Abstract
Background: Paired-like homeodomain transcription factor 1 (PITX1) participates in miscellaneous biological processes including cell growth, development, progression and invasion in various malignant tumors. However, the analysis of the association between PITX1 expression and the survival in breast cancer remains unclear. Methods: Clinical prognostic parameters and survival data related to PITX1 in breast cancer patients were performed using the bioinformatic analysis including Oncomine, Bc-GenExMiner v4.3, PrognoScan and UCSC Xena. Results: We found that PITX1 gene expression was significantly higher in different histological classification of breast cancer. The Scarff–Bloom–Richardson (SBR) grade, Nottingham prognostic index (NPI), estrogen receptor (ER) negative, epidermal growth factor receptor-2 (HER2) positive, lymph node positive, triple-negative status and basal-like status were positively correlated with PITX1 level, except for patients’ age and the progesterone receptor (PR) status. We have found that the increased PITX1 expression correlated with worse relapse-free survival, disease specific survival and overall survival. PITX1 was positively correlated with metastatic relapse-free survival and distant metastasis-free survival. We also confirmed positive correlation between PITX1 and the nucleotide-binding oligomerization domain 2 (NOD2). Conclusion: The lower expression of PITX1 was associated with better clinical prognostic parameters and clinical survival in breast cancer according to the bioinformatic analysis.
Keywords: bioinformation, breast cancer, PITX1, prognosis
Introduction
Breast cancer is the most common malignant tumor in women and the primary cause of female cancer death worldwide [1]. Although the application of locoregional surgery, conventional chemotherapy, precision radiotherapy, endocrine therapy and monoclonal antibody has significant benefits for the prognosis of breast cancer patients, still lots of patients were subjected to the threaten of recurrence and death. Prognosis prediction of breast cancer is related to clinical, pathological and molecular characteristics [2]. Therefore, the identification of new prognostic markers can provide new insight to early detection of breast cancer and decrease the mortality and recurrence.
Paired-like homeodomain transcription factor 1 (PITX1) participates in cell growth and development. It was considered as a binary transcription factor involved in the transcription of proopiomelanocortin gene, which may play a role in the differentiation and formation of pituitary cells [3]. In addition, the role of PITX1 in the development of the hind limbs is mainly found in the areas such as cartilage joints, long bones and skeletal muscles [4]. Haploid deficiency of mice and human PITX1 can lead to talipes equinovarus [5]. Therefore, PITX1 was identified as the key role of the hind limb development.
PITX1 is down-regulated as a tumor suppressor in various malignant tumors and correlated with poor prognosis in gastric carcinoma [6], lung carcinoma [7], head and neck squamous cell carcinoma [8], colorectal carcinoma [9], hepatic carcinoma [10], cutaneous malignant melanoma [11], osteosarcoma [12] and clear cell renal cell carcinoma [13]. PITX1 promotes the expression of RAS negative regulators as a transcription factor [14]. The tumorigenic effect of PITX1 gene was achieved by some signaling pathways, such as EMT and WNT/β-catenin signaling pathway that have a positive regulatory effect on proliferation and invasion in gastric cancer [15], while it was related to KRAS, WNT, NF-κB activation pathway and TGF-β signaling pathway in colon cancer [16]. The important role of PITX1 in the development of malignant tumor can be observed from cancer development, differential expression in cancer and related molecular mechanisms. However, the analysis of PITX1 in breast cancer is rare and the association between PITX1 expression and breast cancer patients’ survival remains unknown.
The present study assessed the correlation between PITX1 expression and the prognostic factor of breast cancer by using various online analysis databases to explore the prognostic significance of PITX1 gene in the treatment of breast cancer.
Methods and materials
Oncomine
Oncomine (http://www.oncomine.org), a cancer microarray database and web-based data mining platform, aims to compare the transcriptome data with normal tissues in most major types of cancer [17]. The gene expression level of PITX1 was analyzed by Oncomine. We compared mRNA level of PITX1 for each microarray data between tissue of normal individual and breast cancer patients with designed parameters including 2-fold change, P value ≤ 1E-4 and top 10% gene rank. T-test was used to analyze the expression difference between different breast cancer pathological types and normal tissues in datasets of PITX1 gene overexpression. In addition, the co-expression gene of PITX1 was analyzed concurrently.
Breast cancer gene-expression miner v4.3 (Bc-GenExMiner v4.3)
Bc-GenExMiner v4.3 (http://bcgenex.centregauducheau.fr/BC-GEM/GEM-Accueil.php?js=1) was analyzed from 36 annotated genomic datasets and three statistical mining functions. Open access database for Bc-GenExMiner v4.3 to analyze the relationship between the expression level of breast cancer specific mRNA level of PITX1 gene and the specific clinicopathological characteristics of breast cancer (including age, Scarff–Bloom–Richardson (SBR) grade, Nottingham prognostic index (NPI), estrogen receptor (ER), progesterone receptor (PR), epidermal growth factor receptor 2 (HER2), nodal status, triple-negative status and basal-like status). Survival analysis was exerted with the software. Moreover, the relationship between co-expression genes of PITX1 was analyzed with the database. Data last updated in July 2019.
PrognoScan
PrognoScan (http://dna00.bio.kyutech.ac.jp/PrognoScan/index.html) is the microarray database of the biological relationship between gene expression and clinical prognosis in various cancers [18]. We utilized PrognoScan database to verify the correlation between mRNA level of PITX1 expression and survival with the adjusted cox P value < 0.05 in breast cancer.
UCSC xena
UCSC Xena (http://xena.ucsc.edu/) is a genome-related database including many tumor research database functions, providing visual analysis for public data centers. Heat map of co-expression gene can be analyzed in the data mining of The Cancer Genome Atlas (TCGA) by UCSC Xena browser.
Results
Overexpression of PITX1 gene in breast cancer patients
The expression of PITX1 gene compared with normal individuals and breast cancer patients was detected in 20 kinds of common cancers with tumor online database Oncomine. Up-regulation of PITX1 gene expression was found in breast cancer, colon cancer, kidney cancer, lung cancer and lymphoma, while down-regulation of PITX1 gene expression being detected in bladder cancer, cervical cancer, esophageal cancer and head and neck cancer. Four out of ten meet the threshold, which are the 11 datasets of overexpression of PITX1 gene level from a total number of 43 breast cancer datasets (Figure 1). Compared with normal individuals, the level of PITX1 gene expression was significantly higher in invasive breast carcinoma, invasive ductal breast carcinoma, invasive lobular breast carcinoma, ductal breast carcinoma, medullary breast carcinoma, invasive ductal and invasive lobular breast carcinoma and tubular breast carcinoma (Figure 2A–K, P=9.96E-28, 3.35E-42, 2.88E-13, 1.18E-7, 2.70E-9, 2.49E-1.4, 1.85E-30, 1.60E-39, 1.48E-76, 1.41E-5 and 3.43E-1.4). In the eleven datasets, three datasets of invasive breast carcinoma appears to be the same trend (Figure 2A, P=9.96E-28, Figure 2D, 1.18E-7, Figure 2J, 1.41E-5, Table 1), while two datasets of invasive ductal breast carcinoma (Figure 2B, 3.35E-4, Figure 2I, 1.48E-76, Table 1) and two datasets of invasive lobular breast carcinoma are the same (Figure 2C, 2.88E-13, Figure 2H, 1.60E-39, Table 1).
Figure 1. Expression of PITX1 gene in 20 common tumors compared with paired normal tissues.

Oncomine database was designed with fold change ≥ 2, P value ≤ 1E-4 and gene rank ≥ top 10%. The graphic represents the numbers of datasets with statistically significant (P<0.01) mRNA over-expression (red) or down-expression (blue) of PITX1 (different types of cancer vs. corresponding normal tissue).
Figure 2. Box plots of normal and tumor differentially expression of PITX1 gene in different subtypes of breast cancer.
(A) Invasive Breast Carcinoma, (B) Invasive Ductal Breast Carcinoma, (C) Invasive Lobular Breast Carcinoma, (D) Invasive Breast Carcinoma, (E) Ductal Breast Carcinoma, (F) Medullary Breast Carcinoma, (G) Invasive Ductal and Invasive Lobular Breast Carcinoma, (H) Invasive Lobular Breast Carcinoma, (I) Invasive Ductal Breast Carcinoma, (J) Invasive Breast Carcinoma and (K) Tubular Breast Carcinoma.
Table 1. Different datasets to analyze PITX1 gene expression in pathological classification of breast cancer.
| Breast cancer subtype | P value | T test | Fold change | Sample |
|---|---|---|---|---|
| Invasive Breast Carcinoma | 9.96E-28 | 14.046 | 4.283 | 137 |
| Invasive Ductal Breast Carcinoma | 3.35E-42 | 19.777 | 4.017 | 450 |
| Invasive Lobular Breast Carcinoma | 2.88E-13 | 9.583 | 3.586 | 97 |
| Invasive Breast Carcinoma | 1.81E-7 | 10.351 | 2.334 | 158 |
| Ductal Breast Carcinoma | 2.70E-9 | 7.431 | 5.787 | 47 |
| Medullary Breast Carcinoma | 2.49E-14 | 11.369 | 6.934 | 176 |
| Invasive Ductal and Invasive Lobular Breast Carcinoma | 1.85E-30 | 14.787 | 6.474 | 234 |
| Invasive Lobular Breast Carcinoma | 1.60E-39 | 15.824 | 5.821 | 292 |
| Invasive Ductal Breast Carcinoma | 1.48E-76 | 29.168 | 6.492 | 1700 |
| Invasive Breast Carcinoma | 1.41E-5 | 5.276 | 4.635 | 165 |
| Tubular Breast Carcinoma | 3.43E-14 | 8.774 | 3.373 | 211 |
PITX1 gene expression with different clinical parameters in breast cancer
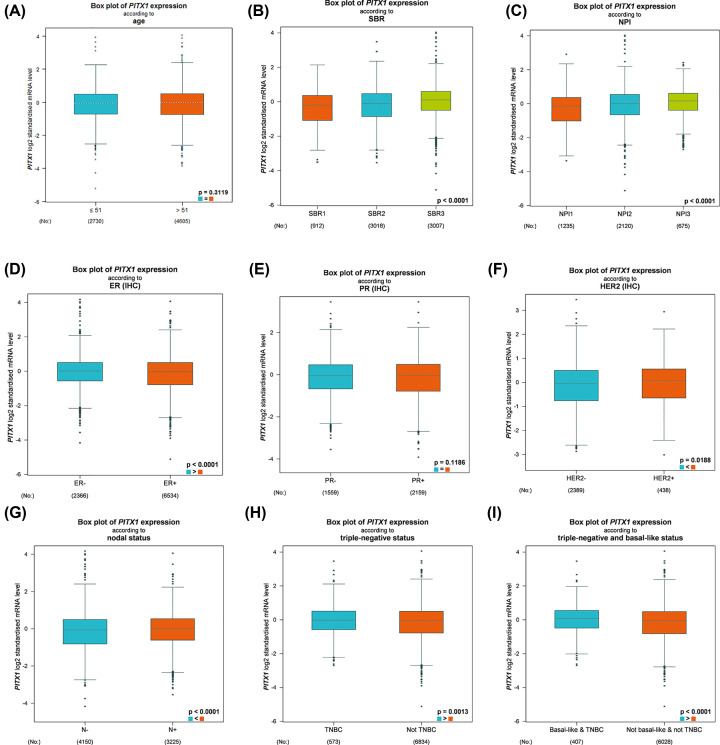
Bc-GenExMiner v4.3 software was used to evaluate the expression of PITX1 gene with several clinical parameters in breast cancer patients. There was no significant difference of PITX1 expression between the <51-year-old group and the >51-year-old group (Figure 3A, P=0.3119, Table 2). The Scarff–Bloom–Richardson grading system (SBR Grade) is the histological grade which based on tumor size (<2 cm, 2–5 cm, ≥5 cm), lymph node status (positive or negative) and vascular invasion status (positive or negative) in breast cancer [19]. The Nottingham Prognostic Index (NPI) is based on histopathological factors (tumor size, lymph node stage and tumor grade) [20]. The expression of PITX1 gene was higher with advanced SBR grade and NPI of breast cancer patients (Figure 3B, P<0.0001, Figure 3C, P<0.0001). The expression of PITX1 gene was higher in ER negative breast cancer patients (Figure 3D, P<0.0001, Table 2). There was no significant difference between the PR positive and PR negative groups (Figure 3E, P=0.1186, Table 2). The expression of PITX1 gene was higher in HER2 positive breast cancer patients (Figure 3F, P=0.0188, Table 2). Compared with lymph node negative patients, the expression of PITX1 gene in lymph node positive patients increased (Figure 3G, P<0.0001, Table 2). In addition, PITX1 was significantly higher in triple negative and basal breast cancer patients than in non-triple negative and non-basal breast cancer patients (Figure 3H, P=0.013, Figure 3I, P<0.0001, Table 2).
Figure 3. Bc-GenExMiner v4.3 to evaluate PITX1 gene expression with box plots according to clinical parameters in breast cancer patients.
(A) Age, (B) SBR grade, (C) NPI, (D) ER, (E) PR, (F) HER-2, (G) nodal status, (H) triple-negative status and (I) basal-like status.
Table 2. PITX1 gene expression analysis in different clinical parameters of breast cancer with Bc-GenExMiner v4.3.
| Variables | No. of patients | PITX1 mRNA | P value* |
|---|---|---|---|
| Age | 0.3119 | ||
| ≤51 | 2730 | – | |
| >51 | 4605 | – | |
| ER | <0.0001 | ||
| Negative | 2366 | Increased | |
| Positive | 6534 | – | |
| PR | – | 0.1186 | |
| Negative | 1559 | – | |
| Positive | 2159 | – | |
| HER-2 | 0.0188 | ||
| Negative | 2389 | – | |
| Positive | 438 | Increased | |
| Nodal status | <0.0001 | ||
| Negative | 4150 | – | |
| Positive | 3225 | Increased | |
| Triple-negative status | 0.0013 | ||
| Non-triple-negative | 6834 | – | |
| Triple-negative | 573 | Increased | |
| Basal-like status | <0.0001 | ||
| Non-basal-like | 7496 | – | |
| Basal-like | 1958 | Increased |
Statistical significance was determined by the Welch’s test.
The effect of the expression of PITX1 gene on prognosis in breast cancer
The survival curves were plotted with different survival information with survival meta-annalistic software PrognoScan, including distant metastasis-free survival, relapse-free survival and disease-specific survival. Breast cancer patients with PITX1 (blue) showed positive related with distant metastasis-free survival (Figure 4A, P=0.024727, Figure 4B, P=0.045388, Figure 4D, P=0.022601, Figure 4E, P=0.040643, Table 3). The Lower PITX1 expression group with blue curve was with preferable relapse-free survival (Figure 4C, P=0.033187, Figure 4F, P=0.048027, Figure 4G, P=0.007207, Table 3). The higher PITX1 gene expression (red) is related with worse disease-specific survival (Figure 4H–J, P=0.008357, P=0.011151, P=0.022379, Table 3). We also analyzed metastatic relapse free survival and overall survival with Bc-GenExMiner v4.3 with the same trend. Expression of PITX1 (purple) presented positive related with metastatic relapse-free survival (Figure 4K, P=0.0105, Table 3) and increased PITX1 expression presented worse overall survival (Figure 4L, P<0.0001, Table 3).
Figure 4. The survival curve of different datasets based on the expression of PITX1 gene was used to analyze the prognostic value in breast cancer.
(A) Distant Metastasis-Free Survival, (B) Distant Metastasis-Free Survival, (C) Relapse-Free Survival, (D) Distant Metastasis-Free Survival, (E) Distant Metastasis-Free Survival, (F) Relapse-Free Survival, (G) Relapse-Free Survival, (H) DiseaseSpecific Survival, (I) Disease-Specific Survival, (J) Disease-Specific Survival, (K) Metastatic relapse-free survival, (L) Overall Survival.
Table 3. Different datasets to analyze the prognosis of PITX1 gene expression in breast cancer.
| Dataset | Probe ID | End point | No. | Cox P-value | HR |
|---|---|---|---|---|---|
| GSE19615 | Distant Metastasis-Free Survival | 209587_at | 115 | 0.024727 | 2.56 [1.13–5.82] |
| GSE19615 | Distant Metastasis-Free Survival | 208502_s_at | 115 | 0.045388 | 1.71 [1.01–2.89] |
| GSE12276 | Relapse-Free Survival | 208502_s_at | 204 | 0.033187 | 1.12 [1.01–1.24] |
| GSE11121 | Distant Metastasis-Free Survival | 209587_at | 200 | 0.022601 | 1.35 [1.04–1.76] |
| GSE11121 | Distant Metastasis-Free Survival | 208502_s_at | 200 | 0.040643 | 1.28 [1.01–1.62] |
| GSE1456-GPL96 | Relapse-Free Survival | 209587_at | 159 | 0.048027 | 1.37 [1.00–1.88] |
| GSE1456-GPL96 | Relapse-Free Survival | 208502_s_at | 159 | 0.007207 | 1.37 [1.09–1.73] |
| GSE1456-GPL96 | Disease-Specific Survival | 208502_s_at | 159 | 0.008357 | 1.46 [1.10–1.94] |
| GSE1456-GPL96 | Disease-Specific Survival | 209587_at | 159 | 0.011151 | 1.65 [1.12–2.42] |
| GSE3494-GPL96 | Disease-Specific Survival | 208502_s_at | 236 | 0.022379 | 1.24 [1.03–1.49] |
Co-expression analysis of PITX1 gene
Oncomine database was analyzed to assess the related co-expression gene with PITX1 gene. A total number of 13,363 samples (111 datasets) were searched for the co-expression gene of PITX1 in Oncomine database. The co-expression profile of PITX1 was identified with a large cluster of 12,624 measured genes across 103 invasive breast carcinomas, the nucleotide-binding oligomerization domain 2 (NOD2) is a principal co-expression gene (Figure 5A). The co-expression gene NOD2 was associated with PITX1 positively according to Bc-GenExMiner v4.3 (Figure 5B, P<0.0001). We analyzed heat map of PITX1 and NOD2 gene expression in 50 gene QPCR analysis (pam50) (Figure 5C) to make sure positive co-expression relationship in breast cancer subtypes of TCGA database generated by UCSC Xena tool.
Figure 5. Co-expression analysis of gene PITX1 and other genes by Oncomine database analysis.
(A) Co-expression analysis of gene PITX1 in 103 invasive breast cancer with Oncomine database. (B). The co-expression gene NOD2 related to PITX1 by bc-GenExMiner software (C). In TCGA database generated by UCSC Xena tool in breast cancer subtypes, the heat map of PITX1 and NOD2 gene expression in 50 gene QPCR analysis (pam50) was analyzed (red represents high expression of gene, blue represents low expression of gene).
Discussion
PITX1 is a member of PITX family member that was considered as development related gene also and tumor suppressor in various carcinoma.
The differential expression of PITX1 was found in many tumors. PITX1 binding to the esterogen receptor α (ERα) acted on the ERα enhancer concentration, and controlled the transcription activity through the binding of ERα target gene and PITX1-binding site in breast cancer cells [21]. However, there was no prognostic association analysis in breast cancer.
The present study demonstrated that the expression of PITX1 was up-regulated in breast cancer patients with respect to normal individuals according to Oncomine database. High PITX1 expression was correlated with poor clinicopathological features in breast cancer. The present results revealed that higher expression of PITX1 was located in different histological classification of breast cancer with respect to normal individuals, including invasive breast carcinoma, invasive ductal breast carcinoma, invasive lobular breast carcinoma, ductal breast carcinoma, medullary breast carcinoma, invasive ductal and invasive lobular breast carcinoma and tubular breast carcinoma. There was no statistical significance in age and PR status in the analysis. It had been reported that SBR grade and NPI are prognostic factors of breast cancer. The expression of PITX1 gene was significantly higher in SBR Grade, NPI, ER negative, HER2 positive, lymph node positive, triple-negative and basal-like status, which were related to poor prognosis of breast cancer respectively. Therefore, overexpression of PITX1 gene may be new biomarker factor related to the poor prognosis in breast cancer. The survival curve of different datasets based on the expression of PITX1 gene was used to analyze the prognostic value in breast cancer with PrognoScan. According to meta-analysis of survival curve data, ten datasets with clinical statistical value were presented. Breast cancer patients with increased PITX1 gene showed worse relapse-free survival, disease-specific survival. PITX1 was positively correlated with distant metastasis-free survival. The same trend also is confirmed that the increased expression of PITX1 gene showed worse overall survival, and PITX1 was positively correlated with metastatic relapse free survival in Bc-GenExMiner v4.3.
We also explored the co-expression relationship gene of PITX1 gene with Oncomine database, which was also verified in Bc-GenExMiner v4.3. NOD2 were regarded co-expression gene of closely related to PITX1 genes positively analyzed by Bc-GenExMiner v4.3. NOD2 increases the risk of breast cancer [22], the research showed that the overexpression of NOD2 inhibited cell proliferation and promoted clone formation in three negative breast cancer cell lines [23]. The incidence of lymph nodes without metastatic is higher in carriers of NOD2 mutations as a prognostic factor [24]. Therefore, the positive correlation between NOD2 and PITX1 gene is reliable.
PITX1 is down-regulated in various malignant tumors and correlated with poor prognosis, may be because it enrichs in promoters of binding genes and regulates gene expression as a transcription factor [25]. PITX1 binds to the target gene PDCD5, which is an apoptosis related gene, so the down-regulation of PITX1 is associated with poor prognosis in gastric cancer [26]. DNA hypermethylation silences PITX1, and the hypermethylation of PITX1 is associated with poor prognosis in esophageal squamous cell carcinoma (ESCC) [27]. Hypermethylation of exon 3 of PITX1 was significantly associated with the risk of death in patients with head and neck squamous cell carcinomas (HNSCC) [28]. There was no prognostic association analysis in breast cancer. High expression of PITX1 in our research may be because gene methylation level of breast cancer differs from other cancers. As a transcription factor binding ERα in breast cancer [21], high PITX1 expression may have a certain impact on poor prognosis, which is worth further study.
In general, under the analysis of various bioinformatic tools, the down-regulation of PITX1 gene was associated with better prognostic clinical parameters such as ER positive, nodal status negative, non-triple-negative, non-basal like status, SBR grade, and NPI. In breast cancer patients, increased PITX1 expression is correlated with worse relapse-free survival, disease-specific survival and overall survival. PITX1 was positively correlated with metastatic relapse-free survival and distant metastasis-free survival. It indicates that breast cancer patients may benefit and have better survival with lower expression of PITX1 gene. Therefore, it may provide some basis for targeted drug therapy related to breast cancer.
Abbreviations
- ER
estrogen receptor
- HER2
human epidermal growth factor receptor-2
- NOD2
nucleotide-binding oligomerization domain 2
- NPI
Nottingham prognostic index
- PITX1
paired-like homeodomain transcription factor 1
- PR
progesterone receptor
- SBR
Scarff–Bloom–Richardson
- TCGA
The Cancer Genome Atlas
Contributor Information
Lei Gan, Email: ganlei19870810@163.com.
Zhixiang Zhuang, Email: 13951106391@139.com.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This research was supported by the National Natural Sciences Foundation of China [grant number 81803553] and the National Natural Science Foudation Pre-research Program of China [grant number SDFEYGJ1608].The research was also be supported by Discipline boosting program of Shanghai Fourth People's Hospital in 2020[grant number SY-XKZT-2020].
Author Contribution
Q.W. designed the experiment and analyzed the data then wrote the paper. S.Z. assisted in analyzing the data and revised the comments. S.Z. and Q.W. are co-first authors with equal contributions. L.G. and Z.Z. corrected the paper.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A. and Jemal A. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Duffy M.J., Walsh S., McDermott E.W. and Crown J. (2015) Biomarkers in Breast Cancer: Where Are We and Where Are We Going? Adv. Clin. Chem. 71, 1–23 10.1016/bs.acc.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 3.Lamonerie T., Tremblay J.J., Lanctot C., Therrien M., Gauthier Y. and Drouin J. (1996) Ptx1, a bicoid-related homeo box transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 10, 1284–1295 10.1101/gad.10.10.1284 [DOI] [PubMed] [Google Scholar]
- 4.Picard C., Azeddine B., Moldovan F., Martel-Pelletier J. and Moreau A. (2007) New Emerging Role of Pitx1 Transcription Factor in Osteoarthritis Pathogenesis. Clin. Orthop. Relat. Res. 462, 59–66 10.1097/BLO.0b013e3180d09d9c [DOI] [PubMed] [Google Scholar]
- 5.Alvarado D.M., McCall K., Aferol H., Silva M.J., Garbow J.R., Spees W.M. et al. (2011) Pitx1 haploinsufficiency causes clubfoot in humans and a clubfoot-like phenotype in mice. Hum. Mol. Genet. 20, 3943–3952, Epub 2011 Jul 20 10.1093/hmg/ddr313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiao F., Gong P., Song Y., Shen X., Su X., Li Y. et al. (2018) Downregulated PITX1 Modulated by MiR-19a-3p Promotes Cell Malignancy and Predicts a Poor Prognosis of Gastric Cancer by Affecting Transcriptionally Activated PDCD5. Cell. Physiol. Biochem. 46, 2215–2231 10.1159/000489590 [DOI] [PubMed] [Google Scholar]
- 7.Chen Y., Knoesel T., Ye F., Pacyna-Gengelbach M., Deutschmann N. and Petersen I. (2007) Decreased PITX1 homeobox gene expression in human lung cancer. Lung Cancer 55, 287–294, Epub 2006 Dec 8 10.1016/j.lungcan.2006.11.001 [DOI] [PubMed] [Google Scholar]
- 8.Takenobu M., Osaki M., Fujiwara K., Fukuhara T., Kitano H., Kugoh H. et al. (2016) PITX1 is a novel predictor of the response to chemotherapy in head and neck squamous cell carcinoma. Mol. Clin. Oncol. 5, 89–94 10.3892/mco.2016.880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knosel T., Chen Y., Hotovy S., Settmacher U., Altendorf-Hofmann A. and Petersen I. (2012) Loss of desmocollin 1-3 and homeobox genes PITX1 and CDX2 are associated with tumor progression and survival in colorectal carcinoma. Int. J. Colorectal Dis. 27, 1391–1399 10.1007/s00384-012-1460-4 [DOI] [PubMed] [Google Scholar]
- 10.Calvisi D.F., Ladu S., Conner E.A., Seo D., Hsieh J.T., Factor V.M. et al. (2011) Inactivation of Ras GTPase-activating proteins promotes unrestrained activity of wild-type Ras in human liver cancer. J. Hepatol. 54, 311–319 10.1016/j.jhep.2010.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osaki M., Chinen H., Yoshida Y., Ohhira T. and Kugoh H. (2013) Decreased PITX1 gene expression in human cutaneous malignant melanoma and its clinicopathological significance. Eur. J. Dermatol. 23, 344–349 [DOI] [PubMed] [Google Scholar]
- 12.Kong G., Liu Z., Wu K., Zhang Y., Deng Z., Feng W. et al. (2015) Strong expression of paired-like homeodomain transcription factor 1 (PITX1) is associated with a favorable outcome in human osteosarcoma. Tumour Biol. 36, 7735–7741 10.1007/s13277-015-3512-1 [DOI] [PubMed] [Google Scholar]
- 13.Yu Z., Chen D., Su Z., Li Y., Yu W., Zhang Q. et al. (2014) miR8863p upregulation in clear cell renal cell carcinoma regulates cell migration, proliferation and apoptosis by targeting PITX1. Int. J. Mol. Med. 34, 1409–1416 10.3892/ijmm.2014.1923 [DOI] [PubMed] [Google Scholar]
- 14.Downward J. (2005) RNA interference libraries prove their worth in hunt for tumor suppressor genes. Cell 121, 813–815 10.1016/j.cell.2005.06.002 [DOI] [PubMed] [Google Scholar]
- 15.Liu L., Tian Y.C., Mao G., Zhang Y.G. and Han L. (2019) MiR-675 is frequently overexpressed in gastric cancer and enhances cell proliferation and invasion via targeting a potent anti-tumor gene PITX1. Cell. Signal. 62, 109352 10.1016/j.cellsig.2019.109352 [DOI] [PubMed] [Google Scholar]
- 16.Watanabe T., Kobunai T., Yamamoto Y., Matsuda K., Ishihara S., Nozawa K. et al. (2011) Differential gene expression signatures between colorectal cancers with and without KRAS mutations: crosstalk between the KRAS pathway and other signalling pathways. Eur. J. Cancer 47, 1946–1954 10.1016/j.ejca.2011.03.029 [DOI] [PubMed] [Google Scholar]
- 17.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D. et al. (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizuno H., Kitada K., Nakai K. and Sarai A. (2009) PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics 2, 18 10.1186/1755-8794-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Doussal V., Tubiana-Hulin M., Friedman S., Hacene K., Spyratos F. and Brunet M. (1989) Prognostic value of histologic grade nuclear components of Scarff-Bloom-Richardson (SBR). An improved score modification based on a multivariate analysis of 1262 invasive ductal breast carcinomas. Cancer 64, 1914–1921 [DOI] [PubMed] [Google Scholar]
- 20.Galea M.H., Blamey R.W., Elston C.E., Ellis I.O., (1992), The Nottingham Prognostic Index in primary breast caner. Breast Cancer Res Treat. 22, 207–219 10.1007/BF00666005 [DOI] [PubMed] [Google Scholar]
- 21.Stender J.D., Stossi F., Funk C.C., Charn T.H., Barnett D.H. and Katzenellenbogen B.S. (2011) The estrogen-regulated transcription factor PITX1 coordinates gene-specific regulation by estrogen receptor-alpha in breast cancer cells. Mol. Endocrinol. 25, 1699–1709 10.1210/me.2011-0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J., He C., Xu Q., Xing C. and Yuan Y. (2014) NOD2 polymorphisms associated with cancer risk: a meta-analysis. PLoS ONE 9, e89340 10.1371/journal.pone.0089340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Velloso F.J., Campos A.R., Sogayar M.C. and Correa R.G. (2019) Proteome profiling of triple negative breast cancer cells overexpressing NOD1 and NOD2 receptors unveils molecular signatures of malignant cell proliferation. BMC Genomics 20, 152 10.1186/s12864-019-5523-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huszno J. and Kolosza Z. (2019) Molecular characteristics of breast cancer according to clinicopathological factors. Mol. Clin. Oncol. 11, 192–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamizu K., Sharov A.A., Piao Y., Amano M., Yu H., Nishiyama A. et al. (2016) Generation and gene expression profiling of 48 transcription-factor-inducible mouse embryonic stem cell lines. Sci. Rep. 6, 25667 10.1038/srep25667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiao F., Gong P., Song Y., Shen X., Su X., Li Y. et al. (2018) Downregulated PITX1 Modulated by MiR-19a-3p Promotes Cell Malignancy and Predicts a Poor Prognosis of Gastric Cancer by Affecting Transcriptionally Activated PDCD5. Cell. Physiol. Biochem. 46, 2215–2231 10.1159/000489590 [DOI] [PubMed] [Google Scholar]
- 27.Otsubo T., Yamada K., Hagiwara T., Oshima K., Iida K., Nishikata K. et al. (2017) DNA hypermethyation and silencing of PITX1 correlated with advanced stage and poor postoperative prognosis of esophageal squamous cell carcinoma. Oncotarget 8, 84434–84448 10.18632/oncotarget.21375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sailer V., Charpentier A., Dietrich J., Vogt T.J., Franzen A., Bootz F. et al. (2018) Intragenic DNA methylation of PITX1 and the adjacent long non-coding RNA C5orf66-AS1 are prognostic biomarkers in patients with head and neck squamous cell carcinomas. PLoS ONE 13, e0192742 10.1371/journal.pone.0192742 [DOI] [PMC free article] [PubMed] [Google Scholar]