Aspergillus fumigatus is an important human-pathogenic fungal species that is responsible for a high incidence of infections in immunocompromised individuals. A. fumigatus high-osmolarity glycerol (HOG) and cell wall integrity pathways are important for the adaptation to different forms of environmental adversity such as osmotic and oxidative stresses, nutrient limitations, high temperatures, and other chemical and mechanical stresses that may be produced by the host immune system and antifungal drugs. Little is known about how these pathways are activated in this fungal pathogen. Here, we characterize four A. fumigatus putative homologues that are important for the activation of the yeast HOG pathway. A. fumigatus SlnASln1p, ShoASho1p, MsbAMsb2p, and OpyAOpy2p are genetically interacting and are essential for the activation of the HOG and cell wall integrity pathways. Our results contribute to the understanding of A. fumigatus adaptation to the host environment.
KEYWORDS: Aspergillus fumigatus, putative receptors, osmotic and cell wall stresses, high-osmolarity glycerol (HOG), caspofungin
ABSTRACT
The high-osmolarity glycerol (HOG) response pathway is a multifunctional signal transduction pathway that specifically transmits ambient osmotic signals. Saccharomyces cerevisiae Hog1p has two upstream signaling branches, the sensor histidine kinase Sln1p and the receptor Sho1p. The Sho1p branch includes two other proteins, the Msb2p mucin and Opy2p. Aspergillus fumigatus is the leading cause of pulmonary fungal diseases. Here, we investigated the roles played by A. fumigatus SlnASln1p, ShoASho1p, MsbAMsb2p, and OpyAOpy2p putative homologues during the activation of the mitogen-activated protein kinase (MAPK) HOG pathway. The shoA, msbA, and opyA singly and doubly null mutants are important for the cell wall integrity (CWI) pathway, oxidative stress, and virulence as assessed by a Galleria mellonella model. Genetic interactions of ShoA, MsbA, and OpyA are also important for proper activation of the SakAHog1p and MpkASlt2 cascade and the response to osmotic and cell wall stresses. Comparative label-free quantitative proteomics analysis of the singly null mutants with the wild-type strain upon caspofungin exposure indicates that the absence of ShoA, MsbA, and OpyA affects the osmotic stress response, carbohydrate metabolism, and protein degradation. The putative receptor mutants showed altered trehalose and glycogen accumulation, suggesting a role for ShoA, MsbA, and OpyA in sugar storage. Protein kinase A activity was also decreased in these mutants. We also observed genetic interactions between SlnA, ShoA, MsbA, and OpyA, suggesting that both branches are important for activation of the HOG/CWI pathways. Our results help in the understanding of the activation and modulation of the HOG and CWI pathways in this important fungal pathogen.
IMPORTANCE Aspergillus fumigatus is an important human-pathogenic fungal species that is responsible for a high incidence of infections in immunocompromised individuals. A. fumigatus high-osmolarity glycerol (HOG) and cell wall integrity pathways are important for the adaptation to different forms of environmental adversity such as osmotic and oxidative stresses, nutrient limitations, high temperatures, and other chemical and mechanical stresses that may be produced by the host immune system and antifungal drugs. Little is known about how these pathways are activated in this fungal pathogen. Here, we characterize four A. fumigatus putative homologues that are important for the activation of the yeast HOG pathway. A. fumigatus SlnASln1p, ShoASho1p, MsbAMsb2p, and OpyAOpy2p are genetically interacting and are essential for the activation of the HOG and cell wall integrity pathways. Our results contribute to the understanding of A. fumigatus adaptation to the host environment.
INTRODUCTION
Aspergillus fumigatus causes aspergillosis, which includes chronic pulmonary aspergillosis (CPA), allergic bronchopulmonary aspergillosis (ABPA), and invasive pulmonary aspergillosis (IPA) (1). IPA has a mortality rate of 50% to 75% when treated, affecting primarily immunocompromised individuals, patients with cancer or hematological neoplasms, or patients undergoing chemotherapy (1–3). A. fumigatus is able to adapt to different forms of environmental adversity such as osmotic and oxidative stresses, nutrient limitations, high temperatures, and other chemical and mechanical stresses, some of which may be produced by the host immune system and antifungal drugs (4). The survival capacity of A. fumigatus under different stress conditions depends on its response and adaptation mechanisms (4).
The high-osmolarity glycerol (HOG) response pathway is a multifunctional signal transduction pathway that specifically transmits ambient osmotic signals (5). The HOG pathway is activated by mitogen-activated protein kinases (MAPKs) during adaptation to environmental stress and morphology regulation (5). The MAPK cascades consist of three kinases, MAPK, MAPK kinase (MAPKK), and MAPKK kinase (MAPKKK), which are highly conserved in fungi (6–8). After the environmental stimulus, the MAPK cascade is activated by sequential phosphorylation, resulting in the activation of transcription factors and expression of target genes that assist in the adaptation to a given condition (5, 9). In Saccharomyces cerevisiae, Hog1p has the following two upstream signaling branches (9–11) (Fig. 1): (i) the Sho1p (synthetic high osmolarity) membrane protein, containing four N-terminal transmembrane domains (TM) and one C-terminal domain (SH3) (9, 11, 12), and (ii) Sln1p (synthetic lethal of N-end rule), a transmembrane histidine phosphotransfer kinase and osmosensor with an intracellular kinase domain forming a phosphorelay system similar to bacterial two-component regulators (9, 11). In yeast, Sho1p is responsible for transmitting signals of osmotic stress by sequentially activating the MAPKKs Pbs2p and Hog1p. In addition, the Sho1p branch includes two other TM proteins, an Msb2p (multicopy suppressor of a budding defect) mucin and an Opy2p (overproduction-induced pheromone-resistant yeast) type 1 TM ancestral protein (13, 14). Under conditions of hyperosmotic stress, these TM proteins form a complex that is composed of two GTPases (Cdc42p and Cdc24p) and three kinases (Ste11p, Ste50p, and Ste20p) and that activates the Hog1p pathway (Fig. 1) (15–17).
FIG 1.
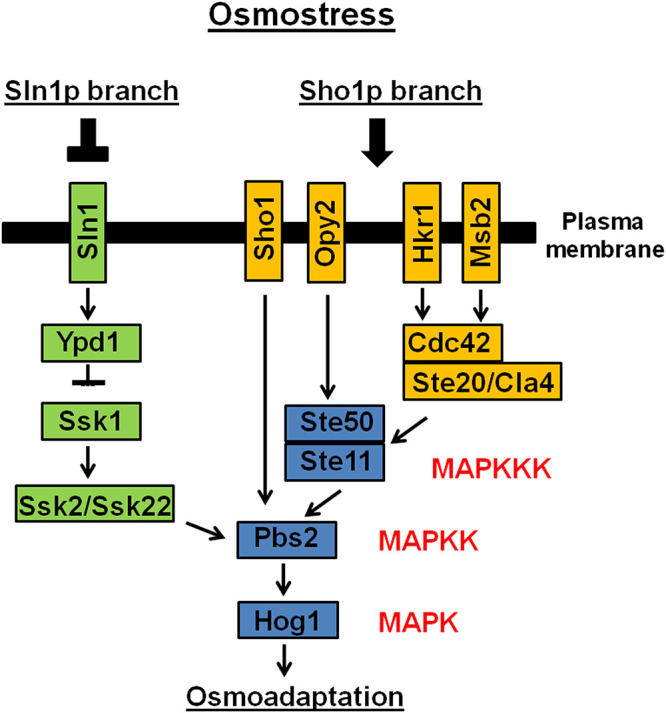
The two branches of Snl1 and Sho1 which are responsible for the activation of the S. cerevisiae high-osmolarity glycerol (HOG) pathway. (Adapted from reference 43).
The Sln1 branch of the yeast HOG pathway is an example of a two-component system (11, 18, 19). The typical organization of a two-component system consists of the following components: (i) a sensor histidine kinase (SHK) that contains an input (or sensor) domain, an HK catalytic domain, and a histidine autophosphorylation site and (ii) a response regulator (RR) that contains a receiver (REC) domain and an output (or effector) domain (20). The sensor domain is modified by a stimulus, a histidine close to the HK domain is phosphorylated (or dephosphorylated), and this phosphoryl group is transferred to the REC domain of the RR (20). Yeast has three REC proteins (Sln1, Ssk1, and Skn7), one SHK (Sln1), and one HPt (Ypd1) (Fig. 1). S. cerevisiae Sln1 is responsible for the coordination of two distinct signaling pathways: the Sln1-Ypd1-Ssk1 phosphorelay pathway, which is important for the regulation of hyperosmolarity responses, and the Sln1-Ypd1-Skn7 pathway, which is important for the regulation of hypo-osmolarity responses (11).
In A. fumigatus, ShoA, the putative homologue of S. cerevisiae Sho1p, has been shown to be important for fungal morphology and oxidative stress; however, a thorough investigation of ShoA function has not been performed (21). The A. fumigatus Sln1p homologue has been previously characterized (22). The ΔtcsBslnA/sln1p mutant was partially sensitive to SDS, and Western blot analysis of both the corresponding wild-type strain and the ΔtcsBslnA/sln1p mutant showed that when stressed with hydrogen peroxide, phosphorylation of Hog1p still occurred in the mutant (22). The A. fumigatus Msb2p mucin homologue, MsbA, has been shown to be important for fungal development, biofilm formation, and cell wall integrity (CWI) (23). The function of the TM Opy2p putative homologue in A. fumigatus (here named OpyA) has not been elucidated, and the interactions of the ShoA branch with the MsbA and OpyA and SlnASln1p branches during HOG MAPK pathway activation are undefined.
Four MAPKs have been identified in A. fumigatus: (i) MpkA, which is important for the CWI pathway and oxidative stress (22, 24, 25); (ii) MpkB, which regulates the pheromone response/filamentous growth pathway, conidiation, and dihydroxynaphthalene (DHN) melanin production (26, 27); and (iii) SakA and (iv) MpkC, which are paralogues that regulate the HOG pathway (28). MpkC and SakA interact physically and play roles in caspofungin tolerance and carbon source utilization, respectively (24, 29–33). We do not have very much information about the upstream mechanisms by which MpkC and SakA are activated and execute their signaling functions.
Here, we investigated in more detail the roles played by both the Sln1 and Sho1 A. fumigatus putative branches during the activation of the MAPK HOG pathway. Our results strongly indicate that these four proteins actively collaborate to promote activation of osmotic stress responses and the cell wall integrity pathway (CWI) MpkA MAPK in A. fumigatus.
RESULTS
The Sho1 branch: functional characterization of three A. fumigatus genes encoding putative receptors.
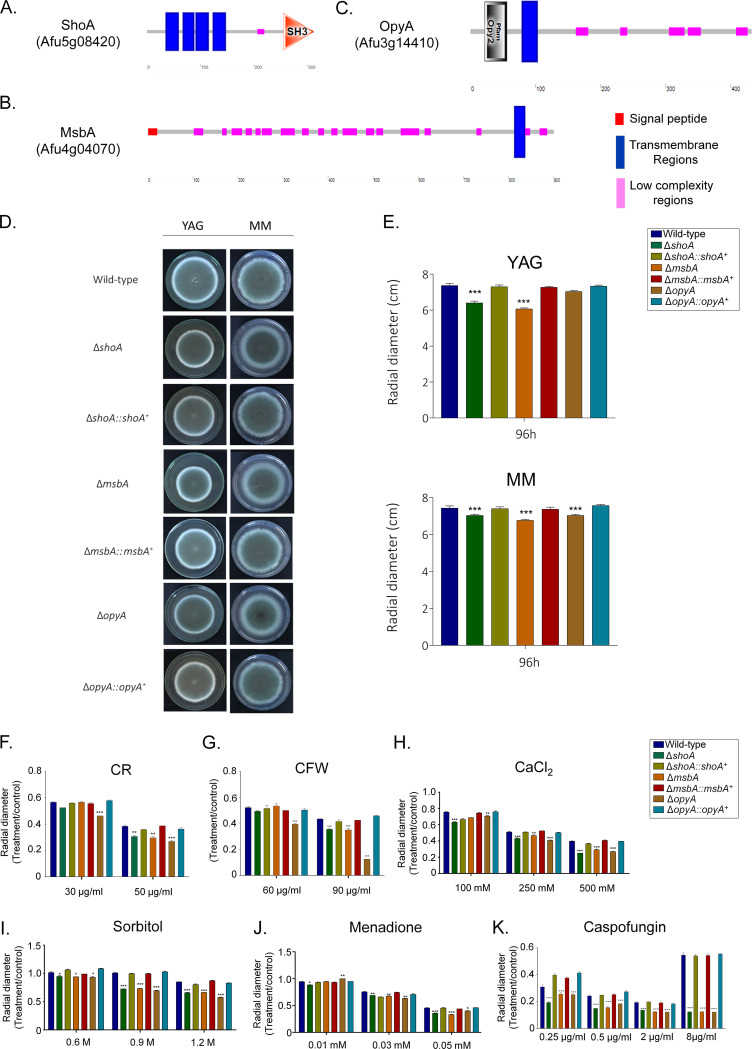
A. fumigatus ShoA and MsbA homologues were previously identified (21, 23). We validated their organization and identified A. fumigatus putative homologues of S. cerevisiae Opy2p. (i) ShoASho1p (A. fumigatus B_055960 [AFUB_055960]) has 311 amino acids and 30% identity and 48% similarity with Sho1p (E value = 4e−39). It has four transmembrane regions and a Src homology 3 (SH3) domain (Interpro IPR001452) that binds to target proteins through sequences containing proline and hydrophobic amino acids (Fig. 2A). (ii) MsbAMsb2p (AFUB_098950) has 901 amino acids and 34% identity and 55% similarity with Msb2p (E value = 8e−12). It has a putative signal peptide (from amino acids 1 to 21) and a single transmembrane region at the C terminus (Fig. 2B). (iii) OpyAOpy2p (AFUB_034820) has 432 amino acids and very low identity with Opy2p that is restricted to the Opy2 domain that acts as a membrane anchor in the HOG signaling pathway (Interpro IPR018571, from amino acids 22 to 56) and a single transmembrane region located at the N terminus (Fig. 2C).
FIG 2.
A. fumigatus ShoA, MsbA, and OpyA. (A to C) Protein organization of A. fumigatus ShoA (A), MsbA (B), and OpyA (C). (D to J) Growth phenotypes of the wild-type strain and the ΔshoA, ΔmsbA, ΔopyA, ΔshoA::shoA+, ΔmsbA::msbA+, and ΔopyA::opyA+ mutants. The strains were grown for 4 days at 37°C (D) on MM or YAG (E), MM plus Congo red (CR) (F), MM plus calcofluor white (CFW) (G), MM plus CaCl2 (H), MM plus sorbitol (I), MM plus menadione (J), and MM plus caspofungin (K), and their radial growth was quantified. The results shown represent the means of measurements of the diameter of 3 colonies for each strain ± standard deviation. The data were analyzed (Prism, GraphPad) using two-way ANOVA followed by Bonferroni posttests. The levels of significance compared to the wild-type results are indicated as follows: *, P < 0.1; **, P < 0.01; ***, P < 0.001.
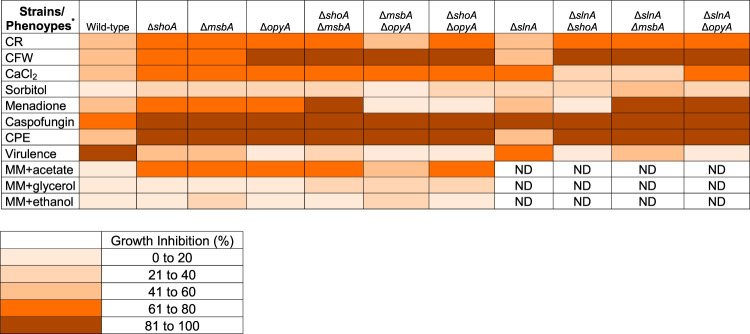
Null mutant strains for shoA, msbA, and opyA were constructed and were functionally complemented by homologous integration (Fig. 2D and E; see also Fig. S1 at https://figshare.com/articles/Membrane_receptors_contribute_to_activation_and_efficient_signaling_of_Mitogen-Activated_Protein_Kinase_cascades_during_adaptation_of_Aspergillus_fumigatus_to_different_stressors_and_carbon_sources/12402125). They had about 15% reduced growth compared to wild-type strain in YAG (except for the ΔopyA mutant, which showed the same radial diameter as the wild-type strain) and minimal medium (MM) (Fig. 2D and E). They were also phenotypically characterized under different stress conditions such as cell wall damage and osmotic and oxidative stresses (Fig. 2F to K). All three mutants were more sensitive to Congo red (CR), calcofluor white (CFW), CaCl2, sorbitol, and menadione than the wild-type and corresponding complementing strains (Fig. 2F to J). All three mutants were more sensitive to caspofungin, an echinocandin that noncompetitively inhibits β-1,3-glucan synthase, impairing fungal cell wall polysaccharide biosynthesis and integrity (Fig. 2K) (24, 34). Caspofungin paradoxical effect (CPE) is described as a phenomenon where high caspofungin concentrations restore the expected inhibition of A. fumigatus growth (35). All three mutants lost the CPE (Fig. 2K). A summary of the phenotypes is shown in Table 1. These results indicate that ShoA, MsbA, and OpyA are important for the activity of the CWI pathway and for responses to osmotic and oxidative stresses.
TABLE 1.
Summary of the growth inhibition of the wild-type strain and slnA, shoA, msbA, and opyA single and double mutants grown under different stressing conditions
*CR, Congo red (50 μg/ml); CFW, calcofluor white (90 μg/ml); CaCl2 (500 mM); Sorbitol 1.2 M; Menadione 0.05 M; Caspofungin (2 μg/ml); CPE, Caspofungin Paradoxical Effect (8 μg/ml); Virulence, virulence in Galleria mellonella; MM+acetate 1%; MM+glycerol 1%; MM+ethanol 1%; ND, not determined.
ShoA, MsbA, and OpyA null mutants have genetic interactions.
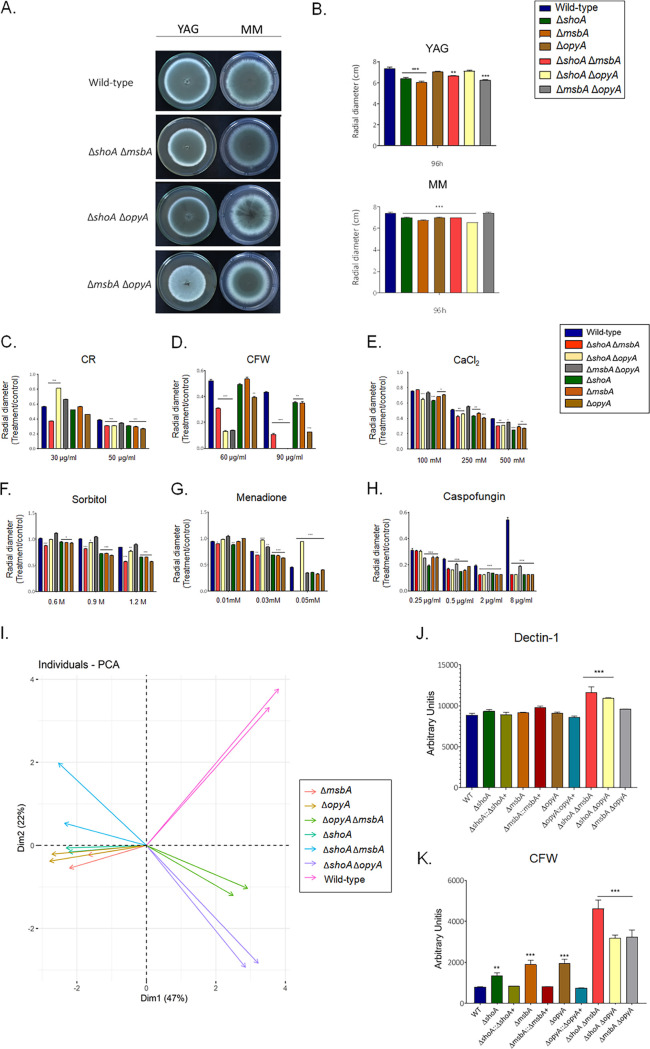
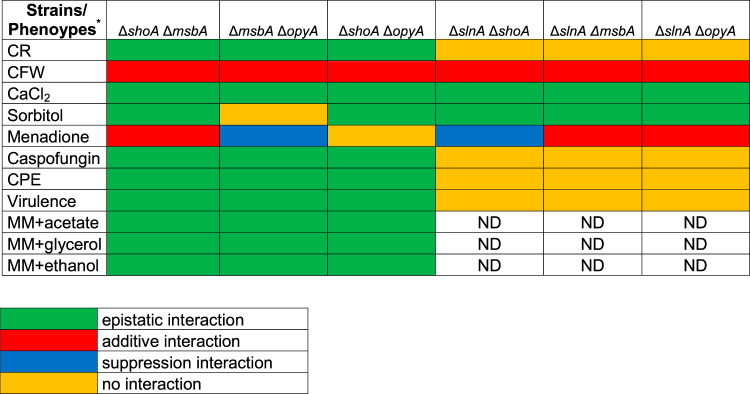
To investigate possible genetic interactions between these putative receptors, we constructed ΔshoA ΔmsbA, ΔshoA ΔopyA, and ΔmsbA ΔopyA double mutants (Fig. 3A and B). The double mutants showed about 15% reduced growth compared to the wild-type strain in YAG and MM (Fig. 3A and B). All of the double mutants showed levels of susceptibility to CR 50 μg/ml similar to those seen with the corresponding single mutants (Fig. 3C), suggesting that shoA, msbA, and opyA are functioning in the same pathway to repair and/or process cell wall damage caused by CR (Tables 1 and 2). In contrast, all the double mutants showed increased susceptibility to CFW 90 μg/ml in comparison to the corresponding single mutants, suggesting an additive interaction (Fig. 3D; see also Tables 1 and 2). The double mutants were also as sensitive to 500 mM CaCl2 as the corresponding single mutants, suggesting again that shoA, msbA, and opyA function in the same pathway for calcium signaling (Fig. 3E). The double mutants, except the ΔmsbA ΔopyA mutant, were as sensitive to osmotic stress caused by 1.2 M sorbitol as the corresponding single mutants (Fig. 3F; see also Tables 1 and 2), suggesting once more that shoA and msbA and shoA and opyA function in the same pathway for osmotic stress signaling. However, the ΔmsbA ΔopyA mutant was more resistant to osmotic stress (Fig. 3F; see also Tables 1 and 2), suggesting that msbA and opyA do not genetically interact. The ΔshoA ΔmsbA mutant was more sensitive to oxidative stress caused by 0.05 mM menadione than the corresponding single mutants, suggesting an additive interaction (Fig. 3G; see also Tables 1 and 2) while the ΔmsbA ΔopyA and ΔshoA ΔopyA mutants were less sensitive than the corresponding single mutants (Fig. 3G; see also Tables 1 and 2), suggesting suppression interactions. All the double mutants were as sensitive to lower caspofungin and CPE concentrations as the single mutants (Fig. 3H; see also Tables 1 and 2), suggesting that they function in the same pathway for caspofungin signaling.
FIG 3.
A. fumigatus double null mutants indicate genetic interactions among ΔshoA, ΔmsbA, and ΔopyA. Growth phenotypes of the wild-type strain and the ΔshoA, ΔmsbA, ΔopyA, ΔmsbA ΔopyA, ΔshoA ΔmsbA, and ΔshoA ΔopyA mutants were determined. (A to H) The strains were grown for 4 days at 37°C (A) on MM or YAG (B) or on MM plus Congo red (CR) (C), MM plus calcofluor white (CFW) (D), MM plus CaCl2 (E), MM plus sorbitol (F), and MM plus menadione (G), and (H) MM plus caspofungin, and their radial growth was quantified. The results shown represent the means of measurements of the diameter of 3 colonies for each strain ± standard deviation. The data were analyzed (Prism, GraphPad) using two-way ANOVA followed by Bonferroni posttests. The levels of significance compared to the wild-type results are indicated as follows: *, P < 0.1; **, P < 0.01; ***, P < 0.001. (I) PCA of the results shown in panels A to E. (J and K) Detection of β-1,3−glucan (Dectin-1; J) and chitin (CFW; K) exposed on the cell surface was performed. WT, wild type. Experiments were performed in triplicate, and the results are displayed as mean values with standard errors (two-way ANOVA followed by Tukey’s P < 0.05).
TABLE 2.
Summary of the genetic interactions between slnA, shoA, and msbA single mutants grown under different stressing conditions
CR, Congo red (50 μg/ml); CFW, calcofluor white (90 μg/ml); CaCl2 (500 mM); Sorbitol 1.2 M; Menadione 0.05 M; Caspofungin (2 μg/ml); CPE, Caspofungin Paradoxical Effect (8 μg/ml); Virulence, virulence in Galleria mellonella; MM+acetate 1%; MM+glycerol 1%; MM+ethanol 1%; ND, not determined.
Considering the paucity of genetic transformation markers for A. fumigatus, we did not complement the double mutants. However, we tested few of the phenotypes described above for two independent candidates of the double mutants (see Fig. S2). They showed exactly the same phenotypes, strongly indicating that there were no additional mutations that were introduced into these double mutants during the transformation process and that could be responsible for the observed phenotypes (see Fig. S2).
The differences between the mutant and wild-type strains were analyzed through a principal-component analysis (PCA) (Fig. 3I). This analysis was performed with the set of all data obtained through measurement of the radial growth of each strain under different stress conditions. In the spatial distribution of the graph, we can see that the single ΔshoA, ΔmsbA, and ΔopyA mutants are grouped, indicating that the single mutants showed similar phenotypes after growth under different stress conditions (CR, CFW, menadione, and sorbitol). These data show that ShoA, MsbA, and OpyA have similar and/or redundant functions.
Looking at the result of the PCA for the double mutants, we can see that the ΔmsbA ΔopyA and ΔshoA ΔopyA double mutants are similar and also grouped apart from the wild-type and the single mutants (Fig. 3H). This suggests that ShoA can fulfill some of the functions performed by MsbA in the ΔmsbA ΔopyA double mutant. Similarly, MsbA can fulfill some of the functions of ShoA in the ΔshoA ΔopyA double mutant. As a consequence, when we subjected the double mutants to stress conditions, we observed that the ΔshoA ΔopyA and ΔmsbA ΔopyA double mutants showed the same behavior.
Finally, we observed that the ΔshoA ΔmsbA double mutant was different from the wild-type strain, the single mutants, and the other two double mutants (the ΔshoA ΔopyA and ΔmsbA ΔopyA mutants). This suggests that ShoA and MsbA have unique functions during signaling caused by stressful conditions. A summary of the phenotypes of and the genetic interactions between the single mutants is shown in Tables 1 and 2. The data set shows that these putative TM proteins can have redundant as well as unique functions during the activation of the signaling pathways in A. fumigatus.
Next, we investigated the cell wall organization using the exposure of different polysaccharides on the cell surface. All the single and double mutants showed a germling length comparable to that seen with the wild-type strains after 16 h of growth at 37°C without shaking. Dectin-1 binding results revealed increased exposure of β-glucans only in the ΔshoA ΔopyA and ΔshoA ΔmsbA cell walls compared with the wild-type strain and all the other mutants (Fig. 3I). However, CFW staining showed that the ΔshoA, ΔopyA, and ΔmsbA mutants exhibited an increase in chitin exposure in comparison to the wild-type and complemented strains (Fig. 3J). The double mutants once more showed a synergistic interaction because their level of chitin exposure was much higher than that of the corresponding single mutants (Fig. 3J).
Taken together, these results strongly indicate a genetic interaction among ShoA, MsbA, and OpyA in response to several stressing conditions that affect the cell wall structure and organization and responses to oxidative and osmotic stresses.
ShoA, MsbA, and OpyA null mutants influence SakA and MpkA phosphorylation.
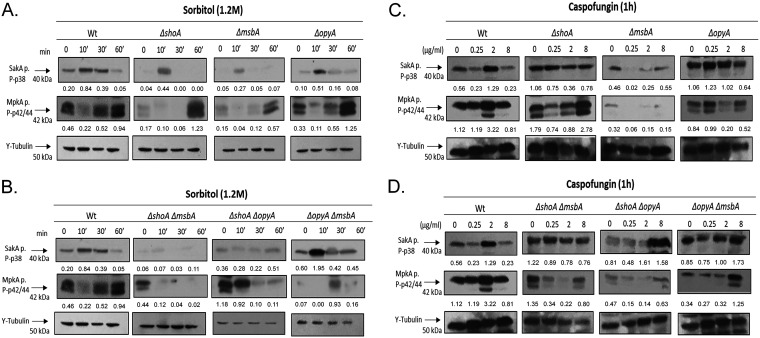
Subsequently, we investigated the impact of the deletion of the three putative receptors on the phosphorylation of MAP kinases SakA and MpkA in the presence of higher osmotic concentrations (1.2 M sorbitol) or different caspofungin concentrations (Fig. 4; for an additional repetition of the Western blot assays, see Fig. S3). These Western blot assays were semiquantitative experiments, and the results were somewhat variable. We focus on the trends that were common to the two independent experiments, though the precise fold changes might have differed between the experiments. The wild-type strain showed increased (about 4-fold) SakA phosphorylation when exposed for 10 min to 1.2 M sorbitol (Fig. 4A). In contrast, the ΔshoA, ΔmsbA, and ΔopyA single mutants showed higher levels of SakA phosphorylation than the wild-type strain, with 11-fold, 5.4-fold, and 5-fold phosphorylation increases at 10 min, respectively (Fig. 4A). The ΔshoA ΔmsbA and ΔshoA ΔopyA double mutants had a synergistic interaction, since both mutants showed much lower SakA phosphorylation than the corresponding single mutants whereas the ΔopyA ΔmsbA double mutant showed about 3-fold SakA phosphorylation at 10 min (Fig. 4B).
FIG 4.
A. fumigatus ShoA, MsbA, and OpyA are important for SakA and MpkA phosphorylation. Western blotting assays of SakA and MpkA phosphorylation in the wild-type strain and single mutants (A and C) or in the wild-type strain and double mutants (B and D) in response to 1.2 M sorbitol for different periods of time (A and B) and in response to caspofungin 0.25, 2.0, or 8.0 μg/ml for 1 h (C and D) were performed. Anti-P-p38 SakA and anti-44/42 MpkA antibodies were used to detect the phosphorylation of SakA and MpkA, respectively, while anti-γ-tubulin was used to detect γ-tubulin. Signal intensities were quantified using ImageJ software, and ratios of (P)-SakA to γ-tubulin or (P)-MpkA to γ-tubulin were calculated.
The wild-type strain exhibited increased (about 2-fold) MpkA phosphorylation when exposed to 1.2 M sorbitol for 60 min (Fig. 4A). All three single mutants show higher induction of MpkA phosphorylation levels than the wild-type strain at 60 min (Fig. 4A). The double mutants ΔshoA ΔmsbA and ΔshoA ΔopyA showed lower MpkA phosphorylation levels than the corresponding single mutants, while the ΔopyA ΔmsbA double mutant showed about 13-fold- and 2-fold-increased MpkA phosphorylation at 30 and 60 min exposure to sorbitol, respectively, compared to the control without sorbitol (Fig. 4B).
When exposed to 2 μg/ml of caspofungin, the wild-type strain showed 2-fold and 2.8-fold more phosphorylation of SakA and MpkA, respectively (Fig. 4C). The ΔshoA, ΔmsbA, and ΔopyA mutants showed no induction of either SakA or MpkA at 2 μg/ml of caspofungin, but the ΔshoA mutant showed about 1.6-fold induction of MpkA phosphorylation at 8 μg/ml of caspofungin (Fig. 4C). The ΔshoA ΔmsbA double mutant showed no increase in SakA and MpkA phosphorylation, while the ΔshoA ΔopyA mutant showed about 2-fold SakA phosphorylation at 2 and 8 μg/ml of caspofungin but no MpkA phosphorylation at any caspofungin concentration (Fig. 4D). The ΔopyA ΔmsbA double mutant showed no SakA phosphorylation in the presence of caspofungin but about 3.7-fold MpkA phosphorylation at 8 μg/ml of caspofungin (Fig. 4D).
Taken together, these results strongly indicate that ShoA, MsbA, and OpyA can affect the levels of MpkA and SakA phosphorylation when A. fumigatus is exposed to osmotic and cell wall stresses.
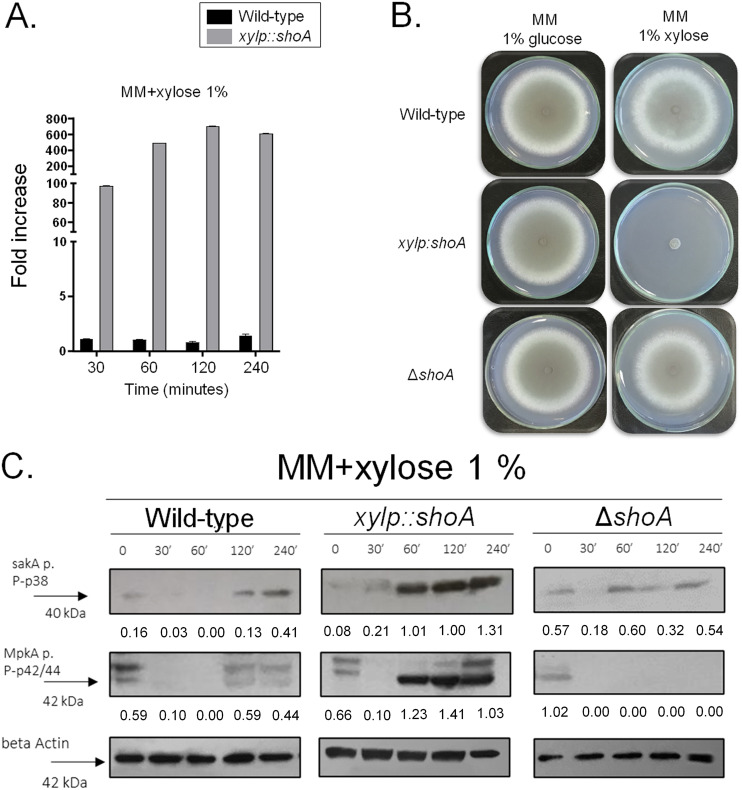
After several attempts, we were unable to construct a ΔshoA ΔmsbA ΔopyA triple mutant. We therefore attempted to construct a conditional xylp::shoA ΔmsbA ΔopyA mutant strain by replacing the shoA endogenous promoter with the xylp promoter from Penicillium chrysogenum, which was induced by xylose and was repressed by glucose (36) in the ΔmsbA ΔopyA strain. However, when this approach failed, we successfully replaced the shoA endogenous promoter with the xylp promoter in the wild-type strain (see Fig. S1). The xylp::shoA strain showed 100-fold and 600-fold increases in the levels of shoA transcripts when exposed for 30 and 120 min to xylose 1%, respectively, compared to the wild-type strain (Fig. 5A). shoA overexpression caused an almost complete reduction of growth rate in the xylp::shoA strain compared to the wild-type strain grown on MM plus 1% xylose (Fig. 5B). Overexpression of shoA also resulted in increased SakA phosphorylation (2.6-fold to 16-fold) and MpkA phosphorylation (about 2-fold) in comparison to the control (strain xylp::shoA grown in 1% glucose) (Fig. 5C). The levels of SakA and MpkA phosphorylation in the wild-type strain grown in xylose showed about a 2.5-fold increase and no increase, respectively, compared to the wild-type strain grown in glucose 1% (Fig. 5C). The levels of SakA and MpkA phosphorylation showed no increase in the ΔshoA mutant grown in xylose 1% compared to glucose 1% (Fig. 5C).
FIG 5.
A. fumigatus shoA overexpression inhibits growth. (A) The wild-type and xylp::shoA strains were grown in MM plus 1% glucose for 16 h at 37°C and transferred to MM plus 1% xylose for 30 to 120 min at 37°C. The results shown represent the means of measurements of the diameter of 3 colonies for each strain ± standard deviation. Gene expression was normalized using tubA (Afu1g10910). Standard deviations present averages of results from three independent biological repetitions (each performed with 2 technical repetitions). (B) The wild-type, xylp::shoA, and ΔshoA strains were grown either on MM plus 1% glucose or on MM plus 1% xylose for 5 days at 37°C. (C) The wild-type, xylp::shoA, and ΔshoA strains were grown in MM plus 1% glucose for 16 h at 37°C and transferred to MM plus 1% xylose for 30 to 120 min at 37°C. Western blotting assays of SakA and MpkA phosphorylation were performed. Anti-P-p38 SakA and anti-44/42 MpkA antibodies were used to detect the phosphorylation of SakA and MpkA, respectively.
Taken together, these results strongly indicate that ShoA, MsbA, and OpyA genetic interactions are important for proper activation of the SakA and MpkA cascade and the response to osmotic and cell wall stresses.
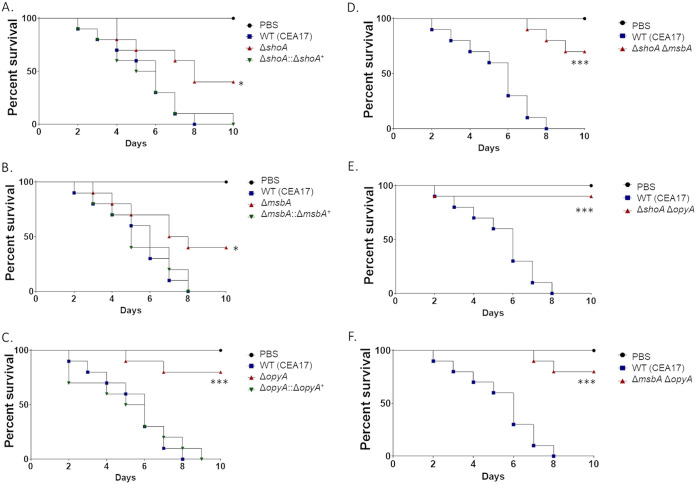
ShoA, MsbA, and OpyA are important for A. fumigatus virulence in Galleria mellonella.
G. mellonella larvae were used to evaluate the importance of ShoA, MsbA, and OpyA in regulating A. fumigatus pathogenicity (Fig. 6). In the G. mellonella model, infection by the wild‐type strain resulted in 100% mortality 8 days postinfection (Fig. 6). However, the ΔshoA and ΔmsbA mutants showed only 60% mortality rates 10 days postinfection, which was statistically significantly different from the results seen with the wild-type strain according to Mantel-Cox and Gehan-Breslow-Wilcoxon tests (P < 0.001; Fig. 6A and B). The ΔopyA mutant caused 20% mortality 10 days postinfection, which was not statistically significantly different from the results seen with the phosphate-buffered saline (PBS) control according to the MantelCox and Gehan-Breslow-Wilcoxon tests (P < 0.001; Fig. 6C). Interestingly, infection by all three double mutants resulted in 10% to 30% mortality rates 10 days postinfection, which was not statistically significantly different from the phosphate buffer saline (PBS) control results according to the Mantel-Cox and Gehan-Breslow-Wilcoxon tests (P < 0.001; Fig. 6D to F), suggesting that shoA, msbA, and opyA function in the same pathway for the establishment of virulence in A. fumigatus (Tables 1 and 2).
FIG 6.
A. fumigatus shoA, msbA, and opyA mutants are important for virulence in Galleria mellonella. Cumulative survival rates of wild-type and shoA (A) msbA (B), and opyA (C) single mutant strains and wild-type and ΔshoA Δmsba (D) ΔshoA ΔopyA (E), and ΔmsbA ΔopyA (F) double mutant strains in the model moth Galleria mellonella are shown. Infection of larvae was carried out via inoculation of 106 conidia. For inoculations, 10 larvae were infected per wild-type or null mutant strain. The levels of significance compared to the wild-type results are indicated as follows: *, P < 0.1; **, P < 0.01; ***, P < 0.001. Phosphate buffered saline (PBS) was used as a negative control.
Taken together, these results clearly demonstrate that ShoA, MsbA, and OpyA play an important role in A. fumigatus virulence.
Proteomic analysis reveals that ShoA, MsbA, and OpyA contribute to the metabolic modulation of A. fumigatus.
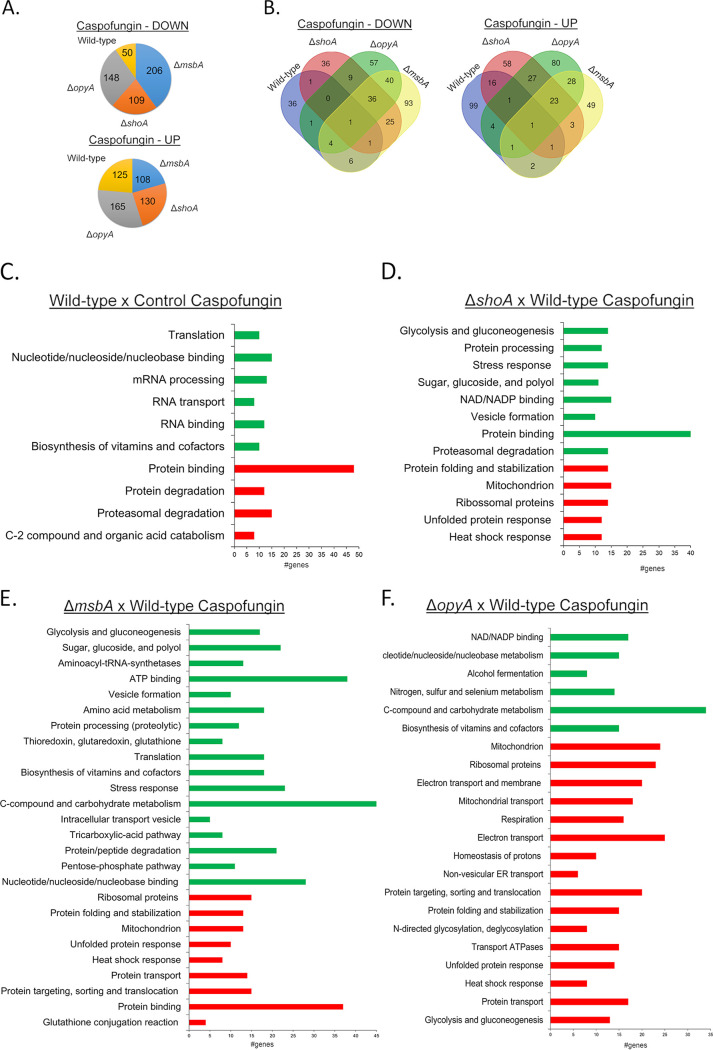
We used label-free quantitative proteomics (spectral counts) to investigate proteins that are differentially abundant in the ΔshoA, ΔmsbA, and ΔopyA mutants upon exposure to caspofungin stress (Fig. 7; see also Table S1). We decided to expose the wild-type and the single mutants to caspofungin (8 μg/h for 1 h) because this concentration and this exposure time are important for CPE, and all the three single mutants lost the CPE (37) (Fig. 2K). Upon exposure to caspofungin (8 μg/h for 1 h), we observed increased abundances of 50, 109, 206, and 148 proteins and decreased abundances of 125, 130, 108, and 165 proteins in the wild-type, ΔshoA, ΔmsbA, and ΔopyA strains, respectively (Fig. 7A). Venn diagrams were generated to correlate the protein abundance profiles for each strain treated with caspofungin (Fig. 7B). Although each strain had a unique set of proteins (135 for the wild-type strain, 94 for the ΔshoA mutant, 142 for the ΔmsbA mutant, and 137 for the ΔopyA mutant), there was extensive overlap among the three mutant strains (Fig. 7B), suggesting independent and common roles for ShoA, MsbA, and OpyA in distinct pathways involved in the caspofungin response. Proteins of significant differential abundances in the ΔshoA, ΔmsbA, and ΔopyA mutants were classified in terms of biological function. Upon caspofungin stress, in the wild-type strain there was an increase in the abundance of proteins involved in but not limited to (i) C-2 compound and organic acid metabolism and (ii) proteasomal degradation. There was also a decrease in the abundance of proteins involved in transcription and translation (Fig. 7C). In the mutant strains, there was an increase in the abundance of proteins involved in but not limited to (i) heat shock response, (ii) unfolded protein response, (iii) mitochondrial function, and (iv) protein folding and stabilization (Fig. 7D to F). In these strains, there was a reduction in the abundance of proteins belonging to categories such as (i) proteasomal degradation; (ii) sugar, glucoside, and polyol; (iii) stress response; (iv) glycolysis and gluconeogenesis; (v) pentose phosphate pathway; and (vi) C-compound and metabolism (Fig. 7D to F). Therefore, this proteomic analysis implies that upon caspofungin exposure, the absence of ShoA, MsbA, and OpyA affects the osmotic stress response, carbohydrate metabolism, and protein degradation.
FIG 7.
Proteomic analysis of the A. fumigatus wild-type strain and shoA, msbA, and opyA null mutants exposed to caspofungin. (A) Number of proteins with increased (UP) and decreased (DOWN) abundance in the wild-type strain and the ΔshoA, ΔmsbA, and ΔopyA mutants when exposed to caspofungin 2 μg/ml for 1 h. (B) Venn diagrams comparing the abundances of proteins in the wild-type and each mutant strain. (C to F) A summary of the FunCat terms overrepresented with respect to increased or decreased abundance (adjusted P value of <0.05) in the wild-type strain exposed to caspofungin and compared to the control (C), in the ΔshoA mutant versus the wild-type strain posttransfer to caspofungin (D), in the ΔmsbA mutant versus the wild-type strain posttransfer to caspofungin (E), and in the ΔopyA mutant versus the wild-type strain posttransfer to caspofungin (F). For the full list, refer to Table S1.
ShoA, MsbA, and OpyA are important for the utilization of several carbon sources.
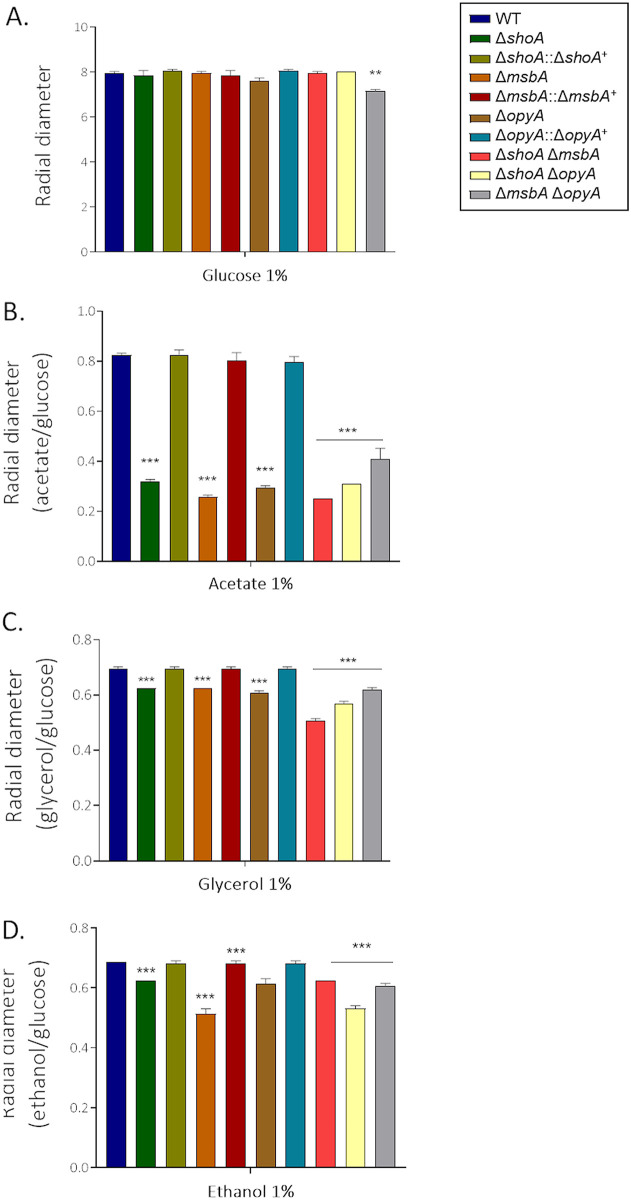
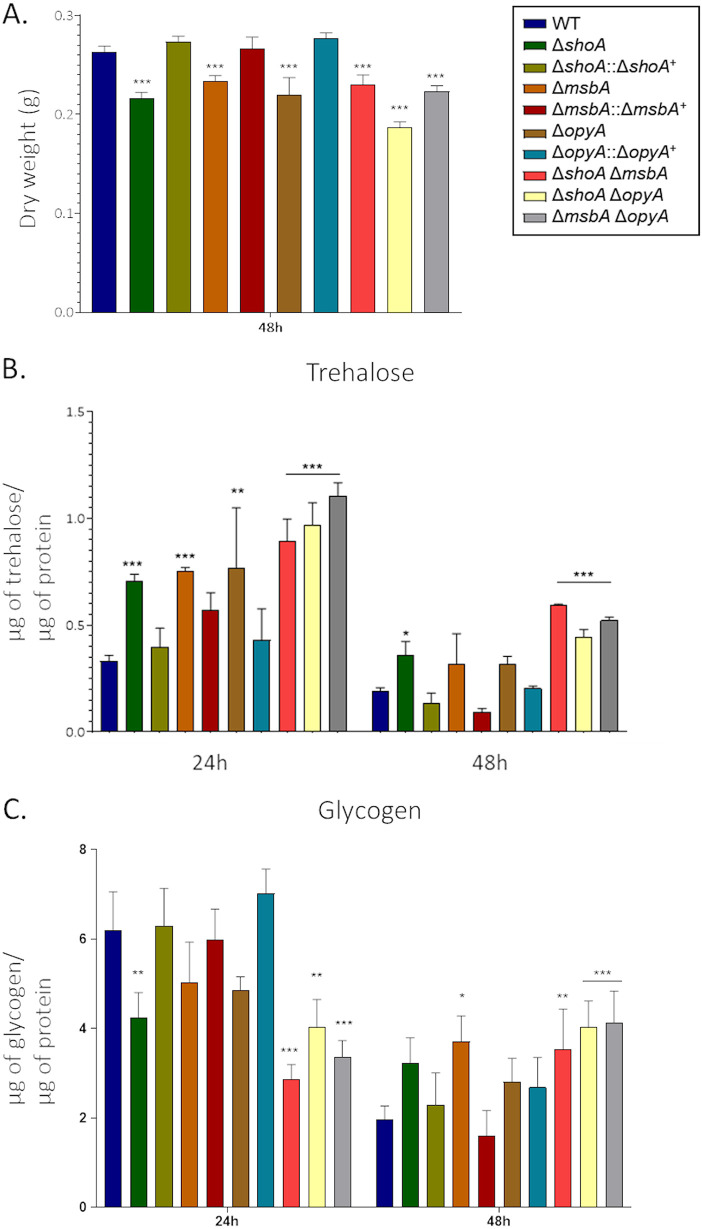
Proteomic data from the ΔshoA, ΔmsbA, and ΔopyA mutants revealed that many proteins involved in the catabolism of glucose and other sugars and amino acids, as well as in glucogenesis, were decreased in abundance (see Table S1). The single and double mutants showed about 10% to 15% growth reduction compared to the wild-type strain on 1% glucose (Fig. 8A), as well as reduced growth on 1% acetate, 1% glycerol, and 1% ethanol (Fig. 8B to D). For each carbon source (acetate, glycerol, or ethanol), the single and double mutants showed about the same level of growth reduction, suggesting that they function in the same pathway for assimilation of these carbon sources (Tables 1 and 2). There was also a reduction in the dry weight of the single and double mutants after 48 h in liquid MM plus 1% glucose (Fig. 9A). Glucose transport was not affected in the mutant strains (see Fig. S4), suggesting that there were no defects in glucose assimilation in these mutants. However, there was increased trehalose accumulation in the single and double mutants at 24 h or 48 h or at both time points (Fig. 9B). We also observed decreased glycogen accumulation in the ΔshoA mutant after 24 h of growth and increased glycogen accumulation in the ΔmsbA mutant at 48 h of growth compared to the wild-type strain (Fig. 9C). Glycogen accumulation was decreased and increased in all three double mutants at 24 and 48 h of growth, respectively (Fig. 9C). These results suggest that glucose metabolism and sugar storage are affected in the putative receptor mutants.
FIG 8.
The ΔshoA, ΔmsbA, and ΔopyA single and double null mutants showed reduced growth on glucose 1% (A), acetate 1% (B), glycerol 1% (C), or ethanol 1% (D) as a single carbon source. The wild-type and null mutant strains were grown for 5 days at 37°C. The results shown represent the means of measurements of the diameter of 3 colonies for each strain ± standard deviation. The data were analyzed (Prism, GraphPad) using two-way ANOVA followed by Bonferroni posttests. The levels of significance compared to the wild-type results are indicated as follows: *, P < 0.1; **, P < 0.01; ***, P < 0.001.
FIG 9.
Altered trehalose and glycogen accumulation in the ΔshoA, ΔmsbA, ΔopyA, and double null mutants. The strains were grown for 24 or 48 h in MM plus 1% glucose. The results shown represent the means of measurements of the diameter of 3 colonies for each strain ± standard deviation. (A) Dry weight. (B) Trehalose accumulation. (C) glycogen accumulation. The data were analyzed (Prism, GraphPad) using two-way ANOVA followed by Bonferroni posttests. The levels of significance compared to the wild-type results are indicated as follows: *, P < 0.1; **, P < 0.01; ***, P < 0.001.
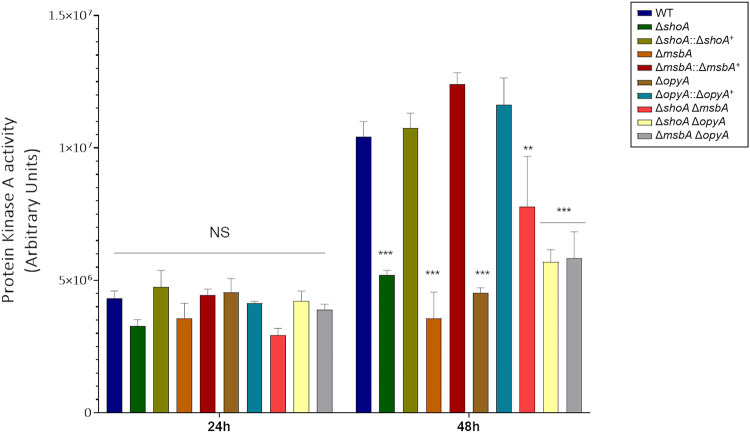
We recently observed that the MAPKs SakA and MpkC are involved in the A. fumigatus cell wall integrity pathway and interact with protein kinase A (PKA) to control carbohydrate mobilization for cell wall remodeling (38). Protein kinase A activity in the single and double mutants was comparable to that seen with the wild-type strain at 24 h of growth but was significantly reduced after 48 h of growth in glucose 1% (Fig. 10). Taken together, these results strongly indicate that the ShoA, MsbA, and OpyA putative receptors can affect carbon source utilization and protein kinase A activity.
FIG 10.
Protein kinase A activity was reduced in the ΔshoA, ΔmsbA, ΔopyA, and double null mutants. The strains were grown for 24 or 48 h in MM plus 1% glucose. The results shown represent the means of measurements of three independent repetitions ± standard deviation. The data were analyzed (Prism, GraphPad) using two-way ANOVA followed by Bonferroni posttests. The levels of significance compared to the wild-type results are indicated as follows: *, P < 0.1; **, P < 0.01; ***, P < 0.001.
The Sln1 branch: genetic interactions between the two branches.
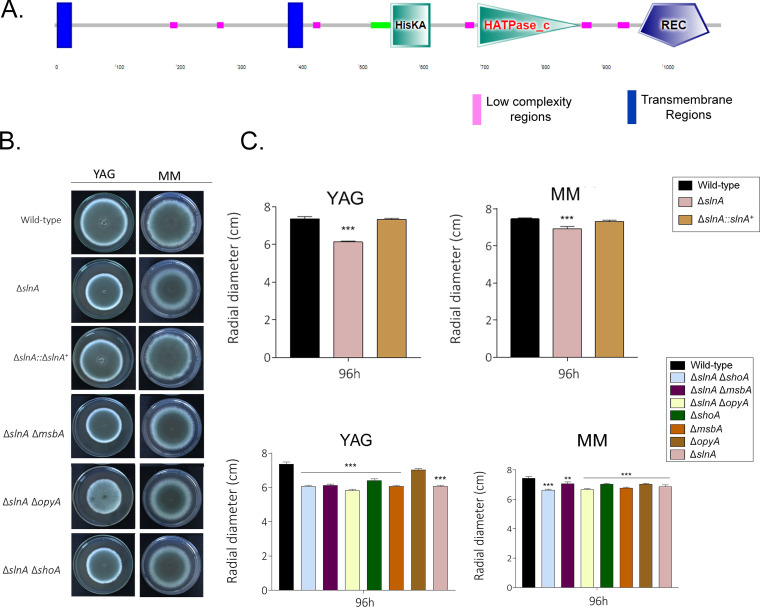
An A. fumigatus Sln1p homologue was previously identified (22). We validated its organization as follows: SlnASln1p (AFUA_2G00660, also called TcsB, www.aspgd.org) has 1,096 amino acids and 43% identity and 63% similarity with Sln1p (E value = 2e−40); it has two transmembrane regions, a HisKA domain (Interpro IPR003661) and a His kinase A phosphoacceptor domain (from amino acid 553 to amino acid 618); HATPase_c (Interpro IPR003594), a histidine kinase-like ATPse (from amino acid 696 to amino acid 863); and a REC domain (Interpro001789), a CheY-homologous receiver domain (from amino acid 960 to amino acid 1077) (Fig. 11A).
FIG 11.
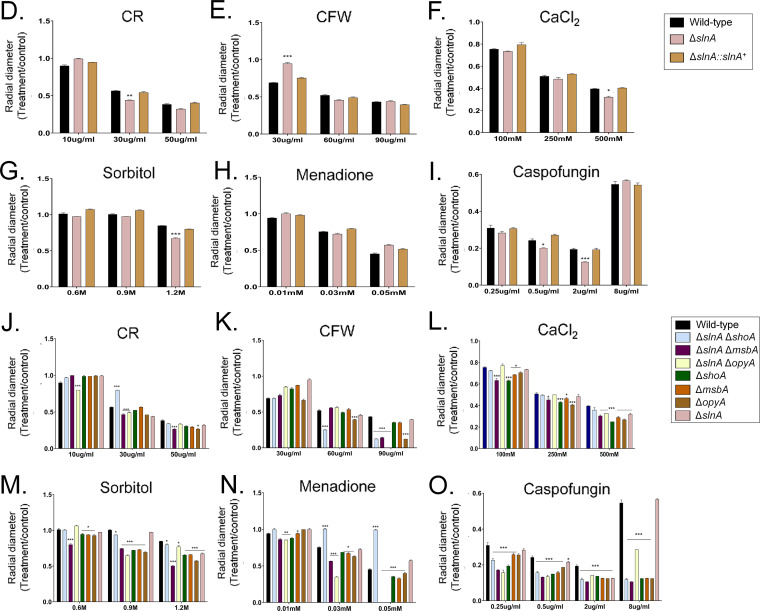
A. fumigatus SlnA and ShoA branches are genetically interacting. (A) Protein organization of A. fumigatus SlnA. (B to O) Growth phenotypes of the wild-type strain and the ΔslnA, ΔshoA, ΔmsbA, ΔopyA, ΔslnA ΔshoA, ΔslnA ΔmsbA, and ΔslnA ΔopyA mutants were determined. The strains were grown for 4 days at 37°C (B) on MM and YAG (C), MM plus Congo red (CR) (D and J), MM plus calcofluor white (CFW) (E and K), MM plus CaCl2 (F and L), MM plus sorbitol (G and M), MM plus menadione (H and N), and MM plus caspofungin (I and O), and their radial growth was quantified. The results shown represent the means of measurements of the diameter of 3 colonies for each strain ± standard deviation. The data were analyzed (Prism, GraphPad) using two-way ANOVA followed by Bonferroni posttests. The levels of significance compared to the wild-type results are indicated as follows: *, P < 0.1; **, P < 0.01; ***, P < 0.001.
The ΔslnA mutant has about 15% reduced growth compared to the wild-type strain in YAG and MM (Fig. 11B and C). The ΔslnA mutant is more sensitive to sorbitol (1.2 M), CaCl2 (500 mM), and caspofungin (0.5 to 2.0 μg/ml) and more resistant to CFW (30 μg/ml) than the wild-type and complemented strains but not to CFW, CaCl2, and menadione (Fig. 11D to I); however, the ΔslnA mutant retained the CPE (Fig. 11I). In the G. mellonella model, infection by the wild‐type and ΔslnA::slnA+ strains resulted in 100% mortality 8 and 9 days postinfection, respectively (see Fig. S5), while the ΔslnA mutant showed a 70% mortality rate 10 days postinfection, which was not statistically significantly different from the rates seen with the wild-type and complemented strains according to Mantel-Cox and Gehan-Breslow-Wilcoxon tests (P < 0.001; see Fig. S5). The double mutants ΔslnA ΔshoA, ΔslnA ΔmsbA, and ΔslnA ΔopyA were as virulent as the single mutants ΔshoA, ΔmsbA, and ΔopyA, suggesting there are no genetic interactions between slnA and shoA, msbA, and opyA for the establishment of virulence in A. fumigatus (Tables 1 and 2).
To investigate possible genetic interactions between the SlnA and ShoA branches, we constructed the double mutants ΔslnA ΔshoA, ΔslnA ΔmsbA, and ΔslnA ΔopyA. They had about 10% to 15% reduced growth compared to the wild-type strain in YAG and MM (Fig. 11B and C). Only the double mutant ΔslnA ΔmsbA was susceptible to 50 μg/ml CR, suggesting there were no genetic interactions with the single mutants in the presence of CR (Fig. 11J; see also Tables 1 and 2). In contrast, all of the double mutants were more sensitive to 90 μg/ml CFW than the corresponding single mutants, suggesting an additive interaction between slnA and shoA, msbA, and opyA (Fig. 11K; see also Tables 1 and 2). All the double mutants were as sensitive to 500 mM CaCl2 and 1.2 M sorbitol as the corresponding single mutants, suggesting that slnA, shoA, msbA, and opyA function in the same pathway for calcium and osmotic stress signaling (Fig. 11L and M; see also Tables 1 and 2). The ΔslnA ΔshoA mutant was less sensitive to 0.05 mM menadione than the corresponding single mutants, suggesting suppression interactions between slnA and shoA, while the slnA and msbA mutants and slnA and opyA mutants were more sensitive to menadione than the corresponding single mutants, suggesting an additive interaction (Fig. 11N; see also Tables 1 and 2). There were no genetic interactions between slnA and shoA, msbA, and opyA, since the ΔslnA double mutants were more resistant to lower and CPE caspofungin concentrations than the corresponding single mutants (Fig. 11O; see also Tables 1 and 2).
We also investigated the impact of slnA deletion on the phosphorylation levels of the MAP kinases SakA and MpkA in the presence of higher osmotic concentrations (1.2 M sorbitol) of CR instead of caspofungin as an alternative agent for mediating cell wall damage (Fig. 12; for an additional repetition of the Western blot assays, see Fig. S3). The wild-type strain showed increased SakA phosphorylation (about 4-fold) when exposed for 10 min to 1.2 M sorbitol (Fig. 12A). The ΔslnA mutant showed a level (about 4-fold at 10 min) of SakA phosphorylation comparable to that seen with the wild-type strain (Fig. 12A). In contrast, there was reduced SakA phosphorylation in the ΔslnA ΔmsbA and ΔslnA ΔopyA strains whereas there was about a 5.4-fold induction of SakA phosphorylation at 10 min of exposure in the ΔslnA ΔshoA strain (Fig. 12A). There was about 2-fold- and 2.3-fold-increased MpkA phosphorylation in the wild-type and ΔslnA strains in the presence of 1.2 M sorbitol, while the ΔslnA ΔmsbA strain showed increases of at least 7.1-fold and 24.8-fold in MpkA phosphorylation (Fig. 12A). The ΔslnA ΔshoA strain exposed to 1.2 M sorbitol for 30 and 60 min showed about 4.1-fold and 13.2-fold phosphorylation (Fig. 12A).
FIG 12.
The A. fumigatus SlnA branch is important for SakA and MpkA phosphorylation. Western blotting assays of SakA and MpkA phosphorylation in response to 1.2 M sorbitol (A) and Congo red 300 μg/ml (B) were performed for different periods of time. Anti-P-p38 SakA and anti-44/42 MpkA antibodies were used to detect the phosphorylation of SakA and MpkA, respectively, while anti-γ-tubulin was used to detect γ-tubulin. Signal intensities were quantified using ImageJ software, and ratios of (P)-SakA to γ-tubulin or (P)-MpkA to γ-tubulin were calculated.
When exposed to CR (300 μg/ml), the wild-type strain showed about 1.7-fold and 1.4-fold more phosphorylation of SakA and MpkA, respectively, at 10 min (Fig. 12B). The increase in SakA phosphorylation in the ΔslnA mutant was comparable to that seen with the wild-type strain (about 1.9-fold) in the presence of CR, but no such increase in MpkA phosphorylation was seen (Fig. 12B). There were increases in SakA phosphorylation in the ΔslnA ΔmsbA mutant of about 2.4-, 2.1-fold, and 3.4-fold in the presence of CR at 10, 30, and 60 min, respectively, and a late increase in SakA phosphorylation (about 1.9-fold at 60 min) in the ΔslnA ΔshoA mutant (Fig. 12B). There was no MpkA phosphorylation induction seen either in the ΔslnA single mutant or in the double mutants (Fig. 12B).
Taken together, these results strongly indicate that the SlnA and ShoA branches have genetic interactions that influence growth under several stressing conditions and are important for the proper activation of the SakA and MpkA cascade and the response to osmotic and cell wall stresses.
DISCUSSION
The ability of pathogenic fungi to survive hostile environments inside and outside the host depends on their ability to respond quickly and robustly to stress (39). MAPK signaling pathways are highly conserved, promoting fungal adaptation to stress by activating the phosphorylation cascades of kinases, which in turn accumulate in the nucleus, triggering adequate cellular responses to different stimuli (40). The S. cerevisiae HOG pathway has already been widely characterized (41, 42). Sho1p and Sln1p are two independent upstream branches that activate the HOG pathway (11). Hkr1, Msb2, and Opy2 interact with Sho1, activating the Ste20-Ste11-Pbs2-Hog1 kinase cascade (9, 11, 43). The Sln1p branch is important for the regulation of hyperosmolarity and hypo-osmolarity responses (11).
Further studies are needed to understand how A. fumigatus activates and controls its HOG pathway. The main objective of this study was to characterize the A. fumigatus putative homologues of the different components of the yeast HOG pathway activation branches Sln1p and Sho1p. Accordingly, we characterized four putative genetic determinants involved in these two branches: SlnASln1p, ShoASho1p, MsbAMsb2p, and OpyAOpy2p. Apparently, there is no A. fumigatus Hrk1p homologue. We have shown that all four genes are not essential and that the ΔslnA mutant is not sensitive to any kind of investigated stress (except osmotic stress) or involved in virulence, while ShoA, MsbA, and OpyA are important for the cell wall integrity pathway and for oxidative stress and virulence. The construction and preliminary phenotypic characterization of ΔshoA, ΔmsbA, and ΔslnA mutants were previously described (21–23). Most of the phenotypes described for the ΔshoA and ΔmsbA mutants (21, 23) were also observed here. However, the growth of the ΔslnA mutant described here was more sensitive to osmotic stress than that of the ΔslnA strain previously described (22). Since the two studies used mutants derived from the same strain, differences could have been due to the growth conditions or medium composition.
The shoA, msbA, and opyA null mutants showed genetic interactions under stressing conditions, indicating that they interact for the activation of stress response mechanisms (Table 2). Although ShoA, MsbA, and OpyA have similar and/or redundant functions, they have unique functions during the activation of the signaling pathways in A. fumigatus. Interestingly, double mutants of the three genes in the Sho1p branch with ΔslnA show genetic interactions to different extents, strongly suggesting the two pathways interact in response to stress conditions. The modulation of MAPK HOG SakA and CWI MpkA phosphorylation levels is altered in these mutants, indicating that these two branches are important for activation of not only HOG but also CWI MAPKs. We cannot discard the possibility that these receptors also affect the MAPK phosphatases, altering the global levels of SakA and MpkA dephosphorylation. We made several unsuccessful attempts to construct triple null mutants with the three putative TM receptors. We were also unable to construct a triple mutant with a conditional xylp::shoA gene in the ΔmsbA ΔopyA background under both repressing and inducing conditions. A single xylp::shoA mutant strain was not able to grow in xylose and induced high SakA and MpkA phosphorylation levels. These results provide direct evidence that the ShoA and SlnA branches are able to modulate activities of MAPKs.
In yeast, genetic analyses showed that the S. cerevisiae Sho1-Opy2 interaction enhances the activation of HOG Pbs2 MAPKKK by Fus3 MAPKK Ste11 but not the activation of Hog1 by Pbs2 (43). The interaction among these putative TM receptors has also been observed in many different human- and plant-pathogenic fungi. In the pathogenic fungus Candida albicans, SLN1 deletion impaired hyphal formation and attenuated virulence (44). C. albicans SHO1 mutants are also sensitive to oxidative stress and to CR and CFW and are defective in morphogenesis (45). C. albicans Msb2p is a global regulator of temperature stress (46), while OPY2 mutants display susceptibility to CR, but it did not play a role in the adaptation to high osmolarity or oxidative stress and virulence in the Galleria mellonella model (47). Sho1 and Msb2 homologues play complementary but distinct roles in stress responses, differentiation, and pathogenicity of the human pathogen Cryptococcus neoformans (48). In the dimorphic plant-pathogenic fungus Ustilago maydis, Sho1 and Msb2-like proteins play a key role during appressorium differentiation and are involved in plant surface sensing (49). In the plant pathogen Verticillium dahliae, Sho1 acts as an osmosensor and is required for plant penetration and melanin biosynthesis (21). A Fusarium oxysporum Δmsb2 mutant was shown to be essential for pathogenicity in tomato and hypersensitive to cell wall targeting compounds (50). F. oxysporum Δsho1 mutants were found to be hypersensitive to CFW, and a Δmsb2 Δsho1 double mutant was even more sensitive. F. oxysporum Sho1 and Msb2 have converging functions activating the Fmk1 MAPK CWI cascade, promoting invasive growth and plant infection, and regulating the CWI pathway (51). These results emphasize the complex roles played by these putative TM receptors and Sln1p homologues in pathogenic fungi.
It is well documented that the response of A. fumigatus to caspofungin involves the combined action of HOG SakA and CWI MpkA MAPK (30, 37, 52). We investigated the protein spectral counts for the wild-type strain and the three putative TM receptor null mutants under unstressed conditions and when exposed to caspofungin. Our results revealed a completely novel aspect of the function of the putative TM receptors: their influence on growth on different carbon sources. The putative TM receptor null mutants showed reduced growth on several carbon sources compared to the wild-type strain. Although they do not have defects in glucose transport, they exhibit aberrant sugar storage metabolism, a strong indicator of problems in carbon assimilation metabolism. Cell wall-related sugars are derived from carbohydrate storage compounds during stress of the cell wall, and this process is dependent on cAMP-dependent protein kinase A (PKA) and on MAPKs SakA and MPkC (38). These protein kinases are important for normal accumulation/degradation of storage sugars through SakA physical interactions with the catalytic and regulatory subunits of protein kinase A PkaC1 and PkaR (38). The putative TM receptor mutants have reduced protein kinase A activity, and the effects on SakA phosphorylation could be the main reason for the reduced growth on several carbon sources.
In summary, we have provided novel information about A. fumigatus SlnAsln1p and ShoASho1p branches and their function in the activation of HOG MAPK pathway. Interestingly, these putative receptors also affect the CWI MpkA pathway. Phosphorylation of CWI MpkA MAPK in response to osmotic and cell wall stress was found to be dependent on HOG MpkC and SakA MAPKs (28, 33). These results indicated that there is a collaboration between the CWI and HOG pathways during cell wall biosynthesis (28, 33). Our work opens new possibilities for understanding the pathways for the activation and maintenance of osmolarity and the cell wall integrity in A. fumigatus. Those pathways are promising targets for drug development and could help to improve the efficacy of antifungal treatment and aspergillosis treatment.
MATERIALS AND METHODS
Strains and culture medium.
The strains were cultured at 37°C in complete yeast glucose (YG, 2% [wt/vol] glucose, 0.5% [wt/vol] yeast extract, trace elements) or minimal medium (MM; 1% [wt/vol] original salts with high nitrate content, trace elements, pH 6.5). For YAG and MM solid media, 1.7% (wt/vol) or 2% (wt/vol) agar were added to YG and MM. Where necessary, urine and uracil (1.2 g/liter) were added. The strains used in this work are described in Table S2 at https://figshare.com/articles/Membrane_receptors_contribute_to_activation_and_efficient_signaling_of_Mitogen-Activated_Protein_Kinase_cascades_during_adaptation_of_Aspergillus_fumigatus_to_different_stressors_and_carbon_sources/12402125.
Construction of A. fumigatus null mutants.
A. fumigatus strain CEA17 was used to generate the following deletion strains used in this study: the ΔmsbA, ΔopyA, and ΔshoA single mutants, with sequences corresponding to msbA (Afu4g04070), opyA (Afu3g14410), and shoA (Afu2g00660) according to the AspGD database (http://www.aspgd.org/). The deletion cassettes were constructed by in vivo recombination in S. cerevisiae as described previously (53). We used approximately 1.0 kb of the 5′ untranslated region (5′-UTR) and the 3′-UTR flanking the target open reading frame (ORF) regions was selected for primer design (see Table S3). Primers 5'Fw and 3'Rv contained a short sequence homologous to plasmid pRS426 and the pyrG gene. Both the 5′-UTR and 3′-UTR fragments were amplified by PCR from A. fumigatus genomic DNA (gDNA). The pyrG gene placed inside the cassette as a prototrophic marker was amplified from plasmid pCDA21. The deletion cassette was generated by transformation into S. cerevisiae SC94721 using the lithium acetate method (54), plus the fragments, together with plasmid pRS426 digested at two sites with enzymes BamHI and EcoRI (New England Biolabs Ltd., United Kingdom). The DNA of the transformants was extracted by a previously described method (55). The deletion cassettes were amplified by PCR from these plasmids using TaKaRa Ex Taq DNA polymerase (Clontech TaKaRa Bio) and were used for the transformation of A. fumigatus. The same protocol was used for the construction of the double mutants. However, the marker gene used for the deletion of the gene was amplified from plasmid pPTRI. The colonies of null mutants were selected by purification of the colonies in 1 μg/ml pyrithiamine hydrobromide (Sigma). The results of Southern blot analysis demonstrated that the transformation cassette was homologously integrated (see Fig. S1). Single gene deletion strains were complemented by cotransformation of approximately 1.0 kb of the 5′-UTR and 3′-UTR plus ORF together with plasmid pPTRI. The complementants were selected by purification of colonies in MM with 1 μg/ml pyrithiamine hydrobromide. Gene complementation was checked by PCR (see Fig. S1). All the primers used in this work are described in Table S3.
Construction of xylP::shoA strain.
The strain of A. fumigatus (CEA17) with pyrG auxotrophic mutant marker was used to generate strain xylP::shoA, whose shoA gene was fused to the xylP promoter, activated by xylose. The cassettes were constructed by in vivo recombination in S. cerevisiae (53), and we used about 1.0 kb of the 5′-UTR flanking region of the region and 3′-UTR of the target ORF regions were selected for the design of the primer. The xylP promoter was amplified from the plasmid pYES-pXyl-hph-devR. The pyrG gene placed inside the cassette as a prototrophic marker was amplified from plasmid pOB430. The fragments necessary for the construction of the cassette were amplified by PCR using the primers listed in Table S3. The construction of the cassette was performed in vivo using S. cerevisiae SC9721 and the lithium acetate method (54), plus the fragments, together with plasmid pRS426 digested in two sites with the enzymes BamHI and EcoRI (New England Biolabs Ltd., United Kingdom). The DNA of the transformants was extracted using a previously described method (55). The xylP::shoA construct cassette was amplified by PCR from these plasmids using TaKaRa Ex Taq DNA polymerase (Clontech TaKaRa Bio) and used for the transformation of A. fumigatus. The colonies of the transformants were selected by purification in MM (1% glucose). Candidates were checked by PCR using 5′-UTR primers external to the cassette used for transformation (see Fig. S1).
Characterization of mutants.
Sensitivity and resistance to several stressing agents were evaluated by measurement of radial growth, comparing the mutant and wild-type strains. A 5-μl volume of a suspension of 2 × 107 conidia was inoculated into MM with or without a corresponding stressing agent, and the plates were incubated for 4 days at 37°C. After this period, the radial diameters were measured and the values were used for statistical analysis. The data were analyzed (Prism, GraphPad) using two-way analysis of variance (ANOVA) followed by Bonferroni posttests. The levels of significance compared to the wild-type results are indicated as follows: *, P < 0.1; **, P < 0.01; ***, P < 0.001.
Immunoblot analysis.
The strains were inoculated (1 × 107 conidia) in 250-ml Erlenmeyer flasks with 50 ml of YAG medium and grown in a rotatory shaker (200 rpm) at 37°C for 16 h before being exposed to different stressing agents, such as Congo red, hydrogen peroxide, sorbitol, and calcium chloride. The proteins were extracted as described previously (56) and quantified according to a previously described method (57). A 60-μg volume of total protein per sample was run on a 12% (wt/vol) SDS-PAGE gel before being transferred to a polyvinylidene difluoride (PVDF) membrane (GE Healthcare). The phosphorylated forms of MpkA and SakA were detected using anti-phospho-p44/42 MAPK and anti-phospho-p38 MAPK (Cell Signaling Technology) antibodies, respectively. Primary antibodies were detected using an anti-rabbit IgG antibody, horseradish peroxidase (HRP) antibody (catalog no. 7074; Cell Signaling Technologies). The detection (chemiluminescence) was performed using a Western ECL Prime (GE Healthcare) blot detection kit according to the manufacturer's instructions. The phosphorylated signal of MpkA and SakA was normalized to the monoclonal antibody IgG γ-tubulin (C-11, sc-17787) ratio. The images generated were submitted for densitometric analysis using ImageJ software (http://rsbweb.nih.gov/ij/index.html).
Enzymatic assays.
Analyses of intracellular trehalose levels (Megazyme) and PKA activity (Promega) were carried out according to the manufacturer’s instructions. Intracellular glycogen levels and extracellular glucose concentrations were quantified as described previously (58).
Cell wall staining.
Cell wall surface polysaccharide straining was performed as described previously (28, 59, 60). Briefly, strains were grown from 2.5 × 103 spores in 200 μl of MM for 16 h at 37°C before the culture medium was removed and germlings were UV irradiated. The hyphal forms were subsequently washed with PBS 1×, and 200 μl of a blocking solution (2% [wt/vol] goat serum, 1% [wt/vol] bovine serum albumin [BSA], 0.1% [vol/vol] Triton X-100, 0.05% [vol/vol] Tween 20, 0.05% [vol/vol] sodium azide, and 0.01 M PBS) was added. Samples were incubated for 30 min at room temperature. For dectin staining, 0.2 μg/ml of Fc-h-dectin-hFc was added to the UV-irradiated germlings and incubated for 1 h at room temperature, followed by the addition of 1:1,000 DyLight 594-conjugated, goat anti-human IgG1 for 1 h at room temperature. The hyphal forms were washed with PBS, and fluorescence was read at 587 nm excitation and 615 nm emission. For chitin staining, 200 μl of a PBS solution with 10 μg/ml of calcofluor white (CFW) was added to the UV-irradiated hyphal forms, incubated for 5 min at room temperature, and washed three times with PBS before fluorescence was read at 380 nm excitation and 450 nm emission. All experiments were performed using 12 repetitions, and fluorescence was read in a microtiter plate reader (SpectraMax i3; Molecular Devices).
Virulence analysis in Galleria mellonella models.
The Galleria mellonella larvae were obtained by breeding adult larvae (61) weighing 275 to 330 mg, kept starving in petri dishes at 37°C in the dark for 24 h prior to infection. All selected larvae were in the final (sixth) stage of larval development. Fresh conidia of each strain of A. fumigatus were counted using a hemocytometer, and the initial concentration of the suspensions of conidia used for the infections was 2 × 108 conidia/ml. A total of 5 μl (1 × 106 conidia/larva) of each suspension was inoculated per larva. The control group was composed of larvae inoculated with 5 μl of PBS to observe death by physical trauma. The inoculation through the last left proleg was performed using a Hamilton syringe (model 7000.5 KH). After infection, the larvae were kept at 37°C in petri dishes in the dark and scored daily. Larvae were considered dead due to lack of movement in response to touch. The viability of the inoculum administered was determined by plating a serial dilution of the conidia on YAG medium and incubating the plates at 37°C for 72 h. All the experiments were performed in parallel and repeated twice.
Purification and identification of proteins by LC-MS/MS.
To precipitate crude protein extracts, they were prepared from ΔshoA, ΔopyA, and ΔmsbA mutant and wild-type cultures that were grown for 24 h and exposed to caspofungin 8 μg/ml (60 min). Crude mycelium protein extracts were obtained by extraction of ground mycelium by the use of B250 buffer (250 mM NaCl, 100 mM Tris-HCl [pH 7.5], 10% glycerol, 1 mM EDTA, and 0.1% NP-40) supplemented with 1.5 ml/liter 1 M dithiothreitol (DTT), 2 tablets/100 ml complete EDTA-free mini-protease inhibitor cocktail (Roche), 3 ml/liter 0.5 M benzamidine, 10 ml/liter phosphatase inhibitors 100× (10 M NaF, 5 M sodium vanadate, 8 M β-glycerol phosphate), and 10 ml/liter of 100 mM phenylmethylsulfonyl fluoride (PMSF). Total protein lysates were centrifuged at 13,000 rpm at 4°C for 10 min, and the supernatant was collected in a new Eppendorf tube. Samples were quantified by the Bradford method (62) and subjected to trypsinization by the use of trypsin (Promega; catalog no. 9PIV511) according to the manufacturer’s instructions. The peptides obtained after trypsinization were bound to a resin (C18 ZipTip pipette tips) according to the manufacturer's instructions (Merck). Digested peptides were separated using reverse-phase liquid chromatography with an Ultimate 3000 NanoLC system (Dionex Corporation, Sunnyvale, CA, USA) followed by mass identification using a Q Exactive mass spectrometer (Thermo Fisher Scientific). Samples were loaded by the use of an autosampler onto a C18 trap column, which was switched on-line with an analytical BioBasic C18 PicoFrit column (C18 PepMap; Dionex) (75-μm inside diameter [i.d.] by 500 mm, 2 μm particle size, 100 Å pore size). Full scans in 300 to 1,700 m/z were recorded with a resolution of 140.000 (m/z 200) with a blocking mass set to 445.12003. Protein identification and label-free quantification (LFQ) normalization were performed using MaxQuant v1.5.2.8 quantitative proteomic software (www.maxquant.org/) in conjunction with Perseus v.1.5.6.0 statistical analysis software (www.maxquant.org/). ANOVA-based multisample t tests were performed using a P value cutoff point of <0.05, allowing the identification of differentially abundant, statistically significant proteins.
RNA extraction and gene expression analysis.
Wild-type and xylP::shoA strains were inoculated (1 × 107 conidia) in 50 ml of MM for 24 h at 37°C. Mycelia were transferred to MM plus 1% xylose for 30 to 240 min. Mycelia were ground to a fine powder in liquid N2, and total RNA was extracted with TRIzol reagent (Thermo Scientific) according to the manufacturer’s protocol. DNA was digested with Turbo DNase I (Ambion Thermo Scientific) according to manufacturer’s instructions. A 2-μg volume of total RNA per sample was subjected to reverse transcription with a High Capacity cDNA reverse transcription kit (Thermo Scientific) using a blend of oligo(dTV) and random primers, according to the manufacturer’s instructions. Quantitative real-time PCRs (qRT-PCRs) were run in a StepOne Plus real-time RT-PCR system (Thermo Scientific) using Power Sybr green PCR master mix (Thermo Scientific). Three independent biological replicates were used, and the mRNA quantity relative fold change values were calculated using standard curves (63). All values were normalized to the expression of the A. fumigatus tubA gene. The primers are described in Table S3.
Data availability.
The proteomic data set can be accessed in Table S1 at https://figshare.com/articles/Membrane_receptors_contribute_to_activation_and_efficient_signaling_of_Mitogen-Activated_Protein_Kinase_cascades_during_adaptation_of_Aspergillus_fumigatus_to_different_stressors_and_carbon_sources/12402125.
ACKNOWLEDGMENTS
We thank the São Paulo Research Foundation (FAPESP; grant no. 2016/07870-9 [G.H.G.], 2016/21392-2 [L.P.S.], and 2014/00789-6 [L.J.D.A.]) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (G.H.G.), both based in Brazil, for financial support. We acknowledge the support of the Irish Research Council (grant no. GOIPG/2018/35 [D.F.]) and Science Foundation Ireland (grant 13/CDA/2142 to O.B.) and the use of the Q-Exactive quantitative mass spectrometer, funded under the Research Infrastructure Call 2012 by Science Foundation Ireland (SFI-12/RI/2346/3).
REFERENCES
- 1.Bassetti M, Giacobbe DR, Grecchi C, Rebuffi C, Zuccaro V, Scudeller L, FUNDICU investigators. 2020. Performance of existing definitions and tests for the diagnosis of invasive aspergillosis in critically ill, adult patients: a systematic review with qualitative evidence synthesis. J Infect 81:131–146. doi: 10.1016/j.jinf.2020.03.065. [DOI] [PubMed] [Google Scholar]
- 2.Kosmidis C, Denning DW. 2015. The clinical spectrum of pulmonary aspergillosis. Postgrad Med J 91:403–410. doi: 10.1136/postgradmedj-2014-206291rep. [DOI] [PubMed] [Google Scholar]
- 3.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 4.Brown NA, Goldman GH. 2016. The contribution of Aspergillus fumigatus stress responses to virulence and antifungal resistance. J Microbiol 54:243–253. doi: 10.1007/s12275-016-5510-4. [DOI] [PubMed] [Google Scholar]
- 5.Posas F, Saito H. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 6.Román E, Arana DM, Nombela C, Alonso-Monge R, Pla J. 2007. MAP kinase pathways as regulators of fungal virulence. Trends Microbiol 15:181–190. doi: 10.1016/j.tim.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Rispail N, Soanes DM, Ant C, Czajkowski R, Grünler A, Huguet R, Perez-Nadales E, Poli A, Sartorel E, Valiante V, Yang M, Beffa R, Brakhage AA, Gow NAR, Kahmann R, Lebrun MH, Lenasi H, Perez-Martin J, Talbot NJ, Wendland J, Di Pietro A. 2009. Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet Biol 46:287–298. doi: 10.1016/j.fgb.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Ma D, Li R. 2013. Current understanding of HOG-MAPK pathway in Aspergillus fumigatus. Mycopathologia 175:13–23. doi: 10.1007/s11046-012-9600-5. [DOI] [PubMed] [Google Scholar]
- 9.Hohmann S. 2009. Control of high osmolarity signalling in the yeast Saccharomyces cerevisiae. FEBS Lett 583:4025–4029. doi: 10.1016/j.febslet.2009.10.069. [DOI] [PubMed] [Google Scholar]
- 10.Hohmann S. 2015. An integrated view on a eukaryotic osmoregulation system. Curr Genet 61:373–382. doi: 10.1007/s00294-015-0475-0. [DOI] [PubMed] [Google Scholar]
- 11.Saito H, Posas F. 2012. Response to hyperosmotic stress. Genetics 192:289–318. doi: 10.1534/genetics.112.140863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maeda T, Takekawa M, Saito H. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 13.O'Rourke SM, Herskowitz I. 2004. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol Biol Cell 15:532–542. doi: 10.1091/mbc.e03-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatebayashi K, Tanaka K, Yang HY, Yamamoto K, Matsushita Y, Tomida T, Imai M, Saito H. 2007. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J 26:3521–3533. doi: 10.1038/sj.emboj.7601796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, Cullen PJ. 2008. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J Cell Biol 181:1073–1081. doi: 10.1083/jcb.200704079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka C, Tan LJ, Mochida K, Kirisako H, Koizumi M, Asai E, Sakoh-Nakatogawa M, Ohsumi Y, Nakatogawa H. 2014. Hrr25 triggers selective autophagy-related pathways by phosphorylating receptor proteins. J Cell Biol 207:91–105. doi: 10.1083/jcb.201402128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C, Jansen G, Zhang J, Thomas DY, Whiteway M. 2006. Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev 20:734–746. doi: 10.1101/gad.1375706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865–870. doi: 10.1016/S0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 19.Saito H. 2001. Histidine phosphorylation and two-component signaling in eukaryotic cells. Chem Rev 101:2497–2509. doi: 10.1021/cr000243+. [DOI] [PubMed] [Google Scholar]
- 20.Bahn YS. 2008. Master and commander in fungal pathogens: the two-component system and the HOG signaling pathway. Eukaryot Cell 7:2017–2036. doi: 10.1128/EC.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang F, Ma D, Wan Z, Liu W, Ji Y, Li R. 2011. The role of sho1 in polarized growth of Aspergillus fumigatus. Mycopathologia 172:347–355. doi: 10.1007/s11046-011-9452-4. [DOI] [PubMed] [Google Scholar]
- 22.Du C, Sarfati J, Latge JP, Calderone R. 2006. The role of the sakA (Hog1) and tcsB (sln1) genes in the oxidant adaptation of Aspergillus fumigatus. Med Mycol 44:211–218. doi: 10.1080/13693780500338886. [DOI] [PubMed] [Google Scholar]
- 23.Gurgel ILDS, Jorge KTOS, Malacco NLSO, Souza JAM, Rocha MC, Fernandes MF, Martins FRB, Malavazi I, Teixeira MM, Soriani FM. 2019. The Aspergillus fumigatus mucin MsbA regulates the cell wall integrity pathway and controls recognition of the fungus by the immune system. mSphere 4:e00350-19. doi: 10.1128/mSphere.00350-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valiante V, Macheleidt J, Föge M, Brakhage AA. 2015. The Aspergillus fumigatus cell wall integrity signaling pathway: drug target, compensatory pathways, and virulence. Front Microbiol 6:325. doi: 10.3389/fmicb.2015.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chelius CL, Ribeiro LFC, Huso W, Kumar J, Lincoln S, Tran B, Goo YA, Srivastava R, Harris SD, Marten MR. 2019. Phosphoproteomic and transcriptomic analyses reveal multiple functions for Aspergillus nidulans MpkA independent of cell wall stress. Fungal Genet Biol 125:1–12. doi: 10.1016/j.fgb.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Manfiolli AO, Siqueira FS, dos Reis TF, Van Dijck P, Schrevens S, Hoefgen S, Föge M, Straßburger M, de Assis LJ, Heinekamp T, Rocha MC, Janevska S, Brakhage AA, Malavazi I, Goldman GH, Valiante V. 2019. Mitogen-activated protein kinase cross-talk interaction modulates the production of melanins in Aspergillus fumigatus. mBio 10:e00215-19. doi: 10.1128/mBio.00215-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frawley D, Stroe MC, Oakley BR, Heinekamp T, Straßburger M, Fleming AB, Brakhage AA, Bayram Ö. 2020. The pheromone module SteC-MkkB-MpkB-SteD-HamE regulates development, stress responses and secondary metabolism in Aspergillus fumigatus. Front Microbiol 11:811. doi: 10.3389/fmicb.2020.00811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruder Nascimento AC, Dos Reis TF, de Castro PA, Hori JI, Bom VL, de Assis LJ, Ramalho LN, Rocha MC, Malavazi I, Brown NA, Valiante V, Brakhage AA, Hagiwara D, Goldman GH. 2016. Mitogen activated protein kinases SakA (HOG1) and MpkC collaborate for Aspergillus fumigatus virulence. Mol Microbiol 100:841–859. doi: 10.1111/mmi.13354. [DOI] [PubMed] [Google Scholar]
- 29.Reyes G, Romans A, Nguyen CK, May GS. 2006. Novel mitogen-activated protein kinase MpkC of Aspergillus fumigatus is required for utilization of polyalcohol sugars. Eukaryot Cell 5:1934–1940. doi: 10.1128/EC.00178-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altwasser R, Baldin C, Weber J, Guthke R, Kniemeyer O, Brakhage AA, Linde J, Valiante V. 2015. Network modeling reveals cross talk of MAP kinases during adaptation to caspofungin stress in Aspergillus fumigatus. PLoS One 10:e0136932. doi: 10.1371/journal.pone.0136932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaimes-Arroyo R, Lara-Rojas F, Bayram Ö, Valerius O, Braus GH, Aguirre J. 2015. The SrkA kinase is part of the SakA mitogen-activated protein kinase interactome and regulates stress responses and development in Aspergillus nidulans. Eukaryot Cell 14:495–510. doi: 10.1128/EC.00277-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrido-Bazán V, Jaimes-Arroyo R, Sánchez O, Lara-Rojas F, Aguirre J. 2018. SakA and MpkC stress MAPKs show opposite and common functions during stress responses and development in Aspergillus nidulans. Front Microbiol 9:2518. doi: 10.3389/fmicb.2018.02518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manfiolli AO, Mattos EC, de Assis LJ, Silva LP, Ulaş M, Brown NA, Silva-Rocha R, Bayram Ö, Goldman GH. 2019. Aspergillus fumigatus high osmolarity glycerol mitogen activated protein kinases SakA and MpkC physically interact during osmotic and cell wall stresses. Front Microbiol 10:918. doi: 10.3389/fmicb.2019.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denning DW. 2002. A new class of antifungal. J Antimicrob Chemother 49:889–891. doi: 10.1093/jac/dkf045. [DOI] [PubMed] [Google Scholar]
- 35.Steinbach WJ, Lamoth F, Juvvadi PR. 2015. Potential microbiological effects of higher dosing of echinocandins. Clin Infect Dis 61(Suppl 6):S669–S677. doi: 10.1093/cid/civ725. [DOI] [PubMed] [Google Scholar]
- 36.Zadra I, Abt B, Parson W, Haas H. 2000. xylP promoter-based expression system and its use for antisense downregulation of the Penicillium chrysogenum nitrogen regulator NRE. Appl Environ Microbiol 66:4810–4816. doi: 10.1128/aem.66.11.4810-4816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ries LNA, Rocha MC, de Castro PA, Silva-Rocha R, Silva RN, Freitas FZ, de Assis LJ, Bertolini MC, Malavazi I, Goldman GH. 2017. The Aspergillus fumigatus CrzA transcription factor activates chitin synthase gene expression during the caspofungin paradoxical effect. mBio 8:e00705-17. doi: 10.1128/mBio.00705-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Assis LJ, Manfiolli A, Mattos E, Fabri JHT, Malavazi I, Jacobsen ID, Brock M, Cramer RA, Thammahong A, Hagiwara D, Ries LNA, Goldman GH. 2018. Protein kinase A and high-osmolarity glycerol response pathways cooperatively control cell wall carbohydrate mobilization in Aspergillus fumigatus. mBio 9:e01952-18. doi: 10.1128/mBio.01952-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown AJP, Cowen LE, di Pietro A, Quinn J. 2017. Stress adaptation, p 463–485. In Heitman J, Howlett BJ, Crous PW, Stukenbrock EH, James TY, Gow NAR (ed), The fungal kingdom. ASM Press, Washington, DC. [Google Scholar]
- 40.Day AM, Quinn J. 2019. Stress-activated protein kinases in human fungal pathogens. Front Cell Infect Microbiol 9:261. doi: 10.3389/fcimb.2019.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. 1993. An osmosensing signal transduction pathway in yeast. Science 259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 42.Krantz M, Ahmadpour D, Ottosson LG, Warringer J, Waltermann C, Nordlander B, Klipp E, Blomberg A, Hohmann S, Kitano H. 2009. Robustness and fragility in the yeast high osmolarity glycerol (HOG) signal-transduction pathway. Mol Syst Biol 5:281. doi: 10.1038/msb.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takayama T, Yamamoto K, Saito H, Tatebayashi K. 2019. Interaction between the transmembrane domains of Sho1 and Opy2 enhances the signaling efficiency of the Hog1 MAP kinase cascade in Saccharomyces cerevisiae. PLoS One 14:e0211380. doi: 10.1371/journal.pone.0211380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada-Okabe T, Mio T, Ono N, Kashima Y, Matsui M, Arisawa M, Yamada-Okabe H. 1999. Roles of three histidine kinase genes in hyphal development and virulence of the pathogenic fungus Candida albicans. J Bacteriol 181:7243–7247. doi: 10.1128/JB.181.23.7243-7247.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Román E, Nombela C, Pla J. 2005. The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol Cell Biol 25:10611–10627. doi: 10.1128/MCB.25.23.10611-10627.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saraswat D, Kumar R, Pande T, Edgerton M, Cullen PJ. 2016. Signalling mucin Msb2 Regulates adaptation to thermal stress in Candida albicans. Mol Microbiol 100:425–441. doi: 10.1111/mmi.13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrero de Dios C, Román E, Diez C, Alonso-Monge R, Pla J. 2013. The transmembrane protein Opy2 mediates activation of the Cek1 MAP kinase in Candida albicans. Fungal Genet Biol 50:21–32. doi: 10.1016/j.fgb.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 48.So YS, Jang J, Park G, Xu J, Olszewski MA, Bahn YS. 2018. Sho1 and Msb2 play complementary but distinct roles in stress responses, sexual differentiation, and pathogenicity of Cryptococcus neoformans. Front Microbiol 9:2958. doi: 10.3389/fmicb.2018.02958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lanver D, Mendoza-Mendoza A, Brachmann A, Kahmann R. 2010. Sho1 and Msb2-related proteins regulate appressorium development in the smut fungus Ustilago maydis. Plant Cell 22:2085–2101. doi: 10.1105/tpc.109.073734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pérez-Nadales E, Di Pietro A. 2011. The membrane mucin Msb2 regulates invasive growth and plant infection in Fusarium oxysporum. Plant Cell 23:1171–1185. doi: 10.1105/tpc.110.075093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pérez-Nadales E, Di Pietro A. 2015. The transmembrane protein Sho1 cooperates with the mucin Msb2 to regulate invasive growth and plant infection in Fusarium oxysporum. Mol Plant Pathol 16:593–603. doi: 10.1111/mpp.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conrad T, Kniemeyer O, Henkel SG, Krüger T, Mattern DJ, Valiante V, Guthke R, Jacobsen ID, Brakhage AA, Vlaic S, Linde J. 2018. Module-detection approaches for the integration of multilevel omics data highlight the comprehensive response of Aspergillus fumigatus to caspofungin. BMC Syst Biol 12:88. doi: 10.1186/s12918-018-0620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schiestl RH, Gietz RD. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 55.Goldman GH, dos Reis Marques E, Duarte Ribeiro DC, de Souza Bernardes LA, Quiapin AC, Vitorelli PM, Savoldi M, Semighini CP, de Oliveira RC, Nunes LR, Travassos LR, Puccia R, Batista WL, Ferreira LE, Moreira JC, Bogossian AP, Tekaia F, Nobrega MP, Nobrega FG, Goldman MHS. 2003. Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: identification of putative homologues of Candida albicans virulence and pathogenicity genes. Eukaryot Cell 2:34–48. doi: 10.1128/ec.2.1.34-48.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fortwendel JR, Juvvadi PR, Perfect BZ, Rogg LE, Perfect JR, Steinbach WJ. 2010. Transcriptional regulation of chitin synthases by calcineurin controls paradoxical growth of Aspergillus fumigatus in response to caspofungin. Antimicrob Agents Chemother 54:1555–1563. doi: 10.1128/AAC.00854-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartree EF. 1972. Determination of protein: a modification of the Lowry method that gives a linear photometric response. Anal Biochem 48:422–427. doi: 10.1016/0003-2697(72)90094-2. [DOI] [PubMed] [Google Scholar]
- 58.Dos Reis TF, Nitsche BM, de Lima PBA, de Assis LJ, Mellado L, Harris SD, Meyer V, Dos Santos RAC, Riaño-Pachón DM, Ries LNA, Goldman GH. 2017. The low affinity glucose transporter HxtB is also involved in glucose signalling and metabolism in Aspergillus nidulans. Sci Rep 7:45073. doi: 10.1038/srep45073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shepardson KM, Ngo LY, Aimanianda V, Latgé JP, Barker BM, Blosser SJ, Iwakura Y, Hohl TM, Cramer RA. 2013. Hypoxia enhances innate immune activation to Aspergillus fumigatus through cell wall modulation. Microbes Infect 15:259–269. doi: 10.1016/j.micinf.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manfiolli AO, dos Reis TF, de Assis LJ, de Castro PA, Silva LP, Hori JI, Walker LA, Munro CA, Rajendran R, Ramage G, Goldman GH. 2018. Mitogen activated kinases (MAPK) and protein phosphatases are involved in Aspergillus fumigatus adhesion and biofilm formation. Cell Surf 1:43–56. doi: 10.1016/j.tcsw.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fuchs BB, O'Brien E, Khoury JB, Mylonakis E. 2010. Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence 1:475–482. doi: 10.4161/viru.1.6.12985. [DOI] [PubMed] [Google Scholar]
- 62.Bradford M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 63.Semighini CP, Marins M, Goldman MH, Goldman GH. 2002. Quantitative analysis of the relative transcript levels of ABC transporter Atr genes in Aspergillus nidulans by real-time reverse transcription-PCR assay. Appl Environ Microbiol 68:1351–1357. doi: 10.1128/aem.68.3.1351-1357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The proteomic data set can be accessed in Table S1 at https://figshare.com/articles/Membrane_receptors_contribute_to_activation_and_efficient_signaling_of_Mitogen-Activated_Protein_Kinase_cascades_during_adaptation_of_Aspergillus_fumigatus_to_different_stressors_and_carbon_sources/12402125.