Sigma B is the alternative sigma factor governing stress response in many Gram-positive bacteria. In C. difficile, a sigB mutant shows pleiotropic transcriptional effects. Here, we determine genes that are likely direct targets of σB by evaluating the transcriptional effects of σB overproduction, provide biochemical evidence of direct transcriptional activation by σB, and show that σB-dependent genes can be activated by antimicrobials. Together, our data suggest that σB is a key player in dealing with toxic radicals.
KEYWORDS: Clostridium difficile, antimicrobial agents, in vitro transcription, luciferase, regulon, sigma factors
ABSTRACT
In many Gram-positive bacteria, the general stress response is regulated at the transcriptional level by the alternative sigma factor sigma B (σB). In C. difficile, σB has been implicated in protection against stressors such as reactive oxygen species (ROS) and antimicrobial compounds. Here, we used an anti-σB antibody to demonstrate time-limited overproduction of σB in C. difficile despite its toxicity at higher cellular concentrations. This toxicity eventually led to the loss of the plasmid used for anhydrotetracycline-induced σB gene expression. Inducible σB overproduction uncouples σB expression from its native regulatory network and allows for the refinement of the previously proposed σB regulon. At least 32% of the regulon was found to consist of genes involved in the response to reactive radicals. Direct gene activation by C. difficile σB was demonstrated through in vitro runoff transcription of specific target genes (cd0350, cd3614, cd3605, and cd2963). Finally, we demonstrated that different antimicrobials and hydrogen peroxide induce these genes in a manner dependent on this sigma factor, using a plate-based luciferase reporter assay. Together, our work suggests that lethal exposure to antimicrobials may result in the formation of toxic radicals that lead to σB-dependent gene activation.
IMPORTANCE Sigma B is the alternative sigma factor governing stress response in many Gram-positive bacteria. In C. difficile, a sigB mutant shows pleiotropic transcriptional effects. Here, we determine genes that are likely direct targets of σB by evaluating the transcriptional effects of σB overproduction, provide biochemical evidence of direct transcriptional activation by σB, and show that σB-dependent genes can be activated by antimicrobials. Together, our data suggest that σB is a key player in dealing with toxic radicals.
INTRODUCTION
Disruption of the normal gastrointestinal flora as a result of antimicrobial treatment can lead to a Clostridioides (Clostridium) difficile infection (CDI) (1). Clostridioides difficile is a Gram-positive, spore-forming obligate anaerobe and the primary cause for nosocomial infectious diarrhea (2). Its highly resistant endospores are usually transmitted via the oral-fecal route and germinate into vegetative cells in the colon upon contact with primary bile acids and other inducing factors (3). In the gut, vegetative C. difficile cells face many environmental stressors, including variations in oxygen tension, pH, osmolarity, nutrient availability, and the inflammatory responses of the immune system (4). The bacteria are also faced with antimicrobial compounds that are produced by the host, the resident microbiota, or given externally during medical therapy (5). The physiological response of C. difficile to these insults and the inflammatory responses triggered by CDI can result in the production of reactive oxygen species (ROS), reactive nitrogen species (RNS), and nitric oxide (NO) (2, 6).
Bacteria need to adapt to changing environmental conditions, including stresses, by adapting their physiology in a timely manner. This is achieved by fast transcriptional reprogramming, followed by briefly delayed changes at the translational level (7). The alternative sigma factor sigma B (σB, encoded by the sigB gene), which regulates the general stress responses in a variety of Gram-positive organisms, is central to the maintenance of cellular homeostasis during stress adaptation (8, 9).
Sigma factor B activity in Firmicutes species is regulated at the protein level by a partner-switching mechanism in which the anti-sigma factor RsbW binds and inhibits σB association with the RNA polymerase under nonstressed conditions. When a σB-activating stress is sensed, the dephosphorylated anti-anti-sigma factor RsbV sequesters RsbW, allowing for the association of free σB with the RNA polymerase core enzyme (8, 10). In C. difficile, the phosphatase RsbZ is responsible for RsbV dephosphorylation (11). The tight regulation of σB activity by a partner-switching mechanism is necessary, as the energy burden associated with σB activity was found to be disadvantageous in several different organisms (12, 13).
Despite the burden associated with its expression, σB is essential for survival for several pathogenic bacterial species in response to host-dependent stressors or antimicrobials. For example, in Listeria monocytogenes, σB is involved in counteracting the effects of the acidic pH encountered in the stomach and upon invasion of intestinal epithelial cells in the lysosome (14, 15). In Staphylococcus aureus, σB overproduction leads to thickening of the cell wall and increased resistance to beta-lactam antimicrobials (16). The sigB homologue sigF of Mycobacterium tuberculosis is induced by small amounts of rifamycin (17). Analogously, Bacillus subtilis σB is involved in resolving a rifampin-induced growth arrest (18). There is also evidence for the involvement of σB in C. difficile in the response to antimicrobial substances. Mutants of sigB show increased susceptibility to rifampin and mitomycin C and are also more sensitive to hydrogen peroxide, nitroprusside, and di‐ethylamine NONOate (19). However, the underlying molecular mechanisms remain unknown. Finally, indirect activation of σB-dependent genes as the result of a gene dosage shift has been demonstrated for C. difficile exposed to DNA polymerase inhibitors such as the phase II drug ibezapolstat/ACX-362E (20).
In this study, we demonstrate that σB overexpression is detectable and is tolerated for short periods of time. This allowed for the experimental identification of a set of genes that is most likely directly regulated by σB by performing transcriptome analyses under conditions of acute σB overexpression. The results obtained show that genes involved in the oxidative and nitrosative stress response form the core of the regulon. Additionally, we show that various antimicrobials and hydrogen peroxide induce the expression of σB-regulated genes in a σB-dependent manner, suggesting a link between the lethal exposure to antimicrobials and oxidative and nitrosative stresses in C. difficile.
RESULTS
C. difficile σB is measurably overproduced upon induction of the sigB gene.
Previous investigations of σB in C. difficile have used a sigB mutant and characterized its gene expression in the stationary growth phase in comparison with that of a wild-type strain (19). Although informative, this method is likely to result in indirect effects of σB due to stationary-phase heterogeneity, prolonged incubation, and possible positive or negative feedback in the σB regulatory circuit. To circumvent these issues and identify genes likely to be regulated by σB directly, we set out to uncouple sigB expression from its native regulatory circuit by expressing it from an inducible promoter.
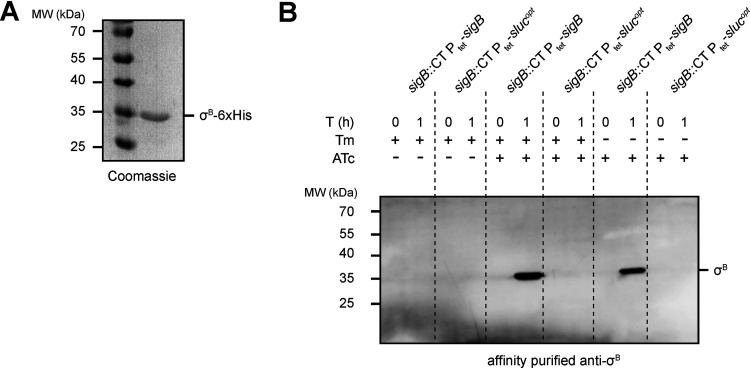
First, in order to confirm overproduction of σB, we measured cellular σB levels using immunoblotting. For this purpose, we heterologously overproduced and purified σB containing a C-terminal His tag (Fig. 1A) and used this protein to raise a polyclonal antiserum. Corresponding polyclonal antibodies were affinity purified to prevent unspecific immune reactions.
FIG 1.
Recombinant σB6×His was used to generate a Clostridiodes difficile σB-specific antibody for intracellular detection. (A) Coomassie blue-stained 12.5% SDS-PAGE gel of purified recombinant σB6×His. (B) Western blot using affinity purified σB antibody (1:500) on strains IB58 (sigB::CT Ptet-sigB) and IB61 (sigB::CT Ptet-slucopt). Cells were grown in lincomycin (20 μg/ml) and in the presence or absence of thiamphenicol (20 μg/ml) until an optical density at 600 nm (OD600) of ≈0.3, after which the indicated samples were induced with 100 ng/ml anhydrotetracycline (ATc). Samples were collected directly after the addition of ATc (or at the time ATc would have been added in the uninduced controls) at T = 0 h and after 1 h of induction (T = 1).
Next, we set out to validate the overproduction of σB in transconjugant C. difficile cells harboring plasmids containing sigB under the control of the anhydrotetracycline (ATc)-dependent promoter Ptet (21). For this purpose, σB was produced in a sigB mutant background (strain IB58; sigB::CT Ptet-sigB). As a control, we introduced a nonrelated expression construct in the same background (IB61; sigB::CT Ptet-slucopt) such that this control strain carries a plasmid with the same replicon, resistance marker, and inducible promoter.
We expected a signal at approximately 30 kDa in Western blot experiments for cells grown in the presence of the inducer ATc for strain IB58, but not for the uninduced cultures of IB58 or the control strain IB61. Additionally, by growing cultures in the presence or absence of thiamphenicol, we investigated whether overproduction of σB required selection for the Ptet-sigB expression plasmid.
When strains were grown in brain heart infusion (BHI) broth supplemented with 0.5% (wt/vol) yeast-extract (BHIY) supplemented with 20 μg/ml lincomycin and induced for 1 h with or without 100 ng/ml ATc in the presence or absence of 20 μg/ml thiamphenicol, we did not detect any signal at the molecular weight expected for σB in the ATc-induced control samples (sigB::CT Ptet-slucopt) or in any of the uninduced samples (Fig. 1B). In contrast, after 1 h of induction, a clear band of the expected molecular weight of σB (≈30 kDa) was observed only in the IB58 (sigB::CT Ptet-sigB) samples (Fig. 1B). Plasmid selection by inclusion of thiamphenicol in the growth medium did not influence σB overproduction in this time frame, which might have occurred as a tradeoff between σB overexpression and cellular toxicity (see further below).
We conclude that the affinity-purified rabbit anti-σB antibody is specific for σB and can be used for its detection in lysates of C. difficile. Furthermore, it is possible to uncouple sigB expression from its tight regulatory network by ATc-inducible overexpression for 1 h in trans.
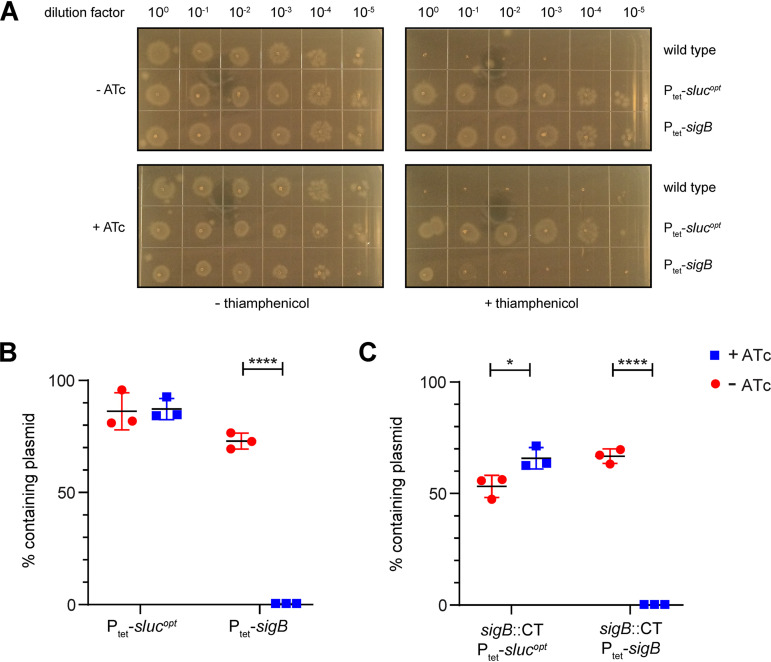
Prolonged overexpression of σB is lethal and leads to a loss of plasmids harboring Ptet-sigB.
Above, we showed that it is possible to overproduce σB in C. difficile and that this is tolerated by the bacterium for 1 h. This observation is somewhat at odds with the previously reported toxic nature of overproduced σB (8, 11). To reconcile these two observations, the effect of long-term overexpression of sigB and the stability of the plasmids used for σB overproduction under such conditions were investigated. First, overnight cultures of 630Δerm (wild-type), AP34 (Ptet-slucopt), and JC096 (Ptet-sigB) strains were adjusted for their optical density at 600 nm (OD600) values and 10-fold serially diluted. Subsequently, 2-μl spots per dilution were made on selective (20 μg/ml thiamphenicol) and nonselective BHIY agar plates, some of which contained 200 ng/ml ATc to induce Ptet-dependent gene expression. All plates were then incubated anaerobically for 24 h. On plates without thiamphenicol, regardless of the presence of the inducer ATc, comparable growth was observed for all three strains (Fig. 2A). As expected, when selecting for the plasmid using thiamphenicol, no growth was observed for the susceptible 630Δerm strain (which lacks the catP gene contained on the expression vector). In the absence of the inducer, no difference in growth was observed for the vector control strain (AP34; Ptet-slucopt) compared to the strain carrying the Ptet-sigB plasmid (JC096). However, upon induction of sigB expression on selective plates, a 3- to 4-log growth defect was observed for the strain carrying Ptet-sigB compared to the vector control strain. We conclude that prolonged induction of sigB expression is toxic when cells are cultured in the presence of thiamphenicol. Our results thus corroborate the finding that σB overproduction is toxic to C. difficile cells in liquid culture (11).
FIG 2.
Overexpression of σB is toxic for C. difficile and leads to plasmid loss. (A) Tenfold serial dilutions on brain heart infusion broth supplemented with 0.5% (wt/vol) yeast-extract (BHIY) agar plates of the 630Δerm (wild-type), AP34 (Ptet-slucopt), and JC096 (Ptet-sigB) strains. Similar results were obtained for strains IB58 and IB61 (data not shown). (B) Percentages of cells retaining the plasmid in AP34 (Ptet-slucopt) and JC096 (Ptet-sigB). (C) Percentages of cells retaining the plasmid in strains IB61 (sigB::CT Ptet-slucopt) and IB58 (sigB::CT Ptet-sigB). Percentages were calculated based on the ratio of CFU/ml of the paired selective (with thiamphenicol) and nonselective (without thiamphenicol) plates. *, P < 0.05; ****, P ≤ 0.0001, as determined by an unpaired Student’s t test (n = 3).
The lethality associated with σB overproduction was not seen when cells were grown without thiamphenicol in our experiment (Fig. 2A). We considered two possible explanations for this observation. As thiamphenicol is used for ensuring plasmid maintenance, its absence might result in plasmid loss due to segregation or negative selection pressure when a toxic protein such as σB is overproduced. The remaining cells that no longer express σB would consequently be susceptible to thiamphenicol (due to the loss of catP) but might outgrow those carrying the plasmid. Alternatively, the combination of σB and thiamphenicol might be toxic to the bacteria. To test whether plasmid loss was the cause of the observed lethality of bacteria overproducing σB in the presence of thiamphenicol, cells from the plates without thiamphenicol (with and without ATc) were resuspended in phosphate-buffered saline (PBS) at 1.0 McFarland turbidity and 10-fold serially diluted in brain heart infusion (BHI) medium. Spots (10 μl) of these dilutions were plated on plasmid-selective (thiamphenicol) and nonselective (no thiamphenicol) plates. Based on the ratio of CFU/ml of the selective and nonselective plates, the percentage of cells which lost their plasmid was calculated. If σB overproduction led to the loss of the plasmid under conditions that do not select for its maintenance (no thiamphenicol), we expected significantly reduced growth on plates containing thiamphenicol. Although some plasmid loss was observed under uninduced conditions, as well as for the negative-control strain AP34 (Ptet-slucopt), all cells originally containing the Ptet-sigB plasmid (strain JC096) completely lost this plasmid upon induction of σB overproduction with ATc (Fig. 2B). Similar results were obtained for sigB mutant strains IB58 (sigB::CT Ptet-sigB) and IB61 (sigB::CT Ptet-slucopt), indicating that the observed effects were solely due to in trans σB overproduction and did not result from an interference of the native sigB regulatory network (Fig. 2C). Together, these results are consistent with a model in which the vector with the low-copy-number pCD6 replicon is rapidly eradicated upon expression of a gene (here sigB) that causes lethal defects (22, 23).
σB primarily activates genes relating to oxidative/nitrosative stress responses.
Above, we have shown that long-term overproduction σB is detrimental and that this leads to loss of the expression plasmid in the absence of thiamphenicol (Fig. 2), but that σB overproduction nevertheless could clearly be demonstrated when induction is limited to 1 h (Fig. 1C). Therefore, we used the time-limited induction to refine the previously proposed regulon (19) in both the presence and absence of thiamphenicol to strike a balance between potential secondary effects due to toxicity associated with σB overproduction (with thiamphenicol), and loss of the expression plasmid from a subpopulation of cells (without thiamphenicol) (Table 1). We compared transcriptome data from strain IB58 (sigB::CT Ptet-sigB) to that of strain IB61 (sigB::CT Ptet-slucopt). IB61 harbors a vector for the inducible expression of a luciferase gene that does not lead to any toxicity or growth phenotype (24).
TABLE 1.
Setup of the DNA array and numbers of differentially expressed genes, including numbers of positively and negatively regulated genes
| Hybridization setup | Control | Target | Conditions | No. of genesa |
||
|---|---|---|---|---|---|---|
| DE | POS | NEG | ||||
| 1 | sigB::CT Ptet-slucopt (IB61) | sigB::CT Ptet-sigB (IB58) | Lincomycin (20 μg/ml), thiamphenicol (20 μg/ml), no ATc | 5 | 4 | 1 |
| 2 | sigB::CT Ptet-slucopt (IB61) | sigB::CT Ptet-sigB (IB58) | Lincomycin (20 μg/ml), thiamphenicol (20 μg/ml), ATc (100 ng/ml) | 183 (178) | 167 (163) | 16 (15) |
| 3 | sigB::CT Ptet-slucopt (IB61) | sigB::CT Ptet-sigB (IB58) | Lincomycin (20 μg/ml), no thiamphenicol, no ATc | 5 | 4 | 1 |
| 4 | sigB::CT Ptet-slucopt (IB61) | sigB::CT Ptet-sigB (IB58) | Lincomycin (20 μg/ml), no thiamphenicol, ATc (100 ng/ml) | 150 (145) | 136 (132) | 14 (13) |
Numbers in brackets correspond to the number of differentially expressed genes after subtracting the differentially expressed genes identified in hybridizations 1 and 3 (that are not dependent on sigB induction). DE, differentially expressed; POS, positively regulated; NEG, negatively regulated.
We expected no genes or a limited number of genes to be differentially expressed (log2 fold change [log2FC] of ≤−1.5 or ≥1.5 and adjusted P value of <0.05) under noninducing conditions. Indeed, we found only five differentially expressed genes in the Ptet-sigB strain (IB58) compared to the Ptet-slucopt control (IB61) strain (hybridizations 1 and 3) (see Data Set S1 in the supplemental material). These genes were similarly positively (CD0583 and CD0584, both GGDEF domain-containing proteins [25], and CD2214 and CD2215, both potential transcriptional regulators [26]) and negatively (CD1616, an EAL domain protein [25]) regulated in all hybridizations, including those where sigB expression was not induced. These results suggest that the basis for the observed differential expression of these genes was vector specific but not dependent on σB induction. These genes were therefore not investigated further and are excluded from the numbers discussed below.
Genes differentially expressed in σB-overproducing cells compared to controls. Cells were harvested from cultures with (induced) or without (uninduced) anhydrotetracycline (ATc), and with (+Tm) or without (−Tm) thiamphenicol. Gene name is the generic gene name (or locus tag if a gene name is not available). log2FC is the log2 of the fold change in gene expression. Four different comparisons are shown, and genes are aligned between comparisons. Upregulated genes are indicated in green. Downregulated genes are indicated in red. Genes not considered not part of the σB regulon are highlighted in yellow. Download Data Set S1, XLSX file, 0.04 MB (37.7KB, xlsx) .
Copyright © 2020 Boekhoud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
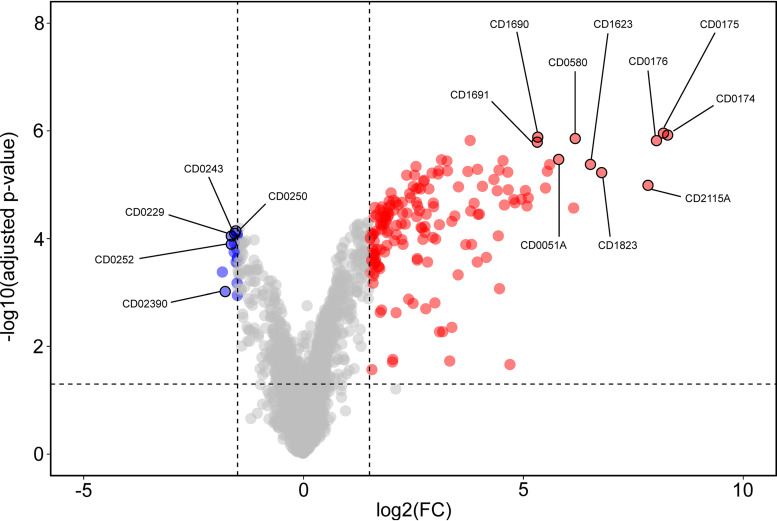
Upon induction of sigB expression, 145 genes were differentially expressed when strains were cultured without thiamphenicol (hybridization 4), and 178 genes were differentially expressed when thiamphenicol was present during cultivation (hybridization 2) (Fig. 3 and Table 1 and Data Set S1). The majority showed an increase in expression upon induction of sigB expression (132 in the samples without thiamphenicol and 163 in the samples with thiamphenicol), consistent with its function as a sigma factor (27), while a minority revealed a decreased expression (13 in the samples without thiamphenicol and 15 in the samples with thiamphenicol). Of note, we observed only a minor difference in the number of differentially expressed genes between the cells grown in the absence and presence of thiamphenicol (33 genes).
FIG 3.
Volcano plot of the transcriptome analysis of the σB regulon. Graphical representation of differential gene regulation upon overproduction of σB. Dashed lines indicated the following significance threshold: |log2FC| of >1.5 and adjusted P value of <0.05. Genes significantly upregulated by σB are indicated in red, and downregulated genes are indicated in blue. The top 10 upregulated genes and 5 selected downregulated genes are annotated in the figure. An interactive version of the graph is available for exploration via the URL provided in Text S1 in the supplemental material.
Link to access an interactive version of the volcano plot based on hybridization 2 (cells harvested from cultures grown in the presence of both anhydrotetracycline and thiamphenicol). Download Text S1, PDF file, 0.05 MB (50.9KB, pdf) .
Copyright © 2020 Boekhoud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Together, these results demonstrate a high level of consistency in the σB regulon despite potential plasmid loss (when grown in the absence of thiamphenicol) or toxic effects (when grown in the presence thiamphenicol). Our results also show that σB primarily activates gene expression.
We focused our further analyses on the data obtained from hybridization 2 (with ATc and thiamphenicol), as this condition provided the broadest data set (178 differentially expressed genes) for the redefinition of the σB regulon under our experimental conditions (Data Set S1).
Of the 163 genes upregulated by σB, the vast majority appeared to be associated with an response to oxidative stress, since they encode various oxidoreductases, peroxidases, and thioredoxin reductases (Table 2). Notably, approximately 51% of the 98 genes previously found to be upregulated under aerobic stress (7) were also positively regulated by σB (Table 2). Five additional genes associated with aerobic/nitrosative stress (cooS [cd0174], iscS2 [cd1279], the cd1280 gene, cysK [cd1594], the cd1823 gene, and msrAB [cd2166]) were also found to be induced by σB, in agreement with previous findings (19).
TABLE 2.
Selected genes differentially expressed upon overexpression of σBa
| Gene group or locus tagb | Gene name | log2FC | Adjusted P value | Predicted function |
|---|---|---|---|---|
| Genes upregulated by aerobic stress and positively regulated by σB | ||||
| CD630DERM_00530 | mrnC | 1.9 | 3.29E-05 | Ribonuclease III domain |
| CD630DERM_01750 | 8.2 | 1.11E-06 | Oxidoreductase, Fe-S subunit | |
| CD630DERM_01760 | 8.0 | 1.52E-06 | Oxidoreductase, NAD/flavin adenine dinucleotide (FAD) binding subunit | |
| CD630DERM_01920 | cls | 2.6 | 2.35E-04 | Cardiolipin synthetase 1 |
| CD630DERM_03500 | 1.9 | 4.63E-05 | Putative hydrolase, HAD superfamily | |
| CD630DERM_03510 | 1.8 | 5.66E-05 | Conserved hypothetical protein | |
| CD630DERM_05600 | nfo | 4.0 | 3.5E-05 | Endonuclease IV |
| CD630DERM_05610 | 2.0 | 2.39E-05 | Putative aldo-/ketoreductase; putative ferredoxin | |
| CD630DERM_05650 | nth | 3.7 | 5.66E-06 | Endonuclease III |
| CD630DERM_05660 | 2.5 | 6.62E-06 | Putative tRNA/rRNA methyltransferase | |
| CD630DERM_05800 | gapN | 6.2 | 1.38E-06 | Glyceraldehyde-3-phosphate dehydrogenase (NADP+) (GAPDH) |
| CD630DERM_11250 | 5.5 | 1.15E-05 | Nitroreductase family protein | |
| CD630DERM_11570 | norV | 4.0 | 5.16E-06 | Anaerobic nitric oxide reductase flavorubredoxin (FlRd) (FlavoRb) |
| CD630DERM_13410 | exoA | 1.8 | 6.76E-05 | Exodeoxyribonuclease |
| CD630DERM_14630 | 1.8 | 1.77E-04 | Conserved hypothetical protein | |
| CD630DERM_15240 | 4.8 | 1.88E-05 | Putative rubrerythrin | |
| CD630DERM_15260 | pyrC | 1.6 | 1.98E-04 | Dihydroorotase |
| CD630DERM_15760 | 1.6 | 2.71E-02 | Putative arylesterase | |
| CD630DERM_16220 | 2.7 | 1.03E-04 | Putative lipoprotein | |
| CD630DERM_16230 | 6.5 | 4.21E-06 | NADH-oxygen oxidoreductase | |
| CD630DERM_16240 | vanR | 1.6 | 8.84E-05 | Two-component response regulator |
| CD630DERM_16900 | trxA | 5.3 | 1.31E-06 | Thioredoxin reductase |
| CD630DERM_16910 | trxB | 5.3 | 1.62E-06 | Putative thioredoxin disulfide reductase |
| CD630DERM_17780 | 2.6 | 1.19E-05 | Conserved hypothetical protein | |
| CD630DERM_17790 | 2.4 | 1.46E-05 | Conserved hypothetical protein | |
| CD630DERM_18180 | ispH | 1.8 | 5.35E-05 | 4-Hydroxy-3-methylbut-2-enyl diphosphate reductase |
| CD630DERM_18220 | bcp | 6.1 | 2.70E-05 | Putative thiol peroxidase |
| CD630DERM_18970 | 1.8 | 3.20E-05 | Conserved hypothetical protein | |
| CD630DERM_19430 | 3.3 | 3.62E-06 | Conserved hypothetical protein | |
| CD630DERM_20460 | 5.5 | 5.60E-06 | Conserved hypothetical protein | |
| CD630DERM_21170 | trxB2 | 5.0 | 1.86E-05 | Thioredoxin reductase |
| CD630DERM_21650 | 2.2 | 1.04E-04 | Transcriptional regulator, helix-turn-helix (HTH)-type | |
| CD630DERM_24760 | 4.6 | 5.84E-06 | Conserved hypothetical protein | |
| CD630DERM_27960 | cwp10 | 5.1 | 2.47E-05 | Cell surface protein |
| CD630DERM_27970 | 5.1 | 1.75E-05 | Putative calcium binding adhesion protein | |
| CD630DERM_29930 | 3.8 | 3.07E-05 | Conserved hypothetical protein | |
| CD630DERM_30380 | 3.9 | 2.21E-05 | Conserved hypothetical protein | |
| CD630DERM_30390 | 4.1 | 1.07E-05 | Putative ATPase | |
| CD630DERM_30400 | 2.1 | 8.46E-05 | Conserved hypothetical protein | |
| CD630DERM_30420 | 2.6 | 2.47E-05 | Putative membrane protein | |
| CD630DERM_33070 | 3.8 | 1.51E-06 | Putative phosphoesterase | |
| CD630DERM_33100 | 3.1 | 6.31E-06 | Putative d-isomer specific 2-hydroxyacid dehydrogenase | |
| CD630DERM_33110 | 2.9 | 1.19E-05 | Conserved hypothetical protein | |
| CD630DERM_34080 | 2.3 | 3.38E-05 | Putative DNA mismatch repair ATPase MutS | |
| CD630DERM_34090 | scoC (hprK) | 2.4 | 1.74E-05 | Phosphotransferase (PTS) system, HPr kinase/phosphorylase |
| CD630DERM_34100 | uvrC | 2.7 | 1.19E-05 | Excinuclease subunit C |
| CD630DERM_34730 | atpC2 | 1.6 | 2.90E-04 | ATP synthase C chain |
| CD630DERM_34760 | atpZ | 1.6 | 2.42E-04 | Putative ATP synthase protein |
| CD630DERM_36100 | 4.6 | 2.38E-05 | Conserved hypothetical protein | |
| CD630DERM_36140 | 2.5 | 1.07E-05 | Conserved hypothetical protein, DUF1130 family | |
| Other genes involved in oxidative/nitrosative stress positively regulated by σB | ||||
| CD630DERM_01740 | cooS | 8.3 | 1.20E-06 | Carbon monoxide dehydrogenase |
| CD630DERM_12790 | iscS2 | 2.8 | 2.00E-03 | Cysteine desulfurase |
| CD630DERM_12800 | 3.0 | 1.56E-03 | Putative NifU-like protein | |
| CD630DERM_14740 | 5.0 | 1.28E-05 | Putative rubrerythrin (Rr) | |
| CD630DERM_15940 | cysK | 3.4 | 4.44E-03 | O-acetyl-serine thiol-lyase A (O-acetyl-sulfhydrylase) (OAS-TL) |
| CD630DERM_18230 | 6.8 | 5.93E-06 | Conserved hypothetical protein, UPF0246 family | |
| CD630DERM_21660 | msrAB | 4.7 | 1.24E-05 | Peptide methionine sulfoxide reductase MsrA/MsrB |
Genes positively regulated by σB and aerobic stress and other genes involved in oxidative/nitrosative stress (7, 19) are highlighted here.
CD numbers corresponding to the published annotation of strain CD630 (52) can be derived by removing “630DERM” and removing the last digit (in case of a 0) or replacing it with an A (in the case of a 1).
Our findings are recapitulated in a volcano plot (28), which clearly shows that genes with lower expression upon sigB induction (in blue) cluster close to the significance threshold, whereas those with increased expression (in red) show a larger fold change (Fig. 3). We calculated the Manhattan distance for each data point (see Data Set S2 in the supplemental material), and discuss the proteins encoded by the top 10 differentially expressed genes below.
Manhattan distance for differentially regulated genes from hybridization 2 (cells harvested from cultures grown in the presence of both ATc and thiamphenicol). Change indicates whether expression is higher (increased) or lower (decreased) upon overproduction of σB. log2FC represents log2 of the fold change in gene expression. Significance is given as −10 · log of the adjusted P value (see Data Set S1). Manhattan distance was calculated by and exported using the online tool VolcaNoseR. Download Data Set S2, XLSX file, 0.02 MB (16.1KB, xlsx) .
Copyright © 2020 Boekhoud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CD0051A is a small hypothetical protein of unknown function. It does not contain any recognizable domains, and a secondary structure prediction using Phyre2 does not give any clues as to its potential function (29). CD0580 (GapN) is annotated as a glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a key glycolytic enzyme, and contains an aldehyde dehydrogenase domain. Interestingly, its activity has been shown to be redox controlled in other bacteria and has been implicated in the response to reactive oxygen and nitrogen species (30–32). CD1623 is a putative oxidoreductase with similarity to FAD flavoproteins and rubredoxins. CD1690 (TrxA) and CD1691 (TrxB) are likely encoded in the same operon (33) and form a thioredoxin/thioredoxin-disulfide reductase couple. CD0174 (CooS; InterPro family IPR010047), CD0175, and CD0176 are likely also encoded in a single operon (33) and function as carbon monoxide dehydrogenase and two putative oxidoreductases. As mentioned above, CD0174 has been implicated in aerobic/nitrosative stress, and it is likely that CD1623, CD1690, and CD1691 also function in this pathway. Finally, CD2115A encodes another small hypothetical protein; as for CD0051A, no function could be assigned on the basis of secondary structure prediction.
As the σB regulon that we define here is substantially smaller than that previously reported, the major conclusion is that at least 32% of the σB regulon is involved in positively regulating oxidative/nitrosative stress responses. In the previous investigation of the σB regulon they were approximately 3.2% (≈32/1,000) (19). Overall, we conclude that the core functions of the σB regulon lie in the regulation of the detoxification response to oxygen and nitro radicals.
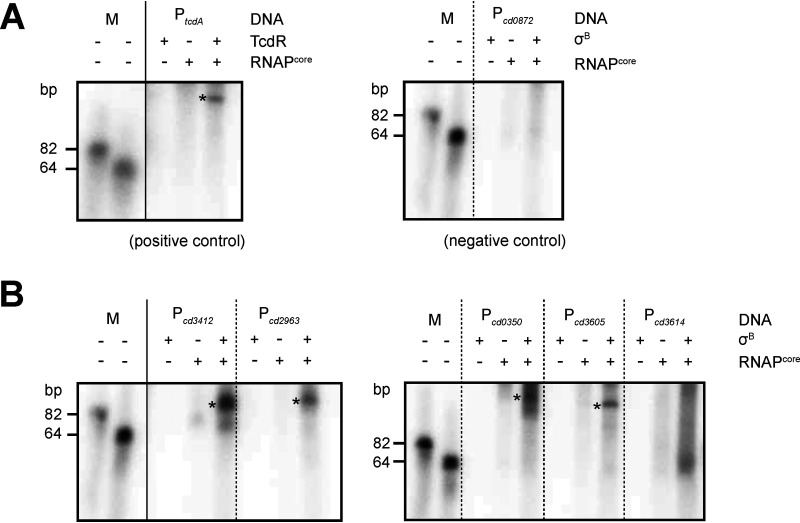
In vitro runoff transcriptions demonstrate direction activation of Pcd0350, Pcd2963, Pcd3412, and Pcd3605 by σB.
Gene expression can directly or indirectly be influenced by σB, and to date no attempts have been made to discriminate these possibilities biochemically (11, 19). Despite the short time of induction and the uncoupling of σB from its normal regulatory network, our analyses could possibly also have picked up indirect effects. To determine if the transcription of selected genes is directly activated by σB, in vitro transcription runoff reactions were performed using purified σB6×His and RNA polymerase core enzyme (RNAPcore) on the upstream regions of a selection of genes. The genes cd0350 (encoding a putative hydrolase involved in oxidative stress; Table 2), cd2963 (encoding an l,d-transpeptidase), cd3412 (encoding UvrB, involved in nucleotide excision repair), cd3605 (encoding a ferredoxin), and cd3614 (encoding a hypothetical protein involved in oxidative stress; Table 2) were selected on the basis of differential expression in our transcriptome analyses (Data Set S1) and those of others (19), availability of reporter constructs that could be used to generate a template for the in vitro transcription reactions (20), and/or the presence of a putative σB consensus upstream in the upstream region (11). The gene cd0872 (encoding maltose O-acetyltransferase) was not differentially expressed in our transcriptome data and was thus included as a negative control. The promoter of the toxin A gene (tcdA) in combination with purified TcdR was used as a positive control for the assay, as previously described (34).
As expected, no in vitro transcript was observed for a linear DNA fragment containing Pcd0872 incubated with purified σB6×His and RNAPcore under our experimental conditions, whereas a specific product was obtained for the positive control PtcdA in the presence of TcdR and RNAPcore (Fig. 4). An RNAPcore- and σB6×His-specific signal was observed for fragments containing the putative promoter regions of the genes cd0350, cd2963, cd3412, and cd3605, demonstrating that expression of these genes was directed by σB. For the fragments containing the putative promoter of cd3614, we did not get a consistent product in the in vitro transcription experiments, although some smearing is visible in the lane with RNAPcore and σB6×His. As cd3614 demonstrates clear differential expression in the DNA array experiments and it upstream region harbors the σB consensus sequence WGWTT-N13-17-(G/T)GGTWA (19), we consider it likely that this gene is directly regulated by σB and that our failure to obtain a discrete signal is due to our experimental conditions or to the lack of an auxiliary factor in our in vitro assays.
FIG 4.
In vitro runoff transcription of selected promoter regions. Samples were run on an 8% urea gel. The two bands corresponding to 82 bp and 64 bp are end-labeled oligonucleotides. Reactions without sigma factor (σB or TcdR, respectively) or RNAPcore were analyzed as controls. Asterisks indicate the presence of distinct transcripts. RNAPcore, Escherichia coli RNA polymerase core enzyme (NEB). (A) Controls for the in vitro runoff transcriptions. Purified TcdR, a sigma factor demonstrated to activate tcdA transcription in vitro (34), was used with PtcdA (from plasmid pCD22) as a positive control for the assay. Pcd0872 (derived from pIB21) shows no altered transcription in the DNA array analysis and was taken along as a negative control. (B) In vitro runoff transcriptions for selected genes induced by σB overproduction.
Overall, we provide the first biochemical evidence for direct σB-dependent activation of several genes identified via transcriptome analyses as part of the σB regulon in C. difficile.
Antimicrobials and hydrogen peroxide activate σB-directed gene transcription.
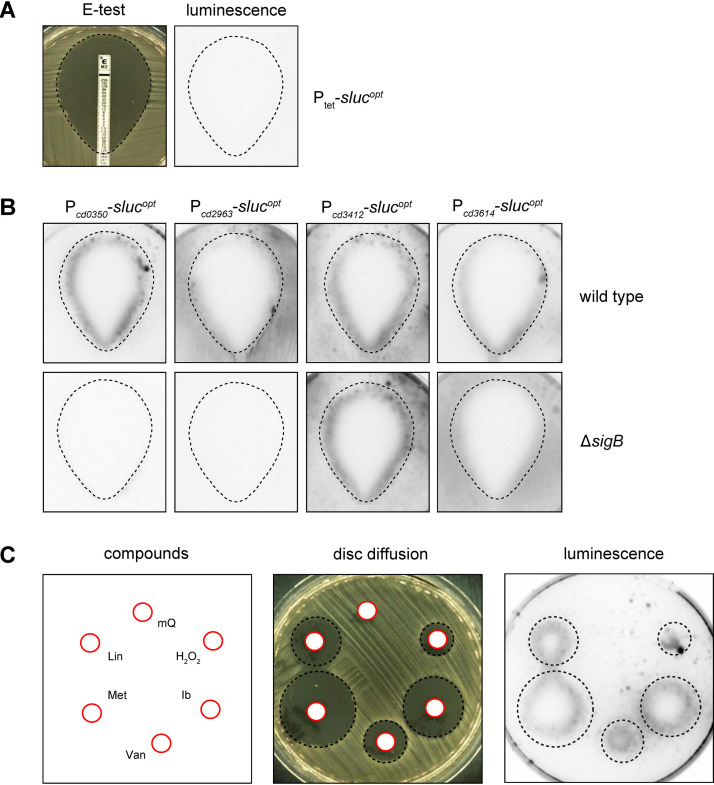
The redefined σB regulon points toward a substantial role for σB in coordinating the oxidative and nitrosative stress response, which could result from antimicrobial treatment. In order to test for the activation of σB-dependent promoters by antimicrobials, we set up a plate-based luciferase reporter assay. In this assay, cells harboring σB-dependent luciferase reporter constructs were plated on BHIY agar to give confluent growth and exposed to antimicrobials either through an epsilometer test (Etest) or through a filter disc. Subsequently, luciferase activity was imaged (for details, see Materials and Methods). A strain harboring Ptet-slucopt (AP34) served as negative control, as this promoter is not expected to respond in a σB-dependent manner (Fig. 5A).
FIG 5.
Plate-based luciferase assay shows σB-dependent promoter activity from antimicrobial and hydrogen peroxide exposure. (A) Setup of the assay. Etest halos (left) were sprayed with luciferase substrate and imaged (right). The dotted line indicates the location of the halo based on the left panel. Strain AP34 (Ptet-slucopt) shows no signal due to the absence of inducer. (B) Luciferase reporters of different σB-regulated promoters were tested for luciferase signal after a metronidazole Etest. Halos are indicated by the dashed lines as in panel A. (C) Luciferase activity of the Pcd0350 luciferase reporter was imaged in the presence of discs containing 10 μl of the following compounds: sterile H2O (mQ), lincomycin (Lin; 3,000 μg/ml), metronidazole (Met; 200 μg/ml), vancomycin (Van; 200 μg/ml), ibezapolstat (Ib; 400 μg/ml) and hydrogen peroxide (H2O2; 1 M). Halos and the location of the different stressors are indicated by red circles (disc) and black dashed circles (halo). Signals outside the halo in these images are representative for the signals across the plate. Experiments were performed at least in triplicate with qualitatively similar results.
First, the σB-dependent response to metronidazole was investigated. Metronidazole, formerly used as a first-line treatment for CDI, is believed to cause DNA damage through the formation of nitro radicals, although its exact mode of action remains unclear (6, 35). To survey a full spectrum of metronidazole concentrations, we evaluated luminescence after 24-h incubation of a metronidazole Etest. If metronidazole treatment results in σB-dependent activation of gene transcription, we expect to see a luciferase signal in the wild type but not in a σB knockout background. In agreement with this, activation of Pcd0350 was observed at the edge of the halo resulting from the metronidazole Etest in the wild-type background but not in the σB knockout strain (Fig. 5B). No signal was observed for the negative control Ptet-slucopt (Fig. 5A). The observed σB-dependent activation of gene expression at the edge of the halo but not further into the plate suggests that the metronidazole-induced, σB-dependent activation of Pcd0350 occurred close to the MIC. Expression of the luciferase from Pcd2963 was found to be strictly dependent on σB, as no luciferase activity was observed in the sigB knockout strain. However, there was limited to no increase in reporter gene expression in the presence of metronidazole. Metronidazole strongly activated transcription from Pcd3412 at MIC levels of metronidazole, but this appeared to be independent of σB in this assay since in the sigB mutant a similar induction was observed. Finally, in a manner comparable to that of Pcd0350, the activation of PCD3614 was strongly induced by metronidazole at values close to the MIC in a σB-dependent manner, but residual activity was observed in the σB knockout strain independent of metronidazole levels. We noted that metronidazole-induced promoter activation appeared to occur on the inside of the Etest halo, which might be attributed to the secretion of the luciferase reporter.
The observed diverse regulatory responses at different tested promoters during the treatment of C. difficile with metronidazole (with respect to basal level, sigB dependence, and induction) pointed toward a more complex regulatory network with the participation of σB but also influenced by other factors. Antimicrobial-driven (and σB-dependent) activation of σB target genes could be specific to metronidazole or represent a more general response to cellular (toxic) stresses. Therefore, we evaluated the effects of different antimicrobial compounds and the radical producer H2O2 as a positive control (19), using the Pcd0350 reporter construct, as this promoter demonstrated the clearest σB-dependent activation in the presence of metronidazole (Fig. 5B). We tested the cell wall biosynthesis inhibitor vancomycin, the protein synthesis inhibitor lincomycin, and the DNA polymerase inhibitor ibezapolstat (formerly known as ACX-362E) (20). We observed clear activation of Pcd0350 in the presence of all added stressors but not for a negative control containing water (Fig. 5C).
We conclude that, at least for the σB-dependent promoter of cd0350, activation does not only occur upon exposure to lethal levels of metronidazole but also occurs with unrelated antimicrobials and toxic stressors such as hydrogen peroxide.
DISCUSSION
In this work, we have demonstrated by Western blotting using an affinity-purified anti-σB antibody that σB can be overproduced for a limited period of time, sufficient for transcriptome analyses. The induced production of σB in a sigB mutant background yielded highly consistent results despite potential toxicity and plasmid loss (Fig. 2), and the results were used to redefine the surprisingly large σB regulon previously proposed (19). As our approach more accurately measures changes in transcription directly related to σB production, the refined regulon described here is much smaller (see Data Set S1 in the supplemental material). Its size is fully in line with that of the σB regulon of other Gram-positive bacteria such as L. monocytogenes (≈130 genes), B. subtilis (≈150 genes), and S. aureus (≈200 genes) (8). The redefined regulon underscores the importance of σB in responding to oxidative/nitrosative stresses, as genes implicated in such processes are significantly enriched in the smaller regulon.
The majority of the genes in our regulon were found to be induced, rather than repressed, by σB. This is in line with sigma factors acting as specificity determinants for transcription initiation (27). Similar observations have been made for the σB regulon of L. monocytogenes (36, 37). For the first time, direct evidence of C. difficile σB-dependent gene activation is provided by the results of the in vitro runoff transcriptions (Fig. 4), which demonstrate that RNAPcore and σB are sufficient to generate transcripts from Pcd0350, Pcd2963, Pcd3412, and Pcd3605. Notably, these experiments pave the way for a further in vitro characterization of this sigma factor in C. difficile, including validation of the σB binding sequence and the interplay with other regulators.
Although the promoters of cd3412 (uvrB) and cd3614 were reported to have a σB consensus sequence and are differentially expressed upon σB overexpression (19, 20), our results clearly demonstrate that they can also be expressed in a σB-independent manner (Fig. 5B). This is most notable for Pcd3412, which is still activated by metronidazole in the absence of σB, in line with results obtained with ibezapolstat in a different study (20). Both metronidazole and ibezapolstat treatment can cause DNA damage, and DNA-damage dependent induction of cd3412 therefore likely depends on a sigB-independent pathway.
The observed σB-dependent gene repression is expected to be indirect (σB induces the transcription of a repressor gene), or the result of competition (σB competes with other sigma factors for RNAP), as sigma factors by their very nature induce gene expression (27). We consider the second scenario more likely for the following reasons. First, little overproduction of σB was detected after 30 min of induction. This leaves only a limited time for indirect effects to occur in our setup. Second, the majority of genes downregulated upon overexpression of σB fall into a single functional group (flagellar motility). These genes are known to be regulated by the dedicated sigma factor, σD (38), supporting the model of sigma factor competition. Strikingly, in L. monocytogenes, σB activity (indirectly) also results in downregulation of flagellar gene expression, but this is mediated by the repressor MogR (39). Protein BLAST analyses revealed that C. difficile does not possess a MogR homologue. Nevertheless, the conserved inverse correlation between the σB-dependent general stress response and bacterial motility could represent a cost-saving strategy for bacterial cells (40). The indirect mechanism underlying the observed σB-dependent downregulation in C. difficile remains to be determined.
There appears to be an intriguing link between σB and the response to toxic compounds, as a sigB mutant was more susceptible to rifampin and mitomycin C (19), and exposure to antimicrobials (metronidazole, vancomycin, lincomycin, and ibezapolstat) or hydrogen peroxide leads to σB-dependent promoter activation (Fig. 5C). The mechanism behind the latter is unclear. It has been suggested that antimicrobials at toxic concentrations can influence metabolism and respiration (41, 42), potentially resulting in the formation of bactericidal concentrations of radical species (43–45). A strong connection between σB and oxidative (and/or nitrosative) stress in C. difficile (Table 2) and other bacteria (7, 18, 19), as well as a recently described radical scavenging strategy that increases tolerance to antimicrobials (46), are consistent with such a model. However, additional research is necessary to determine exactly how these processes occur and are influenced by antimicrobials in anaerobic organisms under anoxic conditions.
In conclusion, we have demonstrated that σB is directly involved in metabolic and oxidative stress responses and that lethal stresses may influence these processes, resulting in activation of σB-targeted genes.
MATERIALS AND METHODS
Construction of σB expression and luciferase reporter vectors.
All oligonucleotides used in this study can be found in Table 3. Plasmids and strains are listed in Table 4. All PCR products used for sequencing or plasmid synthesis were generated with Q5 high-fidelity polymerase (NEB). The PT7-sigB6×His expression vector pIB14 was created by restriction-ligation using the restriction enzymes NdeI and XhoI. Using primers oIB-1 and oIB-2 on C. difficile 630Δerm chromosomal DNA, the sigB coding sequence (CDS) was amplified by PCR. The resulting DNA fragment was digested and ligated into NdeI-XhoI-digested pET21b(−) vector, generating expression vector pIB14. Plasmids pIB27, pIB68, pIB69, and pIB74 have been described previously (20). The cd0872 promoter area was amplified using primers oIB-14 and oIB-15, and the Pcd0872 luciferase-reporter plasmid was created by restriction-ligation using restriction enzymes KpnI and SacI in digested pAP24 backbone, generating plasmid pIB21. The Pcd3605 luciferase reporter plasmid was generated by Gibson assembly as described previously (20) using primers oIB-90 and oIB-99, yielding plasmid pIB73. A plasmid containing Ptet-sigB was generated by cloning the 630Δerm sigB CDS amplified with oWKS-1498 and oWKS-1499 in pMiniT (catalog no. E1202; NEB) per the manufacturer’s instructions. Using restriction enzymes SacI and BamHI, this PCR fragment was cloned into pRPF185, yielding pWKS1760. All plasmids were verified by Sanger sequencing.
TABLE 3.
Oligonucleotides used in this study
| Name | Sequence (5′–3′)a | Description | Source or reference |
|---|---|---|---|
| Cdi-sigB-F | GTAGCTAATGCTACACATTAC | Verification of sigB ClosTron mutant | This study |
| Cdi-sigB-R | CAGTCATCTGTGATATCCCTAG | Verification of sigB ClosTron mutant | This study |
| EBSuniversal | CGAAATTAGAAACTTGCGTTCAGTAAA | Verification of sigB ClosTron mutant | 49 |
| ErmRAM-F | ACGCGTTATATTGATAAAAATAATAGTGGG | Verification of sigB ClosTron mutant | 49 |
| ErmRAM-R | ACGCGTGCGACTCATAGAATTATTTCCTCCCG | Verification of sigB ClosTron mutant | 49 |
| oIB-1 | TAGCCATATGAAAAATGTAGCTAATGCTACAC | Forward primer for sigB cloning in pET21b, contains a NdeI restriction site | This study |
| oIB-2 | ACTGCTCGAGTAAATTTTTTTCATATTCTTTTTTCAG | Reverse primer for sigB cloning in pET21b, contains an XhoI restriction site | This study |
| oIB-14 | GTACGGTACCTTTACATATACTATATATGTTAGAAAAAC | Forward primer for Pcd0872 containing a KpnI restriction site | This study |
| oIB-15 | GGTAGAGCTCATAGTTTACTCCTTTTTGTTATAATTG | Reverse primer for Pcd0872 containing a SacI restriction site | This study |
| oIB-26 | GGAAGGTACCGTTGAATAAAGTATTTATTTTCCATG | Forward primer for Pcd3412 containing a KpnI restriction site | 20 |
| oIB-27 | GGTAGAGCTCAGTATCACTCCTTTTTTCGAAC | Reverse primer for Pcd3412 containing a SacI restriction site | 20 |
| oIB-80 | ctagcataaaaataagaagcctgcatttgcAAATTTACGAAAAGCTTGC | Forward primer for Pcd0350 | 20 |
| oIB-82 | ctagcataaaaataagaagcctgcatttgcTTGTGTTTAAGGGATTTTGAAAG | Forward primer for Pcd2963 | 20 |
| oIB-90 | ctagcataaaaataagaagcctgcatttgcATGTAAAGAAGCCGAAGAAG | Forward primer for Pcd3605 | This study |
| oIB-92 | ctagcataaaaataagaagcctgcatttgcGAATAAAAAAGGTGGTGTC | Forward primer for Pcd3614 | 20 |
| oIB-94 | agctattaataattttttacttggtctcatTTTTACCTCCATGTAACATTTATTG | Reverse primer for Pcd0350 | 20 |
| oIB-95 | agctattaataattttttacttggtctcatAATTAAATCCTTCCTTACATTGTAATTAC | Reverse primer for Pcd2963 | 20 |
| oIB-99 | agctattaataattttttacttggtctcatATTTCAGCCCTCCATATTTG | Reverse primer for Pcd3605 | This study |
| oIB-100 | agctattaataattttttacttggtctcatATAAACACCCTCCTATTCTTTG | Reverse primer for Pcd3614 | 20 |
| oWKS-1498 | GAGCTCCTGCAGTAAAGGAGAAAATTTTATGAAAAATGTAGCTAATGCTACAC | Forward primer for sigB CDS | This study |
| oWKS-1499 | GGATCCTTATAAATTTTTTTCATATTCTTTTTTCAG | Reverse primer for sigB CDS | This study |
| oWKS-1513 | GAGCTCAAATTTGAATTTTTTAGGGGGAAAATACCATGCATCATCACCATCACCACGGTTCCGAAATCGGTACTGGCTTTCC | Oligonucleotide used for end labeling | This study |
| oWKS-1506 | GAGCTCAAATTTGAATTTTTTAGGGGGAAAATACCATGGTTTCAAAAGGAGAAGAATTATTTAC | Oligonucleotide used for end labeling | This study |
Restriction sites are underlined, and 30-bp overlapping regions used in Gibson Assembly are indicated in lowercase letters.
TABLE 4.
Plasmids and strains used in this study
| Name of plasmid or strain | Relevant featuresa | Source or reference |
|---|---|---|
| Plasmids | ||
| pAP24 | Ptet-slucopt; catP | 24 |
| pCD22 | PtcdA; catP | 53 |
| pIB14 | PT7-sigB6×His; amp | This study |
| pIB21 | Pcd0872-slucopt; catP | This study |
| pIB27 | Pcd3412-slucopt; catP | 20 |
| pIB68 | Pcd0350-slucopt; catP | 20 |
| pIB69 | Pcd2963-slucopt; catP | 20 |
| pIB73 | Pcd3605-slucopt; catP | This study |
| pIB74 | Pcd3614-slucopt; catP | 20 |
| pMTL007C-E2_sigB171s::intron_ermB | sigB-retargeted intron (ermB) | This study |
| pWKS1750 | Ptet-sigB; catP | This study |
| Strains | ||
| 630Δerm | MLS-susceptible derivative of Clostridioides difficile strain 630 | 54, 55 |
| AP34 | 630Δerm pAP24; Thiar | 24 |
| DH5α | Escherichia coli F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG purB20 ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK− mK+) λ− | Laboratory stock |
| IB14 | Rosetta (DE3) pLysS pIB14; Ampr, Chlorr | This study |
| IB18 | 630Δerm sigB::CT; Ermr/Lincor | This study |
| IB37 | 630Δerm pIB27; Thiar | 20 |
| IB56 | 630Δerm ΔsigB | 20 |
| IB58 | IB18 pWKS1750; Thiar, Ermr/Lincor | This study |
| IB61 | IB18 pAP24; Thiar, Ermr/Lincor | This study |
| IB95 | 630Δerm pIB68; Thiar | 20 |
| IB96 | 630Δerm pIB69; Thiar | 20 |
| IB98 | IB56 pIB27; Thiar | 20 |
| IB99 | IB56 pIB68; Thiar | 20 |
| IB100 | IB56 pIB69; Thiar | 20 |
| IB108 | 630Δerm pIB74; Thiar | 20 |
| IB111 | IB56 pIB74; Thiar | 20 |
| JC096 | 630Δerm pWKS1750; Thiar | This study |
MLS, macrolides-lincosamides-streptogramin B; Amp, ampicillin; Chlor, chloramphenicol; Erm, erythromycin; Linco, lincomycin; Thia, thiamphenicol; r, resistance.
Bacterial strains and growth conditions.
Strains of Escherichia coli were grown aerobically at 37°C in Luria-Bertani broth (Affymetrix) supplemented with ampicillin (50 μg/ml), kanamycin (50 μg/ml), and/or chloramphenicol (20 μg/ml) when required. Plasmids were maintained in E. coli strains DH5α or MDS42 (Scarab Genomics) under appropriate antimicrobial selection, and cells were transformed using standard procedures (47). For plasmid conjugation into recipient wild-type C. difficile 630Δerm and the isogenic sigB mutant strains, E. coli strain CA434 was used as a donor strain as previously described (48). C. difficile strains were cultured anaerobically at 37°C in either a Don Whitley VA-1000 or A55 workstation. Cells were cultured in brain heart infusion (BHI; Oxoid) broth supplemented with 0.5% (wt/vol) yeast-extract (BHIY) and 20 μg/ml thiamphenicol when appropriate. Unless additional antimicrobials/stressors were added (metronidazole Etest and sterile pads supplemented with different stressors), medium was supplemented with C. difficile selective supplement (CDSS; Oxoid).
The sigB ClosTron mutant described in this study was generated as described previously (49), using pMTL007C-E2_sigB171s::intron_ermB, synthesized by DNA2.0 (now ATUM). Design of the retargeted intron was performed with the Perutka algorithm, via the ClosTron website (http://clostron.com/). The mutant was verified using primers Cdi-sigB-F, Cdi-sigB-R, EBSuniversal, ErmRAM-F, and ErmRAM-R.
Overproduction, purification and affinity purification of σB6×His for synthesis of a polyclonal anti-σB antibody.
(i) Overproduction and purification of σB6×His. Overexpression of σB6×His was performed by using Escherichia coli Rosetta (DE3) pLysS cells (Novagen) harboring the E. coli expression plasmid pIB14. These cells were cultured in Luria-Bertani (LB) broth and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 1 h starting at an optical density of ≈0.6. Cells were collected by centrifugation at 4°C, and the resulting cell pellets were resuspended in lysis buffer (pH = 8.0; 50 mM NaH2PO4, 300 mM NaCl, 5 mM β-mercaptoethanol, 0.1% NP-40, and complete protease inhibitor cocktail [CPIC; Roche Applied Science]). Through the addition of 1 mg/ml lysozyme and sonication (6 × 20 s), cells were lysed. The lysate was drawn through a blunt 1.2-mm needle and was clarified by centrifugation at 13,000 × g at 4°C for 25 min. Recombinant σB6×His was purified from the supernatant on Talon Superflow resin (GE Healthcare) per the manufacturer’s instructions. Proteins were dialyzed and stored in buffer (pH = 8.0) containing 50 mM NaH2PO4, 300 mM NaCl, and 12% glycerol. Protein concentrations were determined using a Bradford assay (Bio-Rad). Two ml of σB6×His protein solution containing 2 mg/ml protein was sent to BioGenes GmbH (Berlin) for generation of a polyclonal rabbit anti-σB antibody.
(ii) Affinity purification of the polyclonal anti-σB antibody. Affinity purification of the antibody was performed to increase specificity of σB detection. Approximately 350 μg of purified σB6×His protein was loaded onto an SDS-PAGE gel. After electrophoresis and transfer of proteins to a polyvinylidene difluoride (PVDF) membrane using standard blotting procedures, purified σB6×His protein was visualized by Ponceau S staining, and the membrane containing the protein was cut as small as possible while retaining the region with the protein. The membrane was destained and washed with Tris-buffered saline with Tween 20 (TBST) buffer (500 mM NaCl, 20 mM Tris base, and 0.05% vol/vol Tween 20 [pH = 7.4]) twice for 5 min at room temperature. The membrane was then preeluted by soaking in acidic glycine solution (100 mM, pH = 2.5) for 5 min prior to washing with TBST twice for 5 min at room temperature. Subsequently, the membrane was blocked in 5% nonfat milk powder solution (Campina Elk, dissolved in TBST buffer) for 1 h at room temperature after again washing twice with TBST for 5 min. Serum containing anti-σB antibody was incubated on the membrane overnight at 4°C. After three 5-min washes with TBST, the membrane was washed twice for 5 min in PBS. Affinity-purified antibody was eluted from the membrane by adding acidic glycine solution and incubating for 10 min at room temperature. The pH of the eluate was adjusted to 7.0 through the addition of 1 M Tris-HCl (pH = 8.0). This step was repeated twice more, and the eluates were pooled and centrifuged (1 min at maximum speed) to remove precipitated protein and membrane particles. Bovine serum albumin (BSA) and sodium azide were added to the affinity-purified anti-σB antibody to end concentrations of 1 mg/ml and 5 mM, respectively, and the affinity-purified antibody was stored at −80°C.
Characterization of the σB regulon.
(i) σB overproduction in C. difficile. Exponentially growing starter cultures of C. difficile strain IB58 and IB61 were diluted to an OD600 of 0.05 in BHIY medium supplemented with 20 μg/ml lincomycin and thiamphenicol (20 μg/ml) where appropriate. Cells were grown until an OD600 of ≈0.3, after which a 1-ml sample was taken for control by Western blotting, and gene expression was induced with 100 ng/ml ATc for 1 h. Subsequently, 1 ml of sample was taken for control by Western blotting, and 50 ml was collected by centrifugation and stored at −20°C until RNA extraction. Noninduced samples were treated and collected identically, except that no ATc was added at an OD600 ≈0.3. All samples were corrected for OD600 prior to analysis by Western blot.
(ii) RNA extraction. Bacterial RNA was extracted and analyzed as previously described (50). Briefly, cell pellets were lysed for 30 min at room temperature in enzymatic lysis buffer consisting of 15 mg/ml lysozyme and Tris-EDTA (TE) buffer. Further disruption of cells was performed by vigorous mechanical lysis for 3 min in RLT buffer to which one spatula of glass beads was added. After samples were centrifuged (3 min at 10,000 rpm at 4°C) and 100% ethanol was added to the supernatant, RNA was purified using the Qiagen RNeasy kit protocol according to manufacturer’s instructions. DNA contamination was removed by using RNase-free DNase I (Qiagen) twice prior to elution of the RNA samples in H2O. RNA quality and integrity numbers (RINs) were assessed with a Bioanalyzer 2100 (Agilent) and RNA 6000 Nano reagents (Agilent). Only samples with an RIN of ≥7 were used for further analysis.
(iii) DNA microarray and data analysis. A customized whole-genome DNA microarray of the 630Δerm strain was used (8 × 15K format; Agilent) (50). Quadruplicate samples were analyzed for the DNA microarray. Using the ULS fluorescent labeling kit for Agilent arrays (Kreatech), 1 μg of total RNA was used for labeling with either Cy3 (Ptet-sigB) or Cy5 (Ptet-slucopt). After pooling and fragmentation, 300 ng of labeled RNA per sample was hybridized according to the two-color microarray protocol from Agilent. DNA microarrays were scanned with an Agilent C scanner and analyzed as described previously (50). A gene was considered differentially expressed if the log2 fold change (log2 FC) was ≤−1.5 or ≥1.5 and the P value was <0.05. Results were visualized in VolcaNoseR (28) and are available as an interactive graph via the URL contained in Text S1 in the supplemental material.
In vitro transcription.
DNA oligonucleotides oWKS-1506 and oWKS-15136 (64 and 82 bp, respectively) were end labeled with ɣ-32P-ATP using T4 polynucleotide kinase (PNK; Invitrogen) and used as a size indicator for the in vitro transcription reactions. For the end labeling reaction, 1 μl ɣ-32P-ATP was incubated together with 200 pmol oligonucleotide and 1 μl (10 U) PNK in Forward reaction buffer (70 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 100 mM KCl, and 1 mM 2-mercaptoethanol) at 37°C for 30 min. For the in vitro runoff transcriptions, sigma factors and RNA polymerase core enzyme were preincubated with PCR-amplified promoter areas (for PCD0350, PCD0872, PCD2963, PCD3412, PCD3605, and PCD3614) or XbaI-linearized pCD22 (PtcdA) for 30 min at 37°C prior to the start of the reaction. PCR products of the promoter areas as used for the in vitro transcription reactions were loaded on and excised from agarose gels and purified using a NucleoSpin gel and PCR clean-up kit (Macherey-Nagel). In vitro transcription reactions mixtures contained 1 μl (1 U) E. coli RNAPcore (catalog no. M0550S; NEB), 16 pmol sigma factor, 0.5 pmol DNA, 10 mM nucleoside triphosphate (NTP) mix, and 0.3 μl α-32P-ATP in reaction buffer (40 mM Tris-HCl, 150 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol [DTT], and 0.01% Triton X-100 [pH = 7.5]) and were incubated for 15 min at 37°C. Transcripts and labeled oligonucleotides to be used as a size indication were purified using P-30 Bio-Gel spin columns (Bio-Rad). All reactions were stopped in gel loading buffer II (Invitrogen) containing 95% formamide, 18 mM EDTA, and 0.025% each of SDS, xylene cyanol, and bromophenol blue at 95°C for 5 min and loaded on 8% monomeric UreaGel (SequaGel; National Diagnostics). Gels were dried and exposed to phosphorimager screens overnight (approximately 17 h) and imaged with a Typhoon 9410 scanner (GE Healthcare).
Spot assay for viability upon σB overproduction in C. difficile and vector stability assay.
C. difficile overnight precultures were corrected for OD600 and were subsequently 10-fold serially diluted in BHI medium. Spots (2 μl) of each dilution were plated on selective (20 μg/ml thiamphenicol) and unselective square (90 × 90 × 15 mm; VWR international) BHI plates with or without 200 ng/ml anhydrotetracycline (ATc). Growth was evaluated after 24 h, and swabs were subsequently taken from all strains grown on unselective BHI agar plates with and without 200 ng/ml ATc for the vector stability assay. These swabs used for the vector stability assay were resuspended in PBS to a McFarland turbidity of 1.0, adjusted for their OD600 values, and 10-fold serially diluted in nonselective BHI medium. Of these serially diluted suspensions, 10-μl spots of each dilution were then plated on selective (20 μg/ml thiamphenicol plus CDSS) and nonselective (BHI plus CDSS) plates, and CFU/ml was counted after 24 to 48 h of growth. The percentage of cells retaining the plasmid was calculated as (CFU/ml)selective/(CFU/ml)nonselective × 100%. If no growth was detected on selective plates containing thiamphenicol, the percentage of plasmid maintained was set as 0%. To calculate statistical significance between percent plasmid maintained in strains induced or not induced by ATc, an unpaired Student’s t test was used.
Plate-based luciferase assay with metronidazole Etest and disk diffusion.
Strains harboring luciferase reporter plasmids were grown on prereduced, selective BHI plates for 24 h. Subsequently, bacterial suspensions corresponding to 1.0 McFarland turbidity were applied on BHI agar supplemented with 0.5% yeast extract, after which a metronidazole Etest or plain disks were applied. Disks were spotted with 10 μl each of sterile H2O, 1 M H2O2, 3,000 μg/ml lincomycin, 200 μg/ml metronidazole, 400 μg/ml ibezapolstat, and 200 μg/ml vancomycin. After 24 h of growth, luciferase activity was visualized by spraying 1:100 reconstituted NanoGlo luciferase substrate (catalog no. N1110; Promega) on the agar plate using a disposable spray flask. One spray corresponded to approximately 250 μl reconstituted NanoGlo luciferase substrate. Luminescence was recorded using a Uvitec Alliance Q9 Advanced imager (BioSPX) after a 10-s exposure time per plate. Luciferase was conjugated into a sigB knockout made by allelic coupled exchange (51), whereas a ClosTron mutant background was used for the DNA arrays. However, no differences between these backgrounds have ever been observed in our assays.
Data availability.
The data used in the VolcaNoseR visualization have been deposited at Zenodo for this purpose (https://doi.org/10.5281/zenodo.3945936). Full transcriptome data have been deposited in the GEO database and can be accessed through the identifier GSE152515.
ACKNOWLEDGMENTS
We thank Bruno Dupuy for kindly providing us with plasmid pCD22 and Annemieke Friggen for technical assistance in strain construction. We thank Nicholas Kint and Isabelle Martin-Verstraete for communicating results prior to publication and Joachim Goedhart for advice on using VolcaNoseR.
REFERENCES
- 1.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 2.Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. 2016. Clostridium difficile infection. Nat Rev Dis Primers 2:16020. doi: 10.1038/nrdp.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paredes-Sabja D, Shen A, Sorg JA. 2014. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol 22:406–416. doi: 10.1016/j.tim.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang FC, Frawley ER, Tapscott T, Vazquez-Torres A. 2016. Bacterial stress responses during host infection. Cell Host Microbe 20:133–143. doi: 10.1016/j.chom.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Kelly C, Loo V, Shaklee Sammons J, Sandora TJ, Wilcox MH. 2018. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 66:987–994. doi: 10.1093/cid/ciy149. [DOI] [PubMed] [Google Scholar]
- 6.Abt MC, McKenney PT, Pamer EG. 2016. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol 14:609–620. doi: 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emerson JE, Stabler RA, Wren BW, Fairweather NF. 2008. Microarray analysis of the transcriptional responses of Clostridium difficile to environmental and antibiotic stress. J Med Microbiol 57:757–764. doi: 10.1099/jmm.0.47657-0. [DOI] [PubMed] [Google Scholar]
- 8.Guldimann C, Boor KJ, Wiedmann M, Guariglia-Oropeza V. 2016. Resilience in the face of uncertainty: sigma factor B fine-tunes gene expression to support homeostasis in Gram-positive bacteria. Appl Environ Microbiol 82:4456–4469. doi: 10.1128/AEM.00714-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Schaik W, Abee T. 2005. The role of σB in the stress response of Gram-positive bacteria – targets for food preservation and safety. Curr Opin Biotechnol 16:218–224. doi: 10.1016/j.copbio.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 10.van Schaik W, Tempelaars MH, Zwietering MH, de Vos WM, Abee T. 2005. Analysis of the role of RsbV, RsbW, and RsbY in regulating σB activity in Bacillus cereus. J Bacteriol 187:5846–5851. doi: 10.1128/JB.187.16.5846-5851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kint N, Alves Feliciano C, Hamiot A, Denic M, Dupuy B, Martin-Verstraete I. 2019. The σB signalling activation pathway in the enteropathogen Clostridioides difficile. Environ Microbiol 21:2852–2870. doi: 10.1111/1462-2920.14642. [DOI] [PubMed] [Google Scholar]
- 12.Guerreiro DN, Wu J, Dessaux C, Oliveira AH, Tiensuu T, Gudynaite D, Marinho CM, Boyd A, García-Del Portillo F, Johansson J, O’Byrne CP. 2020. Mild stress conditions during laboratory culture promote the proliferation of mutations that negatively affect sigma B activity in Listeria monocytogenes. J Bacteriol 202:e00751-19. doi: 10.1128/JB.00751-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boylan SA, Thomas MD, Price CW. 1991. Genetic method to identify regulons controlled by nonessential elements: isolation of a gene dependent on alternate transcription factor sigma B of Bacillus subtilis. J Bacteriol 173:7856–7866. doi: 10.1128/jb.173.24.7856-7866.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garner MR, James KE, Callahan MC, Wiedmann M, Boor KJ. 2006. Exposure to salt and organic acids increases the ability of Listeria monocytogenes to invade Caco-2 cells but decreases its ability to survive gastric stress. Appl Environ Microbiol 72:5384–5395. doi: 10.1128/AEM.00764-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H, Boor KJ, Marquis H. 2004. Listeria monocytogenes σB contributes to invasion of human intestinal epithelial cells. Infect Immun 72:7374–7378. doi: 10.1128/IAI.72.12.7374-7378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morikawa K, Maruyama A, Inose Y, Higashide M, Hayashi H, Ohta T. 2001. Overexpression of sigma factor, σB, urges Staphylococcus aureus to thicken the cell wall and to resist beta-lactams. Biochem Biophys Res Commun 288:385–389. doi: 10.1006/bbrc.2001.5774. [DOI] [PubMed] [Google Scholar]
- 17.DeMaio J, Zhang Y, Ko C, Bishai WR. 1997. Mycobacterium tuberculosis sigF is part of a gene cluster with similarities to the Bacillus subtilis sigF and sigB operons. Tuber Lung Dis 78:3–12. doi: 10.1016/s0962-8479(97)90010-1. [DOI] [PubMed] [Google Scholar]
- 18.Bandow JE, Brotz H, Hecker M. 2002. Bacillus subtilis tolerance of moderate concentrations of rifampin involves the σB-dependent general and multiple stress response. J Bacteriol 184:459–467. doi: 10.1128/jb.184.2.459-467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kint N, Janoir C, Monot M, Hoys S, Soutourina O, Dupuy B, Martin-Verstraete I. 2017. The alternative sigma factor σB plays a crucial role in adaptive strategies of Clostridium difficile during gut infection. Environ Microbiol 19:1933–1958. doi: 10.1111/1462-2920.13696. [DOI] [PubMed] [Google Scholar]
- 20.van Eijk E, Boekhoud IM, Kuijper EJ, Bos-Sanders IMJG, Wright G, Smits WK. 2019. Genome location dictates the transcriptional response to PolC inhibition in Clostridium difficile. Antimicrob Agents Chemother 63:e01363-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fagan RP, Fairweather NF. 2011. Clostridium difficile has two parallel and essential Sec secretion systems. J Biol Chem 286:27483–27493. doi: 10.1074/jbc.M111.263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ransom EM, Ellermeier CD, Weiss DS. 2015. Use of mCherry red fluorescent protein for studies of protein localization and gene expression in Clostridium difficile. Appl Environ Microbiol 81:1652–1660. doi: 10.1128/AEM.03446-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boekhoud IM, Hornung BVH, Sevilla E, Harmanus C, Bos-Sanders IMJG, Terveer EM, Bolea R, Corver J, Kuijper EJ, Smits WK. 2020. Plasmid-mediated metronidazole resistance in Clostridioides difficile. Nat Commun 11:598. doi: 10.1038/s41467-020-14382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira Paiva AM, Friggen AH, Hossein-Javaheri S, Smits WK. 2016. The signal sequence of the abundant extracellular metalloprotease PPEP-1 can be used to secrete synthetic reporter proteins in Clostridium difficile. ACS Synth Biol 5:1376–1382. doi: 10.1021/acssynbio.6b00104. [DOI] [PubMed] [Google Scholar]
- 25.Gao X, Dong X, Subramanian S, Matthews PM, Cooper CA, Kearns DB, Dann CE. 3rd, 2014. Engineering of Bacillus subtilis strains to allow rapid characterization of heterologous diguanylate cyclases and phosphodiesterases. Appl Environ Microbiol 80:6167–6174. doi: 10.1128/AEM.01638-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poquet I, Saujet L, Canette A, Monot M, Mihajlovic J, Ghigo JM, Soutourina O, Briandet R, Martin-Verstraete I, Dupuy B. 2018. Clostridium difficile biofilm: remodeling metabolism and cell surface to build a sparse and heterogeneously aggregated architecture. Front Microbiol 9:2084. doi: 10.3389/fmicb.2018.02084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helmann JD. 2019. Where to begin? Sigma factors and the selectivity of transcription initiation in bacteria. Mol Microbiol 112:335–347. doi: 10.1111/mmi.14309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goedhart J, Luijsterburg MS. 2020. VolcaNoseR—a web app for creating, exploring and sharing volcano plots. bioRxiv doi: 10.1101/2020.05.07.082263. [DOI] [PMC free article] [PubMed]
- 29.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hillion M, Imber M, Pedre B, Bernhardt J, Saleh M, Loi VV, Maaß S, Becher D, Astolfi Rosado L, Adrian L, Weise C, Hell R, Wirtz M, Messens J, Antelmann H. 2017. The glyceraldehyde-3-phosphate dehydrogenase GapDH of Corynebacterium diphtheriae is redox-controlled by protein S-mycothiolation under oxidative stress. Sci Rep 7:5020. doi: 10.1038/s41598-017-05206-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christodoulou D, Link H, Fuhrer T, Kochanowski K, Gerosa L, Sauer U. 2018. Reserve flux capacity in the pentose phosphate pathway enables Escherichia coli’s rapid response to oxidative stress. Cell Syst 6:569–578.e7. doi: 10.1016/j.cels.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Imber M, Huyen NTT, Pietrzyk-Brzezinska AJ, Loi VV, Hillion M, Bernhardt J, Tharichen L, Kolsek K, Saleh M, Hamilton CJ, Adrian L, Grater F, Wahl MC, Antelmann H. 2018. Protein S-bacillithiolation functions in thiol protection and redox regulation of the glyceraldehyde-3-phosphate dehydrogenase gap in Staphylococcus aureus under hypochlorite stress. Antioxid Redox Signal 28:410–430. doi: 10.1089/ars.2016.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, Dubchak IL, Alm EJ, Arkin AP. 2010. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res 38:D396–400. doi: 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matamouros S, England P, Dupuy B. 2007. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol Microbiol 64:1274–1288. doi: 10.1111/j.1365-2958.2007.05739.x. [DOI] [PubMed] [Google Scholar]
- 35.Edwards DI. 1993. Nitroimidazole drugs-action and resistance mechanisms I. Mechanism of action. J Antimicrob Chemother 31:9–20. doi: 10.1093/jac/31.1.9. [DOI] [PubMed] [Google Scholar]
- 36.Mujahid S, Orsi RH, Vangay P, Boor KJ, Wiedmann M. 2013. Refinement of the Listeria monocytogenes σB regulon through quantitative proteomic analysis. Microbiology (Reading) 159:1109–1119. doi: 10.1099/mic.0.066001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmer ME, Chaturongakul S, Wiedmann M, Boor KJ. 2011. The Listeria monocytogenes σB regulon and its virulence-associated functions are inhibited by a small molecule. mBio 2:e00241-11. doi: 10.1128/mBio.00241-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Meouche I, Peltier J, Monot M, Soutourina O, Pestel-Caron M, Dupuy B, Pons JL. 2013. Characterization of the SigD regulon of C. difficile and its positive control of toxin production through the regulation of tcdR. PLoS One 8:e83748. doi: 10.1371/journal.pone.0083748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Regnault B, Coppee JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 40.Mattick JS. 2002. Type IV pili and twitching motility. Annu Rev Microbiol 56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 41.Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CT, Lobritz MA, Braff D, Schwarz EG, Ye JD, Pati M, Vercruysse M, Ralifo PS, Allison KR, Khalil AS, Ting AY, Walker GC, Collins JJ. 2014. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A 111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lobritz MA, Belenky P, Porter CB, Gutierrez A, Yang JH, Schwarz EG, Dwyer DJ, Khalil AS, Collins JJ. 2015. Antibiotic efficacy is linked to bacterial cellular respiration. Proc Natl Acad Sci U S A 112:8173–8180. doi: 10.1073/pnas.1509743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 44.Belenky P, Ye JD, Porter CB, Cohen NR, Lobritz MA, Ferrante T, Jain S, Korry BJ, Schwarz EG, Walker GC, Collins JJ. 2015. Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep 13:968–980. doi: 10.1016/j.celrep.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dingsdag SA, Hunter N. 2018. Metronidazole: an update on metabolism, structure-cytotoxicity and resistance mechanisms. J Antimicrob Chemother 73:265–279. doi: 10.1093/jac/dkx351. [DOI] [PubMed] [Google Scholar]
- 46.Knippel RJ, Wexler AG, Miller JM, Beavers WN, Weiss A, de Crecy-Lagard V, Edmonds KA, Giedroc DP, Skaar EP. 2020. Clostridioides difficile senses and hijacks host heme for incorporation into an oxidative stress defense system. Cell Host Microbe doi: 10.1016/j.chom.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 48.Purdy D, O’Keeffe TA, Elmore M, Herbert M, McLeod A, Bokori-Brown M, Ostrowski A, Minton NP. 2002. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol Microbiol 46:439–452. doi: 10.1046/j.1365-2958.2002.03134.x. [DOI] [PubMed] [Google Scholar]
- 49.Berges M, Michel A-M, Lassek C, Nuss AM, Beckstette M, Dersch P, Riedel K, Sievers S, Becher D, Otto A, Maaß S, Rohde M, Eckweiler D, Borrero-de Acuña JM, Jahn M, Neumann-Schaal M, Jahn D. 2018. Iron regulation in Clostridioides difficile. Front Microbiol 9:3183. doi: 10.3389/fmicb.2018.03183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hofmann JD, Otto A, Berges M, Biedendieck R, Michel A-M, Becher D, Jahn D, Neumann-Schaal M. 2018. Metabolic reprogramming of Clostridioides difficile during the stationary phase with the induction of toxin production. Front Microbiol 9:1970. doi: 10.3389/fmicb.2018.01970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP. 2012. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl Environ Microbiol 78:4683–4690. doi: 10.1128/AEM.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 53.Dupuy B, Sonenshein AL. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol 27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 54.van Eijk E, Anvar SY, Browne HP, Leung WY, Frank J, Schmitz AM, Roberts AP, Smits WK. 2015. Complete genome sequence of the Clostridium difficile laboratory strain 630Δerm reveals differences from strain 630, including translocation of the mobile element CTn5. BMC Genomics 16:31. doi: 10.1186/s12864-015-1252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dannheim H, Riedel T, Neumann-Schaal M, Bunk B, Schober I, Sproer C, Chibani CM, Gronow S, Liesegang H, Overmann J, Schomburg D. 2017. Manual curation and reannotation of the genomes of Clostridium difficile 630Δerm and C. difficile 630. J Med Microbiol 66:286–293. doi: 10.1099/jmm.0.000427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes differentially expressed in σB-overproducing cells compared to controls. Cells were harvested from cultures with (induced) or without (uninduced) anhydrotetracycline (ATc), and with (+Tm) or without (−Tm) thiamphenicol. Gene name is the generic gene name (or locus tag if a gene name is not available). log2FC is the log2 of the fold change in gene expression. Four different comparisons are shown, and genes are aligned between comparisons. Upregulated genes are indicated in green. Downregulated genes are indicated in red. Genes not considered not part of the σB regulon are highlighted in yellow. Download Data Set S1, XLSX file, 0.04 MB (37.7KB, xlsx) .
Copyright © 2020 Boekhoud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Link to access an interactive version of the volcano plot based on hybridization 2 (cells harvested from cultures grown in the presence of both anhydrotetracycline and thiamphenicol). Download Text S1, PDF file, 0.05 MB (50.9KB, pdf) .
Copyright © 2020 Boekhoud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Manhattan distance for differentially regulated genes from hybridization 2 (cells harvested from cultures grown in the presence of both ATc and thiamphenicol). Change indicates whether expression is higher (increased) or lower (decreased) upon overproduction of σB. log2FC represents log2 of the fold change in gene expression. Significance is given as −10 · log of the adjusted P value (see Data Set S1). Manhattan distance was calculated by and exported using the online tool VolcaNoseR. Download Data Set S2, XLSX file, 0.02 MB (16.1KB, xlsx) .
Copyright © 2020 Boekhoud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The data used in the VolcaNoseR visualization have been deposited at Zenodo for this purpose (https://doi.org/10.5281/zenodo.3945936). Full transcriptome data have been deposited in the GEO database and can be accessed through the identifier GSE152515.