Abstract
BACKGROUND
Efforts should be made to develop a deep-learning diagnosis system to distinguish pancreatic cancer from benign tissue due to the high morbidity of pancreatic cancer.
AIM
To identify pancreatic cancer in computed tomography (CT) images automatically by constructing a convolutional neural network (CNN) classifier.
METHODS
A CNN model was constructed using a dataset of 3494 CT images obtained from 222 patients with pathologically confirmed pancreatic cancer and 3751 CT images from 190 patients with normal pancreas from June 2017 to June 2018. We established three datasets from these images according to the image phases, evaluated the approach in terms of binary classification (i.e., cancer or not) and ternary classification (i.e., no cancer, cancer at tail/body, cancer at head/neck of the pancreas) using 10-fold cross validation, and measured the effectiveness of the model with regard to the accuracy, sensitivity, and specificity.
RESULTS
The overall diagnostic accuracy of the trained binary classifier was 95.47%, 95.76%, 95.15% on the plain scan, arterial phase, and venous phase, respectively. The sensitivity was 91.58%, 94.08%, 92.28% on three phases, with no significant differences (χ2 = 0.914, P = 0.633). Considering that the plain phase had same sensitivity, easier access, and lower radiation compared with arterial phase and venous phase , it is more sufficient for the binary classifier. Its accuracy on plain scans was 95.47%, sensitivity was 91.58%, and specificity was 98.27%. The CNN and board-certified gastroenterologists achieved higher accuracies than trainees on plain scan diagnosis (χ2 = 21.534, P < 0.001; χ2 = 9.524, P < 0.05; respectively). However, the difference between CNN and gastroenterologists was not significant (χ2 = 0.759, P = 0.384). In the trained ternary classifier, the overall diagnostic accuracy of the ternary classifier CNN was 82.06%, 79.06%, and 78.80% on plain phase, arterial phase, and venous phase, respectively. The sensitivity scores for detecting cancers in the tail were 52.51%, 41.10% and, 36.03%, while sensitivity for cancers in the head was 46.21%, 85.24% and 72.87% on three phases, respectively. Difference in sensitivity for cancers in the head among the three phases was significant (χ2 = 16.651, P < 0.001), with arterial phase having the highest sensitivity.
CONCLUSION
We proposed a deep learning-based pancreatic cancer classifier trained on medium-sized datasets of CT images. It was suitable for screening purposes in pancreatic cancer detection.
Keywords: Deep learning, Convolutional neural networks, Pancreatic cancer, Computed tomography
Core Tip: We developed a deep learning-based, computer-aided pancreatic ductal adenocarcinoma model trained on computed tomography images with pathologically confirmed pancreatic cancer in this retrospective study. We evaluated the approach used on the datasets in terms of both binary and ternary classifier, with the purposes of detecting and localizing masses, respectively. In the binary classifier, the performance of plain, arterial and venous phase had no difference. Its accuracy on plain scan was 95.47%, sensitivity was 91.58%, and specificity was 98.27%. In the ternary classifier, the arterial phase had the highest sensitivity in detecting cancer in the head of the pancreas among the three phases. Our model is suitable for screening purposes in pancreatic cancer detection.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is the most common solid malignancy of the pancreas. It is aggressive and challenging to treat, which is more commonly called “pancreatic cancer”[1]. Pancreatic cancer is a highly lethal malignancy with a very poor prognosis[2]. Despite recent advances in surgical techniques, chemotherapy, and radiation therapy, the 5-year survival rate remains a dismal 8.7%[3]. Most patients with pancreatic cancer have nonspecific symptoms, and the disease is often found at an advanced stage. Only 10%-20% of patients present at the localized disease stage, at which complete surgical resection and chemotherapy offer the best chance of survival, with a 5-year survival rate of approximately 31.5%. The remaining 80%-90% of patients miss the chance to benefit from surgery because of general or local metastases at the time of diagnosis[4,5].
Currently, effective early diagnosis remains difficult, and it depends mainly on imaging modalities[6]. Compared with ultrasonography, magnetic resonance imaging (MRI), endoscopic ultrasonography, and positron emission tomography, computed tomography (CT) is the most commonly used imaging modality for the initial evaluation of suspected pancreatic cancer[7,8]. CT scans are also used for screening asymptomatic patients at high risk of developing pancreatic cancer. Patients with pancreatic cancer that were incidentally diagnosed during an imaging examination for an unrelated disease have a longer median survival time than those who are already symptomatic[9]. Sensitivity of CT for the pancreatic adenocarcinoma detection ranges from 70% to 90%[10]. The choice for pancreatic cancer diagnosis is a thin section with contrast-enhanced, dual-phase multidetector computed tomography[11].
Recently, due to promising achievements in deep neural networks and increasing medical needs, computer-aided diagnosis (CAD) systems have become a new research focus. There have been some initial successes in applying deep learning to assess radiological images. Deep learning-aided decision-making has been used in support of pulmonary nodule and skin tumor diagnoses[12,13]. Efforts should be made to develop CAD systems to distinguish pancreatic cancer from benign tissue due to the high morbidity of pancreatic cancer. Therefore, developing an advanced discrimination method for pancreatic cancer is necessary. A convolutional neural network (CNN) is a class of neural network models that can extract features from images by exploring the local spatial correlations presented in images. CNN models have been shown to be effective and powerful for addressing a variety of image classification problems[14].
In this study, we demonstrated that a deep learning method can achieve pathologically certified pancreatic ductal adenocarcinoma classification using clinical CT images.
MATERIALS AND METHODS
Data collection and preparation
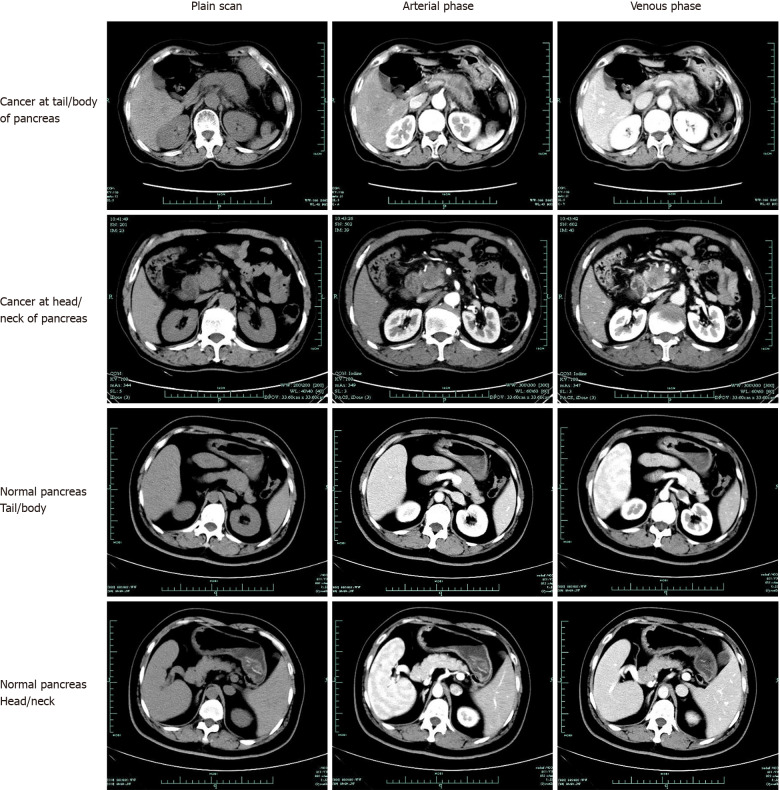
Dataset: Between June 2017 and June 2018, patients with pathologically diagnosed pancreatic cancer in the First Affiliated Hospital, Zhejiang University School of Medicine, China, were eligible for inclusion in the present study. Patients with CT-confirmed normal pancreas were also randomly collected in the same period. All data were retrospectively obtained from patients’ medical records. Images of pancreatic cancers and normal pancreases were extracted from the database. All the cancer diagnoses were based on pathological examinations, either by pancreatic biopsy or by surgery (Figure 1). Participants gave informed consent to allow data collected from them to be published. Because of the retrospective study design, we verbally informed all the participants included in the study. Patients who do not want their information to be shared could opt out. Subject information was anonymized at the collection and analysis stage. All the methods were performed in accordance with the approved guidelines. The Hospital Ethics Committee approved the study protocol. A total of 343 patients were pathologically diagnosed with pancreatic cancer from June 2017 to June 2018. Of these patients, 222 underwent an abdominal enhanced-CT in our hospital before surgery or biopsy. We randomly collected 190 patients who underwent enhanced-CT with normal pancreas. Thus, among the 412 enrolled subjects, 222 were pathologically diagnosed with pancreatic cancer, and the remaining 190 diagnosed with normal pancreas were included as a control group.
Figure 1.
Examples of dataset.
Imaging techniques: Multiphasic CT was performed following a pancreas protocol and using a 256-channel multidetector row CT scanner (Siemens). The scanning protocol included unenhanced and contrast material–enhanced biphasic imaging in the arterial and venous phases after intravenous administration of 100 mL ioversol at a rate of 3 mL/sec using an automated power injector. Images were reconstructed at 5.0-mm thickness. For each CT scan, one to nine pictures of the pancreas were selected from each phase. Finally, datasets of 3494 CT images obtained from 222 patients with pathologically confirmed pancreatic cancer and 3751 CT images from 190 patients with normal pancreas were collected.
Deep learning technique
Data preprocessing: We adopted a CNN model to classify the CT images. A CNN requires the input images to be the same size. Thus, we first cropped each CT image starting at the center to transform it into a fixed 512 × 512 resolution. Each image was stored in the RGB color model, which is a model with red, green, and blue light merged together to reproduce multiple colors, and thus consisted of three-color channels (i.e., red, green, and blue). We normalized each channel of every image using 0.5 as the mean and the standard deviation. This normalization was performed because all the images were processed by the same CNN, and the results might improve if the feature values of the images were scaled to a similar range.
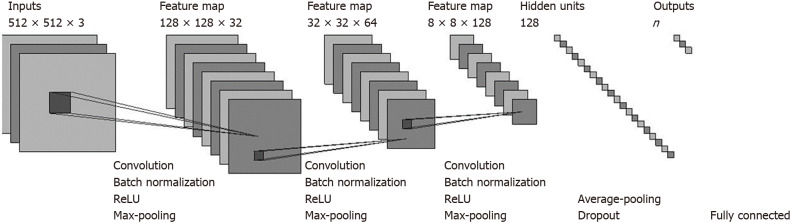
CNN: In this work, we designed a CNN model to classify the pancreatic CT images to assist in pancreatic cancer diagnosis. The architecture of our proposed CNN model is presented in Figure 2. Our model consisted primarily of three convolutional layers and a fully connected layer. Each convolutional layer was followed by a batch normalization (BN) layer that normalized the outputs of the convolutional layer, a rectified linear unit (ReLU) layer that applied an activation function to its input values, and a max-pooling layer that conducted a down-sampling operation. We also adopted an average-pooling layer before the fully connected layer to reduce the dimensions of the feature values input to the fully connected layer. Following the work by Srivastava et al[15], a dropout rate of 0.5 was used between the average-pooling layer and the fully connected layer to avoid overfitting and increase the performance. We also tried Spatial Dropout[16] between each max-pooling layer and its following convolutional layer, but found that such dropouts resulted in performance degradation. Therefore, we did not apply Spatial Dropout. As input, the network takes the pixel values of a CT image, and it outputs the probability that the image belongs to a certain class (e.g., the probability that the corresponding patient has pancreatic cancer). The CT images were fed into our model layer by layer. The input to each layer is the output values of the previous layer. The layers perform specific transformations on the input values and then pass the processed values to the next layer.
Figure 2.
Architecture of our convolutional neural network model.
The convolutional layers and the pooling layers require several hyper-parameters whose settings are shown in Supplementary Material. In the Supplementary Material, we also discuss these layers in sequence: First, the convolutional layer, then, the batch normalization (BN) layer, the Rectified Linear Unit (ReLU) layer, the max-pooling, and average-pooling layers, and finally, the fully connected layer. Then, we present the hyper-parameter settings for our model.
Training and testing the CNN: We collected three types of CT images: Plain scan, venous phase, and arterial phase and built three datasets from the collected images based on the image types. Each dataset may include several images collected from one patient. To divide a dataset into training, validation, and test sets, we first collected the identity documents (IDs) of all the patients in the dataset. Each patient was labeled as follows: The label may be “no cancer (NC)”, “with cancer at the tail and/or body of the pancreas (TC)” or “with cancer at the head and/or neck of the pancreas (HC)”. For each label, e.g., “no cancer”, we randomly placed 10% of the patients with this label into the validation set, 10% into the test set, and the remaining 80% into the training set. Notablly, images of the same patient appear in only one set.
All patients and their CT images were marked by one of the three labels, i.e., “no cancer”, “with cancer in the tail of pancreas” and “with cancer in the head of the pancreas”. For each dataset, we could treat the TC and HC patients as “with cancer (CA)”. Then, we trained a binary classifier to classify all the CT images. We also trained a ternary classifier to determine the specific cancer location. Our proposed approach was flexible enough to be used as either a binary classifier or a multiple-class classifier; we needed only to specify the hyperparameter of the fully-connected layer to control the classifier type.
Given a dataset and the number of target classes (denoted as n), we trained our model on the training set and set the mini-batch size to 32. After each training iteration, we used the cross-entropy loss function to calculate the loss between the predicted results (i.e., the probability distribution P output by the fully connected layer) of our model and the ground truth (denoted as G), computed as Formula 1.
Formula 1.
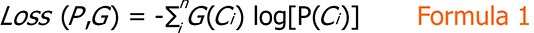
This loss was used to guide the updates of the weights in our CNN model; we used Adam as the optimizer. The statistics of each dataset are presented in Table 1.
Table 1.
Statistics of our datasets
| CT phase |
Number of patients |
Number of images |
|||||||||
| Without cancer cancer |
With cancer |
Total | Without cancer |
With cancer |
Total | ||||||
| At tail1 | At head | Total | At tail | At head | Total | ||||||
| Sets | Plain Scan | 182 | 91 | 123 | 214 | 396 | 1182 | 416 | 496 | 912 | 2094 |
| Arterial phase | 179 | 91 | 129 | 220 | 399 | 1282 | 575 | 735 | 1310 | 2592 | |
| Venous phase | 178 | 93 | 129 | 222 | 400 | 1287 | 573 | 699 | 1272 | 2559 | |
| Total | 539 | 275 | 381 | 656 | 1195 | 3751 | 1564 | 1930 | 3494 | 7245 | |
“At tail” means at tail or body of pancreas, while “At head” means at head or neck of pancreas. CT: Computed tomography.
After updating the model, we calculated the accuracy (see section Evaluation below) of the new model on the validation set to assess the quality of the current model. We trained our model for a maximum of 100 epochs, and the model with the highest accuracy on the validation set was selected as the final model. A 10-fold cross-validation process was used to evaluate our techniques. We randomly divided the images in each phase into 10 folds, 8 of which were used to do the training, 1 fold was the validation set, and the remaining one was used to test the model. The entire process was repeated 10 times, and each fold will be used as the test set once. The average performance was recorded. We evaluated the effectiveness of our CNN model on the test sets in terms of accuracy, precision, and recall (see section Evaluation below).
Evaluation: We evaluated our approach on the three datasets in terms of both binary and ternary classifications and measured the effectiveness of our approach relying on widely adopted metrics of classification aspects: Accuracy, precision, and recall. Accuracy is the proportion of the images that are correctly classified (denoted as TP) among all the images (denoted as All) for all classes. The precision for class Ci is the proportion of images that are correctly classified as class Ci (denoted as TPi) among all images that are classified as class (denoted as TPi + FPi). The recall for class Ci is the proportion of images that are correctly classified as class Ci (denoted as TPi) among all the images that actually belong to class Ci (denoted as Alli). These metrics are calculated as follows:
Accuracy = TP/All;
Precisioni = TPi/( TPi + FPi);
Recalli= TPi/Alli.
We evaluated our approach relying on the accuracy because it measures the overall quality of a classifier on all classes instead of only a specific class Ci, which is shown as follows:
Sensitivity = Recall in cancer detection = (The correctly predicted malignant lesions)/(All the malignant lesions);
Specificity = Recall in detecting noncancer = (The correctly predicted nonmalignant cases)/(All non-malignant cases);
Precision in cancer detection = (The correctly predicted malignant lesions)/(All images classified as malignant).
Evaluation between deep learning and gastroenterologists
Ten board-certified gastroenterologists and 15 trainees participated in the study, and the accuracy of their image classifications was compared with the predictions of the deep learning technique. Each gastroenterologist or trainee classified the same 100-image set in plain scan randomly selected from the test dataset of the deep learning technique. The human response time was approximately 10 s per image. The images accurately classified by the board-certified gastroenterologists and trainees were compared with the results of the deep learning model.
Statistical analysis
We performed statistical analyses using SPSS 13.0 for Windows (SPSS, Chicago, IL, United States). Continuous variables are expressed as mean ± SD and were compared using Student’s t-test. The χ2 test was used to compare categorical variables. A value of P < 0.05 (2-tailed test) was considered statistically significant.
RESULTS
Characteristics of the study participants
Among the 412 enrolled subjects, 222 were pathologically diagnosed with pancreatic cancer, and 190 diagnosed with normal pancreas were included as a control group. The characteristics of the enrolled participants, classified by the presence or absence of pancreatic cancer, are shown in Table 2. The mean age was 63.8 ± 8.7 years for cancer group (range, 39-86 years, 124 men/98 women) and 61.0 ± 12.3 years for non-cancer group (range, 35-83 years, 98 men/92 women). These two groups had no significant differences in age or gender (P > 0.05). For the cancer group, 129 cases were located at the head and neck of pancreas, 93 cases at the tail and body of pancreas. The median tumor size of cancer group was 3.5 cm (range, 2.7-4.3 cm).
Table 2.
Characteristics of study participants
| Variables | With pancreatic cancer (n = 222) | Without cancer (n = 190) | Z value | P value |
| Age (yr) | 63.8 (8.7) | 61.0 (12.3) | 3.391 | 0.66 |
| Gender (male/female) | 124/98 | 98/92 | 0.3852 | 0.54 |
| Diagnosis method surgery/biopsy3 | 161/61 | - | - | - |
| Tumor location; head and neck/tail and body | 129/63 | - | - | - |
| Tumor size4 [median (quartile 1, quartile 3)], cm | 3.5 (2.7-4.3) | - | - | - |
| ≤ 2 | 29 | - | - | - |
| 2-4 | 134 | - | - | - |
| > 4 | 59 | - | - | - |
Data are expressed as the mean ± SD.
t value.
χ2 value.
“Biopsy” includes patients who underwent biopsy only, patients who underwent biopsy before surgery were calculated in “Surgery”, and tumor size was evaluated in greatest dimension.
Tumor size was mainly evaluated by the gross surgical specimen, for patient who only underwent biopsy, the tumor size was calculated by computed tomography image.
Performance of the deep convolutional neural network used as a binary classifier
Datasets of 3494 CT images obtained from 222 patients with pathologically confirmed pancreatic cancer and 3751 CT images from 190 patients with normal pancreas were included, statistics of each dataset are presented in Table 1. We labeled each CT image as “with cancer” or “no cancer”. Then, we constructed a binary classifier using our CNN model by 10-fold cross validation on 2094, 2592, and 2559 images in the plain scan, arterial phase, and venous phase, respectively (Table 1).
The overall diagnostic accuracy of the CNN was 95.47%, 95.76%, and 95.15% on the plain scan, arterial phase, and venous phase, respectively. The sensitivity of the CNN (known as recall in cancer detection - the correctly predicted malignant lesions divided by all the malignant lesions) was 91.58%, 94.08%, and 92.28% on the plain scan, arterial phase, and venous phase images, respectively. The specificity of the CNN (known as recall in detecting non-cancer - the correctly predicted nonmalignant cases divided by all nonmalignant cases) was 98.27%, 97.57% and 97.87% on the three phases, respectively. The results are summarized in Table 3.
Table 3.
Performance of the binary classifiers
| Plain scan | Arterial phase | Venous phase | χ2 value | P value | |
| Accuracy | 0.954747 | 0.957580 | 0.951549 | 0.346 | 0.841 |
| Specificity | 0.982710 | 0.975695 | 0.978692 | 0.149 | 0.928 |
| Sensitivity | 0.915758 | 0.940808 | 0.922756 | 0.914 | 0.633 |
The difference in accuracy, specificity and sensitivity among the three phases were not significant (χ2 = 0.346, P = 0.841; χ2 = 0.149, P = 0.928; χ2 = 0.914, P = 0.633; respectively). Sensitivity of the model is considerably more important than its specificity and accuracy, because the purpose of a CT scan is cancer detection. Compared with arterial and venous phase, plain phase had same sensitivity, easier access, and lower radiation. Thus, these results indicated that the plain scan alone might be sufficient for the binary classifier.
Comparison between CNN and gastroenterologists for the binary classification
Table 4 shows the results of the image evaluation of the test data by ten board-certified gastroenterologists and 15 trainees. The accuracy, sensitivity, and specificity in the plain phase were 81.0%, 84.4%, and 80.4%, respectively. The gastroenterologist group was found to have significantly higher accuracy (92.2% vs 73.6%, P < 0.05), specificity (92.1% vs 79.2%, P < 0.05), and sensitivity (92.3% vs 72.5%, P < 0.001) than trainees.
Table 4.
Diagnostic accuracy of the binary classifiers in plain scan: Convolutional neural network vs gastroenterologists and trainees
| CNN | Doctors |
|||
| Gastroenterologists | Trainees | Total | ||
| No. of doctors | 10 | 15 | 25 | |
| Accuracy | 0.954747 | 0.922 | 0.736 | 0.815 |
| Specificity | 0.982710 | 0.923 | 0.725 | 0.847 |
| Sensitivity | 0.915758 | 0.921 | 0.792 | 0.809 |
CNN: Convolutional neural network.
As described in the methods section, ten board-certified gastroenterologists and 15 trainees participated in the study, and their image classification accuracy was compared with that of the deep learning technique as a binary classifier. The accuracy of the gastroenterologists, trainees, and the CNN was 92.20%, 73.60%, and 95.47%, respectively. The accuracy by CNN and board-certified gastroenterologists achieved higher accuracies than trainees (χ2 = 21.534, P < 0.001; χ2 = 9.524, P < 0.05; respectively). However, the difference between CNN and gastroenterologists was not significant (χ2 = 0.759, P = 0.384). Figure 3 demonstrates the receiver operating characteristic (ROC) curves for the binary classification of the plain scan.
Figure 3.
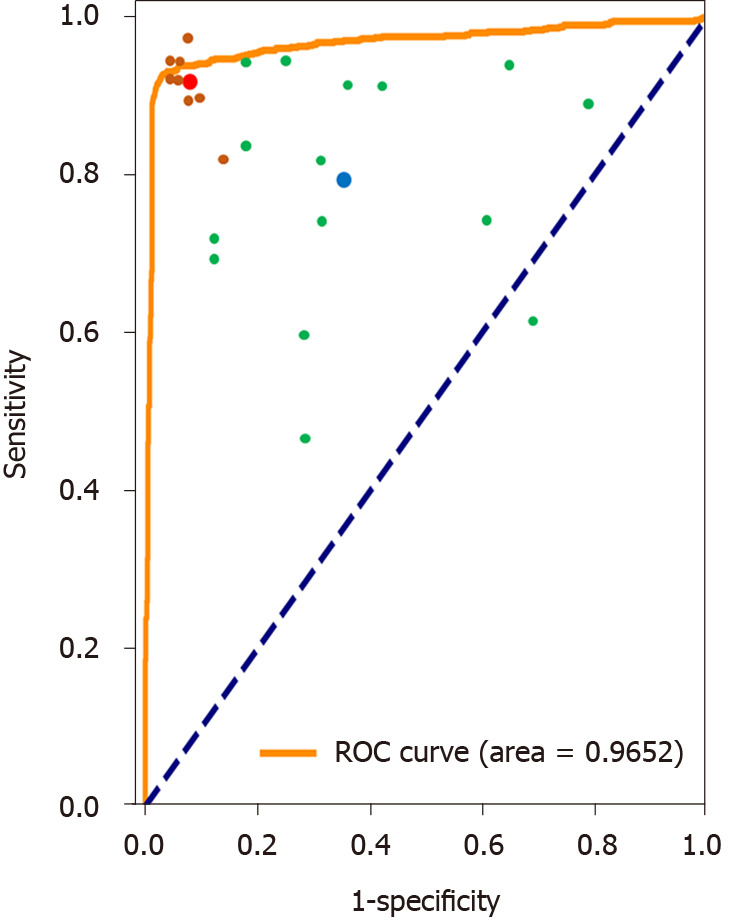
Receiver operating characteristic curves and AUC values for the binary classification of the plain scan using the convolutional neural network model. Each trainee’s prediction is represented by a single green point. The blue point is the average prediction of them. Each gastroenterologist’s prediction is represented by a single brown point. The red point is the average prediction of them. ROC: Receiver operating characteristic.
Performance of the deep convolutional neural network as a ternary classifier
We also trained a ternary classifier using our CNN model and evaluated it by 10-flod cross validation (Table 1). The overall diagnostic accuracy of the ternary classifier CNN was 82.06%, 79.06%, and 78.80% on the plain scan, arterial phase, and venous phase, respectively. The sensitivity scores for detecting cancers in the tail of pancreas were 52.51%, 41.10% and 36.03% on the three phases. The sensitivity scores for detecting cancers in the pancreas head were 46.21%, 85.24%and 72.87% on the three phases, respectively.
The difference in accuracy and specificity among the three phases was not significant (χ2 = 1.074, P = 0.585; χ2 = 0.577, P = 0.749). The difference in the sensitivity scores of cancers in the pancreas head among the three phases was significant (χ2 = 16.651, P < 0.001), with the arterial phase having the highest sensitivity. However, difference in sensitivity in cancers in the pancreas tail among the three phases was not significant (χ2 = 1.841, P = 0.398). The results are summarized in Table 5.
Table 5.
Performance of the ternary classifiers
| Dataset | Plain scan | Arterial phase | Venous phase | χ2 value | P value |
| Accuracy | 0.820568 | 0.790633 | 0.788076 | 1.074 | 0.585 |
| Specificity | 0.985721 | 0.984770 | 0.990305 | 0.577 | 0.749 |
| Sensitivity (cancer at the tail/body of pancreas) | 0.520122 | 0.411098 | 0.360272 | 1.841 | 0.398 |
| Sensitivity (cancer at the head/neck of pancreas) | 0.462148 | 0.852390 | 0.728743 | 16.651 | < 0.001 |
DISCUSSION
In this study, we developed an efficient pancreatic ductal adenocarcinoma classifier using a CNN trained on medium-sized datasets of CT images. We evaluated our approach on the datasets in terms of both binary and ternary classifications, with the purposes of detecting and localizing masses. In the binary classifiers, the performance of plain, arterial and venous phase had no difference, its accuracy on plain scan was 95.47%, sensitivity 91.58%, and specificity 98.27%. In the ternary classifier, the arterial phase had the highest sensitivity in detecting cancer in the head of the pancreas among three phases, but it achieved only moderate performances.
Artificial intelligence has made great strides in bridging the gap between human and machine capabilities. Among the available deep learning architecture, the CNN is the most commonly applied algorithm for analyzing visual images; it can receive an input image, assign weights to various aspects of the image and distinguish one type of image content from another[17]. A CNN includes input, an output layer, and multiple hidden layers. The hidden CNN layers typically consist of convolutional layers, a BN layer, a ReLU layer, pooling layers, and fully connected layers[14]. The CNN acts like a black box, and it can make judgments independent of prior experience or the human effort involved in creating manual features, which is a major advantage. Previous studies showed that CT had a sensitivity of 76%-92%, and an accuracy of 85%-95% for diagnosing pancreatic cancer according to the ability of doctors[18,19]. Our results indicate that our computer-aided diagnostic systems have same detection performance.
The primary goal for a CNN classifier is to detect pancreatic cancer effectively, thus, the model needs to consider sensitivity as a priority over specificity. In the constructed binary classifier, all three phases had high levels of accuracy and sensitivity, with no significant differences among the three phases. This indicates the potential ability of plain scan in tumor screening. Relatively same performance of sensitivity on plain phase can be explained by the size of tumor in our study and redundant information given by arterial or venous phase. In the current study, most tumors were larger than two centimeters, allowing plain scan easier to assess tumor morphology and size. In addition, there are less noisy and unrelated information in the images of the plain scan phase. Thus, it is relatively easy for our CNN model to distill pancreatic-cancer-related features from such images. Currently, the accuracy of the binary classifier on plain scan was 95.47%, its sensitivity 91.58%, and its specificity 98.27%. When compared with the judgments of gastroenterologists and trainees on the plain phase, the CNN model achieved good performance. The accuracy of the CNN and board-certified gastroenterologists was higher than that of the trainees; however, the difference between CNN and gastroenterologists was not significant. We executed our model using a Nvidia GeForce GTX 1080 GPU when performing classifications; its response time was approximately 0.02 seconds per image. Compared with the 10 s average reaction time required by physicians, although our CNN model cannot stably outperform gastroenterologists, the CNN model can process images much faster and is less prone to fatigue. Thus, binary classifiers might be suitable for screening purposes in pancreatic cancer detection.
In our ternary classifier, the accuracy differences among the three phases were also not significant. Regarding sensitivity, the arterial phase had the highest sensitivity in finding malignant lesions among all malignancies in the pancreas head. As the typical appearance of an exocrine pancreatic cancer in CT is a hypoattenuating mass within the pancreas[20], the complex vascular structure around the head and neck of the pancreas could be an explanation for the better performance of CNN classifier in detecting pancreas head and neck lesions in the arterial phase. It is worth noting that unopacified superior mesenteric vein (SMV) at arterial phase may cause confusion in tumor detection. However, SMV has a relatively fixed position in CT image, accompanied by the superior mesenteric artery, which may help the classifier distinguish it from tumor. Further studies in pancreatic segmentation should be carried out to solve this problem. The reason why we also tested a ternary classification is that surgeons choose the surgical approach based on the location of the mass in the pancreas. The conventional operation for pancreatic cancer of the head or uncinate process is pancreaticoduodenectomy. Surgical resection of cancers located in the body or tail of the pancreas involves a distal subtotal pancreatectomy, usually combined with a splenectomy. Compared with gastroenterologists, the performance of the ternary classifier was not as good, because when the physicians judged that a mass existed, they also knew the location of the mass.
Many CNN applications to evaluate organs have been reported, including Helicobacter pylori infection, skin tumors, liver fibrosis, colon polyps, and lung nodules[12,13,21-23], as well as applications for segmenting prostates, kidney tumors, brain tumors, and livers[24-27]. A CNN also has potential applications for pancreatic cancer, mainly focusing on pancreas segmentation by CT[28,29]. Our work concentrates on the detection of pancreatic cancer, and the results demonstrated that on a medium-sized dataset, an affordable CNN model can achieve comparable performance on pancreatic cancer diagnosis and can be helpful as an assistant of the doctors. Another interesting work by Liu et al[30], adopted the faster R-CNN model, which is more complex and harder to train and tune, for pancreatic cancer diagnosis. Their model was mixed images with different phases with an AUC 0.9632, while we trained three classifiers for the plain scan, arterial phase, and venous phase, respectively. Our results indicate that the plain scan, which has easier access and lower radiation, is sufficient for the binary classifier, with an AUC 0.9653.
Our study has several limitations. First, we used only pancreatic cancer and normal pancreas images in this study; thus, our model was not tested with images showing inflammatory conditions of the pancreas, nor was it trained to assess vascular invasion, metastatic lesions and other neoplastic lesions, e.g., intraductal papillary mucinous neoplasm. In the future, we will investigate the performance of our deep learning models on detecting these diseases. Second, our dataset was created using a database with pancreatic cancer/normal pancreas ratio of approximately 1:1; thus, the risk of malignancy in our study cohort was much higher than the normal real-world rate, which made model calculations easier. Therefore, distribution bias might have influenced the entire study, and further studies are needed to clarify this issue. A third limitation is that although the binary classifier achieved the same accuracy as the gastroenterologists, the classifications were based on the information obtained from a single image. We speculate that if the physicians were given additional information, such as the clinical course or dynamic CT images, their classification of the condition would be more accurate. Further studies are needed to clarify this issue.
CONCLUSION
We developed a deep learning-based, computer-aided pancreatic ductal adenocarcinoma classifier trained on medium-sized CT images. The binary classifier may be suitable for disease detection in general medical practice. The ternary classifier could be adopted to localize the mass, with moderate performance. Further improvement in the performance of models would be required before it could be integrated into a clinical strategy.
ARTICLE HIGHLIGHTS
Research background
Pancreatic cancer is a highly lethal malignancy with a very poor prognosis. With promising achievements in deep neural networks and increasing medical needs, computer-aided diagnosis systems have become a new research focus.
Research motivation
Efforts should be made to develop a deep-learning diagnosis system to distinguish pancreatic cancer from benign tissue due to the high morbidity of pancreatic cancer.
Research objectives
To identify pancreatic cancer in computed tomography (CT) images automatically by constructing a convolutional neural network (CNN) classifier.
Research methods
A CNN model was constructed using a dataset of 3494 CT images obtained from 222 patients with pathologically confirmed pancreatic cancer and 3751 CT images from 190 patients with normal pancreas from June 2017 to June 2018. We built three datasets from our images according to the image phases, evaluated our approach in terms of binary classification and ternary classification using 10-fold cross validation, and measured the effectiveness of the model with regard to the accuracy, sensitivity, and specificity.
Research results
In the binary classifiers, the performance of plain, arterial and venous phase showed no difference. Considering that plain phase had relatively same sensitivity, easier access, and lower radiation compared with arterial phase and venous phase, it is more sufficient for the binary classifier. Its accuracy on plain scans was 95.47%, sensitivity was 91.58%, and specificity was 98.27%. In the ternary classifier, the arterial phase had the highest sensitivity in detecting cancer in the head of the pancreas among three phases, but it achieved only moderate performances.
Research conclusions
In this study, we developed a deep learning-based, computer-aided pancreatic ductal adenocarcinoma classifier trained on medium-sized CT images. It was suitable for screening purposes in pancreatic cancer detection.
Research perspectives
Further improvement in the performance of models would be required before it could be integrated into a clinical strategy.
ACKNOWLEDGEMENTS
We would like to thank all the participants and physicians who contributed to the study.
Footnotes
Institutional review board statement: The study was reviewed and approved by the review board of Zhejiang University School of Medicine, Zhejiang Province, China.
Informed consent statement: Patients were not required to give written informed consent to the study because the analysis used anonymous clinical data that were obtained after each patient agreed to treatment by written consent.
Conflict-of-interest statement: We have no financial relationships to disclose.
Manuscript source: Invited manuscript
Peer-review started: April 20, 2020
First decision: May 1, 2020
Article in press: August 26, 2020
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Jurman G, Shichijo S, Wong Y S-Editor: Liu JH L-Editor: MedE-Ma JY P-Editor: Li X
Contributor Information
Han Ma, Department of Gastroenterology, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310003, Zhejiang Province, China.
Zhong-Xin Liu, College of Computer Science and Technology, Zhejiang University, Hangzhou 310058, Zhejiang Province, China.
Jing-Jing Zhang, Department of Gastroenterology, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310003, Zhejiang Province, China.
Feng-Tian Wu, State Key Laboratory for Diagnosis and Treatment of Infectious Disease, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310003, Zhejiang Province, China.
Cheng-Fu Xu, Department of Gastroenterology, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310003, Zhejiang Province, China.
Zhe Shen, Department of Gastroenterology, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310003, Zhejiang Province, China.
Chao-Hui Yu, Department of Gastroenterology, Zhejiang Provincial Key Laboratory of Pancreatic Disease, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310003, Zhejiang Province, China. zyyyych@zju.edu.cn.
You-Ming Li, Department of Gastroenterology, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310003, Zhejiang Province, China.
Data sharing statement
Consent was not obtained, but the presented data are anonymized and risk of identification is low.
References
- 1.Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, Hruban RH. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63:318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 3.Howlader N, Noone AM, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (eds) SEER Cancer Statistics Review, 1975-2016, National Cancer Institute. Bethesda. Available from: https://seer.cancer.gov/csr/1975_2016/, based on November 2018 SEER data submission, posted to the SEER web site, April 2019. [Google Scholar]
- 4.Khorana AA, Mangu PB, Berlin J, Engebretson A, Hong TS, Maitra A, Mohile SG, Mumber M, Schulick R, Shapiro M, Urba S, Zeh HJ, Katz MHG. Potentially Curable Pancreatic Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol. 2017;35:2324–2328. doi: 10.1200/JCO.2017.72.4948. [DOI] [PubMed] [Google Scholar]
- 5.Balaban EP, Mangu PB, Khorana AA, Shah MA, Mukherjee S, Crane CH, Javle MM, Eads JR, Allen P, Ko AH, Engebretson A, Herman JM, Strickler JH, Benson AB, 3rd, Urba S, Yee NS. Locally Advanced, Unresectable Pancreatic Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol. 2016;34:2654–2668. doi: 10.1200/JCO.2016.67.5561. [DOI] [PubMed] [Google Scholar]
- 6.Takhar AS, Palaniappan P, Dhingsa R, Lobo DN. Recent developments in diagnosis of pancreatic cancer. BMJ. 2004;329:668–673. doi: 10.1136/bmj.329.7467.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chari ST. Detecting early pancreatic cancer: problems and prospects. Semin Oncol. 2007;34:284–294. doi: 10.1053/j.seminoncol.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Hawary MM, Francis IR, Chari ST, Fishman EK, Hough DM, Lu DS, Macari M, Megibow AJ, Miller FH, Mortele KJ, Merchant NB, Minter RM, Tamm EP, Sahani DV, Simeone DM. Pancreatic ductal adenocarcinoma radiology reporting template: consensus statement of the Society of Abdominal Radiology and the American Pancreatic Association. Radiology. 2014;270:248–260. doi: 10.1148/radiol.13131184. [DOI] [PubMed] [Google Scholar]
- 9.Zhou B, Xu JW, Cheng YG, Gao JY, Hu SY, Wang L, Zhan HX. Early detection of pancreatic cancer: Where are we now and where are we going? Int J Cancer. 2017;141:231–241. doi: 10.1002/ijc.30670. [DOI] [PubMed] [Google Scholar]
- 10.Chen FM, Ni JM, Zhang ZY, Zhang L, Li B, Jiang CJ. Presurgical Evaluation of Pancreatic Cancer: A Comprehensive Imaging Comparison of CT Versus MRI. AJR Am J Roentgenol. 2016;206:526–535. doi: 10.2214/AJR.15.15236. [DOI] [PubMed] [Google Scholar]
- 11.Vargas R, Nino-Murcia M, Trueblood W, Jeffrey RB., Jr MDCT in Pancreatic adenocarcinoma: prediction of vascular invasion and resectability using a multiphasic technique with curved planar reformations. AJR Am J Roentgenol. 2004;182:419–425. doi: 10.2214/ajr.182.2.1820419. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, Feng X, Chi W, Li Z, Duan W, Liu H, Liang W, Wang W, Chen P, He J, Liu B. Deep learning aided decision support for pulmonary nodules diagnosing: a review. J Thorac Dis. 2018;10:S867–S875. doi: 10.21037/jtd.2018.02.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujisawa Y, Otomo Y, Ogata Y, Nakamura Y, Fujita R, Ishitsuka Y, Watanabe R, Okiyama N, Ohara K, Fujimoto M. Deep-learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses board-certified dermatologists in skin tumour diagnosis. Br J Dermatol. 2019;180:373–381. doi: 10.1111/bjd.16924. [DOI] [PubMed] [Google Scholar]
- 14.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ (eds). Advances in Neural Information Processing Systems 25. Curran Associates Inc. 2012: 1097-1105. [Google Scholar]
- 15.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–1958. [Google Scholar]
- 16.Tompson J, Goroshin R, Jain A, LeCun Y, Bregler C. Efficient object localization using convolutional networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015: 648-656. [Google Scholar]
- 17.Rawat W, Wang Z. Deep Convolutional Neural Networks for Image Classification: A Comprehensive Review. Neural Comput. 2017;29:2352–2449. doi: 10.1162/NECO_a_00990. [DOI] [PubMed] [Google Scholar]
- 18.Scialpi M, Reginelli A, D'Andrea A, Gravante S, Falcone G, Baccari P, Manganaro L, Palumbo B, Cappabianca S. Pancreatic tumors imaging: An update. Int J Surg. 2016;28 Suppl 1:S142–S155. doi: 10.1016/j.ijsu.2015.12.053. [DOI] [PubMed] [Google Scholar]
- 19.Prokesch RW, Chow LC, Beaulieu CF, Bammer R, Jeffrey RB., Jr Isoattenuating pancreatic adenocarcinoma at multi-detector row CT: secondary signs. Radiology. 2002;224:764–768. doi: 10.1148/radiol.2243011284. [DOI] [PubMed] [Google Scholar]
- 20.Yoon SH, Lee JM, Cho JY, Lee KB, Kim JE, Moon SK, Kim SJ, Baek JH, Kim SH, Kim SH, Lee JY, Han JK, Choi BI. Small (≤ 20 mm) pancreatic adenocarcinomas: analysis of enhancement patterns and secondary signs with multiphasic multidetector CT. Radiology. 2011;259:442–452. doi: 10.1148/radiol.11101133. [DOI] [PubMed] [Google Scholar]
- 21.Yasaka K, Akai H, Kunimatsu A, Abe O, Kiryu S. Deep learning for staging liver fibrosis on CT: a pilot study. Eur Radiol. 2018;28:4578–4585. doi: 10.1007/s00330-018-5499-7. [DOI] [PubMed] [Google Scholar]
- 22.Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, Baldi P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology. 2018;155:1069–1078.e8. doi: 10.1053/j.gastro.2018.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shichijo S, Nomura S, Aoyama K, Nishikawa Y, Miura M, Shinagawa T, Takiyama H, Tanimoto T, Ishihara S, Matsuo K, Tada T. Application of Convolutional Neural Networks in the Diagnosis of Helicobacter pylori Infection Based on Endoscopic Images. EBioMedicine. 2017;25:106–111. doi: 10.1016/j.ebiom.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun C, Guo S, Zhang H, Li J, Chen M, Ma S, Jin L, Liu X, Li X, Qian X. Automatic segmentation of liver tumors from multiphase contrast-enhanced CT images based on FCNs. Artif Intell Med. 2017;83:58–66. doi: 10.1016/j.artmed.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Wang S, Chen R, Qu X, Chen Y, Huang S, Liu C. Nested Dilation Networks for Brain Tumor Segmentation Based on Magnetic Resonance Imaging. Front Neurosci. 2019;13:285. doi: 10.3389/fnins.2019.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Dou H, Hu X, Zhu L, Yang X, Xu M, Qin J, Heng PA, Wang T, Ni D. Deep Attentive Features for Prostate Segmentation in 3D Transrectal Ultrasound. IEEE Trans Med Imaging. 2019;38:2768–2778. doi: 10.1109/TMI.2019.2913184. [DOI] [PubMed] [Google Scholar]
- 27.Yu Q, Shi Y, Sun J, Gao Y, Zhu J, Dai Y. Crossbar-Net: A Novel Convolutional Neural Network for Kidney Tumor Segmentation in CT Images. IEEE Trans Image Process. 2019 doi: 10.1109/TIP.2019.2905537. [DOI] [PubMed] [Google Scholar]
- 28.Hussein S, Kandel P, Bolan CW, Wallace MB, Bagci U. Lung and Pancreatic Tumor Characterization in the Deep Learning Era: Novel Supervised and Unsupervised Learning Approaches. IEEE Trans Med Imaging. 2019;38:1777–1787. doi: 10.1109/TMI.2019.2894349. [DOI] [PubMed] [Google Scholar]
- 29.Roth HR, Lu L, Farag A, Shin HC, Liu J, Turkbey EB, Summers RM. DeepOrgan: Multi-level Deep Convolutional Networks for Automated Pancreas Segmentation. In: Navab N, Hornegger J, Wells W, Frangi A (eds), editors. Medical Image Computing and Computer-Assisted Intervention -- MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, vol 9349. Cham: Springer, 2015. [Google Scholar]
- 30.Liu SL, Li S, Guo YT, Zhou YP, Zhang ZD, Li S, Lu Y. Establishment and application of an artificial intelligence diagnosis system for pancreatic cancer with a faster region-based convolutional neural network. Chin Med J (Engl) 2019;132:2795–2803. doi: 10.1097/CM9.0000000000000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Consent was not obtained, but the presented data are anonymized and risk of identification is low.