models. Advantages and typical cases of lncRNAs researches by different models are listed. In most cases, lncRNAs are not essential for whole animals.
Declaration of competing interest
All the authors declared that they have no conflicts of interest to this work. We declare that there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in, or the review of, the manuscript entitled.
Acknowledgement
This work was supported by grants to H. C.: the National Natural Science Foundation of China (31900442); G. S.: the National Key R&D Program of China (2019YFA0802600 and 2018YFC1004500), the National Natural Science Foundation of China (31725016, 31930019, and 91940303).
Contributor Information
He Chen, Email: hechen@ustc.edu.cn.
Ge Shan, Email: shange@ustc.edu.cn.
References
- 1.Mattick J.S., Makunin I.V. Non-coding RNA. Hum. Mol. Genet. 2006;15:17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 2.Eddy S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2(12):919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 3.Ma L., Bajic V.B., Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10(6):925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravasi T., Suzuki H., Pangetal K.C., Katayama S., Furuno M., Okunishi R. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16(1):11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponting C.P., Oliver L.P., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Jeffries M.A. Osteoarthritis year in review 2018: genetics and epigenetics. Osteoarthritis Cartilage. 2019;27(3):371–377. doi: 10.1016/j.joca.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z., Huang C., Bao C., Chen L., Lin M., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2005;22(3):256–364. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 8.Liu X., Wang X., Li J., Hu S., Deng Y., Yin H. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci. China Life Sci. 2020 doi: 10.1007/s11427-020-1631-9. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 9.Clark M.B., Mercer T.R., Bussotti G., Leonardi T., Haynes K.R. Quantitative gene profiling of long noncoding RNAs with targeted RNA sequencing. Nat. Methods. 2015;12(4):339–342. doi: 10.1038/nmeth.3321. [DOI] [PubMed] [Google Scholar]
- 10.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 11.Ponjavic J., Ponting C.P., Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17(5):556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flynn R.A., Chang H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14(6):752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei S., Chen H., Dzakah E.E., Yu B., Wang X., Fu T. Systematic evaluation of C. elegans lincRNAs with CRISPR knockout mutants. Genome Biol. 2019;20(1):7. doi: 10.1186/s13059-018-1619-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amandio A.R., Necsulea A., Joye E., Mascrez B., Duboule D. Hotair is dispensable for mouse development. PLoS Genet. 2016;12(12) doi: 10.1371/journal.pgen.1006232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B., Arun G., Mao Y.S., Lazar Z., Hung G., Bhattacharjee G. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2(1):111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abulwerdi F.A., Xu W., Ageeli A.A., Yonkunas M.J., Arun G., Nam H. Selective small-molecule targeting of a triple helix encoded by the long noncoding RNA, MALAT1. ACS Chem. Biol. 2019;14(2):223–235. doi: 10.1021/acschembio.8b00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauvageau M., Goff L.A., Lodato S., Bonev B., Groff A.F., Gerhardinger C. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2 doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujimoto A., Furuta M., Totoki Y., Tsunoda T., Kato M., Shiraishi Y. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 2016;48(5):500–509. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 24.Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clemson C.M., Hutchinson J.N., Sara S.A., Ensminger A.W., Fox A.H., Chess A. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Standaert L., Adriaens C., Radaelli E., Van K.A., Blanpain C., Hirose T. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA. 2014;20(12):1844–1849. doi: 10.1261/rna.047332.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui Y., Li G., Zhang X., Dai F., Zhang R. Increased MALAT1 expression contributes to cisplatin resistance in non-small cell lung cancer. Oncol. Lett. 2018;16(4):4821–4828. doi: 10.3892/ol.2018.9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fox A.H., Lam Y.W., Leung A.K., Lyon C.E., Andersen J., Mann M. Paraspeckles: a novel nuclear domain. Curr. Biol. 2002;12(1):13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 29.Faulkner G.J., Kimura Y., Daub C.O., Wani S., Plessy C., Irvine K.M. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009;41(5):563–567. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 30.Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Canc. Res. 2011;71(20):6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 31.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schorderet P., Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7(5) doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hung T., Chang H.Y. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 2010;7(5):582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dimitrova N., Zamudio J.R., Jong R.M., Soukup D., Resnick R., Sarma K. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell. 2014;54(5):777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loewer S., Cabili M.N., Guttman M., Loh Y.H., Thomas K., Park I.H. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010;42(12):1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K. Glen Munson, et al., LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atianand M.K., Hu W., Satpathy A.T., Shen Y., Ricci E.P., Alvarez-Dominguez J.R. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016;165(7):1672–1685. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu M.T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 39.Arnberg A.C., van Ommen G.J., Grivell L.A., Van Bruggen E.F., Borst P. Some yeast mitochondrial RNAs are circular. Cell. 1980;19(2):313–319. doi: 10.1016/0092-8674(80)90505-x. [DOI] [PubMed] [Google Scholar]
- 40.Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 41.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu C.Y., Liu H.J., Hung L.Y., Kuo H.C., Chuang T.J. Is an observed non-co-linear RNA product spliced in trans, in cis or just in vitro? Nucleic Acids Res. 2014;42(14):9410–9423. doi: 10.1093/nar/gku643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Umekage S., Kikuchi Y. Production of circular streptavidin RNA aptamer in vivo. Nucleic Acids Symp. Ser. 2007;(51):391–392. doi: 10.1093/nass/nrm196. [DOI] [PubMed] [Google Scholar]
- 44.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9) doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y., Zhang X., Chen T., Xiang J., Yin Q., Xing Y. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 47.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 48.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 50.Guo J.U., Agarwal V., Guo H., P Bartel D. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barbagallo D., Condorelli A., Ragusa M., Salito L., Sammito M., Banelli B. Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2016;7(4):4746–4759. doi: 10.18632/oncotarget.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piwecka M., Glažar P., Hernandez-Miranda L.R., Memczak S., Wolf S.A., Rybak-Wolf A. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357(6357) doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 53.Xu H., Guo S., Li W., Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu L., Gong X., Sun L., Zhou Q., Lu B., Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PloS One. 2016;11(7) doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Xia P., Wang S., Ye B., Du Y., Li C., Xiong Z. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity. 2018;48(4):688–701. doi: 10.1016/j.immuni.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 56.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491(7424):454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 58.Louro R., Smirnova A.S., Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics. 2009;93(4):291–298. doi: 10.1016/j.ygeno.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 59.Yan M., Hong C., Lai G., Cheng A., Lin Y., Chuang S. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum. Mol. Genet. 2005;14(11):1465–1474. doi: 10.1093/hmg/ddi156. [DOI] [PubMed] [Google Scholar]
- 60.Hu S., Wang X., Shan G. Insertion of an Alu element in a lncRNA leads to primate-specific modulation of alternative splicing. Nat. Struct. Mol. Biol. 2016;23(11):1011–1019. doi: 10.1038/nsmb.3302. [DOI] [PubMed] [Google Scholar]
- 61.Huang C., Wang X., Liu X., Cao S., Shan G. RNAi pathway participates in chromosome segregation in mammalian cells. Cell Discov. 2015;1:15029. doi: 10.1038/celldisc.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sulston J.E., Brenner S. The DNA of Caenorhabditis elegans. Genetics. 1974;77(1):95–104. doi: 10.1093/genetics/77.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., C Mello C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 65.Liu H., Wang X., Wang H., Wu J., Ren J., Meng L. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat. Commun. 2012;3:1073. doi: 10.1038/ncomms2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hellwig S., Bass B.L. A starvation-induced noncoding RNA modulates expression of Dicer-regulated genes. Proc. Natl. Acad. Sci. U.S.A. 2008;105(35):12897–12902. doi: 10.1073/pnas.0805118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kiontke K.C., Herrera R.A., Vuong E., Luo J., Schwarz E.M., Fitch D.H.A. The long non-coding RNA lep-5 promotes the Juvenile-to-Adult transition by destabilizing LIN-28. Dev. Cell. 2019;49(4):542–555. doi: 10.1016/j.devcel.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lawson H., Vuong E., Miller R.M., Kiontke K., Fitch D.H., Portman D.S. The Makorin lep-2 and the lncRNA lep-5 regulate lin-28 to schedule sexual maturation of the C. elegans nervous system. Elife. 2019;8 doi: 10.7554/eLife.43660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frezal L., Felix M.A. C. elegans outside the petri dish. Elife. 2015;4 doi: 10.7554/eLife.05849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nam J.W., Bartel D.P. Long noncoding RNAs in C. Elegans. Genome Res. 2012;22(12):2529–2540. doi: 10.1101/gr.140475.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu W., Yu E., Chen S., Ma X., Lu Y., Liu X. Spatiotemporal expression profiling of long intervening noncoding RNAs in Caenorhabditis elegans. Sci. Rep. 2017;7(1):5195. doi: 10.1038/s41598-017-05427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu B., Wang X., Wei S., Fu T., Dzakah E.E., Waqas A. Convergent transcriptional programs regulate cAMP levels in C. elegans GABAergic motor neurons. Dev. Cell. 2017;43(2):212–226. doi: 10.1016/j.devcel.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 73.Howell K., White J.G., Hobert O. Spatiotemporal control of a novel synaptic organizer molecule. Nature. 2015;523(7558):83–87. doi: 10.1038/nature14545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Castle W.E. Inbreeding, cross-breeding and sterility in Drosophila. Science. 1906;23(578):153. doi: 10.1126/science.23.578.153. [DOI] [PubMed] [Google Scholar]
- 75.Morgan T.H. Sex limited inheritance in Drosophila. Science. 1910;32(812):120–122. doi: 10.1126/science.32.812.120. [DOI] [PubMed] [Google Scholar]
- 76.Levine J.D., Funes P., Dowse H.B., Hall J.C. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3:1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Imler J.L., Hoffmann J.A. Toll signaling: the TIReless quest for specificity. Nat. Immunol. 2003;4(2):105–106. doi: 10.1038/ni0203-105. [DOI] [PubMed] [Google Scholar]
- 78.Tupy J.L., Bailey A.M., Dailey G., Evans-Holm M., Siebel C.W., Misra S. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 2005;102(15) doi: 10.1073/pnas.0501422102. 5495-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Willingham A.T., Dike S., Cheng J., Manak J.R., Bell I., Cheung E. Transcriptional landscape of the human and fly genomes: nonlinear and multifunctional modular model of transcriptomes. Cold Spring Harbor Symp. Quant. Biol. 2006;71:101–110. doi: 10.1101/sqb.2006.71.068. [DOI] [PubMed] [Google Scholar]
- 80.Graveley B.R., Brooks A.N., Carlson J.W., Duff M.O., Landolin J.M., Yang L. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471(7339):473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soshnev A.A., Ishimoto H., McAllister B.F., Li X., Wehling M.D., Kitamoto T. A conserved long noncoding RNA affects sleep behavior in drosophila. Genetics. 2011;189(2):455–468. doi: 10.1534/genetics.111.131706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wen K., Yang L., Xiong T., Di C., Ma D., Wu M. Critical roles of long noncoding RNAs in Drosophila spermatogenesis. Genome Res. 2016;26(9):1233–1244. doi: 10.1101/gr.199547.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meller V.H., Rattner B.P. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL Complex. EMBO J. 2002;21(5):1084–1091. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chu C., Qu K., Zhong F.L., Artandi S.E., Chang H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chelmicki T., Dündar F., Turley M.J., Khanam T., Aktas T., Ramírez F. MOF-associated complexes ensure stem cell identity and Xist repression. Elife. 2014;3 doi: 10.7554/eLife.02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Prasanth K.V., Rajendra T.K., Lal A.K., Lakhotia S.C. Omega Speckles - a novel class of nuclear speckles containing hnRNPs associated with noncoding Hsr-Omega RNA in Drosophila. J. Cell Sci. 2000;113(Pt 19):3485–3497. doi: 10.1242/jcs.113.19.3485. [DOI] [PubMed] [Google Scholar]
- 87.Rajendra T.K., Prasanth K.V., C Lakhotia S. Male sterility associated with overexpression of the noncoding Hsromega gene in cyst cells of testis of Drosophila melanogaste. J. Genet. 2001;80(2):97–110. doi: 10.1007/BF02728335. [DOI] [PubMed] [Google Scholar]
- 88.Mallik M., Lakhotia S.C. Improved activities of CREB binding protein, heterogeneous nuclear ribonucleoproteins and proteasome following downregulation of noncoding Hsromega transcripts help suppress poly(Q) pathogenesis in fly models. Genetics. 2010;184(4):927–945. doi: 10.1534/genetics.109.113696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McCorkindale A.L., Wahle P., Werner S., Jungreis I., Menzel P., Shukla C.J. A gene expression atlas of embryonic neurogenesis in Drosophila reveals complex spatiotemporal regulation of lncRNAs. Development. 2019;146(6):dev175265. doi: 10.1242/dev.175265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gummalla M., Galetti S., Maeda R.K., Karch F. Hox gene regulation in the central nervous system of Drosophila. Front. Cell. Neurosci. 2014;8:96. doi: 10.3389/fncel.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li M., Wen S., Guo X., Bai B., Gong Z., Liu X. The novel long non-coding RNA CRG regulates Drosophila locomotor behavior. Nucleic Acids Res. 2012;40(22):11714–11727. doi: 10.1093/nar/gks943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li M., Liu L. Neural functions of long noncoding RNAs in Drosophila. J. Comp. Physiol. A. 2015;201(9):921–926. doi: 10.1007/s00359-014-0937-8. [DOI] [PubMed] [Google Scholar]
- 93.Streisinger G., Walker C., Dower N., Knauber D., Singer F. Production of clones of homozygous diploid zebra fish (brachydanio rerio) Nature. 1981;291(5813):293–296. doi: 10.1038/291293a0. [DOI] [PubMed] [Google Scholar]
- 94.Chakrabarti S., Streisinger G., Singer F., Walker C. Frequency of gamma-Ray induced specific locus and recessive lethal mutations in mature germ cells of the Zebrafish. BRACHYDANIO RERIO, Genetics. 1983;103(1):109–123. doi: 10.1093/genetics/103.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meyer A., Biermann C.H., Orti G. The phylogenetic position of the zebrafish (Danio Rerio), a Model system in developmental biology: an invitation to the comparative method. Proc. Biol. Sci. 1993;252(1335):231–236. doi: 10.1098/rspb.1993.0070. [DOI] [PubMed] [Google Scholar]
- 96.Kimmel C.B. Genetics and early development of Zebrafish. Trends Genet. 1989;5(8):283–288. doi: 10.1016/0168-9525(89)90103-0. [DOI] [PubMed] [Google Scholar]
- 97.Torraca V., Mostowy S. Zebrafish infection: from pathogenesis to cell biology. Trends Cell Biol. 2018;28(2):143–156. doi: 10.1016/j.tcb.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zon L.I., Peterson R.T. In vivo drug discovery in the zebrafish. Nat. Rev. Drug Discov. 2005;4(1):35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
- 99.Fagegaltier D., Lescure A., Walczak R., Carbon P., Krol A. Structural analysis of new local features in SECIES RNA hairpins. Nucleic Acids Res. 2000;28(14):2679–2689. doi: 10.1093/nar/28.14.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Haque S., Kaushik K., Leonard V.E., Kapoor S., Sivadas A., Joshi A. Short stories on zebrafish long noncoding RNAs. Zebrafish. 2014;11(6):499–508. doi: 10.1089/zeb.2014.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147(7):1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pauli A., Valen E., Lin M.F., Garber M., Vastenhouw N.L., Levin J.Z. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22(3):577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kaushik K., Leonard V.E., Kv S., Lalwani M.K., Jalali S., Patowary A. Dynamic expression of long non-coding RNAs (lncRNAs) in adult zebrafish. PloS One. 2013;8(12) doi: 10.1371/journal.pone.0083616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li K., Blum Y., Verma A., Liu Z., Pramanik K., Leigh N.R. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 2010;115(1):133–139. doi: 10.1182/blood-2009-09-242180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cheng Z., Zhang Q., Yin A., Feng M., Li H., Liu H. The long non-coding RNA uc.4 influences cell differentiation through the TGF-beta signaling pathway. Exp. Mol. Med. 2018;50(2):e447. doi: 10.1038/emm.2017.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Basu S., Müller F., Sanges R. Examples of sequence conservation analyses capture a subset of mouse long non-coding RNAs sharing homology with fish conserved genomic elements. BMC Bioinf. 2013;14(7):S14. doi: 10.1186/1471-2105-14-S7-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen W., Zhang X., Li J., Huang S., Xiang S., Hu X. Comprehensive analysis of coding-lncRNA gene co-Expression network uncovers conserved functional lncRNAs in Zebrafish. BMC Genom. 2018;19(2):112. doi: 10.1186/s12864-018-4458-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yuan W., Jiang S., Sun D., Wu Z., Wei C., Dai C. Transcriptome profiling analysis of sex-based differentially expressed mRNAs and lncRNAs in the brains of mature zebrafish (Danio rerio) BMC Genom. 2019;20(1):830. doi: 10.1186/s12864-019-6197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goudarzi M., Berg K., Pieper L.M., Schier A.F. Individual long non-coding RNAs have no overt functions in zebrafish embryogenesis, viability and fertility. Elife. 2019;8 doi: 10.7554/eLife.40815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Briggs J.A., Wolvetang E.J., Mattick J.S., Rinn J.L., Barry G. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. 2015;88(5):861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 112.Dey B.K., Mueller A.C., Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5(4) doi: 10.4161/21541272.2014.944014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moran V.A., Perera R.J., Khalil A.M. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40(14):6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jain S., Thakkar N., Chhatai J., Bhadra M.P., Bhadra U. Long non-coding RNA: functional agent for disease traits. RNA Biol. 2017;14(5):522–535. doi: 10.1080/15476286.2016.1172756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Azzu V., Valencak T.G. Energy metabolism and ageing in the mouse: a mini-review. Gerontology. 2017;63(4):327–336. doi: 10.1159/000454924. [DOI] [PubMed] [Google Scholar]
- 116.Su A.I., Cooke M.P., Ching K.A., Hakak Y., v J.R., Wiltshire T. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 2002;99(7):4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pachnis V., Brannan C.I., Tilghman S.M. The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J. 1988;7(3):673–681. doi: 10.1002/j.1460-2075.1988.tb02862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kitsberg D., Selig S., Brandeis M., Simon I., Keshet I., Driscoll D.J. Allele-specific replication timing of imprinted gene regions. Nature. 1993;364(6436):459–463. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- 119.Brockdorff N., Ashworth A., Kay G.F., McCabe V.M., Norris D.P., Cooper P.J. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71(3):515–522. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 120.Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T. Polycombproteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McHugh C.A., Chen C.K., Chow A., Surka C.F., Tran C., McDonel P. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. doi: 10.1038/nature14443. 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chu C., Zhang Q.C., da Rocha S.T., Flynn R.A., Bharadwaj M., Calabrese J.M. Systematic discovery of xist RNA binding proteins. Cell. 2015;161(2):404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111(2):185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 124.Bassett A.R., Akhtar A., Barlow D.P., Bird A.P., Brockdorff N., Duboule D. Considerations when investigating lncRNA function in vivo. Elife. 2014;3 doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marahrens Y., Panning B., Dausman J., Strauss W., Jaenisch R. Xist-deficient mice are defective in Dosage Compensation but not spermatogenesis. Genes Dev. 1997;11(2):156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 126.Brannan C.I., Dees E.C., Ingram R.S., Tilghman S.M. The product of the H19 gene may function as an RNA. Mol. Cell Biol. 1990;10(1):28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Koerner M.V., Pauler F.M., Huang R., Barlow D.P. The function of non-coding RNAs in genomic imprinting. Development. 2009;136(11):1771–1783. doi: 10.1242/dev.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Thamban T., Agarwaal V., Khosla S. Role of genomic imprinting in mammalian development. J. Biosci. 2020;45:20. [PubMed] [Google Scholar]
- 129.Ripoche M.A., Kress C., Poirier F., Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11(12):1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- 130.Nakagawa S., Naganuma T., Shioi G., Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011;193(1):31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nakagawa S., Ip J.Y., Shioi G., Tripathi V., Zong X., Hirose T. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18(8):1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li L., Liu B., Wapinski O.L., Tsai M.C., Qu K., Zhang J. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5(1):3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shamovsky I., Nudler E. Gene control by large noncoding RNAs. Sci. STKE. 2006. 2006;355:pe40. doi: 10.1126/stke.3552006pe40. [DOI] [PubMed] [Google Scholar]
- 134.Nakagawa S. Lessons from reverse-genetic studies of lncRNAs. Biochim. Biophys. Acta. 2016;1859(1):177–183. doi: 10.1016/j.bbagrm.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 135.Liu W., Ding C. Roles of LncRNAs in viral infections. Front. Cell. Infect. Microbiol. 2016;7:205. doi: 10.3389/fcimb.2017.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xu H., Jiang Y., Xu X., Su X., Liu Y., Ma Y. Inducible degradation of lncRNA Sros1 promotes IFN-γ-mediated activation of innate immune responses by stabilizing Stat1 mRNA. Nat. Immunol. 2019;20(12):1621–1630. doi: 10.1038/s41590-019-0542-7. [DOI] [PubMed] [Google Scholar]
- 137.Gutschner T., Hämmerle M., Eissmann M., Hsu J., Kim Y., Hung G. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Canc. Res. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang P., Xu J., Wang Y., Cao X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science. 2017;358(6366):1051–1055. doi: 10.1126/science.aao0409. [DOI] [PubMed] [Google Scholar]
- 139.Muret K., Désert C., Lagoutte L., Boutin M., Gondret F., Zerjal T. Long noncoding RNAs in lipid metabolism literature review and conservation analysis across species. BMC Genom. 2019;20(1):882. doi: 10.1186/s12864-019-6093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ramos A.D., Attenello F.J., Lim D.A. Uncovering the roles of long noncoding RNAs in neural development and glioma progression. Neurosci. Lett. 2019;625:70–79. doi: 10.1016/j.neulet.2015.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Perry R.B., Hezroni H., Goldrich M.J., Ulitsky I. Regulation of neuroregeneration by long noncoding RNAs. Mol. Cell. 2018;72(3):553–567. doi: 10.1016/j.molcel.2018.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]