Fig. 2.
In vitro paracrine properties and transcriptomic signature of oMSC and sMSC
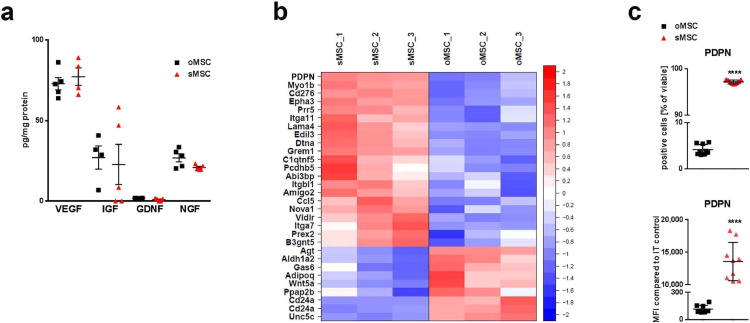
(a) Secreted trophic factors (mRNA clustered in Supplementary Fig.2a) in cell culture supernatants of oMSC and sMSC 48 h after plating of cells analysed by ELISA, n=5, each data point represents one biological replicate per group; replicates analysed collectively by specific ELISAs for GDNF, IGF, NGF and VEGF; oMSC were compared with sMSC by ANOVA with Bonferroni's multiple comparisons test. Error bars: SEM; GDNF, Glial cell-derived neurotrophic factor; IGF, Insulin-like growth factor; NGF, Nerve growth factor; VEGF, vascular endothelial growth factor.
(b) Heatmap of microarray data displaying genes related to cell migration and/or adhesion. Only genes differing significantly and relevantly between murine oMSC and sMSC are displayed. Row-wise z-scores of normalized log2 signal intensities in murine oMSC and sMSC, n=3 per group (3 different passages). Z-scores color-coded as indicated by colour bar on the right.
(c) Quantification of PDPN protein by flow cytometry analysis of o/sMSC to validate PDPN mRNA expression (microarray), n=9, each data point represents one biological replicate per group; two-tailed t-test was used for comparison of percentage and MFI counts of oMSC vs. sMSC (***p<0.001); Error bars: SD. IT, isotype control; MFI, mean fluorescence intensity.