Summary
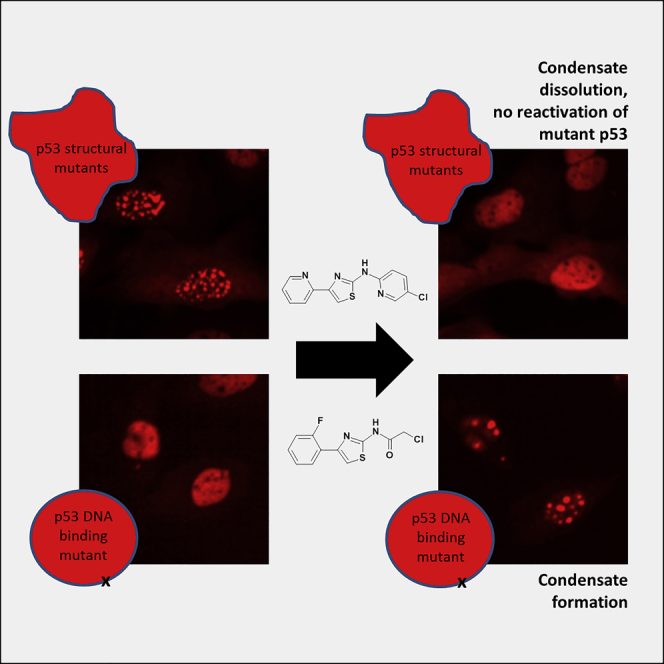
Structural mutants of p53 induce global p53 protein destabilization and misfolding, followed by p53 protein aggregation. First evidence indicates that p53 can be part of protein condensates and that p53 aggregation potentially transitions through a condensate-like state. We show condensate-like states of fluorescently labeled structural mutant p53 in the nucleus of living cancer cells. We furthermore identified small molecule compounds that interact with the p53 protein and lead to dissolution of p53 structural mutant condensates. The same compounds lead to condensation of a fluorescently tagged p53 DNA-binding mutant, indicating that the identified compounds differentially alter p53 condensation behavior depending on the type of p53 mutation.
In contrast to p53 aggregation inhibitors, these compounds are active on p53 condensates and do not lead to mutant p53 reactivation. Taken together our study provides evidence for structural mutant p53 condensation in living cells and tools to modulate this process.
Subject Areas: Biochemistry Methods, Medical Biochemistry, Structural Biology
Graphical Abstract
Highlights
-
•
Fluorescent versions of structural p53 mutants form protein condensates in cells
-
•
We report the identification of p53-interacting small molecules
-
•
Identified compounds differentially alter fluorescent mutant p53 condensation behavior
Biochemistry Methods; Medical Biochemistry; Structural Biology
Introduction
The transcription factor p53 integrates diverse cellular stress events and regulates the expression of stress response genes and can induce apoptotic and anti-proliferative processes. As a potent tumor suppressor, p53 is frequently downregulated or mutated in tumors, a state associated with adverse prognosis in cancer (Donehower et al., 2019; Kastenhuber and Lowe, 2017).
Cancer cells can suppress p53 activity via different mechanisms. Cellular p53 levels are kept low by ubiquitin-mediated protein degradation. Accordingly, MDM2 as one of the relevant E3 ligases is characterized as an oncogene and is frequently upregulated in a variety of human cancers (Wang et al., 2017). Compounds that disrupt MDM2-p53 interaction have been identified and profiled for potential anti-cancer therapies in tumors with wild-type p53 (Wang et al., 2017). However, depending on the cancer type a large amount of tumors contain truncating or inactivating p53 mutations that most likely would not positively respond to MDM2-directed therapies. Indeed, most p53 mutations occur as missense mutations in the DNA-binding domain of p53 and result in inactivated transcription factor function, MDM2 insensitivity, and can confer oncogenic gain of function (Donehower et al., 2019; Muller and Vousden 2013, 2014; Soussi and Wiman, 2015). A majority of p53 missense mutants fall into several hotspot codons. These can be subdivided based on their functional consequences on p53 into DNA contact defective mutants (e.g., mutants at residue 273) that directly interfere with p53-DNA interactions and structural mutants that lead to p53 misfolding and aggregation (Donehower et al., 2019; Muller and Vousden 2013, 2014; Rangel et al., 2014; Silva et al., 2014, 2018; Soussi and Wiman, 2015; Xu et al., 2011).
Structural p53 mutants such as R175H, R282W, and Y220C lead to global protein destabilization and protein misfolding. p53 misfolding in turn is thought to lead to the externalization of an aggregation-prone segment followed by prion-like aggregation and inactivation of mutant p53 (Costa et al., 2016; Rangel et al., 2014; Silva et al., 2014, 2018; Soragni et al., 2016; Wang and Fersht, 2017).
p53 structural mutants form aggregates that co-aggregate with and inactivate interaction partners such as p53 family members p63 and p73 (Costa et al., 2016; Gaiddon et al., 2001; Liu et al., 2011; Muller and Vousden, 2013; Wiech et al., 2012; Xu et al., 2011). This can explain potential oncogenic gain-of-function effects of p53 structural mutants resulting in the development of more invasive tumor phenotypes and poorer prognosis. Furthermore, structural mutant p53 aggregates show prion-like behavior (Silva et al., 2018). As a significant part of human tumors harbor structural mutant forms of the p53 protein, approaches aimed at preventing mutant p53 gain of function and disrupting the pathological interactions among p53 family members as well as preventing amyloidogenic prion-like properties might be of clinical value (Lang et al., 2004; Li and Prives, 2007; Li et al., 2019; Mantovani et al., 2019; Muller and Vousden 2013, 2014; Olive et al., 2004; Silva et al., 2018; Wang and Fersht, 2017; Xu et al., 2011).
Recent work suggests that p53 potentially transitions through a condensate-like state before aggregation (Boija et al., 2018; Kamagata et al., 2020; Safari et al., 2019; Silva et al., 2018). Here, we show that fluorescently labeled structural p53 mutants, but not DNA-binding defective mutants, form liquid-like condensates in the nucleus of living cancer cells.
Furthermore, we report the identification of small molecules that show direct interaction with p53. In contrast to other structural mutant p53 reactivators (Bykov et al., 2018; Li et al., 2019; Silva et al., 2018), the compounds reported here differentially modulate condensation properties of p53 mutants without leading to mutant p53 reactivation. Therefore the identified small molecules constitute a separate class of p53-directed compounds. These can be used as tool compounds to dissect structural p53 loss-of-function and gain-of-function effects and have the potential to be developed into anti-cancer drugs targeting mutant p53 condensation.
Results
Condensate Formation of Mutant p53 Variants
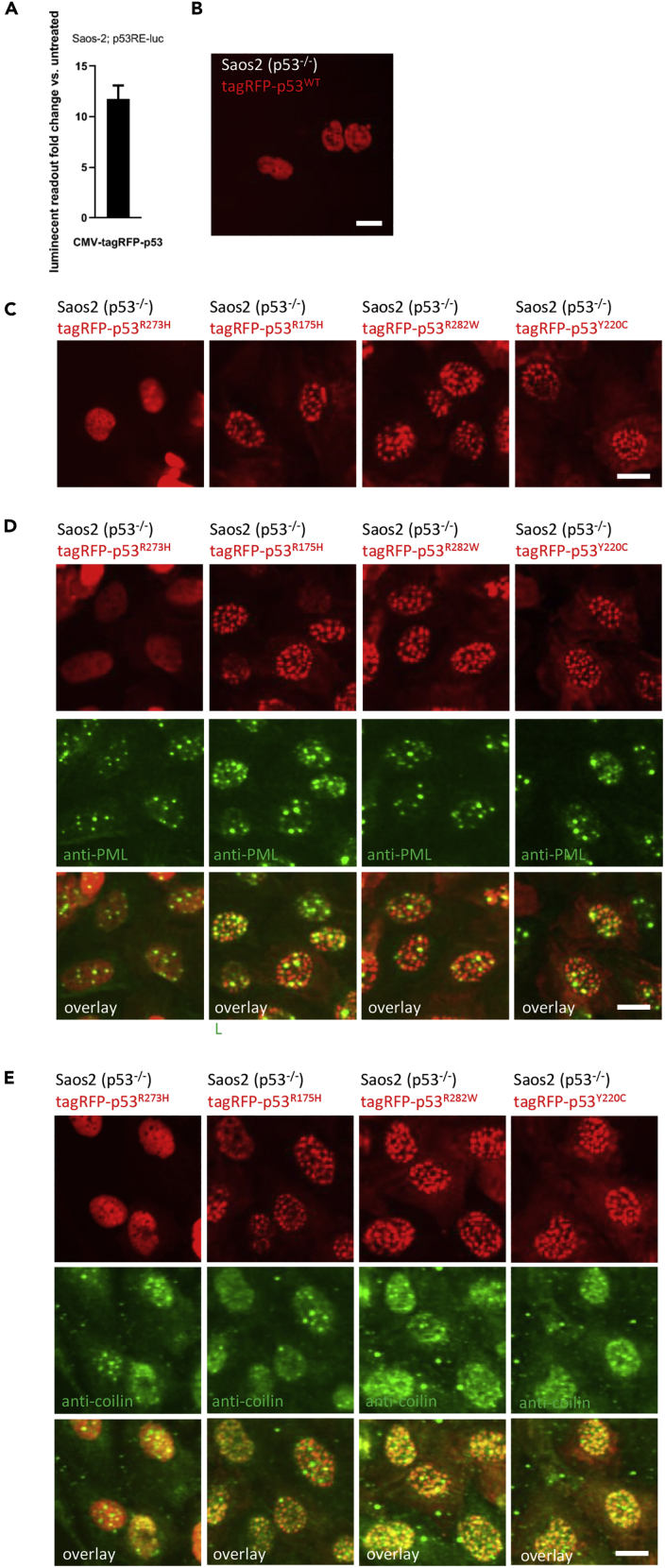
To directly monitor condensation or aggregation of p53 in live cells, we established a series of fluorescently labeled (monomeric tagRFP (Merzlyak et al., 2007)) p53 mutant variants expressed in the otherwise p53−/− Saos2 cell line. Transient expression of fluorescently tagged p53WT activated a p53 activity reporter and confirmed general functionality of fluorescently tagged p53 (Figure 1A). As expected, tagRFP-p53WT is homogenously distributed in the nucleus of Saos2 cells (Figure 1B). According to its role as a tumor suppressor with anti-proliferative activity it was not possible to establish cell lines stably expressing fluorescently labeled p53WT. However, similar to tag-RFP-p53WT, cells stably expressing a fluorescent version of the DNA contact mutant p53R273H showed mainly homogeneous distribution of the fluorescent signal throughout the nucleus (Figure 1C, Video 1D). In stark contrast, stable cell lines expressing fluorescent versions of the structural mutants p53R175H (Figures 1C, Video 1A), p53R282W (Figure 1C, Video 1B), and p53Y220C (Figure 1C, Video 1C) showed dot-like accumulation at several foci throughout the nucleus. Similar condensate-like structures could also be observed in H1299 (p53−/−) and AU565 (p53R175H) cells expressing tagRFP-p53R175H (Figure S1). Protein condensates are characterized by a spherical shape and their ability to fuse (Alberti et al., 2019; Banani et al., 2017; Boija et al., 2018; Cai et al., 2019; Safari et al., 2019; Shin and Brangwynne, 2017). Accordingly, structural mutant tagRFP-p53 foci are spherical and show liquid-like properties and sporadic fusion in live-cell imaging (arrows in Videos 1A, 1B, and 1C). Therefore, based on their condensate-like properties we further refer to the foci as p53 condensates.
Figure 1.
Condensate Formation of Fluorescent Structural Mutant p53 Variants
(A) Saos2 containing a p53 activity reporter showed reporter activation after transduction of a tagRFP-p53 expression vector. Luminescent readout normalized to DMSO controls. Bars show mean with SD (n = 3).
(B) Saos2 cells transiently transfected with a tagRFP-p53WT expression vector show homogeneous distribution of tagRFP-p53 in the nucleus. Scale bar, 10 μm.
(C) Saos2 cells expressing fluorescently tagged structural mutants show p53 condensates in the nuclei. Scale bars, 10 μm.
(D and E) Antibody staining of Saos2 cells expressing fluorescently tagged structural mutants with anti-PML or coilin antibodies. No co-localization of fluorescently tagged mutant p53 condensates with either PML (D) or Cajal bodies (E) in the nucleus is observable. Scale bars, 10 μm.
Saos2 cells expressing fluorescently tagged DNA-binding defective mutant shows homogeneous nuclear fluorescence. Time-lapse video, 3 min per frame. Scale bar, 10 μm.
Saos2 cells expressing fluorescently tagged structural mutants show condensate-like behavior (condensates undergoing fusion events marked by arrows). Time-lapse video, 3 min per frame. Scale bar, 10 μm.
Saos2 cells expressing fluorescently tagged structural mutants show condensate-like behavior (condensates undergoing fusion events marked by arrows). Time-lapse video, 3 min per frame. Scale bar, 10 μm.
Saos2 cells expressing fluorescently tagged structural mutants show condensate-like behavior (condensates undergoing fusion events marked by arrows). Time-lapse video, 3 min per frame. Scale bar, 10μm.
To further characterize structural mutant p53 condensates and evaluate if p53 condensates are recruited as clients to preexisting biomolecular nuclear condensates such as promyelocytic leukemia (PML) or Cajal bodies (Ditlev et al., 2018) or exist as independent condensates we stained Saos2 cells expressing fluorescent mutant p53 with anti-PML or anti-coilin antibodies. We find no colocalization of mutant p53 condensates with either PML (Figure 1D) or Cajal bodies (Figure 1E), indicating that nuclear p53 condensates form independent of PML or Cajal bodies.
Identification of Compounds that Interact with p53 Proteins and Their Effects on Mutant p53 Condensates
A previous high throughput screen (HTS) in the laboratory on an internal compound collection exceeding 4 million compounds (Follmann et al., 2019) identified compounds that potentially interact with mutant p53 (data not shown).
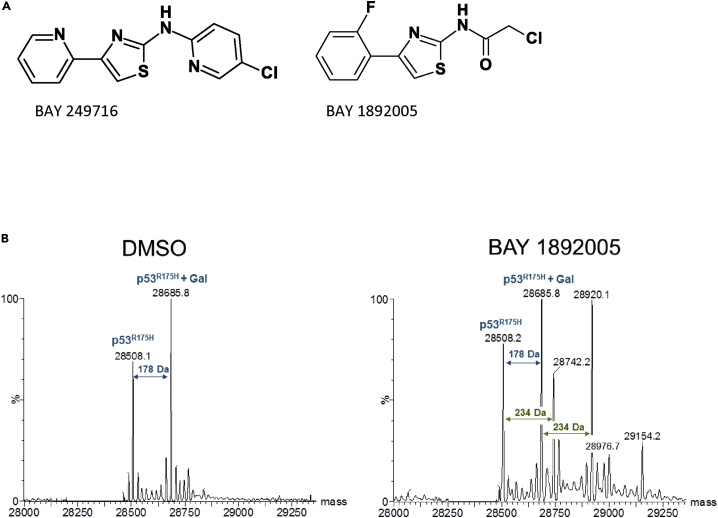
To identify compounds from this hit list that directly interact with the p53 protein, selected substances were tested in nano-differential scanning fluorimetry on recombinantly expressed and purified p53 proteins (comprising the p53 DNA-binding domain, see Material and Methods) of structural mutant p53R175H and p53Y220C as well as p53WT. Generally, p53 containing the structural mutations R175H or Y220C showed significantly lower melting temperatures compared with the WT protein (Tm p53WT = 41.6°C ± 0.01°C, Tm p53R175H = 35.0°C ± 0.086°C, p53Y220C = 34.7°C ± 0.02°C), confirming the destabilizing nature of the mutations (Ang et al., 2006). Compounds from a structural cluster of aminothiazoles showed significant stabilization of p53 (Table 1). BAY 249716 showed significant stabilization of all three p53 protein variants, whereas BAY 1892005 showed stabilization of p53WT and p53Y220C, indicating direct interaction with p53. BAY 1892005 contains a reactive head group (Figure 2A) that could be involved in covalent binding to p53. Indeed, by mass spectrometry, we confirmed that BAY 1892005 binds covalently to mutant p53R175H and p53Y220C (Figures 2B and S2). Taken together we conclude that the identified aminothiazoles potentially directly interact with structural p53 mutant proteins.
Table 1.
nDSF: Stabilization upon Compound Binding
| Compound | nDSF p53R175H D°C | nDSF p53Y220C D°C | nDSF p53WT D°C |
|---|---|---|---|
| BAY 249716 | 1.0 | 0.6 | 1.2 |
| BAY 1892005 | −0.4 | 1.1 | 0.8 |
nDSF, nano-differential scanning fluorimetry
Exemplary data of multiple experiments are shown (n ≥ 2). Ligands, that show a +ΔTm greater than six standard deviations from the mean of the control were considered as hit (stabilizer, bold).
Figure 2.
Identification of Compounds that Interact with p53 Proteins
(A and B) (A) Hit compounds. (B) Deconvoluted spectra of intact mass analysis of p53 shows covalent binding of BAY 1892005 to p53R175H. DMSO control shows two peaks of p53R175H protein, one for the expected mass of 28,508 Da and one of 28,685 Da representing N-terminal gluconoylated (Gal) p53R175 (blue arrows). Incubation with BAY 1892005 showed mass shifts of 234 Da to both the apoprotein and the glyconoylated p53R175H (green arrows), indicative for covalent binding of BAY 1892005. An additional mass shift of 234 Da indicates a partial two-fold binding of BAY 1892005 to p53R175H. Exemplary data of multiple experiments are shown (n ≥ 2).
Compounds that directly bind to and stabilize mutant p53 have the potential to modulate p53 misfolding, condensation, and aggregation (Bykov et al., 2018; Li et al., 2019; Safari et al., 2019; Silva et al., 2018).
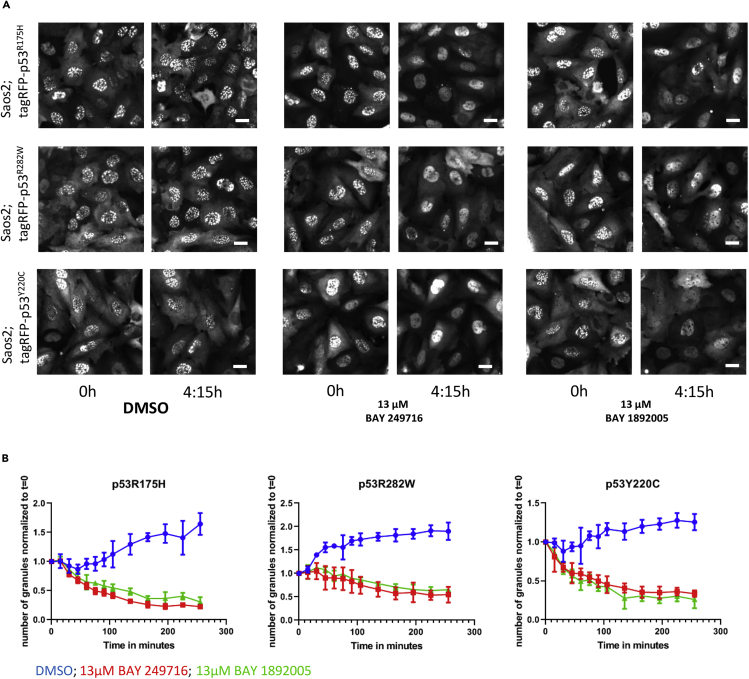
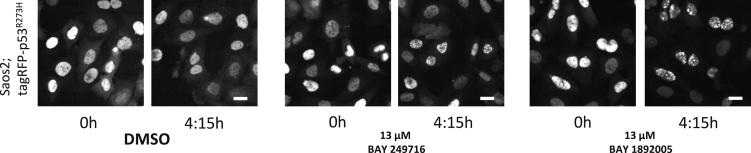
To elucidate if aminothiazoles modulate structural mutant p53 condensates, we treated the cells with BAY 249716 or BAY 1892005 and followed mutant p53 by time-lapse imaging (Videos 2A, 2B, 2C, and 2D). We find that the aminothiazoles lead to a rapid dissolution of structural mutant p53 condensates in most cells of the population (Figures 3A and 3B, Videos 2A, 2B, and 2C). In contrast the same compounds lead to the formation of condensates of the DNA-binding defective mutant p53R273H in some cells of the population (Figure 4, Video 2D). Taken together we conclude that aminothiazoles differentially alter fluorescent mutant p53 condensation behavior depending on the type of p53 mutation (structural vs. DNA-binding mutants). To evaluate if the identified compounds selectively target p53 condensates, treated cells were fixed and stained for PML nuclear bodies. These were not affected by compound treatment, indicating that the identified compounds specifically interfere with mutant p53 condensation (data not shown).
Figure 3.
Condensate Formation of Fluorescent Structural Mutant p53 Variants and Their Dissolution by Aminothiazoles
(A) Saos2 cells expressing fluorescently tagged structural mutants show p53 condensates in the nuclei, which are dissolved after aminothiazole treatment. Stills (0 and 4:15 h) from time-lapse Videos 2A, 2B, and 2C. Scale bars, 10 μm.
(B) Kinetics of condensate dissolution in Saos2 cells expressing structural p53 mutants in response to treatment with aminothiazoles. Bars show mean with SD (n = 4). Exemplary data of multiple experiments are shown (n ≥ 2).
Figure 4.
Aminothiazoles Induce Condensation of Fluorescent DNA-Binding Defective p53R273H Mutant Variant
Saos2 cells expressing a fluorescently tagged DNA-binding domain mutant show homogeneous distribution of the fluorescent signal in the nuclei, and aminothiazole treatment leads to the formation of condensate-like structures in some cells of the population. Stills (0 and 4:15 h) from time-lapse Video 2D. Scale bars, 10 μm. Exemplary data of multiple experiments are shown (n ≥ 2).
Fluorescent structural mutant condensates are dissolved after aminothiazole treatment. Time-lapse video of Saos2 cells expressing fluorescently tagged mutant p53, 15 min per frame for the first 4 frames, afterward 30 min per frame. Scale bar, 10 μm.
Fluorescent structural mutant condensates are dissolved after aminothiazole treatment. Time-lapse video of Saos2 cells expressing fluorescently tagged mutant p53, 15 min per frame for the first 4 frames, afterward 30 min per frame. Scale bar, 10 μm.
Fluorescent structural mutant condensates are dissolved after aminothiazole treatment. Time-lapse video of Saos2 cells expressing fluorescently tagged mutant p53, 15 min per frame for the first 4 frames, afterward 30 min per frame. Scale bar, 10 μm.
Aminothiazole treatment leads to condensation of p53R273H after aminothiazole treatment. Time-lapse video of Saos2 cells expressing fluorescently tagged mutant p53, 15 min per frame for the first 4 frames, afterward 30 min per frame. Scale bar, 10 μm.
Aminothiazoles Lead to Reduction of Nuclear Accumulation of Endogenous Structural p53 Mutants
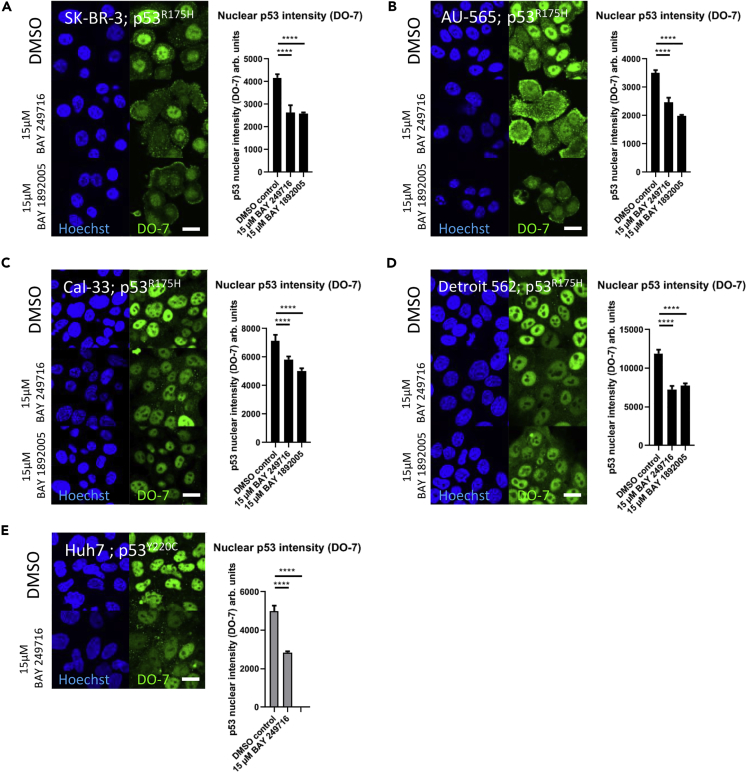
Depending on mutational status, endogenous p53 can show nuclear accumulation in specific nuclear inclusion bodies (De Smet et al., 2017). To evaluate the compounds in cells with endogenous structural p53 mutants, we stained four different p53R175H mutant cell lines and one p53Y220C mutant cell line by immunofluorescence (IF) using different antibodies directed against p53 (Figures 5 and S3). Structural p53R175H or p53Y220C mutants lead to p53 inactivation, misfolding, aggregation, and accumulation. Accordingly all cell lines showed positive staining for p53 in the nucleus (Figures 5A–5E and S3A–S3E). Furthermore we find a statistically significant reduction in nuclear p53 aggregation in all four cell lines carrying p53R175H after 6 h of aminothiazole treatment (Figures 5A–5D and S3A–S3D). In line with aminothiazoles also binding to p53Y220C (Table 1) and dissolving fluorescent tagRFP-p53Y220C condensates (Figures 3A and B) BAY 249716 also showed a clear reduction of p53 staining intensity in the nucleus of the p53Y220C cell line Huh7 (Figures 5E and S3E). Total p53 protein levels were also evaluated by western blot, in which a slight reduction of total p53 levels was observed (Figure S4). Even though p53 aggregates are present in biopsies and also cell culture cell lines, nuclear p53 aggregates are hard to visualize by IF (De Smet et al., 2017; Pedrote et al., 2020). In line with this, cell lines harboring endogenous structural p53 mutants showed mostly homogeneous staining in the nucleus and no clear visualization of p53 condensates. In addition, we did not find significant changes in antibody stainings after compound treatment with antibodies directed against oligomers or fibrils (data not shown), which indicates that either mutant p53 condensate states do not directly translate into p53 aggregation or as the antibodies are not specific for p53 oligomers or aggregates, they may not be able to resolve subtle changes in oligomeric/amyloid states of p53.
Figure 5.
Aminothiazoles Lead to Reduction of Nuclear Accumulation of Endogenous Structural p53 Mutants
(A–E) p53R175H cell lines (A) SK-BR-3, (B) AU-565, (C) Cal-33, and (D) Detroit 562 and (E) p53Y220C cell line Huh7 were treated with aminothiazoles for 6 h and stained by IF for p53 (DO-7), and the intensity of the staining in the nucleus was quantified. ∗∗∗∗p value < 0.0001. Scale bar, 10 μm. Bars show mean with SD (n ≥ 5). Exemplary data of multiple experiments are shown (n ≥ 2).
Identified Hits Do Not Lead to Structural Mutant p53 Reactivation
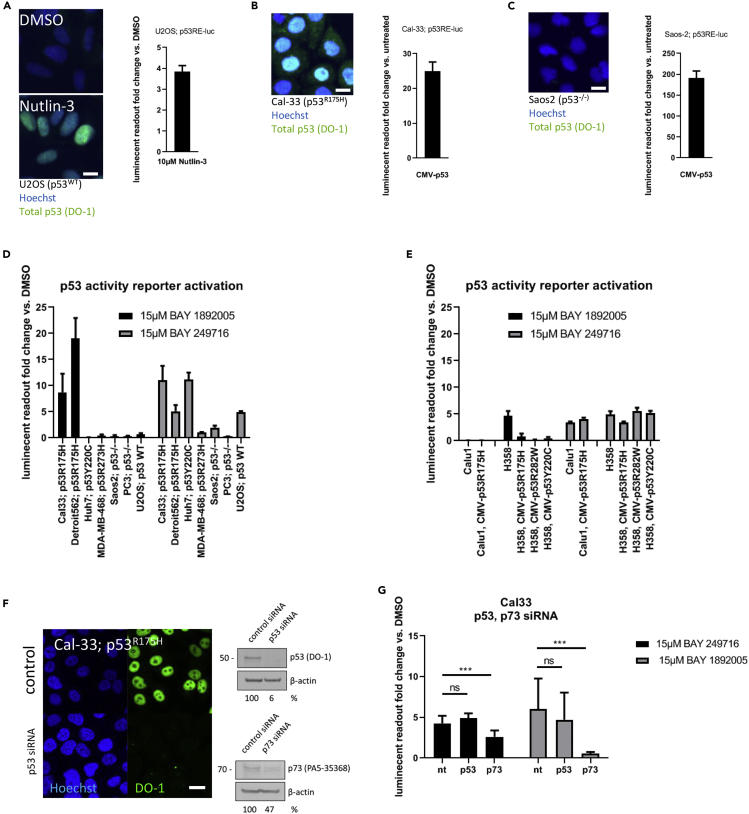
To test for the functional consequence of compound treatment on p53 activity, we generated cell lines containing a p53 response element-driven luciferase reporter (p53 activity reporter, see Material and Methods). Functionality of the reporter was validated in U2OS cells (p53WT) treated with the MDM2 inhibitor Nutlin as well as in Cal-33 (p53R175H) or Saos2 (p53−/−) cells by transduction of a p53 expression vector (Figures 6A–6C).
Figure 6.
Identified Hits Do Not Lead to Structural Mutant p53 Reactivation
(A) U2OS (p53WT) cells containing the p53 activity reporter show nuclear accumulation of p53 upon 10 μM Nutlin-3 treatment and induction of the activity reporter.
(B) Cal-33 (p53R175H) cells containing the p53 activity reporter show nuclear accumulation of p53R175H and no p53 activity reporter induction after Nutlin-3 treatment (data not shown) but induction after transduction of a CMV-driven p53 expression vector.
(C) Saos2 (p53−/−) cells containing the p53 activity reporter showed no staining for p53 and no induction of the p53 activity reporter after Nutlin-3 treatment (data not shown) but induction of the p53 activity reporter after transduction of a CMV-driven p53 expression vector. Luminescent readout normalized to DMSO controls. Exemplary data of multiple experiments are shown (n ≥ 2) Scale bars, 10 μm.
(D) p53 activity reporter activation upon aminothiazole treatment in cells with different p53 mutation status. Luminescent readout normalized to DMSO controls. Exemplary data of multiple experiments are shown (n ≥ 2), Bars show mean with SD (n = 8).
(E) Expression of structural mutant p53 in p53−/− cells does not enhance p53 activity reporter activation. Luminescent readout normalized to DMSO controls. Exemplary data of multiple experiments are shown (n ≥ 2), Bars show mean with SD (n = 8).
(F) p53 knockdown validated by IF and western blot; partial p73 knockdown shown in western blot. Exemplary data of multiple experiments are shown (n ≥ 2); full western blot data show in Figure S6. Scale bar, 10 μm.
(G) p53 knockdown does not prevent p53 activity reporter activation in Cal-33 cells, whereas p73 knockdown shows suppression of p53 activity reporter activation. Luminescent readout normalized to DMSO controls. Exemplary data of multiple experiments are shown (n ≥ 2), Bars show mean with SD (n = 10). ∗∗∗p value < 0.001; ns, not significant.
Next, the compounds were tested for activation of the p53 activity reporter on a set of cell lines with different p53 status. Both aminothiazoles led to weak activation of the p53 activity reporter in Cal-33 and Detroit 562 cells (structural mutant p53R175H), whereas BAY 249716 also showed activity in Huh7 cells (structural mutant p53Y220C) and both showed less activity in MDA-MB-468 cells (DNA contact mutant p53R273H) and in Saos2 or PC3 cells (p53−/−). BAY 249716 also showed p53 activity reporter activation in U2OS cells (p53WT) (Figure 6D), and both compounds showed slight activation of the p53 activity reporter in Calu1 and H358 cells (p53−/−) (Figure 6E). To further elucidate if aminothiazoles lead to the reactivation of structural p53 mutants, we tested if the compounds induce the p53 activity reporter in p53−/− cells stably expressing mutant p53. Treatment with aminothiazoles did not lead to stronger activation of the p53 activity reporter compared with control cells in p53−/− Calu1 cells expressing p53R175H or in p53−/− H358 cells expressing the structural p53 mutants p53R175H, p53R282W, or p53Y220C (Figure 6E) or in Saos2 cells expressing fluorescent versions of mutant p53 (data not shown). This indicates that aminothiazoles do not lead to reactivation of structural mutant p53 protein. Indeed, p53 knockdown by small interfering RNA in Cal-33 (p53R175H) (Figure 6F) led to complete loss of nuclear p53 staining but did not prevent activation of the p53 activity reporter (Figure 6G). Taken together we conclude that aminothiazoles do not lead to direct reactivation of structural p53 mutants.
Structural p53 mutants are thought to show gain of function by co-condensation or co-aggregation of p53WT and other p53 family members such as p73 (de Oliveira et al., 2015; Kehrloesser et al., 2016; Li and Prives, 2007; Stindt et al., 2015). Therefore, we speculated that the weak p53 activity reporter activation seen in cells with structural p53 mutants could be triggered by activation of p73. Indeed, p53 family members share similar response element structures (Luh et al., 2013). To test the influence of p73 on p53 reporter activation, we partially knocked down p73 in Cal-33 cells containing the p53 activity reporter (Figure 6F) and found that reporter activation could be partly suppressed (Figure 6G). Therefore, we conclude that aminothiazoles act on the p53 activity reporter possibly via p73 activation.
Finally, we tested the aminothiazoles for long-term anti-proliferative activity in a set of cell lines with different p53 mutation status. In summary, we demonstrate that both compounds show anti-proliferative activity independent of p53 status (Table 2), indicating p53-independent effects of the aminothiazoles on long-term viability.
Table 2.
Anti-proliferative IC50 of Compounds Tested on Cell Lines with Different p53 Mutation Status
| Anti-proli IC50 [M] | p53R175H |
P53Y220C |
p53−/− |
p53WT |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cal-33 | Detroit 562 | SK-BR-3 | Au −565 | Huh7 | Saos2 | H1299 | H358 | Calu1 | HMC-1-8 | U2OS | MCF7 | |
| BAY 249716 | 1.88 × 10−6 | 1.86 × 10−6 | 2.20 × 10−6 | 6.78 × 10−6 | 7.27 × 10−6 | 4.07 × 10−6 | 2.54 × 10−6 | 7.12 × 10−6 | 2.82 × 10−6 | 7.57 × 10−7 | 4.29 × 10−6 | 8.15 × 10−7 |
| BAY 1892005 | 4.21 × 10−6 | 9.70 × 10−6 | 5.19 × 10−6 | 9.78 × 10−6 | 1.12 × 10−5 | 4.60 × 10−6 | 9.76 × 10−6 | 1.11 × 10−5 | 1.32 × 10−6 | 2.21 × 10−6 | 3.97 × 10−6 | 6.81 × 10−6 |
Taken together we show the visualization of structural mutant p53 condensates in live cells and identify compounds that directly interact with p53 and modulate p53 condensation and nuclear accumulation. As these compounds modulate p53 condensation and do not lead to mutant p53 reactivation, the identified small molecules constitute a separate class of p53-directed compounds.
Discussion
p53 is a transcription factor with a crucial role as a tumor suppressor. p53 controls a wide range of processes including induction of apoptosis and senescence, DNA repair, metabolic and antioxidant responses, and cell-cycle arrest. p53 is inactivated in more than 50% of all cancers. Structural mutants of p53 lead to global p53 protein destabilization (Ang et al., 2006) and aggregation. Co-aggregation of other p53 family members or other interaction partners could explain specific gain-of-function effects of structural p53 mutants. However, it is unclear how the exact process of p53 aggregation occurs. Recent data indicate that p53 can be organized as part of protein condensates and that p53 aggregation could potentially occur through a transient condensate-like state (Boija et al., 2018; Kamagata et al., 2020; Kilic et al., 2019; Safari et al., 2019). Using artificial expression of p53 mutants N-terminally tagged with monomeric tagRFP, we show that p53 variants that carry structural mutants p53R175H, p53R282W, or p53Y220C but not the DNA contact mutant p53R275H or p53WT are localized to punctate structures in the nucleus. Given their spherical shape, dynamic behavior during the cell cycle, and fusion events of single spots, these structures potentially represent structural mutant p53 protein condensates (Alberti et al., 2019). Interestingly, no clearly visible condensate-like structures can be detected in cells with endogenous structural p53 mutants by IF. As condensate-like structures are observed only in cells expressing structural p53 mutants and not in the DNA-binding defective mutant or p53WT, this observation is not an artifact of fluorescent tagging of p53 with monomeric tagRFP but rather a feature of structural p53 mutants. Furthermore, condensate-like structure formation of fluorescently tagged structural p53 mutants is not a cell line-specific effect, as condensate-like structures could also be observed after expression of the fluorescent constructs in H1299 (p53−/−) or AU565 (p53R175H) cell lines. p53 has been shown to form anomalous condensate-like states and to co-condensate with other factors (Boija et al., 2018; Kilic et al., 2019; Safari et al., 2019). Indeed p53 contains several intrinsically disordered regions, which are thought to be driving elements for protein condensation (Anbo et al., 2019; Xue et al., 2013). Therefore, we speculate that artificial expression of fluorescent p53 constructs highlights otherwise sub-microscopic condensates that normally are transient structures in the process of mutant p53 aggregation (de Oliveira et al., 2019; De Smet et al., 2017; Pedrote et al., 2020; Safari et al., 2019). Furthermore, the N-terminal region of p53 is important for p53 aggregation (Kamagata et al., 2020; Melo Dos Santos et al., 2019). Therefore modulation of this region by a fluorescent tag could slow down transitioning through a condensate-like state before aggregation and thereby additionally contributing to highlighting otherwise transient p53 condensate states.
Although several compounds and biomolecules have been identified that reactivate mutant p53 and interfere with p53 aggregation, no compounds have been reported that modulate p53 condensation (Bykov et al., 2018; Li et al., 2019). Mutant p53 condensation, aggregation, and its associated gain-of-function phenotypes as well as their prion-like properties are increasingly viewed as new opportunities for therapeutic intervention. We here identify two compounds that differentially modulate fluorescent mutant p53 condensation behavior. Even though one of the compounds contains a reactive head group and covalently binds to p53, covalent binding is not required for activity, as BAY 249716 does not contain a reactive group for covalent protein modification. Interestingly, the compounds interfere with fluorescent structural mutant p53 condensation but in cells expressing fluorescent DNA contact mutant p53, the same compounds induce p53 condensation in the nucleus. This indicates that the compounds differently interfere with the dynamic behavior of mutant p53 condensation in structural p53 mutants versus DNA-binding defective p53 mutants.
p53 knockdown leads to near-complete reduction of p53 nuclear staining in cells with structural p53 mutants (Figure 2C). Therefore, nuclear accumulation of structural mutant p53 requires constant replenishment of newly synthetized protein and interference with the condensation or aggregation process of structural mutant p53 should lead to decrease in p53 nuclear aggregation. Accordingly, we find that the identified compounds lead to a reduction of p53 accumulation in different cancer cells with endogenous p53 structural mutants. However, we noted that aminothiazoles seem to not affect all cells in a population homogeneously (Figure S5, arrows). A possible explanation for this could be that aminothiazoles target a specific condensate state that is modulated by co-condensates and co-factors. Indeed, several factors have been reported to influence p53 aggregation (Stindt et al., 2015; Wiech et al., 2012) and p53 condensates can co-condensate with other factors (Boija et al., 2018; Kilic et al., 2019). In future studies, it will be interesting to decipher how cellular parameters such as variations in expression level of p53 itself and modulating co-factors as well as the influence of neighboring cells or the cell cycle phase affect the effectivity of aminothiazoles on p53 condensation of cells in a population.
Structural p53 mutants accumulate as amyloid-like protein aggregates (Levy et al., 2011) and have the capability to co-aggregate other p53 family members and other transcription factors (Boija et al., 2018; Kilic et al., 2019; Safari et al., 2019; Wiech et al., 2012; Xu et al., 2011). Indeed, we observed that activation of the p53 activity reporter in cells with structural p53 mutants at least partially depends on p73. Therefore, aminothiazoles could act in part by releasing co-condensed or co-aggregated factors from p53 condensates or aggregates. Furthermore, based on the observation that p53 aggregates can be taken up by surrounding cells and induce further aggregation, p53 is considered to show prion-like properties. It will be interesting to see if by modulating p53 condensation the identified aminothiazoles also influence prion-like effects of p53 mutants. Indeed, 2-aminothiazoles have been proposed as potential leads to treat prion diseases (Gallardo-Godoy et al., 2011).
Given the long-term p53-independent anti-proliferative effects, the molecules reported here are suitable tool compounds for the evaluation of short-term gain-of-function effects of structural p53 mutants. For evaluation of long-term effects, e.g., on prevention of mutant p53 gain-of-function effects on cell migration or invasion or prion-like effects, optimization of the compounds should separate p53-dependent from additional p53-independent effects.
Limitations of the Study
This study identifies condensation-like states of fluorescent structural mutant p53 and identifies tool compounds that differentially modulate p53 condensation, leading to condensate dissolution in fluorescent structural p53 mutant variants and at the same time inducing condensation of a DNA-binding defective fluorescent p53 variant in a time frame 6 h after compound addition. These compounds do not lead to direct mutant p53 reactivation but seem to activate p53 reporter constructs partially by activation of p73. The identified compounds showed p53-independent anti-proliferative behavior in long-term proliferation experiments, indicating additional p53-independent activities. Therefore optimization of the compounds should separate p53-dependent from additional p53-independent effects.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Patrick Steigemann (patrick.steigemann@nuvisan.com).
Materials Availability
Plasmids generated in this study can be obtained from sirion Biotech (https://www.sirion-biotech.com); there are restrictions to the availability of aminothiazoles due to limited stock amounts.
Data and Code Availability
The published article includes all datasets generated or analyzed during this study.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The authors would like to thank Oliver Beutel, Ann Kwong, and Avinash Patel for helpful discussions.
Author Contributions
L.S. and N.Z. generated cell lines, performed luciferase, siRNA, IF, and live cell imaging experiments, P.S analyzed the data. C.L., C.S., and M.L. performed and analyzed cell proliferation and cell mechanistic assays. H.M. performed and analyzed mass spectrometry experiments. J.W., N.Barak., and U.E. performed and analyzed nDSF data. R.L. and A.K. performed and analyzed deep sequencing experiments. K.J. performed and analyzed western blotting. N.Braeuer. supervised Medicinal Chemistry activities. N.W., L.L., T.M.L., J.M., M.S., H.S., K.E., A.E., and P.S. advised on experiments and analysis. This work was conceptualized by C.L. and P.S. P.S. wrote the original draft, which was reviewed by the entire team. P.S. supervised this work.
Declaration of Interests
All authors are or were employees of Bayer AG.
Published: September 25, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101517.
Contributor Information
Clara Lemos, Email: clara.lemos@bayer.com.
Patrick Steigemann, Email: patrick.steigemann@nuvisan.com.
Supplemental Information
References
- Alberti S., Gladfelter A., Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbo H., Sato M., Okoshi A., Fukuchi S. Functional segments on intrinsically disordered regions in disease-related proteins. Biomolecules. 2019;9:88. doi: 10.3390/biom9030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang H.C., Joerger A.C., Mayer S., Fersht A.R. Effects of common cancer mutations on stability and DNA binding of full-length p53 compared with isolated core domains. J. Biol. Chem. 2006;281:21934–21941. doi: 10.1074/jbc.M604209200. [DOI] [PubMed] [Google Scholar]
- Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A., Klein I.A., Sabari B.R., Dall'Agnese A., Coffey E.L., Zamudio A.V., Li C.H., Shrinivas K., Manteiga J.C., Hannett N.M. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018;175:1842–1855 e16. doi: 10.1016/j.cell.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykov V.J.N., Eriksson S.E., Bianchi J., Wiman K.G. Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer. 2018;18:89–102. doi: 10.1038/nrc.2017.109. [DOI] [PubMed] [Google Scholar]
- Cai D., Feliciano D., Dong P., Flores E., Gruebele M., Porat-Shliom N., Sukenik S., Liu Z., Lippincott-Schwartz J. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat. Cell Biol. 2019;21:1578–1589. doi: 10.1038/s41556-019-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa D.C., de Oliveira G.A., Cino E.A., Soares I.N., Rangel L.P., Silva J.L. Aggregation and prion-like properties of misfolded tumor suppressors: is cancer a prion disease? Cold Spring Harb. Perspect. Biol. 2016;8:a023614. doi: 10.1101/cshperspect.a023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira G.A., Rangel L.P., Costa D.C., Silva J.L. Misfolding, aggregation, and disordered segments in c-Abl and p53 in human cancer. Front Oncol. 2015;5:97. doi: 10.3389/fonc.2015.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira G.A.P., Cordeiro Y., Silva J.L., Vieira T. Liquid-liquid phase transitions and amyloid aggregation in proteins related to cancer and neurodegenerative diseases. Adv. Protein Chem. Struct. Biol. 2019;118:289–331. doi: 10.1016/bs.apcsb.2019.08.002. [DOI] [PubMed] [Google Scholar]
- De Smet F., Saiz Rubio M., Hompes D., Naus E., De Baets G., Langenberg T., Hipp M.S., Houben B., Claes F., Charbonneau S. Nuclear inclusion bodies of mutant and wild-type p53 in cancer: a hallmark of p53 inactivation and proteostasis remodelling by p53 aggregation. J. Pathol. 2017;242:24–38. doi: 10.1002/path.4872. [DOI] [PubMed] [Google Scholar]
- Ditlev J.A., Case L.B., Rosen M.K. Who's in and who's out-compositional control of biomolecular condensates. J. Mol. Biol. 2018;430:4666–4684. doi: 10.1016/j.jmb.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L.A., Soussi T., Korkut A., Liu Y., Schultz A., Cardenas M., Li X., Babur O., Hsu T.K., Lichtarge O. Integrated analysis of TP53 gene and pathway Alterations in the cancer genome Atlas. Cell Rep. 2019;28:1370–1384 e5. doi: 10.1016/j.celrep.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follmann M., Briem H., Steinmeyer A., Hillisch A., Schmitt M.H., Haning H., Meier H. An approach towards enhancement of a screening library: the Next Generation Library Initiative (NGLI) at Bayer - against all odds? Drug Discov. Today. 2019;24:668–672. doi: 10.1016/j.drudis.2018.12.003. [DOI] [PubMed] [Google Scholar]
- Gaiddon C., Lokshin M., Ahn J., Zhang T., Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo-Godoy A., Gever J., Fife K.L., Silber B.M., Prusiner S.B., Renslo A.R. 2-Aminothiazoles as therapeutic leads for prion diseases. J. Med. Chem. 2011;54:1010–1021. doi: 10.1021/jm101250y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamagata K., Kanbayashi S., Honda M., Itoh Y., Takahashi H., Kameda T., Nagatsugi F., Takahashi S. Liquid-like droplet formation by tumor suppressor p53 induced by multivalent electrostatic interactions between two disordered domains. Sci. Rep. 2020;10:580. doi: 10.1038/s41598-020-57521-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenhuber E.R., Lowe S.W. Putting p53 in context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrloesser S., Osterburg C., Tuppi M., Schafer B., Vousden K.H., Dotsch V. Intrinsic aggregation propensity of the p63 and p73 TI domains correlates with p53R175H interaction and suggests further significance of aggregation events in the p53 family. Cell Death Differ. 2016;23:1952–1960. doi: 10.1038/cdd.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic S., Lezaja A., Gatti M., Bianco E., Michelena J., Imhof R., Altmeyer M. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 2019;38:e101379. doi: 10.15252/embj.2018101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G.A., Iwakuma T., Suh Y.A., Liu G., Rao V.A., Parant J.M., Valentin-Vega Y.A., Terzian T., Caldwell L.C., Strong L.C. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Levy C.B., Stumbo A.C., Ano Bom A.P., Portari E.A., Cordeiro Y., Silva J.L., De Moura-Gallo C.V. Co-localization of mutant p53 and amyloid-like protein aggregates in breast tumors. Int. J. Biochem. Cell Biol. 2011;43:60–64. doi: 10.1016/j.biocel.2010.10.017. [DOI] [PubMed] [Google Scholar]
- Li Y., Prives C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene. 2007;26:2220–2225. doi: 10.1038/sj.onc.1210311. [DOI] [PubMed] [Google Scholar]
- Li Y., Wang Z., Chen Y., Petersen R.B., Zheng L., Huang K. Salvation of the fallen angel: reactivating mutant p53. Br. J. Pharmacol. 2019;176:817–831. doi: 10.1111/bph.14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Ling S., Lin W.C. TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol. Cell Biol. 2011;31:4464–4481. doi: 10.1128/MCB.05574-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luh L.M., Kehrloesser S., Deutsch G.B., Gebel J., Coutandin D., Schafer B., Agostini M., Melino G., Dotsch V. Analysis of the oligomeric state and transactivation potential of TAp73alpha. Cell Death Differ. 2013;20:1008–1016. doi: 10.1038/cdd.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani F., Collavin L., Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019;26:199–212. doi: 10.1038/s41418-018-0246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo Dos Santos N., de Oliveira G.A.P., Ramos Rocha M., Pedrote M.M., Diniz da Silva Ferretti G., Pereira Rangel L., Morgado-Diaz J.A., Silva J.L., Rodrigues Pereira Gimba E. Loss of the p53 transactivation domain results in high amyloid aggregation of the Delta40p53 isoform in endometrial carcinoma cells. J. Biol. Chem. 2019;294:9430–9439. doi: 10.1074/jbc.RA119.007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzlyak E.M., Goedhart J., Shcherbo D., Bulina M.E., Shcheglov A.S., Fradkov A.F., Gaintzeva A., Lukyanov K.A., Lukyanov S., Gadella T.W., Chudakov D.M. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- Muller P.A., Vousden K.H. p53 mutations in cancer. Nat. Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- Muller P.A., Vousden K.H. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive K.P., Tuveson D.A., Ruhe Z.C., Yin B., Willis N.A., Bronson R.T., Crowley D., Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Pedrote M.M., Motta M.F., Ferretti G.D.S., Norberto D.R., Spohr T., Lima F.R.S., Gratton E., Silva J.L., de Oliveira G.A.P. Oncogenic gain of function in glioblastoma is linked to mutant p53 amyloid oligomers. iScience. 2020;23:100820. doi: 10.1016/j.isci.2020.100820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel L.P., Costa D.C., Vieira T.C., Silva J.L. The aggregation of mutant p53 produces prion-like properties in cancer. Prion. 2014;8:75–84. doi: 10.4161/pri.27776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safari M.S., Wang Z., Tailor K., Kolomeisky A.B., Conrad J.C., Vekilov P.G. Anomalous dense liquid condensates host the nucleation of tumor suppressor p53 fibrils. iScience. 2019;12:342–355. doi: 10.1016/j.isci.2019.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Silva J.L., Cino E.A., Soares I.N., Ferreira V.F., Oliveria G., A.P. Targeting the prion-like aggregation of mutant p53 to combat cancer. Acc. Chem. Res. 2018;51:181–190. doi: 10.1021/acs.accounts.7b00473. [DOI] [PubMed] [Google Scholar]
- Silva J.L., De Moura Gallo C.V., Costa D.C., Rangel L.P. Prion-like aggregation of mutant p53 in cancer. Trends Biochem. Sci. 2014;39:260–267. doi: 10.1016/j.tibs.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Soragni A., Janzen D.M., Johnson L.M., Lindgren A.G., Thai-Quynh Nguyen A., Tiourin E., Soriaga A.B., Lu J., Jiang L., Faull K.F. A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell. 2016;29:90–103. doi: 10.1016/j.ccell.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T., Wiman K.G. TP53: an oncogene in disguise. Cell Death Differ. 2015;22:1239–1249. doi: 10.1038/cdd.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stindt M.H., Muller P.A., Ludwig R.L., Kehrloesser S., Dotsch V., Vousden K.H. Functional interplay between MDM2, p63/p73 and mutant p53. Oncogene. 2015;34:4300–4310. doi: 10.1038/onc.2014.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Fersht A.R. Multisite aggregation of p53 and implications for drug rescue. Proc. Natl. Acad. Sci. U S A. 2017;114:E2634–E2643. doi: 10.1073/pnas.1700308114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zhao Y., Aguilar A., Bernard D., Yang C.Y. Targeting the MDM2-p53 protein-protein interaction for new cancer therapy: progress and challenges. Cold Spring Harb. Perspect. Med. 2017;7:a026245. doi: 10.1101/cshperspect.a026245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiech M., Olszewski M.B., Tracz-Gaszewska Z., Wawrzynow B., Zylicz M., Zylicz A. Molecular mechanism of mutant p53 stabilization: the role of HSP70 and MDM2. PLoS One. 2012;7:e51426. doi: 10.1371/journal.pone.0051426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Reumers J., Couceiro J.R., De Smet F., Gallardo R., Rudyak S., Cornelis A., Rozenski J., Zwolinska A., Marine J.C. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat. Chem. Biol. 2011;7:285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
- Xue B., Brown C.J., Dunker A.K., Uversky V.N. Intrinsically disordered regions of p53 family are highly diversified in evolution. Biochim. Biophys. Acta. 2013;1834:725–738. doi: 10.1016/j.bbapap.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Saos2 cells expressing fluorescently tagged DNA-binding defective mutant shows homogeneous nuclear fluorescence. Time-lapse video, 3 min per frame. Scale bar, 10 μm.
Saos2 cells expressing fluorescently tagged structural mutants show condensate-like behavior (condensates undergoing fusion events marked by arrows). Time-lapse video, 3 min per frame. Scale bar, 10 μm.
Saos2 cells expressing fluorescently tagged structural mutants show condensate-like behavior (condensates undergoing fusion events marked by arrows). Time-lapse video, 3 min per frame. Scale bar, 10 μm.
Saos2 cells expressing fluorescently tagged structural mutants show condensate-like behavior (condensates undergoing fusion events marked by arrows). Time-lapse video, 3 min per frame. Scale bar, 10μm.
Fluorescent structural mutant condensates are dissolved after aminothiazole treatment. Time-lapse video of Saos2 cells expressing fluorescently tagged mutant p53, 15 min per frame for the first 4 frames, afterward 30 min per frame. Scale bar, 10 μm.
Fluorescent structural mutant condensates are dissolved after aminothiazole treatment. Time-lapse video of Saos2 cells expressing fluorescently tagged mutant p53, 15 min per frame for the first 4 frames, afterward 30 min per frame. Scale bar, 10 μm.
Fluorescent structural mutant condensates are dissolved after aminothiazole treatment. Time-lapse video of Saos2 cells expressing fluorescently tagged mutant p53, 15 min per frame for the first 4 frames, afterward 30 min per frame. Scale bar, 10 μm.
Aminothiazole treatment leads to condensation of p53R273H after aminothiazole treatment. Time-lapse video of Saos2 cells expressing fluorescently tagged mutant p53, 15 min per frame for the first 4 frames, afterward 30 min per frame. Scale bar, 10 μm.
Data Availability Statement
The published article includes all datasets generated or analyzed during this study.