Introduction
Many targets of the Wnt/β-catenin signaling pathway are regulated by TCF transcription factors, which play important roles in animal development, stem cell biology and oncogenesis. TCFs can regulate Wnt targets through a “transcriptional switch”, repressing gene expression in unstimulated cells, and promoting transcription upon Wnt signaling. However, it is not clear whether this switch mechanism is a general feature of Wnt gene regulation, or limited to a subset of Wnt targets. Co-repressors of the TLE family are known to contribute to repression of Wnt targets in the absence of signaling, but how they are inactivated or displaced by Wnt signaling is poorly understood. In this mini-review, we discuss several recent reports that address the prevalence and molecular mechanisms of the Wnt transcription switch, including the finding of Wnt-dependent ubiquitination/inactivation of TLEs. Together, these findings highlight the growing complexity of the regulation of gene expression by the Wnt pathway.
The Wnt pathway facilitates short-range signaling during development and in adult tissues. It has important roles in stem cell homeostasis and in the etiology of several cancers and human diseases.[1,2] Transcriptional regulation by the Wnt pathway begins with the stabilization and nuclear accumulation of β-catenin.[3,4] This Wnt/β-catenin pathway (also known as canonical Wnt signaling) regulates gene expression via recruitment of β-catenin to Wnt-responsive cis-regulatory modules (W-CRMs).[5] By far the best-characterized family of transcription factors that bind β-catenin and regulate W-CRMs belong to the T-cell factor/lymphoid enhancer-binding factor 1 (TCF/LEF1) family,[6] hereafter referred to as TCFs.
Shortly after the discovery that transcriptional activation by Wnt signaling was mediated by TCFs through direct binding to β-catenin,[7–9] it was also recognized that TCFs have an additional β-catenin-independent function: repression of Wnt target gene expression in the absence of signaling. This realization came from a couple of key observations. First, in Drosophila and C. elegans embryos, loss of TCF activity can elevate Wnt transcriptional readouts.[10–13] Second, characterization of TCF regulation of the siamois promoter in Xenopus blastomeres reveals that TCF binding sites mediate both basal repression and β-catenin-dependent activation of this element.[14,15] TCF repression of Wnt target gene expression is important for many aspects of animal development across the evolutionary spectrum[11,12,16–18] and is crucial for maintenance of embryonic stem cells.[19–22] In addition, TCFs act as tumor suppressors in colorectal cancer.[23,24] Given the importance of basal TCF repression for the proper regulation of Wnt target genes, understanding the mechanism of this repression and how β-catenin converts TCFs to transcriptional activators has been a major focus of the field.
A key insight into the mechanism of transcriptional inhibition by TCFs was the finding that they can directly bind to members of the transducin-like enhancer of split (TLE) family of transcriptional co-repressors, and this association counteracts TCF/β-catenin transactivation activity.[25,26] These observations favor a model of dual regulation of Wnt targets by TCFs, TLEs and β-catenin (Fig. 1). Targets of other signaling pathways (e.g., Ras and Notch signaling), while regulated by different transcription factors and co-regulators, nonetheless have a similar dual regulation, leading to the idea of a “transcriptional switch” controlling the expression of genes induced by signaling pathways.[27]
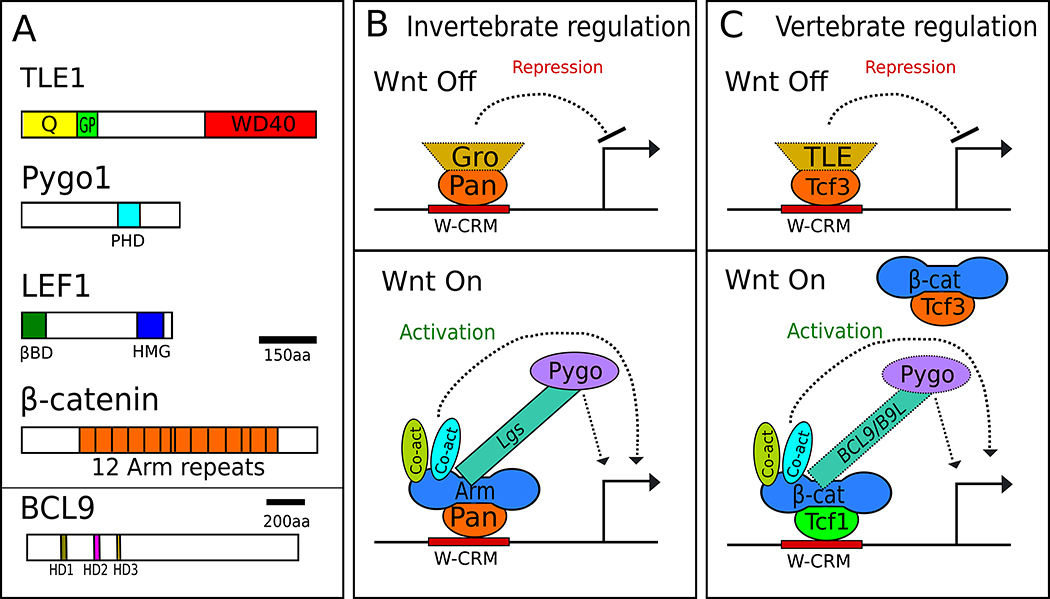
Fig 1. Traditional models of the Wnt transcriptional switch.
A) Domain architecture of the principal components of the switch, using human proteins of each family as archetypes. Colored portions represent interaction domains: Q – glutamine-rich domain, GP – Glycine/Proline-rich domain, PHD – PHD zinc finger domain, βBD – β-catenin binding domain, HMG – High Mobility Group, PBD – Pygopus binding domain, HD1-3 – Homology Domains 1-3 B) The transcriptional switch model in invertebrates. In the absence of Wg signaling in Drosophila, Pan recruits Gro to W-CRMs and represses transcription. When Wg signaling is activated, Gro is displaced and Arm binds to Pan, recruiting Lgs, Pygo and other co-activators, which activate transcription. Lgs acts as an adaptor between Arm and Pygo. C) The TCF switch model in vertebrate Wnt signaling. In the absence of Wnt signaling, repressive TCFs such as TCF3 are bound to W-CRMs. TLEs likely plays a role for repression of many targets. Upon Wnt stimulation, TCF3 is replaced by activating TCFs such as TCF1, which recruits β-catenin and other co-activators. Some factors in panels B and C are depicted with dotted lines to highlight the likelihood that they likely do not regulate all Wnt targets in their respective systems.
TLEs act with many repressive transcription factors in addition to TCFs.[28,29] TLEs form homotetramers through their N-terminal “Q” domains and bind directly to histone deacetylases through the adjacent (GP) domain (Fig. 1A). Both interactions are thought to contribute to the ability of TLEs to repress transcription.[30–33] In addition, the C-terminal WD40 domain is implicated in nucleosome binding and condensation.[34] Drosophila have one TLE, Groucho (Gro), which antagonizes signaling by Wingless (Wg, a fly Wnt).[25,35,36] The Gro homolog UNC-37 plays a similar role in C. elegans embryogenesis.[37] There are four TLEs in mammalian genomes, also known as Groucho related genes (Grgs), along with a fifth family member lacking the WD40 domain that is thought to act as a naturally occurring dominant negative.[38] Several mammalian TLEs have been linked to repression of Wnt target genes.[30,39–43]
In this mini-review we will discuss several recent reports that revise the classic Wnt transcriptional switch model. These reports address the importance of TLEs in repressing Wnt targets, highlight new players and mechanisms by which Wnt signaling inactivates TLE activity, and identify other factors that may contribute to the transcriptional switch.
Is the transcriptional switch a general feature of Wnt gene regulation?
Analysis of TCF mutants in several systems has provided abundant evidence that they can antagonize Wnt gene regulation.[6,40,44–48] Because vertebrates contain four or five TCF genes, loss of function studies cannot easily address whether all Wnt targets are repressed by TCFs, due to potential redundancy. The situation is simpler in nematodes and flies, which contain only one TCF gene. In these invertebrates, there are examples where loss of TCF resulted in no detectable derepression, i.e., increase in expression of Wnt targets.[16,49] In addition, while some transcriptional reporters of Wnt signaling are derepressed by the mutation of TCF binding sites,[14,50,51] many others simply show a loss of Wnt-dependent activation and no evidence of derepression.[52,53] However, these reporters may not fully recapitulate all aspects of their cognate gene’s expression. Thus, the fraction of Wnt targets regulated by a switch-based mechanism remains an open question.
A recent paper addresses this question in Kc167 cells, a Drosophila cell line commonly used to study Wg gene regulation. Using CRISPR/Cas9 editing, the authors isolated cell lines lacking Pangolin (Pan) or Armadillo (Arm), the fly TCF and β-catenin, respectively.[54] RNAseq analysis identified 40 genes activated by Wg treatment, all of which required both Arm and Pan for activation. A fraction (37.5%) of these displayed significant depression when Pan was absent in unstimulated cells.[54] This data set reinforces the view that TCF-mediated repression of Wnt targets in the absence of Wnt signaling is common, but not universally observed (Fig. 2A).
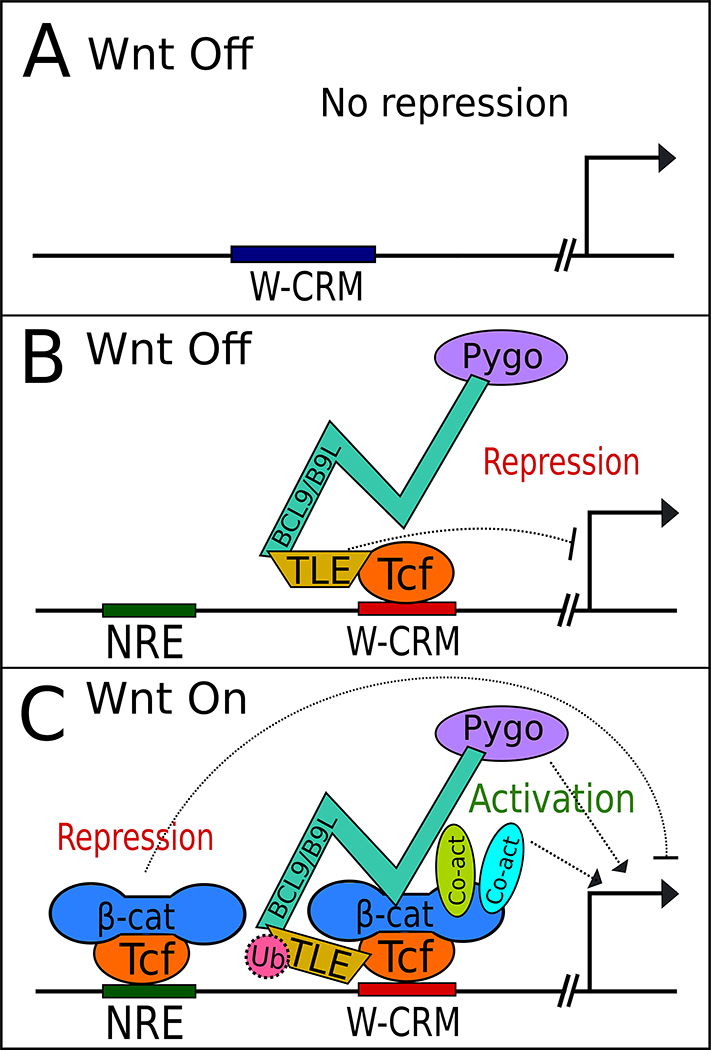
Fig. 2.
Emerging complexities in Wnt target gene regulation covered in this mini-review. A) As described by Franz et al, many Wnt target genes show no evidence of TCF/Pan dependent repression in the absence of Wnt signaling.[54] B) Model proposed in vanTienen et al, where in the absence of Wnt signaling, TLE, BCL9/B9L and Pygo proteins co-occupy W-CRMs.[60] C) This model postulated that these factors are also associated with TCF in Wnt-stimulated cells, along with β-catenin and other co-activators. There is also evidence that TLEs are inactivated by ubiquitylation via UBR5 (not shown).[42] In addition, newly discovered negative regulatory elements (NREs) are proposed to be bound by TCFs and β-catenin, repressing transcription upon Wnt stimulation, fine-tuning the transcriptional response of traditional W-CRMs.[76] Each of these models have important considerations. The work of Franz et al is limited to a single Drosophila cell line, while the “TCF complex reconfiguration” model of vanTienen et al is based on complex composition in solution, not on W-CRM chromatin. The extent of UBR5 modification of TLEs, while evolutionarily conserved, requires further study, as does the functional importance of NREs in regulating Wnt targets. See text for further comment.
One caveat of this study is the possibility of other repressive mechanisms acting redundantly with TCF in this system. For example, the Wnt target naked cuticle (nkd) was derepressed by RNAi depletion of Pan or Gro. Additional depletion of the co-repressor CtBP with Pan or Gro resulted in a synergistic derepression.[35] It would be interesting to see whether the 62.5% of Wnt targets identified by Franz and colleagues that are not derepressed by Pan depletion are sensitive to simultaneous loss of Pan and CtBP.
Does activation of Wnt signaling lead to displacement of TLEs from W-CRMs?
How important are TLEs in regulating Wnt targets? While there are several examples of derepression of Wnt target gene expression in Drosophila gro mutants,[25,35,36,42] the functional evidence implicating these co-repressors in vertebrate systems is largely based on overexpression.[40,43] However, ChIP-seq data from early Xenopus gastrulae, reveals an 86% correlation between β-catenin and TLE-bound sites.[55] Similarly, in mouse hair follicle stem cells, more than half the genes occupied by TCFs are also occupied by TLEs.[40] These studies suggest that TLE-regulation of Wnt targets is widespread.
From the time of the initial discovery of TLEs as repressors of Wnt target gene expression, it has been attractive to propose that the Wnt transcriptional switch is mediated by β-catenin displacing TLEs on TCFs (Fig. 1B,C). In support of this model, ChIP studies indicate a Wnt signaling-dependent loss of TLE from W-CRMs regulating c-myc and cyclin D2 expression.[41,56] In addition, biochemical experiments suggest that β-catenin and TLE1 binding to TCFs are mutually exclusive,[57] but this result has been challenged by more recent results from the same lab, which found that TCF can bind both proteins simultaneously.[39] At present, there is insufficient evidence to fully support β-catenin displacement of TLEs from W-CRMS as a general model.
A corollary of the switch model is that β-catenin recruits transcriptional co-activators to W-CRMs.[46] One such co-activator complex is a heterodimer consisting of B-cell CLL/lymphoma 9 (Bcl9 or the related B9L- known as Legless or Lgs in flies) and Pygopus 1/2 (Pygo). Basler and colleagues have demonstrated the existence of a trimeric complex, with the HD1 domain of Bcl9 bound to β-catenin and the HD2 domain to Pygo.[5,58,59] Interestingly, Pygo also associates with Gro in flies, and analysis of gro, pygo double mutants suggests that a major function of Pygo is to inactivate Gro.[36] A recent study from Bienz and coworkers examined proteins associated with biotin ligase-tagged BCL9 and B9L, in the presence and absence of Wnt signaling using a technique called BioID.[60] Surprisingly, the authors found both BCL9 and B9L associated with TLEs irrespective of the state of Wnt signaling in HEK293 cells. Their co-immunoprecipitation experiments also indicate that BCL9/B9L and TCF are a part of the same complex even in the absence of Wnt signaling. Similar results were obtained when Pygo was used. The authors argue that rather that stimulating an exchange of co-regulators, the addition of β-catenin causes a change in the conformation of the TCF transcriptional complex (Fig. 2B,C).[60]
The results of Bienz and coworkers challenge the β-catenin displacement of TLE model, but some caution is needed, as the BioID and co-IP methodology utilized do not distinguish between complexes in solution and those on W-CRM chromatin. Further ChIP experiments, preferably at the genomic level, will be needed to fully test whether TLE displacement or conformational change of the TCF transcriptional complex is the major mechanism by which the transcriptional switch occurs.
Wnt-dependent ubiquitination of TLEs regulates the transcriptional switch
If β-catenin does not simply displace TLEs at W-CRMs, how does Wnt signaling inactivate these co-repressors? Another recent report from the Bienz group offers an intriguing mechanism.[42] They found that E3 ubiquitin-protein ligase UBR5 (UBR5) is required for efficient Wnt signaling in HEK293 cells. In Drosophila, cells lacking the UBR5 homolog hyperplastic discs (hyd) displayed a classic loss of Wg signaling in wing imaginal discs. UBR5/Hyd functioned downstream of β-catenin stabilization. A mass-spectrometry screen identified an interaction between UBR5 and TLE3, and UBR5 ubiquitinated TLE3 on its C-terminal WD40 domain.[42] Activation of Wnt/β-catenin signaling stimulated UBR5 ubiquitination of TLE3. In flies, the hyd phenotype is partially suppressed by concomitant loss of gro, indicating that TLE/Gro is a major target of Hyd action. Taken together, these results strongly support a model where activation of UBR5 by Wnt signaling inactivates TLEs through ubiquitination.[42]
UBR5 generates K48-linked polyubiquitin chains on TLE3, a modification that is commonly thought to target proteins for proteasomal degradation.[61] However, Bienz and colleagues found no evidence for UBR5/Hyd regulation of TLE3/Gro protein levels or turnover.[42] Rather, they demonstrated that the AAA+ ATPase Valosin-containing protein (VCP, also known as p97) bound to and reduced the level of ubiquitinated TLE3 in Wnt stimulated cells. Genetic or chemical inhibition of VCP/p97 reduced Wnt gene activation.[42] The authors suggest that VCP/p97, which is thought to unfold ubiquitinated proteins via its ATPase activity,[62] disrupts the tetramerization of ubiquitinated TLE3, which is required for its ability to repress Wnt targets.[39]
This work fills an important gap in our understanding of the interactions between the Wnt signaling machinery and TLEs, but additional mechanisms may also exist. For example, Lee and coworkers reported that the E3 ubiquitin ligase X-linked Inhibitor of Apoptosis (XIAP) bound and mono-ubiquitinated TLE3 at its N-terminal Q domain. This modification disrupted TCF4-TLE3 binding.[63] XIAP was recruited to an Axin2 W-CRM upon Wnt stimulation. XIAP knockdown via siRNA in mammalian cell culture or via morpholinos in Xenopus embryos greatly reduced Wnt/β-catenin signaling.[63] It should be noted that XIAP mutant mice are viable and do not exhibit detectable Wnt signaling defects[64] and CRISPR/Cas9 knockout of XIAP in HEK293 cells displayed a modest effect on a Wnt reporter compared to UBR5 knockouts.[42] Nonetheless, given that UBR5 and XIAP modify TLE3 in distinct ways (poly versus mono-ubiquitination) and at different locations on the primary TLE3 sequence, the possibility exists that the two E3 ligases act independently to inactivate TLE activity at W-CRMs. Further research, including examining the phenotype of UBR5, XIAP double knockouts on a variety of Wnt readouts, should help to clarify their respective roles in mediating the Wnt transcriptional switch.
Is TCF3 a dedicated repressor in the transcriptional switch?
While invertebrates contain one TCF, which participates in both sides of the transcriptional switch (Fig. 1B),[46] the presence of multiple TCFs in vertebrates has long raised the question of whether they have specialised transcriptional functions. Many genetic studies support the view that TCF3 (also known as TCF7L1) is a dedicated repressor and LEF1 a dedicated activator of Wnt targets, while TCF1 and TCF4 (also termed TCF7 and TCF7L2, respectively) display either activity depending on the context.[1,6,65] Partially supporting this, TCF3 and TCF4 bind to TLEs with higher affinity than LEF1 or TCF1.[39] In support of a “TCF exchange” model (Fig. 1C), TCF3 is removed from W-CRMs via Wnt-dependent phosphorylation[66,67] or downregulation of its expression.[18,19,68]
One model for investigating the relationship between Wnt signaling and TCF3 are mouse embryonic stem cells (mESCs). TCF3 is the most abundantly expressed TCF in mESCs, where it limits self-renewal through repression of pluripotency genes.[19–22] TCF3 expression and/or function is inhibited by activation of Wnt/β-catenin signaling.[18,19,22,68] TCF3 and TCF1 have been reported to have opposing activities in mESCs, consistent with TCF3 repressing and TCF1 activating the same transcriptional targets.[69]
The antagonistic relationship between TCF1 and TCF3 has been challenged by recent work from Doble and colleagues.[70] They used CRISPR/Cas9 to knockout all four TCF genes in mESCs. While proliferation of these quadruple knockouts (QKO) was unaffected, the activation of Wnt reporters was negligible. When subjected to differentiation conditions, QKO cells could not form mesendoderm and were biased towards neurectoderm formation. Interestingly, this defect could be rescued by knocking in cDNAs of TCF1 or TCF3 at their endogenous loci. Transcriptome profiling of the QKOs and their rescues revealed that the expression profiles of both TCF1 and TCF3 rescued QKOs were similar to each other and were more similar to WT mESCs than the QKO cells.[70]
The results of Doble’s group are at odds with the literature that TCF3 is predominately/exclusively a repressor, but these studies are based on removal of TCF3 in genetic backgrounds where the other three TCFs are present. Perhaps in the QKO/rescue experiments, TCF3 is capable of mediating both sides of the transcriptional switch, taking over the activating function normally carried out by other TCFs. It should be noted that TCF3 does mediate β-catenin-dependent transcriptional activation in overexpression assays.[9,71,72] It would be interesting to examine whether TCF3 can rescue the developmental phenotypes of other TCFs,[71,73] during mouse embryogenesis if it was the sole TCF expressed.
Other potential factors acting in the switch
In addition to TCFs, TLEs, β-catenin and UBR5, there are many other nuclear factors that have been proposed to contribute to the Wnt transcriptional switch,[46] and more factors continue to be identified. For example, O-GlcNAc Transferase (OGT) has been reported to promote TLE repression by direct interaction with TLEs, and this interaction was attenuated by Wnt signaling in mammalian cell culture.[56] The transcription factor Ladybird homeobox 2 (Lbx2) activated Wnt transcriptional readouts in zebrafish by binding to the WD40 domain of TLEs, blocking their ability to bind to TCFs.[74] In another recent report, the selenoenzyme Glutathione peroxidase 4 (GPX4) was shown to bind to TCFs and prevent their association with chromatin. The Wnt antagonistic functions of GPX4 were independent of its catalytic activity.[75] Whether these factors are tissue/cell specific regulators or more broadly required for Wnt transcriptional regulation requires further study.
While these trans-acting factors add more complexity to the Wnt gene regulatory circuit, a new cis-regulatory motif influencing W-CRMs was also recently reported. An 11 bp motif (termed a negative regulatory element, or NRE) was identified upstream of the siamois promoter, a previously characterized direct Wnt target gene.[76] The NRE represses siamois expression, and was specifically bound by TCF in vitro. Similar motifs were identified upstream of a W-CRM regulating expression of the T (Brachyury) gene in mouse embryonic stem cells, and removing them via CRISPR/Cas9 caused an increase in T mRNA levels.[76][73]
NREs have a different consensus than traditional TCF sites, in that way evoking the WGAWAW sites identified in genes repressed by Wnt/β-catenin signaling in fly hematopoietic cells (Fig. 2C).[77,78] β-catenin was detected near the NREs in the T and siamois promoters using ChIP,[76] but whether or not β-catenin regulates gene expression at those locations remains to be determined. Computationally, NREs are enriched near W-CRMs, suggesting that they may be a common feature of Wnt gene regulation.[76]
Conclusions and outlook
While probably not universal, the transcriptional switch model appears to contribute to the regulation of many Wnt/TCF targets, although more functional studies in vertebrate systems would strengthen this claim. Basic mechanistic features of the switch are yet to be clarified, most notably whether Wnt/β-catenin signaling promotes an exchange of factors at W-CRMs or a rearrangement of their configuration. It is also possible that different Wnt targets are regulated by distinct switch mechanisms, and all of the factors discussed in this review likely only regulated a subset of Wnt targets. While the field’s ability to genetically manipulate switch components is at an unprecedented level thanks to CRISRP/Cas9 editing, a lack of ChIP-quality antibodies for many of the key factors is limiting our understanding of what is happening on the chromatin. As these reagents are developed, further investigation of the regulation of biologically important genes, as well as genome-wide studies should provide a greater understanding of how β-catenin alters the transcriptional complexes regulating Wnt target genes.
Acknowledgments
Thanks to Bahaar Chalwa, Juan Ibarra and Kavya Adiga for comments on the manuscript. Supported by a grant from the National Institutes of Health (GM108468) and a University of Michigan Mcube award to KMC.
Bibliography
- 1.Ramakrishnan AB, Cadigan KM. F1000Research. 2017;6:746. doi: 10.12688/f1000research.11034.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nusse R, Clevers H. Cell. 2017;169:985. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 3.Stamos JL, Weis WI. Cold Spring Harb Perspect Biol. 2013;5:a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacDonald BT, He X. Cold Spring Harb Perspect Biol. 2012;4:a007880. doi: 10.1101/cshperspect.a007880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valenta T, Hausmann G, Basler K. EMBO J. 2012;31:2714. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadigan KM, Waterman ML. Cold Spring Harb Perspect Biol. 2012;4:a007906. doi: 10.1101/cshperspect.a007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature. 1996;382:638. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 8.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, Kemler R. Mech Dev. 1996;59:3. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 9.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destrée O, Clevers H. Cell. 1996;86:391. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 10.Brunner E, Peter O, Schweizer L, Basler K. Nature. 1997;385:829. doi: 10.1038/385829a0. [DOI] [PubMed] [Google Scholar]
- 11.Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha YH, Ali M, Priess JR, Mello CC. Cell. 1997;90:707. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- 12.Thorpe CJ, Schlesinger A, Carter JC, Bowerman B. Cell. 1997;90:695. doi: 10.1016/s0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- 13.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Cell. 1997;88:789. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 14.Brannon M, Gomperts M, Sumoy L, Moon RT, Kimelman D. Genes Dev. 1997;11:2359. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan MJ, Grüning W, Walz G, Sokol SY. Proc Natl Acad Sci U S A. 1998;95:5626. doi: 10.1073/pnas.95.10.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schweizer L, Nellen D, Basler K. Proc Natl Acad Sci U S A. 2003;100:5846. doi: 10.1073/pnas.1037533100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gribble SL, Kim HS, Bonner J, Wang X, Dorsky RI. Development. 2009;136:781. doi: 10.1242/dev.027995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shy BR, Wu CI, Khramtsova GF, Zhang JY, Olopade OI, Goss KH, Merrill BJ. Cell Rep. 2013;4:1. doi: 10.1016/j.celrep.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison G, Scognamiglio R, Trumpp A, Smith A. EMBO J. 2016;35:356. doi: 10.15252/embj.201592116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereira L, Yi F, Merrill BJ. Mol Cell Biol. 2006;26:7479. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi F, Pereira L, Merrill BJ. Stem Cells Dayt Ohio. 2008;26:1951. doi: 10.1634/stemcells.2008-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A. Nat Cell Biol. 2011;13:838. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roose J, Huls G, van Beest M, Moerer P, van der Horn K, Goldschmeding R, Logtenberg T, Clevers H. Science. 1999;285:1923. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- 24.Angus-Hill ML, Elbert KM, Hidalgo J, Capecchi MR. Proc Natl Acad Sci. 2011;108:4914. doi: 10.1073/pnas.1102300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Nature. 1998;395:604. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 26.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destrée O, Clevers H. Nature. 1998;395:608. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 27.Barolo S, Posakony JW. Genes Dev. 2002;16:1167. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- 28.Cinnamon E, Paroush Z. Curr Opin Genet Dev. 2008;18:435. doi: 10.1016/j.gde.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 29.Turki-Judeh W, Courey AJ. Curr Top Dev Biol. 2012;98:65. doi: 10.1016/B978-0-12-386499-4.00003-3. [DOI] [PubMed] [Google Scholar]
- 30.Arce L, Pate KT, Waterman ML. BMC Cancer. 2009;9:159. doi: 10.1186/1471-2407-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen G, Nguyen PH, Courey AJ. Mol Cell Biol. 1998;18:7259. doi: 10.1128/mcb.18.12.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen G, Fernandez J, Mische S, Courey AJ. Genes Dev. 1999;13:2218. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song H, Hasson P, Paroush Z, Courey AJ. Mol Cell Biol. 2004;24:4341. doi: 10.1128/MCB.24.10.4341-4350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekiya T, Zaret KS. Mol Cell. 2007;28:291. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM. EMBO J. 2006;25:2735. doi: 10.1038/sj.emboj.7601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mieszczanek J, de la Roche M, Bienz M. Proc Natl Acad Sci. 2008;105:19324. doi: 10.1073/pnas.0806098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calvo D, Victor M, Gay F, Sui G, Luke MP, Dufourcq P, Wen G, Maduro M, Rothman J, Shi Y. EMBO J. 2001;20:7197. doi: 10.1093/emboj/20.24.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jennings BH, Ish-Horowicz D. Genome Biol. 2008;9:205. doi: 10.1186/gb-2008-9-1-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chodaparambil JV, Pate KT, Hepler MRD, Tsai BP, Muthurajan UM, Luger K, Waterman ML, Weis WI. EMBO J. 2014;33:719. doi: 10.1002/embj.201387188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lien WH, Polak L, Lin M, Lay K, Zheng D, Fuchs E. Nat Cell Biol. 2014;16:179. doi: 10.1038/ncb2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sierra J, Yoshida T, Joazeiro CA, Jones KA. Genes Dev. 2006;20:586. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flack JE, Mieszczanek J, Novcic N, Bienz M. Mol Cell. 2017;67:181. doi: 10.1016/j.molcel.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ro H, Dawid IB. EMBO J. 2011;30:2894. doi: 10.1038/emboj.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizumoto K, Sawa H. Trends Cell Biol. 2007;17:465. doi: 10.1016/j.tcb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 45.Phillips BT, Kimble J. Dev Cell. 2009;17:27. doi: 10.1016/j.devcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cadigan KM. In: Curr Top Dev Biol. P S, Payre F, editors. Academic Press; 2012. pp. 1–34. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe K, Dai X. Sci Signal. 2011;4:pe41. doi: 10.1126/scisignal.2002436. [DOI] [PubMed] [Google Scholar]
- 48.Sokol SY. Dev Camb Engl. 2011;138:4341. doi: 10.1242/dev.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lam N, Chesney MA, Kimble J. Curr Biol CB. 2006;16:287. doi: 10.1016/j.cub.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knirr S, Frasch M. Dev Biol. 2001;238:13. doi: 10.1006/dbio.2001.0397. [DOI] [PubMed] [Google Scholar]
- 51.Shetty P, Lo MC, Robertson SM, Lin R. Dev Biol. 2005;285:584. doi: 10.1016/j.ydbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 52.Chang JL, Chang MV, Barolo S, Cadigan KM. Dev Biol. 2008;321:446. doi: 10.1016/j.ydbio.2008.05.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhambhani C, Ravindranath AJ, Mentink RA, Chang MV, Betist MC, Yang YX, Koushika SP, Korswagen HC, Cadigan KM. PLOS Genet. 2014;10:e1004133. doi: 10.1371/journal.pgen.1004133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franz A, Shlyueva D, Brunner E, Stark A, Basler K. PLOS Genet. 2017;13:e1006700. doi: 10.1371/journal.pgen.1006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakamura Y, de P Alves E, Veenstra GJC, Hoppler S. Development. 2016;143:1914. doi: 10.1242/dev.131664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu J, Bowe DB, Sadlonova A, Whisenhunt TR, Hu Y, Rustgi AK, Nie Y, Paterson AJ, Yang X. J Biol Chem. 2014;289:12168. doi: 10.1074/jbc.M114.553859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daniels DL, Weis WI. Nat Struct Mol Biol. 2005;12:364. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 58.Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Züllig S, Basler K. Cell. 2002;109:47. doi: 10.1016/s0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- 59.Städeli R, Basler K. Mech Dev. 2005;122:1171. doi: 10.1016/j.mod.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 60.van Tienen LM, Mieszczanek J, Fiedler M, Rutherford TJ, Bienz M. eLife. 2017;6:e20882. doi: 10.7554/eLife.20882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolf DH, Hilt W. Biochim Biophys Acta. 2004;1695:19. doi: 10.1016/j.bbamcr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 62.Xia D, Tang WK, Ye Y. Gene. 2016;583:64. doi: 10.1016/j.gene.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanson AJ, Wallace HA, Freeman TJ, Beauchamp RD, Lee LA, Lee E. Mol Cell. 2012;45:619. doi: 10.1016/j.molcel.2011.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harlin H, Reffey SB, Duckett CS, Lindsten T, Thompson CB. Mol Cell Biol. 2001;21:3604. doi: 10.1128/MCB.21.10.3604-3608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Archbold HC, Yang YX, Chen L, Cadigan KM. Acta Physiol. 2012;204:74. doi: 10.1111/j.1748-1716.2011.02293.x. [DOI] [PubMed] [Google Scholar]
- 66.Hikasa H, Sokol SY. J Biol Chem. 2011;286:12093. doi: 10.1074/jbc.M110.185280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hikasa H, Sokol SY. Cold Spring Harb Perspect Biol. 2013;5:a007955. doi: 10.1101/cshperspect.a007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Atlasi Y, Noori R, Gaspar C, Franken P, Sacchetti A, Rafati H, Mahmoudi T, Decraene C, Calin GA, Merrill BJ, Fodde R. PLOS Genet. 2013;9:e1003424. doi: 10.1371/journal.pgen.1003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yi F, Pereira L, Hoffman JA, Shy BR, Yuen CM, Liu DR, Merrill BJ. Nat Cell Biol. 2011;13:762. doi: 10.1038/ncb2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moreira S, Polena E, Gordon V, Abdulla S, Mahendram S, Cao J, Blais A, Wood GA, Dvorkin-Gheva A, Doble BW. Cell Rep. 2017;20:2424. doi: 10.1016/j.celrep.2017.08.043. [DOI] [PubMed] [Google Scholar]
- 71.Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Mol Cell Biol. 1998;18:1248. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Merrill BJ, Gat U, DasGupta R, Fuchs E. Genes Dev. 2001;15:1688. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Galceran J, Fariñas I, Depew MJ, Clevers H, Grosschedl R. Genes Dev. 1999;13:709. doi: 10.1101/gad.13.6.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu FI, Sun YH, Wei CY, Thisse C, Thisse B. Nat Commun. 2014;5:5368. doi: 10.1038/ncomms6368. [DOI] [PubMed] [Google Scholar]
- 75.Rong X, Zhou Y, Liu Y, Zhao B, Wang B, Wang C, Gong X, Tang P, Lu L, Li Y, Zhao C, Zhou J. Development. 2017;144:1687. doi: 10.1242/dev.144261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim K, Cho J, Hilzinger TS, Nunns H, Liu A, Ryba BE, Goentoro L. Curr Biol. 2017;27:2357. doi: 10.1016/j.cub.2017.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blauwkamp TA, Chang MV, Cadigan KM. EMBO J. 2008;27:1436. doi: 10.1038/emboj.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang CU, Blauwkamp TA, Burby PE, Cadigan KM. PLOS Genet. 2014;10:e1004509. doi: 10.1371/journal.pgen.1004509. [DOI] [PMC free article] [PubMed] [Google Scholar]