Abstract
The acquisition of metal ions and the proper maturation of holo-metalloproteins are essential processes for all organisms. However, metal ion homeostasis is a double-edged sword. A cytosolic accumulation of metal ions can lead to mismetallation of proteins and cell death. Therefore, maintenance of proper concentrations of intracellular metals is essential for cell fitness and pathogenesis. Staphylococcus aureus, like all bacterial pathogens, uses transcriptional metalloregulator proteins to aid in the detection and the genetic response to changes in metal ion concentrations. Herein, we review the mechanisms by which S. aureus senses and responds to alterations in the levels of cellular zinc, iron, heme, and copper. The interplay between metal ion sensing and metal-dependent expression of virulence factors is also discussed.
Keywords: Staphylococcus aureus, metal ion, metal homeostasis, metalloregulator, iron, manganese
Metal ion acquisition and usage
Transition metals, including iron (Fe), copper (Cu), manganese (Mn), and zinc (Zn), are essential micronutrients (see Glossary) in virtually all biological systems and play vital roles for living organisms, including bacterial pathogens. The acquisition of some metal ions is necessary for survival (i.e. Fe and Zn), while other metal ions are typically cytotoxic and are only adventitiously imported into the cell (i.e. Hg and Cu). Relatively high titers of certain metal ions, like Fe, are necessary for fitness. Others, like Mn, are only required in minute amounts. Importantly, unlike other kinds of essential nutrients, metal ions are not biochemically synthesized and cannot be degraded. In Staphylococcus aureus, the use of metal ion cofactors is required in a diverse array of biochemical processes including carbon transformations, nucleic acid and protein synthesis, DNA replication, regulation of virulence factor expression, and the metabolism of reactive oxidative species (ROS) [1–3].
In response to infection, and to combat invading pathogens like S. aureus, vertebrate hosts restrict the availability of transition metals in a process known as nutritional immunity (reviewed in [4]). A number of studies have identified the S100 family of proteins as metal chelators that can inhibit bacterial growth by competing for metal ion nutrients (reviewed in [5]). Within S. aureus abscesses, neutrophils release calprotectin (S100A8 and S100A9) to inhibit growth of S. aureus by limiting its access to divalent metals, including Zn2+, Mn2+, Fe2+, and Cu2+ [6–8]. Other S100 family members include psoriasin (S100A7), which was demonstrated to ligate Zn2+ and prevent the growth of bacterial pathogens in vitro and on human skin [9], as well as calgranulin C (S100A12), which is a Zn2+- and Cu2+-binding neutrophil protein [10–12]. Staphylococcal access to plasma-associated Fe is also restricted by the host via the secretion of lactoferrin and transferrin, which chelate free iron ions, as well as hemopexin and haptoglobin, which sequester heme and hemoglobin, respectively (reviewed in [13]). The combined effect of these molecules lowers the concentration of metal nutrients available to pathogens.
To oppose host strategies of metal ion limitation, S. aureus synthesizes small molecule metallophores and proteins which promote the acquisition of metal ions from the environment. S. aureus produces two siderophores (staphyloferrin A and staphyloferrin B), which bind to free iron with high affinity and compete with iron-sequestering host proteins like lactoferrin and transferrin [14–17]. Iron ions bound to staphyloferrin A and B are imported by the HtsABC and SirABC transporters, respectively [17–19]. During infection, S. aureus produces staphylopine to compete with the host for Ni, Co, and Zn [20, 21], as well as hemolysins to rupture red blood cell membranes, resulting in hemoglobin release. Hemoglobin is subsequently degraded to heme and free iron via a series of import and processing steps mediated by the staphylococcal Isd system (reviewed in [22]).
Although metals are essential for viability and growth, cytosolic accumulation can result in poisoning. An excess of iron or copper ions can be harmful in aerobic environments due to their roles as catalysts in Fenton reaction chemistry, which leads to the production of damaging oxygen radicals [23, 24]. Transition metals can also be toxic due to their binding to adventitious sites on apo-proteins (known as mismetallation), which can result in protein inactivation or other unintended allosteric effects [25].
In S. aureus and other organisms, metal-dependent regulatory proteins (metalloregulators) work to maintain metal homeostasis by sensing the bioavailability of metal ion(s) and controlling the transcription of genes involved in metal ion import, storage, distribution, and efflux. These metal sensing mechanisms typically involve reversible interaction of the regulator protein with one or more specific metal ions, which alters the affinity of the regulatory protein for specific DNA sequences located in target promoter operons. Alterations in the occupancy of the metalloregulator on the operator modulates transcription of target genes. In the case of two-component regulatory systems, stimulation of a histidine kinase (via association with a ligand and/or a change in physiological state) modulates its kinase or phosphatase activity, thereby altering the phosphorylation status of the response regulator (reviewed in [26, 27]). The phospho-state of the response regulator determines its affinity for promoter DNA sequences, and ultimately regulates transcription of downstream target genes. Altogether, these processes sustain the appropriate levels of transition metal ions and enable S. aureus to adapt to changing conditions within the host environment and/or shifting metabolic demands.
In this review, we describe several of the key metalloregulators involved in the genetic regulation of metal ion homeostasis in S. aureus. We highlight fundamental and recent developments in the understanding of their functions and mechanisms. We also discuss the crosstalk between genetic regulation of metal ion homeostasis and virulence factor production.
The Fur family
The ferric uptake regulator (Fur) family of metalloregulators are widespread in the Staphylococci, as well as other Gram-negative and Gram-positive bacteria [28]. There are three paralogs of the Fur family found in S. aureus: Fur, Zur (zinc uptake regulator), and PerR (peroxide-sensing regulator) [29]. Fur family regulators are homodimers consisting of two structural and functional domains: a N-terminal winged-helix DNA binding domain, and a C-terminal dimerization domain (Figure 1A). Each monomer contains binding sites for two or three metal ions. The key metal sensing site is typically located near the hinge region that links the DNA binding and dimerization domains. Metal ion binding to this site bridges both domains, causing a structural change that repositions the two DNA binding domains in a caliper-like conformation compatible with DNA binding [30, 31]. A non-regulatory metal binding site includes a structural Zn2+ that is required for protein folding and dimerization [32, 33]. Some Fur family members contain a third metal binding site of unclear function [34, 35]. The metal binding specificity of Fur family members is thought to be dictated by both the affinity of the binding site ligands for the cognate metal(s) and the relative availability of each metal ion [36].
Figure 1: Structure and activation mechanism of Fur metalloregulators.
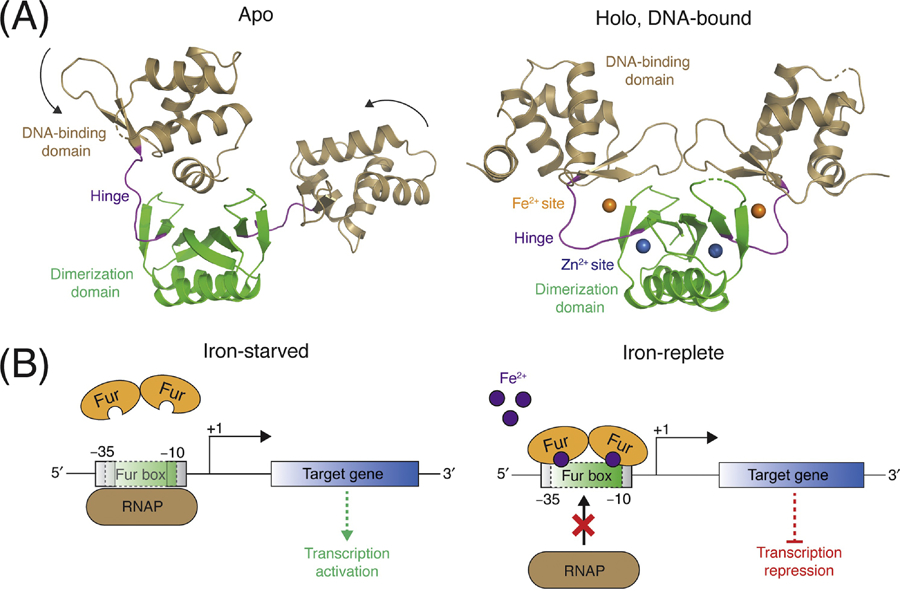
(A) Fur metalloregulators are dimers. Each monomer consists of an N-terminal DNA-binding domain (tan) and a C-terminal dimerization domain (green), which are connected by a hinge region (purple). There are typically two metal binding sites per monomer: the Fe2+ sensing site (orange) is coordinated by residues in the hinge region, and the non-regulatory Zn2+ site (blue) is located within the dimerization domain. In the apo-state (left), the conformations of the two monomers differ. Upon metal and DNA binding (right), Fur adapts a caliper-like conformation with a two-fold axis of symmetry (PDB IDs 4RB0 and 4RB1) [30]. (B) Left: When free iron is limiting, Fe2+ is released from Fur, which triggers dissociation of the apo-Fur dimer from the Fur box, and subsequent binding and transcription initiation by RNA polymerase. Right: During iron-replete conditions, Fur binds Fe2+ and associates with the inverted repeat Fur box within the promoter of the target gene. When bound to the Fur box, the Fur dimer occludes the −35 and/or −10 initiation sites, which inhibits RNA polymerase binding and represses gene transcription.
As the namesake member of the Fur family, the physiological roles and mechanisms of Fur as an Fe-dependent transcriptional repressor are well-established. In iron-replete conditions, Fur is primarily associated with Fe2+ and bound to a conserved operator DNA sequence (Fur box) in the promoter region upstream of target genes [37, 38]. When bound to the Fur box, Fur often partially overlaps the −35 and/or −10 initiation sites, thereby occluding RNA polymerase binding and repressing gene transcription (Figure 1B) [39]. Upon a decrease in cellular iron levels, Fe2+ is released from Fur, triggering dissociation of the Fur dimer from the Fur box, and subsequent binding and transcription initiation by RNA polymerase (Figure 1B) [39]. Recent work has demonstrated in Escherichia coli that Fur also responds to dioxygen tension (via oxidation of intracellular Fe2+ to Fe3+) and that transcription of the Fur regulon is activated under anaerobic growth conditions [40, 41]. While it has been established in S. aureus that Fur responds to media iron concentrations, it is currently unclear if it also responds to alterations in intracellular redox status.
The Fur regulon in S. aureus consists of genes involved in iron import and acquisition, including the siderophore transport system (sir) and the ferrichrome uptake operon (fhu) [42, 43]. A S. aureus fur mutant alters the expression of genes coding for enzymes utilized in central metabolism suggesting that Fur-dependent regulation extends beyond genes utilized for iron acquisition [44]. This includes decreasing flux through the TCA cycle, which is reliant on Fe-dependent enzymes, and increasing glycolytic flux, resulting in increased fermentation and decreased respiration [44]. A fur mutant also has altered production of excreted proteins (exoproteins) [2]. Many of these exoproteins are involved in pathogenesis, including cytotoxins (LukD, LukE, Hla). The fur mutant has increased virulence in murine models of pneumonia and skin abscess [2, 42]. It remains to be determined if these phenotypes are the result of direct Fur regulation or defective Fe homeostasis.
In some bacteria, Fur can function as a transcriptional activator via RNA polymerase recruitment, anti-repressor activity, and transcriptional regulation of small, non-coding RNAs (reviewed in [45]). In B. subtilis, it was recently shown that Fur is a transcriptional activator for the PerR-repressed pfeT iron efflux pump [46]. It is not yet clear if Fur also activates gene expression in S. aureus. However, a number of loci (i.e. aconitase, acnA) have decreased expression in a fur mutant, supporting the possibility. There is also evidence of direct regulation by apo-Fur in Helicobacter pylori [47–49], although it is currently unclear whether this property of Fur is conserved in S. aureus.
Zur is a Zn-dependent metalloregulator [29]. Most biochemical studies of Zur have been conducted in B. subtilis. Like Fur, Zur forms a dimer and contains multiple metal binding sites, which include a structural Zn2+ site and two regulatory metal sensing Zn2+ sites [50]. Interestingly, the two metal sensing sites have different Zn-binding affinities (~1013 and ~1012 M−1, respectively) [50]. This observation indicates that a Zur dimer binds Zn with negative cooperativity, which allows Zur to sense a wider range of cellular Zn concentrations and gradually derepress target operons [50, 51]. It also reveals a role for Zur in buffering cellular Zn levels at very low concentrations, which likely prevents mismetallation of other Fe- or Mn-binding proteins that could bind Zn2+ with greater affinity than their cognate metals based on the relative positions of each ion within the Irving-Williams series [50]. In S. aureus, intracellular levels of free Zn are normally buffered at levels below those of Fe and Mn [35].
During Zn-replete conditions in S. aureus, Zur represses the expression of mreAB, which encodes for Zn importers [29]. However, a zur mutant does not display increased Zn sensitivity, suggesting that other mechanisms exist to protect S. aureus from cytosolic Zn accumulation [29]. Zur has also been shown to regulate transcription of the Zn metallochaperone ZigA [52]. It was recently reported that the cnt locus is dual regulated by Zur and Fur [53]. The cnt operon (previously named opp1) is comprised of three gene clusters which function in the synthesis, transport, and export of staphylopine [21]. Analysis of these gene clusters found that the promoter regions contained bona fide Fur- and Zur-boxes, which are occupied by Fur and Zur, respectively, during metal-replete conditions to repress cnt transcription [53]. These observations describe a previously unknown link between Zur and staphylopine-dependent metal transport, which plays an important role in virulence and infection outcomes [54].
PerR is the third Fur family member present in S. aureus. First characterized in B. subtilis, PerR is a metal-dependent peroxide sensor that regulates inducible peroxide-defense genes [55]. PerR contains two metal-binding sites: a structural Zn2+ site and a regulatory metal binding site that coordinates either Fe2+ or Mn2+ [56]. In the presence of low intracellular concentrations of peroxide, oxidation of PerR is catalyzed by the bound Fe2+ ion, which leads to the incorporation of an oxygen atom into nearby histidine ligand(s) within the PerR metal binding site [33, 57]. Oxidation induces a conformational change in PerR, resulting in decreased affinity for its DNA operator sequence and derepression of target gene transcription [58]. The S. aureus PerR regulon includes genes involved in ROS metabolism (katA, ahpCF, trxB) and iron regulation and storage (fur, bcp, ftn), revealing a key connection between peroxide stress response and Fe homeostasis [42, 59–61]. In S. aureus, PerR is hypersensitive to peroxide and the oxidized Fe3+ bound form predominates under normal growth conditions [62]. Work in B. subtilis demonstrated that most target regulons are repressed by both the Mn and Fe forms of PerR, but some genes (including fur and perR itself) are repressed only when PerR is bound to Mn2+ [63]. In these cases, PerR functions as a sensor of the cellular Fe/Mn ratio. It is unclear what determines the molecular basis for this differential regulation, or if a similar role for PerR is conserved in S. aureus.
Heme sensor system (HssRS)
The concentration of free iron in host tissues is maintained at low concentrations that is likely unable to support the growth of S. aureus [64]. Nonetheless, host tissues are rich in other forms of iron, such as heme, which serves as a key cofactor for hemoproteins found in human blood. Heme is thought to be the preferred Fe source for S. aureus [65]. To satisfy iron nutrient requirements, S. aureus utilizes the iron-regulated surface determinant system (Isd) to acquire, traffic, import, and release of iron ions from heme [22] (see Figure 2). Additionally, it has been proposed that a second locus, known as the heme transport system (htsABC), is also involved in heme import [65, 66]. Several studies have indicated that Hts is a transporter of staphyloferrin A [18, 19]. However, it remains unclear whether Hts also directly imports heme, and if so, what are the mechanism(s) of transport. Expression of both the isd and hts operons is under the transcriptional control of Fur [67, 68]. During infection, the Isd and Hts systems are integral to staphylococcal virulence and pathogenesis [65, 69].
Figure 2: Heme sensing and signaling in S. aureus.
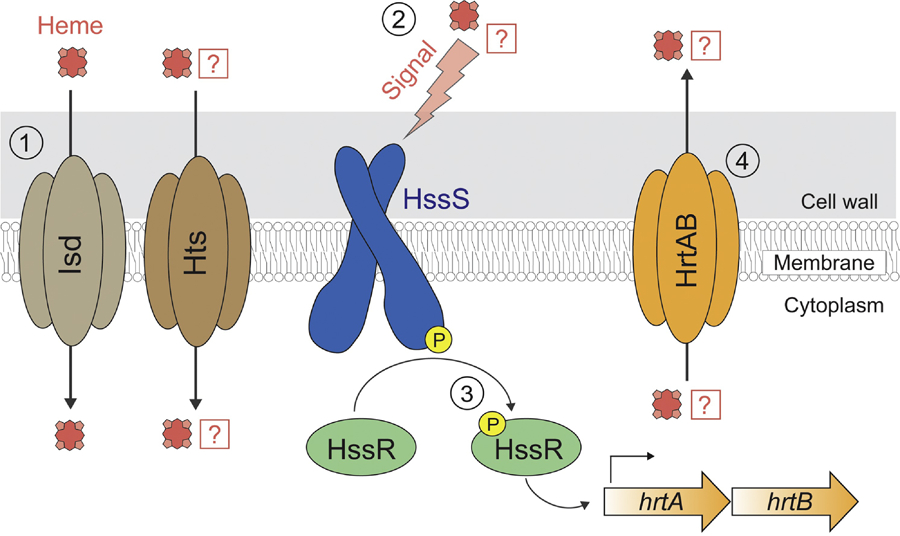
(1) Heme is imported into the cytoplasm via the Isd system. It has been proposed that Hts, which has an established role in siderophore import, may also play a role in heme transport. (2) The HssS histidine kinase is activated by a heme-associated stimulus. Whether HssS directly senses heme or a byproduct of heme metabolism remains unknown. (3) Activation of HssS results in autophosphorylation of HssS and transphosphorylation of the HssR response regulator. Phosphorylated HssR binds to the promoter sequence of hrtAB and stimulates its transcription via recruitment of RNA polymerase. (4) Heme, or a heme-associated byproduct, is effluxed by the HrtAB exporter.
At high concentrations heme can be a toxic molecule capable of generating ROS and cellular damage [70]. To modulate intracellular heme levels and to protect from the toxic side effects of heme, S. aureus encodes for heme detoxification machinery, which is regulated by the heme sensor system, HssRS [71]. HssRS is a two-component regulatory system comprised of a membrane-localized histidine kinase (HssS) and a cytoplasmic response regulator with a DNA-binding domain (HssR) [72] (Figure 2). The generally accepted model for HssRS function is consistent with other two-component systems, in which HssS recognizes a heme-associated stimulus, triggering the phosphorylation of HssR [72]. Phosphorylation increases the binding affinity of HssR for target DNA sequences and stimulates their transcription [72]. HssRS may represent one of only a few described two-component regulatory systems that responds to hemoglobin, a molecular marker of vertebrate tissue [73–75].
Upon stimulation, HssRS increases transcription of the heme-regulated ABC transporter HrtAB [71, 72] (Figure 2). S. aureus strains lacking either hrtA or hrtB fail to adapt to heme toxicity [71]. Perhaps counterintuitively, in murine abscess models of infection an S. aureus hrtA mutant strain exhibited increased virulence in liver tissue [71]. It is suggested that this phenotype arises from a dramatic increase in expression and secretion of immunomodulatory effectors observed in the hrtA mutant, which resulted in inhibited recruitment of host neutrophils to the site of infection [71].
The nature of the signal that is directly sensed by the HssS histidine kinase has remained elusive. It has been speculated that HssS may sense heme by direct binding. However, bioinformatic-based approaches do not reveal a potential heme-binding domain in HssS, given that the predicted sensing domain of HssS shares little sequence similarity to known heme-binding proteins and does not contain conserved ligands typically engaged in iron coordination [4, 72]. One possibility is that rather than binding heme directly, HssS may sense one of the toxic effects that heme has on the cell, such as the buildup up heme metabolites which arise from the breakdown of heme by S. aureus [72]. Similarly, it remains unclear whether HrtAB directly exports heme molecules and/or potentially toxic heme metabolic byproduct(s) [76]. It is unclear if HssRS has other targets beyond hrtAB [72].
Copper-sensitive operon regulator (CsoR)
Unlike the other metal ions discussed herein, copper is not thought to be an essential micronutrient in S. aureus. The only predicted Cu-utilizing enzyme in S. aureus is the QoxABCD aa3 menaquinol terminal oxidase, which functions redundantly with a second, Cu-independent cytochrome bd terminal oxidase, CydAB [77]. Therefore, copper is not essential for dioxygen respiration in S. aureus during growth under standard laboratory conditions. However, a qox mutant strain is defective in liver colonization of a murine infection model, suggesting that Cu acquisition and/or utilization could be an important process in the host [78].
Upon entering S. aureus, intracellular reductants, including low-molecular weight thiols, reduce Cu2+ to Cu1+. Intracellular Cu1+ can be cytotoxic if concentrations are not carefully regulated [79]. The copper-sensitive operon regulator (CsoR) is a Cu1+-responsive regulator of copper homeostasis in S. aureus (see Figure 3) [80]. As a member of the larger CsoR/RcnR family of bacterial transcriptional repressors, CsoR forms a tetrameric structure and uses a WXYZ amino acid fingerprint to coordinate copper ions [81]. There is not a structure of a CsoR family regulator in complex with DNA; however, biophysical studies suggest that two CsoR proteins bind symmetrically per operator sequence [81, 82]. Cu1+ binding by CsoR is predicted to induce a conformational change which compacts the protein structure and allosterically inhibits DNA binding, causing release of operator DNA [82, 83].
Figure 3: Model for copper ion homeostasis in S. aureus USA300.
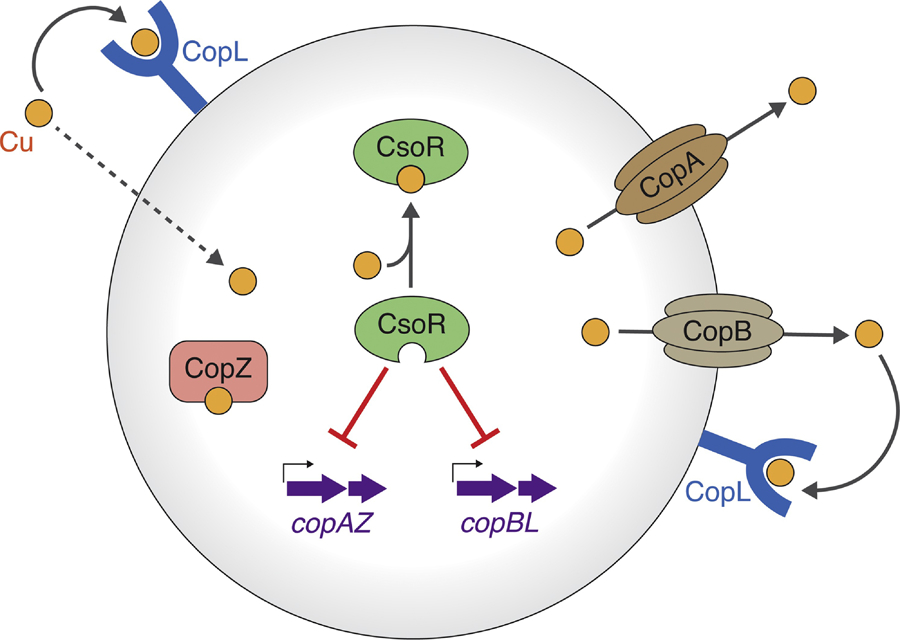
The CsoR transcriptional regulator binds intracellular Cu1+, leading to derepression of the copAZ and copBL operons. copBL (copB has also referred to as copX) is encoded on the ACME mobile genetic element. Cu1+ is effluxed via the CopA and CopB exporters. Extracellular copper binds to CopL lipoprotein on the surface of the cell, where it is prevented from entering or re-entering the cytoplasm. Intracellular copper ions are scavenged and buffered by the CopZ metallochaperone, which can traffic the Cu to CopA. The mechanism(s) of copper import remain unknown.
The CsoR regulon in S. aureus consists of the copper-sensitive operon copAZ, which contains genes encoding proteins involved in Cu1+ copper efflux (CopA) and cytosolic buffering and scavenging (CopZ) [80, 84] (Figure 3). Recently, it was discovered that in addition to copAZ, CsoR regulates the copBL (copB has also been referred to as copX) and copBmco operons, found on mobile DNA, which provide hypertolerance to Cu in some S. aureus strains [79, 84–86]. Genetic evidence suggests that copB encodes a second Cu1+ exporter that is functionally redundant with CopA, while copL encodes for a membrane-bound, surface-exposed, Cu1+-binding lipoprotein [79]. It is proposed that copL functions to prevent copper uptake by binding extracellular copper ions and preventing them from entering or re-entering the cell after export by CopA or CopB [79] (Figure 3). In some S. aureus strains, CsoR also regulates the transcription of mco, which encodes a predicted Cu-dependent multicopper oxidase, although the precise function(s) of Mco remains unknown [87]. S. aureus strains lacking the copAB and copBmco operons have decreased survival in phagocytes [86]. These findings highlight the importance of proper Cu homeostasis for pathogenesis. Moreover, copBL is the only genetic operon shared between the ACME and COMER transposable elements, which were acquired by the USA300 and USA300-LV clones, respectively [88]. These strains have contributed clonal epidemics of community acquired S. aureus in North American (USA300) and South American (USA300-LV) countries [88]. Shared copBL acquisition suggests that the hypertolerance to Cu may have helped facilitate the spread of these clones.
Manganese transport regulator (MntR)
Manganese is an essential micronutrient in S. aureus where it is utilized as a cofactor of superoxide dismutase and plays a key role in defending against the oxidative burst of phagocytes during infection [1]. Two staphylococcal systems are used to import Mn ions from the environment: the MntABC ABC-type transporter and the MntH Nramp family transporter [89] (see Figure 4). During pathogenesis, both the MntABC and the MntH systems compete with calprotectin to acquire Mn in tissue abscesses [90].
Figure 4: Model for manganese ion homeostasis in S. aureus.
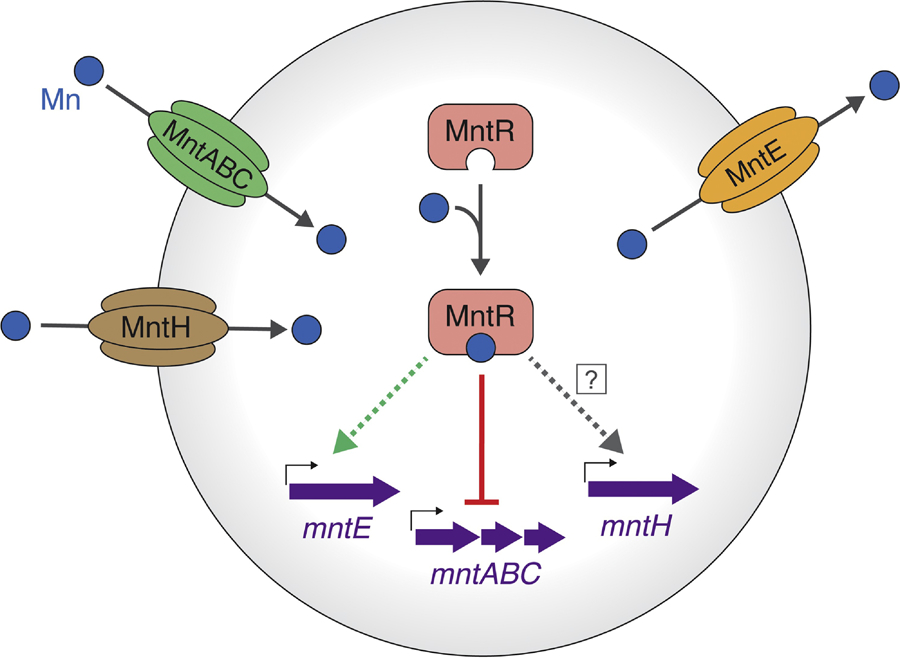
Mn2+ ions are imported into the cell by the MntABC and MntH transporters. The MntR transcriptional regulator binds intracellular manganese and represses transcription of mntABC. It is unclear if transcription of mntH is Mn-dependent and/or regulated by MntR. To prevent cytotoxicity, excess Mn is effluxed by MntE. While upregulation of mntE is dependent on the presence of MntR, it is not known if this effect is a result of direct or indirect regulation.
Transcription of mntABC is regulated by the DtxR family metalloregulator MntR [89] (Figure 4). MntR shares a similar structural architecture to other metalloregulators, consisting of an N-terminal HTH-motif DNA-binding domain and a C-terminal dimerization domain, with the metal sensing site located between the two domains [91]. The metal binding site of each MntR monomer can occupy two metal ions. The coordination geometry determines the selective response of MntR to its cognate metals; larger metal ions (Mn2+ and Cd2+) form a binuclear complex with MntR and are activated, while smaller metal ions (Fe2+, Co2+, and Zn2+) do not fully occupy the site resulting in low activity [92, 93]. The interdomain region of MntR is flexible and can adopt a range of orientations relative to the dimerization domain. Upon Mn2+ or Cd2+ binding, the linker region is rigidified, which restricts the conformation of the protein and is thought to promote DNA binding [94]. There is not yet a structure of MntR in complex with DNA.
The promoters of both mntABC and mntH contain proposed MntR box sequences that are binding sites recognized by MntR. In B. subtilis, MntR acts a repressor of mntABC transcription and an activator of mntH transcription [95]. There are conflicting studies on the role of manganese and MntR in mntH transcription in S. aureus. While one report noted Mn-dependent regulation of mntH transcription, others have found little to no effect ([89, 90] and Al-Tameemi et al. under review). These seemingly discordant observations may be the result of differing culture conditions or different genetic backgrounds.
The gene encoding the RsaC small non-coding RNA is co-transcribed with mntABC and regulated by MntR [96]. RsaC interacts with the superoxide dismutase (sodA) transcript to inhibit its expression [96]. By decreasing sodA expression, RsaC prevents the synthesis of Mn2+-dependent SodA when Mn is limiting. RsaC also interacts with the sufC transcript which encodes an iron-sulfur cluster synthesis enzyme, as well as zur, znuB, and znuC that function in Zn homeostasis [96, 97]. These findings suggest potential crosstalk between Mn, Zn, and Fe homeostasis. mntABC transcription is also negatively regulated by PerR [89].
Like other metals, excess cytosolic Mn is cytotoxic. Until recently, the mechanism(s) of Mn efflux in S. aureus were unclear. In the presence of high Mn, S. aureus expresses the cation diffusion facilitator family transporter MntE to remove excess cytosolic Mn [98] (Figure 4). Upregulation of MntE was shown to be dependent on the presence of MntR, but it remains unknown if MntR directly regulates mntE transcription [98]. An S. aureus strain lacking MntE exhibits accumulated intracellular Mn, but decreased levels of intracellular Fe, highlighting the interplay between Fe and Mn homeostasis [98]. The presence of MntR and MntE were also required for full virulence of S. aureus in murine models of infection, indicating the importance of controlling intracellular Mn levels for pathogenesis [98].
Concluding Remarks
The acquisition of metal ions and prevention of cytosolic metal ion imbalance are essential processes for all microbes. Here, we have discussed how these processes are controlled by a set of metalloregulators that carefully monitor and respond to alterations in the concentrations of cytosolic metal ion pools in S. aureus. These responses vary from the increased import of necessary metals to the removal of unwanted, cytotoxic ions or small molecules. Recently, researchers have also highlighted the roles of these metalloregulators in controlling the transcription of efflux pumps that help restore metal ion homeostasis when titers exceed ideal levels [79, 98]. The study of metal ion homeostasis in S. aureus continues to gain attention because of four key discoveries: (1) hosts protect themselves through nutritional immunity by sequestering specific metal ions [6], (2) hosts use Cu ions to aid bacterial killing in the phagolysosome [99], (3) S. aureus uses metal detoxification genes, including those encoded on mobile DNA, to circumvent the human immune system [85, 97], and (4) metal ion acquisition and homeostasis are promising antimicrobial targets [3]. There is also renewed interest in the roles of metalloregulators in controlling the expression of S. aureus genes that function in pathogenetic actions including toxin production and biofilm formation. Future studies will uncover if these phenotypes are through direct regulation or an indirect consequence of altered cytosolic metal ion pools. As the threat of antibiotic resistant S. aureus increases, metal ion acquisition and homeostasis remain viable targets for antimicrobial development.
Highlights.
The Fe-responsive metalloregulator Fur controls expression of virulence factors, including exoproteins, suggesting that the S. aureus Fur-dependent regulon extends beyond genes utilized in Fe homeostasis.
Fur and Zur co-operate to regulate synthesis of staphylopine, a metallophore used to acquire metal ions by competing with host factors that provide nutritional immunity.
The HssRS two-component regulatory system prevents heme intoxication by sensing cytosolic heme levels and increasing expression of HrtAB, which exports heme or heme metabolites.
The Cu-responsive metalloregulator CsoR controls transcription of the copAZ Cu detoxification genes and the newly described copBL and copBmco operons that provide hypertolerance to Cu.
The MntR metalloregulator preserves Mn homeostasis by regulating transcription of the Mn importer mntABC, as well as the expression of mntE to export excess cytosolic Mn.
Outstanding Questions.
Free cytosolic metal ions can be cytotoxic. Once a metal ion has entered a S. aureus cell, are there factors that act as intracellular metal ion buffers? Do small molecules, such as low molecular weight thiols, play a role in helping to buffer the cytosol and/or facilitate the donation of metal ions to metalloregulators [100]?
The apo-form of the E. coli Fur metalloregulator was found to be proteolytically degraded upon the loss of its metal cofactor [101]. Are S. aureus metalloregulators recycled or degraded if they are not associated with DNA or metal ions?
A large portion of intracellular Fe is used to synthesize iron-sulfur (FeS) clusters in S. aureus and other bacteria. What role(s) do metalloregulators have in regulating this process?
In E. coli, Fur acts as an indirect oxygen sensor by sensing the balance between pools of Fe2+ and Fe3+. Does Fur use a similar mechanism in S. aureus to detect alterations in intracellular redox status?
A number of loci (example: acnA) have decreased expression in an S. aureus fur mutant. Does Fur activate gene expression in S. aureus, and if so, what is the mechanism of activation? Does apo-Fur play a regulatory role in S. aureus?
In B. subtilis, mRNA transcripts corresponding to more than twenty genes have significantly altered abundances in a zur mutant, and eighty direct Zur-binding sites have been identified [102]. What genes are regulated by Zur in S. aureus?
PerR functions as a sensor of the cellular Fe/Mn ratio in B. subtilis. Is this role of PerR conserved in S. aureus?
The HssSR two-component regulatory system responds to cytosolic heme accumulation. Does HssS directly bind heme, or is it activated by a heme-associated byproduct?
The HtsABC transporter imports iron via transport of staphyloferrin A. Does the Hts system also play a direct role in heme transport, and if so, what is the mechanism?
The CsoR metalloregulator aids in preventing intoxication by Cu ions. What is the mechanism(s) of copper toxicity in S. aureus?
In B. subtilis, MntR acts a repressor of mntABC transcription and an activator of mntH transcription [95]. Does MntR play a role in mntH expression in S. aureus?
Glossary
- Fenton reaction
the chemical process in which peroxide (H2O2) is converted to a hydroxyl radical (•OH) via a metal ion catalyst, typically Fe2+
- Fur box
a conserved, inverted repeat sequence of DNA found in the promoter region of Fur target genes; the binding site of typical Fur occupation during iron-replete conditions (also note: MntR box, Per box, and Zur box, which are the sites of MntR, PerR, and Zur binding, respectively)
- Heme
an iron-containing compound of the porphyrin class which functions as the cofactor for hemoglobin (within erythrocytes) and myoglobin (within myocytes)
- Irving-Williams series
a reference order of the relative stabilities of complexes formed between ligands and divalent transition metals (Mn2+ < Fe2+ < Co2+ < Ni2+ < Cu2+ > Zn2+)
- Metallophore
an excreted soluble molecule that transiently binds to metal ions to aid in their sequestration and transport
- Metalloregulator
a DNA-binding protein that alters the transcription of DNA to RNA in direct response to the availability of metal ion(s)
- Micronutrient:
a vitamin or mineral that is required in trace amounts for the normal growth and development of an organism
- Mismetallation
occurs when an inappropriate metal ion has occupied the metal binding site of a holo-metalloprotein
- Nutritional immunity
the process during infection in which a host sequesters nutrients essential to bacterial fitness
- S100 proteins
a family of human proteins produced by the host immune system that tightly bind to metal ions to aid nutritional immunity
- Siderophore
a high-affinity, iron-chelating compound secreted by microorganisms that is used to sequester and transport iron ions from the environment into the cell
- Staphylopine
a nicotinanamine-like metallophore produced by S. aureus that aids in the acquisition and transport of Co2+, Zn2+, and Ni2+
- Two-component regulatory system
a stimulus-response coupling mechanism used by prokaryotes to detect and adapt to changes in environmental conditions; consists of a membrane-localized sensor kinase and a DNA-binding cytosolic response regulator
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Garcia YM et al. (2017) A Superoxide Dismutase Capable of Functioning with Iron or Manganese Promotes the Resistance of Staphylococcus aureus to Calprotectin and Nutritional Immunity. PLoS Pathog 13 (1), e1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres VJ et al. (2010) Staphylococcus aureus Fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect Immun 78 (4), 1618–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts CA et al. (2017) The Suf Iron-Sulfur Cluster Biosynthetic System Is Essential in Staphylococcus aureus, and Decreased Suf Function Results in Global Metabolic Defects and Reduced Survival in Human Neutrophils. Infect Immun 85 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassat JE and Skaar EP (2012) Metal ion acquisition in Staphylococcus aureus: overcoming nutritional immunity. Seminars in Immunopathology 34 (2), 215–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donato R et al. (2013) Functions of S100 proteins. Curr Mol Med 13 (1), 24–57. [PMC free article] [PubMed] [Google Scholar]
- 6.Corbin BD et al. (2008) Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319 (5865), 962–5. [DOI] [PubMed] [Google Scholar]
- 7.Nakashige TG et al. (2015) Human calprotectin is an iron-sequestering host-defense protein. Nature Chemical Biology 11 (10), 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Besold AN et al. (2018) Role of Calprotectin in Withholding Zinc and Copper from Candida albicans. Infect Immun 86 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glaser R et al. (2005) Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat Immunol 6 (1), 57–64. [DOI] [PubMed] [Google Scholar]
- 10.Yang Z et al. (2001) Proinflammatory properties of the human S100 protein S100A12. J Leukoc Biol 69 (6), 986–94. [PubMed] [Google Scholar]
- 11.Dell’Angelica EC et al. (1994) Primary structure and binding properties of calgranulin C, a novel S100-like calcium-binding protein from pig granulocytes. J Biol Chem 269 (46), 28929–36. [PubMed] [Google Scholar]
- 12.Moroz OV et al. (2003) Structure of the human S100A12-copper complex: implications for host-parasite defence. Acta Crystallogr D Biol Crystallogr 59 (Pt 5), 859–67. [DOI] [PubMed] [Google Scholar]
- 13.Weinberg ED (2009) Iron availability and infection. Biochimica et Biophysica Acta - General Subjects 1790 (7), 600–605. [DOI] [PubMed] [Google Scholar]
- 14.Courcol RJ et al. (1997) Siderophore production by Staphylococcus aureus and identification of iron-regulated proteins. Infection and Immunity 65 (5), 1944–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drechsel H et al. (1993) Purification and chemical characterization of staphyloferrin B, a hydrophilic siderophore from staphylococci. Biometals 6 (3), 185–92. [DOI] [PubMed] [Google Scholar]
- 16.Konetschny-Rapp S et al. (1990) Staphyloferrin A: a structurally new siderophore from staphylococci. European Journal of Biochemistry 191 (1), 65–74. [DOI] [PubMed] [Google Scholar]
- 17.Dale SE et al. (2004) Involvement of SirABC in iron-siderophore import in Staphylococcus aureus. J Bacteriol 186 (24), 8356–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beasley FC et al. (2009) Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Molecular Microbiology 72 (4), 947–963. [DOI] [PubMed] [Google Scholar]
- 19.Grigg JC et al. (2010) The Staphylococcus aureus siderophore receptor HtsA undergoes localized conformational changes to enclose staphyloferrin A in an arginine-rich binding pocket. J Biol Chem 285 (15), 11162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grim KP et al. (2017) The Metallophore Staphylopine Enables Staphylococcus aureus To Compete with the Host for Zinc and Overcome Nutritional Immunity. mBio 8 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghssein G et al. (2016) Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science 352 (6289), 1105–9. [DOI] [PubMed] [Google Scholar]
- 22.Skaar EP and Schneewind O (2004) Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes and Infection 6 (4), 390–397. [DOI] [PubMed] [Google Scholar]
- 23.Gunther MR et al. (1995) Hydroxyl radical formation from cuprous ion and hydrogen peroxide: a spin-trapping study. Arch Biochem Biophys 316 (1), 515–22. [DOI] [PubMed] [Google Scholar]
- 24.Haber F and Weiss J (1932) On the catalysis of hydroperoxide. Naturwissenschaften 20 (51), 948–50. [Google Scholar]
- 25.Cotruvo JA Jr. and Stubbe J (2012) Metallation and mismetallation of iron and manganese proteins in vitro and in vivo: the class I ribonucleotide reductases as a case study. Metallomics 4 (10), 1020–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mascher T et al. (2006) Stimulus Perception in Bacterial Signal-Transducing Histidine Kinases. Microbiology and Molecular Biology Reviews 70 (4), 910–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao R and Stock AM (2009) Biological insights from structures of two-component proteins. Annu Rev Microbiol 63, 133–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JW and Helmann JD (2007) Functional specialization within the Fur family of metalloregulators. Biometals 20 (3–4), 485–99. [DOI] [PubMed] [Google Scholar]
- 29.Lindsay JA and Foster SJ (2001) zur: a Zn(2+)-responsive regulatory element of Staphylococcus aureus. Microbiology 147 (Pt 5), 1259–1266. [DOI] [PubMed] [Google Scholar]
- 30.Deng Z et al. (2015) Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nature Communications 6 (1), 7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarvan S et al. (2018) Variation on a theme: investigating the structural repertoires used by ferric uptake regulators to control gene expression. BioMetals 31 (5), 681–704. [DOI] [PubMed] [Google Scholar]
- 32.Althaus EW et al. (1999) The Ferric Uptake Regulation (Fur) Repressor Is a Zinc Metalloprotein. Biochemistry 38 (20), 6559–6569. [DOI] [PubMed] [Google Scholar]
- 33.Lee J-W and Helmann JD (2006) Biochemical Characterization of the Structural Zn2+ Site in the Bacillus subtilis Peroxide Sensor PerR. Journal of Biological Chemistry 281 (33), 23567–23578. [DOI] [PubMed] [Google Scholar]
- 34.Dian C et al. (2011) The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites. Molecular Microbiology 79 (5), 1260–1275. [DOI] [PubMed] [Google Scholar]
- 35.Ma Z et al. (2012) Origins of specificity and cross-talk in metal ion sensing by Bacillus subtilis Fur. Molecular Microbiology 86 (5), 1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helmann JD (2014) Specificity of Metal Sensing: Iron and Manganese Homeostasis in Bacillus subtilis. Journal of Biological Chemistry 289 (41), 28112–28120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bagg A and Neilands JB (1987) Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26 (17), 5471–5477. [DOI] [PubMed] [Google Scholar]
- 38.de Lorenzo V et al. (1987) Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. Journal of bacteriology 169 (6), 2624–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Escolar L et al. (1997) Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Molecular Microbiology 26 (4), 799–808. [DOI] [PubMed] [Google Scholar]
- 40.Beauchene NA et al. (2015) Impact of Anaerobiosis on Expression of the Iron-Responsive Fur and RyhB Regulons. mBio 6 (6), e01947–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beauchene NA et al. (2017) O2 availability impacts iron homeostasis in Escherichia coli. Proc Natl Acad Sci U S A 114 (46), 12261–12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horsburgh MJ et al. (2001) In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J Bacteriol 183 (2), 468–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong A et al. (2000) Molecular characterization of the ferric-uptake regulator, Fur, from Staphylococcus aureus. Microbiology 146 ( Pt 3), 659–68. [DOI] [PubMed] [Google Scholar]
- 44.Friedman DB et al. (2006) Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog 2 (8), e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hassan H and Troxell B (2013) Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Frontiers in Cellular and Infection Microbiology 3 (59). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinochet-Barros A and Helmann JD (2020) Bacillus subtilis Fur is a transcriptional activator for the PerR-repressed pfeT gene encoding an iron efflux pump. J Bacteriol. [DOI] [PMC free article] [PubMed]
- 47.Ernst FD et al. (2005) Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 151 (2), 533–546. [DOI] [PubMed] [Google Scholar]
- 48.Delany I et al. (2001) Iron-dependent transcription of the frpB gene of Helicobacter pylori is controlled by the Fur repressor protein. J Bacteriol 183 (16), 4932–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ernst FD et al. (2005) Iron-responsive regulation of the Helicobacter pylori iron-cofactored superoxide dismutase SodB is mediated by Fur. J Bacteriol 187 (11), 3687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma Z et al. (2011) Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur. Nucleic acids research 39 (21), 9130–9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin J-H and Helmann JD (2016) Molecular logic of the Zur-regulated zinc deprivation response in Bacillus subtilis. Nature Communications 7 (1), 12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jordan MR et al. (2019) Mechanistic Insights into the Metal-Dependent Activation of ZnII-Dependent Metallochaperones. Inorganic Chemistry 58 (20), 13661–13672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fojcik C et al. (2018) Independent and cooperative regulation of staphylopine biosynthesis and trafficking by Fur and Zur. Mol Microbiol 108 (2), 159–177. [DOI] [PubMed] [Google Scholar]
- 54.Remy L et al. (2013) The Staphylococcus aureus Opp1 ABC transporter imports nickel and cobalt in zinc-depleted conditions and contributes to virulence. Molecular Microbiology 87 (4), 730–743. [DOI] [PubMed] [Google Scholar]
- 55.Bsat N et al. (1998) Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol 29 (1), 189–98. [DOI] [PubMed] [Google Scholar]
- 56.Herbig AF and Helmann JD (2001) Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Molecular Microbiology 41 (4), 849–859. [DOI] [PubMed] [Google Scholar]
- 57.Lee J-W and Helmann JD (2006) The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440 (7082), 363–367. [DOI] [PubMed] [Google Scholar]
- 58.Jacquamet L et al. (2009) Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Molecular Microbiology 73 (1), 20–31. [DOI] [PubMed] [Google Scholar]
- 59.Morrissey JA et al. (2004) The Staphylococcal Ferritins Are Differentially Regulated in Response to Iron and Manganese and via PerR and Fur. Infection and Immunity 72 (2), 972–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cosgrove K et al. (2007) Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol 189 (3), 1025–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horsburgh MJ et al. (2001) PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect Immun 69 (6), 3744–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji C-J et al. (2015) Staphylococcus aureus PerR is a Hypersensitive Hydrogen Peroxide Sensor Using Fe-mediated Histidine Oxidation. Journal of Biological Chemistry. [DOI] [PMC free article] [PubMed]
- 63.Fuangthong M et al. (2002) Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J Bacteriol 184 (12), 3276–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bullen JJ et al. (1978) Role of Iron in Bacterial Infection In Current Topics in Microbiology and Immunology: Volume 80 (Arber W et al. eds), pp. 1–35, Springer; Berlin Heidelberg. [DOI] [PubMed] [Google Scholar]
- 65.Skaar EP et al. (2004) Iron-source preference of Staphylococcus aureus infections. Science 305 (5690), 1626–8. [DOI] [PubMed] [Google Scholar]
- 66.Mazmanian SK et al. (2003) Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299 (5608), 906–9. [DOI] [PubMed] [Google Scholar]
- 67.Dryla A et al. (2003) Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Molecular Microbiology 49 (1), 37–53. [DOI] [PubMed] [Google Scholar]
- 68.Hammer ND and Skaar EP (2011) Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu Rev Microbiol 65, 129–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Torres VJ et al. (2006) Staphylococcus aureus IsdB Is a Hemoglobin Receptor Required for Heme Iron Utilization. Journal of Bacteriology 188 (24), 8421–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stojiljkovic I et al. (2001) Antimicrobial properties of porphyrins. Expert Opinion on Investigational Drugs 10 (2), 309–320. [DOI] [PubMed] [Google Scholar]
- 71.Torres VJ et al. (2007) A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1 (2), 109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stauff DL et al. (2007) Signaling and DNA-binding activities of the Staphylococcus aureus HssR-HssS two-component system required for heme sensing. J Biol Chem 282 (36), 26111–21. [DOI] [PubMed] [Google Scholar]
- 73.Bibb LA et al. (2007) The ChrA-ChrS and HrrA-HrrS signal transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae. Infect Immun 75 (5), 2421–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmitt MP (1999) Identification of a two-component signal transduction system from Corynebacterium diphtheriae that activates gene expression in response to the presence of heme and hemoglobin. J Bacteriol 181 (17), 5330–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stauff DL and Skaar EP (2009) Bacillus anthracis HssRS signalling to HrtAB regulates haem resistance during infection. Mol Microbiol 72 (3), 763–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stauff DL et al. (2008) Staphylococcus aureus HrtA is an ATPase required for protection against heme toxicity and prevention of a transcriptional heme stress response. J Bacteriol 190 (10), 3588–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gotz F and Mayer S (2013) Both terminal oxidases contribute to fitness and virulence during organ-specific Staphylococcus aureus colonization. mBio 4 (6), e00976–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hammer ND et al. (2013) Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. MBio 4 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rosario-Cruz Z et al. (2019) The copBL operon protects Staphylococcus aureus from copper toxicity: CopL is an extracellular membrane-associated copper-binding protein. J Biol Chem 294 (11), 4027–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grossoehme N et al. (2011) Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J Biol Chem 286 (15), 13522–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Higgins K and Giedroc D (2014) Insights into Protein Allostery in the CsoR/RcnR Family of Transcriptional Repressors. Chemistry letters 43, 20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tan BG et al. (2014) Conformational and thermodynamic hallmarks of DNA operator site specificity in the copper sensitive operon repressor from Streptomyces lividans. Nucleic Acids Res 42 (2), 1326–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang F-MJ et al. (2014) Cu(I)-mediated Allosteric Switching in a Copper-sensing Operon Repressor (CsoR). Journal of Biological Chemistry 289 (27), 19204–19217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baker J et al. (2011) The Staphylococcus aureus CsoR regulates both chromosomal and plasmid-encoded copper resistance mechanisms. Environ Microbiol 13 (9), 2495–507. [DOI] [PubMed] [Google Scholar]
- 85.Purves J et al. (2018) A horizontally gene transferred copper resistance locus confers hyper-resistance to antibacterial copper toxicity and enables survival of community acquired methicillin resistant Staphylococcus aureus USA300 in macrophages. Environ Microbiol 20 (4), 1576–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zapotoczna M et al. (2018) Mobile-Genetic-Element-Encoded Hypertolerance to Copper Protects Staphylococcus aureus from Killing by Host Phagocytes. MBio 9 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sitthisak S et al. (2005) Characterization of a multicopper oxidase gene from Staphylococcus aureus. Appl Environ Microbiol 71 (9), 5650–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Planet PJ et al. (2015) Parallel Epidemics of Community-Associated Methicillin-Resistant Staphylococcus aureus USA300 Infection in North and South America. The Journal of infectious diseases 212 (12), 1874–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Horsburgh MJ et al. (2002) MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol Microbiol 44 (5), 1269–86. [DOI] [PubMed] [Google Scholar]
- 90.Kehl-Fie TE et al. (2013) MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun 81 (9), 3395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Glasfeld A et al. (2003) Structure of the manganese-bound manganese transport regulator of Bacillus subtilis. Nature Structural & Molecular Biology 10 (8), 652–657. [DOI] [PubMed] [Google Scholar]
- 92.McGuire AM et al. (2013) Roles of the A and C Sites in the Manganese-Specific Activation of MntR. Biochemistry 52 (4), 701–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kliegman JI et al. (2006) Structural Basis for the Metal-Selective Activation of the Manganese Transport Regulator of Bacillus subtilis. Biochemistry 45 (11), 3493–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Golynskiy M et al. (2007) Conformational studies of the manganese transport regulator (MntR) from Bacillus subtilis using deuterium exchange mass spectrometry. JBIC Journal of Biological Inorganic Chemistry 12 (5), 699–709. [DOI] [PubMed] [Google Scholar]
- 95.Que Q and Helmann JD (2000) Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol 35 (6), 1454–68. [DOI] [PubMed] [Google Scholar]
- 96.Lalaouna D et al. (2019) RsaC sRNA modulates the oxidative stress response of Staphylococcus aureus during manganese starvation. Nucleic Acids Res 47 (18), 9871–9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Choby JE et al. (2016) A Small-Molecule Inhibitor of Iron-Sulfur Cluster Assembly Uncovers a Link between Virulence Regulation and Metabolism in Staphylococcus aureus. Cell Chem Biol. [DOI] [PMC free article] [PubMed]
- 98.Grunenwald CM et al. (2019) Manganese Detoxification by MntE Is Critical for Resistance to Oxidative Stress and Virulence of Staphylococcus aureus. MBio 10 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hodgkinson V and Petris MJ (2012) Copper homeostasis at the host-pathogen interface. J Biol Chem 287 (17), 13549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rosario-Cruz Z and Boyd JM (2016) Physiological roles of bacillithiol in intracellular metal processing. Curr Genet 62 (1), 59–65. [DOI] [PubMed] [Google Scholar]
- 101.Mettert EL and Kiley PJ (2005) ClpXP-dependent proteolysis of FNR upon loss of its O2-sensing [4Fe-4S] cluster. J Mol Biol 354 (2), 220–32. [DOI] [PubMed] [Google Scholar]
- 102.Prestel E et al. (2015) Genome-wide identification of Bacillus subtilis Zur-binding sites associated with a Zur box expands its known regulatory network. BMC Microbiology 15 (1), 13. [DOI] [PMC free article] [PubMed] [Google Scholar]


