Summary
Glioblastoma, the predominant adult malignant brain tumor, has been computationally classified into molecular subtypes whose functional relevance remains to be comprehensively established. Tumors from genetically engineered glioblastoma mouse models initiated by identical driver mutations in distinct cells of origin portray unique transcriptional profiles reflective of their respective lineage. Here, we identify corresponding transcriptional profiles in human glioblastoma and describe patient derived xenografts with species-conserved subtype-discriminating functional properties. The oligodendrocyte lineage associated glioblastoma subtype requires functional ERBB3 and harbors unique therapeutic sensitivities. These results highlight the importance of cell lineage in glioblastoma independent of driver mutations and provide a methodology for functional glioblastoma classification for future clinical investigations.
In Brief
Wang et al. identify lineage-specific subtypes in mouse and human glioblastoma that harbor distinct functional properties and therapeutic vulnerabilities, highlighting the role of cell lineage in glioblastoma and providing a classification system for future clinical investigations.
Graphical Abstract

Introduction
Glioblastoma (GBM) is an aggressive and lethal malignant brain tumor characterized by cellular and molecular heterogeneity (Brennan et al., 2013; Frattini et al., 2013; Parsons et al., 2008). Phillips and colleagues first proposed stratification of GBM based on transcriptome relatedness (Phillips et al., 2006). This was expanded into a bioinformatic classification based on gene transcription, mutation, and copy number variation that has dominated the field for a decade (Brennan et al., 2013; Chen et al., 2012b; Verhaak et al., 2010). This widely used tool summarizes the gene expression patterns of large patient cohorts based on graded trends of molecular relatedness and described four separable subtypes: proneural, neural, classical and mesenchymal. These subtypes were further associated with GBM hallmark mutations, like PDGFRA with proneural, EGFR with classical, and NF1 with mesenchymal, although such mutations are not exclusive but rather correlative (Brennan et al., 2013; Verhaak et al., 2010). As the field has applied the bioinformatics based GBM classification, the degree of applied parameter stringency has fluctuated (Bhat et al., 2013; Patel et al., 2014; Segerman et al., 2016). Indeed, a recent updated reclassification by Verhaak and colleagues removed the neural subgroup altogether, reported unstable genomic signatures between spheroid cultures and primary tumors, and noted subtype switching when comparing primary and recurrent gliomas (Wang et al., 2017). Other studies implicate tumor subtype evolution mediated by incremental mutation implying that most TCGA subtypes arise from a proneural-like precursor (Ozawa et al., 2014), or subclass switching of proneural cells undergoing differentiation to a mesenchymal state in a TNF-α/NF-κB-dependent manner (Bhat et al., 2013), or via ASCL1 (Park et al., 2017). Thus, rather than constituting biologically unique and separable tumor subtypes, the purely computational classification may better reflect fluid GBM transcriptional cell states possibly influenced by microenvironment and other variables including cell culture, that can be captured by unsupervised classification algorithms (Bhat et al., 2013; Neftel et al., 2019; Patel et al., 2014; Segerman et al., 2016; Sottoriva et al., 2013). Single cell sequencing studies further support this view with the indication that individual tumors contain a spectrum of GBM subtypes and hybrid cellular states (Neftel et al., 2019; Patel et al., 2014). Thus, while the biological underpinnings of TCGA formulated molecular subtypes continue to be investigated, the clinical relevance remains in question (Chen et al., 2017; Segerman et al., 2016).
Fully penetrant genetically engineered mouse models (GEMMs) of GBM harboring common tumor suppressor mutations (Trp53, Nf1 and Pten) were generated by specifically targeting either adult subventricular zone (SVZ) neural stem cells (NSCs) or adult oligodendrocyte lineage cells (OLCs) (Alcantara Llaguno et al., 2009; Alcantara Llaguno et al., 2015; Chen et al., 2012a; Kwon et al., 2008; Zhu et al., 2005; Zong et al., 2015). Transcriptional profiles for SVZ-derived (Type 1) and OLC-derived (Type 2) GBM are distinct and reflect the tumor cell of origin lineage (Alcantara Llaguno et al., 2015). We therefore hypothesized that the cell lineage constitutes a key determinant of GBM molecular subtype and phenotype.
Results
GBM lineage features are conserved in vitro and in vivo
We previously demonstrated that GBM relevant tumor suppressors mutated in adult SVZ NSCs versus OLCs drive two distinct tumor types, Type 1 and Type 2 GBM, respectively (Figures 1A–B and S1A–D) (Alcantara Llaguno et al., 2015). Transcriptional analysis was performed on 7 Type 1 and 7 Type 2 GEMM tumors, and 394 differentially expressed genes (DEGs) were identified with 189 genes in Type 1 GBM, and 205 in Type 2 GBM (Table S1; adjusted p <0.1 and two-fold change). Predictably, gene ontology (GO) analysis associated Type 1 tumors with astrocyte development genes which relate to NSC lineage while Type 2 DEGs identified enrichment of biological processes that reflect the oligodendrocytic lineage such as myelination and oligodendrocyte differentiation (Table S1). Members of two notable gene families - the ErbB growth factor receptor and SoxE transcription factor families - are differentially expressed, with epidermal growth factor receptor (Egfr) and Sox9 both prominent in Type 1 GBM, and ErbB receptor tyrosine kinase 3 (Erbb3) and Sox10 in Type 2 GBM (Figure 1C–E). The additional ErbB family members Erbb2 and Erbb4 are similarly expressed in both tumor subtypes (Figure S1E) and likewise, the canonical stem cell marker, SOX2, is expressed in all tumors examined (Figure 1D). EGFR has been shown to promote GBM through SOX9 (Liu et al., 2015). In CNS development, EGFR is expressed in actively dividing NSC and transit amplifying cells (Doetsch et al., 2002; Mich et al., 2014), while its relative, ERBB3, has key functions in oligodendrocyte lineage development (Makinodan et al., 2012; Zhang et al., 2014). Similarly, SOX9 is implicated in NSC induction and maintenance (Scott et al., 2010), while SOX10 is a master regulator in the oligodendrocytic lineage (Stolt et al., 2002; Wang et al., 2014). We verified differential gene expression by quantitative real time PCR (qRT-PCR), western blot, and immunohistochemistry (IHC) analyses using GEMM primary tumor tissues (Figures 1D–E and S1F). Thus, murine Type 1 and Type 2 GBM display consistent phenotypic and molecular differences and retain transcriptional signatures reflective of their distinct lineage origins.
Figure 1. Nf1, Trp53, and Pten inactivation in adult NSCs versus OLCs generates distinct GBM subtypes.
(A) Nestin-CreERT2;Rosa-lsl-Yfp and NG2-CreER™;Rosa-lsl-tdTom transgenes were used to drive reporter expression in SVZ NSCs and OLCs, respectively after tamoxifen induction at 1 month of age and analysis 1 week later. YFP expression was revealed by anti-GFP staining while TOMATO expression was endogenous. Scale bars, 1000 μM. (B) Representative images of Type 1 and 2 GBM analyzed by hematoxylin and eosin (H&E) staining. Regions marked by the white box (left, scale bars, 1000 μM) are magnified (right, scale bars, 50 μM). (C) Volcano plot illustrating DEGs (green dots) in Type 1 and 2 GBM (n=7 each for Type 1 and Type 2 GBM; adjusted p<0.1; 2-fold change). (D) Western blot analysis in Type 1 (n=7) and Type 2 (n=7) primary GBM tissue samples. (E) Representative immunofluorescence images in Type 1 and 2 tumors. Scale bars, 50 μM. In A-B and E, results are representative of n≥3 biological replicates. See also Figure S1 and Table S1.
To examine the molecular and histopathological stability of Type 1 and 2 GBM features, primary spheroid and monolayer cultures were established in standard serum-free media with growth factors (Figure S2A–B). Both Type 1 and 2 GBM cultures showed sustained high expression for EGFR/SOX9 or ERBB3/SOX10, respectively, through serial passages (Figure 2A and data not shown). Similar conservation of molecular features was maintained following intracranial transplantation into immunodeficient mice (Figure 2B–C), and western blot analysis showed activated EGFR or ERBB3 signaling in transplanted tumors (Figure 2C). Growth factor response studies showed that Type 1 GBM cells have increased growth in the presence of EGF while Type 2 cells grow best in the presence of the ERBB3 receptor ligand, Neuregulin 1 (NRG1; Figure 2D–E). Consistent with this, ERBB3 knockdown in Type 2 cells blunts neuregulin-mediated cell growth and tumor development (Figure S2C–F). Taken together, these data confirm distinct and stable gene expression profiles, and functional cell growth and tumor properties for GEMM Type 1 and 2 GBM tumors and primary cells.
Figure 2. Type 1 and Type 2 GBM cells exhibit conserved molecular and histological phenotypes as primary cultures and following intracranial transplantation.
(A) Western blot analysis in Type 1 (n=5) and Type 2 (n=6) mouse GBM primary cultures. (B) Representative H&E staining for tumors generated by intracranial transplantation of Type 1 (n=3) and 2 (n=3) cultures into nude mice. Scale bars: 1000 μM (left); 100 μM (right). (C) Representative western blot analysis in transplanted Type 1 and 2 tumors. (D-E) Growth factor dose response for EGF (D) and NRG1 (E) (concentrations from 0–40 ng/ml) of Type 1 and Type 2 cells measured by ATP assays (CellTiter-Glo®; n=3 replicates). Data are presented as mean ± standard deviation (S.D.). See also Figure S2.
Type 1 & 2 GBM transcriptome & methylation profiles segregate human data sets
We examined whether Type 1 and 2 GBM gene expression signatures are represented in human GBM by probing the largest available gene expression data set (TCGA provisional U133 microarray; n=528) that includes 498 GBM patient samples after exclusion of tumors with known IDH1 mutations according to the Brennan et. al. annotation (Brennan et al., 2013). The z-score transformed mRNA expression was directly downloaded from cBioPortal (www.cbioportal.org; (Cerami et al., 2012; Gao et al., 2013). Stringent and stepwise criteria were applied to identify related human GBM transcriptional profiles. First, the 498 human GBM panel was probed for expression of the 4 strong mouse Type 1 and 2 GBM markers, EGFR;SOX9 and ERBB3;SOX10 respectively (Figure 3A). In total, 107 human Type I candidates, were defined as EGFR;SOX9 high (z-score >0.5), and ERBB3;SOX10 low (z-score <0.5 ) (Figures 3A and S3A). Conversely, 68 human Type II candidates, were identified as ERBB3;SOX10 high (z-score >0.5) and EGFR;SOX9 low (z-score <0.5) (Figures 3A and S3A). We also examined the z-score transformed RNAseq data (142 IDH1 wild type GBM in TCGA) curated by cBioPortal and found a similar gene expression distribution among samples as for the microarray platform. Concordantly, there was significant overlap of Type I & II candidates between the microarray and the RNAseq platforms on shared samples (Figure S3B; Table S2). Thus, the z-score standardization does not appear to be specifically biased to Affymetrix data, and independent of the platforms, the correlated expression of all four genes more likely reflects a biological distinction.
Figure 3. Type I & Type II classification of human GBM cases.
(A) Work flow showing stepwise identification of human Type I and Type II GBM core samples. Transcriptional data from 498 IDH1 wild-type TCGA GBM were analyzed (z-score threshold of 0.5) to identify Type I and Type II candidates. GSVA using human homologues of mouse Type 1 versus Type 2 GBM DEG was applied. (B) Hierarchical clustering analysis dendrogram of human and mouse GBM profiles using mouse Type 1 and 2 GBM DEG. (C) Multi-dimensional scaling (MDS) plot using global methylation status of TCGA Type I (n=7) and Type II (n=9) GBM core samples on HM450K platform with 1000 most variable probes. (D) Gene expression heatmap of Type I and Type II GBM signature genes, TIsig and TIIsig, in TCGA core (n=52), extended (n=87), and Rembrandt (n=28) Type I, as well as TCGA core (n=38), extended (n=54) and Rembrandt (n=26) Type II samples. (E) Progression free survival of available data from Type I (n=139), Type II (n=92) and remaining non-Type I&II (n=267) TCGA GBM patients (Type I vs. Type II, p=0.011, likelihood ratio test). See also Figure S3 and Tables S2 and S3.
We next used the human homologs of the previously identified DEG from mouse Type 1 (189 genes) or Type 2 GBM (205 genes; Table S1) and applied gene set variation analysis (GSVA) to obtain enrichment scores of relatedness among the human Type I (T1DEGscore) and Type II (T2DEGscore) candidates, respectively (Table S2). Fifty-two cases among the 107 human Type I candidates scored as TCGA “core Type I” (T1DEGscore>0.1), and 38 cases among the 68 human Type II candidate samples scored as TCGA “core Type II” (T2DEGscore>0.1). Unsupervised hierarchical clustering of GEMM tumors and core TCGA Type I and II GBM samples using GEMM Type 1 and 2 GBM signatures illustrated the similarity among the tumor types by dendrogram co-clustering of mouse Type 1 with human Type I GBM and mouse Type 2 with human Type II GBM (Figure 3B).
Examination of the methylation status for core Type I and Type II GBM also demonstrated subtype separation. The Infinium HumanMethylation450 (HM450) data set that covers more than 450,000 methylation sites was available for 7 Type I and 9 Type II human GBM core samples scored from the TCGA data set. Multi-dimensional scaling plots using the 1,000 most variable probes separated the Type I and Type II GBM core samples into two distinct subgroups (Figure 3C), as did a more comprehensive probe set (n=331,895) (Figure S3C). Similarly, the Infinium HumanMethylation27 (HM27) platform that interrogates more than 27,000 CpG dinucleotides, applying either the 1,000 most variable probes (Figure S3D), or the more comprehensive set of 19,111 probes (Figure S3E), segregated the available 20 Type I and 18 Type II GBM core samples (Bibikova et al., 2011; Brennan et al., 2013). Thus, human Type I and Type II TCGA GBM core samples each aggregate by methylation profiles.
Through comparison of the 52 core Type I GBM samples against the remaining TCGA cases (n=446), we established a list of 72 highly expressed genes (adjusted p <0.05, fold change >2) as a Type I signature (TIsig; Figure 3D; Table S3). Similarly, analysis of the 38 core Type II GBM samples against the remaining cases (n=460) provided an 80-gene Type II signature (TIIsig; adjusted p <0.05, fold change >2; Figure 3D; Table S3). Reevaluation of non-core GBM with TIsig and TIIsig identified an additional 87 samples as Type I, and 54 samples as Type II (Figure 3D; see Methods). Thus, 139 GBM satisfy these criteria and classify as Type I and 92 as Type II, encompassing 46.4% of TCGA samples (Table S3).
To further examine the coherence of this subtyping method, we probed the independently annotated 146 sample GBM Rembrandt data set (Madhavan et al., 2009) with TIsig and TIIsig, and identified 28 Type I and 26 Type II GBM samples under the same criteria as that for TCGA samples, and these consistently showed similar expression patterns with TCGA Type I and II GBM, respectively (Figure 3D; Table S3). Progression free survival (PFS) between stratified Type I and II patients from TCGA was statistically different in multivariate survival analysis considering subtypes, age and the interaction between age and subtypes as variables (Figure 3E), although there was no significant difference in overall survival (data not shown).
Type I and II classification was derived from differential gene expression of tumor tissue samples. A recent large single cell sequencing study of human primary GBM provided an additional layer of tumor data to probe Type I and Type II signatures (Neftel et al., 2019). We first generated pseudo-tissue expression profiles by averaging gene expression for all cells within each individual tumor (20 GBM samples) to determine the Type I, Type II, or non-TypeI/II status for each tumor, as indicated in Figure S3F. Next, we analyzed the entire 4,916 malignant cells from the 20 tumors for TIsig and TIIsig using a rank-based single cell gene set enrichment algorithm (AUCell; see Methods). As depicted by the scatter plot in Figure S3F, each single cell was given a TIsig or TIIsig enrichment score (TIsigScore: X-axis and TIIsigScore: Y-axis), although these scores do not necessarily have equivalent biological meaning as those on the tumor tissue level. The data indicate that cells from each of the 20 tumors are either predominantly high TIsigScore, high TIIsigScore, or low for both scores (non-Type I and II); with only a few cells scoring high for both. Concordantly, the dominant single cell subtype represented in each tumor is in line with the subtype identity of the corresponding GBM. This was observed in a large majority of cells within a given tumor, as exemplified by representative Type I or II tumor scatter plots in Figure S3G. These data show consistent stratification of our signatures at both single cell and entire tumor levels.
Lineage-specific genes and mutational spectrum across Type I and II GBM
Examination of a comprehensive mouse brain single cell RNA sequencing study (Mizrak et al., 2019) further showed that lineage-associated human Type I and II GBM, like their mouse counterparts, are also marked by genes enriched in NSC versus OLC lineage (Figures 4A and S4A). Likewise, ToppCluster and GO analysis of the human Type I and II signatures indicated that the TIsig is enriched in biological process terms like astrocyte differentiation and cell migration, whereas TIIsig enriched in myelination, gliogenesis, and oligodendrocyte differentiation (Figures 4B and S4B). These profiles are in remarkably close alignment with those previously documented for mouse Type 1 and 2 GBM (Table S1). Taken together, the data consistently delineate two human GBM subtypes (Type I & II) that strongly resemble the murine subtypes (Type 1 & 2) originally defined by cell lineage of tumor origin.
Figure 4. Human Type I and II GBM signature genes and mutations.
(A) mRNA expression (shown as log fold change) of representative lineage markers that are differentially expressed in human Type I vs. Type II GBM in TCGA core samples (U133 microarray data), mouse Type 1 vs. Type 2 GBM (microarray data), and NSCs vs. OLCs (Mizrak et al., 2019). mNSC, mouse Neural Stem Cell; mOPC, mouse Oligodendrocyte progenitor cell. (B) Toppcluster analysis (https://toppcluster.cchmc.org/) showing genes and biological processes associated with TIsig and TIIsig. (C) Gene amplification in TCGA Type I and Type II GBM (***p<0.001, Fisher’s exact test for two proportions). (D) Frequency of GBM driver mutations in Type I and Type II GBM (*p<0.05, Fisher’s exact test for two proportions). (E) Frequency of ERBB3 gene amplification in the TCGA dataset. See also Figure S4 and Table S4.
All Type I GBM show high EGFR expression consistent with a preponderance of amplifications at the EGFR locus (7p11.2; 75.7%), a feature that partially aligns Type I with the bioinformatics-based classical (CL) stratification. Likewise, Type II GBM show increased amplification of genes in the vicinity of PDGFRA and KIT locus (4q12; 31.5% and 23.6%, respectively) that partially align with the proneural (PN) classification (Figure 4C; Table S4). GBM driver mutations did not show statistically significant bias among Type I and Type II GBM with the exception of increased EGFR mutations in Type I GBM and mutant TP53 representation in Type II (Figure 4D; Table S4). Notably, Type II tumors show neither ERBB3 mutations nor increased amplification (Figure 4E). Thus, Type I-associated elevated EGFR expression is mostly mediated by gene amplification, whereas increased ERBB3 expression in Type II GBM is likely the result of transcriptional regulation.
Consistent with the above noted partial alignments, direct analysis of the bioinformatics-based subclassification showed statistical overlap of Type I with CL and Type II with PN (Figure S4C–D; (Wang et al., 2017). However, when the Type I and Type II gene signatures (TIsig & TIIsig) were directly compared to the CL and PN signatures respectively, in both cases the number of overlapping genes was fewer than 20% (Figure S4E). Furthermore, both Type I and II subclasses included tumors identified as mesenchymal (MES) by the computational method (Figure S4C; Wang et al., 2017). In aggregate, Type I and Type II GBM signatures show partial convergence with the CL and PN computational signatures, however the overriding transcriptional association with cell lineage dominates and separates their identity.
Type I and II primary GBM cultures maintain core molecular features
To examine functional properties of human Type I and II tumor cells, we established 17 primary GBM cultures from the MSKCC Brain Tumor Center PDX collection limiting the study to tumors derived from treatment naive patient samples. Through a western blot screen, we identified primary cells from 7 primary GBM samples and their corresponding PDX derived cells with high EGFR;SOX9 and low ERBB3;SOX10 protein expression and 7 samples with high ERBB3/SOX10 and low EGFR/SOX9 expression (Figure 5A). The 14 samples collectively contained a range of potential driver mutations (Table S5) and the PDX primary cultures were subjected to mRNA sequencing (RNAseq) for subtyping. To enhance the transcriptional separation between Type I and II primary cultures, we calculated scores (TIDEGScore and TIIDEGScore) using the DEGs that distinguished core Type I and core Type II and encompass TIsig and TIIsig, respectively (Table S5). Four primary cultures scored as Type I, and 7 cultures scored as Type II (Figure 5B; Table S5). Confirmation of EGFR and SOX9 expression in Type I, and ERBB3 and SOX10 expression in Type II tumors was verified by IHC on the original PDX tumor samples (Figure 5C–D). Additionally, SLC1A3, a representative Type I signature gene, and FA2H, a Type II signature gene were also differentially expressed (Figure 5D; Table S5). Like their mouse counterparts, human Type I and II primary cultures also showed differential growth response to EGF and NRG1 (Figure S5A–B).
Figure 5. Type I and II GBM identification in patient-derived cultures and xenografts.
(A) Western blot analysis in a panel of 17 patient-derived GBM cells designated as Type I and II candidates. (B) Heat map showing primary cell enrichment scores for DEG that distinguished core Type I & II GBM (TIDEGScore and TIIDEGScore) in 14 PDXs. (C) Representative images of H&E and IHC staining for EGFR and ERBB3 of Type I and Type II GBM PDX brain sections. Scale bars, 50 μM. (D) Immunofluorescence staining for Type I markers, SOX9 and SLC1A3, and Type II markers, SOX10 and FA2H, on GBM PDX brain sections. Scale bars, 50 μM. In C-D, results are representative of n≥3 biological replicates. See also Figure S5 and Table S5.
Differential drug response functionally distinguishes GBM subtypes
We next investigated a panel of chemotherapeutic agents and pharmacologic inhibitors for activity in Type I versus Type II tumors (Table S6). Among them, the ERBB2 inhibitor Tucatinib (Heffron, 2016) selectively inhibited both mouse Type 2 and human Type II GBM cell growth in vitro (Figure 6A–B) and effectively abolished downstream RAS and phosphoinositide-3-kinase (PI3K) activation while showing no measurable effects on mouse Type 1 or human Type I GBM cells (Figures 6C–D, S6A–B). An additional ERBB2 inhibitor, CP724714 (Heffron, 2016), showed similar specificity (data not shown). Dasatinib, a multi-target tyrosine kinase inhibitor including c-ABL, c-SRC, c-KIT, and PDGFRβ (Heffron, 2016), also showed specific growth inhibition of mouse Type 2 and human Type II cells (Figure 6E–F). We examined known Dasatinib targets for potential roles in the differential sensitivity of GBM subtypes and found that SRC phosphorylation was equivalently inhibited in all GBM cells (Figures 6G–H, S6C–D). Moreover, SRC phosphorylation was unaffected in Type 2/II cells by Tucatinib (Figure S6A–B), thus ruling out a role. Additionally, neither c-KIT nor c-ABL protein tyrosine phosphorylation could be detected, also ruling them out as potential Dasatinib mediators (Figure S6E). In Type II cells, phospho-PDGFRβ was attenuated together with attenuated ERBB2 and ERBB3 tyrosine phosphorylation and downstream signaling. This effect on PDGFRβ was not seen in Type I cells (Figure S6E). However, Imatinib, a potent PDGFRβ inhibitor, did not preferentially inhibit Type II cell growth, ruling out PDGFRβ (Figure S6F). Thus, in mouse Type 1 and human Type I GBM cells, Tucatinib and Dasatinib resistance rests on retained EGFR activity and its ability to maintain effective downstream signaling. In sensitive Type II and Type 2 cells, ERBB2/ERBB3 activity is essential and subject to Dasatinib-mediated inhibition either directly, or indirectly via its downstream signaling (Davis et al., 2011). 5-Azacytidine, a nucleoside analogue that inhibits DNA methylation (Christman, 2002), also exhibits partial inhibitory effect on Type II cells (Figure S6G). Taken together, these species-conserved differential drug responses illustrate the coherent and distinct functional properties between the lineage stratified GBM subtypes.
Figure 6. Differential response to Tucatinib and Dasatinib by Type 2/II GBM.
(A-B) Dose response curves of primary mouse Type 1 & 2 (A) and human Type I & II (B) GBM cells to Tucatinib. Cells were treated with Tucatinib and analyzed 96 h after exposure using ATP assay (CellTiter-Glo®; n=3 replicates). Data are presented as mean ± S.D. (C-D) Western blot analyses for GBM cells from human Type I (C) and human Type II (D) treated with Tucatinib. Primary cells were cultured as monolayer and growth factor starved for 24 h, then treated with Tucatinib at 0.5 μM, 1 μM and 2 μM for 1 h, followed by stimulation with EGF+NRG1 (10 ng/ml each) together. Cells were harvested for analysis 30 min post-stimulation. (E-F) Dose response growth curves of mouse Type 1 & 2 cells (E) and human Type I & II cells (F) to Dasatinib. Cells were treated with Dasatinib and analyzed 96 h after exposure using ATP assay (CellTiter-Glo®; n=3 replicates). Data are presented as mean ± S.D. (G-H) Western blot analyses for GBM cells from human Type I (G) and human Type II (H) treated with Dasatinib. Primary cells were cultured as monolayer and growth factor starved for 24 h, then treated with Dasatinib for 1 h, followed by stimulation with EGF or NRG1 (10 ng/mL) individually. Cells were harvested for analysis 30 min post-stimulation. In C-D and G-H, results are representative of n≥3 biological replicates. See also Figure S6 and Table S6.
Dasatinib inhibits Type 2 & II GBM growth in vivo
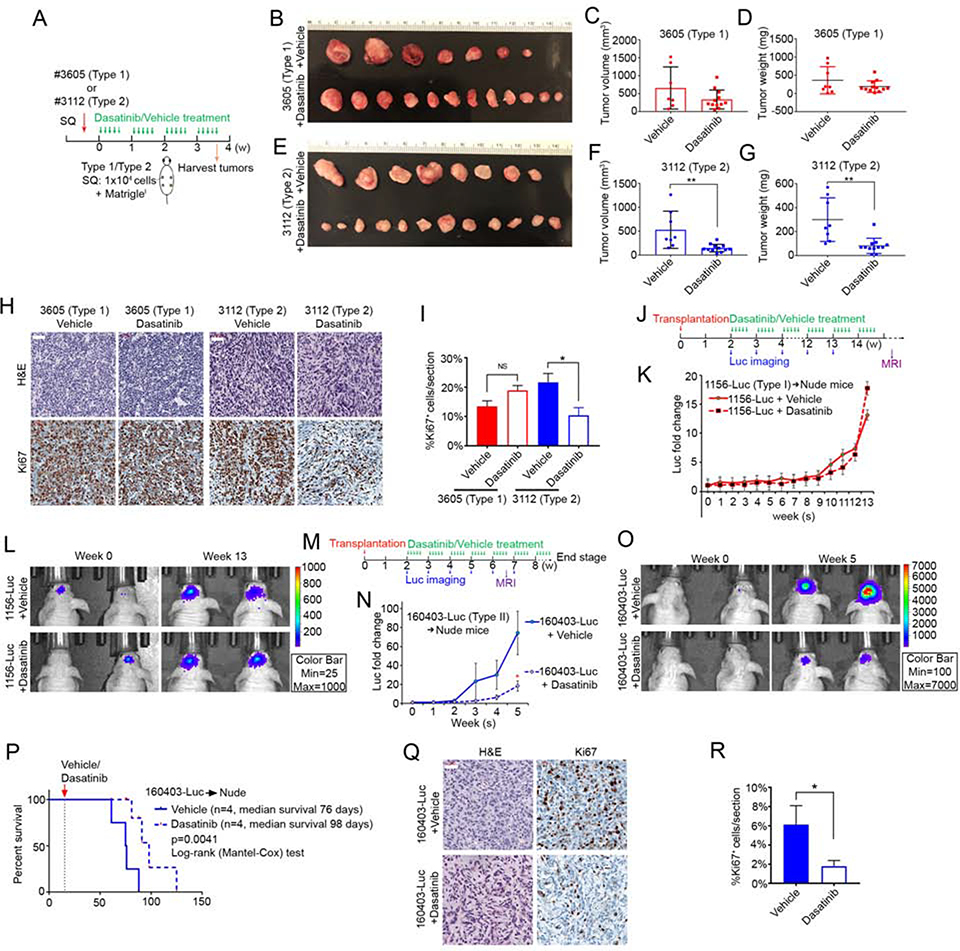
To further test the biological distinction between the species-conserved subtypes and examine a potential clinical application for lineage based GBM stratification, we evaluated the effect of Dasatinib on murine Type 1 & 2 and human Type I & II GBM growth in vivo. First, subcutaneous allografts of murine Type 1 and 2 GBM cells were established and mice were subjected to daily Dasatinib (50 mg/kg) gavage beginning 3 days post-transplantation and continued for 5 weeks (Figure 7A). Type 1 GBM growth qualitatively appeared mildly attenuated but lacked statistical significance compared to controls (Figure 7B–D). In contrast, Type 2 GBM showed clear and statistically significant inhibition in tumor volume and weight (Figure 7E–G). H&E and Ki67 IHC indicated no apparent difference in histology and proliferation in Dasatinib-treated Type 1 tumors (Figure 7H–I). In contrast, Dasatinib-treated Type 2 tumors displayed altered patterns of cellular organization with presence of fibrillary, fascicular, edematous and myxoid changes, and concordant decrease in mitotic index measured by Ki67 IHC (Figure 7H–I). In each case, Type 1 and Type 2 GBM subcutaneous allografts maintained their respective signature expression for Egfr/Sox9 and ErbB3/Sox10 following Dasatinib treatment (Figure S7 A–B).
Figure 7. Dasatinib attenuates Type 2 & II GBM growth in vivo.
(A) Experimental scheme for subcutaneously (SQ) transplanted mouse Type 1 and Type 2 GBM cells. (B) Type 1 GBM (#3605) SQ tumors harvested from vehicle- and Dasatinib-treated mice. (C-D) Tumor volume (C, p=0.12, student’s t-test) and tumor weight (D, p=0.17, student’s t-test) of Type 1 GBM (#3605) SQ tumors from vehicle-treated (n=7) and Dasatinib-treated (n=12) mice. Data are presented as mean ± SD. (E) Type 2 GBM (#3112) SQ tumors harvested from vehicle- and Dasatinib-treated mice. (F-G) Tumor volume (F, **p=0.0035, student’s t-test) and tumor weight (G, **p=0.0011, student’s t-test) of Type 2 GBM (#3112) SQ tumors from vehicle-treated (n=8) and Dasatinib-treated (n=12) mice. (H) Representative images of H&E and anti-Ki67-stained sections of Type 1 GBM (#3605) and Type 2 GBM (#3112) from vehicle- and Dasatinib-treated mice. Scale bars, 50 μM. (I) Quantification of Ki67+ cell densities in Type 1 (#3605) and Type 2 (#3112) GBM SQ tumors after vehicle (n=4 Type 1, n=3 Type 2) or Dasatinib (n=4 Type 1, n=4 Type 2) treatment. Data are presented as mean ± S.E.M. ;N.S., not significant; *p=0.0369, student’s t-test. (J) Experimental scheme for intracranially transplanted human Type I GBM cells (#1156-Luc, 5×105 cells/mouse). Tumor bearing mice were gavaged with vehicle or Dasatinib, once per day, 5 days per week, starting at 2 weeks post-transplantation. Luminescence imaging monitored tumor growth until week 13 and MRI at week 16. (K) Plot of luminescence intensity of transplanted Type I GBM (#1156-luc) mice treated with vehicle (n=4) or Dasatinib (n=4). (L) Representative luminescence images of mice bearing Type I GBM (#1156-luc) at week 0 and week 13 following vehicle and Dasatinib treatment. (M) Experimental scheme for intracranially transplanted human Type II GBM cells (#160403-Luc, 5×105 cells/mouse). Tumor growth was monitored with luminescence imaging weekly and MRI at week 6. (N) Plot of Type II GBM (#160403-luc) cells luminescence intensity over time for vehicle- and Dasatinib-treated mice. Luminescence intensity fold change was normalized to week 0 of individual mouse and presented as average ± S.D. (n=4 at each time point). *p=0.030, student’s t-test. (O) Representative luminescence images of intracranially transplanted Type II GBM (#160403-luc) mice at week 0 and week 5 following vehicle or Dasatinib treatment. (P) Kaplan-Meier survival curve of intracranially transplanted Type II GBM (#160403-luc) mice treated with vehicle (n=4) or Dasatinib (n=4). ***p=0.0041, Log-rank (Mantel-Cox) test. (Q) H&E and anti-Ki67 analysis of Type II tumors with vehicle and Dasatinib treatment. Scale bar, 50 μM. (R) Quantification of Ki67+ cell densities in Type II GBM (#160403-luc) intracranial tumors after vehicle (n=4) and Dasatinib (n=4) treatment. Data are presented as mean ± S.E.M. *p=0.0484, student’s t-test. See also Figure S7 and Table S7.
We next assessed Dasatinib activity on PDX models. Type I and II PDX primary cells were infected with lentivirus expressing Luciferase/GFP (Luc-ZsGreen), sorted for GFP expression (Luc-ZsGreen+), and intracranially transplanted into nude mice. Tumor growth was monitored weekly by bioluminescence imaging (Figure 7J–O). Dasatinib treatment began 2 weeks post-transplantation and continued until animals demonstrated neurological symptoms and morbidity. Type I tumor growth was similar between Dasatinib and vehicle-treated mice (Figure 7J–L). Similar results were obtained by magnetic resonance imaging (MRI) showing no significant difference in tumor size (Figure S7C). In contrast, Type II GBM showed significant response to Dasatinib, manifested by blunted luminescence (Figure 7M–O), reduced tumor size as measured by MRI (Figure S7D), and prolonged median survival (from 76 days to 98 days; p=0.0041, Log-rank (Mantel-Cox) test; Figure 7P). H&E analysis of tumors revealed modified histological features including fibrillary and fascicular patterns, as seen in Dasatinib-treated subcutaneous Type 2 GBM (Figure 7Q) and reduced mitotic index measured by Ki67 analysis (Figure 7R). In sum, the in vivo data demonstrate both murine Type 2 and human Type II GBM have unique, measurable, and reproducible favorable response to Dasatinib. Intratumoral Dasatinib levels in tissues from both intracranial human Type II and subcutaneous mouse Type 2 GBM were assayed by Mass Spectrometry (Table S7). Subcutaneous tumor drug delivery was more effective than intracranial delivery, possibly accounting for the more pronounced inhibitory effects on the former.
Despite significant delay, all Type II Dasatinib treated mice eventually developed tumors (Figure 7P). We therefore examined the status of signaling by western blot analysis of intracranial tumor explant tissue at the study end points. Both Dasatinib and vehicle-treated Type II fresh tumor tissues showed similar Ras downstream signaling indicating a developed evasion of drug pathway inhibition for the former (Figure S7E–F). We next prepared primary cultures from the harvested residual Dasatinib treated Type II tumors and examined their response to drug. Primary cell cultures from Dasatinib-treated intracranial Type II tumors exhibited retained growth sensitivity to both Dasatinib and Tucatinib, equivalent to cells from vehicle-treated Type II tumors (Figure S7G–H). In addition, both tumor-derived cells showed continued response to NRG1 or EGF stimulation (Figure S7I). Thus, despite prolonged in vivo Dasatinib treatment accompanied by significant tumor growth delay, explanted Type II GBM cells apparently did not acquire drug resistance. The cause of in vivo resistance is unclear but may be related to microenvironmental influences or to tumor cell plasticity that does not include loss of the Type II signature since it remains stable in both conditions.
Discussion
The use of modern genomic tools for molecular sub-classification of seemingly related tumors can provide a powerful means for discrimination and evaluation of phenotype and outcome (Northcott et al., 2017). This study assigns specific transcriptional profiles and signatures to two classes of GBM that in autochthonous GEMMs were induced in different lineage cells under identical GBM relevant driver mutations. Strikingly distinct phenotypes emerge and segregate according to the tumor lineage identity (SVZ-derived Type 1 vs. OLC-derived Type 2). The strictly maintained transcriptional signatures remain stable in extended cell culture, following orthotopic transplantation, and are identifiable in archived human GBM databases (TCGA; Rembrandt; Neftel et al. 2019). Human Type I and Type II GBM cultures and orthotopic tumors, like their mouse counterparts, also maintain stable molecular and functional characteristics. It is worth emphasizing that the Type I and II tumor subtypes identified in data sets and PDXs harbor a representative range of glioma relevant driver mutations beyond those used to originally generate Type 1 and 2 GEMMs. The lineage-based GBM stratification could be functionally validated by species-conserved differential response to growth factors and to chemotherapeutic drugs in primary culture and in vivo.
In addition to illustrating the biological difference between the two GBM subtypes, this differential drug susceptibility by stratification presents a potential translational application. Dasatinib has previously failed efficacy tests in phase 2 clinical trials for recurrent GBM (Lassman et al., 2015; Schiff and Sarkaria, 2015). It remains to be determined whether a potential clinical response by prospectively molecularly classified Type II GBM patients, which would represent fewer than 20% of enrolled cases in a random trial, might be masked by overrepresentation of a majority of non-Type II cases. Dasatinib is a broad spectrum tyrosine kinase inhibitor (Hantschel et al., 2008). Our results suggest that Dasatinib may also directly inhibit ERBB3 to mediate Type II tumor inhibition. Reported Dasatinib dissociation constant assays indicate an intermediate ERBB3 binding affinity unique among the ErbB receptor family, that may suffice to mediate functional inhibition (Davis et al., 2011). The similarity of functional activity and signaling profiles with the ERBB2 specific inhibitor Tucatinib further supports this possibility. Additional studies with both compounds will help resolve this issue.
The present study was limited to tumor samples from therapy naïve patients and did not assess therapeutic responsiveness from post radiation and temozolomide treated cases. Additional studies and surveys of anticancer compounds under this stratification may further reveal subtype specific responses. We unexpectedly found that primary cultures harvested from residual tumors from Dasatinib treated PDX mice resumed full drug sensitivity accompanied by renewed inhibition of the RAS and AKT signaling pathways. Given that in vivo treatment substantially delayed tumor growth and blunted downstream signaling, these results suggest that over time the tumor microenvironment (Saggar et al., 2013) may instigate Dasatinib insensitivity. It is also possible that tumor cell plasticity, not reflected by loss of the Type 2/II signature identity, may fluctuate between in vivo and culture conditions (Neftel et al., 2019).
The computational stratification of TCGA based genomic GBM data sets into subtypes has played a dominant role in the field (Brennan et al., 2013; Verhaak et al., 2010). While valuable and widely applied, this approach has lacked a biological foundation and continues to evolve, as emphasized by the recently proposed exclusion of the neural subtype (Wang et al., 2017). The heterogeneity of human tumors in terms of mutational spectra, age of initiation, as well as presence of predisposing factors and/or somatic mutations, may all contribute to computational genomic subgroup discrimination. In contrast, mouse models, with well-defined initiating mutations, cell of origin, and timing of initiation, may clarify focus on fundamental biological processes that are conserved between species, and therefore, are more likely to reveal functional determinants. Nevertheless, prospective patient stratification to assess clinical trial outcome remains an objective of critical importance. A clinical benefit of Bevacizumab has been suggested for newly diagnosed proneural GBM in a retrospective study (Sandmann et al., 2015), although it remains to be validated. Similar studies using the lineage-based subgroups proposed here will assess its power for stratification of clinical trials.
We provide a cogent biological premise for GBM stratification of approximately one half of all GBM that exhibit partial convergent features with the bioinformatics-based transcriptional PN and CL subclasses although with notable differences (Wang et al., 2017). A range of possibilities may help resolve stratification of the remaining non-Type I and II GBM in the future. For example, the adult rodent brain harbors additional distinct stem/progenitor cell populations in the hippocampus, hypothalamus and pons (Goncalves et al., 2016; Lindquist et al., 2016; Paul et al., 2017). Our preliminary data indicate these populations can also produce GBM in genetically engineered mice, thus potentially expanding the possible repertoire of tumor cells of origin and associated molecular subtypes. It is also possible that specific configurations of GBM oncogenic events may override the lineage-associated phenotypes present in Type I and II GBM. Continued scrutiny will be required to yield answers.
Recent reports have fueled controversy concerning the existence of stem cell populations in the adult human brain (Boldrini et al., 2018; Ernst et al., 2014; Sorrells et al., 2018). Our studies, here and elsewhere indicate that in rodents, not only are stem/progenitor cells the principal source of GBM, but in fact more differentiated cell types lack the potential to generate GBM (Alcantara Llaguno et al., 2009; Alcantara Llaguno et al., 2019; Alcantara Llaguno et al., 2015). Given the existence of functional adult CNS stem cell populations in birds, rodents and primates (Gould et al., 1999; Nottebohm, 2005), as well as reports for humans (Boldrini et al., 2018; Ernst et al., 2014; Lee et al., 2012), it is difficult to conceive how exclusion of such plasticity from the most evolved version of the central nervous system (CNS) would afford an evolutionary advantage. Our studies thus best align with those that indicate persistence of stem cells in the human CNS and a fundamental role in GBM development, identity, and phenotype.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Luis F. Parada (paradal@mskcc.org).
Materials Availability
The GEMMs, primary cultures, and PDX materials from the MSKCC Brain Tumor Center will have restrictions according to Institutional Review Board and Material Transfer Agreement institutional policies. Other materials not specified will be made available from the corresponding author on request.
Data and Code Availability
Genomic data generated in this study have been downloaded to the Gene Expression Omnibus (GEO numbers GSE127273 and GSE127274).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mouse experiments were approved and performed according to the guidelines of the Institutional Animal Care and Use Committee of UT Southwestern Medical Center and Memorial Sloan Kettering Cancer Center. Mice with Nestin-creERT2 (Alcantara Llaguno et al., 2009; Chen et al., 2009; Groszer et al., 2001; Lin et al., 2004; Zhu et al., 2001) or NG2-CreER™ (Zhu et al., 2011) transgenes were crossed with conditional Trp53 allele or R26-lsl-Yfp or R26-lsl-tdTom reporters. Nestin-CreERT2;Nf1f/+;Trp53f/f;Ptenf/+;Rosa-lsl-tdTom mice or NG2-CreER™;Nf1f/+;Trp53f/f;Ptenf/+;Rosa-lsl-tdTom mice were generated by breeding Nestin-CreERT2;Trp53f/f or NG2-CreER™;Trp53f/f mice with Nf1f/f;Trp53f/f;Ptenf/f;Rosa-lsl-tdTom mice. All mice were maintained under formal MSKCC IACUC protocols.
Mouse Primary Culture
Mouse primary tumor tissues were freshly dissected from symptomatic mutant mice. Harvested tissues were minced and incubated with Accutase in 37°C water bath for 20 minutes, disassociated, and cultured in serum free medium supplemented with B27 and N2 (SFM), plus EGF, FGF and PDGF-AA (10 ng/ml each) in 5% oxygen, 37°C incubator. Primary cultures were established as tumor spheres at P0, later passages were as spheres in non-coated plates, or as monolayer in laminin pre-coated plates (10 ng/ml Laminin diluted in DPBS with Ca2+/Mg2+).
Patient-derived xenografts
PDXs were established by the Brain Tumor Center at Memorial Sloan Kettering Cancer Center. All patients provided written informed consent for use of specimens and all studies were performed under approval and oversight by the MSKCC Institutional Review Board and IACUC.
Human Primary Culture
Cultures from primary human GBM or PDX tissue were obtained from the Brain Tumor Center at MSKCC. All patients provided written informed consent for use of specimens and all studies were performed under approval and consent of MSKCC’s Institutional Review Board and IACUC. Primary human cultures were first established in SFM with EGF, FGF, PDGF-AA and NRG1 (10 ng/mL each). Designated Type I GBM cells were maintained in SFM with EFP (EGF, FGF and PDGF-AA, each 10 ng/ml), whereas Type II GBM cells were maintained in SFM with NFP (NRG1, FGF and PDGF-AA, each 10 ng/mL).
METHOD DETAILS
Mouse genotyping and induction
PCR genotyping of GEMMs was performed as previously described (Kwon et al., 2008). Mice with or without the Cre transgene were administered with tamoxifen (Sigma) by oral gavage (10 mg/20g body weight) at 6–8 weeks of age and followed until symptomatic or at indicated time points.
Histology and Tumor Analysis
For reporter analysis, mice of Nestin-CreERT2;Rosa-lsl-Yfp, or Nestin-CreERT2;Rosa-lsl-tdTom, and NG2-CreER™;Rosa-lsl-tdTom were tamoxifen induced and harvested at age and time as specified. Mice were transcardially perfused and brains were fixed overnight in 4% paraformaldehyde (PFA), and vibratome sectioned at 50 μm. Staining of floating sections was performed by starting with PBS wash and then sections were permeabilized with 0.4% PBST (PBS with 0.4% TritonX-100) for 4 hours, blocked with 3% donkey serum in 0.4% PBST, and incubated with primary antibodies diluted in 0.4% PBST overnight in 4°C on shaker. Sections were then PBS washed and followed by secondary antibody staining at RT for 2 hours. Sections were applied to slides and air dried in the dark, mounted with mounting medium and air dried before sealing with nail polish. Images were acquired with confocal microscopy (Zeiss).
Tumor bearing brains were harvested with or without perfusion and fixed with 4% PFA overnight, processed and embedded in paraffin. Paraffin brain sections were cut at 5 μm and analyzed by H&E, fluorescence and horseradish peroxidase-based immunostaining, as previously described (Alcantara Llaguno et al., 2009). Please refer to the Key Resources Table for all primary antibodies used in the study. Brain sections were independently examined by D.K.B. and T.A.B, board certified neuropathologists, and tumor diagnosis was determined based on the World Health Organization criteria (Louis et al., 2007). Images were taken using Zeiss confocal (LSM 800) microscopy or Mirax scanning. Quantification of images was done using Fiji/ImageJ.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-EGFR santa cruzsc-03rabbit 1:1000AB_631420 | Abcam | Cat#: ab52894; RRID: AB_869579 |
| Rabbit anti-EGFR | Santa Cruz | Cat#: sc-03; RRID: AB_631420 |
| Rabbit anti-ERBB3 | Cell Signaling | Cat#: 12708; RRID: AB_2721919 |
| Goat anti-Sox10 | Santa Cruz | Cat#: sc-17342; RRID: AB_21953 |
| Goat anti-Sox10 | R&D | Cat#: AF2864-SP; RRID: AB_442208 |
| Rabbit anti-Sox10 | Abcam | Cat#: ab155279; RRID: AB_2650603 |
| Rabbit anti-Sox9 | Cell Signaling | Cat#: 82630S; RRID: AB_2665492 |
| Mouse anti-Sox9 | Novus Biologicals | Cat#:H00006662-M02; RRID: AB_875842 |
| Rabbit anti-Pdgfra | Santa Cruz | Cat#: sc-338; RRID: AB_631064 |
| Rabbit anti-Sox2 | Abcam | Cat#: ab97959; RRID: AB_2341193 |
| Rabbit anti-NG2 | Millipore | Cat#: AB5320; RRID: AB_11213678 |
| Goat anti-Nestin | Santa Cruz | Cat#: sc-21248; RRID: AB_2148925 |
| Rabbit anti-Fa2h | Proteintech | Cat#: 15452–1-AP; RRID: AB_2101886 |
| Sheep anti-Slc1a3 | R&D | Cat#: AF6048-SP; RRID: AB_2044669 |
| Rabbit anti-Olig2 | Millipore | Cat#: Ab9610; RRID: AB_570666 |
| Rabbit anti-Gfap | Cell Signaling | Cat#: 12389T; RRID: AB_2631098 |
| Mouse anti-Tubulin | Cell signaling | Cat#: 3873S; RRID: AB_1904178 |
| Mouse anti-Gapdh | Fisher scientific | Cat#: MA5–15738; RRID: AB_10977387 |
| Rabbit anti-ERBB2 | Santa Cruz | Cat#: sc-284; RRID: AB_632013 |
| Rabbit anti-ERBB4 | Santa Cruz | Cat#: sc-283; RRID: AB_2231308 |
| Rabbit anti-pEGFRY1068 | Cell Signaling | Cat#: 2234s; RRID: AB_331701 |
| Rabbit anti-pERBB2Y1196 | Cell Signaling | Cat#: 6942s; RRID: AB_10860762 |
| Rabbit anti-pERBB3Y1197 | Cell Signaling | Cat#: 4561s; RRID: AB_2099707 |
| Rabbit anti-pAKTSer473 | Cell Signaling | Cat#: 4060s; RRID: AB_2315049 |
| Rabbit anti-pAKTThr308 | Cell Signaling | Cat#: 13038s; RRID: AB_2629447 |
| Rabbit anti-pERK | Cell Signaling | Cat#: 4370s; RRID: AB_2315112 |
| Rabbit anti-ERK | Cell Signaling | Cat#: 4695s; RRID: AB_390779 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma | Cat#: T5648 |
| NRG1 | Lifespan biosciences, Inc | Cat#: LS-G3634 |
| Tucatinib | MedKoo Biosciences | Cat#: 200291 |
| Dasatinib | MedKoo Biosciences | Cat#: 100230 |
| AZD3759 | MedKoo Biosciences | Cat#: 206182 |
| Imatinib | SelleckChem | Cat#: S1026 |
| Mk2206 | SelleckChem | Cat#: S1078 |
| Pictilisib | SelleckChem | Cat#: S1065 |
| Sorafenib | SelleckChem | Cat#: S7397 |
| Erlotinib | SelleckChem | Cat#: S7786 |
| Cp724714 | SelleckChem | Cat#: S1167 |
| KX2–391 | SelleckChem | Cat#: S2700 |
| Sunitnib | SelleckChem | Cat#: SU11248 |
| bAP15 | SelleckChem | Cat#: S4920 |
| 5-Azacytidine | MedKoo Biosciences | Cat#: 100070 |
| Nifedipine | SelleckChem | Cat#: S1808 |
| Pitavastatin | SelleckChem | Cat#: S1759 |
| Salmeterol | SelleckChem | Cat#: S4296 |
| Stattic | SelleckChem | Cat#: S7024 |
| Vinscristine | SelleckChem | Cat#: S1241 |
| 2DG | SelleckChem | Cat#: S4701 |
| SirReal2 | SelleckChem | Cat#: S7845 |
| EGF | Fisher Scientific | Cat#: PHG0311 |
| bFGF | Gemini | Cat#: 300–113P 50ug |
| PDGF-AA | R&D | Cat#: 1055-AA-050 |
| XenoLight D-Luciferin - K+ Salt | PerkinElmer | Cat#: 122799 |
| Critical Commercial Assays | ||
| CellTiter-Glo® ATP assay | Promega | Cat#: G7571 |
| iScript cDNA synthesis kit | Biorad | Cat#: 1708890 |
| RNeasy Extraction Kit | Qiagen | Cat#: 74104 |
| Lipid Tissue RNeasy Mini Kit | Qiagen | Cat#: 74804 |
| Deposited Data | ||
| Gene Expression Analysis of Nestin-creERT2, or NG2-creERTM driven Trp53f/f; Nf1f/+; Ptenf/+ mouse GBM | Gene Expression Omnibus | GSE127273 |
| RNA Sequencing Analysis of human glioblastoma derived cells and xenograft derived primary cells | Gene Expression Omnibus | GSE127274 |
| Rembrandt GBM microarray data set | G-DOC®: A Systems Medicine Platform for Personalized Oncology | Rembrandt GBM https://icbi.georgetown.edu/g-doc/ |
| TCGA GBM RNAseq and microarray data set | Cbioportal https://www.cbioportal.org/ | GBM (TCGA 2003) |
| TCGA GBM Methylation data | https://portal.gdc.cancer.gov/ | GBM |
| Experimental Models: Cell Lines | ||
| Nes-3605; mouse GBM primary tumor stem cell line derived from Nestin-CreERT2;Nf1f/+;Trp53f/f;Ptenf/+;Rosa-lsl-tdTom | Established at MSKCC | N/A |
| Nes-2024; mouse GBM primary tumor stem cell line derived from Nestin-CreERT2;Nf1f/+;Trp53f/f;Ptenf/+;Rosa-lsl-tdTom | Established at MSKCC | N/A |
| Nes-1329; mouse GBM primary tumor stem cell line derived from Nestin-CreERT2;Nf1f/+;Trp53f/f;Ptenf/+;Rosa-lsl-tdTom | Established at MSKCC | N/A |
| Nes-4619; mouse GBM primary tumor stem cell line derived from Nestin-CreERT2;Nf1f/+;Trp53f/f;Ptenf/+;Rosa-lsl-tdTom | Established at MSKCC | N/A |
| NG2–3112; mouse GBM primary tumor stem cell line derived from NG2-CreER™;Nf1f/+;Trp53f/f;Ptenf/+;Rosa-lsl-tdTom | Established at MSKCC | N/A |
| NG2–775; mouse GBM primary tumor stem cell line derived from NG2-CreER™;Nf1f/+;Trp53f/f;Ptenf/+;Rosa-lsl-tdTom | Established at MSKCC | N/A |
| NG2–631; mouse GBM primary tumor stem cell line derived from NG2-CreER™;Nf1f/+;Trp53f/f;Ptenf/+;Rosa-lsl-tdTom | Established at MSKCC | N/A |
| NG2–1870; mouse GBM primary tumor stem cell line derived from NG2-CreER™;Nf1f/+;Trp53f/f;Ptenf/+;Rosa-lsl-tdTom | Established at MSKCC | N/A |
| PDX160929; GBM patient-derived primary tumor stem cell line | Established at MSKCC (Brain Tumor Center) | N/A |
| PDX160412; GBM patient-derived primary tumor stem cell line | Established at MSKCC (Brain Tumor Center) | N/A |
| PDX1156; GBM patient-derived primary tumor stem cell line | Established at MSKCC (Brain Tumor Center) | N/A |
| PDX160324; GBM patient-derived primary tumor stem cell line | Established at MSKCC (Brain Tumor Center) | N/A |
| PDX13050; GBM patient-derived primary tumor stem cell line | Established at MSKCC (Brain Tumor Center) | N/A |
| PDX160927; GBM patient-derived primary tumor stem cell line | Established at MSKCC (Brain Tumor Center) | N/A |
| PDX160721; GBM patient-derived primary tumor stem cell line | Established at MSKCC (Brain Tumor Center) | N/A |
| PDX12111; GBM patient-derived primary tumor stem cell line | Established at MSKCC (Brain Tumor Center) | N/A |
| PDX160225; GBM patient-derived primary tumor stem cell line | Established at MSKCC (Brain Tumor Center) | N/A |
| PDX160403; GBM patient-derived primary tumor stem cell line | Established at MSKCC (Brain Tumor Center) | N/A |
| PDX12178; GBM patient-derived primary tumor stem cell line | Established at MSKCC (Brain Tumor Center) | N/A |
| PDX12224; GBM patient-derived primary tumor stem cell line | Established at MSKCC (Brain Tumor Center) | N/A |
| PDX160812; GBM patient-derived primary tumor stem cell line | Established at MSKCC (Brain Tumor Center) | N/A |
| Experimental Models: Organisms/Strains | ||
| Nestin-CreERT2; Nf1f/+;Trp53f/f;Ptenf/+;Rosa-lsl-tdTom | Alcantara Llaguno et al., 2009 https://www.ncbi.nlm.nih.gov/pubmed/19111880 | N/A |
| NG2-CreER™;Nf1f/+;Trp53f/f;Ptenf/+;Rosa-lsl-tdTom | Alcantara Llaguno, S., et. al., 2009 https://www.ncbi.nlm.nih.gov/pubmed/19111880 | N/A |
| Nestin-CreERT2 | Chen et al., 2009 https://www.ncbi.nlm.nih.gov/pubmed/19117051 | N/A |
| NG2-CreER™ | Zhu et al., 2011 https://www.ncbi.nlm.nih.gov/pubmed/21266410 | N/A |
| Nf1flox | Zhu et al., 2001 https://www.ncbi.nlm.nih.gov/pubmed/11297510 | N/A |
| Trp53flox | Lin et al., 2004 https://www.ncbi.nlm.nih.gov/pubmed/15150107 | N/A |
| Ptenflox | Groszer et al., 2001 https://www.ncbi.nlm.nih.gov/pubmed/11691952 | N/A |
| Rosa-lsl-tdTom | Jackson Laboratories | Stock No:007909 |
| Rosa-lsl-YFP | JacksonLaboratories | Stock No:006148 |
| nu/nu | JacksonLaboratories | Stock No:007850;RRID:IMSR_JAX:007850 |
| Oligonucleotides | ||
| Gapdh-Forward: AGGTCGGTGTGAACGGATTTG | Eton Bioscience | N/A |
| Gapdh-Reverse: TGTAGACCATGTAGTTGAGGT | Eton Bioscience | N/A |
| EGFR-Forward: GCCATCTGGGCCAAAGATACC | Eton Bioscience | N/A |
| EGFR- Reverse: GTCTTCGCATGAATAGGCCAAT | Eton Bioscience | N/A |
| ErbB3-Forward: AAGTGACAGGCTATGTACTGGT | Eton Bioscience | N/A |
| ErbB3-Reverse: GCTGGAGTTGGTATTGTAGTTCA | Eton Bioscience | N/A |
| Sox9- Forward: GAGCCGGATCTGAAGAGGGA | Eton Bioscience | N/A |
| Sox9- Reverse: GCTTGACGTGTGGCTTGTTC | Eton Bioscience | N/A |
| Sox10-Forward: AGGTTGCTGAACGAAAGTGAC | Eton Bioscience | N/A |
| Sox10- Reverse: CCGAGGTTGGTACTTGTAGTCC | Eton Bioscience | N/A |
| Recombinant DNA | ||
| pCW-Cas9 | Addgene | Cat#50661; RRID:RRID:Addgene_50661 |
| pCW-Cas9-sgErbB3 (sgRNA:5’-cctaacctccgagtggtccg-3’ ) | constructed at MSKCC | N/A |
| pHIV-Luc-ZsGreen | Addgene | Cat#39196;RRID:Addgene_39196 |
| Software and Algorithms | ||
| GraphPad Prism v.7 | www.graphpad.com | RRID:SCR_002798 |
| Image J | www.imagej.net | RRID:SCR_003070 |
| Bioconductor | www.bioconductor.org | RRID:SCR_006442 |
| Beadarray | https://www.bioconductor.org/packages/release/bioc/html/beadarray.html | RRID:SCR_001314 |
| inSilicoMerging | https://www.bioconductor.org/packages//2.10/bioc/html/inSilicoMerging.html | RRID:SCR_012829 |
| Limma | https://www.bioconductor.org/packages/release/bioc/html/limma.html | RRID:SCR_010943 |
| biomaRt | https://www.bioconductor.org/packages/release/bioc/html/biomaRt.html | RRID:SCR_002987 |
| fpc | https://cran.r-project.org/web/packages/fpc/index.html | N/A |
| A2R | https://rdrr.io/github/alexjgriffith/CCCA/src/inst/scripts/A2R.r | N/A |
| gplots | https://cran.r-project.org/web/packages/gplots/index.html | N/A |
| cBioPortal | www.cBioPortal.org | RRID:SCR_014555 |
| GSVA | https://bioconductor.org/packages/release/bioc/html/GSVA.html | N/A |
| gcrma | https://bioconductor.org/packages/release/bioc/html/gcrma.html | N/A |
| Virtualarray | https://bioconductor.org/packages/2.11/bioc/html/virtualArray.html | RRID:SCR_001361 |
| AUCell | https://bioconductor.org/packages/release/bioc/html/AUCell.html | N/A |
| STAR 2.4.2.8 | https://www.encodeproject.org/software/star/ | N/A |
| Survival | https://cran.r-project.org/web/packages/survival/index.html | N/A |
| Survcomp | https://www.bioconductor.org/packages/release/bioc/html/survcomp.html | RRID:SCR_003054 |
| ChAMP | https://www.bioconductor.org/packages/release/bioc/html/ChAMP.html | RRID:SCR_012891 |
In vitro inhibitor experiments
To investigate the downstream signaling events upon Tucatinib or Dasatinib inhibition, GBM cells were maintained in serum-free medium (supplemented with B27 and N2) without growth factors for 24 hours, then treated with specified concentrations of inhibitors for one hour, followed by stimulation with EGF and/or NRG1 (10 ng/mL), and harvested 30 minutes post-stimulation.
Transplantations and Dasatinib in vivo experiments
Type 1 and Type 2 GBM cells were transplanted into nude mice subcutaneously (1×104/location) and mice were gavaged once daily with vehicle (80 mM Citric acid, PH 3.1, n=2) or Dasatinib (50 mg/kg, n=3), started 3 days post transplantation, 5 days a week. Treatment continued for 26 days and all tumors were harvested at the same time when the first tumor reached end point (2 cm in diameter).
For in vivo luminescence imaging study using luciferase reporter, human Type I (#1156) and Type II (#160403) cells were infected with lentivirus expressing Luciferase/GFP, FACS-sorted for GFP+ cells, expanded and transplanted intracranially into nude mice (5×105 cells/mouse). Mice were gavaged with vehicle (80 mM Citric acid, PH 3.1; n=4) or Dasatinib (50 mg/kg; n=4), started 2 weeks post transplantation, once daily, 5 days a week. Tumor growth was monitored with luminescence imaging weekly.
For intracranial transplantation, 6-week-old female nu/nu mice were anesthetized and injected with tumor cells at indicated cell numbers into the striatum (coordinates: 1.0 AP, −1.8 ML, 4.5 DV with respect to the bregma). Mice were followed until symptomatic.
Intratumoral Dasatinib levels in intracranial and subcutaneous tumors harvested at the end of the experiments were assayed by mass spectrometry done by the Antitumor Assessment Core Facility at MSKCC.
Erbb3 knockdown experiments
Gene block containing short guide RNA (5’-cctaacctccgagtggtccg-3’) targeting mouse Erbb3 under the control of hU6 promoter was synthesized from IDTDNA and cloned into pCW-cas9 vector (Addgene#50661) at BamHI site. Lentiviral particles containing media was prepared following co-transfection of 293T cells with shuttle vector pCW-Cas9-sgErbb3 and packaging vectors psPAX2 and pMD2.G, and used to infect mouse primary GBM cells. Following puromycin selection, Doxycycline was added to the culture at 1 μg/mL to induce Cas9 expression and Erbb3 knockout. Efficiency of Erbb3 knockout was measured by western blot 7 days after Doxycycline induction, and functionally evaluated by NRG1 growth promoting activity using Cell TiterGlo ATP assay.
Western blot Analysis
Primary tissues or cell pellets harvested from culture were homogenized in cold lysis buffer (50 mM Tris (pH 7.5), 150 mM NaCl, 1% NP40, 10% Glycerol, with Protease inhibitor cocktail and PhosSTOP (Roche), incubated on ice for 30 minutes and centrifuged at 12k for 10 minutes at 4°C. Supernatants were collected and quantified for protein concentration, mixed with 2x Laemmli sample buffer (Biorad), denatured on 65°C heating block for 20 minutes. 4–20% Precast protein gels (Biorad) were used for gel running, nitrocellulose membranes were used for transfer, with constant current at 350 mA for 2h in 4°C. 5% non-fat milk in TBST (TBS with 0.1% tween20) was used for blocking and dilution of primary and second antibodies. Phospho-antibodies were diluted in TBST without milk. Membranes were blocked at room temperature (RT) for 1 hour and incubated with primary antibody overnight in 4°C cold room on shaker. Secondary antibodies were applied after wash and incubated at RT for 1 hour. Membranes were then washed with TBST and signals were developed with an enhanced chemiluminescence detection kit (Amersham) and detected with X ray films or Amersham Imager 600.
Quantitative RT-PCR
Total RNA from tissue/cells was extracted using Qiagen RNeasy extraction kit with trizol. iScript cDNA synthesis kit (Biorad) with random hexamer primers was used for cDNA synthesis. Sybr Green (Applied Biosystems) was used for real time detection of PCR product. GAPDH was used as internal control. Please refer to Key Resources Table for primers used. Quantification and statistics were performed from triplicates.
CellTiter-Glo® assay for cell viability
We used CellTiter-Glo® ATP assay as a surrogate measurement of cell viability. For assay of EGF or NRG1 dependent growth, cells were plated at 4000 cells/well in 96-well plates in SFM supplemented only with N2 and B27, serially diluted EGF or NRG1, then added in each well with triplicates of each condition. For assays of drug sensitivity, cells were plated at 4000 cells/well in 96-well plates with both EGF and NRG1 at 10 ng/ml, drugs were serially diluted and applied to each well with triplicates of each condition. All analyses were performed 96 hours after treatment. CellTiter-Glo® kit was used to measure ATP levels according to manufacturer instructions (Promega). All data are presented as mean ± standard deviation (S.D.).
Mouse GBM gene expression analysis
Mouse primary tumor samples were flash frozen in liquid nitrogen and total RNA was extracted using Qiagen Lipid Tissue RNeasy Mini Kit with trizol. Total RNA samples were submitted to the UT Southwestern Microarray Core for processing and Illumina MouseWG-6 BeadChip was used. The Illumina BeadArray microarray data was summarized using the Illumina Beadstudio software and then processed through Bioconductor in R programming environment by beadarray package (Dunning et al., 2007). Two batches of mouse primary tumor samples were prepared at different times, and the batch effect was removed by package inSilicoMerging using DWD method (Taminau et al., 2012). The same processing was used for batch effect removal in the integrated gene expression analysis between TCGA samples, mouse primary tumors and Rembrandt GBM samples. Differentially expressed genes from microarray platform were identified using the limma package (Ritchie et al., 2015) at probe level and then mapped to official gene symbols using package biomaRt (Durinck et al., 2005). Unless specified, genes with adjusted p values less than 0.05 and more than 2-fold change were considered as differentially expressed. Volcano plot was drawn using the calibrate package. Unsupervised hierarchical clustering was performed using the fpc package and A2R algorithm, and gene expression heatmaps were created in R using the gplots package.
Type I and Type II subtyping for human GBM
We defined the mouse Type 1 and 2 signatures as differentially expressed genes (DEG) from comparison between Type 1 and Type 2 mouse tumors (Table S1). We used gene expression data from the Affymetrix platform of 528 patients and downloaded the z-score transformed data from cBioPortal (Cerami et al., 2012; Gao et al., 2013). IDH1 mutant gliomas were removed from further analysis according to the annotations from the Brennan 2013 Cell paper (Brennan et al., 2013). We preselected patient samples with mutually exclusive criteria: for Type I candidates, z-score is greater than 0.5 for both EGFR & SOX9 expression, and less than 0.5 for both ERBB3 & SOX10 expression; and for Type II candidates, z-score is greater than 0.5 for both ERBB3 & SOX10 expression, and less than 0.5 for both EGFR & SOX9 expression. This step produced 107 Type I candidates and 68 Type II candidates. We also evaluated z-score transformed RNAseq data (142 IDH1 wild type GBM in TCGA) curated by cBioPortal that demonstrated a similar distribution as that of the microarray platform, and further confirmed the significant overlap of the Type I&II candidates between the microarray and the RNAseq platforms (Table S2).
Next, we calculated gene set enrichment scores by GSVA package (Hanzelmann et al., 2013) based on single sample gene expression profiles. T1DEGScore was calculated among the Type I candidates using mouse Type 1 upregulated DEGs, resulting in 52 samples with T1DEGScore >0.1 and were defined as TCGA core Type I samples. T2DEGScore was calculated among the Type II candidates using mouse Type 2 upregulated DEGs, and 38 samples with T2DEGScore >0.1 were defined as TCGA core Type II samples (Table S2). We then compared the gene expression profiles of the 52 core Type I versus the rest of the TCGA GBM samples (n=446), and the 38 core Type II versus the rest of TCGA GBM (n=460) to obtain the signatures for human Type I (TIsig) and Type II (TIIsig) using the limma package (Ritchie et al., 2015) at probe level and then mapped to official gene symbols using package biomaRt Table S3). Resultant human Type I and Type II GBM signatures were used to perform GSVA analysis to identify TCGA extended Type I or II samples in remaining non-core samples. Type I extended samples were defined as TIsigScore >0.3 & TIIsigScore<0; and Type II extended samples were defined as TIIsigScore>0.3 & TIsigScore<0 (Table S3).
To test the human signatures in an independent data set, Rembrandt GBM raw data on Affymetrix U133 platform and the annotations were downloaded from Georgetown Database of Cancer (G-DOC) (https://gdoc.georgetown.edu/gdoc/workflows) (Madhavan et al., 2011). The gene expression profiles were extracted and normalized using the gcrma package and the probe intensities was collapsed into unique gene symbols using virtualarray package. Among all 220 Rembrandt GBM samples claimed with raw Affymetrix U133 gene expression profiles on G-DOC, we accurately located 146 GBM samples with raw data. Type I and Type II GBM were identified using human Type I and II signatures by the GSVA algorithm, and we defined Type I GBM by TIsigScore>0.3 & TIIsigScore<0, and Type II GBM by TIsigScore<0 & TIIsigScore>0.3, which resulted in 28 Type I and 26 Type II GBM (Table S3) samples.
We applied data overlap analysis on 369 TCGA patients which have subtype assignment from this study and the recently described 3 subtypes by Wang et al., 2017. Fisher exact test was used for the statistical analysis implemented through the GeneOverlap package in R.
We analyzed the recent single cell smart-seq data set of 20 primary GBM from Neftel et al. (Neftel et al., 2019). To subtype each of the 4,916 single tumor cells, we calculated the TIsigScore and TIIsigScore with our TIsig and TIIsig using AUCell algorithm (Aibar et al., 2017). AUCell calculates the gene set enrichment score using rank-based algorithm specifically in single cell data, resembling the GSVA method which is designed for microarray or bulk RNAseq data, but was optimized to handle sparse matrix like single cell data transcriptome with many zero in gene counts. To determine the subtype for each of the 20 GBM, we generated a pseudo-tissue transcriptome of individual GBM by averaging all available cells that are defined and sequenced by the original study including malignant cells, immune cells and oligodendrocytes, so that we can have close simulation of the tissue and also avoid the sparse single cell count matrix. We applied the GSVA method to the pseudo-tissue transcriptome the same way as described in handling TCGA data, and the subtypes were identified as labeled in Figure S3.
Lineage marker analysis using single cell sequencing data from adult mouse brain SVZ
Single cell sequencing data were downloaded from Gene Expression Omnibus as GSE109447. Gene expression profiles of 29,319 cells from adult mouse ventricular-subventricular zone in the study of Mizrak were used in our lineage marker analysis (Mizrak et al., 2019). The mapped counts matrix was normalized by the library size using edgeR package. Cell type assignment of 29,319 cells was adapted to the original study that classified 4,789 cells as a population consisting of NSCs, transient activating cells and neuroblasts, and 518 cells as OPC population. Gene expression of selected lineage markers were extracted from specific cells and averaged to calculate the fold changes.
GBM PDX primary cell sequencing and subtyping
Fourteen primary cultures of GBM PDXs were collected and used within 5 passages. Total RNA of primary cells was extracted using Qiagen mini RNA extraction kit with trizol. RNA samples were submitted to Weil Cornell Medicine genomics core for RNA-seq using Hiseq4000 Illumina sequencer. The transcript raw counts were aligned to Refseq related to build GRCh37-hg19 using STAR 2.4.2.8 by default settings on MSKCC high performance computation clusters.
The genes from raw counts matrix of 14 samples were kept only when at least 2 out of 14 samples have normalized counts per million mapped reads more than one calculated by edgeR package. To determine the subtypes of PDX cultures, we applied the same GSVA algorithm to calculate single sample enrichment scores using differentially expressed genes (adjusted p >0.05 and fold change >2) from direct comparison of TCGA Type I and Type II core GBM as gene sets. The TIDEG included 134 upregulated genes in Type I GBM core sample and the TIIDEG had 146 genes (Table S5). In consideration of the differences between in vivo primary tumor tissue and in vitro primary cultures, we applied a less stringent criteria for subtyping PDX cells, where Type I GBM is defined by TIDEGScore>0 & TIIDEGScore<0 and Type II GBM is TIDEGScore<0 and TIIDEGScore>0. We obtained 4 Type I, 7 Type II and 3 non-Type I & II GBM cultures from the PDX cohort.
Patient Survival analysis
Clinical information of TCGA GBM data was downloaded from cBioPortal. The final 498 GBM from TCGA was stratified by our Type I and Type II tumor criteria, the progression free survival comparing Type I and Type II GBM were analyzed using multivariate survival analysis using Cox Regression model considering subtypes, age and the interaction between age and subtypes by survival package and survcomp package (Schroder et al., 2011). The survival progression free data are available for 139 Type I, 91 Type II and 260 Non-TITII characterized in the study according to TCGA annotations (Brennan et al., 2013).
Methylation analysis
Level 3 DNA methylation data of all TCGA GBM samples were downloaded through the NIH data portal at https://portal.gdc.cancer.gov/. Data were processed using ChAMP package (Morris et al., 2014). For both platforms, Infinium Human Methylation 450K BeadChip (HM450K) and Infinium Human Methylation27 BeadChip (HM27K), SNPs that may influence the analysis were removed according to the literature (Zhou et al., 2017). Probes mapped to the x and y chromosomes and multi-hit probes were removed as instructed in ChAMP package. Multidimensional Scaling Plot (MDSplot) was generated based on the top 1000 most variable probes amongst all samples for both HM450K and HM27K. The clustering plot for all samples was generated using the filtered probes, specifically 331,895 probes for HM450K data and 19,111 probes for HM27K data.
Gene copy number alteration (CNA) and mutation analysis
Designated Type I and Type II sample IDs as well as IDs for all 498 patients in the study cohort were used to retrieve CNAs and mutation data files from cBioPortal, and available from supplemental Table S4 (CNAs and mutation). Mutations and CNAs analysis of GBM in PDX are summarized in Table S5. Statistical analysis was performed using Fisher’s exact test of two proportions.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis between groups was performed using two-tailed unpaired Student’s t-test. Fisher’s exact test for two proportions was used to calculate the p values when comparing frequencies of mutation or copy number alterations between TCGA Type I and Type II GBM populations. Kaplan-Meier survival curves were analyzed using log-rank (Mantel-Cox) test. For bar figures presented, the center line represents mean ± S.D. or mean ± S.E.M., as indicated. Data were analyzed using Excel, GraphPad Prism v.7 and Fiji. p values less than 0.05 were considered significant. Assays are representative of ≥3 independent and biological replicates.
Supplementary Material
Table S1, Related to Figure 1. DEGs and selected list of enriched biological process terms of highly expressed genes of GEMM Type 1 vs. Type 2 GBM microarray.
Table S2, Related to Figure 3. Overlap of Type I/II candidates between microarray and RNAseq platforms, and T1DEGScore and T2DEGScore of the Type 1 and Type 2 GBM candidate samples by GSVA analysis.
Table S3, Related to Figure 3. TCGA Type I and II GBM signatures and sample IDs, and Rembrandt GBM GSVA scores with subtype assignment.
Table S4, Related to Figure 4. CNAs and gene mutations in TCGA Type I and II GBM and overall data set.
Table S5, Related to Figure 5. PDX genotyping and TIDEG and TIIDEG from TCGA Type I core vs. Type II core analysis.
Table S6, Related to Figure 6. List of chemotherapeutic agents tested in the study.
Table S7, Related to Figure 7. Intratumoral Dasatinib levels in intracranial Type II and subcutaneous (SQ) Type 2 tumors.
Significance.
Glioblastoma is a highly heterogenous tumor that lacks effective treatment options. To date, previous molecular classification strategies for glioblastoma have not strongly impacted clinical practice. We demonstrate here a species-conserved biological and functional stratification for glioblastoma. This discrimination reveals therapeutic sensitivities in mouse and human tumors that are masked in the absence of classification. Thus, cell-of-origin-based glioblastoma molecular subtyping has the potential to prospectively stratify patients with predicted similar responses to treatment. This could help improve the design, interpretation, and outcome of glioblastoma clinical trials which have so far been disappointing compared to other cancer types.
Highlights.
Tumor suppressor loss in NSCs and OLCs generate distinct murine GBM subtypes
Human GBM subtypes identified based on species-conserved transcriptional profiles
Mouse and human cell lineage subtypes exhibit distinct tumor properties
OLC-associated GBM show ERBB3 dependence and unique therapeutic sensitivity
Acknowledgments
We thank D. K. Burns (UT Southwestern) for pathological consultation and Neal Rosen for helpful discussions. We thank the MSKCC Brain Tumor Center staff for PDX and primary GBM cultures and the Antitumor Assessment Core Facility for mass spectrometry. L.F.P. received funding from NCI R35: CA210100; R01: CA131313; NIH/NCI Cancer Center Support Grant P30 CA008748, and holds the Albert C. Foster Chair in Cancer Research.
Footnotes
Declaration of Interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat Methods 14, 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, and Parada LF (2009). Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer cell 15, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno S, Sun D, Pedraza AM, Vera E, Wang Z, Burns DK, and Parada LF (2019). Cell-of-origin susceptibility to glioblastoma formation declines with neural lineage restriction. Nat Neurosci 22, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno SR, Wang Z, Sun D, Chen J, Xu J, Kim E, Hatanpaa KJ, Raisanen JM, Burns DK, Johnson JE, et al. (2015). Adult Lineage-Restricted CNS Progenitors Specify Distinct Glioblastoma Subtypes. Cancer cell 28, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KPL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD, et al. (2013). Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer cell 24, 331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et al. (2011). High density DNA methylation array with single CpG site resolution. Genomics 98, 288–295. [DOI] [PubMed] [Google Scholar]
- Boldrini M, Fulmore CA, Tartt AN, Simeon LR, Pavlova I, Poposka V, Rosoklija GB, Stankov A, Arango V, Dwork AJ, et al. (2018). Human Hippocampal Neurogenesis Persists throughout Aging. Cell Stem Cell 22, 589–599 e585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. (2013). The somatic genomic landscape of glioblastoma. Cell 155, 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kwon CH, Lin L, Li Y, and Parada LF (2009). Inducible site-specific recombination in neural stem/progenitor cells. Genesis 47, 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, and Parada LF (2012a). A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 488, 522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, McKay RM, and Parada LF (2012b). Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell 149, 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Smith-Cohn M, Cohen AL, and Colman H (2017). Glioma Subclassifications and Their Clinical Significance. Neurotherapeutics 14, 284–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman JK (2002). 5-Azacytidine and 5-aza-2’-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21, 5483–5495. [DOI] [PubMed] [Google Scholar]
- Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, and Zarrinkar PP (2011). Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol 29, 1046–1051. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, and Alvarez-Buylla A (2002). EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron 36, 1021–1034. [DOI] [PubMed] [Google Scholar]
- Dunning MJ, Smith ML, Ritchie ME, and Tavare S (2007). beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 23, 2183–2184. [DOI] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, and Huber W (2005). BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440. [DOI] [PubMed] [Google Scholar]
- Ernst A, Alkass K, Bernard S, Salehpour M, Perl S, Tisdale J, Possnert G, Druid H, and Frisen J (2014). Neurogenesis in the striatum of the adult human brain. Cell 156, 1072–1083. [DOI] [PubMed] [Google Scholar]
- Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, Keir ST, Ji AX, Zoppoli P, Niola F, et al. (2013). The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet 45, 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves JT, Schafer ST, and Gage FH (2016). Adult Neurogenesis in the Hippocampus: From Stem Cells to Behavior. Cell 167, 897–914. [DOI] [PubMed] [Google Scholar]
- Gould E, Reeves AJ, Graziano MS, and Gross CG (1999). Neurogenesis in the neocortex of adult primates. Science 286, 548–552. [DOI] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, and Wu H (2001). Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science 294, 2186–2189. [DOI] [PubMed] [Google Scholar]
- Hantschel O, Rix U, and Superti-Furga G (2008). Target spectrum of the BCR-ABL inhibitors imatinib, nilotinib and dasatinib. Leuk Lymphoma 49, 615–619. [DOI] [PubMed] [Google Scholar]
- Hanzelmann S, Castelo R, and Guinney J (2013). GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron TP (2016). Small Molecule Kinase Inhibitors for the Treatment of Brain Cancer. J Med Chem 59, 10030–10066. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Zhao D, Chen J, Alcantara S, Li Y, Burns DK, Mason RP, Lee EY, Wu H, and Parada LF (2008). Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer research 68, 3286–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassman AB, Pugh SL, Gilbert MR, Aldape KD, Geinoz S, Beumer JH, Christner SM, Komaki R, DeAngelis LM, Gaur R, et al. (2015). Phase 2 trial of dasatinib in target-selected patients with recurrent glioblastoma (RTOG 0627). Neuro Oncol 17, 992–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Clemenson GD, and Gage FH (2012). New neurons in an aged brain. Behav Brain Res 227, 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lee KF, Nikitin AY, Hilsenbeck SG, Cardiff RD, Li A, Kang KW, Frank SA, Lee WH, and Lee EY (2004). Somatic mutation of p53 leads to estrogen receptor alpha-positive and -negative mouse mammary tumors with high frequency of metastasis. Cancer research 64, 3525–3532. [DOI] [PubMed] [Google Scholar]
- Lindquist RA, Guinto CD, Rodas-Rodriguez JL, Fuentealba LC, Tate MC, Rowitch DH, and Alvarez-Buylla A (2016). Identification of proliferative progenitors associated with prominent postnatal growth of the pons. Nat Commun 7, 11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Hon GC, Villa GR, Turner KM, Ikegami S, Yang H, Ye Z, Li B, Kuan S, Lee AY, et al. (2015). EGFR Mutation Promotes Glioblastoma through Epigenome and Transcription Factor Network Remodeling. Mol Cell 60, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, and Kleihues P (2007). The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114, 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan S, Gusev Y, Harris M, Tanenbaum DM, Gauba R, Bhuvaneshwar K, Shinohara A, Rosso K, Carabet LA, Song L, et al. (2011). G-DOC: a systems medicine platform for personalized oncology. Neoplasia 13, 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, and Buetow K (2009). Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res 7, 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinodan M, Rosen KM, Ito S, and Corfas G (2012). A critical period for social experience-dependent oligodendrocyte maturation and myelination. Science 337, 1357–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mich JK, Signer RA, Nakada D, Pineda A, Burgess RJ, Vue TY, Johnson JE, and Morrison SJ (2014). Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. Elife 3, e02669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrak D, Levitin HM, Delgado AC, Crotet V, Yuan J, Chaker Z, Silva-Vargas V, Sims PA, and Doetsch F (2019). Single-Cell Analysis of Regional Differences in Adult V-SVZ Neural Stem Cell Lineages. Cell Rep 26, 394–406 e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, and Beck S (2014). ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics 30, 428–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, et al. (2019). An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 178, 835–849 e821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, Ehrenberger T, Grobner S, Segura-Wang M, Zichner T, Rudneva VA, et al. (2017). The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottebohm F (2005). The neural basis of birdsong. PLoS Biol 3, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K, Charles N, Michor F, and Holland EC (2014). Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer cell 26, 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park NI, Guilhamon P, Desai K, McAdam RF, Langille E, O’Connor M, Lan X, Whetstone H, Coutinho FJ, Vanner RJ, et al. (2017). ASCL1 Reorganizes Chromatin to Direct Neuronal Fate and Suppress Tumorigenicity of Glioblastoma Stem Cells. Cell Stem Cell 21, 411. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. (2008). An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A, Chaker Z, and Doetsch F (2017). Hypothalamic regulation of regionally distinct adult neural stem cells and neurogenesis. Science 356, 1383–1386. [DOI] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. (2006). Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer cell 9, 157–173. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saggar JK, Yu M, Tan Q, and Tannock IF (2013). The tumor microenvironment and strategies to improve drug distribution. Front Oncol 3, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann T, Bourgon R, Garcia J, Li C, Cloughesy T, Chinot OL, Wick W, Nishikawa R, Mason W, Henriksson R, et al. (2015). Patients With Proneural Glioblastoma May Derive Overall Survival Benefit From the Addition of Bevacizumab to First-Line Radiotherapy and Temozolomide: Retrospective Analysis of the AVAglio Trial. J Clin Oncol 33, 2735–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff D, and Sarkaria J (2015). Dasatinib in recurrent glioblastoma: failure as a teacher. Neuro Oncol 17, 910–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder MS, Culhane AC, Quackenbush J, and Haibe-Kains B (2011). survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics 27, 3206–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CE, Wynn SL, Sesay A, Cruz C, Cheung M, Gomez Gaviro MV, Booth S, Gao B, Cheah KS, Lovell-Badge R, et al. (2010). SOX9 induces and maintains neural stem cells. Nat Neurosci 13, 1181–1189. [DOI] [PubMed] [Google Scholar]
- Segerman A, Niklasson M, Haglund C, Bergstrom T, Jarvius M, Xie Y, Westermark A, Sonmez D, Hermansson A, Kastemar M, et al. (2016). Clonal Variation in Drug and Radiation Response among Glioma-Initiating Cells Is Linked to Proneural-Mesenchymal Transition. Cell Rep 17, 2994–3009. [DOI] [PubMed] [Google Scholar]
- Sorrells SF, Paredes MF, Cebrian-Silla A, Sandoval K, Qi D, Kelley KW, James D, Mayer S, Chang J, Auguste KI, et al. (2018). Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 555, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, and Tavare S (2013). Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proceedings of the National Academy of Sciences of the United States of America 110, 4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, and Wegner M (2002). Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes & development 16, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taminau J, Meganck S, Lazar C, Steenhoff D, Coletta A, Molter C, Duque R, de Schaetzen V, Weiss Solis DY, Bersini H, et al. (2012). Unlocking the potential of publicly available microarray data using inSilicoDb and inSilicoMerging R/Bioconductor packages. BMC Bioinformatics 13, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. (2010). Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell 17, 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Pol SU, Haberman AK, Wang C, O’Bara MA, and Sim FJ (2014). Transcription factor induction of human oligodendrocyte progenitor fate and differentiation. Proceedings of the National Academy of Sciences of the United States of America 111, E2885–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, deCarvalho AC, Lyu S, Li P, Li Y, et al. (2017). Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer cell 32, 42–56 e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Laird PW, and Shen H (2017). Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res 45, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Hill RA, Dietrich D, Komitova M, Suzuki R, and Nishiyama A (2011). Age-dependent fate and lineage restriction of single NG2 cells. Development 138, 745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Guignard F, Zhao D, Liu L, Burns DK, Mason RP, Messing A, and Parada LF (2005). Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer cell 8, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, Marth JD, and Parada LF (2001). Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes & development 15, 859–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H, Parada LF, and Baker SJ (2015). Cell of origin for malignant gliomas and its implication in therapeutic development. Cold Spring Harb Perspect Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Related to Figure 1. DEGs and selected list of enriched biological process terms of highly expressed genes of GEMM Type 1 vs. Type 2 GBM microarray.
Table S2, Related to Figure 3. Overlap of Type I/II candidates between microarray and RNAseq platforms, and T1DEGScore and T2DEGScore of the Type 1 and Type 2 GBM candidate samples by GSVA analysis.
Table S3, Related to Figure 3. TCGA Type I and II GBM signatures and sample IDs, and Rembrandt GBM GSVA scores with subtype assignment.
Table S4, Related to Figure 4. CNAs and gene mutations in TCGA Type I and II GBM and overall data set.
Table S5, Related to Figure 5. PDX genotyping and TIDEG and TIIDEG from TCGA Type I core vs. Type II core analysis.
Table S6, Related to Figure 6. List of chemotherapeutic agents tested in the study.
Table S7, Related to Figure 7. Intratumoral Dasatinib levels in intracranial Type II and subcutaneous (SQ) Type 2 tumors.
Data Availability Statement
Genomic data generated in this study have been downloaded to the Gene Expression Omnibus (GEO numbers GSE127273 and GSE127274).