Abstract
Aphids secrete proteins from their stylets that evidence indicates function similar to pathogen effectors for virulence. Here, we describe two small candidate effector gene families of the pea aphid, Acyrthosiphon pisum, that share highly conserved secretory signal peptide coding regions and divergent non-secretory coding sequences derived from miniature exons. The KQY candidate effector family contains eleven members with additional isoforms, generated by alternative splicing. Pairwise comparisons indicate possible four unique KQY families based on coding regions without the secretory signal region. KQY1a, a representative of the family, is encoded by a 968 bp mRNA and a gene that spans 45.7 kbp of the genome. The locus consists of 37 exons, 33 of which are 15 bp or smaller. Additional KQY members, as well as members of the KHI family, share similar features. Differential expression analyses indicate that the genes are expressed preferentially in salivary glands. Proteomic analysis on salivary glands and saliva revealed 11 KQY members in salivary proteins, and KQY1a was detected in an artificial diet solution after aphid feeding. A single KQY locus and two KHI loci were identified in Myzus persicae, the peach aphid. Of the genes that can be anchored to chromosomes, loci are mostly scattered throughout the genome, except a two-gene region (KQY4/KQY6). We propose that the KQY family expanded in A. pisum through combinatorial assemblies of a common secretory signal cassette and novel coding regions, followed by classical gene duplication and divergence.
Keywords: pea aphid, Acyrthosiphon pisum, salivary gland, secretion protein, effector protein, gene family, proteomic analysis
Introduction
Aphids are important pests of plants that can cause economical damage through loss of crop yield and dissemination of plant viruses through their feeding habits (Miles, 1999). There are many different species of aphid that have been found to cause crop damage, including the pea aphid, Acyrthosiphon pisum. The various species of aphid all display a diversity of host ranges, extending from narrow to broad (Jaouannet et al., 2014). An aphid with a narrow host range consumes either one individual species or closely related plants within a single family. An aphid with a broad host range can feed on many different plant species spanning different taxonomic families. During this interaction, aphids extract phloem sap from the leaves and stems of the host plant through stylets, which are inserted into phloem cells. Plants possess both a constitutive and inducible immune response that fights insect consumption (Cook et al., 2015). Once fed upon, a plant can mount a defensive response to thwart parasitic processes. Aphid interactions with non-host plants are hypothesized to fail, in part, due to an immune reaction, while a successful aphid feeding involves suppressing the plant immune response (Jaouannet et al., 2014).
During feeding, aphids secrete saliva, which contains numerous proteins, enzymes, and other compounds, that assist stylet insertion, nutrient extraction, and host tissue interactions (Miles, 1999; Tjallingii, 2006; Will et al., 2007). Upon probing of a potential feeding plant, aphids secrete gelling saliva that acts to surround and protect the stylet. After puncturing the plant, the aphids secrete a watery saliva to thwart plant defenses (Miles, 1999). Components of the salivary proteins are hypothesized to play a role in facilitating the interaction with the host, in analogy to effectors of plant pathogenic bacteria and fungi. In contrast to pathogen effectors, functional evidence for effector action is limited in aphids. Nonetheless, variations in candidate effectors of aphids are hypothesized to contribute to the adaptation of aphid populations to specific host species (biotypes) and genotypes. Ectopic expression and silencing of some candidate aphid effectors have been shown to affect aphid fecundity and growth on host plants (Mutti et al., 2008; Pitino and Hogenhout, 2013).
Effector proteins are often relatively small proteins with no clear function based on relatedness to other proteins and are secreted into the host cell or extracellular milieu. A prominent example of a pea aphid effector is the protein C002. Identified initially from an EST library from the salivary glands, C002 is secreted into the target plant and hypothesized to assist in feeding (Mutti et al., 2008). Reduced expression through inhibitory RNA (RNAi) of the C002 transcript resulted in reduced feeding time of the aphids and, ultimately, premature death. Since the discovery of C002, C002 homologs and additional candidate aphid effectors have been identified (Elzinga and Jander, 2013; Rodriguez and Bos, 2013; Chaudhary et al., 2015; Thorpe et al., 2016; Boulain et al., 2018). One effector of M. persicae, Mp10, has been immunologically localized to the cytoplasm and chloroplasts of plant cells (Mugford et al., 2016).
The pea aphid is a model aphid species that exhibits a narrow host range feeding on legumes exclusively. The pea aphid genome and multiple other aphid genomes are available for analysis and comparisons (Richards et al., 2010; Burger and Botha, 2017; Wenger et al., 2017; Chen et al., 2019; Li et al., 2019; Quan et al., 2019). Additional genomic resources and salivary gland expressed sequence tag (EST) libraries of A. pisum, and other phytophagous aphids, provide numerous effector candidates (International Aphid Genomics Consortium, 2010; Legeai et al., 2010; Shigenobu et al., 2010). Additionally, mass spectrometry proteomic analysis has been used to identify these proteins from salivary glands tissue and saliva secreted into artificial diets (Carolan et al., 2009; Cooper et al., 2010; Carolan et al., 2011; Rao et al., 2013; Chaudhary et al., 2015; Boulain et al., 2018). Despite this progress, much remains to be known about the effectors of aphid salivary proteins in aphid-host plant interactions (Mutti et al., 2006; Carolan et al., 2011; Rao et al., 2013; Boulain et al., 2018). Here, we report the identification of two candidate effector gene families of A. pisum and M. persicae.
Results
Identification of Cassette Gene Families in Pea Aphid
Previously sequenced salivary gland cDNA sequences for A. pisum were retrieved from NCBI, and dataset was analyzed for sequences encoding predicted secreted peptides. Multiple transcripts were identified that encoded relatively short (100–450 aa) proteins and, upon alignment, could be divided into two families based on predicted amino acid sequence similarities ( Supplemental Figures 1 and 2 ). Each family, named KQY and KHI, was composed of multiple genes and, in some cases, two to four isoforms, which were produced by alternative splicing ( Table 1 ). At least one member of the loci, with the exception of KQY2, were found previously to be up-regulated in salivary glands ( Table 1 , Boulain et al., 2018). Three related sequences were also identified in the peach aphid (Myzus persicae) genome ( Table 1 ). The notable feature of the predicted proteins is the conserved signal peptide region, ranging in size from 19 to 28 amino acids, combined with C-terminal divergent sequences ( Figures 1A, B ). The families were, hereafter, referred to as candidate cassette effectors, and the two families were named KQY and KHI after conserved amino acid sequences in the N-terminal region of all or most members ( Figures 1A, B ). The KQY family is comprised of eleven genes and seventeen different isoforms due to splicing variants ( Table 1 ). One member was identified in M. persicae. The KHI family is composed of six genes and 10 isoforms. Two members were identified in M. persicae. The proteins range from 9.2 to 24.4 kDa.
Table 1.
Members of the KQY and KHI families.
| Gene Family | Gene/Isoform a | Gene Locus | Transcript ID | SG Up b | Protein ID | Signal Peptide Predication(TargetP-2.0) c |
|---|---|---|---|---|---|---|
| KQY | KQY1a | LOC100158789 ACYPI000223 |
NM_001162442 | + | NP_001155914 | MIFFKQYSMMITFIVIAVWVMPAITSE (0.8578) |
| KQY1b | LOC100158789 ACYPI000223 |
AK339882 | BAH70584 | MIFFKQYSMMITFIVIAVWVMPAITSE (0.855) | ||
| KQY2a | LOC100302371 | AK342599 | BAH72568 | MVFYKQYLLTITCIVITAWVIPTSA (0.9902) | ||
| KQY2b | LOC100302371 | AK342927 |
BAH72760 NP_001156348 |
MVFYKQYLLTITCIVITAWVIPTSA (0.9902) | ||
| KQY3 | LOC100301916 ACYPI073633 | AK341661 | + |
BAH72003 NP_001153885 |
MVFFKQYLITLTCIVISVWITPVNT (0.9924) | |
| KQY4a | LOC100302370 | AK342948 | + |
BAH72770 NP_001156343 |
MIFFKQYLIILTFIVIAVLVMPVTP (0.9362) | |
| KQY4b | LOC100302370 | AK342242 | BAH72349 | MIFFKQYLIILTFIVIAVLVMPVTP (0.9324) | ||
| KQY4c | LOC100302370 | AK342690 | BAH72627 | MIFFKQYLIILTFIVIAVLVMPVTP (0.9297) | ||
| KQY4d | LOC100302370 | AK342678 | BAH72619 | MIFFKQYLIILTFIVIAVLVMPVTP (0.9537) | ||
| KQY5a | LOC100302375 | AK342406 | + |
BAH72443 NP_001156434 |
MVFFKQYLLTLTCIVIVVQVMPASA (0.9963) | |
| KQY5b | LOC100302376 | AK342378 |
BAH72425 NP_001156435 |
MVFFKQYLLTLTCIVIVVQVMPASA (0.9857) | ||
| KQY6 | LOC100302481 (NV12) | AK340126 | + |
BAH70788 NP_001156817 |
MIFFKQYLIMLTFIIIAVWVMPANT (.9693) | |
| KQY7 | LOC100302485 (NV22) | AK340563 | + |
BAH71149 NP_001156835 |
MSFFKQYLTLTFIVISVWNMSEA (0.9649) | |
| KQY8 | LOC100302439 ACYPI24906 | AK342473 | + |
BAH72486 NP_001156591 |
MVFFKQFLITLTVIIITEA (0.9676) | |
| KQY9 | LOC100302403 | AK342683 | + |
BAH72621 NP_001156509 |
MVFFKLYLLTLTCIVIAVWVMPVSA (0.9981) | |
| KQY10 | LOC100302381 | AK342808 | + |
BAH72693 NP_001156441 |
MFNVLIILSLISYTFEPSYTLYKFKMVFFKQDLLMLTCITIAVWIMPPSASTN (0.8081) | |
| KQY11 | LOC100302480 | AK340121 | + |
BAH70784 NP_001156816 |
MVFFRQFLITLSVILITEA (0.9787) | |
| KQYMp | LOC111039170 (Myzus persicae) |
XM_022322515 | XP_022178207 | MHFFKHYLIVLTYIVISFWFMPSASL (0.9345) | ||
| KHI | KHI1a | LOC100159750 ACYPI001099 |
AK339863 | + |
BAH70570 NP_001155863 |
MFKHIIVLVLCFMAYFVGNLDA (0.998) |
| KHI1b | LOC100159750 ACYPI001099 |
AK339862 | BAH70569 | MFKHIIVLVLCFMAYFVGNLDA (0.9983) | ||
| KHI2a | LOC100302383 | AK341162 | + | BAH71618 | MDKHIIMLALCLMVYIIGNIDA (0.9936) | |
| KHI2b | LOC100302383 | AK341161 |
BAH71617 NP_001156448 |
MDKHIIMLALCLMVYIIGNIDA (0.9937) | ||
| KHI2c | LOC100302383 | AK342769 | BAH72672 | MDKHIIMLALCLMVYIIGNIDA (0.9952) | ||
| KHI3a | LOC100166702 ACYPI007553 |
AK340197 | + |
BAH70850 NP_001156548 |
MLKHIIVLALYLMAYIIGNIDA (0.9965) | |
| KHI3b | LOC100166702 ACYPI007553 |
AK342603 |
BAH72572 NP_001155718 |
MLKHIIVLALYLMAYIIGNIDA (0.9947) | ||
| KHI4 | LOC100570519 ACYPI46154 |
AK341077 | + |
BAH71554 NP_001280397 |
MLKHILLALCFMAYIIENIG (0.9586) | |
| KHI5 | LOC100534636 | AK341390 | + |
BAH71796 NP_001191953 |
MLKHILLALCFMAYIIENIGA (0.9976) | |
| KHI6 | LOC100571631 | AK340760 | + |
BAH71306 NP_001233103 |
MLKHIIVLVLCFMPYIIG (0.9985) | |
| KHIMp1a | LOC111029516 | XM_022308527 | nd | XP_022164219 | MVRHIIMLAICIMFYIIGNAMALTPAERKA | |
| KHIMp1b | LOC111029516 | XM_022308528 | nd | XP_022164220 | MVRHIIMLAICIMFYIIGNAMALTPAERKA | |
| KHIMp1c | LOC111029516 | XM_022308529 | nd | XP_022164221 | MVRHIIMLAICIMFYIIGNAMALTPAERKA | |
| KHIMp2a | LOC111029518 | XM_022308532 | nd | XP_022164224 | MSTMVKHINMLALFIMFYIIGNAMALTPAERKA | |
| KHIMp2b | LOC111029518 | XM_022308533 | nd | XP_022164225 | MSTMVKHINMLALFIMFYIIGNAMALTPAERKA | |
| KHIMp2c | LOC111029518 | XM_022308534 | nd | XP_022164226 | MSTMVKHINMLALFIMFYIIGNAMALTPAERKA | |
| KHIMp2d | LOC111029518 | XM_022308536 | nd | XP_022164228 | MSTMVKHINMLALFIMFYIIGNAMALTPAERKA | |
| C002 | C002Ap | LOC100167863 ACYPI008617 |
XM_001948323 | + | XP_001948358 | MGSYKLYVAVMAIAIAVVQEVRC (0.9704) |
Mp, Myzus persicae, Ap, Acyrthosiphon pisum.
+, Identified as up-regulated in salivary glands in comparison to alimentary tract by Boulain et al. (2018). Up-regulation of locus is indicated, and no differential expression of isoforms is implied. nd, not detected, no salivary gland ESTs from M. persicae were identified by BLAST.
KQY10 and KHIMp isoforms predicted N-terminal peptide, which may be misannotated.
Figure 1.
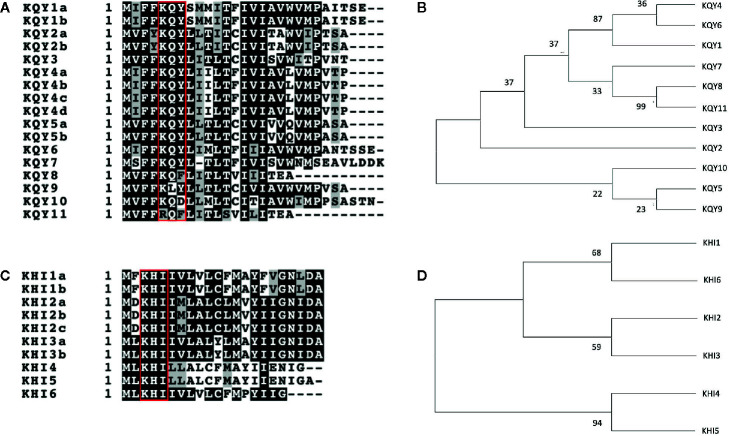
Alignment and phylogeny of KQY and KHI members. (A, B) Amino acid sequence alignment of the N-terminal regions of KQY (A) and KHI (B) gene families, respectively. The alignments were produced with the ClustalW multiple alignment program. The reverse-shaded amino acids represent identical amino acid residues among member of the gene family. The red squares highlight the conserved residues from which the names KQY and KHI were derived. (C, D) Phylogeny based on the nucleic acid sequence of the signal peptides of the KQY (C) and KHI (D) gene families. Maximum-likelihood tree with numbers next to the branches showing bootstrap values as a percentage out of 1,000 replicates.
A maximum likelihood phylogeny was produced, using the N-terminal nucleotides coding sequences for signal peptide region that is unique for each gene ( Figures 1C, D ). Two distinct groups of KQY genes cluster together through high bootstrap values; KQY1, KQY4, KQY6, and KQY8, KQY11. KQY1, KQY4, and KQY6 possess a related bootstrap value of 87, though the KQY4 and KQY6 are more distantly related within this group, only containing a bootstrap value of 36. KQY8 and KQY11 are highly similar, which is related in their bootstrap value of 99. Beyond the secretory signal peptide coding region, pairwise BLAST analysis of the KQY coding sequences indicates four possible gene families (KQY1, 4, 6; KQY2, 5, 9, 10; KQY3, 8, 11, Mp; KQY 7) at the probability level of 1 x 10-5( Supplemental Figure 3 ). KQY7 shares no sequence relatedness beyond the secretory signal region at the DNA or protein level. At the same time, all members share some sequence identity in 3’ region of the transcripts at the nucleotide level, with the exception of KQY7 and KQYMp ( Supplemental Figure 4 ).
Similarly, within the KHI gene family, only two KHI members cluster close together according to bootstrap values, KHI4 and KHI5. KHI4 and KHI5 possess a bootstrap value of 94, indicated high homology. The remaining the KHI gene signal peptides are loosely related with KHI1, KHI6, and KHI2, KHI3 clustering with bootstrap values of 68 and 59, respectively.
KQY10 is annotated with twenty-nine additional N-terminal amino acid residues in comparison to the other family members ( Table 1 ). Both the sequence, as annotated, or a shortened version are predicted to contain a signal peptide. Similarly, the KHIMp2 locus, including all isoforms, are annotated with three additional amino acid residues at the N-terminus ( Table 1 ).
Identification of Candidate Cassette Effectors by Proteomic Analysis of Salivary Gland Proteins
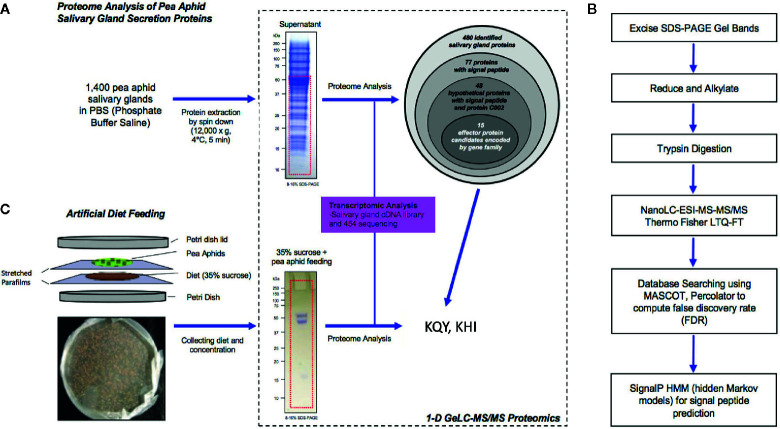
A proteomic analysis was conducted to determine whether member of the two families were present in salivary glands and secreted in salivary fluids ( Figure 2A ). Proteins were extracted from the salivary gland tissues of A. pisum and separated on a SDS-PAGE (1DE) gel ( Figure 2B ). Proteins in 10–60 kDa range were then subjected to 1-D GeLC-MS/MS. Of the 480 proteins, 77 proteins with predicted secretion signals were present ( Supplementary Table 1 ). Notably, 16 of the candidate secreted gland proteins were members of the KQY and KHI families ( Table 2 ; Supplemental Table 1 ). Ten KQY proteins were found, namely KQY1a, KQY2a, KQY2b, KQY3, KQY4a, KQY4c, KQY5b, KQY9, KQY10, and KQY11. KQY2a and b, KQY4a and c, and KHI2a and b were the isoforms found concurrently. Five of the KHI gene family corresponding proteins of the six KHI genes were identified, and six out of the ten protein isoforms were found ( Table 2 , Supplemental Table 1 ). In addition to other candidate effectors, the analysis identified the conserved aphid effector C002 ( Table 2 ).
Figure 2.
Proteomic analysis of pea aphid salivary gland secretion proteins. (A) Schematic representation of secretion proteome analysis of pea aphid salivary gland. (B) 1-D GeLC-MS/MS Proteomics flowchart to identify salivary gland secretion proteins. (C) Schematic of artificial diet feeding experiment.
Table 2.
Cassette effectors from proteomic analysis A. pisum salivary glands.
| Gene Family | Protein Name | NCBI Accession Number | # of Peptides (Unique) | % Coverage |
|---|---|---|---|---|
| KQY | KQY1a ACYPI000223 LOC100158789 |
NP_001155914 | 17(14) | 66 |
| KQY2a LOC100302371 |
BAH72568 | 9(8) | 47 | |
| KQY2b LOC100302371 |
NP_001156348 | 8(6) | 36 | |
| KQY3 LOC100301916 |
NP_001153885 | 2(1) | 13 | |
| KQY4a LOC100302370 |
NP_001156343 | 10(9) | 43 | |
| KQY4c LOC100302370 |
BAH72627 | 10(9) | 45 | |
| KQY5b LOC100302376 |
NP_001156435 | 8(8) | 45 | |
| KQY9 LOC100302403 |
NP_001156509 | 3(3) | 32 | |
| KQY9 LOC100302403 |
NP_001156509 | 3(3) | 24 | |
| KQY10 LOC100302381 |
NP_001156441 | 1(1) | 6 | |
| KQY11 LOC100302480 |
NP_001156816 | 3(3) | 24 | |
| KHI | KHI1a ACYPI001099 LOC100159750 |
NP_001155863 | 4(3) | 33 |
| KHI2a LOC100302383 |
BAH71618 | 2(2) | 14 | |
| KHI2b LOC100302383 |
NP_001156448 | 2(2) | 14 | |
| KHI3a LOC100166702 |
NP_001156548 | 12(9) | 56 | |
| KHI5 LOC100534636 |
NP_001191953 | 6(6) | 38 | |
| KHI6 LOC100571631 |
NP_001233103 | 4(4) | 44 | |
| C002 | C002 | XM_001948323 | 5(4) | 33 |
Proteins were collected from artificial diet media after feeding by A. pisum to determine if any of the family members could be detected in extracellular fluids using an artificial diet ( Figure 2C ). Total protein was analyzed through 1-D GeLC-MS/MS ( Figure 2B ). A total of nine aphid proteins were identified, including KQY protein, KQY1a ( Table 3 ). Additional proteins included amino peptidase and angiotensin converting enzymes and have been previously observed (Boulain et al., 2018).
Table 3.
A. pisum salivary gland proteins detected in synthetic diet using 1-D geLC-MS/MS.
| Protein Name | NCBI Accession | Top Ion E-value | # of Peptides (Unique) | % Coverage | Signal Peptide Probability (D-score) | Mw (kDa) | Predicted Protein Name/Function |
|---|---|---|---|---|---|---|---|
| KQY1a | NP_001155863 | 2.10E-04 | 1(1) | 8 | 0.645 | 23.1 | KQY gene family |
| Aminopeptidase N-like | gi|193636568 | 2.20E-09 | 19(15) | 51 | 0.822 | 70.1 | M1 zinc dependent metalloprotease |
| Angiotensin converting enzyme-like | gi|193669489 | 5.20E-07 | 7(6) | 42.9 | 0.703 | 74.2 | M2 zinc dependent angiotensin converting enzyme (peptidase) |
| Angiotensin converting enzyme-like | gi|209571509 | 1.30E-04 | 4(4) | 9.6 | 0.994 | 74.5 | M2 zinc dependent angiotensin converting enzyme (peptidase) |
| Aminopeptidase N-like | gi|193702193 | 4.00E-06 | 18(9) | 29.2 | 0.265 | 90 | M1 zinc dependent metalloprotease- |
| Leucyl-cystinyl aminopeptidase-like | gi|193643503 | 3.30E-07 | 9(6) | 31.5 | 0.974 | 105 | M1 zinc dependent metalloprotease |
| Aminopeptidase N-like isoform 1 | gi|193634323 | 9.00E-10 | 16(13) | 59.8 | 0.991 | 105.2 | M1 zinc dependent metalloprotease |
| Aminopeptidase N-like | gi|193657504 | 8.30E-06 | 5(3) | 7.8 | 0.753 | 105.4 | M1 zinc dependent metalloprotease |
| Aminopeptidase N-like | gi|193575561 | 2.80E-10 | 30(21) | 31.6 | 0.981 | 105.5 | M1 zinc dependent metalloprotease |
Large Gene Structure and Genomic Location of Candidate Cassette Effector Genes
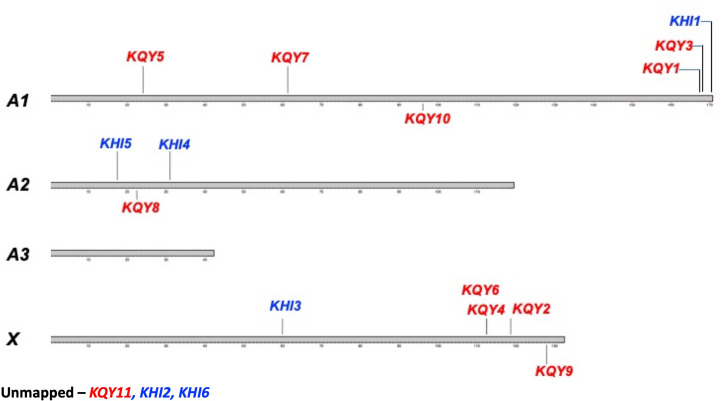
The pea aphid genome consists of four different chromosomes. The gene for KQY1a protein (gi|241896885) is anchored on the A1 chromosome in the A. pisum strain AL4f genome (GCF_005508785.1) ( Table 4 ). Each of the pea aphid KQY genes have been found placed within the genome except for KQY11. KQY genes can be found placed on chromosome A1, A2, and X but not A3 ( Table 4 , Figure 3 ). Two of the KHI genes, KHI2 and KHI6, were unable to be placed within the pea aphid genome, and the remaining KHI genes were also found on chromosome A1, A2, and X, but not A3 ( Table 4 , Figure 3 ).
Table 4.
Genome locations of KQY and KHI families.
| Gene Family | Gene/Isoform Designation | Gene Locus | Chromosome | Location | Size (kb)exons (range bp)/introns (range bp) |
|---|---|---|---|---|---|
| KQY | KQY1a | LOC100158789 | Chr A1 NC_042494.1 | 167,823,999–167,869,655 | 45.7 37(6-416)/36(180-5168) |
| KQY1b | LOC100158789 | ||||
| KQY2a | LOC100302371 | Chr X NC_042493.1 |
118,679,504–118,712,594 | 33.1 | |
| KQY2b | LOC100302371 | 31.2 >13(5-420)/>12(555-9442) |
|||
| KQY3 | LOC100301916 | Chr A1 NC_042494.1 | Complement 168,127,940–168,141,540 |
13,6 15(9-582)/14(281-5949) |
|
| KQY4a | LOC100302370 | Chr X NC_042493.1 |
Complement 112,529,404–112,604,186 |
74.8 32(5-389)/>31(203-10515) |
|
| KQY4b | LOC100302370 | ||||
| KQY4c | LOC100302370 | ||||
| KQY4d | LOC100302370 | ||||
| KQY5a | LOC100302375 | 87.8 >27(6-399)/>26(377-43413) |
|||
| KQY5b | LOC100302376 | Chr A1 NC_042494.1 | 24,100,856–24,188,627 | 87.8 | |
| KQY6 | LOC100302481 (NV12) | Chr X NC_042493.1 |
112,481,406–112,505,954 | 24.5 | |
| KQY7 | LOC100302485 (NV22) | Chr A1 NC_042494.1 | Complement 61,298,211–61,309,480 |
11.3 | |
| KQY8 | LOC100302439 | Chr A2 NC_042495.1 | Complement 22,473,653–22,510,347 Length: 36,695 nt |
36.7 | |
| KQY9 | LOC100302403 | Chr X NC_042493.1 |
Complement 127,899,608–127,978,192 |
78.6 | |
| KQY10 | LOC100302381 | Chr A1 NC_042494.1 | 96,124,545–96,133,787 | 9.2 | |
| KQY11 | LOC100302480 | NW_021761267.1 unplaced | |||
| KHI | KHI1a | LOC100159750 | Chr A1 NC_042494.1 | complement 170,686,182–170,699,132 |
13.0 12.8, 11(27-564)/10(170-2537) |
| KHI1b | LOC100159750 | ||||
| KHI2a | LOC100302383 | NW_021771857.1 unplaced | |||
| KHI2b | LOC100302383 | ||||
| KHI2c | LOC100302383 | ||||
| KHI3a | LOC100166702 | Chr X NC_042493.1 |
59,926,126–59,940,736 | 14.6 13(6-355)/12(173-2296) |
|
| KHI3b | LOC100166702 | 14.6 14(6-355)/13(95-2296) |
|||
| KHI4 | LOC100570519 (ACYPI46154) |
Chr A2 NC_042495.1 | complement 31,024,034–31,037,869 |
13.9 | |
| KHI5 | LOC100534636 | Chr A2 NC_042495.1 | complement 17,449,710–17,453,573 |
18.9 >13(16-254)/>12(67-6096) |
|
| KHI6 | LOC100571631 | NW_021770650.1 unplaced |
Figure 3.
Placement of the KQY and KHI genes on the pea aphid chromosomes. The pea aphid chromosomes A1, A2, A3, and X. KQY genes are colored in red and the KHI genes are colored in blue. KQY11, KHI2, and KHI6 are unmapped.
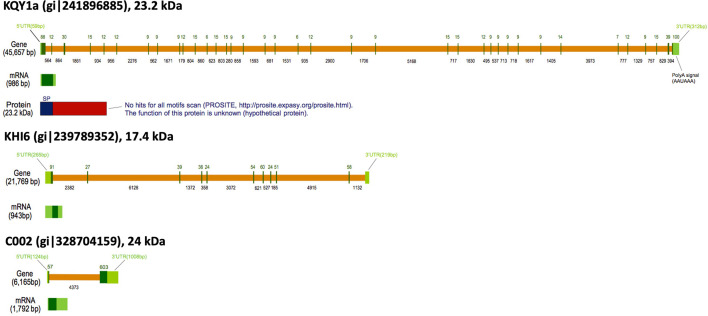
KQY1a covers approximately 45.7 kbp, and the transcript is comprised of 37 relatively small exons (6−416 bp) ( Figure 4 ). This structure of a large gene coding for a small protein using many miniature exons is also observed with other KQY gene family members ( Table 4 ). KHI members are also generated from relatively large genes. The KHI6 transcript (gi|239789352) is 943 bps long and comes from 10 exons in a gene that is 21.789 kbps long ( Figure 4 ). The gene sizes of the mentioned gene families range from 12 kbp to 87 kbp. The first reported pea aphid effector/secretion protein, C002, shown here for contrast, has relatively small gene size (~6 kbp) with only two exons ( Figure 4 ). No significant similar/conserved protein motifs and domains were found. The protein function of the gene family is unknown (hypothetical protein). A separate predicted locus (LOC100569066) can be found within an intron of KQY2. The gene product is highly conserved RAD50-interacting protein 1 (XP_016658051).
Figure 4.
Gene structures of KQY1a and KHI6. C002 is shown for comparison. Diagrams are based on the graphic sequence display generated from the NCBI genome sequence viewer. Orange bars = introns; Green bars = exons.
Discussion
Here, we add to the characterization of candidate effectors of A. pisum, and, by sequence relatedness, possibly, M. persicae with the description of two families of genes, which by several criteria, appear to be variable secreted salivary gland proteins (Carolan et al., 2009; Carolan et al., 2011; Rao et al., 2013; Boulain et al., 2018). Twenty-seven protein candidates based on representative cDNAs could be assigned to either the KQY or KHI families, and most of the cDNA were represented in salivary gland RNAseq libraries. All of the loci, with exception of KQY2, were previously shown to have at least one isoform up-regulated in salivary gland in relation to alimentary tract expression, and all are predicted to encode secreted small molecular weight proteins (~12−28 kDa). Furthermore, peptides from a majority of the loci were detected in protein extractions of washed salivary glands, and one was detected in artificial feeding media. In a previous analysis, unidentified isoforms of three cassette effectors were detected in an artificial diet, including KQY2, which lacked clear evidence for salivary gland expression (Boulain et al., 2018).
The members of the two families were named cassette effectors due to the conserved N-terminal region, which harbors the signal secretion motif, and the divergent coding sequences distal to the secretory signal region. The model implies that novel coding sequences could be swapped on to the signal cassette, generating novel secreted proteins, which, in turn, can then facilitate the adaptation process of the aphid to new hosts or host varieties. The KQY genes can be grouped into three gene subfamilies that have, at least in part, expanded by gene duplication and divergence. KQY7 constitutes a single gene subfamily. Nonetheless, members of different families share sequence similarities beyond the coding regions indicating possible mosaic gene structure. The presence of a single KQY candidate from the related but distant green peach aphid (M. persicae) may be the result of amplification of a single gene during adaptation of pea aphids to various leguminous hosts. Whether cassette swapping was involved in adaption to a new host cannot be definitively stated. Analysis of various biotypes of A. pisum may reveal subspecies cassette gene content. Cassette effectors analogous to KQY and KHI have been previously identified in the Hessian fly genome, where the SSSGP-1 family share a similar structure (Chen et al., 2010), and domain swapping with secretory domains has been proposed, to name a few, to drive complexity in scorpion venom, in the evolution of plastid nuclear encoded proteins, and new virulence in nematodes (Tonkin et al., 2008; Vanholme et al., 2009; Wang et al., 2016). Exon shuffling has long been proposed, in itself, as one benefit of eukaryotic gene structure (Koonin et al., 2013; Smithers et al., 2019). The KQY and KHI genes are represented by varying numbers of mRNAs isoforms. However, definitive conclusions with regards to the levels of individual isoforms or loci remain unclear.
Some of the candidate cassette effector genes are quite large. KQY1a, as an example, is produced from a 986 base mRNA, which, in turn, is spliced from 46 kb of DNA, containing 37 exons and 36 introns. The gene sizes are not the largest, but, given the protein product, they are remarkable. The human gene for type III collagen, for example, is 44 kb and has 52 exons. However, the mRNA is 5460 bases, encoding a protein of 1446 amino acid residues in length, compared to the 986 mRNA and 204 aa products. KQY4 and KQY5 may be nearly twice the size of KQY1a. Further conclusions regarding KQY4 and KQY5 and some other gene of the candidate cassette effectors await improved genome sequencing and assembly. General conclusions regarding the arrangement of the genes may change due to future assembly improvements. The gene that can be mapped are scattered throughout the genome and, at present, only one pair are present in tandem (KQY4 and KQY6), despite the general view that highly evolving loci occur in multigenic loci. The contribution of cassette family genes to aphid adaptation awaits attempts to alter the expression of individual genes.
Materials and Methods
Pea Aphids, Salivary Glands, Proteins Collection
Pea aphid (A. pisum) clone LSR1 was maintained on Vicia faba at 20°C. Salivary glands of feeding adult aphid on the host plants were dissected following a protocol of the previous study (Mutti et al., 2006). For salivary gland protein extraction, the dissected salivary glands of A. pisum were stored in PBS solution with protease inhibitor cocktail (Roche) and centrifuged at 12,000 × g for 15 min at 4°C without tissue homogenization to avoid cellular proteins. After centrifugation and collecting supernatant, salivary gland proteins of the supernatant were precipitated with 20% TCA (v/v) and incubated at -20°C, overnight. The protein pellet was collected by centrifugation (1,500 × g for 10 min, 4°C) and then washed with 100% acetone 3 times and allowed the protein pellet to air dry. The protein pellet was dissolved in SDS-PAGE sample buffer [0.25 M Tris-HCl (pH6.8), 50% glycerol, 5% SDS, and 5% β-mercaptoethanol] for protein separation by 1-D SDS-PAGE for proteome analysis.
Saliva Collection From Artificial Diet
Synthetic diet preparation and saliva collection were conducted under aseptic conditions (Will et al., 2007). Pea aphid saliva collection plates were prepared by stretching sterilized parafilm over the bottom of the 100 by 15 mm plastic petri dishes. Parafilm sheet surface sterilized and exposed to UV light for 30 min and the parafilms were stretched to 50% of the original size. Five milligrams chemically defined synthetic diet (35% sucrose solution) was placed on the stretched parafilm and cover with the other sterilized stretched parafilm ( Figure 1A ). Fifteen aphid saliva collection plates (approximately 1,600 pea aphid on each plate) were prepared for the secreted saliva collection from the synthetic diet. The diet from a 24 h collection period was pooled to give a volume approximately 75 ml, followed by concentration using a Vivaspin concentrator (GE Healthcare) with 3,000 molecular weight cut-off PES membrane at 4°C. The concentrated proteins were separated by 1-D SDS-PAGE and visualized with Coomassie blue R-250.
In Gel Sample Preparation for Mass Spectrometry
For salivary gland proteome analysis, we have identified salivary gland proteins using by 1-D GeLC-MS/MS proteome approach. Proteins from salivary gland tissues and artificial diet were separated on 8%–16% Tris-HCl precast gel (Bio-Rad) in a Mini-Protean Electrophoresis Unit (Bio-Rad) and stained with Coomassie blue R-250 ( Figure 1A ). The stained protein bands of interest were excised using sterile surgical blades and the gel slices (no larger than 2 × 5 mm) were transferred to individual 1.5 ml microcentrifuge tubes with 10 μl HPLC grade water to prevent dehydration and prepared In-gel digestion. Proteins in the gel slices were reduced with 10 mM DTT in 200 mM ammonium bicarbonate at 60°C for 15 min, and then subjected to amidation in 20 mM iodoacetamide in 200 mM ammonium bicarbonate at room temperature in the dark for 30 min. The gel pieces were washed with 200 mM ammonium bicarbonate/50% acetonitrile (v/v) before addition of 250 ml of acetonitrile and incubation at room temperature for 15 min. The remaining solvent was removed, and the gel slices were completely dried using SpeedVac system (Thermo Fisher Scientific). The proteins in the gel slices were digested with 5 ng/ml sequencing grade modified porcine trypsin (Promega) in 200 mM ammonium bicarbonate/10% acetonitrile (v/v) at 55°C for 2 h. Trypsin was inactivated by adding 0.1% trifluoroacetic acid after protein digestion and the supernatant was transferred into 0.5 ml microcentrifuge tube for mass spectrometric analysis.
Capillary Liquid Chromatography-Mass Spectrometry Analysis for Protein Identification
Samples were analyzed by LC-MS/MS using a NanoAcquity chromatographic system (Waters Corp., Milford, MA) coupled to an LTQ-FT mass spectrometer (ThermoFinnigan, Bremen, Germany). Peptides were separated on a reverse-phase C18 column, 5 cm, 500 µm I.D. (CVC Microtech). A gradient was developed from 1% to 40% B (99.9% acetonitrile, 0.1% formic acid) in 50 min, ramped to 95% B in 4 min and held at 95% B for 5 min at a flow rate of 20 µl/min with solvents, A (99.9% H2O, 0.1% formic acid) and B. NanoAquity UPLC Console (Waters Corp., Version 1.3) was used to execute the injections and gradients. The ESI source was operated with spray voltage of 2.8 kV, a tube lens offset of 160 V and a capillary temperature of 200°C. All other source parameters were optimized for maximum sensitivity of the YGGFL peptide MH+ ion at m/z 556.27. The instrument was calibrated using an automatic routine based on a standard calibration solution containing caffeine, peptide MRFA, and Ultramark 1621 (Sigma). Data-dependent acquisition method for the mass spectrometer (configured version LTQ-FT 2.2) was set up using Xcalibur software (ThermoElectron Corp., Version 2.0). Full MS survey scans were acquired at a resolution of 50,000 with an Automatic Gain Control (AGC) target of 5×105. Five most abundant ions were fragmented in the linear ion trap by collision-induced dissociation with AGC target of 2×103 or maximum ion time of 300 ms. The ion selection threshold was 500 counts. The LTQ-FT scan sequence was adapted from the reference (Olsen and Mann, 2004).
Database Searches
MS/MS spectra were analyzed using Mascot (Matrix Science, London, UK; Version 2.3). Mascot was set up to search the SwissProt database and our pea aphid salivary gland transcriptome data of A. pisum assuming the trypsin digestion. Search was performed with a fragment ion mass tolerance of 0.20 Da and a parent ion tolerance of 20 PPM. Iodoacetamide derivative of cysteine was specified as a fixed modification. Oxidation of methionine was specified as a variable modification. Scaffold software (Version 3.6, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identification from the MS/MS data was performed using the MASCOT to correlate the data against NCBI non-redundant database and our salivary gland transcriptome data of A. pisum. To improve peptide identification accuracy, the results of protein identification were validated by multiple search engines (Mascot, Sequest and X! Tandem) using Scaffold software. Peptide identifications were accepted if they could be established at greater than 50.0% probability as specified by the Peptide Prophet algorithm (Keller et al., 2002). Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Protein Sequence and Domain/Motif Analysis
The amino acid sequence of the proteins of the gene family was analyzed with ClustalW alignment program for the gene family protein grouping (https://www.genome.jp/tools-bin/clustalw) using the slow parameters of a 10.0 gap open penalty and a 0.1 gap extension penalty with the BLOSUM (for protein) weight matrix. The amino acid alignment was produced by T-Coffee using default parameters (ebi.ac.uk/Tools/msa/tcoffee/) and illustrated using BoxShade (embnet.vital-it.ch/software/BOX_form.html). The MS-identified protein sequences were analyzed with the ScanProsite and SMART program at the ExPaSy (http://expasy.org/), and EMBL (http://smart.embl-heidelberg.de/) for the domain/motif analysis to predict protein functions. Signal peptide of the all MS-identified proteins was predicted by using SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) with a eukaryote D-cutoff value of 0.6. The pea aphid genome map was produced using karyoploteR (bioconducter.org/packages/release/bioc/html/karyoploteR.html) (Gel and Serra, 2017). Transcript similarity analysis was done using BLASTN comparing two or more sequences (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch). The KQY3 transcript without the secretory peptide and polyA regions was analyzed using BLASTN against single members of the KQY gene families also without their signal peptide and polyA nucleotides.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in Supplemental Table 1 , further inquiries can be directed to the corresponding author/s.
Author Contributions
MD and JO are co-first authors. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SH declared a past co-authorship with one of the authors AS to the handling editor.
Acknowledgments
The authors wish to thank Nadya Galeva at the Mass Spectrometry & Analytical Proteomics Laboratory, The University of Kansas for advice with mass spectrometry analysis. FW and JO wish to thank the Kansas State University Arthropod Genomics Center of Excellance for funds to conduct this project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01230/full#supplementary-material
Full amino acid sequence alignment of the KQY salivary gland secretion protein candidates. The alignment was produced with the ClustalW multiple alignment program.
Full amino acid sequence alignment of the KHI salivary gland secretion protein candidates. The alignment was produced with the ClustalW multiple alignment program.
Pairwise BLASTP analysis of KQY family. Number indicates probability of match by chance (expect value). Cells of the same color indicate member of possible gene family at probability below 1e-05. Only one isoform was used for each gene.
Alignment of KQY3 transcript with other members of the KQY family by BLAST. Colored boxes indicate alignment scores above 40. Red ticks indicate relative location of the stop codon for each gene.
References
- Boulain H., Legeai F., Guy E., Morlière S., Douglas N. E., Oh J., et al. (2018). Fast evolution and lineage-specific gene family expansions of aphid salivary effectors driven by interactions with host-plants. Genome Biol. Evol. 10, 1554–1572. 10.1093/gbe/evy097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger N. F. V., Botha A. M. (2017). Genome of Russian wheat aphid an economically important cereal aphid. Stand Genom. Sci. 28, 90. 10.1186/s40793-017-0307-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carolan J. C., Fitzroy C., II, Ashton P. D., Douglas A. E., Wilkinson T. L. (2009). The secreted salivary proteome of the pea aphid Acyrthosiphon pisum characterised by mass spectrometry. Proteomics 9, 2457–2467. 10.1002/pmic.200800692 [DOI] [PubMed] [Google Scholar]
- Carolan J. C., Caragea D., Reardon K. T., Mutti N. S., Dittmer N., Pappan K., et al. (2011). Predicted effector molecules in the salivary secretome of the pea aphid (Acyrthosiphon pisum): A dual transcriptomic/proteomic approach. J. Proteome Res. 10, 1505–1518. 10.1021/pr100881q [DOI] [PubMed] [Google Scholar]
- Chaudhary R., Atamian H., Shen Z., Briggs S., Kaloshian I. (2015). Potato aphid salivary proteome: Enhance salivation using resorcinol and identification of aphid phosphoproteins. J. Proteome Res. 14, 1792–1778. 10.1021/pr501128k [DOI] [PubMed] [Google Scholar]
- Chen M. S., Liu X., Yang Z., Zhao H., Shukle R. H., Stuart J. J., et al. (2010). Unusual conservation among genes encoding small secreted salivary gland proteins from a gall midge. BMC Evol. Biol. 28 10, 296. 10.1186/1471-2148-10-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Shakir S., Bigham M., Richter A., Fei Z., Jander G. (2019). Genome sequence of the corn leaf aphid (Rhopalosiphum maidis Fitch). Gigascience 8, 1–12. 10.1093/gigascience/giz033. pii: giz033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D. E., Mesarich C. H., Thomma B. P. (2015). Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 53, 541–563. 10.1146/annurev-phyto-080614-120114 [DOI] [PubMed] [Google Scholar]
- Cooper W. R., Dillwith J. W., Puterka G. J. (2010). Salivary proteins of Russian wheat aphid (Hemiptera: Aphididae). Environ. Entomol. 39, 223–231. 10.1603/EN09079 [DOI] [PubMed] [Google Scholar]
- Elzinga D. A., Jander G. (2013). The role of protein effectors in plant-aphid interactions. Curr. Opin. Plant Biol. 16, 451–456. 10.1016/j.pbi.2013.06.018 [DOI] [PubMed] [Google Scholar]
- Gel B., Serra E. (2017). KaryoploteR: An R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 33, 3088–3090. 10.1093/bioinformatics/btx346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Aphid Genomics Consortium (2010). Genome sequence of the pea aphid Acyrthosiphon pisum . PloS Biol. 23, e1000313. 10.1371/journal.pbio.1000313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaouannet M., Rodriguez P. A., Thorpe P., Lenoir C. J. G., MacLeod R., Escudero-Martinez C., et al. (2014). Plant immunity in plant-aphid interactions. Front. Plant Sci. 5, 663. 10.3389/fpls.2014.00663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A., II, Kolker E., Aebersold R. (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392. 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Csuros M., Rogozin I. B. (2013). Whence genes in pieces: reconstruction of the exon-intron gene structures of the last eukaryotic common ancestor and other ancestral eukaryotes. Wiley Interdiscip. Rev. RNA 4, 93–105. 10.1002/wrna.1143 [DOI] [PubMed] [Google Scholar]
- Legeai F., Shigenobu S., Gauthier J. P., Colbourne J., Rispe C., Collin O., et al. (2010). AphidBase: a centralized bioinformatic resource for annotation of the pea aphid genome. Insect Mol. Biol. 19 Suppl 2, 5–12. 10.1111/j.1365-2583.2009.00930.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Park H., Smith T. E., Moran N. A. (2019). Gene family evolution in the pea aphid based on chromosome-level genome assembly. Mol. Biol. Evol. 36, 2143–2156. 10.1093/molbev/msz138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles P. W. (1999). Aphid saliva. Biol. Rev. 74, 41–85. 10.1111/j.1469-185X.1999.tb00181.x [DOI] [Google Scholar]
- Mugford S. T., Barclay E., Drurey C., Findlay K. C., Hogenhout S. A. (2016). An immune-suppressive aphid saliva protein is delievered into the cytosol of the plant mesophyll cells during feeding. Mol. Plant Microbe 29, 854–861. 10.1094/MPMI-08-16-0168-R [DOI] [PubMed] [Google Scholar]
- Mutti N. S., Park Y., Reese J. C., Reeck G. R. (2006). RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum . J. Insect Sci. 6, 1–7. 10.1673/031.006.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutti N. S., Louis J., Pappan L. K., Pappan K., Begum K., Chen M.-S., et al. (2008). A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc. Natl. Acad. Sci. 105, 9965–9969. 10.1073/pnas.0708958105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A., II, Keller A., Kolker E., Aebersold R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658. 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- Olsen J. V., Mann M. (2004). Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc. Natl. Acad. Sci. 101, 13417–13422. 10.1073/PNAS.0405549101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitino M., Hogenhout S. A. (2013). Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol. Plant Microbe 26, 130–139. 10.1094/mpmi [DOI] [PubMed] [Google Scholar]
- Quan Q., Hu X., Pan B., Zeng B., Wu N., Fang G., et al. (2019). Draft genome of the cotton aphid Aphis gossypii . Insect Biochem. Mol. Biol. 105, 25–32. 10.1016/j.ibmb.2018.12.007 [DOI] [PubMed] [Google Scholar]
- Rao S. A. K., Carolan J. C., Wilkinson T. L. (2013). Proteomic profiling of cereal aphid saliva reveals both ubiquitous and adaptive secreted proteins. PloS One 8, 57413. 10.1371/journal.pone.0057413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S., Gibbs R. A., Gerardo N. M., Moran N., Nakabachi A., Stern D., et al. (2010). Genome sequence of the pea aphid Acyrthosiphon pisum . PloS Biol. 8, e1000313. 10.1371/journal.pbio.1000313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez P. A., Bos J. I. (2013). Toward understanding the role of aphid effectors in plant infestation. Mol. Plant Microbe Interact. 26, 25–30. 10.1094/MPMI-05-12-0119-FI [DOI] [PubMed] [Google Scholar]
- Shigenobu S., Richards S., Cree A. G., Morioka M., Fukatsu T., Kudo T., et al. (2010). A full-length cDNA resource for the pea aphid, Acyrthosiphon pisum . Insect Mol. Biol. 19, 23–31. 10.1111/j.1365-2583.2009.00946.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithers B., Oates M., Gough J. (2019). ‘Why genes in pieces?’-revisited. Nucleic Acids Res. 47, 4970–4973. 10.1093/nar/gkz284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe P., Cock P., Bos J. (2016). Comparative transcriptomics and proteomics of three different aphid species identifies core and diverse effector sets. BMC Genomics 12, 172. 10.1186/s12864-016-2496-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjallingii W. F. (2006). Salivary secretions by aphids interacting with proteins of phloem wound responses. J. Exp. Bot. 57, 739–745. 10.1093/jxb/erj088 [DOI] [PubMed] [Google Scholar]
- Tonkin C. J., Foth B. J., Ralph S. A., Struck N., Cowman A. F., McFadden G., II (2008). Evolution of malaria parasite plastid targeting sequences. Proc. Natl. Acad. Sci. U. S. A. 105, 4781–4785. 10.1073/pnas.0707827105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme B., Kast P., Haegeman A., Jacob J., Grunewald W., Gheysen G. (2009). Structural and functional investigation of a secreted chorismate mutase from the plant-parasitic nematode Heterodera schachtii in the context of related enzymes from diverse origins. Mol. Plant Pathol. 10, 189–200. 10.1111/j.1364-3703.2008.00521.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Gao B., Zhu S. (2016). Exon shuffling and origin of scorpion venom biodiversity. Toxins (Basel) 9, E10. 10.3390/toxins9010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger J. A., Cassone B. J., Legeai F., Johnston J. S., Bansal R., Yates A. D., et al. (2017). Whole genome sequence of the soybean aphid, Aphis glycines . Insect Biochem. Mol. Biol. 102917, 1–10. 10.1016/j.ibmb.2017.01.005. pii: S0965-1748(17)30005-X. [DOI] [PubMed] [Google Scholar]
- Will T., Tjallingii W. F., Thö A., Van Bel A. J. E. (2007). Molecular sabotage of plant defense by aphid saliva. Proc. Natl. Acad. Sci. 104, 10536–10541. 10.1073/pnas.0703535104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full amino acid sequence alignment of the KQY salivary gland secretion protein candidates. The alignment was produced with the ClustalW multiple alignment program.
Full amino acid sequence alignment of the KHI salivary gland secretion protein candidates. The alignment was produced with the ClustalW multiple alignment program.
Pairwise BLASTP analysis of KQY family. Number indicates probability of match by chance (expect value). Cells of the same color indicate member of possible gene family at probability below 1e-05. Only one isoform was used for each gene.
Alignment of KQY3 transcript with other members of the KQY family by BLAST. Colored boxes indicate alignment scores above 40. Red ticks indicate relative location of the stop codon for each gene.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in Supplemental Table 1 , further inquiries can be directed to the corresponding author/s.