Summary
Post-translational modification by SUMO is a key regulator of cell identity. In mouse embryonic fibroblasts (MEFs), SUMO impedes reprogramming to pluripotency, while in embryonic stem cells (ESCs), it represses the emergence of totipotent-like cells, suggesting that SUMO targets distinct substrates to preserve somatic and pluripotent states. Using MS-based proteomics, we show that the composition of endogenous SUMOylomes differs dramatically between MEFs and ESCs. In MEFs, SUMO2/3 targets proteins associated with canonical SUMO functions, such as splicing, and transcriptional regulators driving somatic enhancer selection. In contrast, in ESCs, SUMO2/3 primarily modifies highly interconnected repressive chromatin complexes, thereby preventing chromatin opening and transitioning to totipotent-like states. We also characterize several SUMO-modified pluripotency factors and show that SUMOylation of Dppa2 and Dppa4 impedes the conversion to 2-cell-embryo-like states. Altogether, we propose that rewiring the repertoire of SUMO target networks is a major driver of cell fate decision during embryonic development.
Keywords: SUMO, proteomics, embryonic stem cells, somatic cells, cell differentiation, pluripotency, 2C-like cells, totipotency, heterochromatin, Dppa2/4
Graphical Abstract
Highlights
-
•
Endogenous SUMO2/3 proteomics in ESCs and MEFs uncovers drastic SUMOylome rewiring
-
•
In ESCs, SUMO2/3 targets densely interconnected repressive chromatin proteins
-
•
In MEFs, SUMO2/3 targets key determinants of fibroblastic cell identity
-
•
SUMOylation of Dppa2/4 prevents conversion of ESCs to the 2C-like state
Using a comparative proteomic analysis of endogenous SUMO2/3 substrates, Theurillat et al. find a shift in the repertoire of SUMOylated proteins during cell differentiation. SUMO2/3 targets pluripotency and heterochromatin transactions in ESCs but general cellular functions and fibroblastic enhancer activity in MEFs.
Introduction
Precise specification of cell fate within stem cell lineages is critical for ensuring correct cell, tissue, organ, and organism identity. During development, restriction of cell potency over time is governed by specific transcription factors (TFs) that implement particular gene expression programs. Remarkably, pioneer TFs have the ability to direct and reverse cellular identities (Graf and Enver, 2009; Zaret and Carroll, 2011). The most extensively studied model is the conversion of primary fibroblasts into induced pluripotent stem cells (iPSCs) by ectopic expression of a set of core pluripotency-related TFs (Takahashi and Yamanaka, 2006). Recently, we and others have shown that the post-translational modification by SUMO (small ubiquitin-like modifier) acts as a general barrier to cell-fate changes (Borkent et al., 2016; Cheloufi et al., 2015; Cossec et al., 2018). In this process, SUMO functions, at least in part, as a cement to stabilize key chromatin-associated substrates required for the maintenance of cell identity (Cossec et al., 2018). Notably, impairing SUMOylation favors reprogramming of mouse embryonic fibroblasts (MEFs) to iPSCs. Moreover, global suppression of SUMOylation in embryonic stem cells (ESCs) promotes their conversion into cells resembling the 2-cell-stage embryo, called 2C-like cells, which express part of the zygotic genome activation (ZGA) transcriptional program (Cossec et al., 2018).
The SUMO proteins share structural similarities with ubiquitin, and their conjugation to substrates occurs through a related enzymatic cascade involving the sequential action of an E1 activating enzyme (Sae1/Uba2), an E2 conjugating enzyme (Ubc9), and several E3 protein ligases (Pias) (Flotho and Melchior, 2013). The SUMO-specific proteases (Senps) are responsible both for maturation of precursor SUMO proteins and for deconjugation of SUMO from target proteins (Hochstrasser, 2009). The mammalian SUMO protein family consists of three functional paralogs: SUMO1, SUMO2, and SUMO3. Mature SUMO2 and SUMO3 are 95% identical, whereas SUMO1 shares only 43% sequence identity to SUMO2 or SUMO3. Importantly, SUMO2/3 is the main component of SUMO chains, a process greatly enhanced by stress (Saitoh and Hinchey, 2000). Global identification of SUMO substrates showed predominant modification of nuclear targets (Hendriks and Vertegaal, 2016) and SUMOylation is emerging as an important regulator of chromatin structure and function (Cubeñas-Potts and Matunis, 2013). In line with this notion, chromatin immunoprecipitation sequencing (ChIP-seq) analyses revealed that SUMO is widely present all over the genome, where it regulates large gene expression programs (Liu et al., 2012; Neyret-Kahn et al., 2013; Niskanen et al., 2015; Seifert et al., 2015). Surprisingly, the SUMO chromatin landscapes are highly divergent in MEFs and in ESCs (Cossec et al., 2018). Whereas in MEFs, SUMO is present mainly on active enhancers, in ESCs, it is enriched in heterochromatin, suggesting that SUMOylation safeguards somatic and pluripotent cell identities through modification of different protein groups, the nature of which remained to be identified.
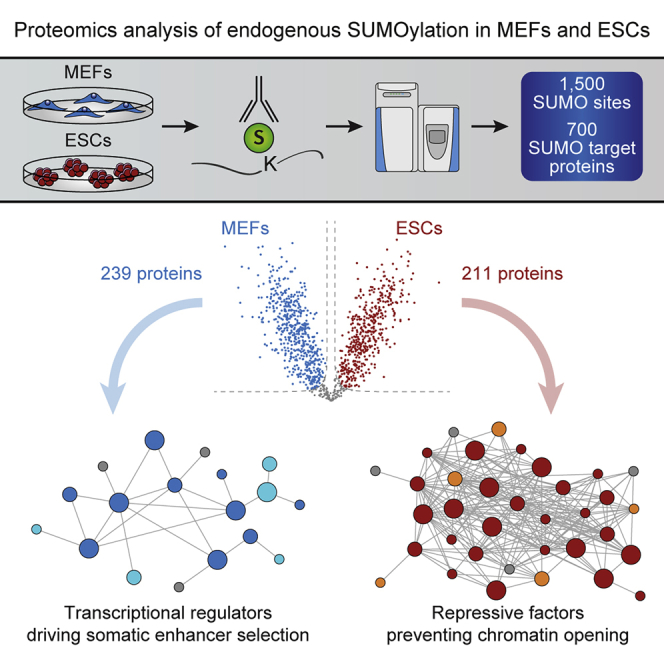
Here we characterized the set of endogenous SUMOylated proteins (SUMOylome) in MEFs and ESCs, taking advantage of a recently developed proteomics approach for quantitative mapping of lysines modified by native SUMO2/3 (Hendriks et al., 2018). Our data reveal strikingly little overlap between the repertoires of SUMO2/3 substrates in differentiated and pluripotent cells. Whereas in MEFs, SUMO2/3 targets proteins involved in general cellular transactions and key determinants of fibroblastic cell identity, in ESCs, SUMO2/3 primarily modifies multiple heterochromatin repressive complexes that are highly interconnected. We moreover mapped the MEF and ESC total proteomes, revealing that the observed heterogeneity in their SUMOylomes cannot be explained by protein expression levels alone. Among the most SUMOylated proteins in ESCs, we identified a number of SUMOylated pluripotency factors, including Dppa2 and Dppa4, which were previously shown to be critical for establishing the 2C-like state (De Iaco et al., 2019; Eckersley-Maslin et al., 2019). Importantly, SUMO-deficient mutants of Dppa2 and Dppa4 favor the conversion of ESCs into 2C-like cells and strengthen their specific transcriptional program. Collectively, our data suggest that concerted SUMOylation of a highly distinct repertoire of critical determinants of cell identity contributes to the establishment and maintenance of somatic and pluripotent states. They also provide a valuable resource for further exploration of the complex SUMO protein network in controlling cellular plasticity.
Results
Site-Specific Profiling of the Endogenous MEF and ESC SUMOylomes
To gain clues regarding the role of SUMO in cell fate determination, we undertook comparative large-scale characterization of endogenous SUMOylated proteins in differentiated cells, MEFs, and pluripotent ESCs. To this aim, we used a recently developed SUMO proteomics strategy that facilitates site-specific profiling of native SUMO2/3 (Hendriks et al., 2018) (Figure 1A). This method uniquely identifies the exact lysine residues that are modified by SUMO2/3, without disturbing the SUMO equilibrium and without the requirement for genetic engineering (Hendriks and Vertegaal, 2016; Sheng et al., 2019).
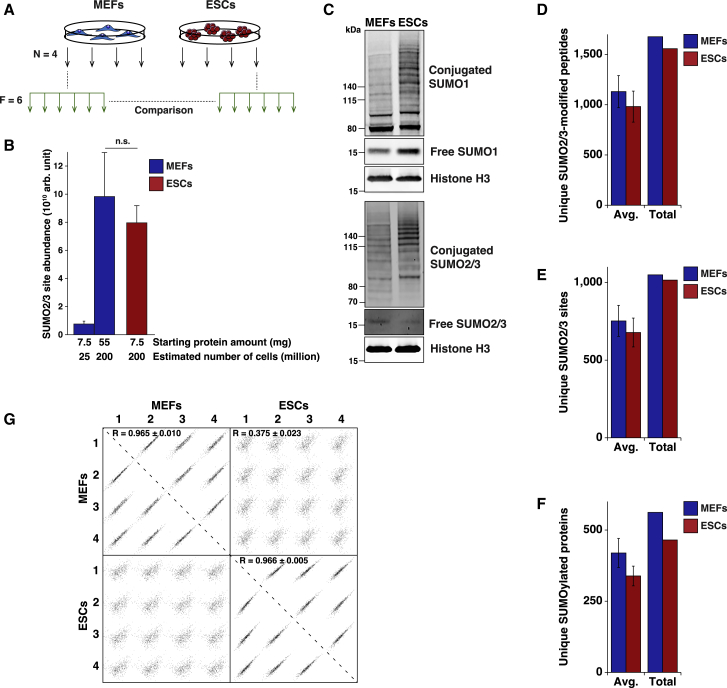
Figure 1.
Experimental Strategy to Characterize Endogenous SUMO2/3 Targets in MEFs and ESCs
(A) Experimental design for the SUMOylome comparison between MEFs and ESCs. All experiments were performed in cell culture quadruplicates and analyzed as six fractions for each replicate.
(B) Comparison of SUMO2/3 site abundance between MEFs and ESCs depending on the amount of protein or the number of cells used for the proteomic analysis. Arb., arbitrary. n = 4.
(C) Immunoblots for SUMO-1 and SUMO2/3 in MEFs and ESCs. Whole-cell lysates from the same number of cells were loaded for the two cell types. Histone H3 was used as a loading control given the comparable histone/chromatin content per cell, regardless of cell type. Representative example, n = 3.
(D) Overview of the average and total numbers of unique SUMO2/3-modified peptides identified from the same number of MEFs or ESCs. Avg., average. n = 4.
(E) Overview of the average and total numbers of unique SUMO2/3 sites identified from the same number of MEFs or ESCs. Avg., average. n = 4.
(F) Overview of the average and total numbers of unique SUMO2/3 target proteins identified from the same number of MEFs or ESCs. Avg., average. n = 4.
(G) Visualization of Pearson correlation between MEF and ESC SUMOylomes. MEF-1 to MEF-4 correspond to the four MEF replicates and ESC-1 to ESC-4 to the four ESCs replicates.
Error bars indicate mean + SD or mean ± SD. See also Tables S1 and S2.
We performed the experiment in quadruplicate to facilitate label-free quantification, while considering the notably larger cellular size of MEFs compared with ESCs. In this regard, we compared both an equal number of cells and an equal amount of post-lysis total protein material. In total, across both experiments and both cell types, 2,480 unique SUMO2/3-modified peptide sequences were identified, corresponding to 1,533 confidently localized SUMO2/3 sites (Table S1), which mapped to 724 SUMO2/3 target proteins (Table S2). Overall, we detected a considerably higher density of SUMO2/3 in ESCs compared with MEFs, with nearly 10-fold more detectable SUMOylated peptides per milligram of total protein (Figure 1B). Intriguingly, when comparing equal numbers of ESCs and MEFs, we did not note a significantly different degree of SUMO2/3 conjugation via mass spectrometry (MS) (Figure 1B). Western blot analysis showed a lower abundance of free SUMO2/3 together with moderately increased SUMO2/3 conjugates in ESCs compared with an equal number of MEFs (Figure 1C). Of note, the amount of both free and conjugated SUMO1 was significantly increased in ESCs (Figure 1C). Specifically, when comparing an equal number of ESC and MEF cells, we observed reproducible and similar numbers of SUMOylation events, with ∼1,500 unique SUMO-modified peptides (Figure 1D), ∼1,000 unique SUMO2/3 sites (Figure 1E), and ∼500 SUMO2/3 target proteins (Figure 1F) identified in each of the two cell types. To avoid bias, the rest of the study was carried out comparing an equal number of cells to ensure a comparable amount of SUMO protein. Quantitative analysis of the SUMOylomes in these conditions allowed quantification of 1,174 (∼77%) SUMO2/3 sites in quadruplicate (Table S1) and quantification of 589 (∼81%) SUMO2/3 target proteins in quadruplicate (Table S2).
A high degree of correlation was observed between experimental replicates (Figure 1G), especially when considering SUMO2/3 target proteins. Contrarily, correlation between MEFs and ESCs was much lower, hinting at a considerable difference in SUMOylome between the two cell types.
The SUMO Equilibrium Is Distinct between Differentiated and Pluripotent Cells
Not all SUMO is conjugated to target proteins, and a pool of free SUMO usually exists in most model systems, which can be indicative of the level of regulatory engagement of the SUMO system (Hendriks et al., 2018). The proteomics methodology we used is uniquely capable of concomitantly quantifying this SUMO equilibrium and can detect free mature and immature SUMO2/3, as well as the relative distribution of conjugated SUMO2/3 across target proteins. In ESCs, we observed 92% of all SUMO2/3 to be conjugated to target proteins (Figure 2A), which is on par with rapidly dividing cell lines (Hendriks et al., 2018). The level of SUMO2/3 conjugation in MEFs was only 72% (Figure 2A), with a considerably larger pool of free SUMO2/3, in line with western blot observations (Figure 1C). The level of immature SUMO2 was significantly lower in ESCs, indicative of a high demand for SUMO conjugation (Figure S1A). In terms of SUMOylation of E1, E2, and E3 enzymes, we observed significantly higher levels of E1 and E2 modification in MEFs, although overall this only accounted for 0.7% of the total SUMO2/3 levels (Figure S1A). More strikingly, we observed high levels of E3 enzyme modification in ESCs, with 11% of the total SUMO pool residing on these enzymes (Figure 2A). Specifically, we observed significantly higher modification levels of Znf451, Pias4, and Pias2 (Tables S1 and S2). Although it remains unclear whether the SUMOylation status of SUMO E3 ligases regulates their activity, this suggests that these three factors may play an active role in defining the SUMO landscape in ESCs. In MEFs, although the overall level of E3 modification was lower, we found Ranbp2 to be significantly more modified (Table S2).
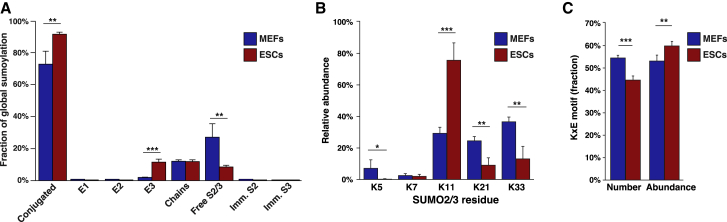
Figure 2.
Comparison of the SUMO Equilibrium between Differentiated and Pluripotent Cells
(A) Quantification of the SUMO2/3 equilibrium in MEFs and ESCs, visualizing the average fraction of total SUMO existing as conjugated to certain target proteins, or as free SUMO. S2, SUMO2; S3, SUMO3; Imm., immature. n = 4.
(B) Quantification of endogenous SUMO2/3 chain architecture in MEFs and ESCs corresponding to endogenous SUMO2 modified by SUMO2/3. n = 4.
(C) Quantification of the SUMO2/3 targeting KxE consensus motifs, either by number of modification sites (number) or in terms of SUMO abundance. n = 4.
Error bars indicate mean + SD. See also Figure S1 and Tables S1 and S2.
Much like ubiquitin, SUMO2/3 can modify itself to form chains, and our data facilitated quantification of SUMO2/3 chain linkages. In terms of global levels of chain formation, we did not find a difference between MEFs and ESCs, with 12% of all SUMO2/3 residing in chains in both cell types. However, the linkage type of the chains was highly distinct between differentiated and pluripotent cells (Figure 2B). In MEFs, SUMO-chain linkages were distributed mainly across three lysines: K21, K33, and K42. Contrarily, in ESCs, the primary linkage was significantly focused on K11, accounting for more than two-thirds of the total SUMO-chain linkages. Of note, acetylation of K11, the prototypical site for SUMO2/3 chain formation (Tatham et al., 2001), was suggested to regulate the formation of non-canonical SUMO2/3 chains (Gärtner et al., 2018). In this regard, it would be interesting to compare K11 acetylation status between the two cell types. We also observed mixed SUMO-ubiquitin chain linkages, with SUMOylation of ubiquitin K48 upregulated in ESCs and SUMOylation of ubiquitin K27 downregulated in ESCs (Table S1). Overall, our proteomics analysis of the SUMO equilibrium in ESCs and MEFs revealed a notably different wiring of the core SUMO enzymatic machinery, which may ultimately be at the root of the highly distinct SUMOylomes and their effect on cell fate.
SUMOylation commonly targets lysines residing in the minimal SUMO consensus motif, KxE, with the glutamic acid at the +2 position facilitating interaction with the E2 enzyme (Bernier-Villamor et al., 2002). Intriguingly, we observed a significantly lower KxE adherence in ESCs, with 44.6% KxE compared with 54.5% KxE in MEFs, with both numbers based on ∼1,000 directly identified SUMO2/3 sites (Figure 2C). Contrarily, when quantifying the relative SUMO occupancy on KxE motifs, we found 59.8% and 53.1% in ESCs and MEFs, respectively. Thus, whereas the overall number of modified sites and quantified SUMO2/3 signal on KxE motifs appeared to match closely in MEFs, the SUMOylome in ESCs was concentrated to a higher degree within a small subset of lysines in KxE motifs. Taken together, this suggests that the core enzymatic regulation of SUMOylation may be differentially orchestrated in pluripotent cells, with focused targeting of much of the SUMO2/3 signal on a small set of KxE motifs while simultaneously also SUMOylating a large number of non-consensus motifs to a lesser degree.
Global SUMOylomes Are Highly Divergent between MEFs and ESCs
Quantitatively, 86% of all SUMO2/3 sites were significantly different (fold change > 2 and false discovery rate [FDR] < 0.05) between MEFs and ESCs (Figures 3A and 3B; Table S1), and 88% of all identified SUMOylated proteins were significantly different between MEFs and ESCs (FDR < 0.05), with 76% of all SUMO2/3 target proteins enriched at least 2-fold in one or the other cell type (239/589 in MEFs and 211/589 in ESCs) (Figure 3B; Table S2). These results demonstrate a dramatic difference in the composition of the SUMOylome between somatic and pluripotent cells. Importantly, among the identified substrates, we found several of the proteins for which SUMOylation was suggested to play an important role in the maintenance of cell identity (Cossec et al., 2018). In MEFs, these include members of the AP-1 functional interaction network (Fosl2, Jdp2, Jund, Cebpb), while in ESCs, we identified members of the PRC1.6 repressive complex (Pcgf6, Mga, L3mbtl2) in addition to proteins involved in the deposition of the heterochromatin mark H3K9me3 (Trim28/Kap1) (Figure 3B). Moreover, several additional relevant substrates, such as Cepba, Setdb1, and Ring1b, were detected in MEFs and/or ESCs, but not quantified across all four replicates (Figure S1B).
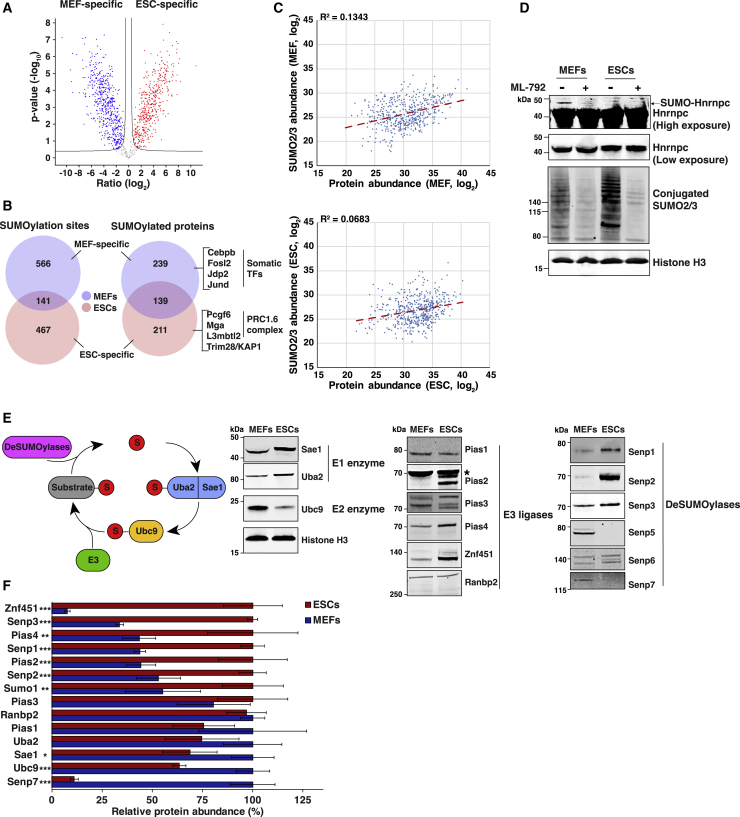
Figure 3.
Global SUMOylome Differences between MEFs and ESCs
(A) Volcano plot displaying differentially modified SUMO2/3 sites in MEFs (blue dots) versus ESCs (red dots). n = 4 cell culture replicates, q < 0.05 with permutation-based FDR calculated at an s0 of 0.5. The black line in the graph represents the FDR cut-off threshold.
(B) Venn diagrams showing the numbers of SUMO2/3 sites (left) and SUMO2/3 target proteins (right) preferentially SUMOylated (log2 fold change [FC] > 1) in MEFs, ESCs, or common to both cell types.
(C) Correlation between protein abundance (x axis) and the SUMO2/3 abundance for the corresponding proteins (y axis) in MEFs (top) and ESCs (bottom). The dotted line corresponds to the regression line.
(D) Immunoblots for Hnrnpc and SUMO2/3 in MEFs and ESCs after 2 days continuous treatment with DMSO or 2 μM ML-792. Histone H3 was used as a loading control given the comparable histone/chromatin content per cell, regardless of cell type. Signals were obtained from the same blot. Two different exposures of the same blot are shown for Hnrnpc. Representative example, n = 2.
(E) Scheme of the SUMOylation cycle (left) and immunoblots (right) for the indicated SUMO enzymes in MEFs and ESCs. Whole-cell lysates from the same number of cells were loaded each time for the two cell types. Fourteen independent blots are shown. Histone H3 was used as a loading control given the comparable histone/chromatin content per cell, regardless of cell type. Loading control for Senp3 is shown; see also Figure S1H. The asterisk indicates a non-specific band. Representative example, n = 2.
(F) Relative protein abundance of members of the SUMOylation machinery identified in the total proteome of MEFs and ESCs, normalized to histone H3 levels. The ratio was fixed to 100% for the cell type with the highest expression for each factor. Error bars indicate mean ± SD.
See also Figures S1 and Tables S1, S2, and S3.
We then asked whether differences in the SUMOylome between the two cell types could be explained by variance of the global proteome in differentiated and pluripotent cells. To address this issue, we performed a quadruplicate total proteome analysis of the exact same samples used for SUMO2/3 enrichment. Overall, we identified 10,189 unique protein groups across MEFs and ESCs (Table S3), with an exceptionally high Pearson correlation of R = 0.992 between cell type replicates (Figure S1C). In MEFs and ESCs, 9,275 and 9,301 proteins, respectively, were detectable in at least duplicate, and 7,355 proteins could be accurately label-free quantified in quadruplicate by at least three peptides. Expression-level correlations between our total proteome (Table S3) and published RNA sequencing (RNA-seq) data (Cossec et al., 2018) were high, with R2 = 0.251 for MEFs and R2 = 0.356 for ESCs (Figure S1D), comparable with what has previously been reported (Vogel and Marcotte, 2012). Out of all SUMO2/3 target proteins identified here (Table S2), 91% were represented in the total proteome analysis, demonstrating good coverage while also highlighting that SUMO can modify proteins that are so low in abundance that total proteome analysis is unlikely to characterize them. Within the subset of SUMO2/3 target proteins, correlations between the total proteome and mRNA levels were R2 = 0.405 for MEFs and R2 = 0.394 for ESCs (Figure S1E). When comparing levels of SUMO2/3-modified proteins to the total proteome, we found only weak correlations of R2 = 0.134 for MEFs and R2 = 0.068 for ESCs (Figure 3C), suggesting that the SUMOylome does not notably follow protein expression levels. Next, we aimed to confirm differential SUMOylation of proteins between MEFs and ESCs, regardless of their expression in the two cell types. Immunoblot analysis of whole lysates from an equal number of MEFs and ESCs revealed that the ribonucleoprotein Hnrnpc is equally expressed in differentiated and pluripotent cells, yet a higher molecular weight form of Hnrnpc was exclusively found in MEFs (Figure 3D). Treatment with ML-792, a specific inhibitor of SUMOylation (He et al., 2017), led to a strong reduction in the intensity of this slower migrating band, thus exemplifying MEF-specific SUMOylation of Hnrnpc in agreement with the MS-based SUMOylome data (Table S2). Altogether, we demonstrate that global proteome differences between differentiated and pluripotent cells are not sufficient to explain the full extent of SUMOylome divergence between the two cell types.
Next, we compared the levels of enzymes involved in the SUMO modification cascade, notably those of E3 ligases and SUMO proteases, which convey some degree of substrate specificity. We validated the specificity of our antibodies by knocking down their respective targets using small interfering RNAs (siRNAs) in either MEFs or ESCs (Figures S1F and S1G). Immunoblot analyses were then performed using equal numbers of MEFs and ESCs to remain consistent with our SUMOylome analysis. This analysis revealed striking differences in the relative amounts of SUMO enzymes across the two cell types (Figures 3E and S1H). We found Pias2 and Pias4 to be more abundant in pluripotent cells. In addition, the amount of Znf451 is greatly increased in ESCs. This is particularly intriguing given the unique ability of Znf451 to trigger SUMO2/3 chain elongation (E4 elongase activity) (Eisenhardt et al., 2015), pointing to a possible important role for Znf451 and elongase activities in pluripotent cells. Moreover, Senp1, Senp2, and Senp3 deSUMOylases were much more abundant in ESCs compared with MEFs, while Senp5 and Senp7 showed the opposite trend. Differential expression was also noted for the unique E1 and E2 enzymes. We ascertained the relative expression levels of SUMO-related enzymes via our total proteome MS data after normalizing to levels of histone H3 (Figure 3F). Except for Sae1, the MS data fully correlated with the western blot data.
Together, these findings indicate that specific combinatorial expression of SUMO enzymes likely participates to the strikingly divergent SUMO proteomes between somatic and pluripotent cells.
Cell Type-Specific SUMO Substrates Assemble into Distinct Functional Networks in Differentiated and Pluripotent Cells
In contrast to other post-translational modifications that are specific for individual proteins, SUMOylation can target whole protein complexes, a process well established in yeast and referred to as protein group modification (Psakhye and Jentsch, 2012). To investigate the propensity of SUMOylation to target functionally interconnected proteins or protein complexes in MEFs and ESCs, we performed protein-protein interaction enrichment analysis using the STRING database (Hendriks et al., 2014). Approximately half (84 of 139) of the common targets between the two cell types were functionally connected (Figure S2), consisting of proteins involved in general cellular processes such as splicing and RNA processing. In addition, SUMO2/3 globally modified general TFs and nucleosome components such as histone variants. Of note, well known SUMO substrates such as PML, Ranbp2, and Top2b, which have been identified in the vast majority of the cell types analyzed so far (Hendriks and Vertegaal, 2016), were also part of the common networks, thus validating the approach.
In MEFs, half (128 of 239) of the preferentially SUMOylated proteins belonged to a diffusely distributed network and were annotated mainly as nucleus localized (Figures 4A and S3). Globally, SUMO2/3 targeted complexes involved in general functions including splicing and translation, as well as components of the cytoskeleton. Moreover, in addition to the TFs that are part of the AP-1 interaction network, we identified members of the Swi/Snf chromatin remodeling complex to be more SUMOylated in MEFs, which could represent other potential candidates for the maintenance of the fibroblastic identity (Vierbuchen et al., 2017).
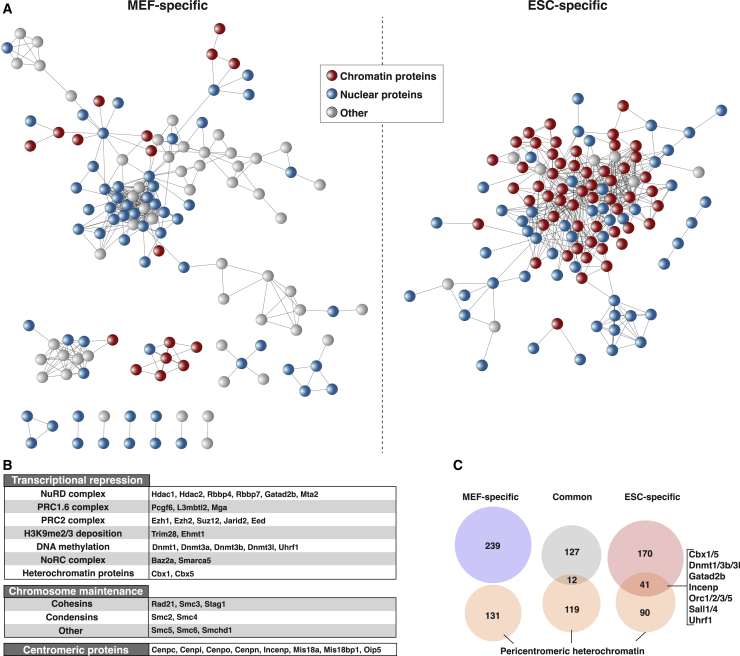
Figure 4.
Comparison of Cell Type-Specific SUMO Substrate Functional Networks
(A) STRING network analysis of proteins preferentially SUMOylated in MEFs (left) and ESCs (right). Each substrate is colored for its cellular localization as annotated by the UniProt protein database.
(B) Examples of ESC-specific SUMO2/3 substrates involved in epigenetic silencing and chromosome maintenance.
(C) Venn diagrams comparing SUMO2/3 substrates with pericentromeric heterochromatin-associated proteins as identified in ESCs (Saksouk et al., 2014).
See also Figures S2–S4.
In contrast, more than half (114 of 211) of the preferentially SUMOylated proteins in ESCs were annotated as chromatin associated and additionally formed a very dense interconnected network (Figures 4A and S4). In addition to members of the PRC1.6 complex and proteins mediating H3K9me2/me3 deposition, SUMO2/3 modified a large number of other repressive factors, including DNA methyltransferases and members of the NuRD, NoRC, and PRC2 repressive complexes (Figure 4B). This latter finding is consistent with the strong enrichment of SUMO at bivalent promoters in ESCs (Cossec et al., 2018). Moreover, other proteins that have been implicated in constitutive heterochromatin formation and maintenance (Cbx1/HP1β, Cbx5/HP1α) were highly modified by SUMO in ESCs. Finally, SUMO2/3 targeted substrates essential for chromosome architecture and mitosis, such as cohesins and condensins, as well as centromeric proteins (Figure 4B), a finding in line with the rapidly cycling state of ESCs and the well-established role for SUMOylation in mitotic chromosome condensation, sister chromatid cohesion and kinetochore function (Mukhopadhyay and Dasso, 2017). To confirm that SUMO is prominently enriched in heterochromatin compartments in pluripotent cells, we cross-correlated our list of SUMO substrates with a published dataset of proteins enriched in pericentromeric heterochromatin regions (Saksouk et al., 2014). We observed that a large number of pericentromeric proteins were represented among ESC-enriched SUMO2/3 substrates, which was not the case for MEF-specific SUMO2/3 targets (31% versus 0%) (Figure 4C).
Overall, in MEFs, SUMO2/3 targets functions that are canonically associated with SUMOylation in somatic cells, such as pre-mRNA splicing and translation (Hendriks and Vertegaal, 2016), as well as fibroblast-specific TFs. In contrast, ESCs display a rather unique SUMO proteome characterized by a highly interconnected set of repressive and structural chromatin proteins, which may suggest increased protein group modification activity.
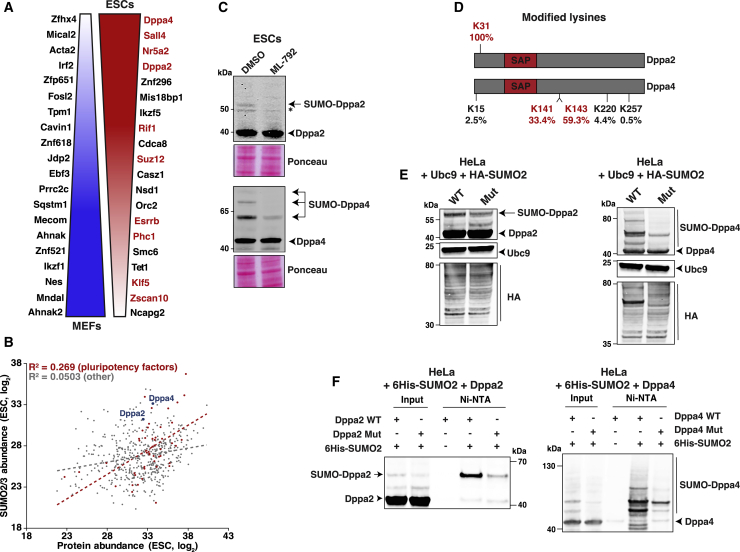
The Pluripotency Factors Dppa2 and Dppa4 Appear as the Top-Scoring SUMO Substrates in ESCs
We next investigated the top 20 most differentially SUMOylated proteins in MEFs versus ESCs (Figure 5A; Table S2). In MEFs, eight of these were cytosolic proteins, including cytoskeletal-associated proteins such as Mical2, Acta2, Tpm1, and Nes, whereas nine substrates corresponded to TFs and TF cofactors, including the AP-1 family members Fosl2 and Jdp2. Of note, the two giant scaffolding proteins Anhak2 and Anhak were among the top hits. In ESCs, all 20 substrates were nuclear proteins, half of which corresponded to pluripotency factors (in red in Figure 5A). We thus investigated the extent to which SUMOylation targets pluripotency factors in ESCs. Correlation between SUMO2/3 modification abundance and protein abundance in ESCs revealed that certain pluripotency factors were among the most highly SUMOylated proteins, far more so than their expression levels would imply (Figure 5B). Of 191 known pluripotency factors detected in the total proteome data, 47 were modified by SUMO2/3, demonstrating a highly significant propensity for SUMO2/3 to specifically target pluripotency factors compared with other proteins (Figure S5A).
Figure 5.
Dppa2 and Dppa4 Are Major Endogenous SUMO Substrates in ESCs
(A) Top 20 substrates differentially modified by SUMO2/3 in MEFs and ESCs. Pluripotency proteins are highlighted in red. SUMO2/3 targets are ranked with the top hits at the bottom for MEFs and at the top for ESCs. Color gradient is for didactic purposes and does not correlate to ratio of change.
(B) Correlation between protein abundance (x axis) and the SUMO2/3 abundance for the corresponding proteins (y axis) in ESCs. Pluripotency factors are highlighted in red. The black dotted line corresponds to the regression line for all SUMO substrates and the red one to pluripotency factors only.
(C) Immunoblots for Dppa2 and Dppa4 in ESCs after 2 day continuous treatment with DMSO or 2 μM ML-792. The star indicates a non-specific band. Representative example, n = 3. Two independent blots are shown. Ponceau staining was used as a loading control.
(D) Schematic representation of Dppa2 and Dppa4 depicting SUMOylation sites identified by mass spectrometry. The percentage indicates the contribution of each lysine to the total SUMOylation of the substrate. In red, the main lysine residues, which were mutated to arginine residues. SAP (SAF-A/B, Acinus, and PIAS) domain is indicated.
(E) SUMOylation of Dppa2 WT or Mut (left) and Dppa4 WT or Mut (right) in vivo. Lysates from HeLa cells transfected with Dppa2 (WT or Mut) or Dppa4 (WT or Mut) together with Ubc9 and HA-SUMO2 and immunoblotted with indicated antibodies
(F) Immunoblot of Dppa2 (left) in HeLa cells overexpressing 6His-SUMO2 together with Dppa2 wild-type (WT) or Dppa2 mutant (Mut). Immunoblot of Dppa4 (right) in HeLa cells overexpressing 6His-SUMO2 together with Dppa4 wild-type (WT) or Dppa4 mutant (Mut). Forty-eight hours post-transfection, cells were harvested, an aliquot of the lysates was directly analyzed (input), and the remaining extracts were used for Ni2+ affinity chromatography to purify 6His-SUMO2 conjugates (Ni-NTA).
Intriguingly, Dppa4 and Dppa2 represented some of the most SUMOylated substrates, while compared with all other proteins in ESCs, they were only moderately expressed (Figure 5B). Notably, in terms of relative enrichment of SUMOylation compared with MEFs, Dppa4 and Dppa2 ranked first and fourth as ESC-specific SUMO2/3 targets, respectively (Figure 5A; Table S2). These two small TFs, which are expressed exclusively in preimplantation embryos, pluripotent cells (Figure S5B), and the germline, were shown recently to activate an early ZGA transcriptional program and to be essential for the transition of ESCs to the 2C-like cells (De Iaco et al., 2019; Eckersley-Maslin et al., 2019). This rare subpopulation of cells, which share some similarities with the 2-cell-stage embryo, arises at very low frequency in ESCs in culture (Macfarlan et al., 2012). Immunoblot analysis of whole lysates revealed the presence of higher molecular weight forms of Dppa2 and Dppa4, which were highly sensitive to ML-792 treatment (Figure 5C). These data strongly suggest that these upper bands correspond to SUMO2/3-modified Dppa2 and Dppa4, and the fact that they are readily visible in whole-cell lysate is indicative for their substantial level of SUMOylation.
To confirm these findings, we generated SUMO-deficient Dppa2 and Dppa4 mutants. Our MS analysis identified a single SUMO2/3 acceptor site at lysine 31 (K31) in Dppa2 (Figure 5D; Table S1) and five SUMOylated lysines in Dppa4, of which the two most prominent sites, at lysines 141 and 143 (K141, K143), accounted for nearly 93% of the protein modification (Figure 5D; Table S1). Mutants for Dppa2 (K31R) and Dppa4 (K141R/K143R) were generated and tested for their effect on SUMOylation in HeLa cells (which do not endogenously express these proteins) upon ectopic expression of the SUMO enzyme Ubc9 together with HA-SUMO2. Both mutants were strongly impaired, though not entirely defective, for SUMO modification (Figure 5E). Similar results were obtained in nickel pull down assays following ectopic expression of His-tagged SUMO2 (Figure 5F).
Together, these data indicate that the developmental proteins Dppa2 and Dppa4 are major endogenous SUMO substrates in ESCs and that K31 and K141/K143 are critical, though not exclusive SUMOylation sites in Dppa2 and Dppa4, respectively.
SUMO-Deficient Dppa2 and Dppa4 Enhance the Conversion to the 2C-like State
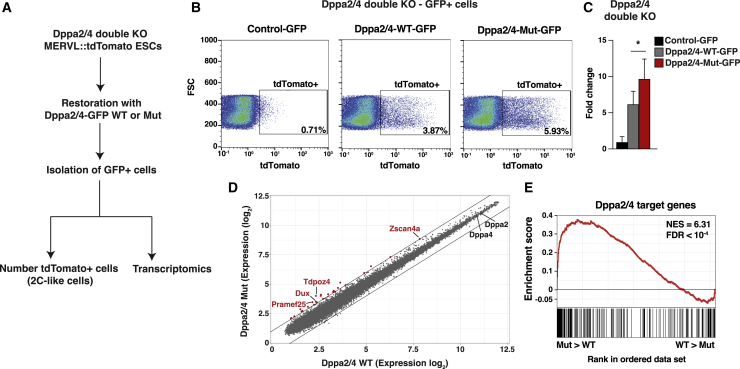
Dppa2 and Dppa4 act in concert in ESCs to establish the 2C-like state through binding and activating the Dux gene, the major driver of the 2C-like transcriptional program (De Iaco et al., 2019; Eckersley-Maslin et al., 2019). Our previous findings that global hypoSUMOylation enhances the conversion of ESCs to 2C-like cells and that SUMO is enriched at the Dux gene and silences its expression (Cossec et al., 2018) prompted us to characterize the functional role of SUMOylation of Dppa2 and Dppa4. We performed rescue experiments to assess the ability of SUMO-deficient Dppa2 and Dppa4 to induce the transition of ESCs to 2C-like cells. We took advantage of a recently developed Dppa2 and Dppa4 double-knockout (KO) Mervl::tdTomato ESC line (Eckersley-Maslin et al., 2019) to follow the appearance of the 2C-like population by tdTomato fluorescence. Dppa2 and Dppa4 wild-type (WT) or mutant (Mut) proteins were fused to green fluorescent protein (GFP) to track complemented cells and we expressed the Dppa2-GFP and Dppa4-GFP constructs in the double-KO ESCs (Figure 6A). Consistent with previous findings (De Iaco et al., 2019; Eckersley-Maslin et al., 2019), simultaneous expression of WT Dppa2 and Dppa4 upregulated the 2C-like fraction (Figure 6B). Interestingly, co-expression of the SUMO-deficient Dppa2 and Dppa4 mutants resulted in a significant increase (1.5-fold) in the number of 2C-like cells in comparison with the WT proteins (Figures 6B and 6C). Similar increases, though less pronounced, were observed upon overproduction of Dppa2 Mut or Dppa4 Mut in the corresponding single-KO ESCs (Figures S6A–S6D).
Figure 6.
SUMOylation of Dppa2 and Dppa4 Inhibits the Transition to 2C-like Cells and Transcript Activation
(A) Experimental procedure used for counting the number of 2C-like cells and for transcriptomic analysis.
(B) Flow cytometry profiles showing expression of the Mervl::tdTomato reporter (x axis) in Dppa2−/− and Dppa4−/− double-knockout (KO) ESCs transfected with GFP (left), Dppa2-WT-GFP + Dppa4-WT-GFP (middle), or Dppa2-Mut-GFP + Dppa4-Mut-GFP (right). The population and percentage of tdTomato-positive cells are shown in the square. Representative example, n = 3.
(C) Percentage of tdTomato-positive cells in Dppa2−/− and Dppa4−/− double-KO ESCs complemented with GFP, Dppa2-WT-GFP + Dppa4-WT-GFP, or Dppa2-Mut-GFP + Dppa4-Mut-GFP. Error bars indicate mean + SD, n = 3.
(D) Scatterplot comparing gene expression of double-KO ESCs complemented with Dppa2-WT-GFP + Dppa4-WT-GFP or Dppa2-Mut-GFP + Dppa4-Mut-GFP. Cells were sorted for GFP expression in both conditions. Red dots indicate overexpressed transcripts (fold change > 2). n = 3.
(E) Gene set enrichment analysis (GSEA) for Dppa2/4-specific target genes in double-KO ESCs complemented with Dppa2-WT-GFP + Dppa4-WT-GFP or Dppa2-Mut-GFP + Dppa4-Mut-GFP. For x axis, genes were ranked on the basis of the ratio of Dppa2/4-Mut-GFP versus Dppa2/4-WT-GFP. NES, normalized enrichment score; FDR, false discovery rate.
To consolidate these observations, we performed a microarray analysis to compare the transcriptome of Dppa2/Dppa4 null ESCs restored with the WT versus Mut versions of Dppa2/Dppa4 (Figure 6A). We verified that WT and Mut proteins were equally expressed as assessed by their mRNA levels (Figure 6D; Table S4). We found that several prominent genes of the 2C-like signature, including Dux and its target genes Zscan4a and Tdpoz4, were slightly, though consistently, upregulated upon expression of the SUMO-deficient proteins compared with the WT forms (Figure 6D). This observation is in line with the finding that artificial fusion of SUMO2 to Dppa2 decreases expression of 2C-like genes (Yan et al., 2019). We next defined a set of genes whose expression is upregulated upon co-expression of the native Dppa2 and Dppa4 proteins in the double-KO ESCs (Figure S6E; Table S5). This set of genes was over-represented among the genes that were upregulated upon expression of Dppa2/Dppa4 mutants, indicating that SUMOylation of these factors restrains their transcriptional activity (Figure 6E).
Collectively, these data indicate that SUMOylation of Dppa2 and Dppa4 plays a direct repressive role on the emergence of the 2C-like cell state and associated transcripts.
Discussion
In this work, we used unbiased MS-based proteomics for in-depth characterization of endogenous SUMO2/3-modified proteins and their SUMO acceptor sites in MEFs and ESCs. Importantly, the proteomics strategy we used does not have a requirement for exogenous expression of SUMO2/3, thereby avoiding perturbation of SUMO homeostasis, which is essential for the maintenance of cell identity (Cheloufi et al., 2015; Cossec et al., 2018). One of our key observations is the dramatic difference between the SUMOylomes of pluripotent and somatic cells, indicating a drastic shift in the repertoire of SUMO2/3 substrates during the process of cell differentiation.
In ESCs, SUMOylated factors are enriched that are associated with gene silencing and heterochromatin structure, including the main repressive complexes, such as PRC1.6, PRC2, NuRD, DNMTs, and NoRC, as well as proteins involved in H3K9me3 deposition and binding. In line with this latter observation, SUMOylation was found to repress endogenous retrovirus expression together with maintaining proper H3K9me3 levels in pluripotent cells (Cossec et al., 2018; Thompson et al., 2015; Yang et al., 2015). Several pericentromeric proteins were similarly enriched in the SUMO2/3 proteome of ESCs, consistent with the reported upregulated expression of major satellite repeats upon loss of SUMOylation in these cells (Cossec et al., 2018). Finally, proteins that control chromosome compaction, such as condensins and cohesins (Skibbens, 2019), were found to be substantially more SUMO modified in ESCs. Remarkably, our screen revealed a high degree of functional interconnectivity of SUMOylated proteins in ESCs compared with MEFs, arguing for potential protein group modification in pluripotent cells. In summary, this MS-based characterization of the SUMO2/3 proteome in ESCs unveiled a predominant repressive chromatin-centric function for SUMOylation, a feature that appears to be unique to this particular cell type.
Developmental progression from totipotency to fully differentiated states is accompanied by a progressive loss in cellular plasticity and gradual compaction of chromatin (Bošković et al., 2014; Gaspar-Maia et al., 2011; Orkin and Hochedlinger, 2011). Notably, pluripotency is characterized by a specific metastable chromatin conformation that, while being more “open” than in somatic cells and thus favoring a transcriptionally permissive state, simultaneously keeps lineage-specific genes in check. In ESCs, SUMO, through concerted modification of a tightly interconnected network of repressive protein complexes, may thus play a major role in contributing to the unique chromatin “milieu” that guides the establishment and maintenance of pluripotency. In this scenario, SUMOylation would prevent chromatin decondensation, and thus reversion to totipotent states, together with avoiding transcriptional permissiveness in this particular open chromatin environment, that could otherwise lead to cell differentiation upon specific developmental signaling.
In MEFs, differentially SUMOylated substrates are associated with biological processes found recurrently across other SUMO proteomic studies, all of which were performed in somatic cells. These processes include splicing, ribosome biogenesis, and cytoskeleton regulation (Hendriks and Vertegaal, 2016). Interestingly, we also identified somatic TFs that are part of the AP-1 complex network, including Fosl2, Jdp2, Jund, and Cebpb. These factors, which are enriched at MEF enhancers, likely contribute to the maintenance of the somatic state, as shown for another AP-1 member, Fosl1 (Chronis et al., 2017). Moreover, several components of the Swi/Snf remodeling complex emerged as MEF-specific SUMO substrates in our study. These results dovetail with a recent report showing that AP-1 together with the Swi/Snf complex play a central role in enhancer selection in MEFs (Vierbuchen et al., 2017). AP-1 TFs select enhancers and further recruit the Swi/Snf complex to establish accessible chromatin at these elements, thus enabling proper expression of the somatic transcriptional program. In MEFs, SUMO is highly enriched on fibroblastic enhancers, and loss in SUMOylation leads to the release of somatic TFs from these elements together with rapid extinction of the fibroblastic program (Cossec et al., 2018). Our data thus suggest that SUMO contributes to MEF-specific gene activation through coordinated modification of key determinants of enhancer selection, likely via stabilization of the factors on these elements, thereby actively guiding cell fate decisions.
The highly distinct composition of the SUMOylome between MEFs and ESCs may simply reflect differences in protein abundance. Notably, in MEFs, TFs that are strongly expressed in somatic cells (Fosl2, Jdp2, Ebf3, and Irf2) are among the most prominent SUMOylation targets, whereas in ESCs, the top hits correspond to pluripotency-associated factors (Dppa4, Dppa2, Sall4, Esrrb, and Nr5a2). However, when comparing absolute levels of protein SUMOylation with the respective abundance of the target proteins in the proteome, we found that protein abundance poorly correlated with SUMOylation levels and accounted for only a minority of the observed difference in the SUMOylomes. Total proteome MS analysis elucidated >6,000 proteins with significantly different expression levels in either MEFs or ESCs, yet only a small fraction (∼10%) of these were found to be SUMOylated, thus highlighting a selectivity in SUMOylation that cannot be extrapolated from protein expression levels alone. Furthermore, whereas the abundant pluripotent factors Oct4 and Sox2 can undergo SUMOylation upon forced expression of the SUMOylation machinery (Tsuruzoe et al., 2006; Wu et al., 2012; Zhang et al., 2007), we failed to detect SUMO-modified forms of the two factors at the endogenous level in ESCs. Although this may be due to modified protein levels below the detection threshold, this observation indicates that highly expressed proteins are not necessarily associated with high levels of SUMOylation, despite the presence of canonical SUMO attachment sites in the protein. Our data indicate that Senp5 and Senp7 are more strongly expressed in MEFs, whereas Pias2, Pias4, Znf451, Senp1, Senp2, and Senp3 are predominant in ESCs. We rather propose that the redistribution of SUMO substrates across the two cell types is mostly dictated by the relative expression of the various components of the SUMO machinery. Of interest, the knockdown of Senp7 in MEFs favors their reprogramming to iPSCs (Zaidan et al., 2018), indicating that changing the ratio between the SUMO proteases may modulate cell fate conversion. Collectively, our data strongly suggest that the observed high divergence in the SUMOylome between ESCs and MEFs, rather than merely reflecting the change in the proteome, is instrumental in establishing the transcriptional programs determinant to cell-type specification.
Suppressing SUMOylation in ESCs was found to trigger the release of the PRC1.6 complex and the H3K9 methyltransferase Setdb1 from Dux, promoting its expression and emergence of 2C-like cells (Cossec et al., 2018). Here, we confirmed that major subunits of the two repressive complexes, such as Mga, L3mbtl2, Pcgf6, and Trim28/Kap1, are extensively SUMOylated at the endogenous level in ESCs. In addition, Dppa2 and Dppa4, which act as upstream activators of Dux (De Iaco et al., 2019; Eckersley-Maslin et al., 2019), emerged as top hits in our ESC proteomics study. The finding that SUMO-deficient Dppa2 and Dppa4 mutants enhance Dux expression and reversion to 2C-like states in rescue settings establishes a direct repressive role of SUMOylation on the transcriptional activity of these factors. Thus, SUMO, as a single modifier for several protein groups, acts locally at multiple levels to repress Dux: by stabilizing repressive complexes onto the Dux locus and by preventing activator-driven transcription. Interestingly, the maternal NELFA/Top2a complex has been shown recently to upregulate Dux and promote 2C-like transition, in part through chromatin decompaction (Hu et al., 2020). Although we did not detect SUMOylated forms of NELFA, with 22 acceptor sites, Top2a appeared as a major SUMO target in ESCs. The exploration of the function of SUMO on Top2a in this context and other Dux regulators to be discovered should shed light on the diverse roles of this modification in regulating the totipotency-to-pluripotency transition.
Finally, future work will be needed to investigate when and where physiological shifts in the repertoire of SUMOylated proteins occur during development and at the adult stage to facilitate rewiring of the regulatory circuitry controlling cell identity. First answers came recently with the observation that the levels of the E3 SUMO ligase Pias4 decrease from the zygote to the 2C embryo stage (Higuchi et al., 2019; Yan et al., 2019). Overexpression of Pias4 results in abnormal ZGA and impairs early mouse embryonic development, underscoring the importance of fine-tuning the balance of SUMOylation/deSUMOylation activities. Finally, our findings may have relevance in cancer, which, in most cases, is associated with hyperactivated SUMOylation (Seeler and Dejean, 2017). Similar proteomics approaches may be used to identify aberrant SUMO substrates in tumors and thus possible cancer cell vulnerabilities to be leveraged to therapeutic ends.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SUMO2/3 (IPs) | Abcam | Cat# ab81371; RRID: AB_1658424 |
| SUMO2/3 (Immunoblots) | MBL | Cat# M114-3; RRID: AB_592769 |
| SUMO1 | Abcam | Cat# ab32058; RRID: AB_778173 |
| Ubc9 | Abcam | Cat# ab75854; RRID: AB_1310787 |
| Dppa2 | Millipore | Cat# MAB4356; RRID: AB_1977389 |
| Dppa4 | R&D systems | Cat# AF3730; RRID: AB_2094166 |
| Hnrnpc | Abcam | Cat# ab133607; RRID: AB_2860560 |
| HA-probe | Santa Cruz | Cat# sc-805; RRID: AB_631618 |
| Sae1 | Abcam | Cat# ab185949; RRID: AB_2827735 |
| Uba2 | Abcam | Cat# ab185955; RRID: AB_2827736 |
| Pias1 | Cell signaling | Cat# 3550; RRID: AB_1904090 |
| Pias2 | Novus Biologicals | Cat# NBP2-19819; RRID: AB_2827737 |
| Pias3 | Santa Cruz | Cat# sc-46682; RRID: AB_628128 |
| Pias4 | Cell signaling | Cat# 4392; RRID: AB_10547884 |
| Znf451 | Sigma | Cat# SAB2108741; RRID: AB_2827738 |
| Ranbp2 | Santa Cruz | Cat# sc-74518; RRID: AB_2176784 |
| Senp1 | Santa Cruz | Cat# sc-271360; RRID: AB_10611042 |
| Senp2 | Santa Cruz | Cat# sc-67075; RRID: AB_2198251 |
| Senp3 | Cell signaling | Cat# 5591; RRID: AB_10694546 |
| Senp5 | Abcam | Cat# ab47631; RRID: AB_882488 |
| Senp6 | Thermo Fisher Scientific | Cat# PA5-69704; RRID: AB_2689788 |
| Senp7 | Thermo Fisher Scientific | Cat# PA5-36089; RRID: AB_2827739 |
| H3 | Abcam | Cat# ab24834; RRID: AB_2553364 |
| Smchd1 | Abcam | Cat# ab31865; RRID: AB_777986 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| ML-792 | Takeda Oncology | N/A |
| Trypsin-EDTA | GIBCO | Cat# 25300-054 |
| FBS | Eurobio | Cat# CVFSVF00-01 |
| GlutaMAX | GIBCO | Cat# 35050-038 |
| NEM-NEAA | GIBCO | Cat# 11140-050 |
| LIF | Miltenyi Biotec | Cat# 130-099-895 |
| KSR | Invitrogen | Cat# 10828028 |
| 2-mercaptoethanol | GIBCO | Cat# 21985-023 |
| TRIzol | Life Technologies | Cat# 15596018 |
| PfuTurbo polymerase | Agilent | Cat# 600255 |
| HIS-Select® Nickel Affinity Gel | Sigma Aldrich | Cat# P6611 |
| Lysyl Endopeptidase, Mass Spectrometry Grade (Lys-C) | Wako | Cat# 125-05061 |
| Protein G Agarose beads | Roche | Cat# 11243233001 |
| Endoproteinase Asp-N Sequencing Grade | Roche | Cat# 11420488001 |
| Critical Commercial Assays | ||
| Affymetrix Mouse Gene 2.0 ST array | Thermo Fisher | Cat# 902119 |
| RNA 6000 Nano kit | Agilent | Cat# 5067-1511 |
| Lipofectamine 2000 | Thermo Scientific | Cat# 301705 |
| Q5® Site-Directed mutagenesis kit | New England Biolabs | Cat# E0554S |
| Gateway BP™ Clonase™ II Enzyme mix | Thermo Scientific | Cat# 11789020 |
| Gateway LR™ Clonase II™ Enzyme mix | Thermo Scientific | Cat# 11791020 |
| Sep-Pak C8 3 cc Vac Cartridge | Waters | Cat# WAT036780 |
| Empore SPE Disks | Sigma Aldrich | Cat# 66883-U |
| ReproSil-Pur 120 C18-AQ, 1.9 μm | Dr. Maisch | Cat# r119.aq. |
| Deposited Data | ||
| Micro-array dataset | This study | NCBI GEO: GSE144881 |
| Raw MS data (SUMOylome) | This study | ProteomeXchange: PXD017697 |
| Raw MS data (total proteome) | This study | ProteomeXchange: PXD020287 |
| Experimental Models: Cell Lines | ||
| R1 mouse ES cells | Cossec et al., 2018 | N/A |
| MERVL::tdTomato Dppa4 KO ES cells | Laboratory of W.Reik | N/A |
| MERVL::tdTomato Dppa2 KO ES cells | Laboratory of W.Reik | N/A |
| MERVL::tdTomato Dppa2/4 double KO ES cells | Laboratory of W.Reik | N/A |
| Mouse embryonic fibroblasts | This study | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | Institut Pasteur | N/A |
| Oligonucleotides and plasmids | ||
| Mutagenesis primers | Eurofins | See Method Details for sequences |
| pCMV6-entry Dppa2 WT plasmid | Origene | Cat# MR224204 |
| pCMV6-entry Dppa2 K31R plasmid | This study | N/A |
| MGC Mouse Dppa4 cDNA | Horizon | Cat# MMM1013-202805602 |
| pCMV Dppa4 WT plasmid | This study | N/A |
| pCMV K141R/K143R plasmid | This study | N/A |
| pDONR221 plasmid | Laboratory of Wolf Reik | N/A |
| pDEST eGFP plasmid | Laboratory of Wolf Reik | N/A |
| pDEST Dppa2 WT-eGFP plasmid | This study | N/A |
| pDEST Dppa2 K31R-eGFP plasmid | This study | N/A |
| pDEST Dppa4 WT-eGFP plasmid | This study | N/A |
| pDEST Dppa4 K141R/K143R-eGFP plasmid | This study | N/A |
| siRNA | Horizon Discovery | Table S6 |
| RT-qPCR primers | Eurofins | Table S6 |
| Software and Algorithms | ||
| STRING 11.0 | Szklarczyk et al., 2019 | N/A |
| FlowJo software | N/A | https://www.flowjo.com/ |
| Graphpad – Prism software | N/A | https://www.graphpad.com/ |
| Cytoscape software | Shannon et al., 2003 | https://cytoscape.org/ |
| MaxQuant version 1.5.3.30 | Cox et al., 2011 | https://www.maxquant.org/ |
| Perseus | Tyanova et al., 2016 | https://maxquant.net/perseus/ |
| Other | ||
| MEF and ESC RNA-seq data | Cossec et al., 2018 | NCBI GEO: GSE99009 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Anne Dejean (anne.dejean@pasteur.fr).
Materials Availability
Plasmids generated in this study are available upon request from the Lead Contact.
Data and Code Availability
The accession number for the micro-array dataset is NCBI GEO: GSE144881. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifiers PXD017697 (SUMOylome data), and PXD020287 (total proteome data).
Experimental Model and Subject Details
Animals
All animal studies were conducted under animal study protocols approved by the French Ministry of Research. C57BL/6J mice were bred at the Pasteur Institute animal facility under specific pathogen–free conditions and housed in a 12-hour light/12-hour dark cycle conditions.
Cell culture
MEF isolation was performed in compliance with the French law and under supervision of the animal core facility (Registration number: A75-15-01, HA0030). Primary MEFs were isolated at 13.5 days post coitum. Pregnant females were sacrificed by cervical dislocation, male and female embryos were removed, and the fetal liver and head were excised. The rest of embryonic tissue was minced and incubated with 0.25% trypsin-EDTA (Sigma-Aldrich) for 15 min at 37°C. MEFs were expanded and frozen at passage 1 for subsequent experiments. MEFs were grown in complete medium: DMEM + GlutaMAX (GIBCO) medium supplemented with 10% FBS and penicillin/streptomycin. HeLa cells were maintained and expanded in complete medium. R1 and MERVL::tdTomato Dppa2/4 knock out embryonic stem cells (ESCs) were maintained in KSR (knock-out serum replacement medium, GIBCO): DMEM + GlutaMAX supplemented with 15% KSR, LIF, 0.1 mM, 2-mercaptoethanol, non-essential amino acid and penicillin/streptomycin on gelatin-coated plates. MERVL::tdTomato Dppa2/4 knock out ESCs were a gift from Wolf Reik (Babraham Institute, Cambridge, UK).
Method Details
Site-specific characterization of endogenous SUMOylation and total proteome
Cell lysates preparation for the SUMO-IP
MEFs and ESCs were amplified to reach approximately 250 million cells per replicate. In the case of MEFs, primary cells had a maximum number of 5 passages. Cells were washed twice with ice-cold PBS and collected on ice by gentle scraping. Collected cells were pelleted by centrifuging at 500g and 4°C for 2 min, and all the PBS was removed. Pellets were resuspended in 2 mL of ice-cold PBS per 100 μL cell pellet. A 100 μL portion was collected, spun down at 500 g and 4°C for 2 min, the PBS was removed and the pellet lysed in 100 μL of SDS NP-40 Tris-buffered saline lysis buffer (2% SDS, 1% NP-40, 150 mM NaCl, and 50 mM TRIS, buffered at pH 7.0) after which it was immediately snap-frozen to serve as an input control. The main batch of cells was spun down at 500g and 4°C for 2 min, PBS was removed and all cells were vigorously lysed using 10 pellet volumes of Lysis Buffer (6 M guanidine, 50 mM TRIS, buffered at pH 8.5), after which they were immediately snap frozen. Lysates were stored at −80°C until further processing.
Peptide preparation for SUMO-IP
In essence, sample preparation and SUMO-IP for native and endogenous mass spectrometry (MS) analysis was performed as described previously (Hendriks et al., 2018). Lysates were thawed at room temperature, after which they were supplemented with 5 mM chloroacetamide (CAA) and 5 mM Tris(2-carboxyethyl)phosphine (TCEP). Subsequently, samples were sonicated using a microtip sonicator, at 30 W and for 1 s per 1 mL of lysate, distributed across 10 s pulses. Samples were incubated for 30 min at 30°C to allow reduction and alkylation to complete, after which endoproteinase Lys-C (Wako) was added in a 1:200 enzyme-to-protein ratio (w/w). Digestion was performed overnight, still, and at room temperature. Samples were diluted with three volumes of 50 mM ammonium bicarbonate (ABC), and a second round of overnight digestion was performed by addition of Lys-C in a 1:200 enzyme-to-protein ratio. Digests were acidified by addition of 0.5% trifluoroacetic acid (TFA), 1:100 vol/vol from a 50% TFA stock, after which they were transferred to 50 mL tubes and centrifuged at 4,250g and at 4°C for 30 min. Clarified digests were carefully decanted into clean 50 mL tubes, after which peptides were purified using C8 Sep-Pak cartridges (Waters), essentially according to the manufacturer’s instructions. Sep-Pak cartridges with 500 mg sorbent were used, with one cartridge used for each ESC replicate, and two cartridges used for each MEF replicate (maximum binding capacity per cartridge is approximately 5% of sorbent amount, e.g., ∼25 mg of digested protein per cartridge). Small and hydrophilic peptides were pre-eluted using 5 mL of 20% ACN in 0.1% TFA, and 3 mL of 25% ACN in 0.1% TFA. SUMOylated peptides were eluted using 1 mL of 35% ACN in 0.1 TFA, 1 mL of 40% ACN in 0.1% TFA, and 2 mL of 45% ACN in 0.1% TFA. For each replicate samples, all elutions were pooled in 50 mL tubes with small holes punctured into the caps, and then frozen overnight at −80°C. Deep-frozen samples were lyophilized to complete dryness for 96 h, with the pressure target set at 0.004 mbar and the condenser coil at −85°C.
Crosslinking of SUMO antibody to beads
In total, 500 μL of Protein G Agarose beads (Roche) were used to capture 300 μL of SUMO-2/3 antibody (8A2, acquired from Abcam, ab81371; ∼5-10 μg/μL antibody). All washing and handling steps were followed by centrifugation of the beads at 500g for 3 min in a swing-out centrifuge with delayed deceleration, and careful aspiration of buffers. Beads were pre-washed 4 times with ice-cold PBS, after which the antibody was added in a 1.5 mL tube, with the tube filled completely with ice-cold PBS. Beads and antibody were incubated at 4°C on a rotating mixer for 1 h, and subsequently washed 3 times with ice-cold PBS. Crosslinking of the antibody to the beads was achieved by addition of 1 mL of 0.2 M sodium borate, pH 9.0, which was freshly supplemented with 20 mM dimethyl pimelimidate (DMP). Crosslinking was performed for 30 min at room temperature on a rotating mixer, after which the crosslinking step was repeated once. SUMO-IP beads where then washed twice with ice-cold PBS, twice with 0.1 M glycine pH 2.8, and three times with ice-cold PBS, after which they were stored until use at 4°C in PBS supplemented with 10 mM sodium azide.
Purification of SUMOylated peptides
Lyophilized peptides were dissolved in 10 mL ice-cold SUMO-IP buffer (50 mM MOPS, 10 mM Na2HPO4, 50 mM NaCl, buffered at pH 7.2). Peptides derived from 7.5 mg of total ESC protein, 7.5 mg of total MEF protein, and 50 mg of total MEF protein, were dissolved separately. The 7.5 mg (equal protein amount compared to ESC) and 50 mg MEF (equal cell count compared to ESC) samples were handled as distinct replicates from this point on. Samples were clarified by centrifugation at 4,250g for 30 min at 4°C in a swing-out centrifuge with delated deceleration. Samples were transferred to new tubes, after which 25 μL SUMO-IP beads were added to the samples based on 7.5 mg protein, and 50 μL SUMO-IP beads were added to the samples based on 50 mg protein. Samples were incubated at 4°C for 3 h in a rotating mixer, after which the beads were washed twice with ice-cold SUMO-IP buffer, twice with ice-cold PBS, and twice with ice-cold MQ water. Upon each first wash with a new buffer, beads were transferred to a clean 1.5 mL LoBind tube (Eppendorf). To minimize loss of beads, all centrifugation steps were performed at 500g for 3 min at 4°C in a swing-out centrifuge with delayed deceleration. Elution of SUMO peptides from the beads was performed by addition of 2 bead volumes of ice-cold 0.15% TFA, and performed for 30 min while standing still on ice, with gentle mixing every 10 min. The elution was transferred to the top of a 0.45 μm spin filter (Millipore), while minimizing carryover of beads. The elution of the beads was repeated once, and the 2nd elution was pooled with the 1st elution on top of the 0.45 μm filter. Next, elutions were clarified of beads by centrifuging at 12,000g for 1 min at 4°C, transferred to a clean 1.5 mL LoBind tube, pH-neutralized by addition of 1/10th volume of 1 M Na2HPO4, and allowed to warm up to room temperature. To shorten the SUMO C terminus to DVFQQQTGG, and to shorten the target peptides to a length feasible for MS analysis, second-stage digestion of SUMOylated peptides was performed with Endoproteinase Asp-N (Roche), using 150 ng of Asp-N for the samples based on 7.5 mg input protein, and 1 μg of AspN for the samples based on 50 mg input protein. Digestion was performed overnight, at 30°C and shaking at 300 rpm, after which samples were frozen at −80°C until further processing.
StageTip purification and high-pH fractionation of SUMO-IP samples
Preparation of StageTips (Rappsilber et al., 2003), and high-pH fractionation of SUMO-IP samples on StageTip, was performed essentially as described previously (Hendriks et al., 2018). Quad-layer StageTips were prepared using four punch-outs of C18 material (Sigma-Aldrich, Empore SPE Disks, C18, 47 mm). StageTips were equilibrated using 100 μL of methanol, 100 μL of 80% acetonitrile (ACN) in 200 mM ammonium, and two times 75 μL 50 mM ammonium. Samples were thawed out, and supplemented with 1/10th volume of 200 mM ammonium, just prior to loading them on StageTip. The StageTips were subsequently washed twice with 75 μL 200 mM ammonium, and afterward eluted as six fractions (F1-6) using 40 μL of 4, 7, 10, 13, 17, and 25% ACN in 50 mM ammonium. All fractions were dried to completion in LoBind tubes, using a SpeedVac for 2 h at 60°C, after which the dried peptides were dissolved using 10 μL of 0.1% formic acid.
Total proteome sample preparation
A small portion (1% of total sample) from the exact peptide mixture from which SUMO2 was immunoprecipitated was used for determination of the total proteome, representing a Lys-C digest eluted off C8 Sep-Paks in the 25%–45% ACN range. The peptides were further digested using modified sequencing grade Trypsin (1:100 w/w; Sigma Aldrich), overnight at 37°C. Tryptic peptides were fractionated on-StageTip at high-pH, essentially as described above for SUMOylated peptides. Peptides were eluted as eight fractions (F1-8) using 80 μL of 2, 4, 7, 10, 13, 17, 22, and 30% ACN in 50 mM ammonium. All fractions were dried to completion in LoBind tubes, using a SpeedVac for 3 h at 60°C, after which the dried peptides were dissolved using 10 μL of 0.1% formic acid. 1.5 μL (0.15% of original total sample) was used for final analysis on the MS.
MS analysis
All samples were analyzed on EASY-nLC 1200 system (Thermo), coupled to either a Q Exactive HF-X Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo) for SUMO samples, or an Orbitrap Exploris 480 mass spectrometer (Thermo) for total proteome samples. Separation of peptides was performed using 15-cm columns (75 μm internal diameter) packed in-house with ReproSil-Pur 120 C18-AQ 1.9 μm beads (Dr. Maisch). Elution of peptides from the column was achieved using a gradient ranging from buffer A (0.1% formic acid) to buffer B (80% acetonitrile in 0.1% formic acid), at a flow of 250 nl/min. Gradient length was 80 min per sample, including ramp-up and wash-out, and an analytical gradient of 50 min. The buffer B ramp for the analytical gradient was as follows for SUMO samples: F1: 13%–24%, F2: 14%–27%, F3-5: 15%–30%, F6: 17%–32%. And as follows for total proteome samples: F1: 5%–38%, F2: 6%–39%, F3: 7%–40%, F4: 8%–41%, F5: 9%–42%, F6: 10%–43%, F7: 12%–45%, F8: 14%–47%. The columns were heated to 40°C using a column oven, and ionization was achieved using either a Nanospray Flex Ion Source (Thermo) for SUMO samples or a NanoSpray Flex NG ion source (Thermo) for total proteome samples. Spray voltage was set at 2 kV, ion transfer tube temperature was set to 275°C, and an RF funnel level of 40% was used. All SUMO samples were measured as two technical replicates, with 5 μL of the sample per injection, and with different technical settings (“Normal” and “Sensitive”) to balance speed versus sensitivity. For “Normal” SUMO analysis, full scan range was set to 400-1,600 m/z, MS1 resolution to 60,000, MS1 AGC target to 3,000,000, and MS1 maximum injection time to 60 ms. Precursors with charges 2-6 were selected for fragmentation using an isolation width of 1.3 m/z, and fragmented using higher-energy collision disassociation (HCD) with normalized collision energy of 25. Precursors were excluded from re-sequencing by setting a dynamic exclusion of 60 s. MS2 resolution was set to 60,000, MS2 AGC target to 200,000, minimum MS2 GC target to 20,000, MS2 maximum injection time to 120 ms, and loop count to 7. For “Sensitive” SUMO analysis all settings were the same except MS1 resolution was set to 120,000, MS1 maximum injection time to 120 ms, MS2 maximum injection time to 500 ms, loop count to 4, and a dynamic exclusion to 120 s. For total proteome analysis, full scan range was set to 300-1,750 m/z, MS1 resolution to 120,000, MS1 AGC target to “200” (2,000,000 charges), and MS1 maximum injection time to “Auto.” Precursors with charges 2-6 were selected for fragmentation using an isolation width of 1.3 m/z, and fragmented using higher-energy collision disassociation (HCD) with normalized collision energy of 25. Monoisotopic Precursor Selection (MIPS) was enabled. Precursors were excluded from re-sequencing by setting a dynamic exclusion of 80 s, with an exclusion mass tolerance of 20 ppm, exclusion of isotopes, and exclusion of alternate charge states for the same precursor. MS2 resolution was set to 15,000, MS2 AGC target to “200” (200,000 charges), MS2 intensity threshold to 430,000, MS2 maximum injection time to “Auto,” and TopN to 18.
Analysis of SUMO MS data
All MS RAW data was analyzed using the freely available MaxQuant software, version 1.5.3.30 (Cox and Mann, 2008; Cox et al., 2011). All SUMO data was processed in a single computational run, and default MaxQuant settings were used, with exceptions specified below. For generation of the theoretical spectral library, the mouse FASTA database was downloaded from Uniprot on the 23rd of July, 2018. The mature sequence of SUMO2 was inserted in the database to allow for detection of free SUMO. In silico digestion of theoretical peptides was performed with Lys-C, Asp-N, and Glu-N, allowing up to 8 missed cleavages. Variable modifications used were protein N-terminal acetylation (default), methionine oxidation (default), peptide N-terminal pyroglutamate, Ser/Thr/Tyr phosphorylation (STY), and Lys SUMOylation, with a maximum of 4 modifications per peptide. The SUMO mass remnant was defined as described previously (Hendriks et al., 2018); DVFQQQTGG, H60C41N12O15, monoisotopic mass 960.4301, neutral loss b7-DVFQQQT, diagnostic mass remnants [b2-DV, b3-DVF, b4-DVFQ, b5-DVFQQ, b6-DVFQQQ, b7-DVFQQQT, b9-DVFQQQTGG, QQ, FQ, FQQ]. Label-free quantification was enabled, with “Fast LFQ” disabled. Maximum peptide mass was set to 6,000 Da. Stringent MaxQuant 1% FDR filtering was applied (default), and additional automatic filtering was ensured by setting the minimum delta score for modified peptides to 20, with a site decoy fraction of 2%. Matching between runs was enabled, with a match time window of 1 min and an alignment window of 20 min. For protein quantification, the same variable modifications were included as for the peptide search. To further minimize false-positive discovery, additional manual filtering was performed at the peptide level. All modified peptides were required to have a localization probability of > 80% and localization delta score of > 6, be supported by diagnostic mass remnants, be absent in the decoy database, and have a delta score of > 40 in case SUMO modification was detected on a peptide C-terminal lysine not preceding an aspartic acid or glutamic acid. All multiply-modified peptides were required to have a delta score of > 40. SUMO target proteins were derived from the “proteinGroups.txt” file, and all post-filtering SUMO sites were manually mapped. Only proteins containing at least one SUMO site were considered as SUMO target proteins, and other putative SUMO target proteins were discarded.
Analysis of total proteome MS data
All total proteome data was processed in a single computational run using MaxQuant 1.5.3.30 and default settings, with exceptions specified below. For generation of the theoretical spectral library, the mouse FASTA database was downloaded from Uniprot on the 23rd of July, 2018. In silico digestion of theoretical peptides was performed with trypsin, allowing up to 3 missed cleavages. Variable modifications used were protein N-terminal acetylation (default), methionine oxidation (default), and Ser/Thr/Tyr phosphorylation (STY), with a maximum of 3 modifications per peptide. Second peptide search was enabled. Label-free quantification was enabled, with “Fast LFQ” disabled, “LFQ min. ratio count” set to 3, and “Skip normalization” enabled. iBAQ was enabled. Stringent MaxQuant 1% FDR filtering was applied at all levels (default). Matching between runs was enabled only within the same (and not neighboring) fractions, with a match time window of 1 min and an alignment window of 20 min. For protein quantification, the same variable modifications were included as for the peptide search.
Quantification of SUMO sites and proteins
All experiments were performed in quadruplicate (n = 4). Quantification of SUMO sites was only performed on SUMO sites that were detected at n = 4/4 in at least one cell type, after which MaxQuant intensity values were median-normalized within experimental conditions. Other detections were considered qualitative. After n = 4/4 filtering and normalization, missing values were globally imputed using Perseus software (Tyanova et al., 2016). SUMO target proteins were processed and quantified analogously to the proteins, but instead using MaxLFQ intensity values from MaxQuant, and requiring at least 2 peptides and at least one SUMOylated peptide per protein. To find significant differences between MEF and ESC, 2-log transformed values were subjected to two-sample testing in Perseus, with a permutation based FDR cut-off of 5% and a p0 value of 0.5.
Quantification of conjugated and free pools of SUMO
The mature sequence of SUMO2 was included as a FASTA file in the MaxQuant search, to allow detection of free mature SUMO2/3. For quantification of pools of SUMO, the “modificationSpecificPeptides.txt” MaxQuant output file was used, and all peptides either modified by SUMO2/3, or peptides derived from SUMO2/3 itself, were considered. Modification of peptides by SUMO was sub-classed into targeting SUMO itself (chain formation), the E1 enzyme subunits (Sae1 and Uba2), the E2 enzyme (Ube2i), the E3 enzymes (Pias, Znf451, Ranbp2), or otherwise conjugation to other targets. Peptides derived from SUMO2/3 were sub-classed as internal, mature free SUMO2/3, immature SUMO2, or immature SUMO3. Peptides ending in QQTGG (predominantly DVFQQQTGG, and to a lesser extent DTIDVFQQQTGG) were considered as mature free SUMO2/3. Peptides containing but not ending with QQTGG were considered as immature SUMO2 (DVFQQQTGGVY), or immature SUMO3 (DVFQQQTGGSASRGSVPTPNRCP). Intensities for each group were summed separately for individual replicates, and fractions were calculated from the summed intensities and used for averages, standard deviations, and Student’s two-tailed t testing.
Quantification of SUMO chains
For SUMO chain quantification, intensity values were directly taken from the “evidence.txt” file. Only confidently localized (> 0.75) evidences were used. SUMO-modified peptides originating from SUMO2/3 were isolated and binned based on modified lysines. In case multiple lysines were simultaneously modified, the peptide intensity was added to each site. Mapped SUMO site positions correspond to SUMO2, the predominantly expressed SUMO family member. Summed site intensities were converted to fractional values within each replicate, and fractional values were averaged across replicates and used for averages, standard deviations, and Student’s two-tailed t testing.
Correlation analysis
For the SUMOylome (n = 4), SUMO target protein LFQ intensity values were used (median CV = 25%–27%). For the total proteome (n = 4), protein intensity values were used (median CV = 15%–18%). For the transcriptome (n = 3) (Cossec et al., 2018), RNA counts were used (median CV = 12%–20%). Low RNA counts (range 1-10) were randomly adjusted by ± 0.5 counts to prevent an artificial lack of noise, and all RNA counts were multiplied by 1 million to shift their magnitude into a range comparable to mass spectrometry intensity values. In all cases, comparisons were only made if non-zero detections were made in at least 2 replicates. Geometric mean values (excluding zeroes) were calculated, 2-log transformed, and correlations were calculated via linear regression.
Protein-protein interaction analyses of the SUMO substrates
A threshold of log2 fold change > 1 was used to specify which substrates were specifically modified in MEFs or ESCs. The protein networks analyses were performed using the online version of STRING (STRING 11.0) (Szklarczyk et al., 2019) with high confidence parameters (0.7), using the sources “Experiments, Databases, and Co-occurrence” and excluding the other sources “Textmining, Neighborhood, Gene Fusion, and Co-expression” for higher stringency. The networks were then visualized and analyzed in Cytoscape software (Shannon et al., 2003).
Directed mutagenesis and cloning
The cDNA for Dppa2 (Origene, MR224204) and Dppa4 (Horizon, MMM1013-202805602) were commercially obtained. First, they were transferred into a PSK vector to perform the mutagenesis using the PfuTurbo polymerase (Agilent) for Dppa2 and the Q5 Site-Directed Mutagenesis kit (NEB) for Dppa4 following the manufacturer’s instructions. The following primers were used for mutagenesis: Dppa2 sense (GTTTGGCTCCTCCTCCTCGCGAAAGGGGACCAGCGTTAAAA), Dppa2 anti-sense (TTTTAACGCTGGTCCCCTTTCGCGAGGAGGAGGAGCCAAAC), Dppa4 sense (GAGGACTGAACCGGGGGAGGAG), Dppa4 anti-sense (ATTCTTTTGTGAGCTGTCTTCAACCTG). The cDNAs were then transferred back in the original commercial vector under CMV promoter for Dppa2 and a home-made expression vector under CMV promoter for Dppa4. For the experiments with Dppa2 and Dppa4 GFP fusions the cDNAs were amplified from the previous plasmids using primers containing AttB1 and AttB2 sequences and cloned into the pDONR221 vector. Gateway cloning (Thermo Scientific) was then used to transfer the cDNA sequences into an in-house-built pDEST vector offered by Wolf Reik (Babraham Institute, Cambridge, UK) containing a CAG promoter and an in-frame C-terminal eGFP-coding sequence and blasticidin resistance by IRES fusion.
Transfection
Transfections were performed using Lipofectamine 2000 (Thermo Fisher) following the manufacturer’s instructions. Cells were plated the day before transfection using 2 μg of plasmid or 50 pmol of siRNAs (Table S6). The medium was changed one day after transfection, and cells were collected two days after transfection for various analyses.
Quantitative PCR
cDNA was generated from 1 μg total RNA purified by Trizol extraction with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative real-time PCR analysis was performed with Power SYBR Green master mix (Thermo Scientific) and the primer sets (Table S6) using cDNA. Quantitative real-time PCR was performed on a CFX96 PCR system (Bio-Rad).
His SUMO-2 immunoprecipitation
The His SUMO-2 immunoprecipitations were performed as described previously (Tatham et al., 2009). Briefly, HeLa wells were seeded one day before transfection with the constructs indicated in the Figure 5F. Two days later, a small fraction of cells was collected in Laemmli buffer to serve as input. The other fraction was washed 3 times in cold PBS and subsequently lysed in cell lysis buffer (6 M Guanidine-HCl, 10 mM Tris, 100 mM sodium phosphate, buffered at pH 8.0). 2-mercaptoepthanol and imidazole were added to the samples to a final concentration of 5 mM. Next, the lysates were sonicated for 30 s at 50% of the full power. Lysates were centrifuged at 3000g to pellet the debris. The supernatant was incubated with 50 μL of Ni2+ NTA beads (QIAGEN) that were pre-equilibrated by washing 3 times in cell lysis buffer. The next day, beads were centrifuged at 750g for 2 min, and washed 5 times in cell lysis buffer. Proteins were eluted using 40 μL of Laemmli buffer.
Immunoblots
Cells were collected and directly lysed in Laemmli buffer. Proteins were quantified using Pierce solution supplemented with Ionic Detergent Compatibility Reagent (Thermo Scientific), following manufacturer’s instructions. Equal amounts of cells were loaded for immunoblotting, and sample loading was assessed by Ponceau staining after membrane transfer. Antibodies against SUMO2 (MBL, M114-3), SUMO1 (Abcam, ab32058), Ubc9 (Abcam, ab75854), Dppa2 (Millipore, MAB4356), Dppa4 (R&D Systems, AF3730), Hnrnpc (Abcam, ab133607), Histone H3 (Abcam, ab24834), Smchd1 (Abcam, ab31865), HA-probe (Santa Cruz, SC805), Sae1 (Abcam, ab185949), Uba2 (Abcam, ab185955), Pias1 (Cell Signaling, 3550), Pias2 (Novus Biologicals, NBP2-19819), Pias3 (Santa Cruz, SC46682), Pias4 (Cell Signaling, 4392), Znf451 (Sigma, SAB2108741), Ranbp2 (Santa Cruz, SC74518), Senp1 (Santa Cruz, SC271360), Senp2 (Santa Cruz, SC67075), Senp3 (Cell Signaling, 5591), Senp5 (Abcam, ab47631), Senp6 (Thermo Fisher Scientific, PA5-69704), and Senp7 (Thermo Fisher Scientific, PA5-36089), were used between 1/1000 and 1/500 concentration according to standard protocols and supplier’s recommendations.
Flow cytometry
For flow cytometry, cells were collected, washed three times in cold PBS, and analyzed on a MACSQuant Analyzer. The results were analyzed in FlowJo. A stringent FSC and SSC gate was used to separate cells from debris. Only cells that were GFP-positive (GFP+) were considered as transfected and used to probe the number of tdTomato-positive (tdTomato+) cells.
Affymetrix micro-arrays
MERVL:tdTomato Dppa2/4 knock out embryonic stem cells were transfected with Dppa2 and Dppa4 WT-GFP plasmids or SUMO deficient Dppa2 and Dppa4 GFP plasmids, using Lipofectamine 2000 according to manufacturer’s instructions. GFP+ cells were selected by flow cytometry and total RNA was prepared by TRIzol extraction (Invitrogen). RNA integrity and concentration were evaluated with a 2100 Bioanalyzer and RNA 6000 Nano kit (Agilent). The preparation of cDNA and its hybridization to an Affymetrix Mouse Gene 2.0 ST Array was performed following the manufacturer’s protocol (Thermo Fisher). A fold change of > 2 was used as the criterion for significance.
Quantification and Statistical Analysis
Statistical analyses were performed using the GraphPad Prism v7 software. Data are represented as Mean + SD or Mean ± SD, statistical significance was assessed by Student’s two-tailed t testing; ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001.
Acknowledgments
We greatly thank Melanie Eckersley-Maslin and Wolf Reik (Babraham Institute) for the Dppa2/4 double- and single-KO ESCs and constructs and for helpful discussions. We are grateful to Alfred Vertegaal (Leiden University) for experimental design advice and to Jacob-Sebastian Seeler for helpful discussions and rereading of the manuscript. We thank Pierre-Henri Commere for fluorescence-activated cell sorting. This work was supported by grants from European Research Council (Advanced Research Grant SUMiDENTITY 832294), Ligue Nationale contre le Cancer (LNCC) (EL2020.LNCC/AnD), Institut National Du Cancer (INCa) (2017-1 PL BIO-05-IP-1), and Agence Nationale de la Recherche (ANR) (SUMOCHROM – ANR-19-CE12-0011-02) to A.D. and by the Novo Nordisk Foundation Center for Protein Research, the Novo Nordisk Foundation (NNF14CC0001 and NNF13OC0006477), and the Danish Council of Independent Research (4002-00051, 4183-00322A, and 8020-00220B) to M.L.N and I.A.H. I.T. was supported by Ministère de l'Enseignement Supérieur et de la Recherche (MESR) and Fondation pour la Recherche Médicale (FRM).
Author Contributions
I.T., I.A.H., J.-C.C., and A.D. conceived the project and designed the experiments. I.A.H. performed the SUMO MS experiments. I.T., I.A.H., J.-C.C., and A.D. interpreted the MS results. I.T., J.-C.C., and A.A. performed cell culture and Dppa2/4 experiments. M.L.N. and A.D. supervised the project. I.T., I.A.H., J.-C.C., and A.D. wrote the paper with input from all co-authors.
Declaration of Interests
The authors declare no competing interests.
Published: September 15, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108146.
Supplemental Information
References
- Bernier-Villamor V., Sampson D.A., Matunis M.J., Lima C.D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- Borkent M., Bennett B.D., Lackford B., Bar-Nur O., Brumbaugh J., Wang L., Du Y., Fargo D.C., Apostolou E., Cheloufi S. A serial shRNA screen for roadblocks to reprogramming identifies the protein modifier SUMO2. Stem Cell Reports. 2016;6:704–716. doi: 10.1016/j.stemcr.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bošković A., Eid A., Pontabry J., Ishiuchi T., Spiegelhalter C., Raghu Ram E.V., Meshorer E., Torres-Padilla M.E. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev. 2014;28:1042–1047. doi: 10.1101/gad.238881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S., Elling U., Hopfgartner B., Jung Y.L., Murn J., Ninova M., Hubmann M., Badeaux A.I., Euong Ang C., Tenen D. The histone chaperone CAF-1 safeguards somatic cell identity. Nature. 2015;528:218–224. doi: 10.1038/nature15749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., Ernst J., Plath K. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 2017;168:442–459.e20. doi: 10.1016/j.cell.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossec J.-C., Theurillat I., Chica C., Búa Aguín S., Gaume X., Andrieux A., Iturbide A., Jouvion G., Li H., Bossis G. SUMO safeguards somatic and pluripotent cell identities by enforcing distinct chromatin states. Cell Stem Cell. 2018;23:742–757.e8. doi: 10.1016/j.stem.2018.10.001. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Cubeñas-Potts C., Matunis M.J. SUMO: a multifaceted modifier of chromatin structure and function. Dev. Cell. 2013;24:1–12. doi: 10.1016/j.devcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A., Coudray A., Duc J., Trono D. DPPA2 and DPPA4 are necessary to establish a 2C-like state in mouse embryonic stem cells. EMBO Rep. 2019;20:e47382. doi: 10.15252/embr.201847382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley-Maslin M., Alda-Catalinas C., Blotenburg M., Kreibich E., Krueger C., Reik W. Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev. 2019;33:194–208. doi: 10.1101/gad.321174.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhardt N., Chaugule V.K., Koidl S., Droescher M., Dogan E., Rettich J., Sutinen P., Imanishi S.Y., Hofmann K., Palvimo J.J., Pichler A. A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly. Nat. Struct. Mol. Biol. 2015;22:959–967. doi: 10.1038/nsmb.3114. [DOI] [PubMed] [Google Scholar]
- Flotho A., Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- Gärtner A., Wagner K., Hölper S., Kunz K., Rodriguez M.S., Müller S. Acetylation of SUMO2 at lysine 11 favors the formation of non-canonical SUMO chains. EMBO Rep. 2018;19:e46117. doi: 10.15252/embr.201846117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A., Alajem A., Meshorer E., Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T., Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- He X., Riceberg J., Soucy T., Koenig E., Minissale J., Gallery M., Bernard H., Yang X., Liao H., Rabino C. Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat. Chem. Biol. 2017;13:1164–1171. doi: 10.1038/nchembio.2463. [DOI] [PubMed] [Google Scholar]
- Hendriks I.A., Vertegaal A.C. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 2016;17:581–595. doi: 10.1038/nrm.2016.81. [DOI] [PubMed] [Google Scholar]
- Hendriks I.A., D’Souza R.C., Yang B., Verlaan-de Vries M., Mann M., Vertegaal A.C. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 2014;21:927–936. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks I.A., Lyon D., Su D., Skotte N.H., Daniel J.A., Jensen L.J., Nielsen M.L. Site-specific characterization of endogenous SUMOylation across species and organs. Nat. Commun. 2018;9:2456. doi: 10.1038/s41467-018-04957-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi C., Yamamoto M., Shin S.-W., Miyamoto K., Matsumoto K. Perturbation of maternal PIASy abundance disrupts zygotic genome activation and embryonic development via SUMOylation pathway. Biol. Open. 2019;8:bio048652. doi: 10.1242/bio.048652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Tan D.E.K., Chia G., Tan H., Leong H.F., Chen B.J., Lau M.S., Tan K.Y.S., Bi X., Yang D. Maternal factor NELFA drives a 2C-like state in mouse embryonic stem cells. Nat. Cell Biol. 2020;22:175–186. doi: 10.1038/s41556-019-0453-8. [DOI] [PubMed] [Google Scholar]
- Liu H.W., Zhang J., Heine G.F., Arora M., Gulcin Ozer H., Onti-Srinivasan R., Huang K., Parvin J.D. Chromatin modification by SUMO-1 stimulates the promoters of translation machinery genes. Nucleic Acids Res. 2012;40:10172–10186. doi: 10.1093/nar/gks819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D., Dasso M. The SUMO pathway in mitosis. In: Wilson V.G., editor. SUMO Regulation of Cellular Processes. Springer International Publishing; Cham: 2017. pp. 171–184. [Google Scholar]
- Neyret-Kahn H., Benhamed M., Ye T., Le Gras S., Cossec J.C., Lapaquette P., Bischof O., Ouspenskaia M., Dasso M., Seeler J. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 2013;23:1563–1579. doi: 10.1101/gr.154872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niskanen E.A., Malinen M., Sutinen P., Toropainen S., Paakinaho V., Vihervaara A., Joutsen J., Kaikkonen M.U., Sistonen L., Palvimo J.J. Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol. 2015;16:153. doi: 10.1186/s13059-015-0717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S.H., Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I., Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151:807–820. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- Saitoh H., Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- Saksouk N., Barth T.K., Ziegler-Birling C., Olova N., Nowak A., Rey E., Mateos-Langerak J., Urbach S., Reik W., Torres-Padilla M.-E. Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol. Cell. 2014;56:580–594. doi: 10.1016/j.molcel.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Seeler J.-S., Dejean A. SUMO and the robustness of cancer. Nat. Rev. Cancer. 2017;17:184–197. doi: 10.1038/nrc.2016.143. [DOI] [PubMed] [Google Scholar]
- Seifert A., Schofield P., Barton G.J., Hay R.T. Proteotoxic stress reprograms the chromatin landscape of SUMO modification. Sci. Signal. 2015;8:rs7. doi: 10.1126/scisignal.aaa2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z., Wang X., Ma Y., Zhang D., Yang Y., Zhang P., Zhu H., Xu N., Liang S. MS-based strategies for identification of protein SUMOylation modification. Electrophoresis. 2019;40:2877–2887. doi: 10.1002/elps.201900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R.V. Condensins and cohesins—one of these things is not like the other! J. Cell Sci. 2019;132:jcs220491. doi: 10.1242/jcs.220491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tatham M.H., Jaffray E., Vaughan O.A., Desterro J.M., Botting C.H., Naismith J.H., Hay R.T. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- Tatham M.H., Rodriguez M.S., Xirodimas D.P., Hay R.T. Detection of protein SUMOylation in vivo. Nat. Protoc. 2009;4:1363–1371. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
- Thompson P.J., Dulberg V., Moon K.M., Foster L.J., Chen C., Karimi M.M., Lorincz M.C. hnRNP K coordinates transcriptional silencing by SETDB1 in embryonic stem cells. PLoS Genet. 2015;11:e1004933. doi: 10.1371/journal.pgen.1004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruzoe S., Ishihara K., Uchimura Y., Watanabe S., Sekita Y., Aoto T., Saitoh H., Yuasa Y., Niwa H., Kawasuji M. Inhibition of DNA binding of Sox2 by the SUMO conjugation. Biochem. Biophys. Res. Commun. 2006;351:920–926. doi: 10.1016/j.bbrc.2006.10.130. [DOI] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T., Ling E., Cowley C.J., Couch C.H., Wang X., Harmin D.A., Roberts C.W.M., Greenberg M.E. AP-1 transcription factors and the BAF complex mediate signal-dependent enhancer selection. Mol. Cell. 2017;68:1067–1082.e12. doi: 10.1016/j.molcel.2017.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo Z., Wu H., Wang X., Yang L., Shi X., Du J., Tang B., Li W., Yang L., Zhang Y. SUMOylation represses Nanog expression via modulating transcription factors Oct4 and Sox2. PLoS ONE. 2012;7:e39606. doi: 10.1371/journal.pone.0039606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.-L., Zhang C., Hao J., Wang X.-L., Ming J., Mi L., Na J., Hu X., Wang Y. DPPA2/4 and SUMO E3 ligase PIAS4 opposingly regulate zygotic transcriptional program. PLoS Biol. 2019;17:e3000324. doi: 10.1371/journal.pbio.3000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B.X., El Farran C.A., Guo H.C., Yu T., Fang H.T., Wang H.F., Schlesinger S., Seah Y.F., Goh G.Y., Neo S.P. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell. 2015;163:230–245. doi: 10.1016/j.cell.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidan N.Z., Walker K.J., Brown J.E., Schaffer L.V., Scalf M., Shortreed M.R., Iyer G., Smith L.M., Sridharan R. Compartmentalization of HP1 proteins in pluripotency acquisition and maintenance. Stem Cell Reports. 2018;10:627–641. doi: 10.1016/j.stemcr.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K.S., Carroll J.S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Liao B., Xu M., Jin Y. Post-translational modification of POU domain transcription factor Oct-4 by SUMO-1. FASEB J. 2007;21:3042–3051. doi: 10.1096/fj.06-6914com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the micro-array dataset is NCBI GEO: GSE144881. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifiers PXD017697 (SUMOylome data), and PXD020287 (total proteome data).