Abstract
The objective of the study was to develop a bio-safe synthetic peptide ELISA for the detection of antibodies against the infectious bronchitis virus (IBV) using a novel multiple antigenic peptide approach (MAP). After initial ELISA optimization, diagnostic sensitivity (DSn) and specificity (DSp) for the linear peptides were determined using receiver operator curve (ROC) analysis. The peptide IBVP1 showed 90.44% DSn and 88.64% DSp at ROC cut off 22.8% while IBVP2 showed 88.24% DSn and 85.23% DSp at ROC cut off 23.05%. The multimerization of linear peptides to MAP design resulted in the improvement of the diagnostic efficiency up to 94.85% DSn and 92.05% DSp for IBVM1 with 19.95% cut off. A similar improvement in the performance was also observed with 92.65% DSn and 90.91% DSp for IBVM2 at 20.72% cut off. All the peptides were tested for diagnostic specificity and did not show the cross-reactivity with Newcastle disease virus and infectious bursal disease virus positive serum samples. In addition, repeatability testing for all linear and multimeric peptide showed that the coefficient of variation for intra-assay was within the expected limits, ranging from 2.4 to 10.4% and inter-assay coefficient of variation was ranging from 5.56 to 14.3%. In a nutshell, the present study used predicted B cell epitope, the synthetic peptide in linear and multimeric design for IBV antibody detection. The study also highlights peptide antigen with modified scaffold design could be a safe alternative to whole virion-based ELISA for IBV antibody detection.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02422-y) contains supplementary material, which is available to authorized users.
Keywords: Antigenic peptides, Gammacoronavirus of poultry, Peptide scaffold, Peptide diagnostics, Respiratory poultry pathogen
Introduction
Infectious bronchitis virus (IBV) is a contagious respiratory pathogen. It is a single stranded, non-segmented RNA virus. The virus belongs to the genus Gammacoronavirus within the family Coronaviridae (Cavanagh 2003). The virus infects domestic fowl chickens of all ages. Infectious bronchitis (IB) disease severely affects the poultry production worldwide leading to major economic losses to the poultry industry. IB is listed as notifiable disease by the office of international des epizootics (OIE)-world organization of animal health, (OIE 2008). Conventionally, IB can be diagnosed by virus neutralization assay, RFLP, RT-PCR, and ELISA (De Wit 2000; Kwon et al. 1993). The RT-PCR and ELISA remains to be the methods of choice for laboratory diagnosis of IB (Nguyen et al. 2013). Several studies have demonstrated the utility of expressed protein subunits in ELISA for the detection of antibodies against IBV (Lei et al. 2017; Lugovskaya et al. 2006; Moneim et al. 2014). These methods provided a valued alternative to the use of whole virus antigen. However, the cumbersome process of protein expression and purification limits the use of expressed protein in ELISA.
The limitations of above mentioned available ELISA protocols underline the need of an alternative method for detection of antibodies against IBV. Use of synthetic peptide ELISA for the detection of antibodies against IBV could be helpful in easing the antigen preparation and easy large-scale antigen production. The peptide is a chemically defined antigen and could be a safe alternative to the whole virus antigen. The IBV encodes four structural proteins; the spike glycoprotein (S), the membrane glycoprotein (M), the internal nucleoprotein (N) and small envelope protein (E). Out of four structural proteins spike glycoprotein and nucleoprotein are preferred antigen for development of the diagnostic assays (Ding et al. 2015; Pradhan et al. 2014). In the present study, we demonstrated a synthetic peptide antigen based bio-safe strategy for detection of antibodies against IBV. In this study, the bioinformatics tools were used to map the linear B cell epitope of IBV spike glycoprotein to predict antigenic peptides. The identified antigenic peptides were used to develop indirect ELISA for the detection of IBV antibodies from chicken serum. Further, the peptides were multimerized to four-armed peptide scaffold and its diagnostic outcome was also studied. The performance of this newly developed linear and multimeric antigenic peptide (MAP) ELISA was evaluated by comparison with the whole virus based commercial ELISA kit (IDEXX, USA). The proposed peptide ELISA could be used as an alternative to screen for antibodies against IBV in the serum samples.
Materials and methods
Antigenic peptide designing
The amino acid sequence of spike glycoprotein of IBV, Massachusetts serotype was retrieved from NCBI. The protein was scanned by Immune Epitope Database Analysis Resource (IEDB) (Vita et al. 2010) to identify the linear B cell epitopic region (Larsen et al. 2006). Bioinformatics analysis was done with the help of various indices viz. Jameson–Wolf antigenic index, surface probability, and hydrophilicity index (Hopp and Wood 1981). Additionally, SOPMA secondary structure prediction tool was used for predictive secondary structure analysis of the peptides (Geourjon and Deleage 1995). After analysis with various indices two peptides namely IBVP1 and IBVP2 with best predictive value were synthesized.
Synthesis of linear and multiple antigenic peptides (MAP)
The peptides were manually synthesized as described earlier with slight modifications (Joshi et al. 2013a). Briefly, the peptides were synthesized on rink amide resin-AM (100–200 mesh, Novabiochem, Merck, Germany) using Fmoc chemistry. Rink amide resin was swollen in dimethylformamide (DMF) overnight followed by removal of resin bound Fmoc protecting group with the treatment of 20% piperidine (v/v) in DMF for 20 min. The first amino acid coupling was carried out by mixing 3 M equivalent of amino acid in DMF, oxyma pure (2.7 M equivalent) and 2.7 M equivalent of N,N,N′,N′-tetramethyl-O-(1H-benzotriazol-1-yl) uronium hexafluorophosphate (HBTU) in presence N,N-diisopropylethylamine (DIEA). Amino acid coupling reactions were carried out on the shaker incubator at 25 °C for 2 h. The resin was then washed with DMF (3 × 1 min), dichloromethane (DCM) (2 × 1 min) and DMF (3 × 1 min). The remaining free amine were capped at each step by treating with the acetylating solution for 10 min. Subsequently, the peptide synthesis was carried out by repeating cycles of deprotection by 20% piperidine (v/v) in DMF and amino acid coupling as described above. Each step of deprotection and amino acid coupling during peptide synthesis was monitored by the Kaiser test. The final synthetic peptide was cleaved from resin in cleavage mixture of trifluoroacetic acid/phenol/triisopropylsilane/water (90:5:2.5:2.5, v/v) for 5 h at 25 °C. Finally, the cleaved peptide was precipitated with excessive prechilled diethyl ether (specially dried). The peptides were then vacuum dried and stored under dry condition until further use. Molecular weight of peptide was analysed by MALDI-TOF (Sandor speciality diagnostics Pvt. Limited, Hyderabad, India). Prior to use, dried peptides were dissolved in HPLC water and purified using reverse phase HPLC on C-8 column using a linear acetonitrile gradient. All the Fmoc amino acids and coupling reagent were purchased from Nova biochem, Merck Germany.
The linear peptide IBVP1 and IBVP2 were synthesized in a multimeric four-armed format. The multiple antigenic peptide form of these linear peptides were named as IBVM1 and IBVM2, respectively. The IBVM1 and IBVM2 were synthesized on inert alanine core with three radially branched lysin residues providing four branching points onto which desired peptide sequence was synthesized. Rink amide-AM resin (100–200 mesh) was used for solid phase peptide synthesis and di-Fmoc-Lys-OH as branching site. The Fmoc amino acid coupling was done as described above to achieve the desired length of multimeric peptide. The Kaiser test was performed at every step of MAP synthesis. The MAP peptide was cleaved from resin in cleavage mixture and precipitated with chilled diethyl ether (specially dried). MAPs were vacuum dried and stored under dry condition until further use. The MAPs were purified by reverse phase-HPLC on C-8 column with a linear acetonitrile gradient.
Optimization of peptide ELISA
The coating buffers, 0.01 M phosphate buffer saline (pH 7.4), 0.15 M citrate phosphate buffer (pH 5.0), 0.05 M carbonate bicarbonate buffer (pH 9.6) were studied for antigen coating. The working peptide antigen concentration (twofold peptide dilutions of 8.0–0.5 µg/ml in coating buffer for linear and 8.0–0.25 µg/ml in coating buffer for MAP) and serum sample dilution were determined for each peptide using checkerboard titrations (twofold serum dilutions from 1/250 to 1/2000 in blocking buffer). Combinations showing a high A492 ratio between positive serum and negative serum (P/N) were selected. Serum from specific pathogen-free (SPF) chicken was used as a negative serum. The working dilutions of blocking buffer (Supplementary Table 1) and HRP anti-chicken antibody (Rabbit anti-chicken HRP conjugate antibody, Merck Life Sciences India) dilution in blocking buffer were optimized.
Peptide ELISA procedure and performance
The 96 microtiters well ELISA plate (High Binding, MICROLONR, Greiner Bio One International, India) was coated with peptide antigen. Antigen coating (100 µl/well) was done in PBS for IBVP1 and citrate phosphate buffer for IBVP2 overnight at 4 °C. The plate was washed three times with phosphate buffer saline with 0.1% Tween 20 (PBST) followed by blocking with blocking buffer for 2 h at 37 °C. After blocking step, the primary antibody/serum samples (100 µl) diluted in blocking buffer was added and incubated for 2 h at 37 °C. Following three washing with PSBT 1 h incubation at 37 °C was done with 100 µl of HRP conjugated secondary antibodies in blocking buffer (1:10,000 dilution). Finally, the plate was washed, and 200 µl of the chromogenic OPD substrate was added (Ready to use OPD tablets dissolved in 20 ml of distilled water with final concentration of 0.4 mg/ml OPD, Sigma-Aldrich, USA). After 15 min reaction was stopped with 50 µl of 3 M H2SO4 and A492 values were recorded. The A492 was recorded using multimode microplate reader (Synergy™ 2, Bio-Tek, USA). A similar procedure was followed for the MAP peptide ELISA.
The performance of each of linear (IBVP1 and IBVP2) and multimeric (IBVM1 and IBVM2) peptide in ELISA was determined in comparison to the whole virus antigen based commercial (IDEXX, USA) ELISA kit. The serum samples were obtained from Disease Investigation Laboratory, Department of Veterinary Public Health and Epidemiology, COVS, LUVAS Hisar, India. A total of 224 chicken serum samples were studied to evaluate peptide ELISA performance. The diagnostic specificity of the proposed ELISA for each peptide was checked using serum from 10 SPF chickens. The cross reactivity of peptides for Newcastle Disease Virus (NDV) and Infectious Bursal Disease Virus (IBDV) in ELISA was evaluated using 15 known positive serum samples for each. Intra-assay and inter-assay repeatability were calculated using 5 negative and 10 positive IBV serum samples. For intra-assay repeatability, three replicates of the same serum samples were tested in the same ELISA plate. The inter-assay repeatability was also performed by two individuals at different time point and place by testing each sample in triplicates.
Data analysis
Based on A492 values, percent positivity (PP) of each sample was calculated using the following formula:
ts test sample, nc negative control, pc positive control.
Using the percent positivity (PP) value, receiver operating characteristic (ROC) curve analysis was done by SPSS version 16.0 software. For ROC analysis IBV positive and negative samples as per commercial ELISA kit were assigned the value of 1 and 0, respectively. The area under curve (AUC) was calculated for samples, AUC value close to 1 indicate the precision of the test. End point cut off in terms of PP value with maximum diagnostic sensitivity (Dsn) and diagnostic specificity (Dsp) was determined for all the peptides.
Results and discussion
Protein scanning, epitope selection, and peptide characterisation
The spike glycoprotein being major structural antigenic protein in IBV, it was used for the development of peptide ELISA (Wang et al. 2002). The whole spike glycoprotein of Massachusetts serotype was used for epitope selection. This selection was based on many previous reports indicating a wide circulation of the Massachusetts serotype in the region (Bayry et al. 2005; Patel et al. 2015; Sumi et al. 2012). The Immune Epitope Database (IEDB) was used for linear B cell epitope scanning. This analysis predicted five antigenic epitope sequences of different lengths based on amino acid sequence character and Hidden Markov Model. The IEDB is the most referred comprehensive resource for experimentally predicted B cell and T cell epitopes of human and other animal species (Potocnakova et al. 2016). The IEDB predicted peptides were further analysed for surface probability score, flexibility index (Supplementary Fig. 1), and secondary structure prediction using SOPMA secondary structure prediction method. Based on predictive values with a high prediction score and physiochemical properties, we selected two peptides namely IBVP1 and IBVP2 (Fig. 1a). Secondary structure analysis of peptide helps to understand the local secondary structure of the peptides. These peptides have a random coil structure with a random coil score of 81.83% for IBVP1 and 84.21% for IBVP2 (Supplementary Fig. 2). We observed the dominant random coil score for both IBVP1 and IBVP2 peptide. The study has suggested that random coil regions are located on the antigen surface and have high probability of forming epitopes (Li et al. 2013). The peptides IBVP1 and IBVP2 were then subjected for multiple sequence alignment to check sequence homology with any other avian pathogen. Based on BLAST results the antigenic sequences found to be specific to spike glycoprotein of IBV.
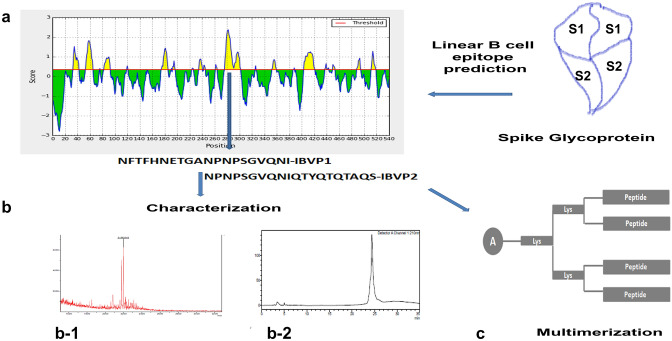
Fig. 1.
Schematic representation of antigenic peptide identification and characterization. a Immune Epitope Database Analysis Resource based Bipred B cell epitope prediction analysis showing antigenicity score of various regions of spike glycoprotein. The Bipred B cell epitope prediction analysis was used for linear B cell epitope identification. This prediction was based on sequence characteristic using Hidden Markov Model (HMM), propensity scale method (Bipred 1.0). A region with high predictive antigen score was selected as antigenic peptides. The amino acid sequence of selected peptide IBVP1 and IBVP2 is indicated with arrow. Using these amino acid sequence peptides synthesized in linear (IBVP1 and IBVP2) and four armed multimeric forms (IBVM1 and IBVM2). b Chemically synthesized peptides were characterized by MALDI-TOF and using RP-HPLC, using C-8 column on linear acetonitrile gradient b-1 the mass spectrum of IBVP1 peptide indicating observed molecular weight 2196.818 Da in MALDI. The calculated average mass of IBVP1 is 2197.08 Da b-2 the peptide chromatogram of IBVP1 peptide at 210 nm wavelength observed under linear acetonitrile gradient from 100% buffer A, to 50–50% of buffer A and buffer B over 30 min (RP-HPLC Buffer: Buffer A: 0.08% TFA in HPLC grade water, Buffer B: 0.08% TFA in HPLC grade acetonitrile) c Antigenic multimerization scheme used in study to synthesize four-armed multiple antigen peptide structure IBVM1 and IBVM2. Di-Fmoc Lysin used as branching point for armed peptide synthesis, A alanine
Once the predicted epitope region was selected the peptides were synthesized and characterized by MALDI TOF for molecular mass and RP-HPLC (Fig. 1b, c).
Development of linear peptide ELISA
ELISA remains to be an assay of choice for IBV antibody detection for many reasons like simplicity, sensitivity, and minimal lab requirements (Zang et al. 2005). Previously, many ELISA studies have described the use of whole virion and subunit protein as antigen for antibody detection (Bronzoni et al. 2005; Lin et al. 2012). Today, the use of both structural and non-structural expressed proteins is widely studied for IBV antibody ELISA (Chen et al. 2003; Finger et al. 2018; Lugovskaya et al. 2006; Wang et al. 1995). However, the use of synthetic peptide molecules can be a safe alternative to both the whole virion antigen and a cumbersome protein expression system (Spencer et al. 2007). This gave impetus to the efforts for alternative antigen preparations. Therefore, we designed the experiment to study the performance of identified predictive peptide epitopes IBVP1 and IBVP2 in ELISA. The peptide ELISA has been used for antibody detection of viral pathogens (Gießauf et al. 2004; Gomara and Haro 2007).
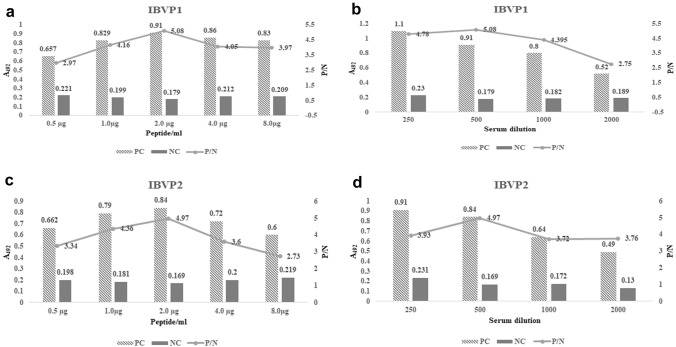
Using checkerboard ELISA, the optimal concentration of peptide antigen and serum dilution was determined for both linear peptide IBVP1 and IBVP2. Twofold peptide dilutions of 8.0–0.5 µg/ml and twofold serum dilutions from 1/250 to 1/2000 were tested in checkerboard ELISA. The antigen and antibody dilution combination with high A492 positive to negative ratio (P/N) was selected. Both the peptide showed better P/N at a combination of 2 µg/ml antigen and 1/500 serum dilution (Fig. 2). We also studied various blocking buffer for peptide ELISA and 5% (w/v) bovine casein in PBS was observed to be effective blocking buffer for all the peptides under study (Supplementary Table 1).
Fig. 2.
Checkerboard ELISA for linear peptide IBVP1 and IBVP2. Checkerboard ELISA for optimization of combination of antigen concentration and antibody dilution for linear peptide IBVP1 and IBVP2. The A492 values of checkerboard ELISA, “PC” positive serum control, “NC” negative control, serum from SPF chickens was used as negative control, P/N indicates ratio between PC and NC values, a and c graphs represents checkerboard ELISA with antigen titration for IBVP1 and IBVP2 linear peptides (0.5, 1, 2, 4 and 8 µg/ml), with 1:250, 1:500, 1:1000, 1:2000 antibody dilutions of PC and NC (b and d). The peptide IBVP1 and IBVP2 shows highest P/N at 2 µg/ml antigen concentration with 1:500 serum dilution combination
After initial ELISA procedure optimization DSn and DSp for IBVP1 and IBVP2 peptides were determined using ROC curve analysis. Peptide IBVP1 shows 90.44% DSn and 88.64% DSp at ROC cut off 22.8% while IBVP2 showed 88.24% DSn and 85.23% DSp at ROC cut off 23.05% (Table 1). The diagnostic accuracy of peptide IBVP1 and IBVP2 with diagnostic performance in comparison with whole virion based commercial ELISA (IDEXX, USA) kit is given in Table 2. The peptide ELISA is a solid phase assay wherein antigenic peptides are immobilized and binding of antibodies to the peptide antigen is recognized as colour signal. The antibody detection ability of peptide depends largely on the antigenicity of epitope present in the peptide (Dubois et al. 2012). This may explain the variation in antibody detection ability of the two peptides under study.
Table 1.
Diagnostic sensitivity, specificity of peptide ELISA to detect antibody against IBV with ROC analysis derived cut off
| Peptide | Cut off (%) | DSn (%) | DSp (%) | ROC-AUC (95% confidence interval) |
|---|---|---|---|---|
| IBVP1 | 22.80 | 90.44 | 88.64 | 0.951 (0.923–0.978) |
| IBVP2 | 23.05 | 88.24 | 85.23 | 0.944 (0.916–0.972) |
| IBVM1 | 19.95 | 94.85 | 92.05 | 0.961 (0.945–0.989) |
| IBVM2 | 20.72 | 92.65 | 90.91 | 0.951 (0.923–0.980) |
A total of 224 serum samples of broiler and layer birds were tested in peptide ELISA and each sample was tested in triplicate. “Cut off%” represents the ROC cut off for each peptide based on which percent diagnostic sensitivity (DSn%) and percent diagnostic specificity (DSp%) was determined
Table 2.
Relative diagnostic outcome with percent accuracy of peptide ELISA for antibody detection on comparison with commercial (IDEXX, USA) ELISA kit
| Peptide ELISA | IDEXX result | Total | Accuracy (%) | ||
|---|---|---|---|---|---|
| + 136 | − 88 | 224 | |||
| IBVP1 | + | 123 | 10 | 133 | 89.73a |
| − | 13 | 78 | 91 | ||
| Total | 136 | 88 | 224 | ||
| IBVP2 | + | 120 | 13 | 135 | 87.05 |
| − | 16 | 75 | 89 | ||
| Total | 136 | 88 | 224 | ||
| IBVM1 | + | 129 | 7 | 136 | 93.75 |
| − | 7 | 81 | 88 | ||
| Total | 136 | 88 | 224 | ||
| IBVM2 | + | 126 | 8 | 134 | 91.96 |
| − | 10 | 80 | 90 | ||
| Total | 136 | 88 | 224 | ||
Using cut off values mentioned in Table 1 diagnostic performance of each peptide ELISA was estimated. “+” Indicates positive and “−” indicates negative
aThe percent accuracy was calculated using formula: % Accuracy IBVP1 = (201/224) × 100 = 89.73%. Results of each peptide ELISA were compared with whole viral antigen based commercial ELISA kit (IDEXX Laboratories, USA)
The repeatability of peptide ELISA was determined by determining the intra-assay and inter-assay coefficient of variance (CV). The intra-assay CV was ranging from 4.5 to 9.6% and 4.9 to 10.4% for IBVP1 and IBVP2 peptide, respectively. The inter-assay CV was observed to 6.6–13.9% (IBVP1) and 5.9–14.3% (IBVP2), (Supplementary Fig. 3). The graph with percent positivity for the serum samples used in the intra-assay is given in Supplementary Fig. 4. The peptides satisfy the prescribed intra-assay and inter-assay coefficient of variance limits. We also examined the specificity of the peptide in ELISA using known NDV and IBDV positive serum samples. No cross reactivity was observed with NDV and IBDV positive serum samples.
Performance of multiple antigenic peptide (MAP) ELISA
As described previously, the antigenicity of peptide sequence can be improved with synthetic branched polymeric scaffold (Kim and Pau 2001). In the present study, we used a divergent four-armed multimeric design known as multiple antigenic peptide (MAP) having multiple copies of the same epitope (Fig. 1c). This design is based on inert core matrix alanine with branched poly-lysine providing the branching points (Kowalczyk et al. 2011). This design of MAP constructs offers many advantages as it provide scaffolding for close packing of antigenic peptide which in turn permit stabilization of the secondary structure (Gomara et al. 2010). The multimeric form of peptides IBVM1 and IBVM2 were studied for its performance in ELISA using the same parameters described earlier for linear peptides.
The checkerboard ELISA of MAP peptides showed better antigen coating and serum reactivity at a lower peptide concentration of 0.25 µg/ml with serum dilution of 1/500 and 1/250 for the IBVM1 and IBVM2, respectively (Fig. 3). The S/P ratio of said antigen–antibody combination was observed to be greater than linear peptides. These improvements might be attributed to the fact that multimeric arrangement helps in better antigen coating and enhances antibody binding even to low affinity antibodies (Tam 1996; Lam et al. 2002). From checkerboard titration, it was clear that the MAP peptide antigen coating was efficient at a lower peptide concentration of 0.25 µg/ml while antigen concentration requirement for the linear peptide was eight times higher i.e. 2 µg/ml. Our finding corroborates with a previous study suggesting peptide multimerization improves the antigen coating and reduces per well antigen concentration (Sarvanan et al. 2004).
Fig. 3.
Checkerboard ELISA for MAP peptide IBVM1 and IBVM2. Optimization of ELISA working combination of antigen concentration and antibody dilution for MAP peptide IBVM1 and IBVM2 using checkerboard ELISA. The A492 values of checkerboard ELISA, “PC” positive serum control, “NC” negative control, serum from SPF chicken was used as negative control, P/N indicates ratio between PC and NC values. The a and c graphs represent antigen titration of IBVM1 and IBVM2 antigen (0.25, 0.5, 1, 2, 4, and 8 µg/ml). b, d Represent antibody dilution for IBVM1 and IBVM2, respectively. The IBVM1 showed highest P/N at 0.25 µg/ml antigen concentration with 1:500 serum dilutions combination while IBVM2 showed highest P/N at 0.25 µg/ml antigen concentration with 1:250 serum dilution combination. MAP peptides showed comparative higher P/N values compared to linear peptides
On performance evaluation, we observed increased diagnostic performance in MAP ELISA. Multimerization leads to an improvement in diagnostic efficiency up to 94.85% DSn and 92.05% DSp for IBVM1. A similar improvement in the performance of ELISA was observed with 92.65% DSn and 90.91% DSp for IBVM2 (Table 1). Our finding confirms previous reports that in immunoassay, MAP antigens shows better antibody-binding capacity compare to linear peptides and improve the diagnostic potential (Nardin et al. 1995; Rai et al. 2017; Tam and Zavara 1989). Apart from the diagnostic performance other reproducibility parameters in ELISA like lower inter-assay intra-assay coefficient of variance showed better compliance in comparison with linear peptides (Supplementary Fig. 3). The relative diagnostic outcome of IBVM1 and IBVM2 along with calculated diagnostic accuracy in comparison with the whole virus based commercial ELISA is given in Table 2. The peptide on multimerization also retained the diagnostic specificity and did not show the cross reactivity with NDV and IBDV positive serum samples. The scheme of peptide multimerization used in study showed the improved diagnostic outcome on comparison with its linear format. With a combination of bioinformatics and the novel antigen design, we could achieve a diagnostic sensitivity up to 94.8% without compromising the diagnostic specificity. Many studies on use of chemically synthesized peptides in viral diagnosis are available (Dechamma et al. 2006; Joshi et al. 2013b; Kumar et al. 2016, 2019), but for IBV, few reports are available on use of synthetic peptides. Jackwood and Hilt (1995) demonstrated synthetic MAP peptides ability to generate peptide specific antibodies and projected use of MAP for IBV diagnosis (Jackwood and Hilt 1995). The Ignjatovic and Sapats (2005), mapped and identified the antigenic and protective peptide epitopes of S1 using seven short synthetic peptides (Ignjatovic and Sapats 2005). However, a complete study on demonstrating the diagnostic potential of synthetic peptide in IBV was lacking.
The present study used predicted B cell epitope, synthetic peptide in linear and multimeric design for IBV antibody detection. The study also highlights that the peptide antigen with modified scaffold design could be a safe alternative to whole virion-based ELISA for IBV antibody detection. Proposed approach may be more helpful for easy detection of antibodies against IBV.
Conclusions
The use of synthetic peptides with appropriate design could provide a safe alternative for antibody detection of IBV. The diagnostic potential of liner peptide epitopes of IBV can be improved impressively with peptide multimeric scaffold designs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Authors are thankful to Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India to fund research project “Development of Peptide and Peptide-nucleic acid Based Diagnostic Assays for Infectious Bronchitis of Poultry-YSS/2014/000406”.
Author contributions
Conceptualization and Supervision: VGJ, RC; Methodology: VGJ, RC, PG; Formal analysis, Experimentation and investigation including peptide synthesis, standardising ELISA protocol: VV, PR, PT; Writing and review and editing: VGJ, VV, PG, NKM; Funding acquisition: VGJ; Resources management and serum samples: NKM, PT.
Funding
Science and Engineering Research Bord, Department of Science and Technology, Government of India, Grant number: YSS/2014/000406.
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Contributor Information
Vikas Verma, Email: vikasverma8051@gmail.com.
Vinay G. Joshi, Email: vinaygjoshi18@luvas.edu.in
Puneet Ranjan, Email: pntrnjn@gmail.com.
Piyush Tomar, Email: tomarvety@gmail.com.
Rajesh Chhabra, Email: rchhabra59@rediffmail.com.
N. K. Mahajan, Email: nkmahajan15@gmail.com
Parveen Goel, Email: parveengoel.hsr@gmail.com.
References
- Bayry J, Goudar MS, Nighot PK, Kshirsagar SG, Ladman BS, Gelb J, Ghalsasi GR, Kolte GN. Emergence of a nephropathogenic avian infectious bronchitis virus with a novel genotype in India. J Clin Microbiol. 2005;43(2):916–918. doi: 10.1128/JCM.43.2.916-918.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronzoni RV, Fatima M, Montassier S, Pereira GT, Gama NM, Sakai V, Montassier HJ. Detection of infectious bronchitis virus and specific anti-viral antibodies using a Concanavalin A-Sandwich-ELISA. Viral Immunol. 2005;18:569–578. doi: 10.1089/vim.2005.18.569. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus. Avian Pathol. 2003;32:567–582. doi: 10.1080/03079450310001621198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Coote B, Attree S, Hiscox JA. Evaluation of a nucleoprotein-based enzyme-linked immunosorbent assay for the detection of antibodies against infectious bronchitis virus. Avian Pathol. 2003;32:519–526. doi: 10.1080/0307945031000154125. [DOI] [PubMed] [Google Scholar]
- De Wit JJ. Detection of infectious bronchitis virus. Avian Pathol. 2000;29:71–93. doi: 10.1080/03079450094108. [DOI] [PubMed] [Google Scholar]
- Dechamma HJ, Dige V, Kumar CA, Singh RP, Jagadish M, Kumar S. Identification of T-helper and linear B epitope in the hypervariable region of nucleocapsid protein of PPRV and its use in the development of specific antibodies to detect viral antigen. Vet Microbiol. 2006;118:201–211. doi: 10.1016/j.vetmic.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Ding MD, Wang HN, Cao HP, Fan WQ, Ma BC, Xu PW, Zhang AY, Yang X. Development of a multi-epitope antigen of S protein-based ELISA for antibodies detection against infectious bronchitis virus. Biosci Biotechnol Biochem. 2015;79:1287–1295. doi: 10.1080/09168451.2015.1025692. [DOI] [PubMed] [Google Scholar]
- Dubois ME, Hammarlund E, Slifka MK. Optimization of peptide-based ELISA for serological diagnostics: a retrospective study of human monkeypox infection. Vector Borne Zoo Dis. 2012;12(5):400–409. doi: 10.1089/vbz.2011.0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger PF, Pepe MS, Dummer LA, Magalhaes CG, de Castro CC, Hubner SO, et al. Combined use of ELISA and Western blot with recombinant N-protein is a powerful tool for the immunodiagnosis of avian infectious bronchitis. Virol J. 2018;15(1):18. doi: 10.1186/s12985-018-1096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geourjon C, Deleage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics. 1995;11(6):681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- Gießauf A, Letschka T, Walder G, Dierich MP, Würzner R. A synthetic peptide ELISA for the screening of rubella virus neutralizing antibodies in order to ascertain immunity. J Immunol Methods. 2004;287(1–2):1–1. doi: 10.1016/j.jim.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Gomara MJ, Haro I. Synthetic peptides for the immunodiagnosis of human diseases. Curr Med Chem. 2007;14(5):531–546. doi: 10.2174/092986707780059698. [DOI] [PubMed] [Google Scholar]
- Gomara MJ, Fernández L, Pérez T, Ercilla G, Haro I. Assessment of synthetic chimeric multiple antigenic peptides for diagnosis of GB virus C infection. Anal Biochem. 2010;396(1):51–58. doi: 10.1016/j.ab.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignjatovic J, Sapats S. Identification of previously unknown antigenic epitopes on the S and N proteins of avian infectious bronchitis virus. Arch Virol. 2005;150:1813–1831. doi: 10.1007/s00705-005-0541-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood MW, Hilt DA. Production and immunogenicity of multiple antigenic peptide (MAP) constructs derived from the S1 glycoprotein of infectious bronchitis virus (IBV) Adv Exp Med Biol. 1995;380:213–219. doi: 10.1007/978-1-4615-1899-0_35. [DOI] [PubMed] [Google Scholar]
- Joshi VG, Singh AK, Kantaraj C, Bais MV, Tiwari AK, Kumar S. Conformational analysis of Infectious Bursal Disease Virus (IBDV) derived cell penetrating peptide (CPP) analogs. Vet World. 2013 doi: 10.5455/vetworld.2013.307-312. [DOI] [Google Scholar]
- Joshi VG, Dighe VD, Thakuria D, Malik YPS, Kumar S. Multiple antigenic peptide (MAP): a synthetic peptide dendrimer for diagnostic, antiviral and vaccine strategies for emerging and re-emerging viral diseases. Indian J Virol. 2013;24(3):312–320. doi: 10.1007/s13337-013-0162-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P, Pau CP. Comparing tandem repeats and multiple antigenic peptides as the antigens to detect antibodies by enzyme immunoassay. J Immunol Methods. 2001;257(1–2):51–54. doi: 10.1016/S0022-1759(01)00444-6. [DOI] [PubMed] [Google Scholar]
- Kowalczyk W, Monso M, de la Torre BG, Andreu D. Synthesis of multiple antigenic peptides (MAPs)-strategies and limitations. J Pept Sci. 2011;17(4):247–251. doi: 10.1002/psc.1310. [DOI] [PubMed] [Google Scholar]
- Kumar N, Malik YPS, Kumar S, Sharma K, Sircar S, Saurabh S, Gulati BR, Singh N, Singh AK, Joshi VG, Banyai K, Dhama K. Peptide-recombinant VP6 protein-based enzyme immunoassay for detection of group A rotavirus in multiple host species. PLoS ONE. 2016;11(7):89. doi: 10.1371/journal.pone.0159027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Yadav K, Kumar R, Chaudhary N, Kumar S. Glycoprotein D peptide-based diagnostic approach for the detection of avian infectious laryngotracheitis antibodies. Avian Pathol. 2019;48:602–609. doi: 10.1080/03079457.2019.1631444. [DOI] [PubMed] [Google Scholar]
- Kwon HM, Jackwood MW, Gelb J. Differentiation of infectious bronchitis virus serotypes using polymerase chain reaction and restriction fragment length polymorphism analysis. Avian Dis. 1993;37:194–202. doi: 10.2307/1591474. [DOI] [PubMed] [Google Scholar]
- Lam LL, Pau CP, Dollard SC, Pellett PE, Spira TJ. Highly sensitive assay for human herpesvirus 8 antibodies that uses a multiple antigenic peptide derived from open reading frame K8. 1. J Clin Microbiol. 2002;40(2):325–329. doi: 10.1128/JCM.40.02.325-329.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen JE, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;24(2):2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Shia T, Suna D, Moa K, Yana Y, Jina Y, Liao ZJ. Development and application of nsp5-ELISA for the detection of antibody to infectious bronchitis virus. J Virol Methods. 2017;243:182–189. doi: 10.1016/j.jviromet.2017.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu X, Zhu Y, Zhou X, Cao C, Hu X, Ding JB. Bioinformatic prediction of epitopes in the Emy162 antigen of Echinococcus multilocularis. Exp Ther Med. 2013;6(2):98. doi: 10.3892/etm.2013.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KH, Lin CF, Chiou SS, Hsu AP, Lee MS, Chang CC, Chang TJ, Shien JH, Hsu WL. Application of purified recombinant antigenic spike fragments to the diagnosis of avian infectious bronchitis virus infection. Appl Microbiol Biotechnol. 2012;95:233–242. doi: 10.1007/s00253-012-4143-8. [DOI] [PubMed] [Google Scholar]
- Lugovskaya NN, Scherbakov AV, Yakovleva AS, Tsyvanyuk MA, Mudrak NS, Drygin VV, Borisov AV. Detection of antibodies to avian infectious bronchitis virus by a recombinant nucleocapsid protein-based enzyme-linked immunosorbent assay. J Virol Methods. 2006;135:292–296. doi: 10.1016/j.jviromet.2006.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moneim ASA, Giesow K, Keil GM. High-level protein expression following single and dual gene cloning of infectious bronchitis virus N and S genes using baculovirus systems. Viral Immunol. 2014;72(2):75–81. doi: 10.1089/vim.2013.0114. [DOI] [PubMed] [Google Scholar]
- Nardin EH, Oliveira GA, Calvo-Calle JM, Nussenzweig RS. The use of multiple antigen peptides in the analysis and induction of protective immune responses against infectious diseases. Adv Immunol. 1995;60:105. doi: 10.1016/S0065-2776(08)60585-4. [DOI] [PubMed] [Google Scholar]
- Nguyen TT, Kwon HJ, Kim IH, Hong SM, Seong WJ, Jang JW, Kim JH. Multiplex nested RT-PCR for detecting avian influenza virus, infectious bronchitis virus and Newcastle disease virus. J Virol Methods. 2013;188:41–46. doi: 10.1016/j.jviromet.2012.12.004. [DOI] [PubMed] [Google Scholar]
- OIE . Avian infectious bronchitis. Manual of standards diagnostic tests and vaccine. Paris: Office International des Epizooties; 2008. pp. 700–710. [Google Scholar]
- Patel BH, Bhimani MP, Bhanderi BB, Jhala MK. Isolation and molecular characterization of nephropathic infectious bronchitis virus isolates of Gujarat state, India. Virus Dis. 2015;26(1–2):42–47. doi: 10.1007/s13337-015-0248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocnakova L, Bhide M, Pulzova LB. An introduction to B-cell epitope mapping and in silico epitope prediction. J Immunol Res. 2016 doi: 10.1155/2016/6760830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan SK, Kamble NM, Pillai AS, Gaikwad SS, Khulape SA, Reddy MR, Mohan CM, Kataria JM, Dey S. Recombinant nucleocapsid protein based single serum dilution ELISA for the detection of antibodies to infectious bronchitis virus in poultry. J Virol Methods. 2014;209:1–6. doi: 10.1016/j.jviromet.2014.08.015. [DOI] [PubMed] [Google Scholar]
- Rai R, Dubey S, Santosh KV, Biswas A, Mehrotra V, Rao DN. Design and synthesis of multiple antigenic peptides and their application for dengue diagnosis. Biologicals. 2017;49:81–85. doi: 10.1016/j.biologicals.2017.08.005. [DOI] [PubMed] [Google Scholar]
- Sarvanan P, Kumar S, Kataria JM. Use of multiple antigenic peptides related to antigenic determinants of infectious bursal disease virus (IBDV) for detection of anti-IBDV-specific antibody in ELISA-quantitative comparison with native antigen for their use in serodiagnosis. J Immunol Methods. 2004;293:61–70. doi: 10.1016/j.jim.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Spencer KA, Osorio FA, Hiscox JA. Recombinant viral proteins for use in diagnostic ELISAs to detect virus infection. Vaccine. 2007;25:5653–5659. doi: 10.1016/j.vaccine.2007.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi V, Singh SD, Dhama K, Gowthaman V, Barathidasan R, Sukumar K. Isolation and molecular characterization of infectious bronchitis virus from recent outbreaks in broiler flocks reveals emergence of novel strain in India. Trop Anim Health Prod. 2012;44(7):1791–1795. doi: 10.1007/s11250-012-0140-2. [DOI] [PubMed] [Google Scholar]
- Tam JP. Recent advances in multiple antigen peptides. J Immunol Methods. 1996;196:17. doi: 10.1016/0022-1759(96)00066-X. [DOI] [PubMed] [Google Scholar]
- Tam JP, Zavala F. Multiple antigen peptide: a novel approach to increase detection sensitivity of synthetic peptides in solid-phase immunoassays. J Immunol Methods. 1989;124:53. doi: 10.1016/0022-1759(89)90185-3. [DOI] [PubMed] [Google Scholar]
- Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N. The immune epitope database 2.0. Nucleic Acids Res. 2010;38:D854–D862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Parr RL, King DJ, Collisson EW. A highly conserved epitope on the spike protein of infectious bronchitis virus. Arch Virol. 1995;140:2201–2213. doi: 10.1007/BF01323240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CH, Hong CC, Seak JC. An ELISA for antibodies against infectious bronchitis virus using an S1 spike polypeptide. Vet Microbiol. 2002;85:333–342. doi: 10.1016/S0378-1135(01)00525-9. [DOI] [PubMed] [Google Scholar]
- Zang DY, Zhou JY, Fang J, Hu JQ, Wu JX, Mu AX. An ELISA for antibodies to infectious bronchitis virus based on nucleocapsid protein produced in Escherichia coli. Vet Med. 2005;50:336–344. doi: 10.17221/5632-VETMED. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.