Abstract
The pharmacological targeting of polyamine metabolism is currently under the spotlight for its potential in the prevention and treatment of several age-associated disorders. Here, we report the finding that triethylenetetramine dihydrochloride (TETA), a copper-chelator agent that can be safely administered to patients for the long-term treatment of Wilson disease, exerts therapeutic benefits in animals challenged with hypercaloric dietary regimens. TETA reduced obesity induced by high-fat diet, excessive sucrose intake, or leptin deficiency, as it reduced glucose intolerance and hepatosteatosis, but induced autophagy. Mechanistically, these effects did not involve the depletion of copper from plasma or internal organs. Rather, the TETA effects relied on the activation of an energy-consuming polyamine catabolism, secondary to the stabilization of spermidine/spermine N1-acetyltransferase-1 (SAT1) by TETA, resulting in enhanced enzymatic activity of SAT. All the positive effects of TETA on high-fat diet-induced metabolic syndrome were lost in SAT1-deficient mice. Altogether, these results suggest novel health-promoting effects of TETA that might be taken advantage of for the prevention or treatment of obesity.
Subject terms: Enzyme mechanisms, Metabolic pathways
Introduction
Like spermidine (IUPAC name: N-(3-aminopropyl)-1,4-butanediamine), triethylenetetramine (TETA, IUPAC name: N,N′-Bis(2-aminoethyl)-1,2-ethanediamine) is a polyamine. Spermidine is a natural compound produced by a biosynthetic pathway present in human cells, but is also produced by the intestinal microbiota and contained in multiple food items [1], while TETA is a fully synthetic product. Spermidine supplementation has the remarkable capacity to extend the lifespan of model organisms (yeast, nematodes, flies, and mice) [2, 3], and a spermidine-rich diet has been linked to reduced mortality in epidemiological studies, in two independent human cohorts from Austria and Northern Italy [4]. The longevity-extending and cardioprotective effects of spermidine have been attributed to its capacity to induce autophagy [5], since they are lost in nonmammalian model organisms lacking essential autophagy genes [3], as well as in autophagy-deficient mice lacking the microtubule-associated protein 1S (Map1s) gene [6]. Similarly, the deficiency of the autophagy-related gene 5 (Atg5) abrogated the cardioprotective effects of spermidine in mice [2]. Mechanistically, the capacity of spermidine to induce autophagy has been attributed to several, possibly tissue-specific effects, namely, (i) the inhibition of acetyltransferase EP300 activity [7] and (ii) the increase in eukaryotic translation initiation factor 5A-1 (eIF5A) hypusination, resulting in the activation of the pro-autophagic transcription factor EB (TFEB) [8].
Beyond its longevity-extending effects, spermidine has rather broad health-promoting effects. Thus, it improves diastolic function of the aging heart, prevents heart failure driven by salt-induced hypertension [2], improves the efficacy of anticancer immunotherapies [9, 10], increases cognition [11], halts inflammation [12], reduces liver fibrosis and nonalcoholic fatty liver disease (NAFLD) [6, 13], and prevents high-fat diet (HFD)-induced obesity and diabetes [14]. Many of these effects depend on the induction of autophagy [5, 6, 9, 10, 14].
TETA is currently used for the treatment of Wilson disease, in patients that have developed adverse effects against the first-line drug d-penicillamine [15, 16]. Wilson disease is caused by the mutation of adenosine triphosphatase copper-transporting β gene (ATP7B), a copper-extruding P-type ATPase that normally avoids excessive copper accumulation in hepatocytes and other cell types, leading to progressive liver failure, typically with an adult onset [17]. TETA is considered as a copper chelator that prevents the intestinal absorption and favors the urinary excretion of excessive copper [18]. TETA can be administered for decades for the long-term treatment of Wilson disease with minor side effects [15, 19]. Interestingly, TETA is rapidly metabolized by two acetyltransferases, thialysine Nε-acetyltransferase and spermidine/spermine N1-acetyltransferase-1 (SAT1) [20, 21], the latter being the rate-controlling enzyme of polyamine catabolism.
Given the structural similarities between spermidine and TETA, as well as the fact that both polyamines are metabolized by the same enzyme, SAT1, we addressed the question whether TETA might be used to induce autophagy, and/or to improve the course of aging or age-related diseases. Here, we report the unexpected finding that TETA can stimulate the activity of SAT1 in vivo, thereby setting a biochemical cascade that protects against obesity and diabetes in mouse models.
Experimental model and subject details
Mouse experiments and tissue processing
Wild-type C57BL/6 (Envigo and Janvier Laboratories, France), ob/ob mice (Charles River laboratories, France), Atg4b−/− C57BL/6 mice (gift of Dr. Carlos Lopez-Otin, University of Oviedo, Spain), and Sat1−/− mice (gift from Prof. Leena Alhonen, University of Eastern Finland, generated as described earlier [22] and bred in Cordeliers Research Center), were bred and maintained according to the FELASA guidelines and local guidelines from the Animal Experimental Ethics Committee (#8216-2016121516216070v3, #2315-2015101617138161v1, #5333-2016050509281672v3, #5272-2016042112271931v2), and approved by the Austrian Government (BMWFW-66.007/0029-WF/V/3b/2017) for lifespan analysis of TETA-supplemented mice. Mice were housed in specific pathogen-free conditions and in a temperature-controlled environment with 12-h light/dark cycles and received chow diet (R04, Safe, Augy, France) or HFD (260HF, Safe) and water ad libitum. Starting from 7 weeks of age, mice (from the reported genetic strain) were divided into several groups by simple randomization and fed with [1] chow diet [2], TETA (triethylenetetramine dihydrochloride, Sigma Aldrich #T5033-25G), 3% weight/volume in the drinking water ad libitum [3], HFD (20% protein, 36% lipids, and 36.7% carbohydrate) [4], HFD and TETA [5], chow diet + 30% sucrose in the drinking water ad libitum [6], and chow diet + TETA (daily intraperitoneal injection, i.p., 100 mg/kg) + 30% sucrose in the drinking water ad libitum. In accordance with ARRIVE guidelines, experiments with mice were conducted with a minimum number of animals. Sample sizes were selected based on prior experience of the laboratory in metabolic studies. A minimum of three biological replicates was used for each experiment. Experiments were not conducted in blind conditions. Mice were weighed once a week, to monitor their weight gain, and subjected to metabolic tests around the fifth/sixth week of treatment. Then, based on the different settings, animals were sacrificed after 2–20 weeks of treatment by cervical dislocation, and organs collected and processed. For the lifespan analysis, 18-month-old C57BL/6JRj mice (Janvier Laboratories) were divided into two groups, housed in a specific pathogen-free condition with 14-/10-h light/dark cycles and cage enrichment as well as group sizes as published in ref. [2], and respectively treated, or not, with TETA-free base (Sigma Aldrich #90460), at 13.7 mM final concentration in the drinking water ad libitum (late-in-life feeding), and their lifespan was monitored until 26 months of age according to published strategies [2]. In this experiment, TETA-free base was used at a concentration of 13.7 mM in drinking water, which equals 3000 ppm referred to TETA*2HCl (TETA dihydrochloride). TETA-free base-supplemented drinking water was freshly prepared every 3–4 days from a 1 M aqueous stock solution titrated with HCl to pH 7.2–7.3 (similar to spermidine stock solutions according to ref. [3]). For the short-term autophagy induction studies, mice were treated with TETA (i.p., 100 mg/kg); 2 h before sacrifice, mice were injected with leupeptin (i.p., 15 mg/kg), and 8 h after TETA administration, mice were sacrificed. Tissues were snap-frozen in liquid nitrogen after extraction, and homogenized in two cycles for 20 s at 5500 rpm using a Precellys 24 tissue homogenator (Bertin Technologies, Montigny-le-Bretonneux, France) in 20 mM Tris buffer (pH 7.4) containing 150 mM NaCl, 1% Triton X-100, 10 mM EDTA, and Complete® protease inhibitor cocktail (Roche Applied Science). Tissue extracts were centrifuged at 12,000 g at 4 °C and supernatants were collected. Protein concentration in the supernatants was evaluated by the bicinchoninic acid technique (#23225, BCA protein assay kit). Then samples were subjected to western blot analysis. For acetyl-lysine staining, mice were sacrificed, and the tissues collected and immediately fixed in 4% paraformaldehyde phosphate-buffered saline (PBS) solution. Fixed tissues were then kept for 24 h in 15% sucrose solution, and subsequently moved to a 30% sucrose solution for another 24 h. Tissues were embedded with tissue-TEK OCT compound (Sakura Fine Technical Co., Ltd) and frozen at −80 °C. In total, 6-μm-thick sections were obtained from the frozen tissue samples by using a cryostat (CM3050S, Leica); it was cut into four slices of tissue for each sample. The slices were then subjected to the immunohistochemistry assay.
Immunohistochemistry
Nonspecific binding sites were blocked with 5% bovine serum albumin in PBS, followed by incubation with primary antibodies, 1:200 in BSA 2% PBS solution (Acetylated-Lysine antibody, Cell Signaling Technology, 9441S), overnight at 4 °C. Later, the sections were incubated with appropriate Alexa Fluor 568 anti-mouse (Invitrogen, A11031), 1:300 in BSA 2% PBS solution (Molecular Probes-Invitrogen, Eugene, OR, USA). Finally, the slices were mounted with 4′,6-diamidin-2-fenilindolo (DAPI) Fluoromount-G (SouthernBiotech, 0100-20) and viewed under Axio Observer inverted fluorescence microscope (Carl Zeiss). Images were acquired using an ImageXpress Micro Confocal microscope (Molecular devices, Sunnyvale, CA, USA) using a CFI PlanApo 10× objective (Leica, Wetzlar, Germany) and built-in DAPI and CY3 filters. For each of the three slices from each organ, the adequate number of viewfields was automatically selected for quantification, when at least 65% of the image was presenting tissue without major fold. Nuclei were segmented using the EBImage package from R based on DAPI staining, defining the region of interest (ROI) for acetylation-intensity measurement. After removing artifacts based on nuclear intensity and nuclear area, the median pixel-intensity value outside of the ROI was then subtracted to assess values for background correction. The median value of all corrected nuclear intensities was finally computed for each mouse. At rare occasions when technical issues with staining occurred, slides were excluded from IHC analysis.
Immunoblotting
For immunoblotting, proteins extracts obtained by tissues lysis in radioimmunoprecipitation assay buffer, were separated on 4–12% Bis-Tris acrylamide precast gels (Invitrogen) and electrotransferred to 0.2 μM polyvinylidene fluoride membranes (#1620177, Bio-Rad). Nonspecific binding sites were saturated by incubating membranes for 1 h in 0.05% Tween 20 (#P9416, Sigma Aldrich) v–v in Tris-buffered saline (TBS) (#ET220, Euromedex) supplemented with 5% nonfat powdered milk (w:v in TBS), followed by an overnight incubation with primary antibodies specific for LC3B (#2775 Cell Signaling Technology). Membranes were cut in order to allow simultaneous detection of different molecular weight proteins. Equal protein loading was monitored by probing membranes with actin-specific antibody (anti-beta Actin antibody [AC-15] (HRP), Abcam, ab49900). Membranes were developed with suitable horseradish peroxidase conjugates followed by chemiluminescence-based detection with the Amersham ECL Prime (#RPN2232, GE Healthcare) and the ImageQuant LAS 4000 software-assisted imager (GE Healthcare, Piscataway, NJ, USA). Quantification was performed by densitometry by means of ImageJ software (National Institutes of Health, Bethesda, Maryland, USA). Autophagy was quantified through evaluation of LC3-II/actin ratio according to ref. [23].
Histological analysis
After treatment, mice were sacrificed by cervical dislocation followed by immediate fixation of the liver tissue and the visceral fat in 4% paraformaldehyde PBS solution. Fixed samples were embedded in paraffin, and 3-µm-thick sections were stained with hematoxylin–eosin–safranin (H&E). Each slide was examined using a Zeiss Axiophot microscope. Histological slides were acquired with a Virtual Slide microscope VS120-SL (Olympus, Tokyo, Japan), 20× air objective (0.75 NA). An in-house algorithm was developed with ImageJ, in order to quantify the hepatic steatosis damage. The method calculates the proportion of steatosis in each sample by computing both the whole surface of the hepatic parenchyma and that occupied by the adipocytes. Tissue detection was performed at low resolution (image decimated four times) on the green component. The “Percentile” automatic segmentation was used followed by mathematical morphology operators (closing and then geodesic opening of size 3) in order to remove the artifacts. Calculation of the surface occupied by the adipocytes was carried out in two stages. Since the full-resolution image was large, it was processed in parts. It was therefore necessary to calculate initially an optimal threshold for the adipocyte segmentation, and then to apply it to each selected area of the original image. The calculation of the adipocytes’ optimal segmentation threshold was carried out on a selected area measuring 2000 × 2000 pixels, of the image at full resolution, in the center of the sample. H&E color deconvolution was computed on this detail image, and the threshold parameters of the automatic thresholding “Triangle dark” applied to the second color component were stored. To process the whole image, H&E color deconvolution was applied to each part of the original image, and then the previously computed threshold parameters were applied to the second color component. Small artifacts were then removed by a size 4 geodesic opening. Coalescent adipocytes were separated by means of a watershed. Finally, only adipocytes with a size in-between ~6 and 400 μm2 were taken into account. The size of adipocytes present within the white adipose tissue was determined with ImageJ at low resolution. In order to visualize adipocytes’ wall, an automatic “Triangle” thresholding of each color component was performed. To prevent artifacts, a Gaussian blurring of size 0.5 of each color component was applied prior to the thresholding. The result of each thresholding was then combined (logical OR) to obtain the adipocyte wall image. Large artifacts (extrahepatic tissues) were removed using mathematical morphology operators and size filter. Small artifacts (such as fragmented adipocyte walls) were removed too by size filtering. For correct cell size analysis, apparently large cells resulting from the coalescence of several cells with broken walls have to be discarded. Therefore, a size filter cannot be used because “normal” large adipocytes may have the same size as a cluster of adipocytes with broken membranes. Watershed was used to characterize the gathered cells. Since these gathered cells usually keep remains of walls, watershed should split them. So only cells that had not been split by watershed were selected and taken into account for the size analysis. Finally, to avoid artifacts, only adipocytes with a size in the range of 1500–30,000 µm2 were selected. For the analysis of the adipocyte size distribution a Kolmogorov–Smirnov test was performed as the following: after calculating the D statistic by applying the empirical distribution function to compared groups, the associated p value was computed by using the R pkolmim package. At rare occasions when technical issues with staining occurred, slides were excluded from histological analysis.
Cytokine detection in plasma
Plasma was harvested from blood collection tubes by centrifugation at 15,000 rpm for 30 min, and stored at −80 °C until use. Leptin (EZML-82K), ghrelin (EZRGRT-91), C-peptide and GIP (MMHMAG-44K), insulin (MMHMAG-44K, MADKMAG-71K), plasminogen activator inhibitor-1 (PAI-1) Total and resistin (MADKMAG-71K), insulin growth factor-binding protein (IGFBP-1), IGFBP-2, IGFBP-3, IGFBP-6, and IGFBP-7 (MIGFBPMAG-43K), KC, eotaxin, G-CSF, IP-10, IL-1a and LIX (MCYTOMAG-70K), LH, FSH, TSH, ACTH and GH (MPTMAG-49K), and adiponectin (MADPNMAG-70K-01) and IL-22 (MTH17MAG-47K) levels were measured using a mouse serum adipokine immunoassay kit (Cytokine multiplexing, Luminex assay), following the protocol provided by the manufacturers (Merck, Darmstadt, Germany, and EMD Millipore, Temecula, CA, USA). The control, chow diet-fed group, of this experiment, is shared with the experiment reported in ref. [24], in Fig. S5.
Tolerance tests
Mice were fasted for 6 h before glucose-tolerance (GTT) and insulin-resistance (ITT) tests. In GTT, non-anesthetized mice were injected i.p. glucose (2 g/kg, Sigma Aldrich, G8270). In the ITT, mice were injected i.p. 0.75 U/kg of insulin (Lilly, HI0210), prepared at 0.1 U/ml in advance. In both GTT and ITT, fasting plasma glucose levels were determined at time 0, and then after injection of glucose or insulin. Glucose levels were determined by a glucometer (Accu-Chek Performa) in blood from the tail vein, at specified time points.
Analysis of whole-body composition
Noninvasive determination of lean tissue mass, fat mass, and free fluid was performed on non-anesthetized mice using Time Domain-NMR technology (minispec LF90II, Bruker BioSpin) in Centre de Recherche des Cordeliers. In other experiments, magnetic resonance imaging (MRI) was performed with a dedicated small-animal 4.7 Tesla MR system (Biospec 47/40 USR Bruker) using a quadrature transmit/receive body coil with a 7-cm inner diameter (Plateforme Imageries du Vivant, INSERM UMR 970). Mice were anesthetized with air and isoflurane (4% for induction and 1% during MRI). Experiments were performed with respiratory gating to avoid movement artifacts. We used a spin-echo 3D sequence of the entire mouse with the following parameters: TR/TE = 750/65 ms and 260 mm of resolution in the three dimensions, to bring out the signal of the fat of the mice.
Food-intake analysis
Mice food intake was analyzed using metabolic cages. Mice were individually housed and acclimated to the metabolic cages for 24 h before experimental measurements. Subsequently, the food consumption was measured daily for 5 days. Mice body weight has been measured before, during, and after the experiments.
Metabolomics analysis
Sample preparation tissue
About 30 mg of biological material for each condition was first weighted and solubilized into 1.5-mL polypropylene microcentrifuge tubes with ceramic beads with 1 mL of cold lysate buffer (MeOH/water/chloroform with stable isotope-labeled internal standard ISTD, 9/1/1, −20 °C). They were then homogenized three times for 20 s at 5500 rpm using Precellys 24 tissue homogenator (Bertin Technologies, Montigny-le-Bretonneux, France), followed by a centrifugation (10 min at 15000 g, 4 °C). Then, the upper phase of the supernatant was split into two parts: the first 270 µL was used for the gas chromatography coupled by mass spectrometry (GC/MS) experiment, the other 250 µL was used for the ultra-high-pressure liquid chromatography coupled by mass spectrometry (UHPLC/MS) experimentations. Concerning the GC–MS aliquots, samples were evaporated, and 50 µL of methoxyamine (20 mg/mL in pyridine) was added on dried extracts, then stored at room temperature in the dark, during 16 h. The day after, 80 µL of N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) was added, and the final derivatization occurred at 40 °C during 30 min. Samples were then directly injected into GC–MS. Concerning the LC–MS aliquots, the collected supernatant was evaporated at 40 °C in a pneumatically assisted concentrator (Techne DB3, Staffordshire, UK). The LC–MS dried extracts were solubilized with 450 µL of Milli-Q water. Samples were aliquoted (100 µL) for LC methods and backup. Biological samples and QC aliquots were kept at −80 °C until injection, or transferred in vials for direct analysis by UHPLC/MS. Concerning the rest of the supernatant and the pellet, 340 µl of methanol with 2% of sulfosalicylic acid (SSA) was added before vortexing and centrifugation (10 min at 15,000 g, 4 °C). In all, 500 µl of the supernatant was transferred in a microtube and evaporated. The dried sample was spiked with 200 µl of Milli-Q water before injection in UHPLC/MS of the polyamine’s method.
Sample preparation plasma (lithium heparin)
A volume of 50 µL of plasma was mixed with 500 µL of a cold solvent mixture (MeOH/water/chloroform with stable isotope-labeled internal standard ISTD, 9/1/1, −20 °C), into 1.5-mL Eppendorf tubes, vortexed, and centrifuged (10 min at 15,000 g, 4 °C). Then, the upper phase of the supernatant was split in two parts: the first 220 µL was used for the GC coupled by MS experiment, the other 200 µL was used for the UHPLC coupled by MS experimentations. Concerning the GC–MS aliquots, samples were evaporated, and 50 µL of methoxyamine (20 mg/mL in pyridine) was added on dried extracts, and then stored at room temperature in the dark, for 16 h. The day after, 80 µL of N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) was added, and the final derivatization occurred at 40 °C for 30 min. Samples were then directly injected into GC–MS. Concerning the LC–MS aliquots, the collected supernatant was evaporated at 40 °C in a pneumatically assisted concentrator (Techne DB3, Staffordshire, UK). The LC–MS dried extracts are solubilized with 400 µL of Milli-Q water. Samples are aliquoted (100 µL) for LC methods and backup. Biological samples and QC aliquots were kept at −80 °C until injection, or transferred in vials for direct analysis by UHPLC/MS. Concerning the rest of the supernatant and the pellet, 285 µl of methanol with 2% of SSA was added before vortexing and centrifugation (10 min at 15,000 g, 4 °C). In all, 500 µl of the supernatant was transferred into a microtube and evaporated. The dried sample was spiked with 200 µl of Milli-Q water before injection in UHPLC/MS of the polyamine’s method.
Sample preparation polyamine-labeling experiment
Tissues were solubilized into 1.5-mL polypropylene microcentrifuge tubes with ceramic beads with 1 mL of cold lysate buffer (MeOH with 1% SSA, without internal standards, −20 °C) to be homogenized and centrifugated (10 min at 15,000 g, 4 °C). Regarding plasma, 500 µl was added with the same extraction buffer, and samples were then vortexed and centrifuged. Respectively, 600 µl and 300 µl of the supernatant were transferred and evaporated. The dried sample was spiked with 300 µl of Milli-Q water before injection in UHPLC/MS of the polyamine’s method.
Targeted analysis of CoAs and nucleoside phosphates by ion-pairing UHPLC coupled to a triple-quadrupole (QQQ) mass spectrometer
Targeted analysis was performed on a RRLC 1260 system (Agilent Technologies, Waldbronn, Germany) coupled to a Triple Quadrupole 6410 (Agilent Technologies) equipped with an electrospray source operating in positive mode. The gas temperature was set to 350 °C with a gas flow of 12 L/min. The capillary voltage was set to 3.5 kV. About 10 μL of sample was injected on a Column XDB-C18 (100 mm × 2.1 mm particle size, 1.8 µm) from Agilent Technologies, protected by a guard column XDB-C18 (5 mm × 2.1 mm particle size, 1.8 μm), and heated at 40 °C by a Pelletier oven. The gradient mobile phase consists of water with 2 mM of dibutylamine ammonium acetate (DBAA) (A) and acetonitrile (B). The flow rate was set to 0.2 mL/min, and gradient as follows: the initial condition is 90% phase A and 10% phase B, maintained during 4 min. Molecules were then eluted using a gradient from 10% to 95% phase B over 3 min. The column was washed using 95% mobile phase B for 3 min and equilibrated using 10% mobile phase B for 3 min. The autosampler was kept at 4 °C. The collision gas was nitrogen. The scan mode used was the MRM for biological samples. Peak detection and integration of the analytes were performed using the Agilent MassHunter quantitative software (B.07.01).
Widely targeted analysis of intracellular metabolite GC coupled to a QQQ mass spectrometer
The GC–MS/MS method was performed on a 7890B gas chromatography coupled to a triple quadrupole 7000 C (Agilent Technologies, Waldbronn, Germany) equipped with a high-sensitivity electronic impact source (EI) operating in positive mode. The front inlet temperature is 250 °C; the injection was performed in splitless mode. The transfer line and the ion-source temperature were 250 °C and 230 °C, respectively. The septum purge flow was fixed at 3 mL/min, the purge flow to split vent operated at 80 mL/min during 1 min, and gas saver mode was set to 15 mL/min after 5 min. The helium gas flows through the column (J&W Scientific HP-5MS, 30 m × 0.25 mm, i.d. 0.25 mm, 25 µm d.f., Agilent Technologies Inc.) at 1 mL/min. Column temperature was held at 60 °C for 1 min, then raises to 210 °C (10 °C/min), followed by a step to 230 °C (5 °C/min), and reached 325 °C (15 °C/min), to be held at this temperature for 5 min. The collision gas was nitrogen. The scan mode used was the MRM for biological samples. Peak detection and integration of the analytes were performed using the Agilent MassHunter quantitative software (B.07.01).
Targeted analysis of bile acids by UHPLC coupled to a QQQ mass spectrometer
Targeted analysis was performed on a RRLC 1260 system (Agilent Technologies, Waldbronn, Germany) coupled to a Triple Quadrupole 6410 (Agilent Technologies) equipped with an electrospray source operating in positive mode. The gas temperature was set to 325 °C with a gas flow of 12 L/min. The capillary voltage was set to 4.5 kV. Ten microliters of sample were injected on a Column Poroshell 120 EC-C8 (100 mm × 2.1 mm particle size, 2.7 µm) from Agilent Technologies, protected by a guard column XDB-C18 (5 mm × 2.1 mm particle size, 1.8 μm), and heated at 40 °C by a pelletier oven. The gradient mobile phase consisted of water with 0.2% of formic acid (A) and acetonitrile/isopropanol (1/1, v/v) (B) freshly made. The flow rate was set to 0.3 mL/min, and gradient as follows: the initial condition is 70% phase A and 30% phase B, maintained during 1.5 min. Molecules were then eluted using a gradient from 30% to 60% phase B over 9 min. The column was washed using 98% mobile phase B for 2 min, and equilibrated using 30% mobile phase B for 2 min. After each injection, the needle was washed twice with isopropanol and thrice with water. The autosampler was kept at 4 °C. The collision gas was nitrogen. The scan mode used was the MRM for biological samples. Peak detection and integration of the analytes were performed using the Agilent MassHunter quantitative software (B.07.01).
Targeted analysis of polyamines by ion-pairing UHPLC coupled to a QQQ mass spectrometer
Targeted analysis was performed on a RRLC 1260 system coupled to a Triple Quadrupole 6410 equipped with an electrospray source operating in positive mode. The gas temperature was set to 350 °C with a gas flow of 12 L/min. The capillary voltage was set to 3.5 kV. Ten microliters of sample were injected on a Column Kinetex C18 (150 mm × 2.1 mm particle size, 2.6 µm) from Phenomenex, protected by a guard column C18 (5 mm × 2.1 mm), and heated at 40 °C by a Pelletier oven. The gradient mobile phase consists of water with 0.1% of heptafluorobutyric acid (HFBA) (A) and acetonitrile with 0.1% of HFBA (B) freshly made. The flow rate was set to 0.2 mL/min, and gradient as follows: the initial condition is 95% phase A and 5% phase B. Molecules were then eluted using a gradient from 5% to 40% phase B over 10 min. The column was washed using 90% mobile phase B for 2.5 min, and equilibrated using 5% mobile phase B for 4 min. The autosampler was kept at 4 °C. The collision gas was nitrogen. The scan mode used was the MRM for biological samples. Peak detection and integration of the analytes were performed using the Agilent MassHunter quantitative software (B.07.01).
The same method was applied to analyze the labeling experiment. Acquisition includes isotopomers, to monitor enrichment of the polyamine pathway. Peak detection and integration of the isotopomers were performed using the Agilent MassHunter quantitative software (B.07.01).
Untargeted analysis of intracellular metabolites by UHPLC coupled to a quadrupole-time-of-flight (QTOF) mass spectrometer
Profiling of intracellular metabolites was performed on a liquid chromatography (LC) 1260 system coupled to a QTOF 6520 equipped with an electrospray source operating in both positive and negative modes, successively, and full-scan mode from 50 to 1000 Da. The gas temperature was set to 350 °C with a gas flow of 12 l/min. The capillary voltage was set to 3.5 kV, the fragmentor to 175 V, and the skimmer to 65 V. Two reference masses were used to maintain the mass accuracy during analysis: m/z 121.050873 and m/z 922.009798 in positive mode, and m/z 112.985587 and m/z 980.016375 in negative mode. Ten microliters of sample were injected on a SB-Aq column (100 mm × 2.1 mm particle size, 1.8 μm) from Agilent Technologies, protected by a guard column XDB-C18 (5 mm × 2.1 mm particle size, 1.8 μm), and heated at 40 °C. The gradient mobile phase consists of water with 0.2% of acetic acid (A) and acetonitrile (B). The flow rate was set to 0.3 mL/min. The initial condition is 98% phase A and 2% phase B. Molecules were then eluted using a gradient from 2% to 95% phase B in 7 min. The column was washed using 95% mobile phase B for 3 min, and equilibrated using 2% mobile phase B for 3 min. The autosampler was kept at 4 °C. Peak detection and integration of the analytes were performed using the Agilent MassHunter quantitative software (B.07.01).
Untargeted analysis of intracellular metabolites by UHPLC coupled to a Q-Exactive mass spectrometer: reversed-phase acetonitrile method
Profiling of intracellular metabolites was performed on a Dionex Ultimate 3000 UHPLC system (Thermo Scientific) coupled to a Q-Exactive (Thermo Scientific) equipped with an electrospray source operating in both positive and negative modes and full-scan mode from 100 to 1200 m/z. The Q-Exactive parameters were sheath gas flow rate 55 au, auxiliary gas flow rate 15 au, spray voltage 3.3 kV, capillary temperature 300 °C, and S-Lens RF level 55 V. About 10 μL of sample were injected on a SB-Aq column (100 mm × 2.1 mm particle size, 1.8 μm) from Agilent Technologies, protected by a guard column XDB-C18 (5 mm × 2.1 mm particle size, 1.8 μm), and heated at 40 °C by a Pelletier oven. The mass spectrometer was calibrated with sodium acetate solution dedicated to low mass calibration. The gradient mobile phase consisted of water with 0.2% of acetic acid (A) and acetonitrile (B). The flow rate was set to 0.3 mL/min. The initial condition is 98% phase A and 2% phase B. Molecules are then eluted using a gradient from 2% to 95% phase B in 22 min. The column was washed using 95% mobile phase B for 2 min, and equilibrated using 2% mobile phase B for 4 min. The autosampler was kept at 4 °C. Peak detection and integration were performed using the Thermo Xcalibur quantitative software (3.1).
Quality control policy
A daily qualification of the instrumentation was set up with automatic tune and calibration processes. These qualifications were completed with double injections of standard mixes, at the beginning and at the end of the run, as for a blank-extracted sample to control the background impurities. Mixtures were adapted for each chromatographic method. After the extraction, the pool of QC sample was used to passivize the column before the analysis with the proper biological matrix, and reinjected during the batch to monitor and correct analytical bias occurring during the batch (m/z, retention time and sensitivity drifts) during post-acquisition treatment signal.
Chemical products
Acetonitrile and Methanol are from HoneyWell. Isopropanol and Acetic acid were from VWR. Chloroform is from Arcos Organics. Formic acid, Methoxyamine hydrochloride, N-Methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA), N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide (MSTBFA), O-Ethylhydroxylamine hydrochloride, Pyridine, DBAA, SSA, and HFBA are from Merck. Detailed references are included in the key resources table, as well as the stable isotopes labeled internal standard references.
Heat-map analyses
The data were first normalized by dividing all the values by the average value for each metabolite. Using R software (R version 3.5.0), it was fitted through the linear models on the normalized data for each metabolite. The output coefficients represent the difference between the average value of one condition and the average value of the control condition. The p values result from two-tailed unpaired Student’s t test applied on coefficients.
Analysis of SAT1 activity
Tissues were homogenized using TissueLyser (30 Hz for 2–4 min, Qiagen) to an ice-cold buffer containing 25 mM Tris-HCl, pH 7.4, 1 mM DTT, 0.1% Triton X-100, and 1× Complete EDTA-free protease inhibitor cocktail (Roche Diagnostics). Lysates were centrifuged at 12,000 g for 30 min at 4 °C, and the supernatant was used for SAT1 activity assays. SAT1 activity was determined by an assay that measures the amount of incorporation of radioactivity from 14C-acetyl-CoA into 8-methylspermidine (8-MeSpd) in 10 min at 37 °C as described previously in ref. [25]. A standard assay mixture contained 100 mM Tris-HCl, pH 7.8, 1 mM semicarbazide, 5 mM EDTA, 3 mM 8-MeSpd [26], and 50 nCi (50–60 mCi/mmol) of 14C-acetyl-CoA (Moravek Inc.) in a total volume of 100 μl. 8-MeSpd has higher affinity for SAT1 in comparison to its natural substrate spermidine (Km 78 ± 3 µM and Vmax of 7.35 ± 0.10 µmol/min/mg vs. Km 151 ± 15 µM and Vmax of 4.28 ± 0.13 µmol/min/mg). Furthermore, 8-MeSpd cannot be enzymatically acetylated at N8-position, thus allowing feasible SAT1-specific activity assay in crude enzyme preparations.
SAT1 protein half-life measurement in liver lysates
Mouse recombinant SAT1 was produced and purified as described earlier [27]. The degradation mixture contained 10 mM Tris-HCl, pH 7.8, 5 mM MgCl2, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 100 μM cycloheximide, 2 mM ATP, 50% liver lysate (a source of proteasomes) from Sat1−/− mice, and 5 ng of mouse recombinant SAT1 in a total volume of 20 μl. After incubation at 37 °C for 0–30 min, SAT1 activity assay was performed using 30 mM spermidine as a substrate. Without added liver lysate, the degradation of SAT1 was only ~97% in 30 min.
Cu chelation
Tissue metal analyses were done as recently reported in ref. [28]. Briefly, around 100 mg of stored frozen heart, liver, or muscle tissue were subjected to wet ashing by incubation in 1 ml of nitric acid and 5–10 min of heating in a microwave. Thereafter, the total volumes were adjusted with distilled water to 8 ml, and metal content was analyzed by Inductively Coupled Plasma Optical Emission Spectrometry (ICP-OES, Ciros Vision, SPECTRO Analytical Instruments GmbH, Kleve, Germany).
Gene expression analysis
Total RNA from murine tissues was isolated by QIAzol (QIAGEN) trituration with Precellys 24 tissue homogenator (Bertin Technologies, Montigny-le-Bretonneux, France), followed by column purification with RNeasy Mini Kit (QIAGEN). Following the manufacturer’s instructions, Superscript III Reverse Transcriptase (Invitrogen) was used with random hexamers (Promega) for generation of cDNA. Quantitative PCR was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems) using TaqMan Gene Expression Master Mix (Applied Biosystems) and the following TaqMan Gene Expression Assays: murine Sat1 (Mm00485911_g1) and the murine Ppia (Mm02342430_g1), purchased by ThermoFisher Scientific.
Statistical analysis
Data are reported as box-and-whisker plots, which show median, first and third quartiles, and maximum and minimum values or as means ± s.e.m., as specified. Circles indicate each mouse used in the experiment, unless otherwise stated in the paper. For statistical analyses, p values were calculated by one-way ANOVA non-corrected or by two-tailed unpaired Student’s t test. Longitudinal statistical comparisons, for mice weight gain, were performed with https://kroemerlab.shinyapps.io/TumGrowth/ [29]; Wald test was used to compute p values, by testing jointly that both slopes and intercepts were the same between the groups of interest. Kolmogorov test was used to compare adipocytes, by comparing their size distributions; acetyl-lysine cell intensities were compared by means of a Wilcoxon test, evaluating sample median differences. Differences were considered statistically significant when p values are *(p < 0.05), **(p < 0.01), ***(p < 0.001).
Results
Metabolic effects of TETA are independent of copper depletion
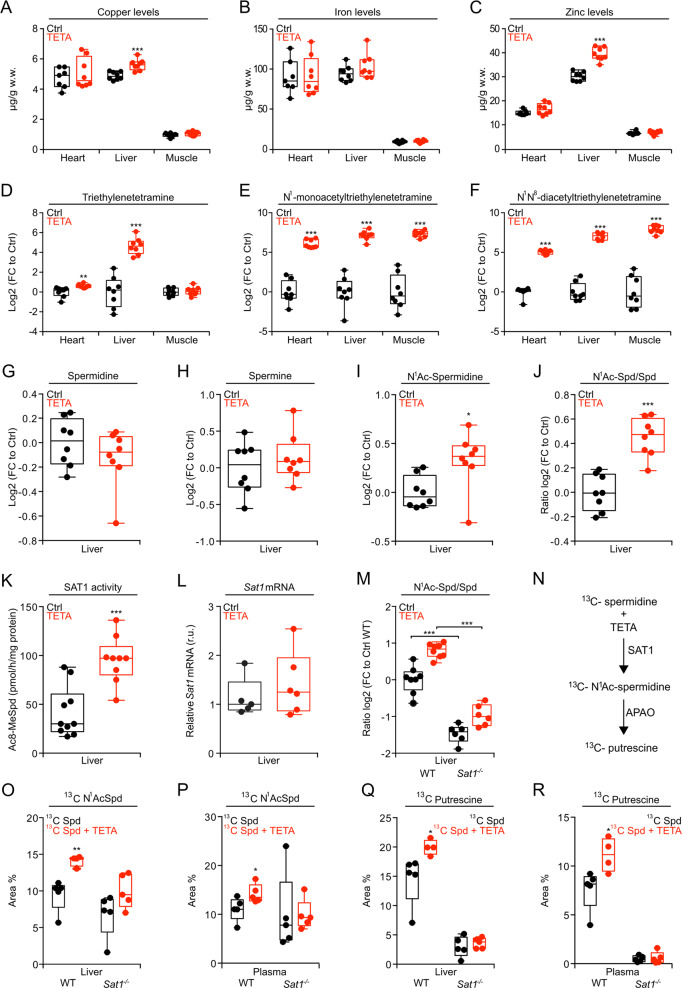
Chronic administration of TETA (3000 ppm in the drinking water) to 18-month-old wild-type (WT) C57Bl/6J:Rj mice (late-in-life feeding), reared under standard conditions, had no major toxic effects, as indicated by the assessment of longevity (Fig. S1a), body weights (Fig. S1b), as well as food and water uptake (Fig. S1c). Of note, at this low dose, and in a non-pathological condition, TETA was unable to deplete heavy metals, including copper (Fig. 1a), iron (Fig. 1b), and zinc (Fig. 1c) in heart, liver, and muscle. In concordance with the available literature, we confirmed that TETA was widely distributed across these organs, as indicated by its mass spectrometric detection (Fig. 1d), as well as that of its principal metabolites, N1-monoacetyltriethylenetetramine (MAT) and N1N10-diacetyltriethylenetetramine (DAT) (Fig. 1e, f) [30]. Commensurate with the structural similarities of the synthetic agent TETA and the natural polyamine spermidine (Fig. S1d), the effects of both compounds on the liver metabolome were largely convergent, though not identical (Fig. S1e, f). The complete list of detected metabolites is reported in Table S1.
Fig. 1. Metabolic effects of TETA on mice.
Wild-type (WT) C57BL/6JOlaHsd male mice fed a chow diet, were treated with TETA (3000 ppm dissolved in drinking water) starting at 7 weeks of age. After 2 weeks of treatment, the organs (heart, liver, and muscle) were collected for the determination of metals (a–c) (n = 7/8 mice/group), TETA, and its mono- or diacetylated metabolites (d–f) (n = 8 mice/group). g–j In addition, in the liver of mice, the endogenous levels of polyamines were measured (n = 8 mice/group). WT male mice, fed a chow diet, were treated with TETA and, after 5 weeks, livers were collected, and the hepatic SAT1 activity (k) (n = 9/10 mice) and Sat1 mRNA expression (l) (n = 5/6 mice) were determined. m WT and Sat1−/− male mice were treated with TETA (3000 ppm) for 2 weeks. The ratio N1-acetylspermidine over spermidine was then assessed in the liver (n = 6/8 mice/group). n Schematic representation of polyamine flux. WT and Sat1−/− male mice were treated with TETA (3000 ppm) for 2 weeks. 13C-spermidine was injected intraperitoneally (i.p., 50 mg/kg) 3 h before the recovery of livers and plasma for the mass spectrometric quantitation of 13C N1-acetylspermidine (o, p) and 13C putrescine (q, r) (n = 4/5 mice/group). In this figure, the results are displayed as box-and-whisker plots, which show median, first and third quartiles, and maximum and minimum values (a–m, o–r). Circles indicate each mouse used in the experiment. For statistical analyses, p values were calculated by two-tailed unpaired Student’s t test (a–l, o–r), comparing TETA-treated with -untreated mice (*p < 0.05, **p < 0.01, ***p < 0.001), or in (m) comparing WT with Sat1−/− mice in control and TETA-treated group (***p < 0.001). APAO acetylpolyamineoxidase, Ctrl control, FC fold change, r.u. relative units, SAT1 spermidine/spermine N1-acetyltransferase-1, Spd spermidine, w.w. wet weight.
TETA increases the activity of spermidine/SAT1
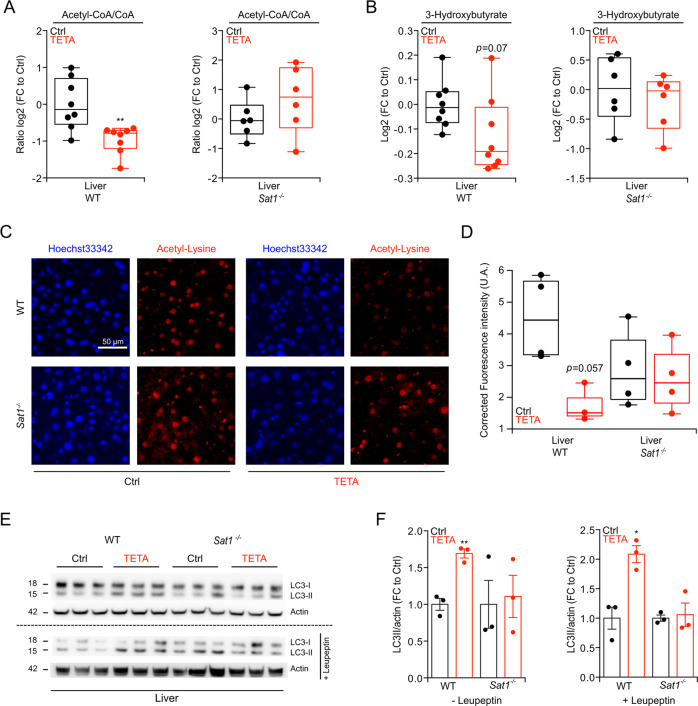
TETA administration did not affect the concentrations of spermidine and spermine in the liver (Fig. 1g, h), yet enhanced that of N1-acetylspermidine (Fig. 1i), as well as the ratio of N1-acetylated over non-acetylated spermidine (Fig. 1j), as compared with untreated controls. The N1 acetylation of spermidine is mostly catalyzed by SAT1 [31], and the enzymatic activity of SAT1 was indeed increased by TETA treatment (Fig. 1k) although Sat1 mRNA was not increased (Fig. 1l and S2a). This may be related to stabilization of the SAT1 protein by TETA, as polyamines and many of their structural analogs have been shown to bind to SAT1 protein and protect it against proteasomal degradation [31]. Indeed, we observed that the enzymatic activity of recombinant SAT1 protein, time-dependently decreased in the presence of liver extracts (a source of proteasomes), an effect that was reduced by addition of TETA (Fig. S2b, c), causing a significant increase in the half-life of SAT1 (Fig. S2d). The ratio of N1- acetylspermidine over spermidine was significantly reduced in Sat1−/− mice and barely elevated by TETA treatment of such mice (Fig. 1m). 13C-stable isotope-labeled spermidine, injected intraperitoneally, was converted into 13C N1-acetylspermidine, and then to putrescine (Fig. 1n). This enzymatic reaction, leading from spermidine to N1-acetylspermidine (catalyzed by SAT1) and then to putrescine (catalyzed by acetylpolyamineoxidase, APAO, official gene name PAOX) [32], was stimulated by TETA in WT but not in Sat1−/− mice (Fig. 1o–r). Commensurate with the increase in SAT1 activity (which can be expected to consume acetyl-CoA) [31], livers from TETA-treated WT mice exhibited a reduced acetyl-CoA/CoA ratio (Fig. 2a, left panel), as well as a reduced level of 3-hydroxybutyrate (Fig. 2b, left panel), which usually correlates with acetyl-CoA abundance [33]. Moreover, the level of Nε lysine acetylation, which is known to be in equilibrium with acetyl-CoA [7], was reduced in the liver of TETA-treated WT mice, as determined by means of a quantitative immunofluorescence assay (Fig. 2c, d). Nε lysine deacetylation of cytoplasmic proteins can stimulate autophagy [7], and, in WT mice, TETA indeed induced an increase in the autophagy-related variant II of the microtubule-associated protein 1A/1B light chain 3B (hereafter referred to as LC3), a lipidated variant of the protein that can be detected by its increased electrophoretic mobility, especially when the lysosomal destruction of LC3-II was blocked by the injection of leupeptin (Fig. 2e, f). The TETA-induced reduction in acetyl-CoA/CoA ratio, 3-hydroxybutyrate and Nε lysine acetylation, coupled to enhanced autophagic flux, was not detectable in Sat1−/− mice, meaning that they require SAT1 function (Fig. 2a–f). Altogether, these results support the contention that TETA acts as a classical caloric restriction mimetic (CRM) [34] inducing protein deacetylation, autophagy, and positive health effects. In mice, the TETA-induced protein deacetylation and autophagy depended on SAT1 activation.
Fig. 2. Acetyl-CoA depletion, deacetylation, and autophagy induction by TETA.
WT or Sat1−/− male mice (7 weeks old) were treated with TETA (3000 ppm in drinking water) for 2 weeks, and livers were collected to determine the acetyl-CoA/CoA ratio (a) and the abundance of 3-hydroxybutyrate (b) by mass spectrometry analysis (n = 6/8 mice/group). c, d TETA was injected i.p. (100 mg/kg), into WT or Sat1−/− male mice to determine, 8 h later, the level of Nε lysine acetylation of hepatic proteins by indirect immunofluorescence (representative images in c, quantitation in d) (n = 3/4 mice/group). e, f In the same experimental conditions described in (c, d), the autophagy- associated level of LC3 lipidation was evaluated in the absence or presence of leupeptin (i.p., 15 mg/kg) administered 2 h before the recovery of livers (representative blots in e, quantitation in f) (n = 3 mice/group in each experiment. N = 3 and N = 2 different experiments were performed without and with leupeptin injection, respectively). In this figure, the results are displayed as box-and-whisker plots, which show median, first and third quartiles, and maximum and minimum values (a, b and d) or mean ± s.e.m. (f). Circles, in the graphs, indicate each mouse used in the experiment. Statistical comparisons were done by two-tailed unpaired Student’s t test (a, b and f) comparing TETA-treated with -untreated mice (*p < 0.05, **p < 0.01). Statistical comparisons in d were done by applying a Wilcoxon test (*p < 0.05), comparing TETA-treated with control mice. Ctrl control, FC fold change, UA arbitrary units.
SAT1-dependent anti-obese and antidiabetic effects of TETA
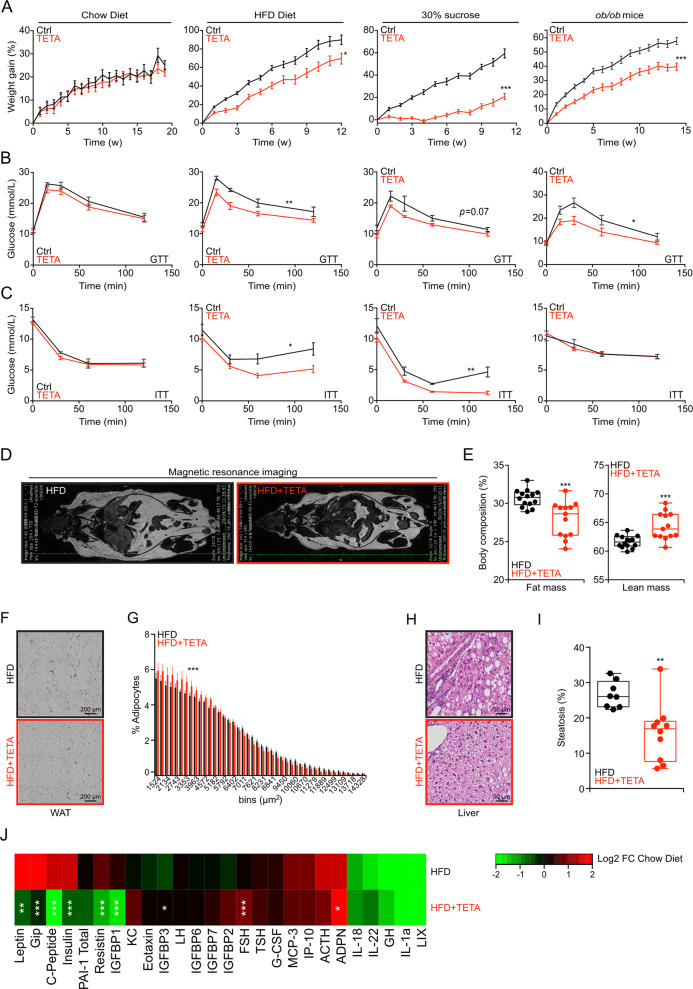
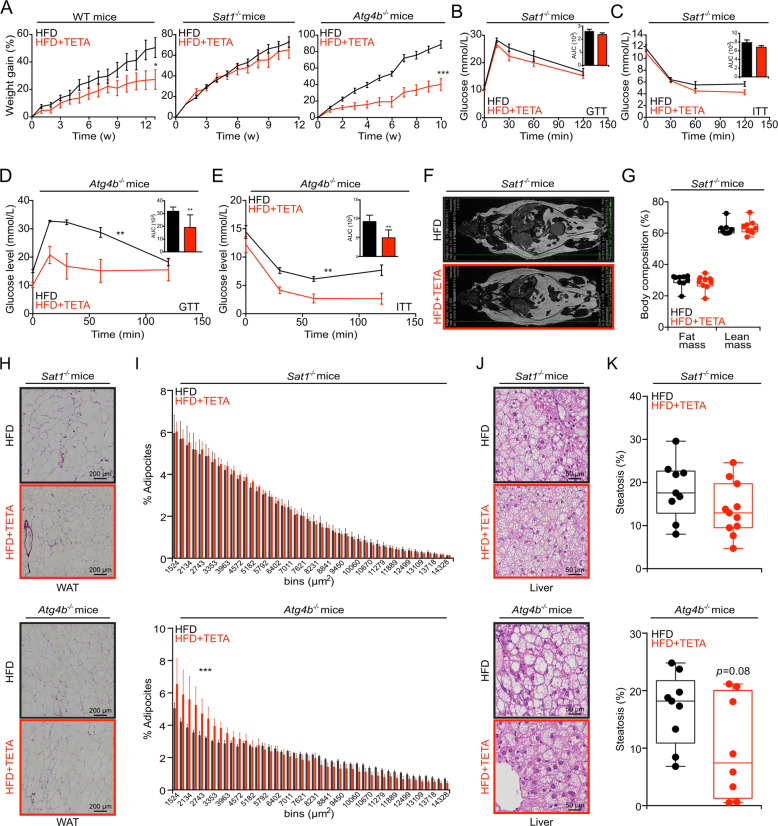
Continuous treatment of mice with TETA, from 7 weeks of age, did not affect weight gain in WT mice fed with normal chow, yet reduced weight gain of WT mice fed with HFD or 30% sucrose in the drinking water (Fig. 3a). TETA also reduced the weight gain of leptin-deficient ob/ob mice fed a normal diet (Fig. 3a). Alongside these anti-obesity effects, TETA ameliorated the glucose- and insulin-tolerance tests in obesogenic conditions (Fig. 3b, c, S3a, b), but failed to improve insulin tolerance in ob/ob mice (Fig. 3c). In the context of HFD, TETA reduced adiposity and increased lean mass (Fig. 3d, e), reduced the diameter of visceral adipocytes (Fig. 3f, g), and limited the extent of hepatic steatosis (Fig. 3h, i). At the biochemical level, TETA also prevented the HFD-induced increase in the plasma levels of, among others, leptin, gastrointestinal peptide (Gip), insulin, PAI-1, resistin, and insulin growth factor-binding protein (IGFBP-3) (Fig. 3j). TETA failed to affect food intake (Fig. S3c, d), and did not reduce the abundance of copper (Fig. S3e), iron (Fig. S3f), and zinc (Fig. S3g) even in conditions of HFD. Importantly, TETA lost its anti-obese effects in Sat1−/− but not in partially autophagy-deficient Atg4b−/− mice (Fig. 4a) [35, 36]. Under HFD, Sat1−/− (but not Atg4b−/−) mice also became resistant to the antidiabetic effects of TETA (Fig. 4b–e), failed to reduce their fat mass (Fig. 4f, g), and ameliorated the histological correlates of visceral adiposity (Fig. 4h, i, upper panels) nor those of hepatosteatosis (Fig. 4j, k, upper panels). However, these effects were preserved in Atg4b−/− mice (Fig. 4h–k, lower panels). In conclusion, all favorable effects of TETA on metabolic health depended on SAT1. This is true for spermidine as well, since spermidine failed to ameliorate HFD-induced phenotype in Sat1−/− mice, with the exception of improved glucose tolerance (Fig. S4a–i). Thus, the activity of Sat1 is required for the positive metabolic effects of both TETA and spermidine.
Fig. 3. Metabolic effects of TETA in the context of obesogenic diets or leptin deficiency.
a–c WT mice received standard chow diet (Ctrl = 10 and TETA = 9 mice), a high-fat diet (HFD) (n = 10 mice/group), or normal chow diet with 30% sucrose in the drinking water (n = 10 mice/group), for at least 12 weeks in the presence, or not, of TETA administration (3000 ppm in drinking water or daily i.p. injection, 100 mg/kg). Similarly, leptin-deficient ob/ob male mice (Ctrl = 9 and TETA = 10 mice), receiving a normal diet, were treated, or not, with TETA (3000 ppm in drinking water). Body weight was monitored weekly (a), and glucose- (b) or insulin-tolerance (c) tests were performed after 6–7 weeks of treatment, except for ob/ob male mice to whom the metabolic tests were performed after 11–12 weeks of treatment (chow diet: GTT and ITT, n = 5 mice/condition; HFD: GTT, n = 10 mice/group, ITT Ctrl = 9 and TETA = 10 mice; sucrose 30%: GTT 5 mice/group and ITT Ctrl = 5 and TETA = 4 mice; ob/ob mice: GTT Ctrl = 9 and TETA = 8 mice, ITT Ctrl = 9 and TETA = 7 mice; one representative experiment). d, e In addition, at 12 weeks of TETA treatment in HFD-fed mice, body composition was determined by magnetic resonance imaging (representative images in d and quantitation in e) (n = 13/14 mice/group). f–i Moreover, visceral white adipose tissues (representative images in f and quantification in g) or livers (representative images in h and quantification in i) were removed and subjected to hematoxylin and eosin staining, and image analysis for quantification of the surface of individual adipocytes (g) (Ctrl = 10 and TETA = 8 mice), or the occupancy of the liver by lipid droplets (i) (n = 8/10 mice per condition), was performed. j In addition, at 12 weeks of treatments, plasma was drawn and subjected to a multiplexed quantification of multiple hormones and cytokines, plotting a heat map (HFD vs. normal diet) in otherwise untreated or TETA-treated mice (n = 5 mice/condition). In this figure, the results are displayed as box-and-whisker plots, which show median, first and third quartiles, and maximum and minimum values (e, i) or mean ± s.e.m. (g). Circles, in the graphs, indicate each mouse used in the experiment. For statistical analysis, longitudinal statistical comparisons for mice weight gain, were performed by Wald test (a) (**p < 0.01, ***p < 0.001); p values were determined by two-tailed unpaired Student’s t test (for b, c, areas under the curve in Fig. S3a, b, and e, i) comparing TETA-treated with -untreated mice (*p < 0.05, **p < 0.01, ***p < 0.001); p values in g were done by means of a Kolmogorov–Smirnov test comparing TETA-treated with -untreated mice (***p < 0.001). In j statistical comparisons were done by Wilcoxon test (*p < 0.05, **p < 0.01, ***p < 0.001). Ctrl control, FC fold change, GTT glucose-tolerance test, HFD high-fat diet, min minutes, ITT insulin-tolerance test, w weeks, WAT white adipose tissues. Cytokines: ACTH adrenocorticotropic hormone, ADPN adiponectin, FSH follicle-stimulating hormone, G-CSF granulocyte colony-stimulating factor, GH growth hormone, Gip gastric-inhibitory polypeptide, IGFBP-1/2/3/6/7 insulin-like growth factor-binding protein 1/2/3/6/7, IL-18/22/1a interleukin, IP-10 interferon gamma-induced protein 10, KC keratinocyte chemoattractant, LH luteinizing hormone, LIX lipopolysaccharide-inducible CXC chemokine, MCP-3 monocyte chemotactic protein-3, PAI-1 total plasminogen activator inhibitor-1, TSH thyroid-stimulating hormone.
Fig. 4. SAT1-dependent metabolic effects of TETA.
a Male mice with the indicated genotypes (WT, Sat1−/−, or Atg4b−/−) were subjected to a HFD diet in the presence, or not, of TETA administration (3000 ppm in drinking water) for at least 10 weeks of treatment, and the body weight was monitored weekly (WT: Ctrl = 9 and TETA = 10 mice; Sat1−/−: Ctrl = 8 and TETA = 7 mice; Atg4b−/−: Ctrl = 10 and TETA = 8 mice). Glucose-tolerance tests (b, d) and insulin-tolerance tests (c, e) were performed after 6–11 weeks of treatment in Sat1−/− (Ctrl = 8 and TETA = 7 mice) and Atg4b−/− mice (Ctrl = 8 and TETA = 5 mice), respectively. After 12 weeks, Sat1−/− mice were subjected to magnetic resonance imaging, representative image in f, and quantitation in g (n = 9 mice/group). Histological analysis of visceral white adipose tissue (h, i), liver histopathology (j, k), was performed and quantified in Sat1−/− (upper panels; for visceral white adipose tissue analysis: Ctrl =9 and TETA = 12 mice; for liver histopathology: Ctrl = 9 and TETA = 11 mice) and Atg4b−/− (lower panels; for visceral white adipose tissue analysis: Ctrl = 10 and TETA = 8 mice; for liver histopathology: Ctrl = 9 and TETA = 8 mice). Representative images are shown in h, j, and quantifications are reported in i, k, respectively. In this figure, the results are displayed as box-and-whisker plots, which show median, first and third quartiles, and maximum and minimum values (g, k) or mean ± s.e.m. (i). Circles, in the graphs, indicate each mouse used in the experiment. Longitudinal statistical comparisons for mice weight gain, were performed by Wald test (a) (*p < 0.05, ***p < 0.001). For statistical analysis, p values were calculated by two-tailed unpaired Student’s t test (b–e calculating the related AUC, and g, k) comparing TETA-treated with -untreated mice. Statistical comparisons in i were done by means of a Kolmogorov–Smirnov test comparing TETA-treated with control group. The HFD control group of Sat1−/− mice in a–c is shared with Fig. S4a–c; HFD control group of Sat1−/− mice in f is shared with the control group in Fig. S4d, as well as the control group in h–k is shared with Fig. S4f–i. AUC area under the curve, Ctrl control, GTT glucose-tolerance test, HFD high-fat diet, min minutes, ITT insulin-tolerance test, min minutes, w weeks, WAT white adipose tissues.
Discussion
Although TETA is generally considered as a copper chelator, justifying its therapeutic use for the treatment of Wilson disease [15, 16, 19, 37], the present report suggests that even prolonged treatment with TETA is unable to deplete copper and other heavy metals from the organs of mice in non-pathological conditions. Moreover, we demonstrated that TETA activates autophagy in vivo; this might suggest that TETA improves the Wilson disease outcome through alternative mechanisms, including the induction of autophagy, which has been suggested to increase the turnover of copper-damaged mitochondria [37, 38]. From a broader perspective, the therapeutic indications of TETA may be extended to pathological settings (i.e., cardiovascular diseases) [39], characterized by an aberrant accumulation of dysfunctional mitochondria and excessive rate of cell death [40] or senescence [41]. It is hence temping to speculate that TETA may restore detrimental mitochondrial dynamics via autophagy-dependent [5] or -independent mechanisms.
Interestingly, TETA, which is metabolized by SAT1, enhanced the activity of SAT1 in the mouse liver, through a mechanism that is difficult to be elucidated due to the absence of suitable SAT1-specific antibodies. It appears clear, however, that TETA does not increase the expression of the Sat1 mRNA, and that it may stabilize the SAT1 enzyme. Irrespective of the precise mechanism of SAT1 stabilization/activation by TETA, there are multiple signs that SAT1 is overactivated in TETA-treated mice, causing enhanced acetylation of spermidine with a commensurate consumption of acetyl-CoA that results in Nε lysine deacetylation of cellular proteins and stimulation of autophagy. This TETA-triggered cascade of biochemical and cellular events is entirely lost in Sat1−/− mice, implying that SAT1 activation is epistatic to the reduction of acetyl-CoA and the induction of autophagy. Driven by the aforementioned observation that the autophagy inducer spermidine, a SAT1 substrate that is structurally related to TETA, can prevent obesity and diabetes [13, 14], we investigated whether TETA might have similar beneficial effects on whole-body metabolism. Indeed, TETA mitigated weight gain in obesogenic conditions (though without reducing food intake) and reduced obesity-related comorbidities such as diabetes and hepatosteatosis. In contrast to the pro-healthy and antidiabetic effects of spermidine, which require autophagy induction (and hence are lost in partially autophagy-deficient Atg4b−/− mice) [1, 14], the beneficial effects of TETA appeared to be Atg4b independent, supporting the notion that autophagy is dispensable for the anti-obese properties of TETA. As a disclaimer, we cannot discard the possibility that Atg4b-independent autophagy might occur in this context [42].
Nonetheless, both TETA and spermidine require SAT1 to mediate their anti-obese effects, meaning that both polyamines lose their anti-obesity, antidiabetic, and anti-steatotic properties in Sat1−/− mice. As a possible scenario, the exacerbated metabolism of spermidine by SAT1 (that is stimulated by TETA, which stabilizes SAT1, and perhaps also by spermidine when it is overabundant) may cause a futile cycle consuming acetyl-CoA, thereby altering cellular metabolism and/or consuming energy to prevent lipo-anabolism and its consequences. Indeed, Sat1−/− mice accumulate more fat, especially on HFD, while transgenic overexpression of SAT1 has previously been shown to yield a lean phenotype [43–46] arguing in favor of this conjecture. Moreover, activation of SAT1 has been shown to contribute to differentiation of white into beige adipocytes [47], suggesting yet another anti-obesity action for this enzyme.
In this scenario, it remains to be determined in which tissues SAT1 activation is particularly important for mediating the salutary effects of TETA, although it may be speculated that tissues that actively participate in the regulation of systemic glucose homeostasis (i.e., white adipose tissue, pancreas, and skeletal muscle) would be particularly important for these TETA effects. Of note, we observed that TETA could enhance the activity of SAT1 in the colon (a major site of polyamine production) [48], leaving open the possibility that TETA could mediate metabolic benefit via the modulation of intestinal polyamine flux as well.
Based on the above-mentioned findings, TETA may be considered as a new type of CRM not falling in any of the previously known categories [34, 49], namely (i) agents that deplete acetyl-CoA by inhibiting its synthesis (examples: hydroxycitrate, SB-204990, two inhibitors of ATP citrate lyase), (ii) agents that inhibit acetyltransferases (examples: aspirin and spermidine that inhibit EP300), and (iii) agents that activate deacetylases (examples: resveratrol and nicotinamide that activate sirtuin-1). Indeed, TETA would be an agent that causes the overconsumption (rather than reduced generation) of acetyl-CoA secondary to the activation of SAT1, thereby acting as a CRM. Given that TETA lacks any major toxicity (as indicated by preclinical experiments, as well as by its long-term use for the treatment of humans with Wilson disease) [15, 16, 19], it is tempting to speculate that this property may be taken advantage of for repurposing TETA for novel clinical applications, including the prevention or treatment of obesity with its comorbidities.
Supplementary information
Acknowledgements
We thank Professor Carlos Lopez-Otin (University of Oviedo, Spain) for helpful discussion and Ms Tuula Reponen (University of Eastern Finland, Finland) for technical help. The authors thank the CRC Core Facilities. GK is supported by the Ligue contre le Cancer (équipe labellisée); Agence National de la Recherche (ANR)—Projets blancs; ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases; Association pour la recherche sur le cancer (ARC); Association “Le Cancer du Sein, Parlons-en!”; Cancéropôle Ile-de-France; Chancelerie des universités de Paris (Legs Poix), Fondation pour la Recherche Médicale (FRM); a donation by Elior; European Research Area Network on Cardiovascular Diseases (ERA-CVD, MINOTAUR); Gustave Roussy Odyssea, the European Union Horizon 2020 Project Oncobiome; Fondation Carrefour; High-end Foreign Expert Program in China (GDW20171100085), Institut National du Cancer (INCa); Inserm (HTE); Inserm Transfert; Institut Universitaire de France; LeDucq Foundation; the LabEx Immuno-Oncology (ANR-18-IDEX-0001); the RHU Torino Lumière; the Seerave Foundation; the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); the SIRIC Cancer Research and Personalized Medicine (CARPEM). MTH and TAK are supported by the Academy of Finland (grants 292574 and 315487). MCM is supported by Fondation ARC (RAC11159LLA). FM is grateful to the Austrian Science Fund FWF (SFB LIPOTOX F3007 & F3012, W1226, P29203, P29262, P27893, and P31727, the Austrian Federal Ministry of Education, Science, and Research, and the University of Graz for grants “Unkonventionelle Forschung-InterFast” and “flysleep”) (BMWFW-80.109/0001-WF/V/3b/2015), as well as the field of excellence program BioHealth. We acknowledge support from NAWI Graz and the BioTechMed-Graz flagship project “EPIAge”. FP is supported by a Karolinska Institute Starting Grant, Starting Grant from the Swedish Research Council (2019_02050_3).
Author contributions
FC, SAM, EEB, EV, SL, MCM, and FP performed the in vivo experiments. TE, CD, TP, and FM conducted the lifespan in vivo experiment. PO and NS conducted the analysis of white adipose tissue and liver hepatosteatosis. AS performed the statistical analysis of white adipose tissues and acetyl-lysine immunohistochemistry. SD, FA, DL, and NB conducted metabolomics analysis. MK performed the metabolome statistical analysis. MTH and TAK performed the SAT1 enzymatic activity and half-life experiments. CE, BM, and HZ conducted the analysis of heavy metals in organs. MCM, FP, FM, TAK, and MTH helped to design the study. FC, FP, MCM, and GK conceived the study, analyzed the data, and wrote the paper.
Compliance with ethical standards
Conflict of interest
GK and FM are co-founders of Samsara Therapeutics. GK and FP hold a patent protecting new medical use for TETA. FM is a co-founder of The Longevity Labs. TE has equity interests in The Longevity Labs. GK is a consultant for The Longevity Labs.
Footnotes
Edited by M. Piacentini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Federico Pietrocola, Guido Kroemer
Contributor Information
Federico Pietrocola, Email: federico.pietrocola@ki.se.
Guido Kroemer, Email: kroemer@orange.fr.
Supplementary information
The online version of this article (10.1038/s41418-020-0550-z) contains supplementary material, which is available to authorized users.
References
- 1.Madeo F, Eisenberg T, Pietrocola F, Kroemer G. Spermidine in health and disease. Science. 2018;359:pii: eaan2788. doi: 10.1126/science.aan2788. [DOI] [PubMed] [Google Scholar]
- 2.Eisenberg T, Abdellatif M, Schroeder S, Primessnig U, Stekovic S, Pendl T, et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med. 2016;22:1428–38.. doi: 10.1038/nm.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–14. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 4.Kiechl S, Pechlaner R, Willeit P, Notdurfter M, Paulweber B, Willeit K, et al. Higher spermidine intake is linked to lower mortality: a prospective population-based study. Am J Clin Nutr. 2018;108:371–80.. doi: 10.1093/ajcn/nqy102. [DOI] [PubMed] [Google Scholar]
- 5.Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yue F, Li W, Zou J, Jiang X, Xu G, Huang H, et al. Spermidine prolongs lifespan and prevents liver fibrosis and hepatocellular carcinoma by activating MAP1S-mediated autophagy. Cancer Res. 2017;77:2938–51.. doi: 10.1158/0008-5472.CAN-16-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015;21:805–21. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Alsaleh G, Feltham J, Sun Y, Napolitano G, Riffelmacher T, et al. Polyamines control eIF5A hypusination, TFEB translation, and autophagy to reverse B cell senescence. Mol Cell. 2019:748–60. [DOI] [PMC free article] [PubMed]
- 9.Lévesque S, Le Naour J, Pietrocola F, Paillet J, Kremer M, Castoldi F, et al. A synergistic triad of chemotherapy, immune checkpoint inhibitors, and caloric restriction mimetics eradicates tumors in mice. Oncoimmunology. 2019;8:e1657375. [DOI] [PMC free article] [PubMed]
- 10.Pietrocola F, Pol J, Vacchelli E, Rao S, Enot DP, Baracco EE, et al. Caloric restriction mimetics enhance anticancer immunosurveillance. Cancer Cell. 2016;30:147–60.. doi: 10.1016/j.ccell.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Signor C, Girardi BA, Lorena Wendel A, Fruhauf PKS, Pillat MM, Ulrich H, et al. Spermidine improves the persistence of reconsolidated fear memory and neural differentiation in vitro: Involvement of BDNF. Neurobiol Learn Mem. 2017;140:82–91. doi: 10.1016/j.nlm.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Gabande-Rodriguez E, M MGdLH, Mittelbrunn M. Control of inflammation by calorie restriction mimetics: on the crossroad of autophagy and mitochondria. Cells. 2019;9:82. [DOI] [PMC free article] [PubMed]
- 13.Gao M, Zhao W, Li C, Xie X, Li M, Bi Y, et al. Spermidine ameliorates non-alcoholic fatty liver disease through regulating lipid metabolism via AMPK. Biochem Biophys Res Commun. 2018;505:93–8. doi: 10.1016/j.bbrc.2018.09.078. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez AF, Barcena C, Martinez-Garcia GG, Tamargo-Gomez I, Suarez MF, Pietrocola F, et al. Autophagy couteracts weight gain, lipotoxicity and pancreatic beta-cell death upon hypercaloric pro-diabetic regimens. Cell Death Dis. 2017;8:e2970. doi: 10.1038/cddis.2017.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohr I, Weiss KH. Current anti-copper therapies in management of Wilson disease. Ann Transl Med. 2019;7:S69. doi: 10.21037/atm.2019.02.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walshe JM. Management of penicillamine nephropathy in Wilson’s disease: a new chelating agent. Lancet. 1969;2:1401–2. doi: 10.1016/s0140-6736(69)90940-4. [DOI] [PubMed] [Google Scholar]
- 17.Czlonkowska A, Litwin T, Dusek P, Ferenci P, Lutsenko S, Medici V, et al. Wilson disease. Nat Rev Dis Prim. 2018;4:21. doi: 10.1038/s41572-018-0018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedera P. Clinical management of Wilson disease. Ann Transl Med. 2019;7:S66. doi: 10.21037/atm.2019.03.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Litwin T, Dziezyc K, Czlonkowska A. Wilson disease-treatment perspectives. Ann Transl Med. 2019;7:S68. doi: 10.21037/atm.2018.12.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerrada-Gimenez M, Weisell J, Hyvonen MT, Park MH, Alhonen L, Vepsalainen J, et al. Complex N-acetylation of triethylenetetramine. Drug Metab Dispos. 2011;39:2242–9. doi: 10.1124/dmd.111.041798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyvonen MT, Weisell J, Khomutov AR, Alhonen L, Vepsalainen J, Keinanen TA. Metabolism of triethylenetetramine and 1,12-diamino-3,6,9-triazadodecane by the spermidine/spermine-N(1)-acetyltransferase and thialysine acetyltransferase. Drug Metab Dispos. 2013;41:30–2. doi: 10.1124/dmd.112.047274. [DOI] [PubMed] [Google Scholar]
- 22.Niiranen K, Keinanen TA, Pirinen E, Heikkinen S, Tusa M, Fatrai S, et al. Mice with targeted disruption of spermidine/spermine N1-acetyltransferase gene maintain nearly normal tissue polyamine homeostasis but show signs of insulin resistance upon aging. J Cell Mol Med. 2006;10:933–45.. doi: 10.1111/j.1582-4934.2006.tb00536.x. [DOI] [PubMed] [Google Scholar]
- 23.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bravo-San Pedro JM, Sica V, Martins I, Pol J, Loos F, Maiuri MC, et al. Acyl-CoA-binding protein is a lipogenic factor that triggers food intake and obesity. Cell Metab. 2019;30:754–67. e9. doi: 10.1016/j.cmet.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Hyvönen MT, Keinänen T, Alhonen L. Assay of ornithine decarboxylase and spermidine/spermine N1-acetyltransferase activities. Bio-Protocol. 2014;4.
- 26.Hyvonen MT, Keinanen TA, Khomutov M, Simonian A, Weisell J, Kochetkov SN, et al. The use of novel C-methylated spermidine derivatives to investigate the regulation of polyamine metabolism. J Med Chem. 2011;54:4611–8. doi: 10.1021/jm200293r. [DOI] [PubMed] [Google Scholar]
- 27.Jarvinen A, Grigorenko N, Khomutov AR, Hyvonen MT, Uimari A, Vepsalainen J, et al. Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural polyamines. J Biol Chem. 2005;280:6595–601. doi: 10.1074/jbc.M412788200. [DOI] [PubMed] [Google Scholar]
- 28.Einer C, Leitzinger C, Lichtmannegger J, Eberhagen C, Rieder T, Borchard S, et al. A high-calorie diet aggravates mitochondrial dysfunction and triggers severe liver damage in Wilson disease rats. Cell Mol Gastroenterol Hepatol. 2019;7:571–96.. doi: 10.1016/j.jcmgh.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enot DP, Vacchelli E, Jacquelot N, Zitvogel L, Kroemer G. TumGrowth: An open-access web tool for the statistical analysis of tumor growth curves. Oncoimmunology. 2018;7:e1462431. doi: 10.1080/2162402X.2018.1462431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J, Chan YK, Poppitt SD, Cooper GJ. Determination of triethylenetetramine (TETA) and its metabolites in human plasma and urine by liquid chromatography-mass spectrometry (LC-MS) J Chromatogr B Anal Technol Biomed Life Sci. 2007;859:62–8. doi: 10.1016/j.jchromb.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Pegg AE. Spermidine/spermine-N(1)-acetyltransferase: a key metabolic regulator. Am J Physiol Endocrinol Metab. 2008;294:E995–1010. doi: 10.1152/ajpendo.90217.2008. [DOI] [PubMed] [Google Scholar]
- 32.Pegg AE. Mammalian polyamine metabolism and function. IUBMB Life. 2009;61:880–94. doi: 10.1002/iub.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry RJ, Peng L, Cline GW, Petersen KF, Shulman GI. A non-invasive method to assess hepatic acetyl-CoA in vivo. Cell Metab. 2017;25:749–56.. doi: 10.1016/j.cmet.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madeo F, Pietrocola F, Eisenberg T, Kroemer G. Caloric restriction mimetics: towards a molecular definition. Nat Rev Drug Discov. 2014;13:727–40. doi: 10.1038/nrd4391. [DOI] [PubMed] [Google Scholar]
- 35.Maciel M, Hernandez-Barrientos D, Herrera I, Selman M, Pardo A, Cabrera S. Impaired autophagic activity and ATG4B deficiency are associated with increased endoplasmic reticulum stress-induced lung injury. Aging. 2018;10:2098–112.. doi: 10.18632/aging.101532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marino G, Fernandez AF, Cabrera S, Lundberg YW, Cabanillas R, Rodriguez F, et al. Autophagy is essential for mouse sense of balance. J Clin Investig. 2010;120:2331–44. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zischka H, Lichtmannegger J, Schmitt S, Jagemann N, Schulz S, Wartini D, et al. Liver mitochondrial membrane crosslinking and destruction in a rat model of Wilson disease. J Clin Investig. 2011;121:1508–18. doi: 10.1172/JCI45401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polishchuk EV, Merolla A, Lichtmannegger J, Romano A, Indrieri A, Ilyechova EY, et al. Activation of autophagy, observed in liver tissues from patients with wilson disease and from ATP7B-deficient animals, protects hepatocytes from copper-induced apoptosis. Gastroenterology. 2019;156:1173–89. e5. doi: 10.1053/j.gastro.2018.11.032. [DOI] [PubMed] [Google Scholar]
- 39.Bonora M, Wieckowski MR, Sinclair DA, Kroemer G, Pinton P, Galluzzi L. Targeting mitochondria for cardiovascular disorders: therapeutic potential and obstacles. Nat Rev Cardiol. 2019;16:33–55. doi: 10.1038/s41569-018-0074-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Del Re DP, Amgalan D, Linkermann A, Liu Q, Kitsis RN. Fundamental mechanisms of regulated cell death and implications for heart disease. Physiol Rev. 2019;99:1765–817. doi: 10.1152/physrev.00022.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Childs BG, Li H, van Deursen JM. Senescent cells: a therapeutic target for cardiovascular disease. J Clin Investig. 2018;128:1217–28. doi: 10.1172/JCI95146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galluzzi L, Green DR. Autophagy-independent functions of the autophagy machinery. Cell. 2019;177:1682–99.. doi: 10.1016/j.cell.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jell J, Merali S, Hensen ML, Mazurchuk R, Spernyak JA, Diegelman P, et al. Genetically altered expression of spermidine/spermine N1-acetyltransferase affects fat metabolism in mice via acetyl-CoA. J Biol Chem. 2007;282:8404–13. doi: 10.1074/jbc.M610265200. [DOI] [PubMed] [Google Scholar]
- 44.Koponen T, Cerrada-Gimenez M, Pirinen E, Hohtola E, Paananen J, Vuohelainen S, et al. The activation of hepatic and muscle polyamine catabolism improves glucose homeostasis. Amino Acids. 2012;42:427–40. doi: 10.1007/s00726-011-1013-0. [DOI] [PubMed] [Google Scholar]
- 45.Mandal S, Mandal A, Park MH. Depletion of the polyamines spermidine and spermine by overexpression of spermidine/spermine N(1)-acetyltransferase 1 (SAT1) leads to mitochondria-mediated apoptosis in mammalian cells. Biochem J. 2015;468:435–47. doi: 10.1042/BJ20150168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pirinen E, Kuulasmaa T, Pietila M, Heikkinen S, Tusa M, Itkonen P, et al. Enhanced polyamine catabolism alters homeostatic control of white adipose tissue mass, energy expenditure, and glucose metabolism. Mol Cell Biol. 2007;27:4953–67. doi: 10.1128/MCB.02034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan F, Zhang L, Cao Y, Gao W, Zhao C, Fang Y, et al. Spermidine/spermine N1-acetyltransferase-mediated polyamine catabolism regulates beige adipocyte biogenesis. Metabolism. 2018;85:298–304. doi: 10.1016/j.metabol.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsumoto M, Kibe R, Ooga T, Aiba Y, Kurihara S, Sawaki E, et al. Impact of intestinal microbiota on intestinal luminal metabolome. Sci Rep. 2012;2:233. doi: 10.1038/srep00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madeo F, Carmona-Gutierrez D, Hofer SJ, Kroemer G. Caloric restriction mimetics against age-associated disease: targets, mechanisms, and therapeutic potential. Cell Metab. 2019;29:592–610. doi: 10.1016/j.cmet.2019.01.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.