Abstract
Background
In cardiac gene therapy to improve contractile function, achieving gene expression in the majority of cardiac myocytes is essential. In preventing cardiac arrhythmias, however, this goal may not be as important, since transduction efficiencies as low as 40% suppressed ventricular arrhythmias in genetically-modified mice with polymorphic catecholaminergic ventricular tachycardia (CPVT).
Methods
Using computational modeling, we simulated 1D, 2D and 3D tissue under a variety of conditions to test the ability of genetically-engineered non-arrhythmogenic “stabilizer cells” to suppress triggered activity (TA) due to delayed (DAD) or early (EAD) afterdepolarizations.
Results
Due to source-sink relationships in cardiac tissue, a minority (20–50%) of randomly distributed “stabilizer” cells engineered to be non-arrhythmogenic can suppress the ability of arrhythmogenic cells to generate DAD- and EAD-related arrhythmias. Stabilizer cell gene therapy strategy can be designed to correct a specific arrhythmogenic mutation, as in the CPVT mice studies, or more generally to suppress DADs or EADs from any cause by overexpressing the inward rectifier K channel Kir2.1 in stabilizer cells.
Conclusions
This promising antiarrhythmic strategy warrants further testing in experimental models to evaluate its clinical potential.
Keywords: arrhythmia, death, sudden, gene therapy, catecholaminergic polymorphic ventricular tachycardia, long QT syndrome, afterdepolarizations
Journal Subject Terms: Arrhythmias, Ventricular Fibrillation, Basic Science Research, Gene Therapy, Computational Biology
Graphical Abstract

Introduction
A current limitation of cardiac gene therapy is achieving gene expression in a high enough percentage of myocytes to reverse the disease phenotype at the whole organ level1. However, partial gene transduction may be less of a limitation for gene therapy of cardiac arrhythmias, particularly those arising from spontaneous TA (TA) due to delayed (DADs) or early afterdepolarizations (EADs). In several recent gene therapy studies in catecholaminergic polymorphic ventricular tachycardia (CPVT) mouse models, gene transduction in a minority of ventricular myocytes in the range of 40–66% was shown to be surprisingly effective at suppressing CPVT DAD-mediated ventricular arrhythmias 2–5. The explanation may lie in the fact that the heart is an electrical syncytium such that a single myocyte generating a DAD or EAD has no chance of initiating a propagating action potential (AP) in normally well-coupled tissue because the surrounding normal myocytes act as current sinks that prevent its voltage from deviating significantly from theirs 6–10. To overcome this source-sink mismatch, it has been estimated that the number of adjacent pacemaking, DAD- or EAD-generating myocytes required to trigger an AP ranges from the tens in 1D tissue, to thousands in 2D tissue and to hundreds of thousands in 3D tissue 11, 12, although factors such as reduced gap junction coupling and fibrosis can significantly lower these numbers.
In the present computational study, we had two objectives. The first was to investigate systematically the gene transduction efficiency required to suppress DAD-mediated TA in CPVT by simulated correction of the molecular defect. Accordingly, we incorporated spontaneous sarcoplasmic reticulum (SR) Ca release, the defect precipitating DAD-mediated arrhythmias in CPVT, into simulated cardiac tissue. We then randomly dispersed stabilizer cells, rendered non-arrhythmogenic by correcting their arrhythmogenic molecular defect with simulated gene therapy, into the tissue. The percentage of stabilizer cells required to suppress DAD-TA was then characterized under a variety of conditions and compared to experimental mouse CPVT studies. Our second objective was to explore through predictive modeling whether a more generalized stabilizer cell gene therapy strategy could be developed to suppress arrhythmias triggered by either DADs or EADs regardless of the underlying molecular causes. We show that overexpression the inward rectifier K current Kir 2.1 is a promising candidate to target the general dynamical mechanisms of DAD and EAD-mediated TA irrespective of the specific underlying molecular drivers.
Based on this analysis, stabilizer cell gene therapy shows promise as an antiarrhythmic strategy and warrants further testing in experimental arrhythmia models to evaluate its potential clinical relevance.
Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request. No IRB approval was necessary since only simulations were performed.
Experimental Design
Tissue simulations in 1D cables (100 myocytes), 2D tissue (100×100 myocytes) and 3D tissue (100×100×100 myocytes) were carried out using the UCLA rabbit ventricular AP cell model previously described in Mahajan et al 13, 14. The partial differential equation governing voltage is:
| [1] |
where V is the membrane voltage and Cm is the membrane capacitance (1 μF/cm2). The diffusion term in Eq.1 is for 1D; for 2D; and for 3D. Further details are provided in the supplementary material.
DADs were generated by commanding spontaneous release of SR Ca as previously described in Liu et al 15. Spontaneous SR Ca release strength was determined by the parameter Gspon which controls spontaneous Ca release flux from the SR into the cytoplasm. The timing of Ca release was set to follow a Gaussian distribution with standard deviation of 50 ms unless otherwise specified. EADs were generated as described in Xie et al 11. Further details on the DAD and EAD models are provided in the supplementary material.
Stabilizer cells were generated using two different strategies. In the first strategy, Gspon was set to 0 to prevent any spontaneous SR Ca release from occurring. In the second strategy, the conductance of the inward rectifier K current (GK1) in the UCLA model13, 14 was enhanced, and also compared to alternative GK1 formulations from the Ten-Tuscher16 and O’Hara-Rudy17 models. Alternatively, an additional current from a weak inward rectifier K conductance from Kir1.1 channels (GKir1.1) was formulated as: and added to model’s control strong inward rectifier K current. Stabilizer cells were interspersed in tissue at various percentages using a uniformly random distribution of either single cells, 3×3 cell clusters, or 5×5 cell clusters. In the simulations testing heterogenous transduction, a central circular region devoid of stabilizer cells was created.
Fibrosis was simulated by randomly removing a percentage of cell-to-cell connections from a central circular region of the tissue. Cell-to-cell diffusion term was modified for each dimension x to be , where is a random number between 0 and 1 for each dimension to set the desired % fibrosis and Vi,j,k is the cell voltage. Breakthrough TA was defined as successful propagation into the surrounding regions with normal cell-to-cell connections.
Statistical methods
Statistical probability of TA was assessed by performing 100 trials of randomized stabilizer cell distributions (or removal of cell-to-cell connections in fibrosis simulations) and reporting the percentage of randomized trials with TA for each parameter set.
Results
Modeling experimental CPVT gene therapy trials
CPVT is caused by mutations in either ryanodine receptors (CPVT1) or calsequestrin (CPVT2) that result functionally in inappropriate spontaneous SR Ca release. During diastole, spontaneous SR Ca release activates inward currents such as the Na-Ca exchange current that cause membrane depolarization, resulting in a DAD that can trigger an AP. To simulate the functional effects of CPVT mutations, we modified the UCLA rabbit ventricular cell model14 to permit spontaneous diastolic SR Ca release on command by activating the model parameter Gspon controlling SR Ca release flux into the cytoplasm (see Methods). The resulting intracellular Ca release activated inward Na-Ca exchange current to generate a DAD with amplitude proportionate to Gspon, which triggered an AP at a critical Gspon value (Fig. 1A). Since the timing of spontaneous SR Ca release events from myocyte to myocyte is variable in cardiac tissue18, the timing of Gspon activation in each cell was randomly selected from a Gaussian distribution with a standard deviation of 50 ms, based on experimental measurements in rabbit and rat ventricular myocytes18, 19. Using this cell model, we first simulated a 1D cable of arrhythmogenic cells in which the commanded Gspon was set above the threshold value required for the DAD to trigger an AP (Fig. 1B, left panel, red traces).
Figure 1.
Suppression of DAD-mediated TA by stabilizer cells. (A) Simulations of a single uncoupled myocyte exhibiting a DAD using a commanded spontaneous Ca release by activating Gspon. Voltage (left) and Ca transient (right) traces are shown for different Gspon values controlling the strength of Ca release. For a single uncoupled cell, TA occurs at a Gspon of 0.07 ms−1 or higher. (B) A homogeneous 1D cable with Gspon = 0.07 ms−1 in all cells results in TA (left, red traces). TA is suppressed when 20% of the cells are replaced with non-arrhythmogenic stabilizer cells with Gspon = 0 to attenuate the tissue DAD amplitude (right, black traces). Note that in B and C, only every 10th cell trace is plotted, so not all stabilizer cells are shown.
To model the experimental gene therapy studies, we assumed that transduced myocytes in those studies had been converted into non-arrhythmogenic stabilizer cells by expressing sufficient gene product to suppress spontaneous SR Ca release. Accordingly, we modeled stabilizer cells by disabling the commanded Gspon activation to prevent spontaneous SR Ca release. Stabilizer cells were then interspersed randomly among arrhythmogenic cells. Fig. 1B (right panel) shows an example of a 1D cable with 20% stabilizer cells (black traces) interspersed among 80% of arrhythmogenic cells (red traces, same parameters as in the left panel). The 20% of randomly distributed non-arrhythmogenic stabilizer cells without DADs created a sufficient sink to prevent the 80% of arrhythmogenic cells with DADs from triggering an AP anywhere in the cable.
Effects of tissue dimension
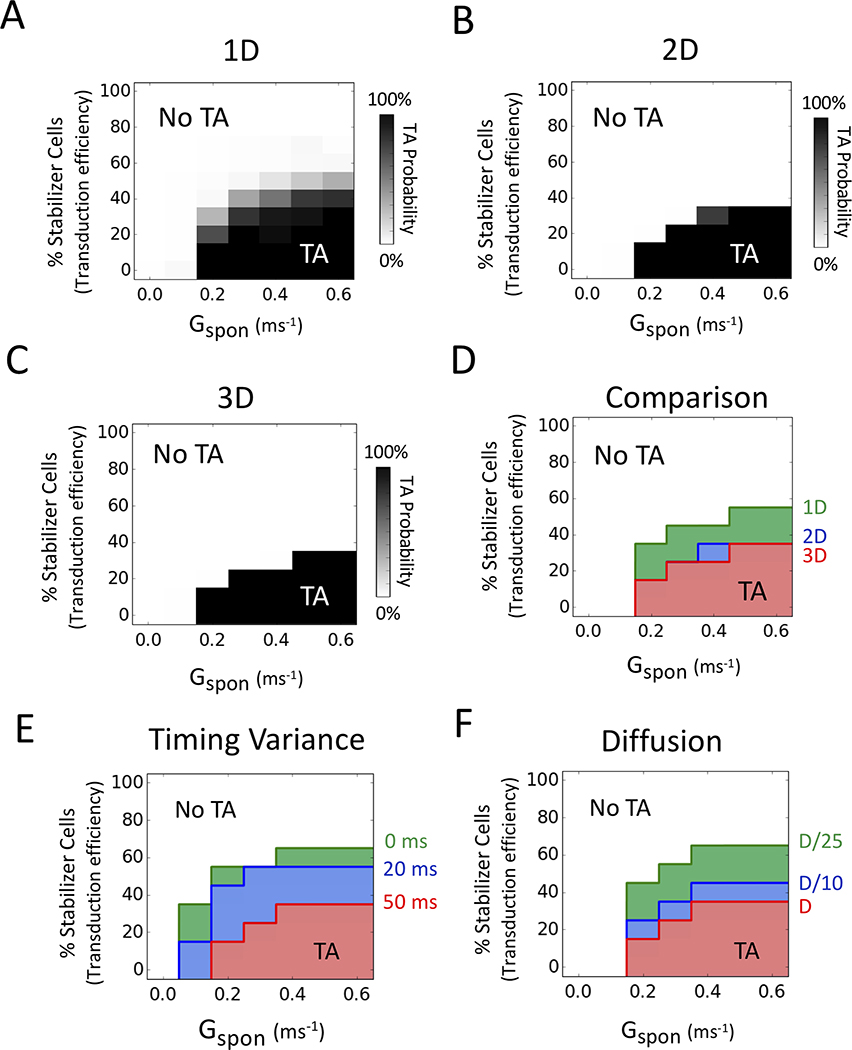
The results from 1D cables over a wide range of Gspon values (to vary the DAD amplitude in the arrhythmogenic cells) are summarized in Fig. 2A. For each Gspon value, 100 random spatial distributions of non-arrhythmogenic stabilizer cells were tested, and the percentage of those 100 trials triggering AP’s are shown on a gray scale as a function of the percentage of stabilizer cells (equivalent to transduction efficiency) in the tissue. Fig. 2 also shows the results for 2D (Fig. 2B) and 3D tissue (Fig. 2C) when stabilizer and arrhythmogenic cells were intermixed in a randomly uniform distribution. The percentage required was higher in 1D (30–50%) than in 2D or 3D tissue (15–30%) (Fig. 2D). Higher values of Gspon corresponding to increased intrinsic DAD amplitude modestly increased the percentage of randomly distributed stabilizer cells required to prevent TA, but still remained below 50%.
Figure 2.
Effects of tissue dimension, DAD timing and reduced diffusion coefficient. Stabilizer cells (with Gspon = 0) were interspersed randomly throughout the tissue. DAD timing had a standard deviation of 50 ms unless otherwise specified. 100 simulations were performed at each parameter combination to obtain the TA probability. (A) Probability of TA in 1D cables (100 cells) versus % stabilizer cells in the tissue (as a surrogate for transduction efficiency) over a range of Gspon values controlling the spontaneous SR Ca release strength. (B) Same in 2D tissue (100×100 cells). (C) Same in 3D tissue (100×100×100 cells). (D) Comparison plot of 1D (green), 2D (blue), and 3D (red) tissue. The envelopes delineating TA probability >10% are superimposed here and in E-F. (E) Effect of the standard deviation of DAD timing (timing variance) on the probability of TA versus % stabilizer cells and Gspon, for standard deviations of 50 ms (red), 20 ms (blue), and 0 ms (green). (F) Effect of reduced diffusion coefficient D (reflecting gap junction coupling) on the probability of TA versus % stabilizer cells and Gspon, for the control value D = 0.0005 cm2/ms (red), D/10 (blue) and D/25 (green).
Effects of synchrony of spontaneous Ca release events
In the above simulations, we assumed that spontaneous Ca release triggered by Gspon activation did not occur synchronously in all arrhythmogenic cells, but followed a Gaussian distribution with standard deviation of 50 ms19, reflecting the stochastic nature of spontaneous SR Ca release. In coupled tissue, the effect of this asynchrony is to reduce the DAD amplitude and broaden its duration in tissue compared to a single cell. To examine how the variance in Gspon activation time affects the ability of randomly-distributed stabilizer cells to suppress TA by the arrhythmogenic cells, we performed 2D tissue simulations in which the standard deviation of spontaneous SR Ca release was reduced from 50 ms to 20 ms or to the nonphysiological extreme of 0 ms. As shown in Fig. 2E, reducing the standard deviation to 20 ms or 0 ms increased the percentage of stabilizer cells required to suppress TA. However, even in the extreme case of zero standard deviation, the percentage of randomly distributed stabilizer cells required to suppress TA remained well below 100%, ranging from 30–60% depending on the Gspon value.
Effects of reduced gap junction conductance (diffusion coefficient)
We also studied the effects of the diffusion coefficient reflecting gap junction conductance between cells. Reducing the diffusion coefficient 10-fold or 25-fold, corresponding to 3.1-fold and 5-fold decreases in conduction velocity respectively, modestly increased the percentage of stabilizer cells required to suppress TA, as shown in Fig. 2F.
Effects of spatial heterogeneity of stabilizer cell distribution
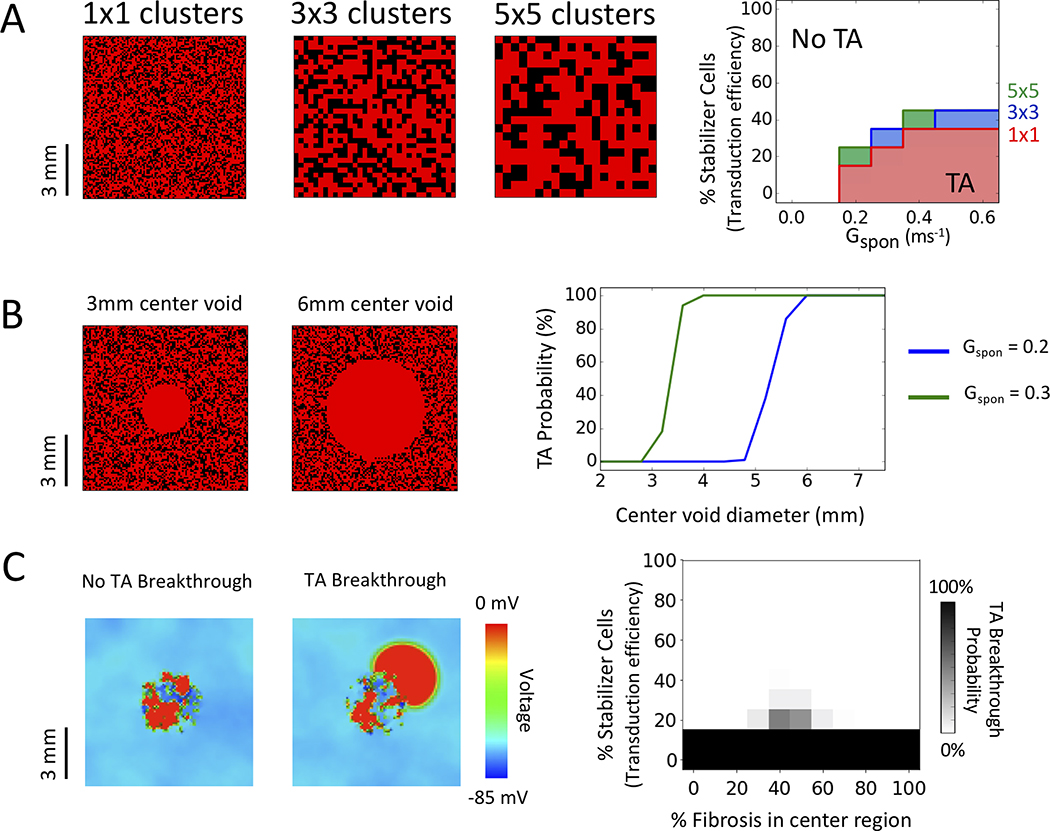
In the above simulations, we interspersed non-arrhythmogenic stabilizer cells among arrhythmogenic cells in a randomly uniform distribution. However, in experimental gene therapy, transduction efficiency can vary widely throughout the tissue, at both microscopic and macroscopic spatial scales20. To examine how microscopic variation in transduction efficiency impacts the ability of stabilizer cells to suppress TA, we compared simulated 2D tissue in which stabilizer cells were randomly distributed singly or in 3×3 or 5×5 cell clusters (Fig. 3A, left panels). As cluster size increased, the percentage of stabilizer cells required to suppress TA increased mildly from 15–30% for single cells to 20–40% for 3×3 or 5×5 clusters (Fig. 3A, right panel).
Figure 3.
Effects of clusters, macroscopic defects and fibrosis. All results are for 2D tissue (100×100 cells, with Gspon=0.2 ms−1, a standard deviation of DAD timing of 50 ms for arrhythmogenic cells and Gspon = 0 for stabilizer cells). (A) Stabilizer cell clustering. Left panels: Patterns of stabilizer cells (40%) randomly distributed singly or in clusters of 3×3 cells or 5×5 cells. DAD-generating cells are red, stabilizer cells black. Right panel: Corresponding probability of TA as a function of % stabilizer cells (transduction efficiency) for randomly distributed single (red), 3×3 (blue) or 5×5 clusters (green) in 2D tissue. Envelopes delineating TA probability >10% are superimposed. (B) Macroscopic defects. Left: Central regions devoid of stabilizer cells, with 40% randomly distributed stabilizer cells elsewhere. Right panel: TA probability as a function of the central region diameter, for the two different Gspon values indicated. (C) Fibrosis. Cell-to-cell connections were removed from a central circular region (diameter 4 mm, 40 cells) to simulate fibrosis, with 40% stabilizer cells distributed randomly throughout the entire tissue. Left panels: Typical voltage snapshots with 50% of cell-to-cell connections randomly removed from the central region (i.e. 50% fibrosis). Depending on the randomization pattern of removed connections, TA in the fibrotic region either did not (No TA Breakthrough) or did (TA Breakthrough) propagate into the surrounding normally-coupled tissue. Right panel: TA breakthrough probability versus % stabilizer cells and % fibrosis.
To assess macroscopic heterogeneities, we simulated 2D tissue containing 40% stabilizer cells sufficient to suppress DAD-mediated TA, and tested how large of a region devoid of stabilizer cells could be tolerated without TA returning. Fig. 3B shows the probability of TA as the diameter of the central region devoid of stabilizer cells was gradually increased. For a Gspon value of 0.20 (just above the threshold for TA in unstabilized tissue), TA reappeared when the diameter exceeded 48 cells (approximately 4.8 mm). For a larger Gspon value of 0.30, TA reappeared when the diameter exceeded 28 cells (approximately 2.8 mm).
Effects of simulated fibrosis
By disrupting cell-to-cell communication between stabilizer cells and arrhythmogenic cells, tissue fibrosis may reduce the sink effect of stabilizer cells required to suppress TA. On other hand, severe fibrosis may also block propagation of that TA into surrounding non-fibrotic tissue. To investigate the interplay between these factors, we simulated 2D tissue (100×100 cells) with an adequate percentage of stabilizer cells to suppress TA. Cell-to-cell connections were then progressively removed from a central circular region (4 mm diameter or 40 cells across) to determine: 1) whether TA recurred in the central region and 2) if so, whether it could propagate outwards and break through into the surrounding normally-coupled tissue. We started with tissue in which 15% of randomly-distributed stabilizer cells were sufficient to suppress TA in a central region. Random removal of 50% of cell-to-cell connections from the central region allowed TA to emerge in the central region, but propagation into the surrounding tissue failed (Fig. 3C, left voltage snapshot). With a different randomized removal of 50% of cell-to-cell connections in the central region, however, propagation into the non-fibrotic normal region succeeded (Fig. 3C, right voltage snapshot). Fig. 3C plots the percentage of stabilizer cells required to prevent TA breakthrough versus the percentage of cell-to-cell connections removed from the central region (“% fibrosis”). For each % fibrosis value in the central region, 100 randomized spatial distributions of removing cell-to-cell connections were tested, and the percentage of trials in which TA successfully propagated into the surrounding normally-coupled tissue (breakthrough TA) is plotted on a gray scale. With 40 to 60% fibrosis in the central region, a modestly increased percentage of stabilizer cells was required to suppress TA breakthrough (from 10–15% to 25–30%). Below 30% fibrosis, the sink effect of stabilizer cells was sufficient to suppress almost all TA in the central region (except for completely disconnected arrhythmogenic cells). Above 60% fibrosis, TA in the central region occurred but was unable to propagate into the surrounding normally-coupled tissue.
A general stabilizer cell gene therapy target to prevent both DAD and EAD-mediated triggered arrhythmias
Although correcting the underlying molecular defect promoting spontaneous SR Ca release is the most direct method to suppress DADs in monogenic diseases like CPVT, DAD-mediated ventricular arrhythmias are also common in acquired diseases such as heart failure and ischemic heart disease. In these conditions, the etiology of spontaneous SR Ca release is multifactorial, involving transcriptional and post-translational alterations in multiple proteins 21, making identification of appropriate molecular targets for gene therapy difficult. An alternative to suppressing spontaneous SR Ca release directly is to reduce the sensitivity of the diastolic membrane potential to depolarization by intracellular Ca. To explore this strategy, we created a second type of stabilizer cell in which spontaneous Ca release was left intact, but the inward rectifier K channel conductance GK1, a strong determinant of the dynamic threshold for a DAD to trigger an AP 22, was enhanced. In addition to attenuating DAD amplitude, we reasoned that by increasing repolarization reserve during the AP, enhanced GK1 might also suppress EADs and EAD-mediated TA. In conditions like heart failure in which altered intracellular Ca dynamics concomitantly promotes DADs and reduced repolarization reserve promotes EADs 23, a single gene therapy target suppressing both DAD- and EAD-mediated arrhythmias would be highly desirable.
Enhancing GK1 to suppress DADs and EADs
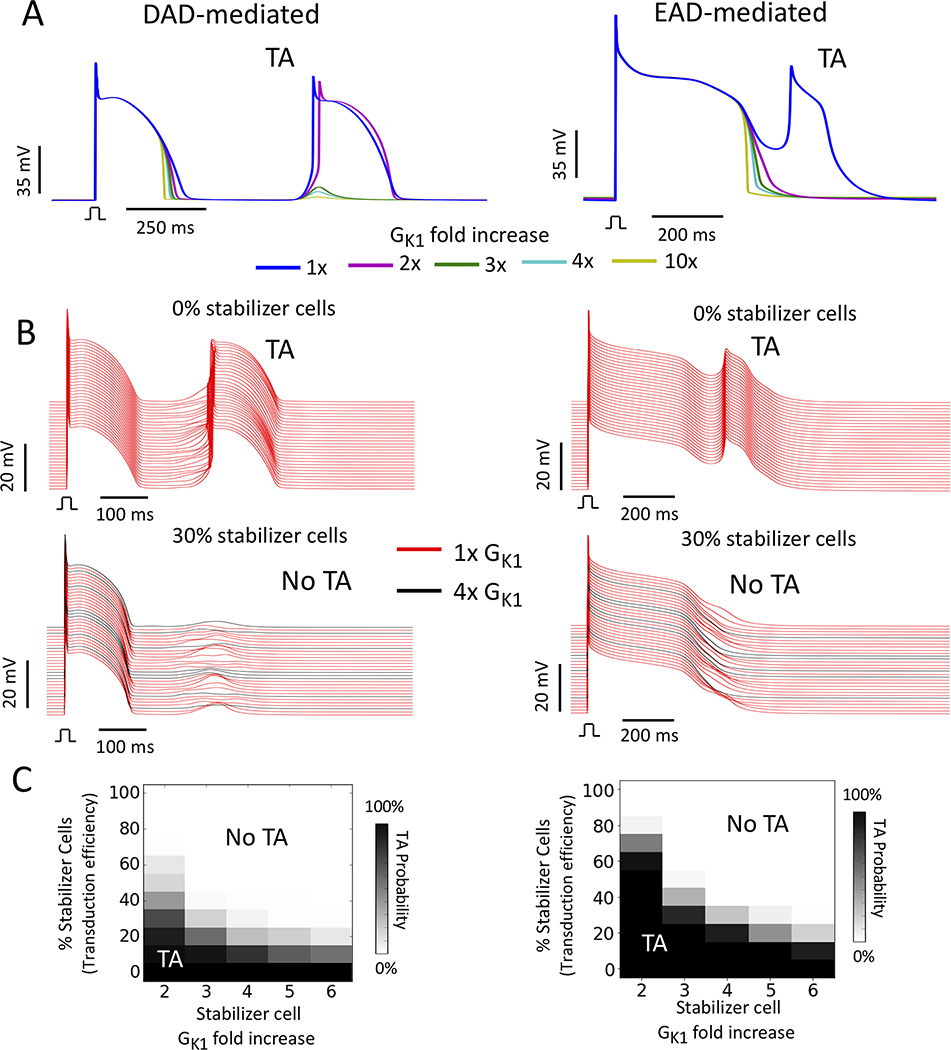
Fig. 4A (left panel) shows the effect of increasing GK1 on a suprathreshold DAD caused by commanding spontaneous Ca release in a single cell with Gspon = 0.2 ms-1. In this example, the DAD amplitude became too small to trigger an AP with a 3-fold or greater increase in GK1. Enhancing GK1 up to 10-fold accelerated phase 3 repolarization and modestly shortened APD without significantly affecting resting membrane potential (Fig 4A, left panel).
Figure 4.
Suppression of DAD- and EAD-mediated TA by stabilizer cells with enhanced GK1. (A) Simulations of single uncoupled myocytes exhibiting a suprathreshold DAD (left, Gspon=0.2 ms−1) or EAD (right) as the control value of GK1 was increased from 2- to 10-fold. DAD- and EAD-mediated TA was suppressed by 3-fold and 2-fold increases in GK1, respectively. (B) Upper: Corresponding 1D cables of 100 coupled cells exhibiting DAD- (left) and EAD-mediated (right) TA (red traces). Lower: Suppression of DAD- (left) and EAD-mediated (right) TA by 30% stabilizer cells with GK1 enhanced 4-fold (black traces) randomly distributed among the arrhythmogenic cells (red traces). (Note: not all stabilizer cell traces are shown since only every 10th cell is plotted). (C) Percent of randomly-distributed stabilizer cells required to suppress DAD- (left) or EAD-mediated (right) TA (TA) in 1D cables versus the fold-increase in GK1 in stabilizer cells.
To examine the effect of enhancing GK1 on EADs, we modified the UCLA cell model13, 14 to generate supra-threshold EADs as in Xie et al11. Doubling GK1 was sufficient to eliminate EADs and shorten APD to ∼400 ms, with further GK1 increases up to 10-fold shortening APD to ∼300 ms (Fig 4A, right panel).
Fig. 4B shows examples from 1D cables in which 30% stabilizer cells, created by enhancing GK1 4-fold, were effective at suppressing TA when randomly dispersed (black traces) among arrhythmogenic cells (red traces) generating DADs (left panels) or EADs (right panels). The fraction of stabilizer cells required to suppress DAD- or EAD-mediated TA in 1D cables as a function of the fold-increase in GK1 is shown in Fig. 4C. Both DAD- and EAD-mediated TA were suppressed by <40% randomly distributed stabilizer cells with a 4-fold increase in GK1, with minimal slowing of conduction velocity (from 0.32 m/s to 0.31 m/s) and a minimal increase in the tissue stimulation threshold (from 16A/F to 18A/F).
Effects of tissue dimension, diffusion coefficient, synchrony of spontaneous SR Ca release, clustering of stabilizer cells, macroscopic regions devoid of stabilizer cells and fibrosis
The ability of stabilizer cells with enhanced GK1 to suppress DAD- and EAD-mediated TA under a variety of conditions are summarized in Fig. 5. For DAD-prone tissue (Fig. 5A), the percentage of stabilizer cells with GK1 enhanced 4-fold required to suppress TA was slightly lower with respect to tissue dimension, diffusion coefficient, clustering of stabilizer cells, macroscopic regions devoid of stabilizer cells and fibrosis, and slightly higher with respect to the timing variance of spontaneous SR Ca release, when compared to stabilizer cells in which spontaneous SR Ca release was directly inhibited by inactivating Gspon (compare to Figs. 2 and 3).
Figure 5.
Effectiveness of stabilizer cells with GK1 enhanced 4-fold at suppressing (A) DAD-mediated and (B) EAD-mediated TA. Upper panels in (A) and (B): Envelopes for TA probability >10% are superimposed as labeled to show the effects of tissue dimension (1D, 2D, 3D) and the effects of diffusion coefficient (D = 0.0005 cm2/ms, D/10, D/25) and clustering of stabilizer cells (1×1, 3×3, 5×5) in 2D tissue (100×100 cells) as a function of Gspon. Lower panels in (A) and (B): Same for the timing variance of DAD onset (standard deviation of 50 ms, 20 ms and 0, for DADs only), as well as the effects of macroscopic central defects devoid of stabilizer cells (TA probability as a function of the central region diameter) and fibrosis (TA breakthrough probability versus % stabilizer cells and % fibrosis) in 2D tissue (100×100 cells). See Figs. 2 and 3 for further details and comparison to stabilizer cells with Gspon=0.
For EAD-prone tissue, the effects of tissue dimension, diffusion coefficient, clustering of stabilizer cells, macroscopic regions devoid of stabilizer cells and fibrosis are summarized in Fig. 5B for different levels of GK1 enhancement in the stabilizer cells. Similar to the DAD protocols, 100 randomly uniform spatial distributions of interspersed non-arrhythmogenic stabilizer cells were tested for each GK1 value. A 4- to 5-fold increase in GK1 in the stabilizer cells was sufficient to keep the percentage of stabilizer cells required to prevent TA in the 40% range in 1D tissue and in the 25% range in 2D and 3D tissue. Decreasing the diffusion coefficient 10-fold or 25-fold in 2D tissue had little effect on the percentage of stabilizer cells required to suppress TA. Clustering of stabilizer cells in 3×3 or 5×5 groups only mildly increased the percentage of stabilizer cells required to suppress TA. In 2D tissue with 40% randomly distributed stabilizer cells with GK1 enhanced 4-fold, the size of a macroscopic heterogeneity devoid of stabilizer cells could reach 5.6 mm (56 cells) before TA reappeared. A central region of fibrosis (simulated by removal of cell-to-cell connections as in Fig. 3C) had almost no effect of the percentage of stabilizer cells required to prevent TA from propagating into surrounding non-fibrotic tissue.
Effects of GK1-enhanced stabilizer cells on APD dispersion
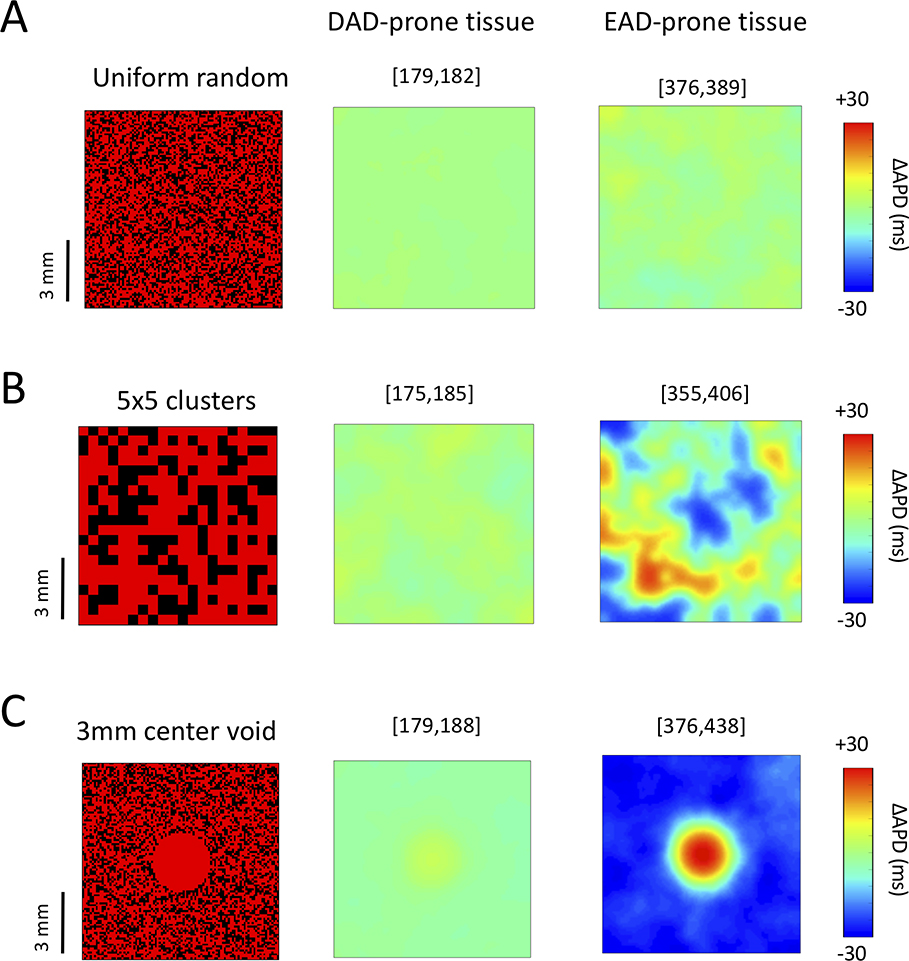
If the stabilizer cells were to increase electrophysiological dispersion significantly in cardiac tissue, susceptibility to initiation of reentry might increase and offset the antiarrhythmic benefits of suppressing triggers, as exemplified by the CAST Trial 24. In our model, creating stabilizer cells by preventing spontaneous Ca release during diastole (i.e. setting Gspon=0) had no effect on the APD preceding a DAD. However, stabilizer cells created by enhancing GK1 did modestly shorten APD (Fig. 4A). To assess the effects on APD dispersion, we performed 2D tissue simulations in which 40% of stabilizer cells with GK1 enhanced 4-fold were randomly distributed to prevent DAD- or EAD-mediated TA (Fig. 6). For DAD-prone tissue, stabilizer cells caused up to 10 ms of APD dispersion in the tissue, whether they were distributed singly, in 5×5 clusters or absent from a 3 mm defect in the center of the tissue (Fig. 6A–C, middle panels).
Figure 6.
Effect of GK1-enhanced stabilizer cells on APD dispersion. Stabilizer cells with GK1 enhanced 4-fold (40% of all cells) were randomly distributed in 2D tissue (100×100 cells) containing either DAD-generating cells (Gspon = 0.2 ms−1 and 50 ms standard deviation in DAD timing, middle panels) or EAD-generating cells (right panels). ΔAPD maps (difference from the mean tissue APD) are shown for: (A) Uniform random spatial distribution of single stabilizer cells; (B) Random distribution of 5×5 stabilizer cell clusters: (C) A 3 mm central defect devoid of stabilizer cells. Numbers in brackets above each panel indicate the tissue APD range in ms.
For EAD-prone tissue, APD dispersion was mild (13 ms) when stabilizer cells were distributed singly, but increased significantly (to 51–62 ms) when stabilizer cells were distributed in 5×5 clusters or when a central region was devoid of stabilizer cells (Fig. 6A–C, right panels).
Effects of modified GK1 properties
To examine how specific biophysical characteristics of GK1 (peak conductance and strength of inward rectification) affect the ability of stabilizer cells to suppress DAD- and EAD-mediated TA, we compared the GK1 formulation in the UCLA model with those in the Ten-Tuscher16 and O’Hara-Rudy17 models, whose peak conductances are right-shifted and left-shifted, respectively (Fig. S1A). Neither alternative GK1 formulation had a significant effect on the DAD-mediated TA suppression (Fig. S1B), but the left-shifted peak in O’Hara-Rudy GK1 model did require a greater fold increase in GK1 to suppress EAD-mediated arrhythmias (Fig. S1C).
To examine the effects of inward rectification strength, instead of increasing GK1 in the stabilizer cells, we added a weak inward rectifying K conductance (GKir1.1) simulating Kir1.1 channels. When added to stabilizer cells, GKir1.1 suppressed DADs and EADs effectively, but APD dispersion was markedly increased compared to GK1-enhanced stabilizer cells, even when stabilizer cells were distributed singly in a uniformly random manner (Fig. S2). This was due to prominent effects of Kir1.1 current on phase 2 of the AP. In contrast, Kir2.1 primarily affects phase 3, which has a much weaker influence on APD (see Discussion).
Discussion
In the present study, we have demonstrated that it is possible to suppress TA arising from DADs or EADs in simulated cardiac tissue by converting a minority of myocytes into “stabilizer cells” engineered to be incapable of developing these arrhythmogenic triggers. Two strategies for engineering stabilizer cells were tested. In the first method, we inhibited DADs by preventing spontaneous SR Ca release during diastole, and recapitulated the results of recent gene therapy experiments in CPVT mouse models that successfully eliminated DAD-mediated arrhythmias despite failing to achieve gene expression in the majority of ventricular myocytes 2–5. In the second more general method, we enhanced the inward rectifier K conductance GK1 to suppress both DADs and EADs. This method has not yet been tested experimentally by gene therapy, but has a broader potential to be useful for treating both DAD- and EAD-mediated arrhythmias in either genetic (e.g. CPVT and long QT syndromes) or acquired (e.g. heart failure) arrhythmogenic disorders.
Comparison to previous experimental gene therapy studies
In the Denegri et al 2 study in which the human CPVT-causing calsequestrin mutation R33Q was recapitulated in a mouse model, the CPVT phenotype was rescued by gene therapy with an AAV9 vector expressing wild-type calsequestrin to inhibit spontaneous SR Ca release, despite transducing only 40% of ventricular myocytes. Since then, three additional studies have been reported in which RYR2 mutations causing CPVT in humans were recapitulated in mice and successfully treated by AAV9-based gene therapy despite transducing only 45–66% of ventricular myocytes 3–5. In all of these studies, the intended effect of gene therapy was to restore normal RyR function in the transduced myocytes so that they would no longer be susceptible to spontaneous Ca release underlying DAD-mediated TA. Our simulations using the first method of creating stabilizer cells by inactivating Gspon to prevent spontaneous SR Ca release had the same functional effects targeted in these mouse gene therapy studies. Consistent with the experimental results, we found that a 40–66% transduction efficiency was more than adequate to account for the suppression of DAD-mediated arrhythmias observed in treated CPVT mice.
For our second more general method of creating stabilizer cells, we enhanced the inward rectifier K conductance GK1 to increase the dynamic threshold required for a DAD to trigger an AP 22. Using formulations of GK1 from three different models (Fig. S1), we found that this method was comparably effective at suppressing DAD-mediated TA as the first method in simulated cardiac tissue. This strategy has not yet been evaluated experimentally in the above-mentioned mouse CPVT models, but is straightforward to test with a viral vector incorporating the KCNJ2 gene that encodes Kir2.1 channels underlying GK1 in heart.
Although there are no experimental gene therapy studies suitable for comparison, GK1-enhanced stabilizer cells were also effective at suppressing simulated EAD-mediated TA. Among the three GK1 formulations tested, the O’Hara-Rudy formulation17 with most left-shifted peak conductance required a significantly higher percentage of stabilizer cells (Fig. S1). For gene therapy, modestly right-shifting the GK1 peak conductance by genetic engineering could potentially lower the minimum GK1 fold-increase needed for effective stabilization.
Mechanism of the stabilizer cell effect
The mechanism whereby a minority of non-arrhythmogenic cells can suppress TA by a majority of arrhythmogenic cells with DADs or EADs is related to the fact that when cells are coupled by gap junctions, any voltage difference between two adjacent cells causes current to flow through the gap junctions to try to equalize their voltages 6–10. In the case of DADs, the DAD amplitude of the arrhythmogenic cells is attenuated by nearby stabilizer cells, making it more difficult to reach the threshold required to trigger an AP (Fig. 1). If the DAD amplitude in the arrhythmogenic myocytes is very close to AP threshold (i.e. low safety factor, corresponding to low Gspon values in the model), then the percentage of stabilizer cells without DADs required to suppress TA is relatively low (Fig. 2). Conversely, when the safety factor is large (corresponding to high Gspon values), a higher percentage is required. In the CPVT mouse gene therapy experiments 2–5, it is unknown whether DADs have a low, high or variable safety factor, but the finding that 40–66% transduction was effective at suppressing DAD-mediated arrhythmias in those experiments is consistent with our simulations showing that even for a high value of Gspon to create a DAD well-above the threshold to trigger an AP, the percentage of stabilizers cells required to suppress TA did not exceed these percentages (Fig. 2). Moreover, the simulation results suggest that if DADs have a low safety factor (up to 1.5 times the Gspon threshold), the percentage of stabilizer cells required to suppress TA may be as low of 15–25% (Fig. 2). It will be interesting to perform dose-response experiments in the CVPT mouse gene therapy models to determine whether transduction rates lower than 40–45% remain effective at suppressing DAD-mediated arrhythmias, as well as to compare the already reported strategies with a gene therapy vector incorporating the KCNJ2 gene encoding Kir2.1 to create GK1-enhanced stabilizer cells.
Our computational results indicate stabilizer cell gene therapy is robust at suppressing DAD- and EAD-mediated TA irrespective of tissue dimension or diffusion coefficient (gap junction coupling). In experimental cardiac gene therapy, transduction efficiency can vary significantly throughout tissue, with both microscopic clusters and macroscopic regions with variable densities of transduced cells (e.g. Fig. 2A in Guo et al 20). To estimate the consequences of heterogeneous gene expression at the microscopic scale, we randomly distributed cells in clusters of up to 5×5 cells throughout 2D tissue (Fig. 3A, 5C, S1C), and found that it had only mild effects on the percent of non-arrhythmogenic stabilizer cells required to suppress TA. This is not unexpected, since the electrical space constant of the tissue (1–2 mm) smooths out microscopic voltage heterogeneities at the submillimeter scale. Macroscopic defects devoid of stabilizer cells larger than the electrical space constant, however, did result in reemergence of DAD- or EAD-mediated TA due to the weakened sink effect in those regions (Figs. 3B and 5).
Finally, it is important to point out that during sinus rhythm, the stabilizer cells have a similar voltage waveform as surrounding unmodified myocytes, such that their own AP triggers a normal Ca transient and contraction; that is, stabilizer cells contribute normally to and do not impair cardiac contractility.
Potentially undesirable effects of stabilizer cell gene therapy
Stabilizer cell gene therapy that selectively targets spontaneous SR Ca release during diastole, such as in CPVT, does not significantly affect the AP during systole. However, gene therapy targeting ionic currents that directly shape the cardiac AP has the potential to exacerbate electrophysiological heterogeneity in a proarrhythmic fashion, especially if the gene expression is variable and spatially heterogeneous. In this regard, GK1-enhanced stabilizer cells created by enhancing GK1 were very effective at suppressing TA without adversely affecting either propagation or intracellular Ca cycling (Fig. 1A & 4A). For suppression of DAD-mediated TA, APD dispersion was also not significantly increased, even when GK1-enhanced stabilizer cells were present in 5×5 clusters or absent in a 3 mm region (Fig. 6, 2nd column). For suppression of EAD-mediated TA, however, APD dispersion was minimal with uniformly random distributed stabilizer cells, but could increase to ∼60 ms when stabilizer cell distribution was spatially non-uniform (Fig. 6B), either with 5×5 clusters or areas devoid of stabilizer cells. Thus, non-uniform transduction efficiency could be a limitation for using stabilizer cells with enhanced GK1 to treat EAD-related arrhythmias, although increased APD dispersion would be restricted to regions with active EADs and not necessarily widespread throughout the heart.
It is notable that stabilizer cells created by enhancing the weak inward rectifier K channel conductance GK1.1 caused much greater APD dispersion than stabilizer cells with enhanced GK1, even when stabilizer cell distribution was randomly uniform (Fig. S2). This was due to the prominent GKir1.1-induced increase in outward K current during phase 2 of the AP, which is the major determinant of APD. Enhancing GK1, on the other hand, has minimal effects on phase 2 and becomes influential only at voltages well below 0 mV where it accelerates rapid repolarization during phase 3 (Fig. 4A right panel). GK1 remains effective at suppressing EAD-mediated TA in this voltage range because EAD propagation requires EAD takeoff potentials well below 0 mV in order to generate a rapid enough EAD upstroke for regenerative propagation. These results make it unlikely that overexpressing other K conductances that increase outward K current during phase 2, such as rapidly-activating (GKr) or slowly-activating (GKs) voltage-dependent K conductances or the small Ca-activated K conductance, would be effective strategies to create stabilizer cells, since they would also be likely to markedly exacerbate APD dispersion. Finally, because of its modest effects on APD, overexpression of wild-type Kir2.1 channels is unlikely to result in short QT Syndromes from Kir2.1 mutations with weakened inward rectification that dramatically shorten APD 25–27.
Limitations
Although our simulations provide a mechanistic explanation for the antiarrhythmic efficacy of gene therapy that transduced less than <50% myocytes in mouse models of CPVT, whether the same strategy would scale to the larger human heart or to diseased hearts is uncertain and requires further experimental evaluation. Off-targets effect of gene expression could also create adverse effects, such as expression of Kir2.1 channels in the sino-atrial node or atrio-ventricular node impairing pacemaker function and/or atrio-ventricular conduction. Our simulations of fibrosis did not fully separate the cell internal resistance from the gap junctional resistance which could have a quantitative effect on TA breakthrough. Tissue anisotropy was also not considered, but we expect the effects would be small given that a 10x overall reduction in diffusion constant still did not have a significant effect. Finally, the success of stabilizer cell gene therapy relies on suppression of triggers rather than reducing tissue vulnerability to reentry. The efficacy of stabilizer cell gene therapy therefore cannot be assessed by programmed electrical stimulation, which assesses tissue substrate vulnerability to reentry. However, programmed electrical stimulation would remain useful for evaluating potential proarrhythmic effects of stabilizer cell gene therapy due to inhomogeneous gene delivery, to ensure that the reduction in triggers is not offset by increased vulnerability to reentry.
Conclusions
Converting a minority of cells in cardiac tissue into stabilizer cells was effective at suppressing DAD- or EAD-triggered arrhythmias regardless of tissue dimension, diffusion coefficient, fibrosis, or submillimeter clumping of stabilizer cells. Stabilizer cell gene therapy shows promise as an antiarrhythmic strategy and deserves further experimental evaluation.
Supplementary Material
What Is Known.
A current limitation of cardiac gene therapy is achieving gene expression in a high enough percentage of myocytes to reverse the disease phenotype at the whole organ level.
However, partial gene transduction may be less of a limitation for gene therapy of cardiac arrhythmias, as demonstrated in several recent genetic mouse models of catecholaminergic polymorphic ventricular tachycardia (CPVT).
What the Study Adds.
In this computational study, we demonstrate that due to source-sink relationships in cardiac tissue, a minority (20–50%) of randomly distributed “stabilizer” cells engineered to be non-arrhythmogenic can suppress the ability of arrhythmogenic cells to generate afterdepolarization-related arrhythmias.
Stabilizer cell gene therapy strategy can be designed to correct a specific arrhythmogenic mutation, as in the CPVT mice studies, or more generally to suppress delayed or early afterdepolarizations from any cause by overexpressing inward rectifier K channels in stabilizer cells.
This promising antiarrhythmic strategy warrants further testing in experimental models to evaluate its clinical potential.
Acknowledgments
We thank our colleagues Peng-Sheng Chen, Alan Garfinkel, Scott A. John, Hrayr S. Karagueuzian, Thao Nguyen and Riccardo Olcese for helpful discussions.
Source of Funding: NIH grants P01 HL078931, R01 HL134709, F30 HL132449 and T32 GM008042, R25GM067110, AHA-WSA Innovative Science Award 12PILT12560004, the Laubisch and Kawata Endowments, and in part by NSF grant PHY17-48958 and the Gordon and Betty Moore Foundation Grant No. 2919.01.
Nonstandard Abbreviations and Acronyms
- AP
action potential
- APD
action potential duration
- CPVT
catecholaminergic polymorphic ventricular tachycardia
- DAD
delayed afterdepolarization
- EAD
early afterdepolarization
- GK1
conductance of the inward rectifier K channel
- GKir1.1
conductance of the inward rectifier Kir1.1 channel
- Gspon
conductance of the SR Ca release channels in the AP model
- SR
sarcoplasmic reticulum
- TA
triggered activity
Footnotes
Disclosures: None.
References
- 1.Hajjar RJ, Ishikawa K. Introducing genes to the heart: All about delivery. Circ Res. 2017;120:33–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denegri M, Bongianino R, Lodola F, Boncompagni S, De Giusti VC, Avelino-Cruz JE, Liu N, Persampieri S, Curcio A, Esposito F et al. Single delivery of an adeno-associated viral construct to transfer the CASQ2 gene to knock-in mice affected by catecholaminergic polymorphic ventricular tachycardia is able to cure the disease from birth to advanced age. Circulation. 2014;129:2673–81. [DOI] [PubMed] [Google Scholar]
- 3.Bongianino R, Denegri M, Mazzanti A, Lodola F, Vollero A, Boncompagni S, Fasciano S, Rizzo G, Mangione D, Barbaro S et al. Allele-specific silencing of mutant mRNA rescues ultrastructural and arrhythmic phenotype in mice carriers of the R4496C mutation in the ryanodine receptor gene (RYR2). Circ Res. 2017;121:525–536. [DOI] [PubMed] [Google Scholar]
- 4.Pan X, Philippen L, Lahiri SK, Lee C, Park SH, Word TA, Li N, Jarrett KE, Gupta R, Reynolds JO, et al. In vivo Ryr2 editing corrects catecholaminergic polymorphic ventricular tachycardia. Circ Res. 2018;123:953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bezzerides VJ, Caballero A, Wang S, Ai Y, Hylind RJ, Lu F, Heims-Waldron DA, Chambers KD, Zhang D, Abrams DJ, et al. Gene therapy for catecholaminergic polymorphic ventricular tachycardia by inhibition of Ca/Calmodulin-dependent kinase II. Circulation. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joyner RW, Sugiura H, Tan RC. Unidirectional block between isolated rabbit ventricular cells coupled by a variable resistance. Biophys J. 1991;60:1038–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaw RM, Rudy Y. Ionic mechanisms of propagation in cardiac tissue. Roles of the sodium and L-type calcium currents during reduced excitability and decreased gap junction coupling. Circ Res. 1997;81:727–41. [DOI] [PubMed] [Google Scholar]
- 8.Spach MS, Boineau JP. Microfibrosis produces electrical load variations due to loss of side-to-side cell connections: a major mechanism of structural heart disease arrhythmias. Pacing & Clinical Electrophysiology. 1997;20:397–413. [DOI] [PubMed] [Google Scholar]
- 9.Rohr S, Kucera JP, Fast VG, Kleber AG. Paradoxical improvement of impulse conduction in cardiac tissue by partial cellular uncoupling. Science. 1997;275:841–844. [DOI] [PubMed] [Google Scholar]
- 10.Wang YG, Kumar R, Wagner MB, Wilders R, Golod DA, Goolsby WN, Joyner RW. Electrical interactions between a real ventricular cell and an anisotropic two-dimensional sheet of model cells. Am J Physiol Heart Circ Physiol. 2000;278:H452–60. [DOI] [PubMed] [Google Scholar]
- 11.Xie Y, Sato D, Garfinkel A, Qu Z, Weiss JN. So little source, so much sink: requirements for afterdepolarizations to propagate in tissue. Biophys J. 2010;99:1408–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tveito A, Lines GT. A condition for setting off ectopic waves in computational models of excitable cells. Math Biosci. 2008;213:141–50. [DOI] [PubMed] [Google Scholar]
- 13.Mahajan A, Sato D, Shiferaw Y, Baher A, Xie L-H, Peralta R, Olcese R, Garfinkel A, Qu Z, Weiss JN. Modifying L-type calcium current kinetics: Consequences for cardiac excitation and arrhythmia dynamics. Biophys J. 2008;94:411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahajan A, Shiferaw Y, Sato D, Baher A, Olcese R, Xie LH, Yang MJ, Chen PS, Restrepo JG, Karma A, et al. A rabbit ventricular action potential model replicating cardiac dynamics at rapid heart rates. Biophys J. 2008;94:392–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu MB, de Lange E, Garfinkel A, Weiss JN, Qu Z. Delayed afterdepolarizations generate both triggers and a vulnerable substrate promoting reentry in cardiac tissue. Heart Rhythm. 2015;12:2115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ten Tusscher KH, Panfilov AV. Alternans and spiral breakup in a human ventricular tissue model. Am J Physiol Heart Circ Physiol. 2006;291:H1088–100. [DOI] [PubMed] [Google Scholar]
- 17.O’Hara T, Virag L, Varro A, Rudy Y. Simulation of the undiseased human cardiac ventricular action potential: Model formulation and experimental validation. PLoS Comput Biol. 2011;7:e1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasserstrom JA, Shiferaw Y, Chen W, Ramakrishna S, Patel H, Kelly JE, O’Toole MJ, Pappas A, Chirayil N, Bassi N, et al. Variability in timing of spontaneous calcium release in the intact rat heart is determined by the time course of sarcoplasmic reticulum calcium load. Circ Res. 2010;107:1117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko CY, Liu MB, Song Z, Qu Z, Weiss JN. Multiscale determinants of delayed afterdepolarization amplitude in cardiac tissue. Biophys J. 2017;112:1949–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y, VanDusen NJ, Zhang L, Gu W, Sethi I, Guatimosim S, Ma Q, Jardin BD, Ai Y, Zhang D, et al. Analysis of cardiac myocyte maturation using CASAAV, a platform for rapid dissection of cardiac myocyte gene function in vivo. Circ Res. 2017;120:1874–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo M, Anderson ME. Mechanisms of altered Ca handling in heart failure. Circ Res. 2013;113:690–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu MB, Ko CY, Song Z, Garfinkel A, Weiss JN, Qu Z. A dynamical threshold for cardiac delayed afterdepolarization-mediated triggered activity. Biophys J. 2016;111:2523–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomaselli GF, Zipes DP. What causes sudden death in heart failure? Circ Res. 2004;95:754–763. [DOI] [PubMed] [Google Scholar]
- 24.Moss AJ, CAST Investigators. Effect of encainide and flecainide on mortality in a random trial of arrhythmia suppression after myocardia infarction. N Engl J Med. 1989;321:406–412. [DOI] [PubMed] [Google Scholar]
- 25.Priori SG, Pandit SV, Rivolta I, Berenfeld O, Ronchetti E, Dhamoon A, Napolitano C, Anumonwo J, di Barletta MR, Gudapakkam S, et al. A novel form of short QT syndrome (SQT3) is caused by a mutation in the KCNJ2 gene. Circ Res. 2005;96:800–7. [DOI] [PubMed] [Google Scholar]
- 26.Hattori T, Makiyama T, Akao M, Ehara E, Ohno S, Iguchi M, Nishio Y, Sasaki K, Itoh H, Yokode M, et al. A novel gain-of-function KCNJ2 mutation associated with short-QT syndrome impairs inward rectification of Kir2.1 currents. Cardiovasc Res. 2012;93:666–73. [DOI] [PubMed] [Google Scholar]
- 27.Deo M, Ruan Y, Pandit SV, Shah K, Berenfeld O, Blaufox A, Cerrone M, Noujaim SF, Denegri M, Jalife J, et al. KCNJ2 mutation in short QT syndrome 3 results in atrial fibrillation and ventricular proarrhythmia. Proc Natl Acad Sci U S A. 2013;110:4291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.