Abstract
The pervasive spread of infectious diseases and pandemics, such as the 2019 coronavirus disease (COVID-19), are becoming increasingly serious and urgent threats to human health. Preventing the spread of such diseases prioritizes the development of sensing devices that can rapidly, selectively, and reliably detect pathogens at minimal cost. Paper-based analytical devices (PADs) are promising tools that satisfy those criteria. Numerous paper-based biosensors have been established that rival conventional pathogen detection methods. Among them, colorimetric strategies are promising since results can be interpreted by eye, and are simple to operate, which is advantageous for point-of-care testing (POCT). Particularly, the application of nanomaterials on paper-based biosensors has become important as these materials are capable of converting signals from pathogens through unique mechanisms to yield an amplified colorimetric readout. To highlight the research progress on using nanomaterials in colorimetric paper-based biosensor for pathogen detection, we discuss the sensing mechanisms of how they work, structural and analytical characteristics of the devices, and representative recent applications. Current challenges and future directions of using PADs and nanomaterial-mediated strategies are also discussed.
Keywords: Paper-based analytical devices, Colorimetric biosensors, Pathogen detection, Nanomaterials, Point-of-care testing
Graphical abstract
1. Introduction
Infectious diseases derived from numerous disease-causing agents, such as pathogenic foodborne and waterborne bacteria and viruses, have become increasingly serious global threats to human health and the economy [1]. On March 19, 2020, the World Health Organization (WHO) declared that the on-going 2019 coronavirus disease (COVID-19) situation was a global pandemic. The COVID-19 pandemic has been spreading rapidly around the world and has widespread community transmission in more than 150 countries including China, Italy, United States, and South Korea. Approximately 100,000 confirmed cases quickly reached 209,839 confirmed cases worldwide within 12 days, with 8778 deaths reported over the 3 months since cases were first reported in Wuhan, China on December 31, 2019 [2,3]. COVID-19 is a variant of a large group of viruses that cause the common cold and other severe illnesses in humans such as Middle East respiratory syndrome (MERS-CoV) and severe acute respiratory syndrome (SARS-CoV) which caused outbreaks in 2002 and 2012, respectively [[4], [5]]. It was reported that by August 2003, there were 8422 infected cases of SARS-CoV which led to more than 900 deaths worldwide while there have been only 2494 cases of MERS-CoV since 2012. However, the number of MERS-associated deaths were roughly comparable to that of SARS-CoV, which were 858 cases since September 2012, with a relatively high mortality rate of 34.4% [6]. Compared to SARS-CoV, COVID-19 has a stronger transmission capacity and its pathogenicity is considered to be comparable to that of MERS-CoV [7]. Additionally, the symptoms of COVID-19 infection are nonspecific and are difficult to distinguish from the common cold. These symptoms include fever, cough, headache, breathing difficulties (dyspnea), and viral pneumonia. Considering these nonspecific symptoms and widespread transmission of this disease, diagnostic examination is critical for reliably confirming suspected cases, screening probable cases, as well as surveilling the spread of the virus [5]. Apart from viral epidemics, pathogenic microorganisms from unsafe foods were also considered as causative agents of morbidity and mortality [1]. The WHO reported that there are 600 million cases of foodborne diseases that cause 420,000 deaths each year across the world [8]. In addition, pathogenic bacteria in contaminated water are also a pressing issue due to environmental pollution, and is a concern even in industrialized nations. For instance, annual deaths in the United States caused by acute gastrointestinal illness have been increasing from consuming unsafe water, rising from 4.26 million to more than 30 million. In addition, approximately 2.2 million people die each year from diarrhea caused by contaminated water intake [9,10]. Thus, it is important to develop rapid, precise, and sensitive approaches for early detection of pathogens to prevent worldwide spread of infectious diseases, and to optimize medical care to reduce mortality and overall cost [1,11].
Several conventional methods have been developed to identify and monitor pathogenic bacteria and viruses. These techniques consist of standard culture-based methods, enzyme-linked immunosorbent assay (ELISA), and polymerase chain reaction (PCR). Culture-based methods are laborious and take a long time. ELISA and PCR are highly sensitive and accurate methods, but are expensive and depend on complex equipment and trained technicians that are only feasible in centralized labs [1,12]. To overcome such shortcomings, the WHO strongly advocates for diagnostic approaches that are affordable, sensitive, specific, user-friendly, robust, rapid, equipment-free, and deliverable to end-users (ASSURED). Point-of-care (POC) analytical devices are potential candidates that satisfy these ASSURED requirements [13]. Paper-based analytical devices (PADs) emerged as critical POC tools due to their simplicity, low fabricating costs, easy storage, portability, and disposability. These advantageous features make PADs important for medical diagnostics, particularly in resource-limited areas, emergencies, and in home healthcare, without relying on external devices and reagents [14,15]. There are diversified signal readouts that have been utilized in paper-based sensors that involve electrochemistry, conductivity, fluorescence, and colorimetry [13]. Among them, colorimetric paper-based biosensors are in demand and are the most attractive because the presence of a specific pathogen can be conveniently monitored by a simple change in color, which can be distinguished easily with the naked eye without expensive and complex instruments [16]. The major limitation of colorimetric assays is low sensitivity since it is often difficult to transform detectable signals into a color readout [1,17].
To resolve the limitation, enzymes were utilized to produce selective and sensitive colorimetric signal on PADs [[18], [19], [20]]; however, they have several intrinsic problems such as instability in diverse environments and high production cost, which critically hinders their practical utilizations [21]. To strengthen the sensitivity of colorimetric assays, a variety of nanomaterials, such as noble metal nanoparticles including gold nanoparticles (AuNPs), magnetic nanoparticles (MNPs), carbon-based nanostructures, conjugated polymeric nanovesicles, and other enzyme-mimicking nanomaterials (nanozymes), have been used [22]. These materials exhibit distinctive physical and chemical properties, making them important signal indicators for colorimetric paper-based biosensors [[22], [23], [24]]. Besides, nanomaterials also support diverse functionalizations with other biological materials, thus, facilitating target capture and detection as well as signal amplification in colorimetric detection of pathogens [1]. The large surface area of nanomaterials can increase the amount of receptors conjugated on their surface [25], consequently increasing the amount of recognition events and improving the detection performances. Thanks to their high surface-to-volume ratio, nanomaterials also generally exhibit remarkably high physicochemical properties such as catalytic activity, conductivity, and magnetic property, all of which could be essentially utilized to enhance signal readout for colorimetric pathogen detection [1,22,26].
Despite these advantages accompanied with nanomaterials, PADs have several limitations that need to be resolved for providing more reliable disease diagnosis to end-users. First, their diagnostic performances such as specificity and sensitivity are usually highly affected and can be varied by the kinds of paper substrate, and thus, the right choice of paper is required to achieve optimal detection performances [27]. Generally, higher speed of capillary flow, which can be varied by the kinds of paper, results in higher specificity but lower sensitivity [28]. Besides, patterning step to construct microchannel on paper substrate should be elaborately designed and considered since it can determine the reaction pathway and efficiency on PADs. There have been several patterning methods on paper substrate, including photolithography and printing methods like inkjet and wax printing [29]. Among them, wax printing is generally considered as the most popular method for creating hydrophobic barriers for patterning in PADs, due to its relatively low cost, simple and rapid printing procedures, easy post-printing steps, and high capacity to create flexible patterns [30]. However, it also has limitations including sample leakages caused by the surfactants in biological samples [14]. Therefore, considering the detection efficiency, cost-effectiveness, and kinds of employed reagent and reaction, appropriate selection of paper substrate and patterning method as well as improvements in the fabrication of PADs are required for the widespread utilizations in diagnostic fields.
In this review, we focus on recent developments in PADs for colorimetric detection of pathogenic microorganisms and viruses using nanomaterials. We describe representative types of paper substrates for POC analytical devices, utilization of noble metal nanomaterials, nanozymes that have peroxidase-like activity, and conjugated polymeric nanovesicles. We also describe prospective nanotechnology that can be used to enhance colorimetric methods to detect pathogens.
2. Representative types for paper-based biosensors
Paper substrates have historically been used in analytical and chemical applications. In particular, paper chromatography is often used for separating and detecting molecules such as amino acids, proteins, antibodies, and small molecules from matrices [14]. The utilization of paper substrates in POCT has become increasingly important [14,15]. Compared to sensors made out of other materials such as glass, silicon, and plastic, PADs are cheaper and easier to fabricate. In addition, PADs are also easy to store and transport, and can be disposed without cost in a safe and environmentally-friendly manner. The intrinsic hydrophilic property of paper, due to its porous structure, is highly advantageous since it can facilitate fluid flow by capillary action without the need for external pumping [15].
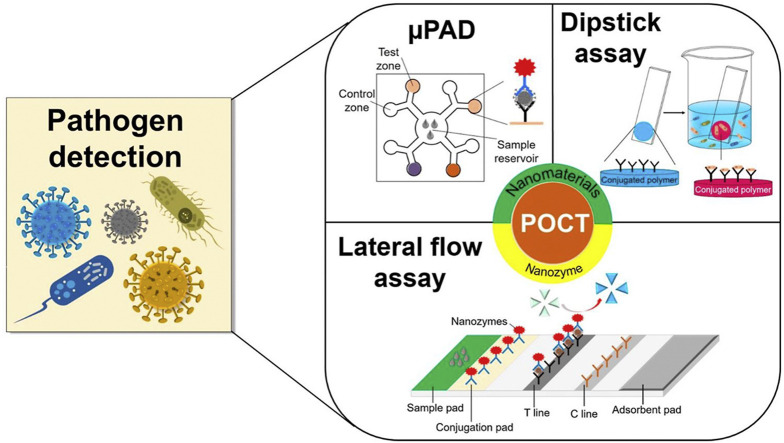
In practice, PADs can be classified into three main types: dipstick assay, lateral flow assay (LFA), and microfluidic paper-based analytical device (μPAD) (Fig. 1 ) [27]. LFA assays are frequently used to detect pathogens since they provide many advantages for practical pathogen detection including fast-response, single-step operation, cost effectiveness, user-friendly format, high sensitivity, and sufficient selectivity [12]. The two major types of paper substrates are cellulose and nitrocellulose membranes. Cellulose membranes are widely used in dipstick and μPAD applications, while nitrocellulose membranes are extensively used in LFA assays [27]. Urine test strips, the most common dipstick assay, are utilized to evaluate urinary tract infection caused by nitrites. The presence of nitrites is not common in urine, and is mainly attributed to the presence of bacteria, in most cases Gram-negative bacteria, that can reduce urinary nitrates into nitrites. The urine test strip is specific, but not sensitive, needing more than 104 cells/mL to present a positive result [31]. The urine test strips are made up of a piece of water-resistant plastic film containing many absorbent pads with immobilized chemical reagents. Depending on the chemical composition of the urine sample, the change in color of the different absorbent pads indicate urinary health [32]. LFA strips are primarily comprised of four integrated fragments: (1) sample pad where the analysis sample is added, (2) conjugation pad where signal tag is linked to the bio-receptor, (3) nitrocellulose membrane where the capture molecules are immobilized to form the test line (T line) and control line (C line), and (4) absorbent pad to absorb excessive fluid and prevent reversed flow of liquid. Regarding to μPAD, hydrophobic boundaries patterning is a principal process as the fluid was driven not only by the capillary force of hydrophilic paper but also the hydrophobic microchannel network [13]. The μPADs are generally constructed into 2D or 3D configurations. The 2D method primarily relies on horizontal or passive fluid movement via capillary force, whereas the 3D method exploits both horizontal and vertical fluid transportation via connected patterned layers.
Fig. 1.
Three main types of PADs for pathogen detection: (a) μPAD, (b) LFA, and (c) dipstick.
For fabricating μPADs, there have been several techniques for 2D configuration such as photolithography, inkjet printing, wax screen printing, and flexographic printing, as well as other fabrication methods for 3D configuration such as stacking of patterned papers, origami method, and 3D wax printing [29]. In 2007, Whiteside's group first applied the photolithography technique to generate microfluidic patterns on paper substrate [33]. This method utilized UV light to transfer patterns of a photomask on photoresist-coated paper substrate. It created hydrophilic channels on the paper by forming hydrophobic surroundings using polymeric materials [11,29]. Other printing methods such as inkjet and wax printing have also been widely utilized to construct μPADs. Inkjet printing can be simply performed with commercially-available inkjet printer, to precisely deposit patterning agent such as alkyl ketene dimer (AKD), to form hydrophobic barriers on the paper [29,34]. AKD can hydrophobize the cellulose component of the paper by esterification; however, the inevitable use of organic solvent to dissolve AKD has destructive effect on printer cartridge [34]. To circumvent this disadvantage, UV-curable acrylate ink was utilized in place of AKD [35]. Wax printing utilized a non-toxic and hydrophobic solid wax to create microfluidic patterns on the paper with a solid-ink printer. After printing, heating treatment should be performed to melt the solid wax as well as enable the liquid wax to spread vertically and horizontally into the entire paper [36]. This method enables rapid and inexpensive patterning on the paper and does not depend on sophisticated instrumentation and harmful organic solvents [29]. Flexographic printing utilized polystyrene solution as hydrophobic patterning ink, and has been utilized to produce a large quantity of μPADs [29,34].
Compared with the above examples describing 2D microfluidic patterning on paper, 3D patterning on the paper has greater potential to enhance detection performances by allowing the integration of many functional layers or reservoirs with controllable fluid flow whose direction can be either horizontal or vertical [34,37]. To fabricate 3D μPADs, stacking method to vertically connect multiple 2D patterned papers using adhesive tape was reported. This method could transfer the fluid more easily and freely as required; however, it has limitations of complicated alignment, bonding, and punching processes, which can negatively affect the fluid flow [29,30,34]. Origami method utilized simple folding process of 2D patterned papers to construct 3D configuration, and thus it is quick and easy to perform, and allows various 3D shapes for diverse operations [34]. Furthermore, 3D wax printing was reported by printing wax on both sides of paper [38]. By elaborately controlling the density of wax as well as heating period, the penetration depth of solid wax could be manipulated and thus several layers of channels on a single paper sheet were constructed. These 3D patterning methods allowed effective integration of many fluid flows, enabling more complexed or multiplexed assays on a single μPAD.
3. Noble metal nanoparticle-mediated colorimetric pathogen detection
Noble metal nanoparticles are interesting in POC biosensing research because they can be directly utilized as signal reporters due to their intrinsic physical and optical features. These features consist of high surface-to-volume ratio which enables suitable surface modification with bioactive compounds, excellent capacity for reaction catalysis, electrical conductance, good biocompatibility, particularly high characteristic extinction coefficient in visible light, as well as visual color transition from the shift of surface plasma absorption by varying size, shape, interparticle distance through the aggregation, and dielectric environment [13,26]. The deployment of noble metal nanoparticles in colorimetric biosensors can significantly amplify signal intensity and thus enhance the sensitivity for target biological molecules such as pathogenic bacteria and viruses, DNA, toxins, proteins, and others [26,39]. Among noble metal nanoparticles, gold and silver nanoparticles are particularly interesting due to their simplicity in producing a color response with high sensitivity [39]. Gold nanoparticles (AuNPs) are extensively used in colorimetric biomedical assays since they are easy to synthesize, are chemically and physically stable, have good biocompatibility, have unique optoelectronic behavior, and are easily modified with bioactive and organic compounds [13,40].
One important property of AuNPs is localized surface plasmon resonance (LSPR). This phenomenon involves collective oscillation of free electrons positioned in the conduction bands of AuNPs that become activated by interacting with a resonant frequency of incident light [26]. This interaction leads to electrical polarization that generates an oscillating dipole, resulting in a unique change of light absorption or scattering. For example, small and symmetrical AuNPs usually absorb light, whereas larger and less symmetrical particles tend to scatter light. The effects of LSPR are easily adjusted by changing structural factors such as size, shape, inter-particle distance, and dielectric value of its surroundings. Monodispersed AuNPs in solution are red due to LSPR-induced absorption peaks at green wavelengths, while the aggregated state induces a color transformation to blue or purple. Thus, size-induced LSPR effects of AuNPs have been used for colorimetric detection of targets [40].
One of the main issues in biosensor development is specific detection of targets from a medium containing several interfering components. The choice of bio-receptor is critical since it can determine detection sensitivity and selectivity for target pathogens. Several types of bio-receptors, such as oligonucleotide aptamers, antibodies, proteins, and other small biomolecules, can be functionalized on nanomaterials to enhance the affinity of these materials towards target pathogenic bacteria and viruses in complex matrices. Among these types of bio-receptors, antibodies and aptamers are most often used for pathogenic detection [1,40].
3.1. Antibody-functionalized noble metal nanoparticles for colorimetric pathogen detection
The high affinity between antigens and their corresponding antibodies is of great interest and has been widely used to detect many types of pathogens [12]. For example, Lei et al. showed that two specific capture antibodies for influenza A subtypes H1N1 and H3N2 were functionalized with AuNPs for rapid colorimetric screening of pathogenic viruses [41]. In this study, capture antibodies were adsorbed onto each well of 48-well microfluidic paper. Samples were then loaded into each well to induce the interaction between virus particles and antibodies. Subsequently, primary antibodies were added, followed by horseradish peroxidase (HRP)- or AuNPs-coupled secondary antibodies to perform sandwich immunoassays. Signal detection by AuNPs was optimized by enhancing gold growth to physically enlarge and fuse the AuNPs to intensify colorimetric output, resulting in better signal response compared with the HRP-mediated enzymatic assay. As a result, the limit of detection (LOD) of H1N1 and H3N2 were lowered to 2.7 × 103 and 2.7 × 104 plaque forming units (PFU)/mL, respectively. The sensitivity and reliability of this device were comparable with ELISA and real-time (RT)-PCR assays, demonstrating its potential for rapid screening of infectious diseases. However, due to many washing steps needed after each reagent was added, the assay was time consuming, requiring at least 1 h to complete. A gold enhancement strategy was used to nucleate gold particles through the reaction between chloroauric acid (HAuCl4) and a reducing agent such as hydroxylamine hydrochloride (NH2OH·HCl). This allowed for in situ reduction of Au3+ ions on the surface of primary AuNPs to increase their size, resulting in enhanced signal intensity. Based on this strategy, Bu et al. also established an LFA strip to detect Salmonella enteritidis within 20 min by visual observation [42]. In this assay, a traditional LFA strip (10 min) was used, and then further dipped into an enhancer solution for another 10 min to boost signal intensity. The LOD of this assay was 104 colony forming units (CFU)/mL, which was 100-times more sensitive than a traditional strip without enhancement. Although higher sensitivity was achieved, an extra manual procedure to apply enhancer solution should be considered for its practical utilization.
In another study, Pan et al. developed an AuNP-enhanced LFA strip for sensitive POC detection of Cronobacter sakazakii (C. sakazakii) in powdered, non-sterile infant formula [43]. Two types of AuNPs were employed; the first was used as a signal indicator and was conjugated to detection antibodies, while the other was used as a signal intensifier and was conjugated to capture antibodies. The AuNP-modified detection and capture antibodies were immobilized onto the conjugation pad and printed onto the test zone, respectively. The use of AuNP-modified capture antibodies increased the number of antibodies at the T line, thus, increasing the interaction between capture antibodies and C. sakazakii. In the presence of C. sakazakii, the AuNP-modified detection antibodies were trapped by AuNP-modified capture antibodies in the test zone to increase the amount of AuNPs available for enhancing signal intensity. As a result, the LOD of the LFA was lowered to 103 CFU/mL, which is about 100-fold more sensitive than the non-amplified strip. Yen et al. reported rapid and multiplexed colorimetric detection of dengue, yellow fever, and Ebola viruses in a single lateral flow strip [44]. This study exploited optical properties of silver nanoparticles (AgNPs), where shape and size variations trigger color transformations in the visible range. In other words, common colloidal AgNPs are yellow, but can change to orange, red, and green depending on the size and/or shape of AgNPs. In this study, each specific antibody type corresponding to yellow fever, Ebola, and dengue viruses were labeled with orange AgNPs, red AgNPs, and green AgNPs, respectively. When the target analyte is present, the sandwich platform allows the pathogen to be trapped by the AgNP-modified antibodies, that become further bound to capture antibodies printed on the test line. The strips were effective in detecting levels as low as 150 ng/mL for dengue, yellow fever, and Ebola viruses. The reported LOD of dengue viral protein (NS1) was about 100 to 300-fold lower than the maximal concentration of NS1 in serum.
An immunological dipstick strip was developed to detect pathogenic Vibrio parahaemolyticus directly from oyster hemolymph (oyster circulatory fluid) [45]. Antibody was electrostatically conjugated on AuNPs, followed by passivation using thiolated polyethylene glycol (PEG) to prevent non-specific interaction. The resulting antibody-conjugated AuNPs were immobilized on test area of dipstick, and then sample fluid moved sequentially through the strip to facilitate proper antigen-antibody interaction with minimizing AuNPs' aggregation and non-specific interaction. With the assay, the LOD was determined to be 4.66 × 105 CFU/mL, which is lower than the reported V. parahaemolyticus dose with a 50% probability to cause a foodborne disease.
3.2. Aptamer-functionalized noble metal nanoparticles for colorimetric pathogen detection
Conventionally, antibodies are used to recognize the presence of pathogens. However, the use of oligonucleotides, particularly aptamers, are becoming increasingly interesting since they have tunable specificity, are easy to synthesize, have prolonged stability, and have high capacity for further functionalization [46]. Aptamers are single-stranded oligonucleotides including DNA and RNA that can form specific patterns such as stems, purine-rich bulges, and guanidine-quadruplexes [12]. These single-stranded nucleic acids can be repeatedly selected towards desired targets via systematic evolution of ligands by exponential (SELEX) enrichment. Aptamers have become a promising class of bioreceptors for pathogenic detection as their performance can be improved by advancing SELEX procedures. Furthermore, several studies have reported that integration of aptamers and nanomaterials promotes signal intensity, which leads to increased sensitivity in diagnosing pandemic and infectious diseases at early stages [12,22].
For example, AuNPs were coupled with aptamers in an LFA strip for visual and rapid monitoring of Salmonella typhimurium (S. typhimurium), Escherichia coli (E. coli) O157:H7, and Staphylococcus aureus (S. aureus) within 10 min [47]. The mechanism relies on the aptamer-based sandwich model. A red line is observed when the target pathogen in the sample solution migrates up the strip to form a complex with AuNP-labeled aptamer 1, where the target pathogen then associates with aptamer 2 in the strip. The validity of the strip was confirmed by another red band, the C line, in addition to the T line. After the pathogen is trapped on the T line to generate a red line, the remaining AuNP-aptamer 1 complex continues to travel to the C line and hybridizes with complementary DNA to produce another red line. Without the target, only the C line is detected. Using this strategy, a simple, fast, and reliable method was established for detecting S. typhimurium, E. coli, and S. aureus, with LODs as low as 103, 104, and 104 CFU/mL, respectively.
Another study used a pair of aptamers that specifically bound to avian influenza H5N2 viruses at multiple sites simultaneously [48]. This study was one of the few studies that used a homologous pair of aptamers to detect whole H5N2 virus particles instead of specific viral proteins, such as hemagglutinins. In order to select pairs of aptamers that specifically bind to whole H5N2 virus particles, a graphene-oxide based SELEX (GO-SELEX) procedure was used. This strategy is based on π-π stacking between single-stranded DNA (ssDNA) and GO. In the presence of target pathogens, ssDNA that can bind the pathogen is released from GO due to a structural change, whereas those that are not specific for the target pathogen remain stable on GO. After screening with GO-SELEX, a pair of aptamers was chosen where one aptamer was used for capturing and was immobilized on the T line, while the other was modified with AuNPs and functioned as the reporter aptamer. When a sample containing virus particles travels along the strip, the interaction between virus cells and AuNP-functionalized aptamers creates a complex that further binds to the capturing aptamer in the test zone. The accumulation of AuNPs at the T line generates a red band that can be observed by eye. By successfully applying a double aptamer sandwich on the LFA strip, the paper-based biosensor was able to detect H5N2 virus particles in concentrations as low as 6 × 105 50% egg infection dose (EID50/mL) in buffer and 1.2 × 106 EID50/mL in duck feces. Although this study showed comparable results with commercial kits for rapid detection of diverse subtypes of influenza A virus, the sensitivity can be further improved for monitoring of influenza viruses.
3.3. Utilization of other receptors with noble metal nanoparticles for colorimetric pathogen detection
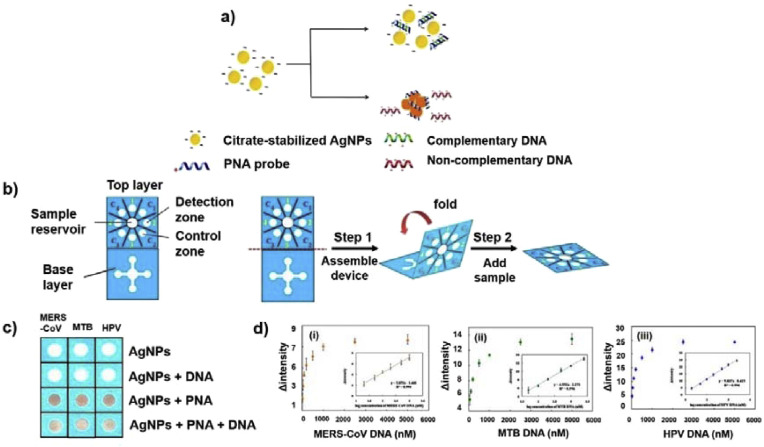
In addition to antibodies and aptamers, other receptor molecules including specific glycoprotein and peptide nucleic acid (PNA), were utilized with noble metal nanoparticles to identify target pathogens via paper-based devices. Shafiee et al. utilized lipopolysaccharide binding protein (LBP), which can recognize and bind the lipid moiety of lipopolysaccharide on the outer cell membrane of Gram-negative bacteria, to detect pathogenic E. coli [49]. AuNPs were functionalized with LBP and the conjugates were applied to detect E. coli, that caused an aggregation of AuNPs on the bacterial surface and subsequent color shift from red to blue. This color-changing strategy was realized on cellulose paper and enabled sensitive colorimetric detection of E. coli infected in blood serum with a LOD of 8 CFU/mL. The entire detection process was completed within 10 min through observing the clear color change with either naked eyes or smartphone camera. An origami-type multiplex 3D PAD was also reported to colorimetrically detect MERS-CoV, Mycobacterium tuberculosis (MTB), and human papilloma virus (HPV) (Fig. 2 ) [50]. In this method, PNA, which is an artificial DNA analog having polypeptide backbone rather than sugar phosphate, was utilized as a probe to detect target pathogenic DNA via the color change induced by the aggregation of AgNPs. Compared to DNA, PNA has many superiorities such as high stability in harsh environments and efficient hybridization with its complementary DNA strands. The origami structure consisted of two layers, which were prepared by wax printing. The top layer contained four detection and control zones, each of them contained AgNPs with PNA probe, while the base layer contained four channels extending outward from a center sample inlet. After the device was folded and stacked together, the channels of the base layer were connected to four detection zones of the top layer. Upon sample addition, the solution moved outward through the channels of the base layer to move into the detection zones of the top layer, leading to color change. In the absence of target DNA, the PNA probe caused an aggregation of citrate-stabilized AgNPs, which led to red color. On the other hand, in the presence of target DNA, DNA-PNA duplex was formed, resulting in dispersion of the AgNPs with their native yellow color. This 3D PAD was used for specifically detecting MERS-CoV, MTB, and HPV, by measuring the color change and the LODs were found to be 1.53, 1.27, and 1.03 nM, respectively.
Fig. 2.
3D μPAD for detecting Middle East respiratory syndrome coronavirus (MERS-CoV), Mycobacterium tuberculosis (MTB), and human papillomavirus (HPV). (a) Schematic illustration representing pathogenic DNA detection by the color change from peptide nucleic acid (PNA)-induced AgNP aggregation, (b) design and operation of origami-type 3D μPAD, (c) real images of visible color transformation from the DNA of MERS-CoV, MTB, and HPV, and (d) color intensities according to concentrations of target DNA with the corresponding calibration plots for (i) MERS-CoV, (ii) MTB, and (iii) HPV. Reproduced from Refs. [50] with permission of ACS Publications.
Furthermore, gold-coated paper coupled with peptide-functionalized MNPs were utilized to detect pathogenic S. aureus [51]. The sensing method was based on the protease activity toward specific peptide, sandwiched between MNPs and gold surface on paper substrate. An external magnet was fixed on the back of the paper sensor to facilitate the cleavage of the MNPs away from the paper upon the sample addition. Due to the cleavage and MNPs removal from the paper strip, the original black color was changed into gold, which was visible by naked eyes and analyzed down to 7 CFU/mL in pure broth culture during just 1 min incubation. With similar detection principle, E. coli O157:H7 was also successfully determined with a LOD of 12 CFU/mL [52].
4. Nanozyme-mediated colorimetric pathogen detection
Enzymes are protein molecules that act as biocatalysts to speed up the reaction rates, as well as facilitate diversified biological reactions in living systems [53]. Because of their mild reaction conditions with high substrate specificity and activity with less toxicity to physiology and environment, they have been widely used in pharmaceutical, food, and beverage industries [54]. Although enzymes have been extensively used in practical applications, they are prone to activity loss in harsh environmental conditions due to changes in pH, temperature, or solvents. They are also laborious to produce and purify, are inefficient to recycle, and are expensive to produce, which significantly hinders their usage [21]. Therefore, emerging “artificial enzymes” that address these limitations of natural enzymes are very desirable [21,54].
Nanozymes are a type of artificial enzymes that are potential alternatives to traditional enzymes since they are highly stable in harsh conditions, cost-effective, easy to mass produce, and have highly tunable catalytic activity [21,55]. In 2007, the first peroxidase-mimicking Fe3O4 MNPs were shown to oxidize peroxidase substrates, 3,3′,5,5′-tetramethylbenzidine (TMB), diazo-aminobenzene (DAB), and o-phenylenediamine (OPD), in response to hydrogen peroxide (H2O2) to their corresponding blue, brown, and orange colors, respectively [56]. Accordingly, many studies have reported enzyme-like activity of metals, metal oxide nanoparticles, nanoclusters, quantum dots, carbon nanostructures, nanowires, and several composites, including organic-inorganic hybrid nanoflowers and metal-organic frameworks [[57], [58], [59]]. Current nanozyme researches demonstrate that the catalytic activity of nanozymes can be significantly enhanced, that is comparable or even higher than that of natural enzyme counterpart, by optimizing their size, shape, composition, and surface functionalities [60,61]. Substrate specificity of nanozymes is still a challenging issue to overcome; however, several potent strategies to engineer the surface of nanozymes, including the growth of molecular imprinted polymer to create substrate binding pockets or the conjugation of active site-mimicking single atom site, were recently devised [62,63]. Compared to conventional enzymes, nanozymes have been utilized as robust catalytic indicators for immunoassays and other biosensing systems, demonstrating the significance and rapid advancement of this research field.
Due to their promising catalytic abilities, nanozymes have been used as a model for POC monitoring platforms, which have been used for clinical diagnosis and detection of infectious pathogens [64]. Nanozyme-mediated paper-based LFAs are considered as a particularly effective strategy [65]. Previous studies have demonstrated that colorimetric readouts of LFAs could be achieved by direct utilization of nanomaterials due to plasmonic properties of AuNPs. However, visual detection of low pathogen concentrations could not be achieved since the concentration of accumulated nanoparticles is too low [42]. Therefore, nanozymes have been integrated to improve the performance and sensitivity of the paper-based biosensors [66,67]. Although nanozyme-based LFAs resemble conventional AuNP-based LFAs, an additional chromogenic substrate is needed which can be placed onto the test site of the strip. Accordingly, in the presence of target analytes, the reaction between the nanozyme complex and chromogenic substrate eventually generates a very strong colorimetric signal from the respective color-producing reaction in comparison with that of conventional LFAs [68].
4.1. Metal nanozymes for colorimetric pathogen detection
Peroxidase-mimicking nanozymes, owing to their catalytic ability to immensely enhance colorimetric signals, have been adopted in PADs for disease assessment [66,67,69,70]. Nanozyme-mediated LFA strips for detecting E. coli O157:H7 have improved colorimetric detection. Jiang et al. prepared porous platinum-gold (Pt–Au) bimetal nanoparticles conjugated to anti–E. coli O157:H7 polyclonal antibodies that were immobilized onto the strip [69]. Han et al. also successfully designed concave palladium-platinum (Pd–Pt) nanoparticles that could be immobilized onto LFA strips [65]. Both Pt–Au and Pd–Pt nanoparticles were unique composite structures that enhanced surface-to-volume ratios compared to single metal nanoparticles. This improvement in surface-to-volume ratio increases the number of catalytic sites, thus these nanoparticles exhibited excellent peroxidase-like activity. Accordingly, the signal intensity was enhanced with Pt–Au and Pd–Pt-based strips with LODs as low as 100 CFU/mL (particularly within 1 min) and 87 CFU/mL, respectively [65,69]. In addition, they also showed high practicality for on-site pathogen monitoring since the strip was demonstrated to be applied in real milk sample for E. coli O157:H7 detection and the lowest detectable limit was determined as 900 CFU/mL, validating them for practical applications considering the infectious dose of this pathogen [65]. In another study, a novel LFA paper-based strip was designed for ultrasensitive detection of E. coli O157:H7 with an LOD as low as 1.25 × 101 CFU/mL [67]. This was achieved using conventional colloidal Au-based strips with a two-step process to intensify the signal. Development of AuNPs was promoted, followed by intrinsic peroxidase-mimicking activity of the newly-developed AuNPs. Synergetic effects from this signal amplification strategy allowed for ultrasensitive detection of E. coli O157:H7, even at a single molecule level. These peroxidase-mimicking nanozyme-based LFA strips exhibited higher sensitivity compared to that of conventional AuNPs-based LFAs; however, nanozymes required additional experimental steps including the addition of colorimetric substrate and H2O2 with prolonged reaction time to produce color signal, which are not friendly to end-users.
Nanozyme-mediated LFA strips have also been used for on-line detection of Campylobacter jejuni (C. jejuni) in contaminated food [71], which is often responsible for gastroenteritis. In this research, gold nanorods coated with thin layers of platinum (AuNR@Pt) were incorporated onto LFA strips. This allowed for a simultaneous colorimetric readout, based on peroxidase–like activity, and surface enhanced Raman scattering (SERS)-mediated readout due to robust electromagnetic forces of the nanoparticles. For the colorimetric readout, AuNR@Pt was tagged with monoclonal antibodies to form a sandwich assay to detect C. jejuni on the test zone of the LFA strip. In the presence of TMB and H2O2, the peroxidase-like activity of AuNR@Pt disproportionates H2O2, and further oxidize the TMB substrate to create a blue-colored band on the strip. Using this approach, the strip can detect target bacteria in concentrations as low as 75 CFU/mL. AuNR@Pt was also functionalized with a Raman reporter (4–MBA) to lower the LOD to 50 CFU/mL. The LFA strip in this study was also used to detect bacteria in milk and chicken samples. The integration of both colorimetric and SERS strategies validate the use of this bimetallic nanozyme-based LFA strip for detection of C. jejuni.
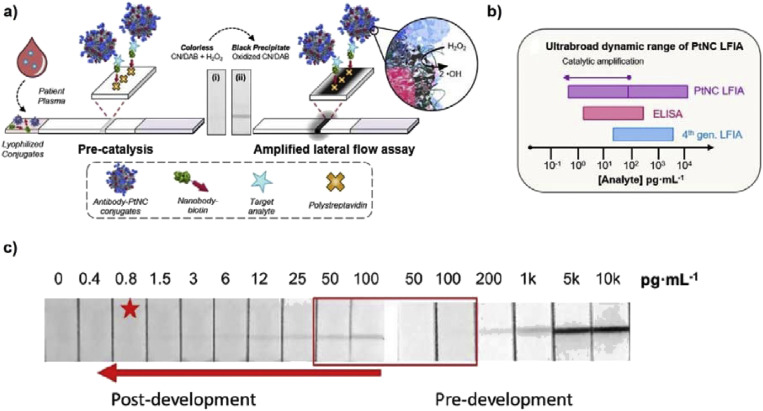
Human immunodeficiency virus (HIV) is responsible for acquired immunodeficiency syndrome (AIDS) which led to 940,000 deaths worldwide in 2017 [72]. Since AIDS is an ongoing public health concern, various analysis kits have been generated for detecting different portions of the virus [66,72]. Detecting biomarkers such as p24 capsid protein allows for early diagnosis, where false–negative results are likely to appear by antibody analysis. Clinical values for p24 during acute infection periods is much lower than LODs established in pioneering colorimetric LFA tests (approximately 10–15 pg/mL). Accordingly, Loynachan et al. developed a rapid and ultrasensitive paper-based LFA strip which can detect femtomolar levels of p24 biomarker within 20 min (Fig. 3 ) [66]. In this paper, a porous Au–Pt core-shell nanozyme was synthesized by overgrowing the Pt layer on the surface of 15 nm diameter AuNP seeds with polyvinylpyrrolidone (PVP) and ascorbic acid as anti-aggregation substance (stabilizers) and reducing agent, respectively. The nanoscopic surface area of AuNPs plays a crucial part in increasing peroxidase-like activity since the surface is accessible to small molecules such as H2O2 through the nanosized pores embedded in the Pt shell. In contrast, larger molecules such as antibodies, albumin, and other serum proteins cannot travel through the pores, consequently stabilizing the nanoparticle from protein-rich medium. Au–Pt core-shell nanoparticles strikingly extend the detection range for p24, ranging from 10,000 pg/mL to less than 1 pg/mL. Between 100 and 10,000 pg/mL of p24, Au–Pt nanozymes appeared as black on the T line, and furthermore, the smaller spectrum (100 to less than 1 pg/mL) could be detected by signal amplification strategy via the reaction with peroxidase-like activity of the probe. By employing the uniquely-designed Au–Pt nanozymes with tailored optimizations, an ultrasensitive method via LFA platform for detecting the HIV biomarker p24 was established with an LOD of 0.8 pg/mL.
Fig. 3.
Platinum nanozyme-mediated LFA strip to enhance signal intensity in colorimetric detection of p24 biomarker (HIV detection). (a) Platinum nanozyme – mediated LFA strip. The platinum nanocatalysts (PtNCs) immobilized onto test line oxidize CN/DAB (4-chloro-1-naphthol/3,3′-diaminobenzidine, tetrahydrochloride) substrate in the presence of H2O2 to form an insoluble black product which can be observed obviously by naked eyes, (b) comparison of dynamic linear ranges among LFA, ELISA, and PtNCs LFA, and (c) representative images of LFA strips before and after signal amplification (red box emphasizes the pre- and post-amplification for 50 and 100 pg/mL sample), corresponding to the varying p24-spiked levels in sera. The star indicates the lowest concentrations which can be recognized by naked eyes. Reproduced from Refs. [66] with permission of ACS Publications.
4.2. Peroxidase-mimicking MNPs for colorimetric pathogen detection
As an another potent approach to improve the sensitivity for detecting pathogen, magnetic separation-coupled assays has usually been employed for target enrichment. This strategy exploits inherent magnetic characteristics of MNPs to isolate target bacterial cells from fluid matrices such as food samples or biofluids. In fact, the use of MNPs in LFA assays is for both their magnetic properties and peroxidase-like activity for colorimetric immunological detection of targets with high sensitivity [17,64]. For instance, MNPs were successfully adopted to efficiently detect Ebola viruses within 30 min [70]. MNP nanoprobes are conjugated with antibodies to perform several functions, including identifying, enriching, as well as signaling. Due to peroxidase-like activity, MNPs improve sensitivity by oxidizing the peroxide substrate DAB to show a brown band. The nanozyme-mediated strip could detect a trace level of specific glycoprotein of Ebola virus as low as 1 ng/ml, showing 100-fold higher sensitivity than a typical LFA strip. The MNP-based LFA strip could provide early diagnosis of Ebola infection, as levels of virus in the bloodstream of infected patients is usually much higher.
In another study, MNP-based paper strips were used to detect Enterobacter sakazakii (E. sakazakii), a common pathogenic food-borne bacteria. E. sakazakii belongs to the Enterobacteriaceae family, is Gram-negative, rod-shaped, and does not form spores [73]. These bacteria are responsible for symptoms that can affect the bloodstream and central nervous system such as sepsis, meningitis, and necrotizing enterocolitis in infants. Therefore, developing methods to detect viable E. sakazakii is necessary. The sample was treated with propidium monoazide to remove dead cells to selectively monitor live bacterial cells only, prior to loop-mediated isothermal amplification (LAMP). The amplified DNA can then be visually detected on the MNP-mediated strip. The use of MNPs instead of colloidal AuNPs strikingly increased the colorimetric signal with a LOD of 10 CFU/mL.
5. Conjugated polymeric nanovesicle-mediated colorimetric pathogen detection
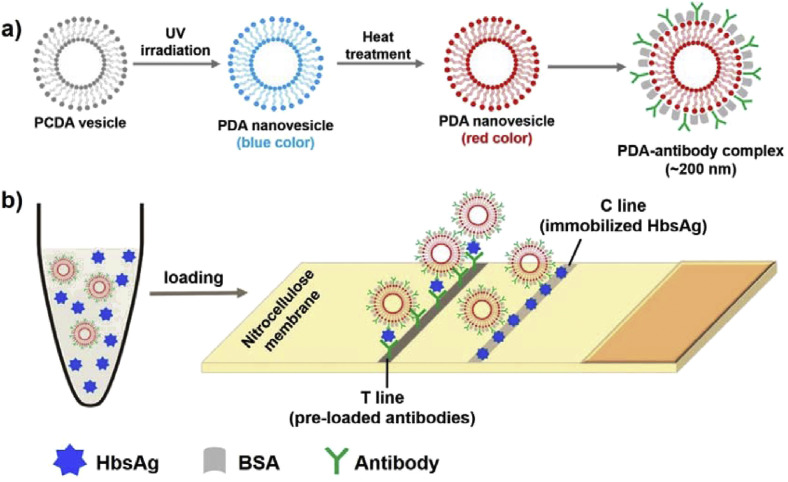
Conjugated polymers have unique chromatic properties that can be utilized for sensitive optical transduction. Polydiacetylene (PDA) is a commonly used conjugated polymer that is synthesized by 1,4–addition polymerization of diacetylene monomers under UV irradiation [74]. In a typical synthesis, nanoscopic vesicles or liposomes were formed through spontaneous self-assembly, having several hundreds of nanometers in their size [75]. Through simple amine modification on the monomers [76] or addition of specific ligands such as PEG [77] involved in the synthesis of PDA vesicles, the particle size was further decreased below 100 nm, which is advantageous to achieve higher sensitivity due to their much larger surface area for the reaction. PDA undergoes a color transformation from blue to red upon changes in various external conditions such as pH, temperature, organic solvent, and ligand-receptor interaction [74,78]. The interaction between PDA and the target induces steric strength that can alter PDA backbone arrangement to initiate the blue–to–red color transition. The distinct optical properties of PDA are appealing for pathogenic detection research [1,79].
PDA-based sensors are classified into solution-based and solid substrate-based platforms. The former utilizes PDA liposome solutions, but are prone to aggregation during long-term storage due to its unstable nature in liquid phase. In contrast, solid substrate-based sensors do not have the same limitation as PDA molecules are immobilized onto solid supports, which facilitate the utilization as POCT devices due to its simplicity, flexibility, and portability [79,80]. For example, an LFA strip utilizing antibody-conjugated red-colored PDA nanovesicles (~200 nm in diameter) was reported to selectively trace hepatitis B antigen (Fig. 4 ) [78]. Hepatitis B is a highly prevalent infectious disease, which is ranked as one of the top 10 leading causes of human death and caused by hepatitis B virus (HBV). Patients with this disease do not suffer from any pain during the initial phases until it further develops into chronic cancers if not properly treated. Thus, early diagnosis for detecting HBV infection is crucially important. Hepatitis B antibody, which specifically binds to surface antigen of HBV, was conjugated to PDA vesicles through covalent linkage, and the prepared PDA vesicle-antibody conjugate was proven to have sizes of ~200 nm. The PDA vesicle-antibody conjugates were then loaded on nitrocellulose membrane as a replacement of conventional AuNPs-antibody conjugates, enabling colorimetric detection of target antigen via antigen-antibody interaction and the concomitant red band on the strip from the bound PDA conjugates. The LOD was reported to be 1 ng/mL. PDA nanovesicles were also immobilized onto a polyvinylidene fluoride (PVDF) dipstick strip for quick and rapid detection of influenza A (H1N1) virus antigen [74]. In the presence of target antigen, the blue color of PDA nanovesicles immobilized on the strip changed into red due to the disruption of vesicle structure by the immunoreaction. In another recent study describing similar PDA vesicle-based paper strip for influenza A virus, smartphone software was utilized for image analysis to enable simple and quantitative analysis of low concentrations of influenza A virus [79]. The LOD of this PDA paper-based device ranged between 103 to 5 × 103 TCID50 (TCID50: 50% tissue culture infective dose).
Fig. 4.
(a) Structural design and formation of PDA-antibody complexes from 10, 12-pentacosadiynoic acid (PCDA) vesicle and (b) schematic illustration how to detect target hepatitis B surface antigen (HBsAg) on paper substrate. Redrawn from Ref. [78].
6. Conclusion and future prospects
The ongoing COVID-19 pandemic has put a heavy burden on healthcare services as well as economic growth. Widespread transmission of this disease has raised a question about early diagnostic methods for detecting viral infection since this can reduce the number of infected patients. Even developed countries have found huge obstacles in treating infectious patients since the number of patients is likely to outweigh healthcare infrastructures. Until now, the most reliable and sensitive methods for detecting pathogenic viruses and bacteria are based on PCR and ELISA. However, these techniques are time-consuming and require skilled technicians that are only feasible in advanced laboratories. PADs are powerful tools for providing fast, sensitive, and reliable assays, a key step in controlling and preventing the spread of diseases (Table 1 ). Further development of novel PADs exhibiting higher sensitivity, selectivity, and reliability for detecting disease targets is in high demand.
Table 1.
Summary of recent developments on PADs for colorimetric detection of pathogens using various nanomaterials.
| Materials | Type | Probe | Target | Sample Type | Receptor | Assay Time | LOD | Ref |
|---|---|---|---|---|---|---|---|---|
| Noble metal nanoparticles | μPAD | AuNPs + GE | Influenza A H1N1 virus | clinical sample | Antibody | 1 h | 2.7 × 103 PFU/mL | [41] |
| μPAD | AuNPs + GE | Influenza A H3N2 virus | clinical sample | Antibody | 1 h | 2.7 × 104 PFU/mL | [41] | |
| LFA | AuNPs + GE | S. enteritidis | milk | Antibody | 20 min | 104 CFU/mL | [42] | |
| LFA | Dual AuNPs | C. sakazakii | infant powder | Antibody | 15 min | 103 CFU/mL | [43] | |
| LFA | AgNPs | Yellow Fever virus | human serum | Antibody | _ | 150 ng/mL | [44] | |
| LFA | AgNPs | Ebola virus | human serum | Antibody | _ | 150 ng/mL | [44] | |
| LFA | AgNPs | Dengue virus | human serum | Antibody | _ | 150 ng/mL | [44] | |
| Dip stick | AuNPs | V. parahaemolyticus | oyster hemolymph | Antibody | 2 h | 4.66 × 105 CFU/mL | [45] | |
| μPAD | AuNPs + GE | GII.4 norovirus | Human fecal sample | Antibody | 10 min | 9.5 × 104 copies/mL | [81] | |
| LFA | AuNPs | S. typhimurium | milk, chicken, food | Aptamer | 10 min | 103 CFU/mL | [47] | |
| LFA | AuNPs | E. coli O157:H7 | milk, chicken, food | Aptamer | 10 min | 104 CFU/mL | [47] | |
| LFA | AuNPs | S. aureus | milk, chicken, food | Aptamer | 10 min | 104 CFU/mL | [47] | |
| LFA | AuNPs | Influenza A H5N2 virus | Ducks' feces | Aptamer | _ | 6 × 105 EID50/mL | [48] | |
| Dip stick | AuNPs | E. coli | whole blood serum | LBP | 10 min | 8 CFU/mL | [49] | |
| 3D μPAD | AgNPs | MERS-CoV | _ | PNA | _ | 1.53 nM, | [50] | |
| 3D μPAD | AgNPs | M. tuberculosis | _ | PNA | _ | 1.27 nM | [50] | |
| 3D μPAD | AgNPs | HPV | _ | PNA | _ | 1.03 nM | [50] | |
| Nanozymes | LFA | Pt–Au NPs | E. coli O157:H7 | _ | Antibody | 1 min | 100 CFU/mL | [69] |
| LFA | Pd–Pt NP | E. coli O157:H7 | milk | Antibody | _ | 0.87 × 102 CFU/mL | [65] | |
| LFA | AuNPs + GE | E. coli O157:H7 | milk | Antibody | _ | 1.25 × 101 CFU/mL | [67] | |
| LFA | AuNR@Pt | C. jejuni | milk, chicken | Antibody | _ | 75 CFU/mL | [71] | |
| LFA | Au–Pt | HIV | human blood serum | Antibody | Under 20 min | 0.8 pg/mL | [66] | |
| LFA | MNP | Ebola virus | human serum | Antibody | 30 min | 240 PFU/mL | [70] | |
| LFA | MNP | E. sakazakii | infant powder | Antibody | 1 h | 10 CFU/mL | [73] | |
| PDA nanovesicles | LFA | PDA | Hepatitis B virus | _ | Antibody | _ | 1 ng/mL, | [78] |
| Dip stick | PDA | Influenza A H1N1 virus | _ | Antibody | _ | _ | [74] |
AuNP: gold nanoparticle; AuNR: gold nanorod; AgNP: silver nanoparticle; CFU: colony-forming units; EID50: 50% egg infection dose; GE: gold enhancement; MNP: magnetic nanoparticle; PDA: polydiacetylene; PFU: plaque-forming units.
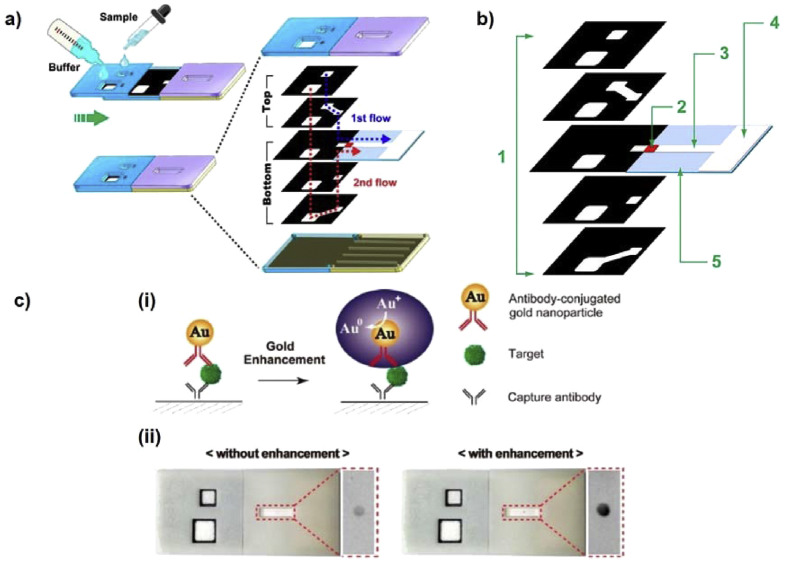
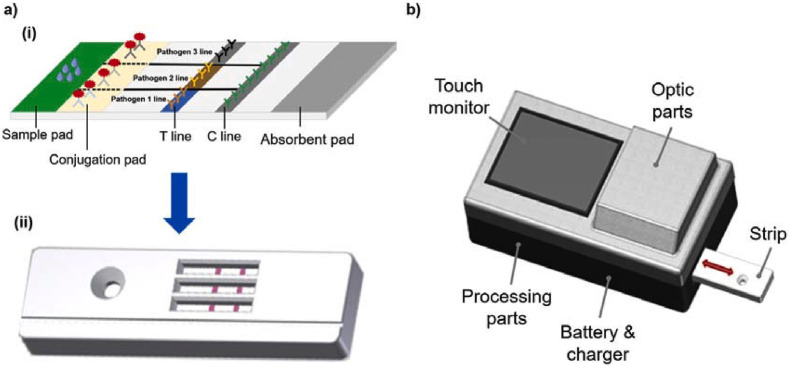
The increasing pressure of preventing spread of infectious diseases and stopping this pandemic has triggered a need to optimize and improve PADs. We believe that there are technical issues with nanomaterial-mediated PADs for colorimetric detection of pathogens that should be urgently addressed. First, the reliability and reproducibility of a device are the main criteria needed for commercialization. Optimizing a protocol for generating the devices can significantly reduce batch variations. For example, using nanoparticles with negligible size variation can remarkably strengthen the reliability of the strip. Conjugation of nanomaterials with antibodies and aptamers should be consistent to ensure that each strip has similar signal enhancement, leading to similar readouts. Second, the sensitivity of PADs has been a major barrier for commercialization. Many nanomaterials have been implemented in PADs to enhance diverse detection capabilities, particularly promoting colorimetric signal intensity. Compared with optical properties of noble metal nanoparticles, nanozymes can catalyze colorimetric reactions, amplify signal intensity, and yield higher sensitivity in PADs. However, employment of nanozymes require additional procedures, including chromogenic substrate addition and washing steps, which are tedious and inconvenient for end-users. To overcome this issue, multi-layered PADs could be used to support broader utilization of nanozyme-mediated PADs (Fig. 5 ) [81]. Integration of hydrophobic patterns and multiple layered 3D structures can be employed to eliminate multiple steps needed for signal amplification, allowing for fluid flow by passive capillary force. Optimizing paper geometry in combination with highly-active nanozymes can improve sensitivity, as well as practicality of using PADs. Third, multiplex approaches should be considered for simultaneous detection of multiple targets using PADs. Many infectious diseases are caused by co-existence of several pathogens and serotypes [82,83]. In many cases, detection of single pathogens is not sufficient for medical diagnoses, thus, rapid and simultaneous monitoring of multiple pathogens is desirable. Several strategies have been identified for developing multiplex LFA strips, such as single-array lateral flow biosensors with multiple dots, and parallel multiple-array lateral flow biosensors. Many multiple-array type assays have been commercialized [84], comprising of many single lateral flow strips in a parallel fashion within one plastic container to detect multiple targets. Nonetheless, the increase in device size is inevitable, resulting in higher amount of reagent utilization as well as overall cost. Thus, we believe that improving the design by incorporating multiple hydrophobic channels in a single lateral strip could be a potential resolution for reducing strip size and reagent usage. Simple wax printing or other methods for printing hydrophobic boundaries can therefore produce single LFA strips that can detect multiple pathogens with multiple specific antibodies and bioreceptors that are immobilized in separate regions (Fig. 6 ). Compared to LFAs, μPADs are highly advantageous for multiplex detection. The construction of 2D and 3D μPADs provides a network of microchannels that enables multiple functions. The design of 3D μPADs is better than 2D μPADs for multiplex assays since they are constructed from a stack of patterned layers, connected by a folding process [37]. By creating microchannels in each layer and stacking multiple layers together, the surface area for sample detection is increased. We predict that using nanomaterials in colorimetric, paper-based analytical devices can be broadly employed in healthcare for rapid, reliable, sensitive, and convenient detection of pathogenic viruses and bacteria in the near future.
Fig. 5.
(a) A schematic showing the design and working principles of 3D slip-PADs. A movable slipping design is used in conjunction with multiple layers stacked on top of one another. The device enables a one-step process for sequential fluid movement to the detection site. The black and white areas illustrate the hydrophobic wax boundary and hydrophilic fluid route, respectively. (b) Structural components of 3D slip-PADs. (1: 3D layers, 2: conjugate pad, 3: nitrocellulose membrane, 4: wicking pad, and 5: adhesive pad). (c) (i) representative scheme of gold enhancement for signal amplification on the 3D slip-PADs. The size-enlarged AuNPs resulted in signal intensification, and (ii) photographs of the PAD obtained before and after gold enhancement at a target IgG (500 ng/mL). Reproduced from Refs. [81] with permission of Nature Research.
Fig. 6.
(a) (i) A schematic showing a multiplexed version of an LFA strip by utilizing hydrophobic lines to separate different regions for various functionalities. In each region, one type of detection antibodies is immobilized onto test line and is able to specifically detect target pathogen, and (ii) expected multiplex colorimetric readouts of LFA device. (b) Expected point-of-care strip reader for rapid and convenient quantification of colorimetric signal obtained from LFA strip.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science and ICT (NRF- 2019R1A2C1087459) and by the Gachon University research fund of 2019 (GCU-2019-0812). This research was also supported and funded by the Korean National Police Agency [Project name: Development of visualization technology for biological evidence in crime scenes based on nano-bio technology/Project Number: PA-K000001-2019-401].
References
- 1.Choi Y., Hwang J.H., Lee S.Y. Recent trends in nanomaterials-based colorimetric detection of pathogenic bacteria and viruses. Small Methods. 2018;2:1700351. doi: 10.1002/smtd.201700351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National Authorities Coronavirus disease 2019 (COVID–19) Situation Report – 59. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- 3.The Visual and Data Journalism Team, Coronavirus: A visual guide to the pandemic. https://www.bbc.com/news/world-51235105 (accessed 19 March 2020).
- 4.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020:1–4. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu D.K., Pan Y., Cheng S., Hui K.P., Krishnan P., Liu Y., Ng D.Y., Wan C.K., Yang P., Wang Q. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikaeen G., Abbaszadeh S., Yousefinejad S. Application of nanomaterials in treatment, anti-infection and detection of coronaviruses. Nanomedicine. 2020;15:1501–1512. doi: 10.2217/nnm-2020-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhuang J., Yin J., Lv S., Wang B., Mu Y. Advanced “lab-on-a-chip” to detect viruses–Current challenges and future perspectives. Biosens. Bioelectron. 2020:112291. doi: 10.1016/j.bios.2020.112291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Organization W.H. World Health Organization; 2015. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015. [Google Scholar]
- 9.Bhardwaj N., Bhardwaj S.K., Bhatt D., Lim D.K., Kim K.-H., Deep A. Optical detection of waterborne pathogens using nanomaterials. Trends Anal. Chem. 2019;113:280–300. [Google Scholar]
- 10.M. P. Miagostovich, F.F. Ferreira, F.R. Guimarães, T.M. Fumian, L. Diniz-Mendes, S.L.B. Luz, L.A. Silva, J.P.G. Leite, Molecular Detection and Characterization of Gastroenteritis Viruses Occurring Naturally in the Stream Waters of Manaus, Central Amazonia, Brazil. [DOI] [PMC free article] [PubMed]
- 11.Nasseri B., Soleimani N., Rabiee N., Kalbasi A., Karimi M., Hamblin M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018;117:112–128. doi: 10.1016/j.bios.2018.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kant K., Shahbazi M.-A., Dave V.P., Ngo T.A., Chidambara V.A., Than L.Q., Bang D.D., Wolff A. Microfluidic devices for sample preparation and rapid detection of foodborne pathogens. Biotechnol. Adv. 2018;36:1003–1024. doi: 10.1016/j.biotechadv.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Liu L., Yang D., Liu G. Signal amplification strategies for paper-based analytical devices. Biosens. Bioelectron. 2019;136:60–75. doi: 10.1016/j.bios.2019.04.043. [DOI] [PubMed] [Google Scholar]
- 14.Mahato K., Srivastava A., Chandra P. Paper based diagnostics for personalized health care: emerging technologies and commercial aspects. Biosens. Bioelectron. 2017;96:246–259. doi: 10.1016/j.bios.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Hristov D.R., Rodriguez-Quijada C., Gomez-Marquez J., Hamad-Schifferli K. Designing paper-based immunoassays for biomedical applications. Sensors. 2019;19:554. doi: 10.3390/s19030554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang Y., Ramasamy R.P. Current and prospective methods for plant disease detection. Biosensors. 2015;5:537–561. doi: 10.3390/bios5030537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majdinasab M., Hayat A., Marty J.L. Aptamer-based assays and aptasensors for detection of pathogenic bacteria in food samples. Trends Anal. Chem. 2018;107:60–77. [Google Scholar]
- 18.Sun J., Huang J., Li Y., Lv J., Ding X. A simple and rapid colorimetric bacteria detection method based on bacterial inhibition of glucose oxidase-catalyzed reaction. Talanta. 2019;197:304–309. doi: 10.1016/j.talanta.2019.01.039. [DOI] [PubMed] [Google Scholar]
- 19.Srisa-Art M., Boehle K.E., Geiss B.J., Henry C.S. Highly sensitive detection of Salmonella typhimurium using a colorimetric paper-based analytical device coupled with immunomagnetic separation. Anal. Chem. 2018;90:1035–1043. doi: 10.1021/acs.analchem.7b04628. [DOI] [PubMed] [Google Scholar]
- 20.Adkins J.A., Boehle K., Friend C., Chamberlain B., Bisha B., Henry C.S. Colorimetric and electrochemical bacteria detection using printed paper-and transparency-based analytic devices. Anal. Chem. 2017;89:3613–3621. doi: 10.1021/acs.analchem.6b05009. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y., Ren J., Qu X. Catalytically active nanomaterials: a promising candidate for artificial enzymes. Acc. Chem. Res. 2014;47:1097–1105. doi: 10.1021/ar400250z. [DOI] [PubMed] [Google Scholar]
- 22.Sharifi S., Vahed S.Z., Ahmadian E., Dizaj S.M., Eftekhari A., Khalilov R., Ahmadi M., Hamidi-Asl E., Labib M. Detection of pathogenic bacteria via nanomaterials-modified aptasensors. Biosens. Bioelectron. 2020:111933. doi: 10.1016/j.bios.2019.111933. [DOI] [PubMed] [Google Scholar]
- 23.Cheon H.J., Adhikari M.D., Chung M., Tran T.D., Kim J., Kim M.I. Magnetic nanoparticles-embedded enzyme-inorganic hybrid nanoflowers with enhanced peroxidase-like activity and substrate channeling for glucose biosensing. Adv. Healthcare Mater. 2019;8:1801507. doi: 10.1002/adhm.201801507. [DOI] [PubMed] [Google Scholar]
- 24.Cho S., Lee S.M., Shin H.Y., Kim M.S., Seo Y.H., Cho Y.K., Lee J., Lee S.P., Kim M.I. Highly sensitive colorimetric detection of allergies based on an immunoassay using peroxidase-mimicking nanozymes. Analyst. 2018;143:1182–1187. doi: 10.1039/c7an01866e. [DOI] [PubMed] [Google Scholar]
- 25.Chen J., Andler S.M., Goddard J.M., Nugen S.R., Rotello V.M. Integrating recognition elements with nanomaterials for bacteria sensing. Chem. Soc. Rev. 2017;46:1272–1283. doi: 10.1039/c6cs00313c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malekzad H., Zangabad P.S., Mirshekari H., Karimi M., Hamblin M.R. Noble metal nanoparticles in biosensors: recent studies and applications. Nanotechnol. Rev. 2017;6:301–329. doi: 10.1515/ntrev-2016-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu J., Wang S., Wang L., Li F., Pingguan-Murphy B., Lu T.J., Xu F. Advances in paper-based point-of-care diagnostics. Biosens. Bioelectron. 2014;54:585–597. doi: 10.1016/j.bios.2013.10.075. [DOI] [PubMed] [Google Scholar]
- 28.Millipore E. vol. 29. EMD Millipore Corporation; Billerica, MA, USA: 2013. pp. 702–707. (Rapid Lateral Flow Test Strips: Considerations for Product Development). [Google Scholar]
- 29.Bhattacharya S., Kumar S., Agarwal A.K. Springer Nature; Singapore: 2019. Paper Microfluidics Theory and Applications. [Google Scholar]
- 30.Yamada K., Shibata H., Suzuki K., Citterio D. Toward practical application of paper-based microfluidics for medical diagnostics: state-of-the-art and challenges. Lab Chip. 2017;17:1206–1249. doi: 10.1039/c6lc01577h. [DOI] [PubMed] [Google Scholar]
- 31.Simerville J.A., Maxted W.C., Pahira J.J. Urinalysis: a comprehensive review. Am. Fam. Physician. 2005;71:1153–1162. [PubMed] [Google Scholar]
- 32.Richard Kock, Urinalysis Test Strips: How They Work, Which To Buy. https://puriapp.com/urinalysis-test-strips/(accessed 5 April 2020).
- 33.Whitesides G.M. Cool, or simple and cheap? Why not both? Lab Chip. 2013;13:11–13. doi: 10.1039/c2lc90109a. [DOI] [PubMed] [Google Scholar]
- 34.Lim H., Jafry A.T., Lee J. Fabrication, flow control, and applications of microfluidic paper-based analytical devices. Molecules. 2019;24:2869. doi: 10.3390/molecules24162869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maejima K., Tomikawa S., Suzuki K., Citterio D. Inkjet printing: an integrated and green chemical approach to microfluidic paper-based analytical devices. RSC Adv. 2013;3:9258–9263. [Google Scholar]
- 36.Carrilho E., Martinez A.W., Whitesides G.M. Understanding wax printing: a simple micropatterning process for paper-based microfluidics. Anal. Chem. 2009;81:7091–7095. doi: 10.1021/ac901071p. [DOI] [PubMed] [Google Scholar]
- 37.Chen C.-A., Yeh W.-S., Tsai T.-T., Chen C.-F. Three-dimensional origami paper-based device for portable immunoassay applications. Lab Chip. 2019;19:598–607. doi: 10.1039/c8lc01255e. [DOI] [PubMed] [Google Scholar]
- 38.Li X., Liu X. Fabrication of three-dimensional microfluidic channels in a single layer of cellulose paper, Microfluid. Nanofluidics. 2014;16:819–827. [Google Scholar]
- 39.Yu L., Li N. Noble metal nanoparticles-based colorimetric biosensor for visual quantification: a mini review. Chemosensors. 2019;7:53–75. [Google Scholar]
- 40.Cordeiro M., Ferreira Carlos F., Pedrosa P., Lopez A., Baptista P.V. Gold nanoparticles for diagnostics: advances towards points of care. Diagnostics. 2016;6:43. doi: 10.3390/diagnostics6040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei K.F., Huang C.-H., Kuo R.-L., Chang C.-K., Chen K.-F., Tsao K.-C., Tsang N.-M. Paper-based enzyme-free immunoassay for rapid detection and subtyping of influenza A H1N1 and H3N2 viruses. Anal. Chim. Acta. 2015;883:37–44. doi: 10.1016/j.aca.2015.02.071. [DOI] [PubMed] [Google Scholar]
- 42.Bu T., Huang Q., Yan L., Huang L., Zhang M., Yang Q., Yang B., Wang J., Zhang D. Ultra technically-simple and sensitive detection for Salmonella enteritidis by immunochromatographic assay based on gold growth. Food Contr. 2018;84:536–543. [Google Scholar]
- 43.Pan R., Jiang Y., Sun L., Wang R., Zhuang K., Zhao Y., Wang H., Ali M.A., Xu H., Man C. Gold nanoparticle-based enhanced lateral flow immunoassay for detection of Cronobacter sakazakii in powdered infant formula. J. Dairy Sci. 2018;101:3835–3843. doi: 10.3168/jds.2017-14265. [DOI] [PubMed] [Google Scholar]
- 44.Yen C.-W., de Puig H., Tam J.O., Gómez-Márquez J., Bosch I., Hamad-Schifferli K., Gehrke L. Multicolored silver nanoparticles for multiplexed disease diagnostics: distinguishing dengue, yellow fever, and Ebola viruses. Lab Chip. 2015;15:1638–1641. doi: 10.1039/c5lc00055f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez-Quijada C., Lyons C., Santamaria C., Quinn S., Tlusty M., Shiaris M., Hamad-Schifferli K. Optimization of paper-based nanoparticle immunoassays for direct detection of the bacterial pathogen V. parahaemolyticus in oyster hemolymph. Anal. Methods. 2020;12:3056–3063. doi: 10.1039/d0ay00725k. [DOI] [PubMed] [Google Scholar]
- 46.Yoo S.M., Kim D.-K., Lee S.Y. Aptamer-functionalized localized surface plasmon resonance sensor for the multiplexed detection of different bacterial species. Talanta. 2015;132:112–117. doi: 10.1016/j.talanta.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Lu C., Gao X., Chen Y., Ren J., Liu C. Aptamer-based lateral flow test strip for the simultaneous detection of Salmonella typhimurium, Escherichia coli O157:H7 and Staphylococcus aureus. Anal. Lett. 2019:1–4. [Google Scholar]
- 48.Kim S.H., Lee J., Lee B.H., Song C.-S., Gu M.B. Specific detection of avian influenza H5N2 whole virus particles on lateral flow strips using a pair of sandwich-type aptamers. Biosens. Bioelectron. 2019;134:123–129. doi: 10.1016/j.bios.2019.03.061. [DOI] [PubMed] [Google Scholar]
- 49.Shafiee H., Asghar W., Inci F., Yuksekkaya M., Jahangir M., Zhang M.H., Durmus N.G., Gurkan U.A., Kuritzkes D.R., Demirci U. Paper and flexible substrates as materials for biosensing platforms to detect multiple biotargets. Sci. Rep. 2015;5:8719. doi: 10.1038/srep08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teengam P., Siangproh W., Tuantranont A., Vilaivan T., Chailapakul O., Henry C.S. Multiplex paper-based colorimetric DNA sensor using pyrrolidinyl peptide nucleic acid-induced AgNPs aggregation for detecting MERS-CoV, MTB, and HPV oligonucleotides. Anal. Chem. 2017;89:5428–5435. doi: 10.1021/acs.analchem.7b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suaifan G.A., Alhogail S., Zourob M. Rapid and low-cost biosensor for the detection of Staphylococcus aureus. Biosens. Bioelectron. 2017;90:230–237. doi: 10.1016/j.bios.2016.11.047. [DOI] [PubMed] [Google Scholar]
- 52.Suaifan G.A., Alhogail S., Zourob M. Paper-based magnetic nanoparticle-peptide probe for rapid and quantitative colorimetric detection of Escherichia coli O157: H7. Biosens. Bioelectron. 2017;92:702–708. doi: 10.1016/j.bios.2016.10.023. [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Viloca M., Gao J., Karplus M., Truhlar D.G. How enzymes work: analysis by modern rate theory and computer simulations. Science. 2004;3:186–195. doi: 10.1126/science.1088172. [DOI] [PubMed] [Google Scholar]
- 54.Chapman J., Ismail A.E., Dinu C.Z. Industrial applications of enzymes: recent advances, techniques, and outlooks. Catalysts. 2018;8:238–248. [Google Scholar]
- 55.Huang Y., Ren J., Qu X. Nanozymes: classification, catalytic mechanisms, activity regulation, and applications. Chem. Rev. 2019;119:4357–4412. doi: 10.1021/acs.chemrev.8b00672. [DOI] [PubMed] [Google Scholar]
- 56.Gao L., Zhuang J., Nie L., Zhang J., Zhang Y., Gu N., Wang T., Feng J., Yang D., Perrett S. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat. Nanotechnol. 2007;2:577–583. doi: 10.1038/nnano.2007.260. [DOI] [PubMed] [Google Scholar]
- 57.Nath I., Chakraborty J., Verpoort F. Metal organic frameworks mimicking natural enzymes: a structural and functional analogy. Chem. Soc. Rev. 2016;45:4127–4170. doi: 10.1039/c6cs00047a. [DOI] [PubMed] [Google Scholar]
- 58.Batule B.S., Park K.S., Gautam S., Cheon H.J., Kim M.I., Park H.G. Intrinsic peroxidase-like activity of sonochemically synthesized protein copper nanoflowers and its application for the sensitive detection of glucose. Sensor. Actuator. B Chem. 2019;283:749–754. [Google Scholar]
- 59.Sun H., Zhou Y., Ren J., Qu X. Carbon nanozymes: enzymatic properties, catalytic mechanism, and applications. Angew. Chem. Int. Ed. 2018;57:9224–9237. doi: 10.1002/anie.201712469. [DOI] [PubMed] [Google Scholar]
- 60.Wu J., Wang X., Wang Q., Lou Z., Li S., Zhu Y., Qin L., Wei H. Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes (II) Chem. Soc. Rev. 2019;48:1004–1076. doi: 10.1039/c8cs00457a. [DOI] [PubMed] [Google Scholar]
- 61.Gooding J.J. Can nanozymes have an impact on sensing? ACS Sens. 2019;4:2213–2214. doi: 10.1021/acssensors.9b01760. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Z., Zhang X., Liu B., Liu J. Molecular imprinting on inorganic nanozymes for hundred-fold enzyme specificity. J. Am. Chem. Soc. 2017;139:5412–5419. doi: 10.1021/jacs.7b00601. [DOI] [PubMed] [Google Scholar]
- 63.Kim M.S., Lee J., Kim H.S., Cho A., Shim K.H., Le T.N., An S.S.A., Han J.W., Kim M.I., Lee J. Heme cofactor-resembling Fe–N single site embedded graphene as nanozymes to selectively detect H2O2 with high sensitivity. Adv. Funct. Mater. 2020;30:1905410. [Google Scholar]
- 64.Niu X., Cheng N., Ruan X., Du D., Lin Y. Nanozyme-based immunosensors and immunoassays: recent developments and future trends. J. Electrochem. Soc. 2019;167 [Google Scholar]
- 65.Han J., Zhang L., Hu L., Xing K., Lu X., Huang Y., Zhang J., Lai W., Chen T. Nanozyme-based lateral flow assay for the sensitive detection of Escherichia coli O157: H7 in milk. J. Dairy Sci. 2018;101:5770–5779. doi: 10.3168/jds.2018-14429. [DOI] [PubMed] [Google Scholar]
- 66.Loynachan C.N., Thomas M.R., Gray E.R., Richards D.A., Kim J., Miller B.S., Brookes J.C., Agarwal S., Chudasama V., McKendry R.A. Platinum nanocatalyst amplification: redefining the gold standard for lateral flow immunoassays with ultrabroad dynamic range. ACS Nano. 2018;12:279–288. doi: 10.1021/acsnano.7b06229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu J., Zhou Y., Huang X., Zhang W., Wu Y., Fang H., Zhang C., Xiong Y. Dramatically enhanced immunochromatographic assay using cascade signal amplification for ultrasensitive detection of Escherichia coli O157: H7 in milk. J. Agric. Food Chem. 2020;68:1118–1125. doi: 10.1021/acs.jafc.9b07076. [DOI] [PubMed] [Google Scholar]
- 68.Huang L., Sun D.W., Pu H., Wei Q. Development of nanozymes for food quality and safety detection: principles and recent applications. Compr. Rev. Food Sci. 2019;18:1496–1513. doi: 10.1111/1541-4337.12485. [DOI] [PubMed] [Google Scholar]
- 69.Jiang T., Song Y., Wei T., Li H., Du D., Zhu M.-J., Lin Y. Sensitive detection of Escherichia coli O157: H7 using Pt–Au bimetal nanoparticles with peroxidase-like amplification. Biosens. Bioelectron. 2016;77:687–694. doi: 10.1016/j.bios.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 70.Duan D., Fan K., Zhang D., Tan S., Liang M., Liu Y., Zhang J., Zhang P., Liu W., Qiu X. Nanozyme-strip for rapid local diagnosis of Ebola. Biosens. Bioelectron. 2015;74:134–141. doi: 10.1016/j.bios.2015.05.025. [DOI] [PubMed] [Google Scholar]
- 71.He D., Wu Z., Cui B., Xu E., Jin Z. Establishment of a dual mode immunochromatographic assay for Campylobacter jejuni detection. Food Chem. 2019;289:708–713. doi: 10.1016/j.foodchem.2019.03.106. [DOI] [PubMed] [Google Scholar]
- 72.Farzin L., Shamsipur M., Samandari L., Sheibani S. Talanta; 2019. HIV Biosensors for Early Diagnosis of Infection: the Intertwine of Nanotechnology with Sensing Strategies; p. 120201. [DOI] [PubMed] [Google Scholar]
- 73.Zhang L., Chen Y., Cheng N., Xu Y., Huang K., Luo Y., Wang P., Duan D., Xu W. Ultrasensitive detection of viable Enterobacter sakazakii by a continual cascade nanozyme biosensor. Anal. Chem. 2017;89:10194–10200. doi: 10.1021/acs.analchem.7b01266. [DOI] [PubMed] [Google Scholar]
- 74.Jeong J.-p., Cho E., Yun D., Kim T., Lee I.-S., Jung S. Label-free colorimetric detection of influenza antigen based on an antibody-polydiacetylene conjugate and its coated polyvinylidene difluoride membrane. Polymers. 2017;9:127. doi: 10.3390/polym9040127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qian X., Städler B. Recent developments in polydiacetylene-based sensors. Chem. Mater. 2019;31:1196–1222. [Google Scholar]
- 76.Thongmalai W., Eaidkong T., Ampornpun S., Mungkarndee R., Tumcharern G., Sukwattanasinitt M., Wacharasindhu S. Polydiacetylenes carrying amino groups for colorimetric detection and identification of anionic surfactants. J. Mater. Chem. 2011;21:16391–16397. [Google Scholar]
- 77.Guo C., Zeng L., Liu S., Chen Q., Dai Z., Wu X. In vitro evaluation and finite element simulation of drug release from polydiacetylene-polyethylene glycol stearate nanovesicles. J. Nanosci. Nanotechnol. 2012;12:245–251. doi: 10.1166/jnn.2012.5136. [DOI] [PubMed] [Google Scholar]
- 78.Roh J., Lee S.Y., Park S., Ahn D.J. Polydiacetylene/anti-HBs complexes for visible and fluorescent detection of hepatitis B surface antigen on a nitrocellulose membrane. Chem. Asian J. 2017;12:2033–2037. doi: 10.1002/asia.201700769. [DOI] [PubMed] [Google Scholar]
- 79.Son S.U., Seo S.B., Jang S., Choi J., Lim J.-W., Lee D.K., Kim H., Seo S., Kang T., Jung J. Naked-eye detection of pandemic influenza a (pH1N1) virus by polydiacetylene (PDA)-based paper sensor as a point-of-care diagnostic platform. Sensor. Actuator. B Chem. 2019;291:257–265. [Google Scholar]
- 80.Yoon B., Lee S., Kim J.-M. Recent conceptual and technological advances in polydiacetylene-based supramolecular chemosensors. Chem. Soc. Rev. 2009;38:1958–1968. doi: 10.1039/b819539k. [DOI] [PubMed] [Google Scholar]
- 81.Han K.N., Choi J.-S., Kwon J. Three-dimensional paper-based slip device for one-step point-of-care testing. Sci. Rep. 2016;6:25710. doi: 10.1038/srep25710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao Y., Wang H., Zhang P., Sun C., Wang X., Wang X., Yang R., Wang C., Zhou L. Rapid multiplex detection of 10 foodborne pathogens with an up-converting phosphor technology-based 10-channel lateral flow assay. Sci. Rep. 2016;6:21342. doi: 10.1038/srep21342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Safenkova I.V., Panferov V.G., Panferova N.A., Varitsev Y.A., Zherdev A.V., Dzantiev B.B. Alarm lateral flow immunoassay for detection of the total infection caused by the five viruses. Talanta. 2019;195:739–744. doi: 10.1016/j.talanta.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 84.Li J., Macdonald J. Multiplexed lateral flow biosensors: technological advances for radically improving point-of-care diagnoses. Biosens. Bioelectron. 2016;83:177–192. doi: 10.1016/j.bios.2016.04.021. [DOI] [PubMed] [Google Scholar]