Abstract
Background
To develop a high-efficiency pulmonary nodule computer-aided detection (CAD) method for localization and diameter estimation.
Methods
The developed CAD method centralizes a novel convolutional neural network (CNN) algorithm, You Only Look Once (YOLO) v3, as a deep learning approach. This method is featured by two distinct properties: (I) an automatic multi-scale feature extractor for nodule feature screening, and (II) a feature-based bounding box generator for nodule localization and diameter estimation. Two independent studies were performed to train and evaluate this CAD method. One study comprised of a computer simulation that utilized computer-based ground truth. In this study, 300 CT scans were simulated by Cardiac-torso (XCAT) digital phantom. Spherical nodules of various sizes (i.e., 3–10 mm in diameter) were randomly implanted within the lung region of the simulated images—the second study utilized human-based ground truth in patients. The CAD method was developed by CT scans sourced from the LIDC-IDRI database. CT scans with slice thickness above 2.5 mm were excluded, leaving 888 CT images for analysis. A 10-fold cross-validation procedure was implemented in both studies to evaluate network hyper-parameterization and generalization. The overall accuracy of the CAD method was evaluated by the detection sensitivities, in response to average false positives (FPs) per image. In the patient study, the detection accuracy was further compared against 9 recently published CAD studies using free-receiver response operating characteristic (FROC) curve analysis. Localization and diameter estimation accuracies were quantified by the mean and standard error between the predicted value and ground truth.
Results
The average results among the 10 cross-validation folds in both studies demonstrated the CAD method achieved high detection accuracy. The sensitivity was 99.3% (FPs =1), and improved to 100% (FPs =4) in the simulation study. The corresponding sensitivities were 90.0% and 95.4% in the patient study, displaying superiority over several conventional and CNN-based lung nodule CAD methods in the FROC curve analysis. Nodule localization and diameter estimation errors were less than 1 mm in both studies. The developed CAD method achieved high computational efficiency: it yields nodule-specific quantitative values (i.e., number, existence confidence, central coordinates, and diameter) within 0.1 s for 2D CT slice inputs.
Conclusions
The reported results suggest that the developed lung pulmonary nodule CAD method possesses high accuracies of nodule localization and diameter estimation. The high computational efficiency enables its potential clinical application in the future.
Keywords: Computer-aided detection (CAD), pulmonary nodule, deep learning
Introduction
Automated pulmonary nodule detection has been a longstanding topic for lung cancer diagnosis. The implementation of computer-aided detection (CAD) systems for nodule detection is a hallmark endeavor used to optimize efficiencies and cost during routine clinical practice. Since the 1980s, conventional CAD studies have investigated multiple methods to detect nodule candidates [Hessian matrix (1-3); Stable 3D Mass-Spring Models (4); thresholding (5); 3D template matching (6)] and minimizing positive detection rates [support vector machine (SVM) (1); neural network (7)]. However, the high false-positive rates and feature annotation costs have remained as hurdles to their clinical implementation.
Recently, deep learning techniques have become popular in academia and industry. Convolutional neural networks (CNN) is the most common deep learning technique in imaging science, with the ability to perform pixel-wise feature extraction (8). Their multiple applications are evidenced by their use in self-driving cars, image segmentation, and facial recognition. Several studies have applied CNN architectures to nodule classification challenges (9-13). However, as they are typically constrained by their binary output (i.e., exist/absence, benign/malignant), these classification systems have not been able to provide detailed nodule information, such as anatomic location and diameters. As such, preliminary two-stage CNN-based CAD systems were developed for pulmonary nodule detection (14,15). In general, these methods consisted of two sub-systems, responsible for (I) the detection of suspicious nodules and (II) the reduction of false-positive rates. Due to the two-stage design, these CAD systems have complex detection procedures and are therefore prone to exceedingly high computational costs. As an attempt to address the issues associated with two-stage CAD systems, a few single-stage CAD systems for lung nodule detection have been reported with reduced computational costs (16,17). However, these single-stage CAD systems were associated with high false-positive rates compared to the two-stage systems. Also, computational costs are high for 3D volumetric detection (18). Furthermore, the ability of these CAD systems to determine nodule anatomic localization and diameter measurement has not been investigated.
In this study, we developed a single-stage pulmonary nodule CAD method based on a novel CNN algorithm—You Only Look Once (YOLO) (19). YOLO has been used in multiple diagnostic modalities, including digital mammograms (20,21), lung X-ray and computed tomography (CT) (22,23), and electroencephalography (EEG) (24). The proposed method customized the latest YOLO v3 algorithm as a CNN implementation (25). The developed method can simultaneously achieve nodule localization and diameter measurements with streamlined computational efficiency using a light-weight architecture. The accuracy of the developed method was first evaluated in a computer simulation study, using a digital phantom. It was subsequently examined in a patient study where data was extracted from a public database, and compared with 9 current lung nodule detection methods.
Methods
CAD method design
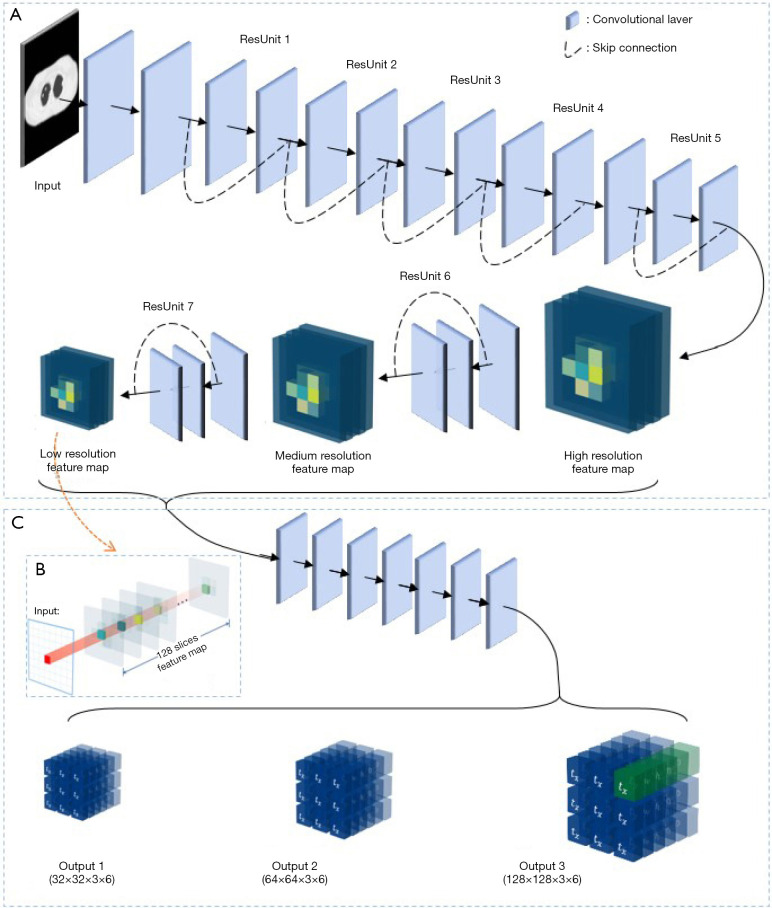
The developed CAD method customized a YOLO v3 CNN algorithm for the detection of pulmonary nodules. Figure 1 demonstrates a schematic illustration of the CNN architecture. In general, the network consists of two major components: (I) a feature extractor that screens nodule characteristics among the input data, and (II) a bounding box generator that determines nodule coordinates and diameter. This CAD method takes 2D axial CT slices as input and yields nodule-specific quantitative values (i.e., number, existence confidence, central coordinates, and diameter) as output.
Figure 1.
Convolutional neural networks (CNN) structure illustration of the developed computer-aided detection (CAD) method. (A) Feature extractor; (B) extension version of the feature map; (C) bounding box generator.
As shown in Figure 1A, the feature extractor is, more specifically, a residual network that contains seven residual units (ResUnit) (26). Each ResUnit has two or three convolutional layers with a skip connection design. A series of pooling layers in the ResUnits (ResUnit 5, 6, and 7) allow the feature extractor to screen potential nodules across three spatial scales. Three feature maps are subsequently generated by the feature extractor with 1/4, 1/8, and 1/16 of the input image resolution. The information stored at each site within the correlating feature space is responsible for a confined input range (i.e., it is spatially dependent). In the low-, medium-, and high-resolution feature maps, the voxel range coverage is 4×4, 8×8, and 16×16 pixels of the input image, respectively. Each feature map has 128 feature slices, as demonstrated in Figure 1B.
The bounding box design is then utilized to describe the nodule location and diameter. As demonstrated in Figure 1C, a seven-layer generator network utilized feature maps as inputs to predict the parameters of the bounding box. These encompassed central coordinates (), height and width (), and nodule score (). Bounding boxes were derived from anchor boxes, which were pre-defined bounding boxes at each position in the feature spaces. By associating the feature information at each map position, the bounding box generator predicted the values of coordinate shifts () and length changes () from the anchor box to bounding boxes. Therefore, the central coordinates () were predicted by,
| [1] |
and
| [2] |
where, and were the coordinates of the anchor boxes in the feature space. The bounding box lengths () were determined by,
| [3] |
and
| [4] |
where, and were the lengths of the anchor boxes. The nodule score () was calculated by the multifaction of two variables: the object score () and the nodule class score (c), which indicated the presence of an object in a feature position and the category of the detected object independently. For each feature map, 3 anchor boxes are designed at each position in the feature space. As seen in Table 1, they have different sizes to measure nodules across a wide range of diameters. As multiple anchor boxes at each voxel position may create detection redundancy, a non-maximum suppression algorithm was used when a nodule was detected by several bounding boxes simultaneously (27). This algorithm calculated the overlap among all detected bounding boxes. Bounding boxes that exceeded an overlap threshold were excluded.
Table 1. Size of anchor boxes in high-/medium-/low-resolution feature maps.
| Feature map resolution | Anchor boxes | Heights (pixels) | Width (pixels) |
|---|---|---|---|
| High | 1 | 8 | 8 |
| 2 | 10 | 10 | |
| 3 | 12 | 12 | |
| Medium | 4 | 15 | 15 |
| 5 | 20 | 20 | |
| 6 | 25 | 25 | |
| Low | 7 | 30 | 30 |
| 8 | 35 | 35 | |
| 9 | 40 | 40 |
The loss function used for network training includes three terms (i.e., object loss (), class loss (), and bounding box loss (), and was mathematically defined as,
| [5] |
where referred to the weighting factor for bounding box loss. Object loss () was defined by,
| [6] |
where S was the feature map resolution, and B represented the number of bounding boxes in each feature map. was a binary value that indicated the existence of an object inside the jth bounding box of ith feature position. is a binary function and is triggered when was bigger than the threshold (τ). Class loss () was defined by,
| [7] |
where BCE represented binary cross-entropy function. represented the predicted class score of category m inside the jth bounding box of ith feature position and was the corresponding ground truth. Bounding box loss () evaluated the difference between the predicted bounding box parameters () and the ground truth (). It was mathematically defined as,
| [8] |
where SE is the square error function.
Experiment design
The proposed CAD method was trained and evaluated via two independent studies: (I) a computer simulation study and (II) a patient study from a public database. These two studies assessed the CAD performance in the computer-based ground truth and human-based ground truth, separately.
In the computer simulation study, 300 3D CT scans containing detailed anatomical information were simulated using the Cardiac-torso (XCAT) digital phantom environment (28). Spherical nodules of various sizes (i.e., 3–10 mm in diameter) were randomly implanted within the lung region of these simulated images. Transverse CT slices that intersect with the center of these spherical nodules were extracted to form the dataset, and a 10-fold cross-validation procedure was implemented to evaluate network hyper-parameterization and generalization.
In the patient study, patient data from the lung image database consortium and image database resource initiative (LIDC–IDRI) (29) were used. The LIDC–IDRI database has 1,018 thoracic CT scans with corresponding nodule information. In this study, CT images with a slice thickness greater than 2.5 mm were excluded, leaving 888 CT images for analysis. Nodule information was marked by four experienced radiologists into one of three groups: (I) no nodule, (II) nodules <3 mm, and (III) nodules ≥3 mm. Among these CT images, 1,186 nodules were considered as positive examples by the criteria of nodule sizes above 3 mm and marked by at least three out of four radiologists. The central transverse CT slices between each nodule’s upper and lower boundary in axial direction were used for 10-fold cross-validation.
The simulated data was generated by digital phantom and the patient data was obtained from a publicly available open-source database, namely LIDC-IDRI database (29). Thus, no ethics approval of an institutional review board or ethics committees was required for this study. The authors acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health, and their critical role in the creation of the free publicly available LIDC/IDRI Database used in this study.
Evaluation method
The average performance among the 10 cross-validation folds was used for evaluation in both the computer simulation study and the patient study. Evaluation metrics included nodule detection accuracy, nodule localization accuracy, and nodule diameter measurement accuracy.
Detection accuracy was assessed based on the sensitivity of nodule identification. Under different nodule score thresholds, nodule detection sensitivities, and the corresponding average false positives (FPs) per image were calculated in the 10 testing folds. Seven sensitivity results (FPs = 1/8, 1/4, 1/2, 1, 2, 4, 8) were reported in two studies. Also, the detection accuracy in the patient study was compared against 9 recently published CAD studies which were developed using the LIDC-IDRI database. The techniques used in these methods included feature-based conventional techniques (30,31), two-dimensional (2D) CNN-based techniques [e.g., regional-based CNN (16), U-NET (32)], and three-dimensional (3D) CNN-based techniques (14,17). Free-receiver response operating characteristic (FROC) curve analysis was used in this comparison study (33). Specifically, detection sensitivities under a wide range of FPs (0<FPs≤8) were acquired by evaluating the proposed method with multiple nodule score thresholds (). As for the methods used in the comparison study, 7 sensitivity results covering the same FPs range (FPs = 1/8, 1/4, 1/2, 1, 2, 4, 8) were acquired from their original reports.
Localization accuracy was quantified by central coordinate shifts between the predicted nodule bounding boxes and the ground truth bounding boxes, expressed as the mean value of shifts in x/y direction. Also, 2D histograms were plotted to visualize the spatial deviation in nodule localization.
The standard error was used in evaluating the diameter measurement accuracy,
| [9] |
where, was the diameter prediction of ith nodule, and was the corresponding ground truth value. n was the total number of nodules.
Activation maps from the two studies were produced to investigate the performance difference between the simulation database and the patient database. To align with the page space requirements of the manuscript, only activation maps generated by the first 10 convolutional layers were reported.
Results
Table 2 summarizes the nodule detection sensitivity results under different false positives (FPs), per image. In the computer simulation study, the developed CAD method achieved an average sensitivity of 99.5%. In the patient study, the developed CAD method reached high detection sensitivities in high FPs settings (FP =1, 2, 4, 8). Sensitivity results were found to be suboptimal when FP <1.
Table 2. 10-fold cross-validation results of detection sensitivities in the computer simulation study and the patient study.
| Study | Sensitivity of false positives (FPs) per image | Averaged sensitivity | ||||||
|---|---|---|---|---|---|---|---|---|
| 1/8 | 1/4 | 1/2 | 1 | 2 | 4 | 8 | ||
| Computer simulation study | 98.9% | 99.3% | 99.3% | 99.3% | 99.5% | 100% | 100% | 99.5% |
| Patient study | 69.0% | 79.7% | 86.0% | 90.0% | 93.5% | 95.4% | 97.7% | 87.3% |
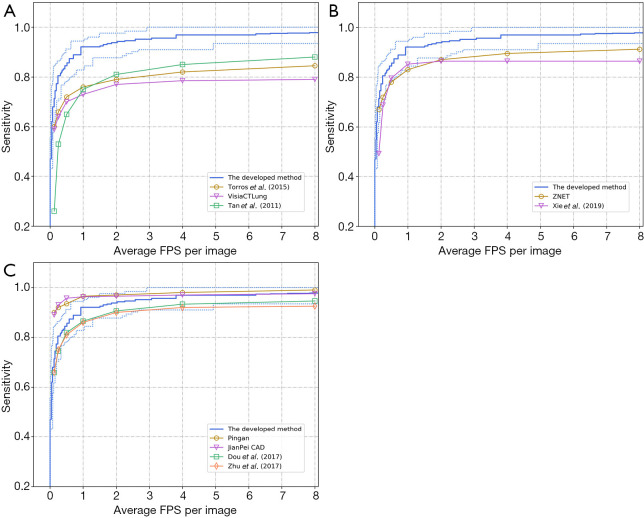
Comparison results are shown in Figure 2 concerning FROC curve analysis. Here, the FROC curve of the developed CAD method is presented as the blue line. The upper/lower dotted blue curves represent the best/poorest testing performance across the 10-fold cross-validation procedure. In Figure 2A, the developed CAD method demonstrated superiority over 3 conventional CAD methods. Similarly, Figure 2B demonstrates improved performance in the developed method, compared to 2 2D-CNN-based detection methods. Figure 2C reveals comparable results between the developed method and computationally-expensive 3D-CNN-based CAD methods. However, two 3D CNN methods (Pingan and JianPei CAD) have higher sensitivity than the developed method when FP <2.
Figure 2.
FROC curve comparison between the developed and (A) conventional CAD methods, (B) 2D-CNN-based method, (C) 3D-CNN-based method. FROC, free-receiver response operating characteristic; CAD, computer-aided detection; CNN, convolutional neural network.
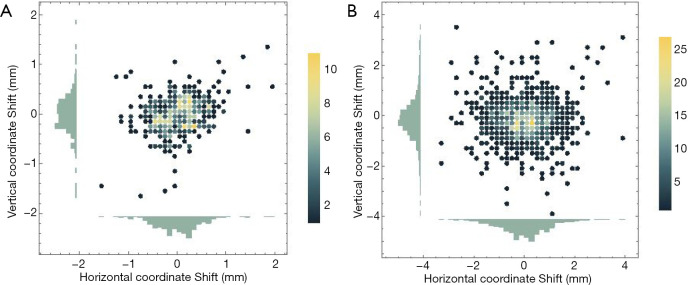
Table 3 summarizes the central coordinate shifts between the predicted nodule and the ground truth bounding boxes. As shown, the average shifts in the x/y direction were less than 1 mm in both studies. The shift in the x-direction was slightly higher than the shift in the y-direction. Figure 3 illustrates the 2D histogram of central coordinate shifts. No apparent spatial deviation in nodule localization was observed, and most shifts were close to the origin.
Table 3. 10-fold cross-validation results of the coordinate shifts.
| Study | Coordinate shifts (mm) | |
|---|---|---|
| x | y | |
| Computer simulation study | 0.345±0.328 | 0.267±0.272 |
| Public patient database study | 0.736±0.657 | 0.575±0.608 |
Figure 3.
2D histograms of the coordinate shifts in (A) the computer simulation study, and (B) the patient study.
Compared to the ground truth, the standard error of diameter measurements was 0.26 mm in the computer-simulated study. The corresponding standard error in the patient study was 0.99 mm. In terms of implementation efficiency, screening a 2D image required 0.07 seconds, when using the developed CAD method.
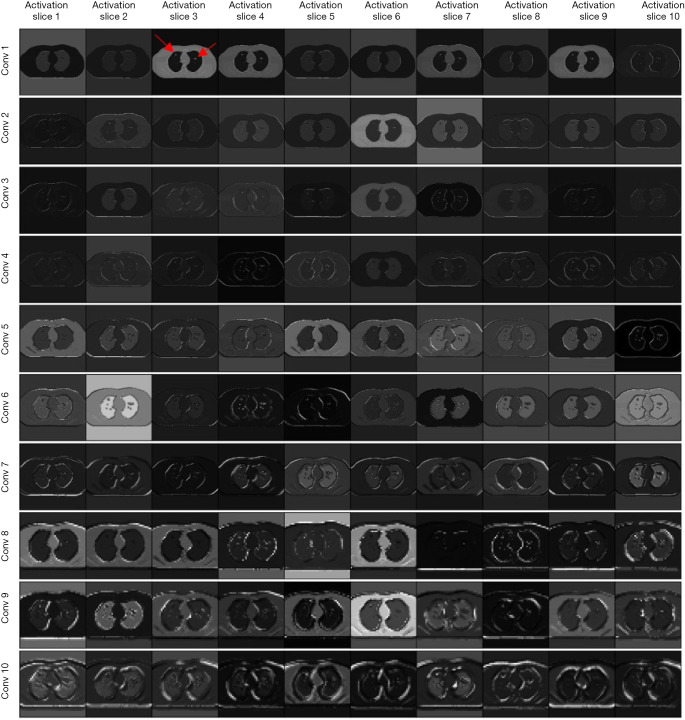
Figure 4 presents the activation maps in the computer simulation study. Activation slices 1 to 10 represent the results of the first 10 filters in each convolution layer. As indicated by the red arrow (Conv 1 – Activation slice 3), two nodules were implanted in this simulated CT image. Figure 5 demonstrates the activation maps of a patient image. A nodule was indicated by the red arrow (Conv 1 – Activation slice 1).
Figure 4.
Activation maps in the simulation study. Red arrow indicates two nodules were implanted in the simulated CT image.
Figure 5.
Activation maps in the patient study. A nodule was indicated by the red arrow.
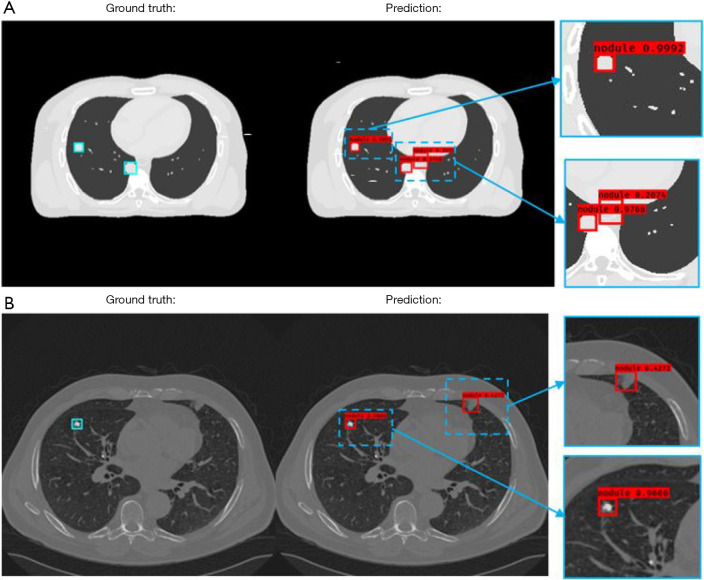
Figure 6 presents 2 independent test results from the simulation and the patient study, where ground truth bounding boxes are indicated by the blue squares on the left column. The detected bounding boxes are illustrated in the middle column, and bounding boxes at a magnified scale is indicated by the right column. The nodule scores () are attached to the detected bounding boxes. In the simulation study results presented in Figure 6A, the nodule score threshold was set to 0.2. Three nodules were detected by the developed CAD method using this threshold. Two true positive detections had higher nodule scores (0.9992/0.9768) than false positive detection (0.2074). For the false positive detection, the CAD misinterpreted the descending aorta as nodules. Figure 6B illustrates the patient study results under a threshold value of 0.4, where the developed CAD method is shown to successfully detect a nodule with a high nodule score (0.9666). Although the ground truth of this detection had a speculated perimeter, the developed method defined the boundary of the nodule accurately. In contrast, the false-positive detection was associated with a much lower nodule score (0.4272).
Figure 6.
Computer-aided detection (CAD) method results of two test patients from (A) the computer simulation study, and (B) the patient study.
Discussion
In this study, we developed a CAD method for the detection of pulmonary nodules in diagnostic CT images. The goal of this CAD method was to achieve accurate nodule localization and diameter estimation. Our approach centralized a YOLO v3 CNN design, which, to the best of our knowledge, has not been used for pulmonary nodule detection. One possible reason that has delayed its application is the reported low accuracy in detecting small objects (34). In our study, we approached this problem by reducing the down-sampling scale in the feature extractor to increase the detection accuracy for small nodules. Quantitative evaluation results demonstrated improved performances in nodule localization and diameter measurements. Mainly, the CAD method was able to localize central nodule coordinates in a range of 1-pixel width (<1 mm) without angular dependence for both studies. Also, this CAD method achieved clinically acceptable precision in nodule diameter estimation. The error in diameter measurement was less than 1 mm, which is smaller than the basic dimensional unit (1 mm) in clinical measurement guidelines and inter-/intra- reader variability (1.73/1.32 mm) (35,36).
The reliability of the developed CAD method was evaluated concerning the computer-based ground truth of the computer simulation study. These simulated images could be used as a digital phantom for quality testing and assurance in future nodule detection studies (37). However, the performance of the developed CAD method is slightly different between the simulated database and the patient database (i.e., the computer simulation study achieved a higher detection sensitivity than patient study). The superior performance of the simulation study may result from the simplicity of the images (i.e., noise-free, well-circumscribed simulated nodules, and homogeneous lung tissue).
The performance difference observed between the two studies may be reflected by the activation maps in Figures 4 and 5. As demonstrated in Figure 4, the edges of the patient’s body and multiple internal organs were highlighted in activation slices of the first five columns (conv 1–5). It can be inferred that the filters in the shallow convolutional layers were responsible for the detection of edges in multiple directions. The information inside the lung region was highlighted in the subsequent deep layers. After that, two nodules were detected. Figure 5 indicates that the filters in the patient database study. With the assistance of edge filters in the shadow layers, the information outside the lung region (e.g., heart, bone) is greatly inhibited, while the information within the lung is enhanced. However, the deep filters in the patient study focused on highlighting the bronchus and pulmonary vessels instead of the nodules directly in the simulation study. This difference may be caused by the complexity inside the lung region, which may be associated with complex functional information associated with imaging data (38). Initial extraction of the bronchus and vessel features may be required in the CAD method before excluding their interference for patient nodule detection.
This CAD method is most prominently characterized by its computationally efficient design. The developed CNN structure consists of 19 convolutional layers in the feature extractor, which was primarily reduced, compared with its original algorithm (i.e., 53 convolutional layers) (25). Also, our proposed approach did not require the computationally-intensive false positive reduction procedure compared to other two-stage methods. Computation consumption is reflected by the number of parameters in any CNN model. As such, our method reduced the total number of parameters compared to other CNN approaches (i.e., 3D U-net or 3D Resnet) due to a lower number of layers and 2D convolution operations. A low-performance GPU, such as GTX1060Ti 6GB used in this study, has adequate memory for loading the full set of parameters needed for model training. However, it is noted that a quantitative comparison of parameters was not feasible due to the limited availability of the source code of other CAD methods.
Finally, we note that the improved computational efficiency did not compromise detection sensitivity. As demonstrated in Figure 2, the developed CAD method out-performed all conventional methods and 2D CNN-based methods. It also achieved comparable results at high FPs settings, compared with computationally expensive 3D CNN-based CAD systems. In low FP settings, however, the developed CAD method did not achieve comparable results to the PATECH and JianPei CAD methods. Future works of task-specific developments of the 3D version of the presented method would emphasize a balance between detection accuracy and implementation cost (39). For example, on-board (i.e., when a patient is under treatment), lung nodule detection for lung radiotherapy using a linear accelerator (LINAC) requires rapid implementation from a light-weight CNN design to achieve real-time detection. Clinical practice preference and evaluation would be the guidelines for future development works towards 3D CNN architecture.
While this study presents a novel approach to nodule localization and diameter estimation, it possesses limitations due to the simplicity of phantom images. Simulating more realistic phantom images (i.e., extra noise, morphological variations) will be essential to fully understand the rationale and robustness of the developed CAD method (40). More sophisticated digital phantoms could customize the simulated image database with specific nodule texture, location, size, and density. This way, further investigation of filter preference (i.e., texture, size, density, location) in feature extraction could be conducted by such simulated images.
Conclusions
In this work, a novel deep-learning CAD method was developed for lung nodule detection with improving computational efficiency and reducing false-positive rates. Preliminary results demonstrated that the developed method achieved nodule localization and diameter estimation with sub-millimeter accuracy. With promising nodule detection accuracy and reduced computation power cost, the developed CAD method has an excellent potential for clinical application.
Acknowledgments
Funding: None.
Ethical Statement: The simulated data was generated by digital phantom and the patient data was obtained from a publically available open-source database, namely LIDC-IDRI database (29). Thus, no ethics approval of an institutional review board or ethics committees was required for this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-19-883). The authors have no conflicts of interest to declare.
References
- 1.Choi WJ, Choi TS. Automated pulmonary nodule detection based on three-dimensional shape-based feature descriptor. Comput Methods Programs Biomed 2014;113:37-54. 10.1016/j.cmpb.2013.08.015 [DOI] [PubMed] [Google Scholar]
- 2.Santos AM, de Carvalho Filho AO, Silva AC, de Paiva AC, Nunes RA, Gattass M. Automatic detection of small lung nodules in 3D CT data using Gaussian mixture models, Tsallis entropy and SVM. Eng Appl Artif Intell 2014;36:27-39. 10.1016/j.engappai.2014.07.007 [DOI] [Google Scholar]
- 3.Chen B, Kitasaka T, Honma H, Takabatake H, Mori M, Natori H, Mori K. Automatic segmentation of pulmonary blood vessels and nodules based on local intensity structure analysis and surface propagation in 3D chest CT images. Int J Comput Assist Radiol Surg 2012;7:465-82. 10.1007/s11548-011-0638-5 [DOI] [PubMed] [Google Scholar]
- 4.Cascio D, Magro R, Fauci F, Iacomi M, Raso G. Automatic detection of lung nodules in CT datasets based on stable 3D mass–spring models. Comput Biol Med 2012;42:1098-109. 10.1016/j.compbiomed.2012.09.002 [DOI] [PubMed] [Google Scholar]
- 5.Wu S, Wang J. Pulmonary Nodules 3D Detection on Serial CT Scans. Wuhan: 2012 Third Global Congress on Intelligent Systems, 2012, 257-260. doi: 10.1109/GCIS.2012.46. [DOI] [Google Scholar]
- 6.Ozekes S, Osman O, Ucan ON. Nodule Detection in a Lung Region that's Segmented with Using Genetic Cellular Neural Networks and 3D Template Matching with Fuzzy Rule Based Thresholding. Korean J Radiol 2008;9:1-9. 10.3348/kjr.2008.9.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin JS, Lo SB, Hasegawa A, Freedman MT, Mun SK. Reduction of false positives in lung nodule detection using a two-level neural classification. IEEE Trans Med Imaging 1996;15:206-17. 10.1109/42.491422 [DOI] [PubMed] [Google Scholar]
- 8.LeCun Y, Kavukcuoglu K, Farabet C. editors. Convolutional networks and applications in vision. Proceedings of 2010 IEEE International Symposium on Circuits and Systems; 30 May 2010 - 2 June 2010. [Google Scholar]
- 9.Hua KL, Hsu CH, Hidayati SC, Cheng WH, Chen YJ. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. Onco Targets Ther 2015;8:2015-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Setio AA, Ciompi F, Litjens G, Gerke P, Jacobs C, van Riel SJ, Wille MM, Naqibullah M, Sanchez CI, van Ginneken B. Pulmonary Nodule Detection in CT Images: False Positive Reduction Using Multi-View Convolutional Networks. IEEE Trans Med Imaging 2016;35:1160-9. 10.1109/TMI.2016.2536809 [DOI] [PubMed] [Google Scholar]
- 11.Gruetzemacher R, Gupta A. Using deep learning for pulmonary nodule detection & diagnosis. 2016.
- 12.Kang G, Liu K, Hou B, Zhang N. 3D multi-view convolutional neural networks for lung nodule classification. PLoS One 2017;12:e0188290. 10.1371/journal.pone.0188290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin H, Li Z, Tong R, Lin L. A deep 3D residual CNN for false-positive reduction in pulmonary nodule detection. Medical Physics 2018;45:2097-107. 10.1002/mp.12846 [DOI] [PubMed] [Google Scholar]
- 14.Dou Q, Chen H, Jin Y, Lin H, Qin J, Heng PA. Automated Pulmonary Nodule Detection via 3D ConvNets with Online Sample Filtering and Hybrid-Loss Residual Learning. ArXiv e-prints. 2017 August 01, 2017. Available online: https://ui.adsabs.harvard.edu/#abs/2017arXiv170803867D
- 15.Ding J, Li A, Hu Z, Wang L. Accurate Pulmonary Nodule Detection in Computed Tomography Images Using Deep Convolutional Neural Networks. ArXiv e-prints. 2017 June 01, 2017. Available online: https://ui.adsabs.harvard.edu/#abs/2017arXiv170604303D
- 16.Xie H, Yang D, Sun N, Chen Z, Zhang Y. Automated pulmonary nodule detection in CT images using deep convolutional neural networks. Pattern Recognition 2019;85:109-19. 10.1016/j.patcog.2018.07.031 [DOI] [Google Scholar]
- 17.Zhu W, Liu C, Fan W, Xie X. DeepLung: 3D Deep Convolutional Nets for Automated Pulmonary Nodule Detection and Classification. ArXiv e-prints. September 01, 2017. Available online: https://ui.adsabs.harvard.edu/#abs/2017arXiv170905538Z
- 18.Huang X, Shan J, Vaidya V. editors. Lung nodule detection in CT using 3D convolutional neural networks. 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017); 18–21 April 2017. [Google Scholar]
- 19.Redmon J, Divvala S, Girshick R, Farhadi A. You Only Look Once: Unified, Real-Time Object Detection. ArXiv e-prints. June 01, 2015. Available online: https://ui.adsabs.harvard.edu/#abs/2015arXiv150602640R
- 20.Al-Masni MA, Al-Antari MA, Park JM, Gi G, Kim TY, Rivera P, Valarezo E, Han SM, Kim TS. Detection and classification of the breast abnormalities in digital mammograms via regional Convolutional Neural Network. Conf Proc IEEE Eng Med Biol Soc 2017;2017:1230-3. [DOI] [PubMed] [Google Scholar]
- 21.Al-Antari MA, Al-Masni MA, Kim TS. Deep Learning Computer-Aided Diagnosis for Breast Lesion in Digital Mammogram. Adv Exp Med Biol 2020;1213:59-72. 10.1007/978-3-030-33128-3_4 [DOI] [PubMed] [Google Scholar]
- 22.Kim YG, Cho Y, Wu CJ, Park S, Jung KH, Seo JB, Lee HJ, Hwang HJ, Lee SM, Kim N. Short-term Reproducibility of Pulmonary Nodule and Mass Detection in Chest Radiographs: Comparison among Radiologists and Four Different Computer-Aided Detections with Convolutional Neural Net. Sci Rep 2019;9:18738. 10.1038/s41598-019-55373-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramachandran SS, George J, Skaria S, V. V. V, editors. Using YOLO based deep learning network for real time detection and localization of lung nodules from low dose CT scans. Medical Imaging 2018: Computer-Aided Diagnosis; 2018 February 01, 2018. [Google Scholar]
- 24.Chambon S, Thorey V, Arnal PJ, Mignot E, Gramfort A. DOSED: A deep learning approach to detect multiple sleep micro-events in EEG signal. J Neurosci Methods 2019;321:64-78. 10.1016/j.jneumeth.2019.03.017 [DOI] [PubMed] [Google Scholar]
- 25.Redmon J, Farhadi A. YOLOv3: An Incremental Improvement. ArXiv e-prints. April 01, 2018. Available online: https://ui.adsabs.harvard.edu/#abs/2018arXiv180402767R
- 26.He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. ArXiv e-prints. December 01, 2015. Available online: https://ui.adsabs.harvard.edu/#abs/2015arXiv151203385H
- 27.Rothe R, Guillaumin M, Van Gool L. editors. Non-maximum suppression for object detection by passing messages between windows. Asian Conference on Computer Vision; 2014: Springer. [Google Scholar]
- 28.Segars WP, Sturgeon G, Mendonca S, Grimes J, Tsui BM. 4D XCAT phantom for multimodality imaging research. Med Phys 2010;37:4902-15. 10.1118/1.3480985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armato SG, 3rd, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA, Kazerooni EA, MacMahon H, Van Beeke EJ, Yankelevitz D, Biancardi AM, Bland PH, Brown MS, Engelmann RM, Laderach GE, Max D, Pais RC, Qing DP, Roberts RY, Smith AR, Starkey A, Batrah P, Caligiuri P, Farooqi A, Gladish GW, Jude CM, Munden RF, Petkovska I, Quint LE, Schwartz LH, Sundaram B, Dodd LE, Fenimore C, Gur D, Petrick N, Freymann J, Kirby J, Hughes B, Casteele AV, Gupte S, Sallamm M, Heath MD, Kuhn MH, Dharaiya E, Burns R, Fryd DS, Salganicoff M, Anand V, Shreter U, Vastagh S, Croft BY. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 2011;38:915-31. 10.1118/1.3528204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres EL, Fiorina E, Pennazio F, Peroni C, Saletta M, Camarlinghi N, Fantacci ME, Cerello P. Large scale validation of the M5L lung CAD on heterogeneous CT datasets. Med Phys 2015;42:1477-89. 10.1118/1.4907970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan M, Deklerck R, Jansen B, Bister M, Cornelis J. A novel computer-aided lung nodule detection system for CT images. Med Phys 2011;38:5630-45. 10.1118/1.3633941 [DOI] [PubMed] [Google Scholar]
- 32.Arindra Adiyoso Setio A, Traverso A, de Bel T, Berens MSN, van den Bogaard C, Cerello P, Chen H, Dou Q, Fantacci ME, Geurts B, van der Gugten R, Ann Heng P, Jansen B, de Kaste MMJ, Kotov V, Lin JYH, Manders JTMC, Sonora-Mengana A, Garcia-Naranjo JC, Papavasileiou E, Prokop M, Saletta M, Schaefer-Prokop CM, Scholten ET, Scholten ET, Scholten ET, Scholten L, Snoeren MM, Torres EL, Vandemeulebroucke J, Walasek N, Zuidhof GCA, van Ginneken B, Jacobs C. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge. ArXiv e-prints. December 01, 2016. Available online: https://ui.adsabs.harvard.edu/#abs/2016arXiv161208012A
- 33.Receiver Operating Characteristic Analysis in Medical Imaging: Contents. J ICRU 2008;8:NP.
- 34.Du Z, Yin J, Yang J. Expanding Receptive Field YOLO for Small Object Detection. J Phys Conf Ser 2019;1314:012202 10.1088/1742-6596/1314/1/012202 [DOI] [Google Scholar]
- 35.Bankier AA, MacMahon H, Goo JM, Rubin GD, Schaefer-Prokop CM, Naidich DP. Recommendations for Measuring Pulmonary Nodules at CT: A Statement from the Fleischner Society. Radiology 2017;285:584-600. 10.1148/radiol.2017162894 [DOI] [PubMed] [Google Scholar]
- 36.Revel M-P, Bissery A, Bienvenu M, Aycard L, Lefort C, Frija G. Are Two-dimensional CT Measurements of Small Noncalcified Pulmonary Nodules Reliable? Radiology 2004;231:453-8. 10.1148/radiol.2312030167 [DOI] [PubMed] [Google Scholar]
- 37.Chang Y, Lafata K, Wang C, Duan X, Geng R, Yang Z, Yin FF. Digital phantoms for characterizing inconsistencies among radiomics extraction toolboxes. Biomed Phys Eng Express 2020;6:025016. [DOI] [PubMed] [Google Scholar]
- 38.Lafata KJ, Zhou Z, Liu JG, Hong J, Kelsey CR, Yin FF. An Exploratory Radiomics Approach to Quantifying Pulmonary Function in CT Images. Sci Rep 2019;9:11509. 10.1038/s41598-019-48023-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng D, Hong JC, Wang C, Zhu X. Radiotherapy Treatment Planning in the Age of AI: Are We Ready Yet? Technol Cancer Res Treat 2019;18:1533033819894577. 10.1177/1533033819894577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang Y, Lafata K, Segars PW, Yin FF, Ren L. Development of realistic multi-contrast textured XCAT (MT-XCAT) phantoms using a dual-discriminator conditional-generative adversarial network (D-CGAN). Phys Med Biol 2020;65:065009. 10.1088/1361-6560/ab7309 [DOI] [PMC free article] [PubMed] [Google Scholar]