Abstract
Seed traits present important breeding targets for enhancing grain yield and quality in various grain legume crops including pigeonpea. The present study reports significant genetic variation for six seed traits including seed length (SL), seed width (SW), seed thickness (ST), seed weight (SWT), electrical conductivity (EC) and water uptake (WU) among Cajanus cajan (L.) Millspaugh acc. ICPL 20340 and Cajanus scarabaeoides (L.) Thouars acc. ICP 15739 and an F2 population derived from this interspecific cross. Maximum phenotypic values recorded for the F2 population were higher than observed in the parent ICPL 20340 [F2 max vs ICPL 20340: SW (7.05 vs 5.38), ST (4.63 vs 4.51), EC (65.17 vs 9.72), WU (213.17 vs 109.5)], which suggested contribution of positive alleles from the wild parent, ICP 15739. Concurrently, to identify the QTL controlling these seed traits, we assayed two parents and 94 F2 individuals with 113 polymorphic simple sequence repeat (SSR) markers. In the F2 population, 98 of the 113 SSRs showed Mendelian segregation ratio 1:2:1, whereas significant deviations were observed for 15 SSRs with their χ2 values ranging between 6.26 and 20.62. A partial genetic linkage map comprising 83 SSR loci was constructed. QTL analysis identified 15 marker-trait associations (MTAs) for seed traits on four linkage groups i.e. LG01, LG02, LG04 and LG05. Phenotypic variations (PVs) explained by these QTL ranged from 4.4 (WU) to 19.91% (EC). These genomic regions contributing significantly towards observed variability of seed traits would serve as potential candidates for future research that aims to improve seed traits in pigeonpea.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02423-x) contains supplementary material, which is available to authorized users.
Keywords: Genetic linkage map, Genotyping, Mapping population, Phenotypic variation, QTL, Seed traits, SSR, Wild relative
Introduction
Pigeonpea [Cajanus cajan (L.) Millspaugh] is the sixth most important grain legume grown across the globe (FAOSTAT 2018). Annually, a total of 5.99 million tons of pigeonpea is harvested from worldwide area of 6.99 mha (FAOSTAT 2018). India and Myanmar are the two major countries contributing 83.6% of the total pigeonpea to the global pigeon pea basket. Nutritionally, pigeonpea serves as a rich source of protein, starch, fat and important minerals to the malnourished human population across the globe, especially in the developing countries (Obala et al. 2019; Bohra et al. 2020).
Seed traits are key to improving seed yield and quality of grain legume crops like pigeonpea. However, limited attention has been paid to improve seed traits in pigeonpea breeding programmes. A variety of seed traits such as number of seeds per plant, 100-seed weight are reported to determine the crop productivity. Seed traits in general receive limited attention in view of the fact that our key breeding targets include developing high-yielding disease- and pest-resistant cultivars. In recent years; however, growing consumer preferences and market requirements have caused a renewed focus on seed traits like seed size and shape (Liang et al. 2005). Niu et al. (2013) have reported that 100-seed weight is positively influenced by seed size, which is a function of its length, width and thickness. Rate of water uptake by seed is also known to influence seed quality in chickpea (Lamichaney et al. 2016), soybean (Singh et al. 2008), flax (Saeidi 2008) fababean (Peksen 2007) and cowpea (Peksen et al. 2004), which is attributed to embryo damage and subsequently leaching out of electrolytes necessary for the growth and development of embryo. Entry of water into seed during imbibition is regulated by the physical and chemical constituents of seed coat (Lamichaney et al. 2017). Negative association of hard seed (inability of seeds to imbibe water) with seed weight has also been reported in soybean (Hirota et al. 2005). The seed coat or testa acts as a barrier between the embryo and external environment, which regulates the transport of metabolites, air and water to the embryo (Smykal et al. 2014).
Wild relatives serve as great reservoir for genetic variability for various traits of breeding importance in various legume crops including pigeonpea (Bohra et al. 2010; Khoury et al. 2015; Smýkal et al. 2015). Incorporation of crop wild relatives for improving seed traits has been demonstrated in various legume species (Abbo et al. 1992; Porch et al. 2013; Bohra et al. 2014). Genetic diversity studies elucidate narrow genetic base of the cultivated pigeonpea (Kassa et al. 2012; Yang et al. 2006). Experimental populations based on the wide crosses have great potential to reveal novel variations/alleles for the important traits including seed traits (Li et al. 2019). Recent advances in DNA marker technologies have allowed molecular mapping of various seed traits in different legume crops such as groundnut (Zhang et al. 2019), soybean (Xu et al. 2011; Han et al. 2012), common bean (Pérez-Vega et al. 2010; Yuste-Lisbona et al. 2014), lentil (Fedoruk et al. 2013; Verma et al. 2015; Jha et al. 2017; Khazaei et al. 2018). In pigeonpea, limited studies have been performed so far on analysis of seed traits using DNA marker technology (Yadav et al. 2019). Availability of robust marker-trait associations could play important role in fast-track improvement of seed traits. The present investigation was carried out with the aim to capture genetic variability in a broad base F2 population segregating for various seed traits and to identify the underlying QTL/genomic regions to enable rapid and targeted improvement of these traits in pigeonpea.
Materials and methods
Plant materials
We followed standard hybridization technique suggested by Sharma and Green (1980) for pigeonpea. Developing flower buds on ICPL 20340 [Cajanus cajan (L.) Millspaugh] were hand emasculated and pollen source buds were collected from ICP 15739 [Cajanus scarabaeoides (L.) Thouars] in the morning hours. After pollination of the emasculated buds, the plant (ICPL 20340) was covered with fine-mesh nylon cloth bags. Coloured threads were used to identify cross-pollinated flowers. The four F1 seeds resulting from the cross were planted and a single true F1 was identified based on the visual inspection (Fig. 1). Ninety-four F2 individuals resulting from self-pollination of true F1 plant were grown at main farm of Indian Institute of Pulses Research (IIPR), India during 2017–18.
Fig. 1.
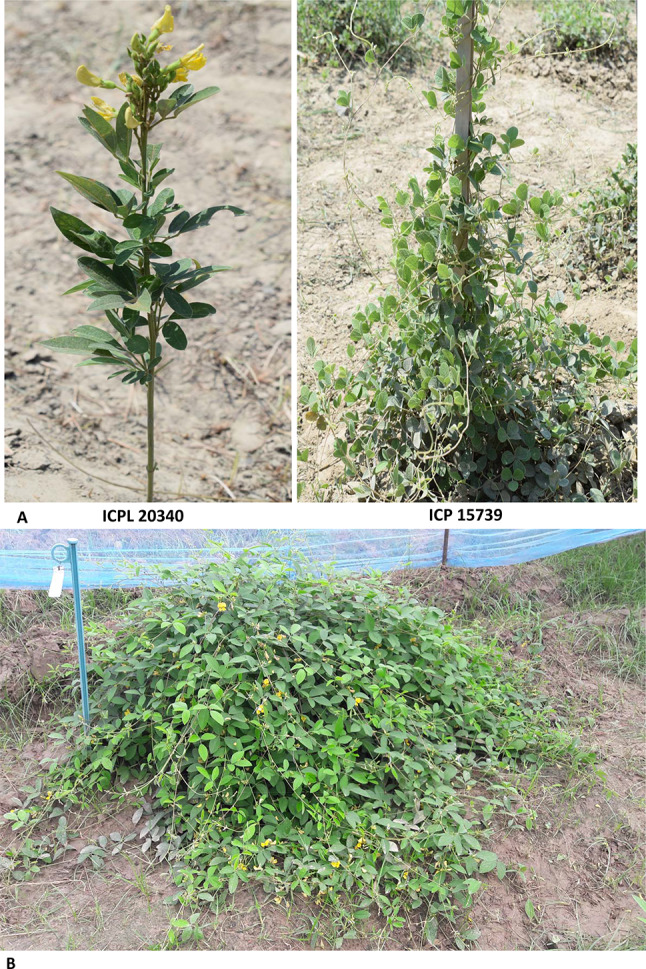
Images of parents and F1 hybrid a Parents ICPL 20340 and ICP 15739 and b Interspecific F1 obtained from the cross ICPL 20340 × ICP 15739, which was self-pollinated to develop an F2 population
Phenotyping of the F2 population was done for six seed traits viz. seed length (SL), seed width (SW), seed thickness (ST), seed weight (SWT), electrical conductivity of seed leachate (EC) and water uptake (WU). For SL, SW and ST, data were taken from 10 individual seed using digital Vernier caliper having an accuracy of ± 0.02 mm and the average data were used for analysis. Owing to shortage of seeds, ten seeds of each sample were weighed and were allowed to soak for 24 h in 50 ml of distill water at 20 °C. After 24 h of soaking the seeds were re-weighed and the percentage increase in water uptake was calculated. The seed leachate remained after 24 h of soaking was used to measure EC using conductivity meter and expressed in μS cm−1 g−1 of seed. The data on all seed traits were analyzed using SAS v. 9.4.
Genomic DNA extraction
In order to genotype the entire F2 population, plant leaves were taken from each individual. Genomic DNA was extracted from all 94 F2 individuals along with the two parents. Genomic DNA was isolated from young leaves by using CTAB method of DNA extraction (Murray and Thompson 1980). Both quantity and quality of DNA were estimated through electrophoresis using 0.8% agarose gel (Sigma-Aldrich, St. Louis, USA).
SSR analysis
A reaction mixture of 10 μl volume was prepared using 4.8 μl of sterilized distilled water, 2.0 μl template DNA (25 ng), 0.5 μl of forward and 0.5 μl of reverse primer (5 μM), 1.0 μl 10 × PCR buffer (10 mM Tris–HCl, 50 mMKCl, pH 8.3), 1.00 μl dNTP mix (0.2 mM each of dATP, dGTP, dCTP and dTTP) and 0.2 μl Taq polymerase (5 U/μl) (Thermo Scientific, Mumbai, India) for the amplification of genomic DNA with SSR primers. The reaction mixture was polymerized in G-40402 thermo cycler (G-STORM, Somerset, UK) in a touchdown PCR profile by using the following amplification profile: initial denaturation at 94 °C for 5 min followed by 10 cycles of touchdown 55–45 °C, 20 s at 94 °C, annealing for 20 s at 55 °C (the annealing temperature for each cycle being reduced by 1 °C per cycle) and extension for 30 s at 72 °C. This was accompanied by 40 cycle of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s, elongation at 72 °C for 45 s and 10 min of final extension at 72 °C. Amplified products were resolved in 3% agarose gel using 0.5 × TBE running buffer and images were analyzed in Quantity one software (Bio-Rad, CA 94,547, USA).
Marker genotyping and construction of genetic linkage map
A set of 113 polymorphic SSR markers comprising Cajanus cajan genomic microsatellites (CcGMs; Bohra et al. 2017; Varshney et al. 2012) and Arahar hypervariable simple sequence repeat (AHSSR; Singh et al. 2012) were used for genotyping mapping parents and F2 individuals. The amplicons were scored as A, B and H. ICIM v. 4.2 was run to build genetic linkage map by using parameters LOD 4, nnTwoOpt algorithm, criterion SARF and window size of 5 cM.
QTL analysis
Genotypic and phenotypic data of parents and F2 population were used for QTL analysis using ICIM v. 4.2 by applying parameters LOD threshold of 2.5, window size 5.0 cM, missing phenotype: deletion, mapping method ICIM-ADD and PIN 0.001.
Results
Measuring variation for seed physical traits
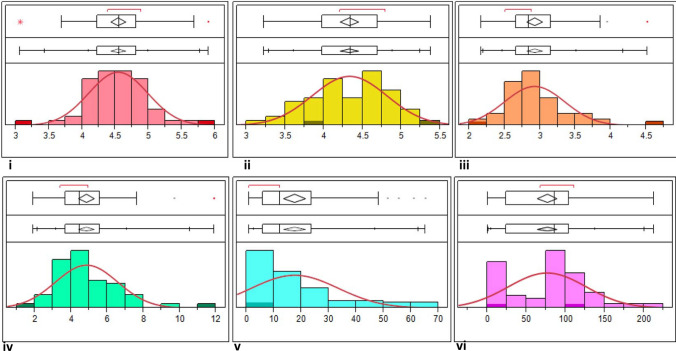
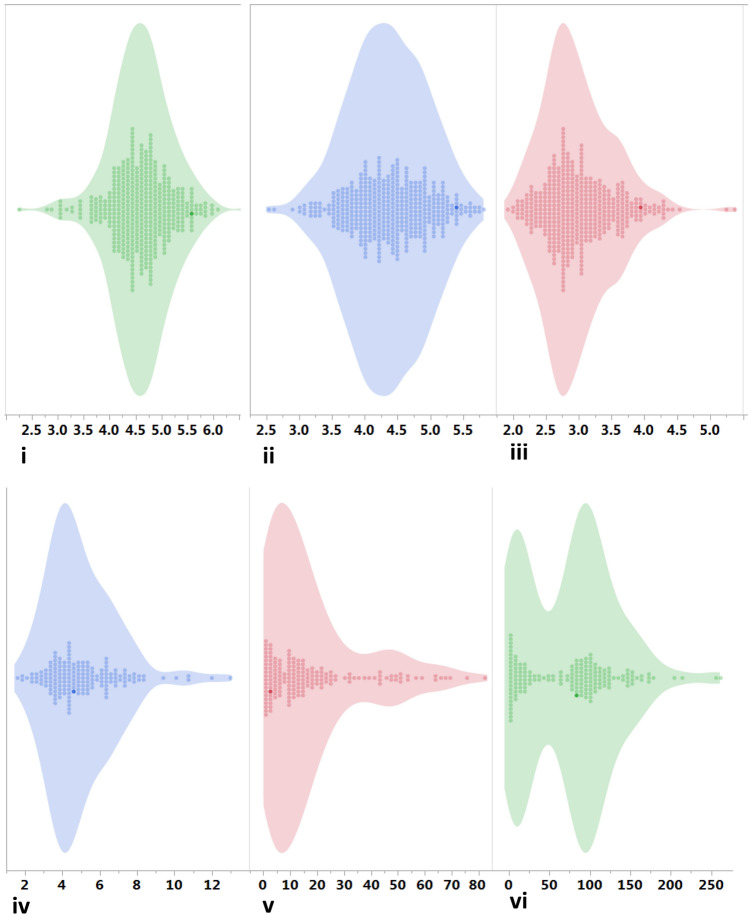
The parents (ICPL 20340 and ICP 15739) were crossed to generate F1s (Fig. 1) and F2 population was obtained by self-pollinating the true F1 plant. The parents and F2 population displayed significant variation for these quantitative traits based on the t test and ANOVA, respectively. The differences in seed and hilum morphology of seeds from parents and random F2 plants are shown in Fig. 2. ICPL 20340 (P1) showed higher SL of 5.89 mm as compared to 3.06 mm for ICP 15739 (P2). Among all individuals, values for SL ranged from 3.69 mm to 5.69 mm with an average value of 4.54 mm (Table 1, Fig. 3a, b). Mapping parents were highly contrasting for SW with ICPL 20340 exhibiting greater width (5.38 mm) than ICP 15739 (3.88 mm), while F2 individuals had SW values in the range of 3.23–7.05 mm. Regarding ST, ICPL 20340 had ST of 4.51 mm which was greater than the other parent ICP 15739 having ST of 2.22 mm. Concerning mapping population, wide variation was observed among F2 individuals for ST varying from 2.17 to 4.63 mm. Similarly, the parent ICPL 20340 displayed SWT of 11.90 g, whereas ICP 15739 had a SWT of 1.90 g. Likewise, F2 population showed significant variation for SWT ranging from 2.30 g to 9.70 g. ICPL 20340 had an EC value of 9.72 μS cm−1 g−1 of seed as compared to the other parent ICP 15739 with an EC value of 0.93 μS cm−1 g−1 of seed. Variation was also observed in the F2 individuals having a minimum EC value of 0.99 and maximum EC of 65.17 μS cm−1 g−1. Mapping parents had contrasting values for WU, i.e. 1.4% (ICP 15739) and 109.57% (ICPL 20340) and the range of WU in the F2 population remained between 0.62 and 213.17%.
Fig. 2.
Variation in seed (upper half) and hilum (lower half) morphology among seeds from parents and F2 plants
Table 1.
Range of trait variation observed between parents and in the F2 population
| S. no. | Traits | P1 | P2 | F2 individuals | *Average (including parents) | Skewness | Kurtosis | |
|---|---|---|---|---|---|---|---|---|
| (ICPL 20340) | (ICP 15739) | |||||||
| Minimum | Maximum | |||||||
| 1 | Seed length (mm) | 5.89 | 3.06 | 3.69 | 5.69 | 4.54 ± 0.22 | 0.09 | 2.24 |
| 2 | Seed width (mm) | 5.38 | 3.88 | 3.23 | 7.05 | 4.33 ± 0.24 | − 0.22 | − 0.45 |
| 3 | Seed thickness (mm) | 4.51 | 2.22 | 2.17 | 4.63 | 2.92 ± 0.21 | 1.20 | 2.48 |
| 4 | 100-seed weight (g) | 11.9 | 1.9 | 2.3 | 9.7 | 4.85 ± 0.87 | 1.49 | 3.94 |
| 5 | Electrical conductivity (μS cm−1 g−1) | 9.72 | 0.93 | 0.99 | 65.17 | 17.57 ± 7.97 | 1.39 | 1.32 |
| 6 | Water uptake (%) | 109.5 | 1.4 | 0.62 | 213.17 | 76.31 ± 25.15 | 0.17 | − 0.20 |
*95% confidence interval
Fig. 3.
Distribution of seed traits among parents and population. Distributions of i) seed length ii) seed width iii) seed thickness iv) 100 seed weight v) electrical conductivity and vi) water uptake are illustrated by a bar plots and b contour and count plots. Both figures elucidate almost normal distribution of seed width, seed thickness, seed length and 100-seed weight, whereas electrical conductivity is highly skewed. The areas with dark colour in bar plots contain data points of parents. In contour and count plot, each data point is expressed as dot showing the frequency of each value in the distribution. The highest frequency or modal value is presented by the longest vertical line joined by dots
Construction of a coarse genetic linkage map
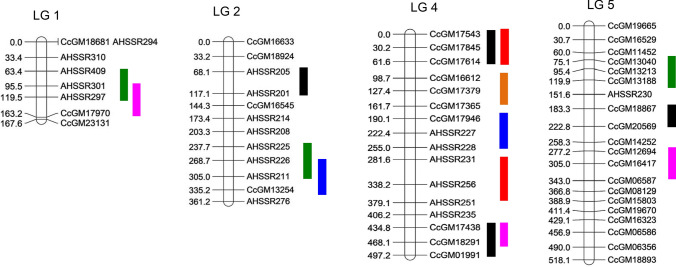
A total of 457 hypervariable SSR markers were used for polymorphism survey between mapping parents. These hypervariable SSR markers were developed from the pigeonpea genome by targeting SSR tract length of ≥ 20 bp (Bohra et al. 2017; Singh et al. 2012; Varshney et al. 2012). Eleven hundred thirteen polymorphic SSR markers were identified following polymorphism survey of parental genotypes. Genotyping data were assembled for these 113 SSR markers in 94 F2 individuals. We then evaluated segregation patterns of 113 SSRs in F2 population using goodness-of-fit test with alpha threshold 0.05 and 2 degree of freedom (Supplementary Table 1). Ninety-eight of the 113 SSRs showed Mendelian segregation ratio 1:2:1, whereas deviations were observed for 15 SSRs with their χ2-values ranging between 6.26 and 20.62. Linkage analysis could provide map positions to 83 loci, resulting in a coarse genetic linkage map spanning a total map length of 2113.4 cM with individual LGs varying between 167.60 cM and 518.10 cM (Table 2). Owing to low number of markers, we could obtain six LGs in the current analysis instead of eleven LGs of pigeonpea (2n = 2x = 22). Nine loci showing segregation distortion were also mapped onto LGs 1, 3, 5 and 6. The number of mapped loci per LG ranged from 8 (LG01) to 20 (LG05). The average inter marker distance was found to be 25.46 cM. The distribution of DNA markers across different LGs was unequal and the size of the LG did not necessarily reflect the number of mapped loci. For example, LG05 with 20 markers had a length of 518.10 cM, whereas LG01 with eight loci spanned a distance of 167.6 cM.
Table 2.
Salient features of the genetic linkage map based on the interspecific population
| Linkage group | Number of mapped loci | Length (cM) | Average marker spacing (cM) |
|---|---|---|---|
| LG1 | 8 | 167.6 | 20.95 |
| LG2 | 12 | 361.2 | 30.1 |
| LG3 | 11 | 305 | 27.72 |
| LG4 | 16 | 497.2 | 31.07 |
| LG5 | 20 | 518.1 | 25.9 |
| LG6 | 16 | 264.3 | 16.51 |
| Total markers | 83 | 2113.4 | 25.46 |
Identification of marker-trait associations (MTAs) for seed traits
The genotyping and phenotyping data were combined to find significant marker-trait associations controlling variation for seed traits (Fig. 4). Consequently, two QTL detected in LG04 showed significant association with SL explaining about 16 and 10% of the total phenotypic variation (Table 3). Likewise, three QTL on LG05, LG02 and LG01 showed significant association with SW, accounting for 16, 12 and 8% PV for the trait. One major QTL, QTL6 (flanked by AHSSR301 and CcGM17970) with 10.47% PV was detected for ST on LG01. One QTL, QTL10 (flanked by CcGM17946 and AHSSR228) for SWT explaining 13.37% PV was detected on LG02. QTL11 (flanked by CcGM16612–CcGM17365) was detected on LG04 that is associated with EC explaining PV of 19.91%. We obtained four QTL for rate of WU on LGs 02, 04, 05 and PVs explained by these QTL ranged from 4.4 to 5%.
Fig. 4.
Linkage groups showing marker intervals of significant QTL. Total 15 QTL were identified for six seed traits from analysis of genotyping and phenotyping data. QTL for SW, ST, SL, SWT, EC, WU are shown with green, magenta, red, blue, orange and black colour, respectively
Table 3.
QTLs for seed-related traits detected in the F2 population (ICPL 20340 × ICP 15739)
| Traits | LGs | QTL | Position (cM) | Marker interval | LOD value | *Additive effect | % PV |
|---|---|---|---|---|---|---|---|
| Seed length (SL) | 4 | QTL1 | 30.2 | CcGM17543–CcGM17614 | 4.67 | − 0.0141 | 15.75 |
| 4 | QTL2 | 338.2 | AHSSR231–AHSSR251 | 3.86 | − 0.2291 | 10.48 | |
| Seed width (SW) | 5 | QTL3 | 95.4 | CcGM13040–CcGM13188 | 4.05 | 0.0537 | 16.07 |
| 2 | QTL4 | 268.7 | AHSSR225–AHSSR211 | 2.75 | − 0.1005 | 12.2 | |
| 1 | QTL5 | 95.5 | AHSSR409–AHSSR297 | 2.59 | − 0.2952 | 7.67 | |
| Seed thickness (ST) | 1 | QTL6 | 119.5 | AHSSR301–CcGM17970 | 3.86 | − 0.2467 | 10.47 |
| 4 | QTL7 | 468.1 | CcGM17438–CcGM18291 | 2.77 | − 0.0886 | 6.89 | |
| 5 | QTL8 | 305 | CcGM12694–CcGM06587 | 2.7 | 0.1856 | 6.75 | |
| Seed weight (SWT) | 4 | QTL9 | 222.4 | CcGM17946–AHSSR228 | 2.97 | − 0.1326 | 8.46 |
| 2 | QTL10 | 305 | CcGM13254–AHSSR226 | 2.58 | 0.4516 | 13.37 | |
| Electrical conductivity (EC) | 4 | QTL11 | 127.4 | CcGM16612–CcGM17365 | 2.71 | 0.9112 | 19.91 |
| Water uptake (WU) | 2 | QTL12 | 96 | AHSSR205–AHSSR201 | 3.3 | 0.0633 | 5 |
| 4 | QTL13 | 44 | CcGM17543–CcGM17614 | 3.8 | − 0.5861 | 5 | |
| 4 | QTL14 | 452 | CcGM17438–CcGM18291 | 2.8 | 2.9773 | 4.4 | |
| 5 | QTL15 | 202 | CcGM18867–CcGM20569 | 6.6 | 3.5851 | 4.9 |
*The positive or negative signs of additive effect of a QTL indicate that, the favourable allele is contributed by the wild parent (ICP 15739) and cultivated parent (ICPL 20340), respectively
Discussion
Seed traits viz., SW, SWT and seed size remain one of the important breeding targets for determining market class and value and consumer preference of grain legume crops like pigeonpea. However, limited efforts have been invested for genetic analysis of seed traits in pigeonpea. The present study is the first attempt to report the genetic variation and identify the QTL for important seed traits in an inter-specific population of pigeonpea. In the present investigation, we analyzed variations in seed traits (SL, SW, ST, SWT, EC and WU) of two contrasting parents and F2 individuals. Mapping parents registered significant difference for the traits examined here. For instance, ICPL 20340 had bold seed with higher seed coat permeability as it recorded higher rate of WU and EC values. Whereas ICP 15739 was small seeded with lower values of WU and EC. The value of all seed traits in F2 individuals roughly spanned between the values recorded in the parents.
The recent advancements in genomic technologies in concert with availability of mapping populations have allowed construction of genetic linkage maps and QTL mapping of various traits of breeding importance in pigeonpea; however, the reported number of QTL controlling seed traits remains dismally low in pigeonpea (Yadav et al. 2019; Obala et al. 2020). In the present study, we observed segregation distortion for 16.95% of the 113 SSRs genotyped in F2 population. Segregation distortion is a common phenomenon in populations resulting from interspecific crosses. In a previous study in pigeonpea, authors found segregation distortion for 63.5% of the SSR loci scored in an interspecific F2 population (Cajanus cajan acc. ICP 28 × Cajanus scarabaeoides acc. ICPW 94) and authors attributed this deviation to occurrence of low parental alleles and overabundance of heterozygous alleles in population (Bohra et al. 2011).
Out of 11 QTL mapped in the current research, seven QTL namely QTLs 1, 2, 3, 4, 6, 10 and 11 could be considered as major QTL as these explained more than 10% PV, while remaining eight QTL (QTLs 5, 7, 8, 9, 12, 13, 14, 15) had minor effects since these accounted for less than 10% PV. Two QTL were mapped onto LG04 for SL, while LG04 and LG02 had one QTL each for SWT. Three QTL each were detected for SW (located at LG01, LG02 and LG05) and ST (located at LG01, LG04 and LG05), while one QTL on LG04 was found controlling variation in EC. Earlier, Yadav et al. (2019) have reported one major QTL (qSS6.1) for seed size on CcLG06 that explained 29.5% PV. Similarly, QTL related to SL, ST and SW have been reported in rice (Qi et al. 2017), wheat (Williams and Sorrells 2014), maize (Liu et al. 2014), lentil (Fedoruk et al. 2013; Khazaei et al. 2018), soybean (Teng et al. 2018), groundnut (Zhang et al. 2019).
Earlier researchers have noted wider genetic variation for SWT in pigeonpea (Zavinon et al. 2019; Obala et al. 2019; Yadav et al. 2019). Obala et al. (2020) reported one QTL flanked by SNP markers S2_11771536 and S2_10960200 for SWT on LG02. In our study, four QTL were detected for WU. Parents ICPL 20340 and ICP 15739 differed in the seed coat permeability as they registered contrasting values for WU and EC. Higher EC value of the seed leachate is an indicator of reduced membrane integrity. Hence, the seeds of ICPL 20340 having higher WU also recorded higher EC of seed leachate. Negative correlation is reported between seed hardness with 100-seed weight and rate of water uptake in soybean (Hirota et al. 2005). Significant MTAs have been reported for seed coat permeability (assessed by rate of water uptake) in other legume crops; such as Singh et al. (2008) identified four QTLs associated with seed coat permeability in soybean, of which one (satt281) showed association with EC.
Wide range of genetic variability revealed in the inter-specific population followed by molecular mapping of the underlying genomic regions reinforces the significance of wild relatives and offers great scope for targeted and accelerated improvement of these important traits. In the present study, traits SW, ST, SWT, EC and WU had QTL with positive additive effects suggested the contribution of favourable allele of these traits from the wild parent ICP 15739. Earlier, Gnanesh et al. (2011) also reported contribution of favourable alleles from low-value parent, i.e. susceptible parent towards observed phenotypic variability in pigeonpea. Once validated in diverse genetic background/after saturation of the candidate genomic regions, the MTAs can be deployed in breeding programs for fast-track improvement of seed traits.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Financial support from Indian Council of Agricultural Research (ICAR), New Delhi is gratefully acknowledged. Dr C.V. Sameer Kumar is highly acknowledged for providing seeds of ICPL 20340 and ICP 15739.
Author contributions
AB conceived the idea, planned experiments and wrote manuscript with RJ, UCJ and NPS. RJ, AT performed genotyping experiments. RJ performed QTL analysis. AL along with AKM, VY recorded measurements related to seed traits. AB, RJ, DS analyzed the data. FS, IPS and DD helped in developing mapping population and interpretation of results. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
Authors declare that there is no conflict of interest.
References
- Abbo S, Ladizinsky G, Weeden NF. Genetic analysis and linkage studies of seed weight in lentil. Euphytica. 1992;58:259–266. [Google Scholar]
- Bohra A, Mallikarjuna N, Saxena KB, Upadhyaya H, Vales I, Varshney R. Harnessing the potential of crop wild relatives through genomics tools for pigeonpea improvement. J Plant Biol. 2010;37:83–98. [Google Scholar]
- Bohra A, Dubey A, Saxena RK, Penmetsa RV, Poornima KN, Kumar N, Farmer AD, Srivani G, Upadhyaya HD, Gothalwal R, Ramesh R, Singh D, Saxena KB, Kavi Kishor PB, Singh NK, Town CD, May GD, Cook DR, Varshney RK. Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea. BMC Plant Biol. 2011;11:56. doi: 10.1186/1471-2229-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohra A, Jha UC, Kavi Kishor PB, Pandey S, Singh NP. Genomics and molecular breeding in lesser explored pulse crops: current trends and future opportunities. Biotechnol Adv. 2014;32:1410–1428. doi: 10.1016/j.biotechadv.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Bohra A, Jha R, Pandey G, Patil PG, Saxena RK, Singh IP, Singh D, Mishra RK, Mishra A, Singh F, Varshney RK, Singh NP. New Hypervariable SSR markers for diversity analysis, hybrid purity testing and trait mapping in pigeonpea [Cajanus cajan (L.) Millspaugh] Front Plant Sci. 2017;8:377. doi: 10.3389/fpls.2017.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohra A, Saxena KB, Varshney RK, Saxena RK. Genomics assisted breeding for pigeonpea improvement. Theor Appl Genet. 2020;133:1721–1737. doi: 10.1007/s00122-020-03563-7. [DOI] [PubMed] [Google Scholar]
- FAOSTAT (2018). https://faostat.fao.org/beta/en/#data/QC/visualize. Accessed 5 Mar 2020
- Fedoruk MJ, Vandenberg A, Bett KE. Quantitative trait loci analysis of seed quality characteristics in lentil using single nucleotide polymorphism markers. Plant Genome. 2013;6:1–10. doi: 10.3835/plantgenome2017.06.0051. [DOI] [PubMed] [Google Scholar]
- Gnanesh BN, Bohra A, Sharma M, Byregowda M, Pande S, Wesley V, Saxena RK, Saxena KB, KaviKishor PB, Varshney RK. Genetic mapping and quantitative trait locus analysis of resistance to sterility mosaic disease in pigeonpea [Cajanus cajan (L.) Millsp.] Field Crops Res. 2011;123:53–61. [Google Scholar]
- Han Y, Li D, Zhu D, Li H, Li X, Teng W, Li W. QTL analysis of soybean seed weight across multi-genetic backgrounds and environments. Theor Appl Genet. 2012;125:671–683. doi: 10.1007/s00122-012-1859-x. [DOI] [PubMed] [Google Scholar]
- Hirota TK, Takahata T, Ogawa M, Iwai IY. Quality of soybean seeds grown in Hyogo prefecture. Bull Hyogo Pre Tech Cent Agric Forest Fish (Agric) 2005;53:6–12. [Google Scholar]
- Jha R, Bohra A, Jha UC, Rana M, Chahota RK, Kumar S, Sharma TR. Analysis of an intraspecific RIL population uncovers genomic segments harbouring multiple QTL for seed relevant traits in lentil (Lens culinaris L.) Physiol Mol Biol Plants. 2017;23:675–684. doi: 10.1007/s12298-017-0438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassa MT, Penmetsa RV, Carrasquilla-Garcia N, Sarma BK, Datta S, Upadhyaya HD, Varshney RK, Von Wettberg EJB, Cook DR. Genetic patterns of domestication in pigeonpea (Cajanus cajan (L,) Millsp.) and wild Cajanus relatives. PLoS ONE. 2012;7:e39563. doi: 10.1371/journal.pone.0039563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazaei H, Fedoruk M, Caron CT, Vandenber A, Bett KE. Single nucleotide polymorphism markers associated with seed quality characteristics of cultivated lentil. Plant Genome. 2018;11:170051. doi: 10.3835/plantgenome2017.06.0051. [DOI] [PubMed] [Google Scholar]
- Khoury CK, Castaeda-Alvarez NP, Achicanoy HA, Sosa CC, Bernau V, Kassa MT, Norton SL, von der Maesen LJG, Upadhyaya HD, Ramirez-Villegas J, Jarvis A, Struik PC. Crop wild relatives of pigeonpea [Cajanus cajan (L.) Millsp.]: distributions, ex situ conservation status, and potential genetic resources for abiotic stress tolerance. Biol Conserv. 2015;184:259–270. [Google Scholar]
- Lamichaney A, Katiyar PK, Natarajan S, Sripathy KV. Relationship among some seed characters, laboratory germination and field emergence in chickpea (Cicer arietinum L.) genotypes differing in testa colour. J Food Legume. 2016;29(1):29–32. [Google Scholar]
- Lamichaney A, Kudekallu S, Kamble U, Sarangapany N, Katiyar PK, Bohra A. Differences in seed vigour traits between desi (pigmented) and kabuli (non-pigmented) ecotypes of chickpea (Cicer arietinum L.) and its association with field emergence. J Environ Biol. 2017;38:735–742. [Google Scholar]
- Li J, Zhao J, Li Y, Gao Y, Hua S, Nadeem M, Sun G, Zhang W, Hou J, Wang X. Identification of a novel seed size associated locus SW9-1 in soybean. Crop J. 2019;7:548–559. [Google Scholar]
- Liang HZ, Li WD, Wang H, Fang XJ. Genetic effects on seed traits in soybean. Acta Genet Sin. 2005;32:1199–1204. [PubMed] [Google Scholar]
- Liu Y, Wang L, Sun C, Zhang Z, Zheng Y, Qiu F. Genetic analysis and major QTL detection for maize kernel size and weight in multi-environments. Theor Appl Genet. 2014;127:1019–1037. doi: 10.1007/s00122-014-2276-0. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res. 1980;8:4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Xu Y, Liu XF, Yang SX, Wei SP, Xie FT, Zhang YM. Association mapping for seed size and shape traits in soybean cultivars. Mol Breed. 2013;31:785–794. [Google Scholar]
- Obala J, Saxena RK, Singh VK, et al. Development of sequence-based markers for seed protein content in pigeonpea. Mol Genet Genom. 2019;294:57–68. doi: 10.1007/s00438-018-1484-8. [DOI] [PubMed] [Google Scholar]
- Obala J, Saxena RK, Singh VK, et al. Seed protein content and its relationships with agronomic traits in pigeonpea is controlled by both main and epistatic effects QTLs. Sci Rep. 2020;10:214. doi: 10.1038/s41598-019-56903-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peksen E. Relationships among electrical conductivity of seed leakage, germination, field emergence percentage and some seed traits in faba bean (Vicia faba) Asian J Chem. 2007;19:3178–3184. [Google Scholar]
- Peksen A, Peksen E, Bozoglu H. Relationships among some seed traits, laboratory germination and field emergence in cowpea genotypes. Pak J Bot. 2004;36:311–320. [Google Scholar]
- Pérez-Vega E, Pañeda A, Rodríguez-Suárez C, et al. Mapping of QTLs for morpho-agronomic and seed quality traits in a RIL population of common bean (Phaseolus vulgaris L.) Theor Appl Genet. 2010;120:1367–1380. doi: 10.1007/s00122-010-1261-5. [DOI] [PubMed] [Google Scholar]
- Porch TG, Beaver JS, Debouck DG, Jackson SA, Kelly JD, Dempewolf H. Use of wild relatives and closely related species to adapt common bean to climate change. Agronomy. 2013;3:433–461. [Google Scholar]
- Qi L, Sun Y, Li J, Su L, Zheng XM, Wang XN, et al. Identify QTLs for grain size and weight in common wild rice using chromosome segment substitution lines across six environments. Breed Sci. 2017;67:472–482. doi: 10.1270/jsbbs.16082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeidi G. Genetic variation and heritability for germination, seed vigour and field emergence in brown and yellow-seeded genotypes of flax. Int J Plant Prod. 2008;2:15–22. [Google Scholar]
- Sharma D, Green JM. Pigeonpea. In: Fehr WR, Hadley HH, editors. Hybridization of crop plants. American Society of Agronomy Crop Science. Wisconsin: Society of America; 1980. pp. 471–481. [Google Scholar]
- Singh RK, Raipuria RK, Bhatia VS, Rani A, Husain SM, Satyavathi CT, Chauhan GS, Mohapatra T. Identification of SSR markers associated with seed coat permeability and electrolyte leaching in soybean. Physiol Mol Biol Plant. 2008;14:173–177. doi: 10.1007/s12298-008-0016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NK, Gupta DK, Jayaswal PK, Mahato AK, Dutta S, Singh S, Bhutani S, Dogra V, Singh BP, Kumawat G, Pal JK, Pandit A, Singh A, Rawal H, Kumar A, Rama Prashat G, Khare A, Yadav R, Raje RS, Singh MN, Datta S, Fakrudin B, Wanjari KB, Kansal R, Dash PK, Jain PK, Bhattacharya R, Gaikwad K, Mohapatra T, Srinivasan R, Sharma TR. The first draft of the pigeonpea genome sequence. J Plant Biochem Biotechnol. 2012;21:98–112. doi: 10.1007/s13562-011-0088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smykal P, Vernoud V, Blair MW, Soukup A, Thompson RD. The role of the testa during development and in establishment of dormancy of the legume seed. Front Plant Sci. 2014;5:351. doi: 10.3389/fpls.2014.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smýkal P, Coyne CJ, Ambrose MJ, et al. Legume crops phylogeny and genetic diversity for science and breeding. Crit Rev Plant Sci. 2015;34:43–104. [Google Scholar]
- Teng WL, Sui MN, Li W, Wu DP, Zhao X, Li HY, Han YP, Li WB. Identification of quantitative trait loci underlying seed shape in soybean across multiple environments. J Agric Sci. 2018;156:3–12. [Google Scholar]
- Varshney RK, Chen W, Li Y, Bharti AK, Saxena RK, Schlueter JA, Donoghue MTA, Azam S, Fan G, Whaley AM, Farmer AD, Sheridan J, Iwata A, Tuteja R, Penmetsa RV, Wu W, Upadhyaya HD, Yang SP, Shah T, Saxena KB, Michael T, McCombie WR, Yang B, Zhang G, Yang H, Wang J, Spillane C, Cook DR, May GD, Xu X, Jackson SA. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol. 2012;30:83–89. doi: 10.1038/nbt.2022. [DOI] [PubMed] [Google Scholar]
- Verma P, Goyal R, Chahota RK, Sharma TR, Abdin MZ, Bhatia S. Construction of a genetic linkage map and identification of QTLs for seed weight and seed size traits in lentil (Lens culinaris Medik.) PLoS ONE. 2015;10:e0139666. doi: 10.1371/journal.pone.0139666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Sorrells ME. Three-dimensional seed size and shape QTL in hexaploid wheat (Triticum aestivum L.) populations. Crop Sci. 2014;54:98–110. [Google Scholar]
- Xu Y, Li HN, Li GJ, Wang X, Cheng LG, Zhang YM. Mapping quantitative trait loci for seed size traits in soybean (Glycine max L. Merr.) Theor Appl Genet. 2011;122:581–594. doi: 10.1007/s00122-010-1471-x. [DOI] [PubMed] [Google Scholar]
- Yadav P, Saxena KB, Hingane A, Kumar CS, Kandalkar VS, Varshney RK, Saxena RK. An “Axiom Cajanus SNP Array” based high density genetic map and QTL mapping for high-selfing flower and seed quality traits in pigeonpea. BMC Genomics. 2019;20:35. doi: 10.1186/s12864-019-5595-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Ash G, Harper J, Varling J, Wenzl P, et al. Low level of genetic diversity in cultivated pigeonpea compared to its wild relatives is revealed by diversity arrays technology. Theor Appl Genet. 2006;113:585–595. doi: 10.1007/s00122-006-0317-z. [DOI] [PubMed] [Google Scholar]
- Yuste-Lisbona FJ, González AM, Capel C, et al. Genetic variation underlying pod size and color traits of common bean depends on quantitative trait loci with epistatic effects. Mol Breed. 2014;33:939–952. [Google Scholar]
- Zavinon F, Adoukonou-Sagbadja H, Bossikponnon A, Dossa H, Ahanhanzo C. Phenotypic diversity for agro-morphological traits in pigeon pea landraces [(Cajanus cajan L.) Millsp.] cultivated in southern Benin. Open Agric. 2019;4:487–549. [Google Scholar]
- Zhang S, Hu X, Miao H, Chu Y, Cui F, Yang W, Wang C, Shen Y, Xu T, Zhao L, Zhang J, Chen J. QTL identification for seed weight and size based on a high-density SLAF-seq genetic map in peanut (Arachis hypogaea L.) BMC Plant Biol. 2019;3:19. doi: 10.1186/s12870-019-2164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.