Abstract
We report on the first case of multiresistant Pseudomonas aeruginosa harbouring the metallo-β-lactamase IMP-15 isolated in Switzerland from a patient repatriated from Cambodia. The laboratory diagnosis of IMP-15 was hampered by two negative tests for carbapenemase detection. The carbapenemase gene was subsequently detected by whole genome sequencing and the isolate further characterised by various phenotypic and genotypic analyses.
Keywords: blaIMP-15, Cabapenemase-producing Pseudomonas aeruginosa, Metallo-β-lactamase, Switzerland, Cambodia, Whole genome sequencing (WGS)
Introduction
The worldwide emergence of multidrug resistant bacteria is a major threat to public health. Pseudomonas aeruginosa is one of the major pathogens causing nosocomial infections like bacteremia and pneumonia. Carbapenemases, especially the metallo-β-lactamases (MBLs) such as IMP, VIM, SPM, GIM, NDM, and FIM are one of the most important mechanisms responsible for resistance to β-lactams including carbapenems in P. aeruginosa. Moreover, such resistance genes can be transferred to further bacterial species on mobile genetic elements [1]. In 1988, the blaIMP-1 was first documented in Japan on a conjugative plasmid [2]. During the last years, multiple case reports and hospital epidemics have reported blaIMP variants worldwide. Here, we present the first case of a P. aeruginosa harbouring a blaIMP-15 isolated from a patient in Switzerland repatriated from Cambodia.
Case
We report the case of a 33-year-old male patient from the north-eastern part of Switzerland, who was successfully resuscitated after a cardiac arrest of unknown cause in his holiday apartment in Cambodia on December 24, 2019. After initial supportive treatment at a provincial hospital in Cambodia, he was transferred to an intensive care unit in Bangkok, Thailand, still unconscious and in a critical state, intubated and on vasopressors. Chest radiograph showed bilateral patchy infiltration. Findings on electro-encephalography were compatible with profound diffuse encephalopathy. Repatriation took place on January 3, 2020. On admission onto intensive care unit in Switzerland, the patient was placed under preventive contact isolation measures due to high risk of colonization with multi-drug resistant organisms. A sputum culture from Bangkok revealed growth of Pseudomonas aeruginosa resistant against tigecycline and Acinetobacter baumanni with intermediate susceptibility to ceftazidime and resistance against tazobactam. Susceptibility testing was otherwise unremarkable. Based on these microbiology results, broad-spectrum parenteral antibiotic treatment with meropenem 500 mg every 12 h (adapted to impaired renal function) was initiated for bilateral pneumonia as per internal guideline. Unfortunately, the patient’s condition deteriorated and he deceased on January 6, 2020. After his death, culture from bronchial aspirate revealed the presence of Burkholderia cepacia complex, being phenotypically susceptible to ceftazidime and trimethoprim/sulfamethoxazole only, and Pseudomonas aeruginosa resistant to all regularly tested antimicrobials except gentamicin. In addition, a rectal swab performed during routine admission screening showed growth of Burkholderia cepacia complex and Pseudomonas aeruginosa as well but was otherwise negative for vancomycin resistant enterococci.
The P. aeruginosa isolated from bronchial aspirate was sent to the microbiology laboratory of the University Hospital Basel for identification of underlying resistance mechanism. The antimicrobial susceptibility testing perfomed with Vitek® 2 (bioMérieux) revealed resistance to all antimicrobial agents tested except meropenem, interpreted as intermediate, and tobramycin, amikacin and colistin, interpreted as susceptible. The isolate was tested with Xpert® Carba-R (Cepheid) PCR and exhibited negative results for the following carbapenemase gene targets: KPC, NDM, VIM, OXA-48-variants and IMP. In order to screen for MBL phenotype, combined Etest® strip using imipenem with and without EDTA (Etest® MBL IP/IPI; bioMérieux) was performed. Since MBL was excluded with this phenotypic test, we presumed that mechanisms other than carbapenemase production were responsible for carbapenem resistance in this P. aeruginosa. Later on, the isolate was submitted to whole genome sequencing (WGS) as part of a study to validate WGS for routine purposes. DNA from isolate 400128-20 was sequenced on both a NextSeq500 platform (Illumina) after Nexteraflex library production (Illumina), resulting in 64x mean coverage and GridION platform (Oxford Nanopore), resulting in 10x mean coverage to aid scaffolding. All reads have been submitted to the ENA under project PRJEB37060. The genome was hybrid assembled using Unicycler v0.3.0b [3], resulting in 18 contigs, annotated with Prokka v1.13 [4] and visualized in Artemis [5]. The isolate belongs to MLST sequence type 155 (https://pubmlst.org/paeruginosa)/ [6]. The 741-bp-long metallo-beta-lactamase blaIMP-15 gene was identified with 100 % identity, using ABRicate (https://github.com/tseemann/abricate) and the NCBI AMRFinderPlus database (doi: 10.1128/AAC.00483-19).
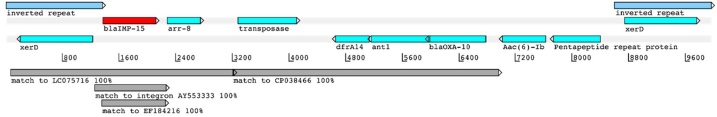
The blaIMP-15 gene is located on a contig of 323Kb, part of the chromosome. The 10Kb region around the blaIMP-15 gene seems to have derived from several plasmids, including LC075716 from P. aeruginosa in Vietnam (3.1Kb, 100 %) and CP038466 from Aeromonas hydrophila in China (3.8Kb, 100 %). This 10Kb section, flanked by two identical tyrosine recombinase genes, carries a transposase and several other antimicrobial resistance markers (Fig. 1; accession number ERZ1303326; Supplementary file). The identity with the blaIMP-15 class I integron from P. aeruginosa in Thailand (accession number AY553333) covers only 997 bp (100 % identity) over the blaIMP-15 gene itself; similarly the match to EF184216 from Mexico [8] covers 889bp (100 % identity) is also over the blaIMP-15 gene (Table 1).
Fig. 1.
blaIMP-15 containing 10Kb region. The region is flanked by 1355bp inverted repeats covering genes encoding tyrosine recombinase XerD. The blaIMP-15 is shown in red, along with other annotated genes in the region in forward or reverse frames. Matches to database sequences are shown below in grey. Figure was adapted from data visualized in Artemis [5].
Table 1.
Overview of carbapenemase test results performed on IMP-15 producing P. aeruginosa isolate.
| Carbapenemase test | Result |
|---|---|
| Phenotypic tests | |
| Etest® MBL IP/IPI | – |
| RAPIDEC® CARBA NP | + |
| β-CARBA Test | + |
| Carbapenem inactivation method mCIM | + |
| Genotypic tests | |
| Xpert® Carba-Ra | – |
| IMP-15 PCR [8] | + |
| IMP PCR [9,10] | + |
Detects KPC, NDM, VIM, OXA-48-variants and IMP (not all IMP subtypes are covered by the assay).
We performed two conventional IMP-targeted PCR tests to confirm the presence of blaIMP-15 gene. The first PCR (A) was described in a study to characterize a hospital epidemic in Mexico in 2008 with blaIMP-15 containing P. aeruginosa [8]. The second IMP PCR (B) was performed according to references [9,10]. Both PCRs were performed on freshly extracted DNA and showed the expected DNA fragment length of 275bp for PCR (A) and 232bp for PCR (B) on the 4200 TapeStation system (Agilent Technologies). Nucleotide sequencing of both PCR fragments using Sanger sequencing in both directions resulted in 100 % identity to blaIMP-15. In order to further characterize our isolate, the following phenotypic tests for detection of carbapenemase production were performed: RAPIDEC® CARBA NP (bioMérieux), β-CARBA Test (BIO-RAD) and modified Carbapenem Inactivation Method mCIM according CLSI [11]. Interestingly, our isolate showed positive results with all three above mentioned assays. The results of all genotypic and phenotypic tests perfomed are shown in summary in Table 1.
Discussion
The MBL variant blaIMP-15 was described for the first time in 2005 in Thailand (GenBank accession no. AY553333) and was associated with a class I integron, genes encoding dihydrofolate reductase (dhfr) and aminoglycoside 6′-N-acetyltransferase (aac(6′)). Further studies reported on IMP-15-producing multiresistant P. aeruginosa: three hospital outbreaks in Mexico in 2003–2005 and one case from USA related to one of the Mexican clones in 2005 [7,8,12,13]. The first blaIMP-15 cases in Europe were communicated in 2013 with two P. aeruginosa and two P. putida strains [14]. In a retrospective study published in 2014, one case from Germany isolated in 2009 was identified [15]. Recently, four P. aeruginosa strains containing blaIMP-15 were detected in Lebanon [16,17].
Due to their high transmissibility and potential for causing outbreaks in healthcare settings, timely recognition of patients colonized or infected with carbapenem-resistant gram-negative bacteria (GNB) is key to implementing appropriate preventive measures in order to stop these bacteria from further spread. Active surveillance for detection of asymptomatic colonization is therefore one of several strongly recommended infection control measures and the target population should include patients with recent hospitalization in countries with high prevalence of carbapenem-resistant GNB [18,19]. The carbapenemase activity of this P. aeruginosa strain could not be detected with our routine diagnostic scheme for carbapenemase production using Xpert® Carba-R PCR and Etest® MBL IP/IPI. However, other phenotypic carbapenemase screening tests performed (Table 1) succeded in detection of this rare carbapenemase type. Concerning the negative result in Xpert® CARBA-R, the manufacturer indicates that the following IMP variants are not covered with this assay: IMP-7, IMP-13, and IMP-14. According to our present data, IMP-15 has to be added to this list until the manufacturer is able to provide a PCR able to detect all IMP variants. The challenging identification of MBL blaIMP-15 in our strain is worrying. Conventional testing with Etest® MBL IP/IPI was not able to detect IMP-15 production in this P. aeruginosa isolate in our laboratory, suggesting that such strains could be overlooked and therefore require a high degree of alertness from clinical microbiologists. If the presence of carbapenamase is suspected despite the negative screening test, a second screening test should be considered, or the isolate should be referred to a reference laboratory for further analyses.
Since the development of antimicrobial resistance in GNB is a constantly moving target, the absence of a genetic resistance determinant should be approached with caution, as organisms can harbour resistance mechanisms for which tests were not developed or are false negative.
In this case, the initial microbiology report from Bangkok evoked a false security in the treating physicians. It is therefore of major importance to expand empiric antibiotic therapy in a seriously ill patient if presence of a multi-drug resistant pathogen such as a carbapenemase producer has to be assumed based on epidemiological data or relevant exposure history.
WGS proved to be a powerful tool for revealing mechanism of resistance which could not be detected with conventional, routinely used methods. Unfortunately, this is a relatively novel technology which is still not widely available in the common microbiology laboratory practice.
Authors’ contributions
DG collected and interpreted laboratory results and wrote the manuscript; DVG collected and interpreted clinical and epidemiological data and wrote the manuscript; KH collected and interpreted laboratory results; VH interpreted laboratory results, supervised phenotypic laboratory testing and reviewed the manuscript; HSS analysed WGS data and wrote the manuscript; AE supervised the study and reviewed the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, ornot-for-profit sectors.
Consent
Written informed consent was obtained from the patient for publication of this case report. A copy of the written consent is available for review by the Editor-in-Chief of this journal on request.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
We thank Berfin Kocyigit, Benjamin Zahm, Daniel Gander and Elisabeth Schultheiss for excellent technical assistance.
Assemblies and searches were performed at sciCORE (http://scicore.unibas.ch/) scientific computing center at University of Basel.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.idcr.2020.e00933.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Hong D.J., Bae I.K., Jang I.H., Jeong S.H., Kang H.K., Lee K. Epidemiology and characteristics of metallo-beta-lactamase-producing Pseudomonas aeruginosa. Infect Chemother. 2015;47(2):81–97. doi: 10.3947/ic.2015.47.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watanabe M., Iyobe S., Inoue M., Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35(1):147–151. doi: 10.1128/aac.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb Genom. 2017;3(10):e000132. doi: 10.1099/mgen.0.000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prokka Seemann T. Rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 5.Carver T., Harris S.R., Berriman M., Parkhill J., McQuillan J.A. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28(4):464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jolley K.A., Bray J.E., Maiden M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garza-Ramos J.U., Sanchez-Martinez G., Barajas J.M., Suarez S., Sanchez-Perez A., Rojas-Moreno T. Variability of the bla(IMP-15)-containing integrons, highly related to In95, on an endemic clone of Pseudomonas aeruginosa in Mexico. Microb Drug Resist. 2010;16(3):191–195. doi: 10.1089/mdr.2010.0017. [DOI] [PubMed] [Google Scholar]
- 8.Garza-Ramos U., Morfin-Otero R., Sader H.S., Jones R.N., Hernandez E., Rodriguez-Noriega E. Metallo-beta-lactamase gene bla(IMP-15) in a class 1 integron, In95, from Pseudomonas aeruginosa clinical isolates from a hospital in Mexico. Antimicrob Agents Chemother. 2008;52(8):2943–2946. doi: 10.1128/AAC.00679-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellington M.J., Kistler J., Livermore D.M., Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007;59(2):321–322. doi: 10.1093/jac/dkl481. [DOI] [PubMed] [Google Scholar]
- 10.Poirel L., Walsh T.R., Cuvillier V., Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 11.CLSI . Clinical and Laboratory Standards Institute; Wayne, PA: 2020. M100 Performance Standards for Antimicrobial Susceptibility Testing; 30th ed. Table 3C. Modified Carbapenem Inactivation Methods for Suspected Carbapenemase Production in Enterobacterales and Pseudomonas aeruginosa. [Google Scholar]
- 12.Martin C.A., Morita K., Ribes J.A., Deshpande L.M., Sader H.S., Castanheira M. IMP-15-producing Pseudomonas aeruginosa strain isolated in a U.S. medical center: a recent arrival from Mexico. Antimicrob Agents Chemother. 2008;52(6):2289–2290. doi: 10.1128/AAC.00299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinones-Falconi F., Galicia-Velasco M., Marchiaro P., Mussi M.A., Ballerini V., Vila A.J. Emergence of Pseudomonas aeruginosa strains producing metallo-beta-lactamases of the IMP-15 and VIM-2 types in Mexico. Clin Microbiol Infect. 2010;16(2):126–131. doi: 10.1111/j.1469-0691.2009.02780.x. [DOI] [PubMed] [Google Scholar]
- 14.Gilarranz R., Juan C., Castillo-Vera J., Chamizo F.J., Artiles F., Alamo I. First detection in Europe of the metallo-beta-lactamase IMP-15 in clinical strains of Pseudomonas putida and Pseudomonas aeruginosa. Clini Microbiol Infect. 2013;19(9):E424–427. doi: 10.1111/1469-0691.12248. [DOI] [PubMed] [Google Scholar]
- 15.Castanheira M., Deshpande L.M., Costello A., Davies T.A., Jones R.N. Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonas aeruginosa collected during 2009-11 in 14 European and Mediterranean countries. J Antimicrob Chemother. 2014;69(7):1804–1814. doi: 10.1093/jac/dku048. [DOI] [PubMed] [Google Scholar]
- 16.Al Bayssari C., Diene S.M., Loucif L., Gupta S.K., Dabboussi F., Mallat H. Emergence of VIM-2 and IMP-15 carbapenemases and inactivation of oprD gene in carbapenem-resistant Pseudomonas aeruginosa clinical isolates from Lebanon. Antimicrob Agents Chemother. 2014;58(8):4966–4970. doi: 10.1128/AAC.02523-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yaghi J., Fattouh N., Akkawi C., El Chamy L., Maroun R.G., Khalil G. Unusually high prevalence of cosecretion of ambler class A and B carbapenemases and nonenzymatic mechanisms in multidrug-resistant clinical isolates of Pseudomonas aeruginosa in Lebanon. Microb Drug Resist. 2020;26(2):150–159. doi: 10.1089/mdr.2019.0040. [DOI] [PubMed] [Google Scholar]
- 18.Magiorakos A.P., Burns K., Rodriguez Bano J., Borg M., Daikos G., Dumpis U. Infection prevention and control measures and tools for the prevention of entry of carbapenem-resistant Enterobacteriaceae into healthcare settings: guidance from the European Centre for Disease Prevention and Control. Antimicrob Resist Infect Control. 2017;6:113. doi: 10.1186/s13756-017-0259-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO . 2017. Guidelines for the prevention and control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in health care facilities. Licence: CC BY-NC-SA 30 IGO. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.