Kunjin and West Nile viruses belong to the arthropod-borne flaviviruses, which can result in severe symptoms, including encephalitis, meningitis, and death. Flaviviruses have expanded into new populations and emerged as novel pathogens repeatedly in recent years, demonstrating that they remain a global threat. Currently, there are no approved antiviral therapeutics against either Kunjin or West Nile viruses. Thus, there is a pressing need for understanding the pathogenesis of these viruses in humans. In this study, we investigated the role of the Kunjin virus helicase on infection in cell culture and in vivo. This work provides new insight into how flaviviruses control pathogenesis and mosquito transmission through the nonstructural protein 3 helicase.
KEYWORDS: cytopathic effect, flavivirus, helicase, mosquito
ABSTRACT
The unwinding of double-stranded RNA intermediates is critical for the replication and packaging of flavivirus RNA genomes. This unwinding activity is achieved by the ATP-dependent nonstructural protein 3 (NS3) helicase. In previous studies, we investigated the mechanism of energy transduction between the ATP and RNA binding pockets using molecular dynamics simulations and enzymatic characterization. Our data corroborated the hypothesis that motif V is a communication hub for this energy transduction. More specifically, mutations T407A and S411A in motif V exhibit a hyperactive helicase phenotype, leading to the regulation of translocation and unwinding during replication. However, the effect of these mutations on viral infection in cell culture and in vivo is not well understood. Here, we investigated the role of motif V in viral replication using West Nile virus (Kunjin subtype) T407A and S411A mutants (T407A and S411A Kunjin, respectively) in cell culture and in vivo. We were able to recover S411A Kunjin but unable to recover T407A Kunjin. Our results indicated that S411A Kunjin decreased viral infection and increased cytopathogenicity in cell culture compared to wild-type (WT) Kunjin. Similarly, decreased infection rates in surviving S411A Kunjin-infected Culex quinquefasciatus mosquitoes were observed, but S411A Kunjin infection resulted in increased mortality compared to WT Kunjin infection. Additionally, S411A Kunjin infection increased viral dissemination and saliva positivity rates in surviving mosquitoes compared to WT Kunjin infection. These data suggest that S411A Kunjin increases viral pathogenesis in mosquitoes. Overall, these data indicate that NS3 motif V may play a role in the pathogenesis, dissemination, and transmission efficiency of Kunjin virus.
IMPORTANCE Kunjin and West Nile viruses belong to the arthropod-borne flaviviruses, which can result in severe symptoms, including encephalitis, meningitis, and death. Flaviviruses have expanded into new populations and emerged as novel pathogens repeatedly in recent years, demonstrating that they remain a global threat. Currently, there are no approved antiviral therapeutics against either Kunjin or West Nile viruses. Thus, there is a pressing need for understanding the pathogenesis of these viruses in humans. In this study, we investigated the role of the Kunjin virus helicase on infection in cell culture and in vivo. This work provides new insight into how flaviviruses control pathogenesis and mosquito transmission through the nonstructural protein 3 helicase.
INTRODUCTION
Kunjin virus, a West Nile virus (WNV) subtype, causes encephalitis epidemics in horses that are localized to Australia (1–4), whereas WNV has a much larger global impact, is present on almost every major continent except for South America and Antarctica (4, 5), and regularly results in encephalitis in humans as well as horses (6). Within the United States alone, approximately 3 million people are thought to have been infected with West Nile virus between 1999 and 2010 (7–9). Kunjin virus and WNV share a natural transmission cycle between Culex mosquito vectors and bird reservoir hosts (2). Humans and horses are considered dead-end hosts because they do not contribute to viral perpetuation. In humans, about 80% of WNV-infected individuals are asymptomatic, and the majority of symptomatic individuals experience a mild febrile illness. However, approximately 1:150 infections result in severe symptoms, including meningitis and/or encephalitis, and ∼9% of these cases are fatal (6, 10). Currently, there are vaccines against WNV for horses, but not for humans; no vaccines against Kunjin virus are available (5). Thus, there is a need for the development of vaccines and/or antiviral therapies against Kunjin virus and WNV infections. Developing a fundamental understanding of how Kunjin virus and WNV replicate within hosts, including the mosquito vector, is essential to the development of interventional strategies.
Kunjin virus and WNV belong to the Flavivirus genus within the Flaviviridae family. Flaviviridae is a group of single-stranded positive-sense RNA viruses with genomes of approximately 11 kb in length (11–13). Kunjin virus is a subtype of WNV, with the viruses having nucleotide and amino acid sequence identities of 82% and 93%, respectively (14–16). However, Kunjin virus infection results in a lower morbidity than WNV infection in humans, making it an excellent tool to study WNV replication with well-established molecular tools while minimizing risk (17). Additionally, Kunjin virus is less cytopathic than WNV, allowing differences in virus-induced cell viability to be more easily visualized. The proteins and processes involved in viral replication are conserved across the Flavivirus genus, including for Kunjin virus, WNV, and dengue, yellow fever, Japanese encephalitis, and Zika viruses (12, 18). Initially, the viral RNA genome is translated into a single polyprotein, which is cleaved by host and viral proteases into three structural proteins (C, prM, and E) and eight nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, 2K, NS4B, and NS5) (12, 18, 19). The viral NS replication proteins then generate a negative-sense antigenomic RNA that is in complex with the positive-sense genomic RNA, forming the double-stranded RNA (dsRNA) intermediate complex (20, 21). The negative-sense antigenomic RNA serves as a template for positive-strand synthesis (20); therefore, unwinding of the dsRNA intermediate is required for replication. Unwinding is achieved by the C-terminal helicase domain of NS3 (22–24).
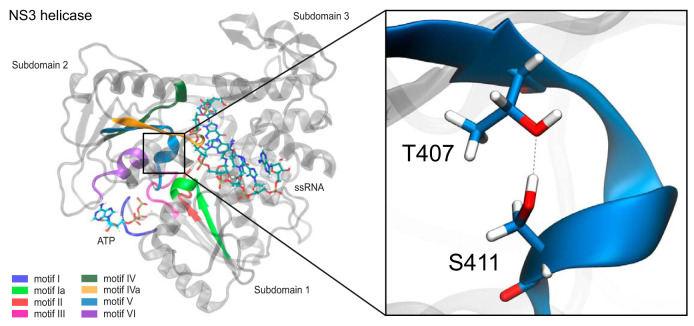
The NS3 helicase domain is a multifunctional viral protein that houses three enzymatic activities: RNA helicase, nucleoside triphosphatase (NTPase), and RNA 5′-triphosphatase (RTPase) (25–28). NS3 helicase is a member of the superfamily 2 (SF2) helicases (29). The helicase domain consists of three subdomains (subdomains 1, 2, and 3). Subdomains 1 and 2 are RecA-like structures that are highly conserved across all SF2 helicases, while subdomain 3 is unique to the viral/DEAH-like group of SF2 helicases (30). Additionally, there are eight structural motifs (motifs I, Ia, II, III, IV, IVa, V, and VI) that are highly conserved across all viral/DEAH-like subfamilies with the SF2 helicases (29). These structural motifs are responsible for both substrate binding and enzymatic function within the helicase. The helicase domain is responsible for the translocation and unwinding of the double-stranded RNA intermediate in an ATP-dependent manner during viral replication (31). Previous studies further identified motif V to be potentially critical for the translocation and unwinding of the double-stranded RNA intermediate (32, 33). Motif V was described to be a potential link between the ATP binding pocket and the RNA binding cleft through a strong correlation between residues within motif V and both binding pockets (32). The strongly correlated movements between ATP binding pocket and RNA binding cleft residues in our simulations suggest a physical linkage between the two sites that may be important for the ATP-driven helicase function. Additionally, the T407A and S411A mutations in motif V increased the unwinding activity and decreased viral genome replication compared to those for the wild type (WT), suggesting that the hydrogen bond between these two residues in the WT inhibits helicase unwinding activity in vitro and in vivo (33). These data suggest that motif V may serve as a molecular throttle on the NS3 helicase function, but what effect these residues play on the larger viral replication cycle was not clear.
To better understand the effects that NS3 motif V mutations have on flavivirus replication, we sought to investigate the role of the motif V T407 and S411 residues on helicase function in cell culture and in vivo by introducing alanine mutations in full-length infectious Kunjin virus with the T407A and S411A mutations (referred to here as T407A Kunjin and S411A Kunjin, respectively). Only S411A Kunjin was recovered, and it resulted in reduced viral yields compared with those of wild-type Kunjin virus (WT Kunjin). Additionally, S411A Kunjin showed an increased cytopathic effect in comparison to that of WT Kunjin virus in cell culture. Similarly, when WT or S411A Kunjin viruses were intrathoracically injected into Culex quinquefasciatus mosquitoes, S411A Kunjin resulted in increased mortality compared with that after injection of WT Kunjin. Upon further investigation of mosquito infection, S411A Kunjin viruses were found to disseminate and transmit more effectively than WT Kunjin viruses, even though the overall rate of infection with S411A Kunjin was lower than that with WT Kunjin. Overall, our data suggest that flaviviruses may use the NS3 motif V to help control the cytotoxicity induced by NS3 during infection and limit virus-induced mortality in mosquito vectors.
RESULTS
S411A Kunjin virus increases the cytopathic effect in cell culture.
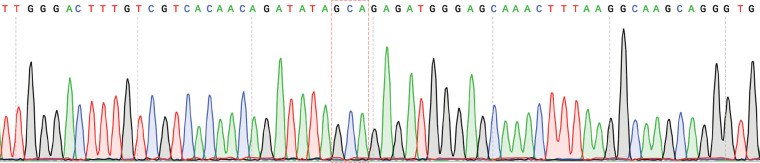
Previously, motif V residues T407 and S411 were mutated to alanine to disrupt a hydrogen bond that potentially stabilizes the motif V secondary structure of the NS3 helicase during viral replication (Fig. 1). These mutations were shown to decrease viral genome replication in a replicon-based system while increasing helicase unwinding activity biochemically (33). In the present study, we introduced these mutations into the full-length infectious Kunjin virus to investigate the effects of these mutations on infectivity compared to that of WT Kunjin both in cell culture and in mosquito infections. We utilized a novel mutagenesis and bacterium-free viral launch system to generate the T407A Kunjin and S411A Kunjin viruses in Vero cells. The first generation of S411A Kunjin was recovered from infected cells, and the presence of the alanine mutation was verified by sequencing (Fig. 2). The sequencing results provided insight into the stability of the alanine mutation, in that the mutation did not revert to the WT Kunjin sequence. On the other hand, we were unable to recover T407A Kunjin, despite repeated attempts, which was consistent with our previously reported decrease in T407A Kunjin viral genome replication in replicon assays (33). Second-generation stocks of WT Kunjin and S411A Kunjin were generated, and the titers of the viruses were determined for further experiments. We noted the plaque morphology for both WT Kunjin and S411A Kunjin (Fig. 3). WT Kunjin showed large, faint plaques with a diameter of 3.3 ± 0.7 mm (Fig. 3A), while S411A Kunjin showed small but distinctly clear plaques with a diameter of 1.5 ± 0.3 mm (Fig. 3B). The S411A Kunjin plaque sizes were significantly different from the WT Kunjin plaque sizes, suggesting a potential decrease in viral cell-to-cell spread and an increase in the cytopathic effect for S411A Kunjin-infected cells compared to that for WT Kunjin-infected cells. Since these results suggest that S411A Kunjin may be more toxic to cells than WT Kunjin during infection, we further investigated the effect of S411A Kunjin on cell viability.
FIG 1.
S411 and T407 interaction within motif V in NS3 helicase. NS3 helicase consists of three subdomains: subdomain 1, subdomain 2, and subdomain 3. All highly conserved structural motifs (motifs I, Ia, II, III, IV, IVa, V, and VI) are located within subdomains 1 and 2. These structural motifs are highlighted. (Inset) Within motif V, residues T407 and S411 interact with each other through a hydrogen bond. ssRNA, single-stranded RNA.
FIG 2.
Verification of the alanine mutation in the S411A Kunjin virus via Sanger sequencing. Results from Sanger sequencing verified the presence of the alanine mutation at position 411 through the presence of the alanine codon (highlighted in a red box). The original serine codon within the red box was TCT. Two nucleotides were changed to introduce the alanine mutation. Refer to GenBank accession number AY274504.1 for the sequence of wild-type Kunjin virus clone FLSDX.
FIG 3.
Plaque morphology suggests an increased cytopathic effect for S411A Kunjin. Viral titers for second-generation WT and S411A Kunjin viruses were obtained via 4-day incubation plaque assays with Vero cells, and the plaque morphology is shown for WT Kunjin (A) and S411A Kunjin (B). The average diameter (∅avg) is reported for both WT Kunjin and S411A Kunjin. The measured diameters were statistically analyzed via a t test (P = 0.000003).
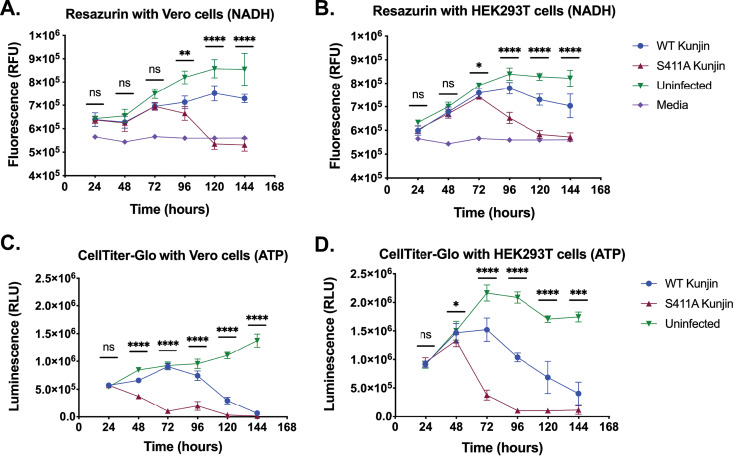
S411A Kunjin reduces NADH and intracellular ATP levels, leading to increased cellular death.
We utilized resazurin and CellTiter-Glo assays to quantify the virus-induced cell killing in HEK293T and Vero cells infected with either WT Kunjin or S411A Kunjin at a multiplicity of infection (MOI) of 5 PFU/cell. Both of these assays estimate cell viability through the measurement of metabolically active cells using fluorescence and luminescence, respectively. In the resazurin assay, resazurin, a nonfluorescent dye, converts to resorufin, a highly fluorescent dye, in response to the reducing environment of heathy, growing cells (34–36). We measured the number of relative fluorescence units (RFU) of resazurin in uninfected, WT Kunjin-infected, or S411A Kunjin-infected Vero and HEK293T cells every 24 h for 6 days (Fig. 4A and B). We also measured the number of RFU of the medium as a negative control to determine the baseline medium fluorescence. The cell viability measurements of uninfected Vero and HEK293T cells increased gradually over the duration of the experiment, suggesting that the cells were healthy and growing for the entirety of the experiment. The cell viability measurements during the first 72 h for WT Kunjin-infected Vero and HEK293T cells were similar to those for uninfected cells. However, the fluorescent signals of the cell viability measurements were lower for infected cells than for uninfected cells. After 72 h postinfection (p.i.), cell viability measurements for WT Kunjin infections continued to increase in fluorescence, reaching 7.5 × 105 ± 0.3 × 105 RFU at 120 h p.i. for Vero cells and 7.8 × 105 ± 0.2 × 105 RFU at 96 h p.i. for HEK293T cells. After these times, the cell viability measurements decreased in fluorescence by 144 h p.i., suggesting that the WT Kunjin-induced cell toxicity overtook cellular replication. In the case of S411A Kunjin-infected Vero and HEK293T cells during the first 72 h, cell viability measurements demonstrated levels of fluorescence similar to those for uninfected cells, although the cell viability measured for S411A Kunjin-infected cells was decreased compared to that measured for uninfected cells. As the S411A Kunjin infection continued, cell viability measurements significantly reduced in fluorescence between 96 and 144 h p.i., ending with 5.3 × 105 ± 0.3 × 105 RFU for Vero cells and 5.7 × 105 ± 0.2 × 105 RFU for HEK293T cells. Together, these data suggest that Kunjin virus-infected Vero and HEK293T cells are relatively healthy for at least the first 72 h, after which point the viability of the S411A Kunjin-infected cell population is negatively affected immediately in both cell lines, whereas 24-h and 48-h delays in decreased cell viability measurements are observed with WT Kunjin infection for HEK293T and Vero cells, respectively.
FIG 4.
S411A Kunjin decreases cell viability. The cellular metabolism of WT Kunjin- and S411A Kunjin-infected Vero cells (A) and HEK293T cells (B) was measured by the resazurin assay. Similarly, intracellular ATP levels in WT Kunjin- and S411A Kunjin-infected Vero cells (C) and HEK293T cells (D) were measured by the CellTiter-Glo assay. All infections were performed at an MOI of 5 PFU/cell in triplicate. Cell viability curves were statistically analyzed by an unpaired t test, and the significance of the difference between WT Kunjin- and S411A Kunjin-infected cells is reported (****, P ≤ 0.0001; ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, not significant [P > 0.05]).
Another way to infer that cells are metabolically active or viable is through the detection of intracellular ATP levels. We utilized the CellTiter-Glo assay, which uses the luciferase reaction, an ATP-dependent reaction, to convert luciferin to oxyluciferin and several by-products, including light (34). The amount of the by-product light was measured in relative luminescence units (RLU) for uninfected, WT Kunjin-infected, or S411A Kunjin-infected Vero and HEK293T cells every 24 h for 6 days (Fig. 4C and D). Over the course of the experiment, uninfected Vero cells progressively increased in luminescence from 5.5 × 105 ± 0.3 × 105 to 1.4 × 106 ± 0.1 × 106 RLU (Fig. 4C), suggesting that the uninfected cells were healthy and metabolically active for the 6-day experiment. However, cell viability measurements of uninfected HEK293T cells increased linearly for the first 72 h, after which point the cell viability measurements decreased and then leveled off at 1.7 × 106 ± 0.07 × 106 RLU (Fig. 4D), suggesting that uninfected HEK293T cells become less metabolically active after 96 h than the Vero cells. As for infection with WT Kunjin, the cell viability measurements steadily increased for the first 72 h for Vero cells and for the first 48 h for HEK293T cells, similar to the observed cell viability measurements of uninfected Vero and HEK293T cells. At 96 h p.i. in Vero cells and 72 h p.i. in HEK293T cells, the cell viability measurements of WT Kunjin-infected cells decreased compared to those of uninfected cells. The viability of the WT Kunjin-infected cell populations continued to decrease, with cell viability measurements reaching 6.5 × 104 ± 3.0 × 104 RLU in Vero cells and 4.0 × 105 ± 2.0 × 105 RLU in HEK293T cells at 144 h. These data suggest that infection with WT Kunjin negatively affected cell viability after 72 h p.i. compared to the effect of no virus infection on cell viability. On the other hand, cell viability measurements with S411A Kunjin infection decreased after 24 h p.i. for Vero cells and after 48 h p.i. for HEK293T cells. For the remainder of the experiment, the viability of the S411A Kunjin-infected Vero and HEK293T cell populations continued to decrease, suggesting that both Vero and HEK293T cells are extremely sensitive to S411A Kunjin and, thus, that cell viability is significantly reduced in the presence of the mutated virus. Together, these results suggest that infection with S411A Kunjin in either Vero or HEK293T cells negatively affected cell viability more quickly than infection with WT Kunjin.
S411A Kunjin results in decreased and delayed viral replication kinetics.
The results presented in the previous section indicated that S411A Kunjin induced increased cellular death during infection. This prompted the following question: how does increased cellular death resulting from infection with S411A Kunjin affect the replication kinetics of the virus? Therefore, we performed a multistep replication kinetics experiment with WT Kunjin- or S411A Kunjin-infected HEK293T cells at an MOI of 0.01 PFU/cell over a 5-day period. Every 12 h the media containing viruses were collected and viral titers were determined via focus-forming assays (Fig. 5). At 12 h postinfection, the WT and S411A Kunjin viral titers were not significantly different. At 24 h p.i., S411A Kunjin remained in the lag phase, while WT Kunjin had entered the exponential replication phase, demonstrating delayed replication with S411A Kunjin infection. Over the last 4 days of infection, S411A Kunjin maintained and expanded the initial delay in exponential replication and reached an ∼1-log lower peak viral titer than WT Kunjin. Overall, these data suggest that S411A Kunjin does not replicate as efficiently as WT Kunjin. These results are consistent with data reported by Du Pont et al., suggesting that the increased helicase unwinding activity seen with the recombinant S411A Kunjin NS3 helicase negatively affects viral replication in fully infectious S411A Kunjin virus (33). Considering the observations that S411A Kunjin resulted in decreased viral replication and increased cellular death, we next investigated the effects of the S411A mutation on Kunjin virus infection in vivo.
FIG 5.
S411A Kunjin decreases and delays viral replication kinetics. Replication kinetics experiments were performed in triplicate for WT and S411A Kunjin viruses. HEK293T cells were infected at an MOI of 0.01 PFU/cell. Viral samples (media) were collected every 12 h for 5 days and processed to obtain viral titers using Vero cells in focus-forming assays. Viral replication curves were compared by the F test, and the significance of the differences between the WT Kunjin- and S411A Kunjin-infected cells is reported (****, P ≤ 0.0001; ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, not significant [P > 0.05]). ffu, focus-forming units.
S411A Kunjin results in increased mortality in mosquitoes compared to WT Kunjin when IT injected but not when blood fed.
For the in vivo studies, we did not have access to a colony of C. annulirostris mosquitoes, the primary vector for Kunjin virus, but we had an established colony of C. quinquefasciatus mosquitoes that were infectible by Kunjin virus. C. quinquefasciatus mosquitoes were fed defibrinated calf blood diluted by half, with the titer being equilibrated for WT Kunjin, S411A Kunjin, or medium alone as a negative control. Similarly, female C. quinquefasciatus mosquitoes were subjected to intrathoracic (IT) injection of 345 PFU per mosquito of WT Kunjin or S411A Kunjin or conditioned medium. Mosquito mortality was recorded daily for 15 or 9 days for mosquitoes receiving the blood meal and subjected to IT injection, respectively. Overall, virus-exposed mosquito mortality was low in both the blood-fed and IT injected cohorts (Fig. 6), consistent with previous observations of Kunjin virus in C. quinquefasciatus mosquitoes (37). When blood fed, no difference in mortality rates was observed between mosquitoes exposed to WT Kunjin and mosquitoes exposed to S411A Kunjin. However, the low rate of mortality for virus-exposed mosquitoes (∼10%) was significantly different from that for mosquitoes exposed to medium alone (Fig. 6A). In contrast to the data for blood-fed mosquitoes but consistent with the cell culture and replication kinetics data, when virus was introduced through IT injection, to bypass the midgut barrier, only exposure to S411A Kunjin resulted in increased mortality (Fig. 6B). Together these data suggest that S411A Kunjin is more lethal to mosquitoes than WT Kunjin, once the virus has been able to establish an infection and/or transverse through the mosquito midgut barrier. This result led us to further investigate the specifics of infection of C. quinquefasciatus mosquitoes by WT and S411A Kunjin viruses.
FIG 6.
S411A Kunjin viruses are more lethal to C. quinquefasciatus mosquitoes than WT Kunjin. Female C. quinquefasciatus mosquitoes were exposed to WT or S411A Kunjin virus either through infectious blood meals (A) or by IT injection (B). Control mosquitoes were exposed to blood meals containing medium or injected with medium alone. Mortality was recorded daily for 15 days (A) or 9 days (B). Survival curves were compared by the log-rank test for trend (****, P < 0.0001; *, P < 0.05). The data are for 425 mosquitoes per condition (A) and 40 mosquitoes per condition (B). In panel B, the traces for WT and S411A Kunjin overlap.
S411A Kunjin has a lower infection rate but disseminates more efficiently than WT Kunjin.
Similar to the mortality experiments, C. quinquefasciatus mosquitoes were infected with either WT Kunjin or S411A Kunjin by a blood meal. Mosquito legs/wings, saliva, and bodies were collected after 7 days and determined to be positive or negative for infection by plaque assay. While ∼58% of mosquitoes infected with WT Kunjin were positive for the virus at day 7, only ∼8% of mosquitoes infected with S411A Kunjin were positive (Fig. 7A). Dissemination was inefficient for WT Kunjin, with only 6% of mosquitoes being positive for virus in the legs and wings, demonstrating a strong barrier to escape from the midgut. Similarly, less than 2% of infected mosquitoes had positive saliva samples (Fig. 7A). Despite low infection rates for mosquitoes infected with S411A Kunjin, positive legs/wings and saliva were identified across multiple replicate experiments, with nearly 50% of infected mosquitoes having disseminated virus and 50% of those with disseminated virus having positive saliva. These data led to the question: does S411A Kunjin allow for higher relative rates of dissemination?
FIG 7.
S411A Kunjin is less capable than WT Kunjin at infecting mosquitoes but disseminates and transmits more efficiently once established. Engorged female C. quinquefasciatus mosquitoes exposed to infectious blood meals containing either WT or S411A Kunjin virus were housed for 7 days (A) or 14 days (B to E) after the blood feed. Mosquitoes were dissected, and their legs/wings, saliva, and bodies were collected and tested for the presence or absence of Kunjin virus by plaque assay on Vero cells of undiluted samples. Data are shown as the percentage of total exposed infected cells (A and B), the total number of mosquitoes with negative and positive bodies (C), the number of mosquitoes with positive legs/wings from all infected mosquitoes (D), or the total number of mosquitoes with positive saliva from all mosquitoes in which virus disseminated. Data are for 64 mosquitoes per condition (A) and for 60 WT Kunjin-infected mosquitoes and 390 S411A Kunjin-infected mosquitoes (B). (A) Error bars represent SEM. (A and C to E) Infection, dissemination, and transmission were compared using Fisher’s exact test (****, P < 0.0001; ***, P < 0.001; *, P < 0.05).
To answer this question, a second, much larger cohort of C. quinquefasciatus mosquitoes was infected by a blood meal with WT Kunjin or S411A Kunjin. Enough mosquitoes were dissected to generate an estimated 30 infected mosquitoes per condition: 60 exposed to WT Kunjin and 390 exposed to S411A Kunjin. Since mosquitoes continue to die up to 14 days after a blood feed, mosquitoes were collected at 14 days instead of 7 days after a blood meal in an attempt to ensure sufficient numbers of S411A Kunjin-infected mosquitoes. Again, WT Kunjin was observed to infect a larger percentage of exposed mosquitoes than S411A Kunjin (∼30% versus ∼15%) (Fig. 7B and C), whereas S411A Kunjin demonstrated higher rates of dissemination than WT Kunjin (Fig. 7B and D). No legs/wings or saliva samples from WT Kunjin-infected mosquitoes were found to be positive at 14 days after the blood meal (Fig. 7B, D, and E). In contrast to and supporting these data from smaller cohorts collected at 7 days after the blood meal, 48% of S411A Kunjin-infected mosquitoes had infected legs/wings and 61% of mosquitoes with S411A Kunjin-infected legs/wings had positive saliva samples. These data demonstrate that S411A Kunjin was less capable of infecting C. quinquefasciatus via a blood meal than WT Kunjin. However, these data also suggest that when S411A Kunjin was able to establish infection in C. quinquefasciatus mosquitoes, it was able to escape the midgut barrier more efficiently than WT Kunjin, resulting in dissemination, infection of the salivary glands, and delivery to the saliva. Finally, when considered in combination with the survival data, these data further support the suggestion that when S411A Kunjin is able to establish infection in C. quinquefasciatus mosquitoes, it is more lethal.
DISCUSSION
Previous work by our group has supported the hypothesis that motif V in flavivirus NS3 helicase is a communication hub for the translocation and unwinding of the dsRNA intermediate during flavivirus replication (32, 33). More specifically, we found that NS3 motif V residues T407 and S411 exhibit an increased helicase unwinding activity in biochemical assays when mutated to alanine residues, while we observed a reduction in the replication of T407 and S411 mutant replicons. These previous results suggest that the T407 and S411 residues are responsible for regulating the NS3 helicase function during flavivirus replication. In this study, we further investigated the role of the T407 and S411 helicase residues in the full-length infectious Kunjin virus in cell culture and in vivo experiments. S411A Kunjin was successfully recovered, and the presence of the mutation was confirmed via sequencing (Fig. 2). However, T407A Kunjin was not recovered, which was consistent with previous results indicating ablated viral genome replication activity (33). We utilized WT Kunjin and S411A Kunjin in several cell culture experiments, including viral replication, resazurin, and CellTiter-Glo assays. Additionally, we compared WT Kunjin and S411A Kunjin in several in vivo experiments, including experiments evaluating infection, dissemination, and transmission within C. quinquefasciatus mosquitoes. We observed that S411A Kunjin reduced cell viability during infection, leading to observation of an increased cytopathic effect in the plaque morphology and several metabolic assays in cell culture. Additionally, the results demonstrated a lower initial infection rate for S411A Kunjin than for WT Kunjin within mosquitoes. However, once infection was established, the more efficient dissemination of S411A Kunjin compared to that of WT Kunjin occurred, potentially causing the observed increased mortality rates in S411A Kunjin-infected mosquitoes compared to that in WT Kunjin-infected mosquitoes. Overall, our data suggest that the S411 residue in motif V of NS3 influences infection-induced cellular death and subsequent mortality in mosquito vectors.
The plaque morphology of viruses is a classical indicator of the effects of a mutation on the viral cytopathic effect in cells and spread between cells. We observed large and fuzzy plaques with WT Kunjin, while S411A Kunjin plaques were small and clearly defined (Fig. 3), suggesting that S411A Kunjin is more toxic to cells than WT Kunjin but is not able to spread as rapidly as WT Kunjin. Our previous work had indicated that the S411A mutation in a replicon-based system reduced viral genome replication (33), so the small plaque size was expected. However, the formation of clearer plaques was not. Therefore, we performed a more quantitative investigation of the S411A Kunjin effect on cell viability using two assays (the resazurin and CellTiter-Glo assays) that probed for different aspects of metabolically active cells, the NADH content and the ATP content. The results from both assays indicated that infection with S411A Kunjin results in a larger decrease in metabolic activity than infection with WT Kunjin within both HEK293T and Vero cells (Fig. 4). Previously, studies have shown that reduced intracellular ATP levels lead to proteasome inhibition, which induces apoptosis, leading to cellular death (38–43). Therefore, our metabolic activity data are consistent with our plaque morphology data, in that infection with S411A Kunjin resulted in reduced intracellular ATP levels and an increased cytopathic effect through increased cell death. S411A Kunjin exhibited delayed and decreased viral replication kinetics compared to WT Kunjin (Fig. 5), suggesting that even though the mutated Kunjin virus is more toxic to cells, it does not replicate as efficiently as WT Kunjin. These data are consistent with those from previous studies reporting a decrease in viral genome replication seen with the S411A helicase replicon (33). The previously reported replicon data are a few orders of magnitude different from the viral replication kinetics data due to the lack of spread from the replicon-infected cells. The experiment with the Kunjin virus replicon is an endpoint experiment, so once the replicon is transfected into the cells, it cannot exit the cells since it does not have the structural proteins that are required for viral assembly, whereas the experiment evaluating viral replication kinetics is a continual experiment using infectious virus that can spread between cells. As subsequent infections occur, the reduced replication rates that accumulate cause a higher reduction in replication compared to that in the replicon system.
An interesting but different hypothesis is that hyperactive NS3 helicase affects cellular mRNA. Studies on NS3 helicase function have focused primarily on its effect on genome replication and packaging (44), but our finding that a hyperactive NS3 helicase mutant increases cell death opens up the possibility that NS3 has roles in altering cellular physiology as well. Previously observed results indicated that a recombinant NS3 S411A helicase mutant had a higher rate of helicase activity than the WT but did not have a significantly higher rate of ATPase activity than the WT (33), so it is unlikely that the reduction of cell viability was due to decreased amounts of ATP from NS3 ATP degradation. However, it is possible that increased cytotoxicity is due to another effect of helicase activity on cellular physiology. The hyperactive NS3 helicase may interact with cellular RNAs, leading to the dysregulation of cellular homeostasis. NS3 could bind to cellular mRNAs and unwind their secondary structures, causing a disruption in RNA stability and the recruitment of translational factors. This unwinding of cellular mRNAs would result in an imbalance within the cell, inducing cellular apoptosis. We are currently exploring if NS3 affects cellular RNAs.
The observed reductions in cell viability led us to investigate the effect of S411A on infection in mosquitoes. Generally, the longevity of mosquitoes infected with flaviviruses is similar to that of uninfected mosquitoes (45, 46). During mosquito infection, flaviviruses must overcome four barriers: (i) the midgut infection barrier, (ii) the midgut escape barrier, (iii) the salivary gland infection barrier, and (iv) the salivary gland escape barrier (47). For the first barrier, the virus must successfully infect and replicate in the midgut epithelial cells (47, 48). Infection is dependent on the arbovirus-specific interactions with the midgut epithelial cell receptors (49). If the virus cannot establish an infection in the midgut epithelial cells, then the mosquito cannot be infected by the virus. If the virus can establish infection in the midgut, then the next barrier is escaping the midgut by crossing the basal lamina, which surrounds the midgut epithelium (47). After escaping the midgut, the virus can disseminate throughout the rest of the mosquito tissues. If the virus is able to penetrate into the salivary gland, the virus must replicate and be deposited into the apical cavities of acinar cells for the mosquito to transmit the virus to other hosts (47). Not all mosquitoes are able to transmit virus due to unknown reasons. The Culex mosquitoes in our study were blood fed or submitted to intrathoracic (IT) injection with either WT or S411A Kunjin. Mosquito mortality was recorded for 15 days for blood-fed mosquitoes or 9 days for IT injected mosquitoes. The results indicated no significant difference in mortality between mosquitoes blood fed with either WT or S411A Kunjin viruses. However, when combined with the low infection rates observed for S411A Kunjin (Fig. 7), these results suggest that S411A Kunjin is more lethal than the WT if infection is established after being taken up by a blood meal. Supporting this, mosquitoes that were intrathoracically injected with S411A Kunjin exhibited an increase in mortality compared to mosquitoes that were intrathoracically injected with WT Kunjin. Interestingly, WT lethality was observed in mosquitoes only when administered by a blood feed, suggesting that Kunjin virus may result in death most commonly as a result of midgut infection. Together, our data suggest that S411A Kunjin viruses are inefficient at crossing the midgut infection barrier to establish infection (Fig. 7). However, upon bypassing the midgut infection and midgut escape barriers through IT injection, S411A Kunjin was more lethal (Fig. 6B). The basis for the observed increased mortality is not yet clear but could be due to an increased cytopathic effect in infected cells, similar to what was observed in cell culture.
To further investigate the distribution of WT Kunjin and S411A Kunjin infection within the C. quinquefasciatus mosquitoes, bodies, legs/wings, and saliva were collected 7 or 14 days after a blood feed and analyzed for the presence of virus. Thirty percent (day 14) to 50% (day 7) of mosquito bodies were positive for WT Kunjin infection, whereas less than 15% of bodies were positive for S411A Kunjin infection on either collection day. These data suggest that WT Kunjin was able to routinely establish infection within midgut epithelial cells, while S411A Kunjin did so less effectively. However, when legs/wings and saliva were analyzed, WT Kunjin was found at extremely low levels, while S411A Kunjin was found in over half of infected mosquitoes, suggesting that, once S411A Kunjin was able to cross the midgut escape barrier, it was able to replicate more efficiently in peripheral tissues than WT Kunjin. One interesting finding was that at 7 days p.i. some mosquitoes infected with WT Kunjin were observed to have virus present in their legs/wings and saliva, but none were found to have virus present in their legs/wings and saliva at 14 days p.i. Since dissemination for WT Kunjin was rare, it is possible that mosquitoes that experience dissemination by WT Kunjin are more likely to die than those in which the infection is maintained within the midgut. This phenomenon is not typical in flavivirus infections of mosquitoes (50, 51).
Previous studies have suggested that arboviruses may require apoptosis to escape the midgut and infect the salivary glands of Culex mosquitoes (48, 52–55). Thus, taking into account the cell culture results suggesting that S411A Kunjin induces increased cellular death, S411A Kunjin viruses may be able to exit the midgut more effectively than WT Kunjin due to the increased induction of apoptosis. Even though S411A Kunjin has a lower initial infection rate than WT Kunjin, the mutant virus was more toxic to infected cells, and thus, the mutant virus may be able to induce apoptosis and disseminate into the rest of the body, leading to a higher potential transmission rate with increased salivary gland infection.
In conclusion, this study provides insight into how a mutation producing a hyperactive NS3 helicase contributes to Kunjin virus replication and the effect on cellular responses during infection. S411A Kunjin negatively affected the overall replication of the virus and increased the cytopathic effect in cells, potentially resulting in increased mosquito mortality. Infection with S411A Kunjin resulted in less metabolic activity in cells and, ultimately, cellular death. When considering the increased mortality of mosquitoes IT injected with S411A Kunjin, it seems likely that cells within mosquitoes undergo a cytopathic effect similar to that observed in cell culture. Cellular death in mosquitoes could allow S411A Kunjin to disseminate into the legs/wings and saliva more efficiently than WT Kunjin and result in increased mosquito death. Virus-induced mortality is not ideal for the long-term maintenance of virus in mosquitoes, so flaviviruses appear to have evolved mechanisms to reduce their helicase activity to reduce virus-induced cell killing. Overall, these data indicate that NS3 helicase activity may have significant roles during viral infection in cell culture and in vivo and that NS3 motif V may play a central role in controlling virus-induced mortality in mosquito vectors to allow for efficient viral transmission.
MATERIALS AND METHODS
Cell culture and viruses.
HEK293T and Vero (African Green Monkey kidney epithelial) cells were maintained in HyClone Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 mM HEPES (pH 7.5), 5% penicillin-streptomycin, and 5% l-glutamine. All cells were grown in humidified incubators at 37°C with 5% CO2. The West Nile virus (Kunjin subtype) infectious clone was generously provided by Alexander Khromykh (University of Queensland) (56).
Virus mutagenesis.
To produce the T407A Kunjin and S411A Kunjin NS3 mutant viruses, a novel bacterium-free virus launch system was used. The system was based on in vitro NEBuilder assembly of PCR-amplified DNAs containing a eukaryotic polymerase II promoter with PCR fragments containing viral genome sequences and the direct transfection of assembled DNAs into Vero cells. Three PCR fragments were produced using the Q5 DNA polymerase system (New England Biolabs) according to the manufacturer’s instructions (56). PCR fragment 1 contained the cytomegalovirus (CMV) immediate early promoter (612 bp), with pcDNA-3.1 used as the PCR template. PCR fragment 2 (5,867 bp) contained the 5′ region of the Kunjin virus genome. PCR fragment 3 (5,309 bp) contained the 3′ end of the Kunjin virus genome, in addition to a hepatitis delta virus ribozyme. The Kunjin virus infectious clone plasmid FLSDXHDVr was used as the PCR template for fragments 2 and 3 (57). The primer sequences used to produce PCR fragments with overlapping 5′ and 3′ ends for NEBuilder assembly were designed using the NEBuilder assembly tool and are listed in Table 1.
TABLE 1.
NEBuilder primers used for detection of T407A and S411A Kunjin viruses
| Fragmenta | NEBuilder primerb | Primer sequenceb,c |
|---|---|---|
| Fragment 1 | CMV forward | atcggaatctGATTATTGACTAGTTATTAATAGTAATCAATTACG |
| CMV reverse | gcgaactactCGGTTCACTAAACGAGCTC | |
| Fragment 2 | ||
| 5′ T407A virus | 5′ Kunjin forward | tagtgaaccgAGTAGTTCGCCTGTGTGAG |
| Kunjin virus | 5′ Kunjin (T407A) reverse | atatatctgtGGCGACGACAAAGTCCCAATC |
| Fragment 3 | ||
| 3′ T407A virus | 3′ Kunjin (T407A) forward | tgtcgtcgccACAGATATATCTGAGATGGG |
| Kunjin virus | 3′ Kunjin reverse | gtcaataatcTTCCGATAGAGAATCGAG |
| Fragment 2 | ||
| 5′ S411A virus | 5′ Kunjin forward | tagtgaaccgAGTAGTTCGCCTGTGTGAG |
| Kunjin virus | 5′ Kunjin (S411A) reverse | ctcccatctcTGCTATATCTGTTGTGACGAC |
| Fragment 3 | ||
| 3′ S411A virus | 3′ Kunjin (S411A) forward | agatatagcaGAGATGGGAGCAAACTTTAAG |
| Kunjin virus | 3′ Kunjin reverse | gtcaataatcTTCCGATAGAGAATCGAG |
The mutant Kunjin viruses were generated from three fragments, fragments 1, 2, and 3. The product of fragment 2 from the NEBuilder assembly reaction contains the specified mutation.
The underlined primers for fragments 2 and 3 contain the alanine mutation at either position 407 or position 411 (underlined).
The sequences indicate the 5′ overlap sequence (lowercase nucleotides)/spacer sequence/and the sequence annealed at the 3′ end (uppercase nucleotides).
The NS3 T407A and S411A mutations (33) were separately engineered into the fragment 2 reverse primer and the fragment 3 forward primers. PCR products were gel extracted with a Qiagen gel extraction kit and quantified by UV spectrophotometry and agarose gel electrophoresis. To assemble the WT Kunjin, T407A Kunjin, or S411A Kunjin fragments, equal molar amounts of each fragment were mixed in a total DNA mass of 200 ng for each virus in ultrapure water in a final volume of 15 μl. An equal volume of New England Biolabs NEBuilder 2× master mix was added to the DNAs, and the reaction mixture was incubated at 50°C for 4 h. The assembled DNAs were transfected directly into Vero cells by adding 1 μl of JetPrime transfection reagent (PolyPlus) to the assembly mixture, the mixture was incubated at 22°C for 15 min, and the transfection mixture was added to 50% confluent Vero cells. The DMEM containing 10% fetal bovine serum and 50 mM HEPES (pH 7.5) was changed 24 h after transfection, and the cells were incubated for 6 additional days and monitored for a cytopathic effect. Medium was collected on day 6 as the passage 0 (P0) stock. Virus was amplified in a T75 flask that had been seeded at 50% confluence for 7 additional days, and clarified medium was collected as the P1 stock. Finally, the P1 stock was used to infect a T150 flask of 50% confluent Vero cells for 7 days, the medium was collected and clarified of cellular debris, and the clarified medium was frozen at −80°C and used as the P2 stock. The P2 stocks were quantified for infectivity via a focus-forming assay. T407A Kunjin was unrecoverable from the infections. The presence of S411A Kunjin was verified by extracting RNA from the P2 stock and reverse transcribing and PCR amplifying the NS3 region of Kunjin virus using the Kunjin virus NS3 sequence forward (5′-ATGCACCAATATCCGACTTACA) and reverse (5′-TGGCCTCAGAATCTTCCTTTC) primers, and the sequence of the PCR 794-bp amplicon was determined by Sanger sequencing.
Viral infectivity.
HEK293T cells were plated into 12-well plates at 20,000 cells/well and allowed to adhere to the plates overnight. On the next day, the cells were infected at an MOI of 0.01 PFU/cell with either WT Kunjin or S411A Kunjin in triplicate under biosafety level 2 (BSL2) conditions. Both intracellular and extracellular viral samples were collected every 12 h for 5 days. The extracellular viral samples were processed through focus-forming assays to determine the viral titer at each time point. The growth curves were plotting using matplotlib software (58).
Resazurin assay.
HEK293T cells were plated into 96-well plates at 10,000 cells/well. Additionally, DMEM with 10% FBS was plated into one row for each plate as a negative control for resazurin. On the following day, cells were either not infected or infected with either WT or S411A Kunjin at an MOI of 5 PFU/cell. The DMEM was not infected. Every 24 h over the course of 6 days, the cells as well as the negative control were treated with resazurin (0.15 mg/ml). The treated plate was then incubated for 1 h at 37°C with 5% CO2 before measuring the fluorescence at an excitation wavelength of 560 nm and an emission wavelength of 590 nm on a Victor X5 multilabel plate reader (Perkin Elmer).
CellTiter-Glo assay.
Vero and HEK293T cells were plated into 96-well plates at 10,000 cells/well. On the following day, the cells in each plate were either not infected or infected with WT or S411A Kunjin at an MOI of 5 PFU/cell. Every 24 h for the next 6 days, the cells were treated with 1× CellTiter-Glo reagent and incubated at room temperature for 10 min, before measuring the luminescence with an exposure time of 0.5 s on a Victor X5 multilabel plate reader.
Mosquitoes.
C. quinquefasciatus mosquito larvae (59) were propagated on a 1:1 mix of powdered tetra food and powdered rodent chow. Adult mosquitoes were kept on a 16-h light, 8-h dark cycle at 28°C with 70% to 80% humidity. Water and sugar were provided ad libitum, and citrated sheep blood was provided to maintain the colony. Mosquito infection experiments with Kunjin virus were performed exclusively on female mosquitoes and under BSL3 conditions.
Infection of mosquitoes with Kunjin virus and analysis.
C. quinquefasciatus mosquitoes were either fed infectious blood meals or intrathoracically injected to introduce Kunjin virus. Blood-fed mosquitoes were fed an infectious blood meal of defibrillated calf blood diluted by half with 2.5 × 106 PFU/ml Kunjin virus or medium alone as a negative control. The blood meals also contained 2 mM ATP. For the IT injection experiments, the mosquitoes were injected with 138 nl WT or S411A Kunjin virus (∼345 PFU/mosquito) using a Nanoject II microinjector (Drummond Scientific). Engorged female mosquitoes were maintained for up to 15 days under the conditions described above but in the BSL3 insectary, and the mortality rate was determined daily. For infection, dissemination, and transmission experiments, after 7 or 14 days of incubation, mosquitoes were anesthetized with cold and kept on ice while their legs and wings were removed, the mosquitoes were salivated for 30 min in a capillary tube filled with immersion oil, and the bodies were collected. Legs/wings and bodies were homogenized at 24 Hz for 1 min in 500 μl mosquito diluent with a stainless steel bead, and saliva samples were stored in 250 μl mosquito diluent as previously described (60). All mosquito samples were clarified by centrifugation at 15,000 × g for 5 min at 4°C and then determined to be positive or negative by infection with undiluted samples by a tragacanth gum overlay plaque assay on Vero cell monolayers as previously described (61).
Visual analysis of NS3.
The dengue virus 4 NS3 helicase crystal structure (Protein Data Bank accession number 2JLV) was obtained for visual presentation. All structural images were generated using VMD software (62). The interaction highlighted between residues 407 and 411 is the measured distance between the two residue side chains.
ACKNOWLEDGMENTS
We acknowledge the support of NIH grants R01 AI132668 to B.J.G. and R01 AI067380 to G.D.E. We also acknowledge helpful discussion with Erin R. Lynch.
K.E.D. developed all of the biochemical and in vitro replication data. N.R.S. developed all of the in vivo mosquito data. Both were involved in the development of the manuscript, with K.E.D. taking the lead in manuscript development.
REFERENCES
- 1.Frost MJ, Zhang J, Edmonds JH, Prow NA, Gu X, Davis R, Hornitzky C, Arzey KE, Finlaison D, Hick P, Read A, Hobson-Peters J, May FJ, Doggett SL, Haniotis J, Russell RC, Hall RA, Khromykh AA, Kirkland PD. 2012. Characterization of virulent West Nile Virus Kunjin strain, Australia, 2011. Emerg Infect Dis 18:792–800. doi: 10.3201/eid1805.111720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray TJ, Burrow JN, Markey PG, Whelan PI, Jackson J, Smith DW, Currie BJ. 2011. West Nile virus (Kunjin subtype) disease in the northern territory of Australia—a case of encephalitis and review of all reported cases. Am J Trop Med Hyg 85:952–956. doi: 10.4269/ajtmh.2011.11-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solomon T. 2004. Flavivirus encephalitis. N Engl J Med 351:370–378. doi: 10.1056/NEJMra030476. [DOI] [PubMed] [Google Scholar]
- 4.Weaver SC, Reisen WK. 2010. Present and future arboviral threats. Antiviral Res 85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. 2017. West Nile virus key facts. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 6.Byas AD, Ebel GD. 2020. Comparative pathology of West Nile virus in humans and non-human animals. Pathogens 9:48. doi: 10.3390/pathogens9010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciota AT. 2017. West Nile virus and its vectors. Curr Opin Insect Sci 22:28–36. doi: 10.1016/j.cois.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Petersen LR, Carson PJ, Biggerstaff BJ, Custer B, Borchardt SM, Busch MP. 2013. Estimated cumulative incidence of West Nile virus infection in US adults, 1999–2010. Epidemiol Infect 141:591–595. doi: 10.1017/S0950268812001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busch MP, Wright DJ, Custer B, Tobler LH, Stramer SL, Kleinman SH, Prince HE, Bianco C, Foster G, Petersen LR, Nemo G, Glynn SA. 2006. West Nile virus infections projected from blood donor screening data, United States, 2003. Emerg Infect Dis 12:395–402. doi: 10.3201/eid1205.051287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flynn LM, Coelen RJ, Mackenzie JS. 1989. Kunjin virus isolates of Australia are genetically homogeneous. J Gen Virol 70:2819–2824. doi: 10.1099/0022-1317-70-10-2819. [DOI] [PubMed] [Google Scholar]
- 11.Grubaugh ND, Ebel GD. 2016. Dynamics of West Nile virus evolution in mosquito vectors. Curr Opin Virol 21:132–138. doi: 10.1016/j.coviro.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindenbach BD, Heinz-Jurgen T, Rice CM. 2007. Flaviviridae: the viruses and their replication, p 1101–1152. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 13.Mastrangelo E, Milani M, Bollati M, Selisko B, Peyrane F, Pandini V, Sorrentino G, Canard B, Konarev PV, Svergun DI, de Lamballerie X, Coutard B, Khromykh AA, Bolognesi M. 2007. Crystal structure and activity of Kunjin virus NS3 helicase; protease and helicase domain assembly in the full length NS3 protein. J Mol Biol 372:444–455. doi: 10.1016/j.jmb.2007.06.055. [DOI] [PubMed] [Google Scholar]
- 14.Scherret JH, Poidinger M, Mackenzie JS, Broom AK, Deubel V, Lipkin WI, Briese T, Gould EA, Hall RA. 2001. The relationships between West Nile and Kunjin viruses. Emerg Infect Dis 7:697–705. doi: 10.3201/eid0704.010418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall RA, Scherret JH, Mackenzie JS. 2001. Kunjin virus: an Australian variant of West Nile? Ann N Y Acad Sci 951:153–160. [PubMed] [Google Scholar]
- 16.Coia G, Parker MD, Speight G, Byrne ME, Westaway EG. 1988. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol 69:1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- 17.Liu WJ, Sedlak PL, Kondratieva N, Khromykh AA. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J Virol 76:10766–10775. doi: 10.1128/jvi.76.21.10766-10775.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R. 2018. Rewiring cellular networks by members of the Flaviviridae family. Nat Rev Microbiol 16:125–142. doi: 10.1038/nrmicro.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chambers TJ, Hahn CS, Galler R, Rice CM. 1990. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol 44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 20.Brand C, Bisaillon M, Geiss BJ. 2017. Organization of the flavivirus RNA replicase complex. Wiley Interdiscip Rev RNA 8:e1437. doi: 10.1002/wrna.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saeedi BJ, Geiss BJ. 2013. Regulation of flavivirus RNA synthesis and capping. Wiley Interdiscip Rev RNA 4:723–735. doi: 10.1002/wrna.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo D, Xu T, Watson RP, Scherer-Becker D, Sampath A, Jahnke W, Yeong SS, Wang CH, Lim SP, Strongin A, Vasudevan SG, Lescar J. 2008. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J 27:3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mastrangelo E, Bolognesi M, Milani M. 2012. Flaviviral helicase: insights into the mechanism of action of a motor protein. Biochem Biophys Res Commun 417:84–87. doi: 10.1016/j.bbrc.2011.11.060. [DOI] [PubMed] [Google Scholar]
- 24.Pérez-Villa A, Darvas M, Bussi G. 2015. ATP dependent NS3 helicase interaction with RNA: insights from molecular simulations. Nucleic Acids Res 43:8725–8734. doi: 10.1093/nar/gkv872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C-C, Huang Z-S, Chiang P-L, Chen C-T, Wu H-N. 2009. Analysis of the nucleoside triphosphatase, RNA triphosphatase, and unwinding activities of the helicase domain of dengue virus NS3 protein. FEBS Lett 583:691–696. doi: 10.1016/j.febslet.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Warrener P, Tamura JK, Collett MS. 1993. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J Virol 67:989–996. doi: 10.1128/JVI.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wengler G, Wengler G. 1991. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology 184:707–715. doi: 10.1016/0042-6822(91)90440-M. [DOI] [PubMed] [Google Scholar]
- 28.Wengler G, Wengler G. 1993. The NS 3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology 197:265–273. doi: 10.1006/viro.1993.1587. [DOI] [PubMed] [Google Scholar]
- 29.Fairman-Williams ME, Guenther UP, Jankowsky E. 2010. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol 20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Story RM, Steitz TA. 1992. Structure of the recA protein-ADP complex. Nature 355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 31.Swarbrick CMD, Basavannacharya C, Chan KWK, Chan S-A, Singh D, Wei N, Phoo WW, Luo D, Lescar J, Vasudevan SG. 2017. NS3 helicase from dengue virus specifically recognizes viral RNA sequence to ensure optimal replication. Nucleic Acids Res 45:12904–12920. doi: 10.1093/nar/gkx1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidson RB, Hendrix J, Geiss BJ, McCullagh M. 2018. Allostery in the dengue virus NS3 helicase: insights into the NTPase cycle from molecular simulations. PLoS Comput Biol 14:e1006103. doi: 10.1371/journal.pcbi.1006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du Pont KE, Davidson RB, McCullagh M, Geiss BJ. 2020. Motif V regulates energy transduction between the flavivirus NS3 ATPase and RNA-binding cleft. J Biol Chem 295:1551–1564. doi: 10.1074/jbc.RA119.011922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilbert DF, Friedrich O. 2017. Cell viability assays. Springer, New York, NY. [Google Scholar]
- 35.O'Brien J, Wilson I, Orton T, Pognan F. 2000. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem 267:5421–5426. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- 36.Rampersad SN. 2012. Multiple applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors (Basel) 12:12347–12360. doi: 10.3390/s120912347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kay BH, Fanning ID, Carley JG. 1982. Vector competence of Culex pipiens quinquefasciatus for Murray Valley encephalitis, Kunjin, and Ross River viruses from Australia. Am J Trop Med Hyg 31:844–848. doi: 10.4269/ajtmh.1982.31.844. [DOI] [PubMed] [Google Scholar]
- 38.Huang H, Zhang X, Li S, Liu N, Lian W, McDowell E, Zhou P, Zhao C, Guo H, Zhang C, Yang C, Wen G, Dong X, Lu L, Ma N, Dong W, Dou QP, Wang X, Liu J. 2010. Physiological levels of ATP negatively regulate proteasome function. Cell Res 20:1372–1385. doi: 10.1038/cr.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Latta M, Künstle G, Leist M, Wendel A. 2000. Metabolic depletion of ATP by fructose inversely controls CD95- and tumor necrosis factor receptor 1-mediated hepatic apoptosis. J Exp Med 191:1975–1986. doi: 10.1084/jem.191.11.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eguchi Y, Shimizu S, Tsujimoto Y. 1997. Intracellular ATP levels determine cell death fate by apoptosis or necrosis. Cancer Res 57:1835–1840. [PubMed] [Google Scholar]
- 41.Leist M, Single B, Castoldi AF, Kühnle S, Nicotera P. 1997. Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. J Exp Med 185:1481–1486. doi: 10.1084/jem.185.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Zheng H, Tang M, Ryu Y-C, Wang X. 2008. A therapeutic dose of doxorubicin activates ubiquitin-proteasome system-mediated proteolysis by acting on both the ubiquitination apparatus and proteasome. Am J Physiol Heart Circ Physiol 295:H2541–H2550. doi: 10.1152/ajpheart.01052.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwong JQ, Henning MS, Starkov AA, Manfredi G. 2007. The mitochondrial respiratory chain is a modulator of apoptosis. J Cell Biol 179:1163–1177. doi: 10.1083/jcb.200704059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patkar CG, Kuhn RJ. 2008. Yellow fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J Virol 82:3342–3352. doi: 10.1128/JVI.02447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samuel GH, Adelman ZN, Myles KM. 2018. Antiviral immunity and virus-mediated antagonism in disease vector mosquitoes. Trends Microbiol 26:447–461. doi: 10.1016/j.tim.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chamberlain RW, Sudia WD. 1961. Mechanism of transmission of viruses by mosquitoes. Annu Rev Entomol 6:371–390. doi: 10.1146/annurev.en.06.010161.002103. [DOI] [PubMed] [Google Scholar]
- 47.Rückert C, Ebel GD. 2018. How do virus-mosquito interactions lead to viral emergence? Trends Parasitol 34:310–321. doi: 10.1016/j.pt.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franz A, Kantor A, Passarelli A, Clem R. 2015. Tissue barriers to arbovirus infection in mosquitoes. Viruses 7:3741–3767. doi: 10.3390/v7072795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mercado-Curiel RF, Black WC, de L Muñoz M. 2008. A dengue receptor as possible genetic marker of vector competence in Aedes aegypti. BMC Microbiol 8:118. doi: 10.1186/1471-2180-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moudy RM, Meola MA, Morin L-LL, Ebel GD, Kramer LD. 2007. A newly emergent genotype of West Nile virus is transmitted earlier and more efficiently by Culex mosquitoes. Am J Trop Med Hyg 77:365–370. doi: 10.4269/ajtmh.2007.77.365. [DOI] [PubMed] [Google Scholar]
- 51.Robison A, Young MC, Byas AD, Rückert C, Ebel GD. 18 May 2020. Comparison of Chikungunya virus and Zika virus replication and transmission dynamics in Aedes aegypti mosquitoes. Am J Trop Med Hyg doi: 10.4269/ajtmh.20-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H, Gort T, Boyle DL, Clem RJ. 2012. Effects of manipulating apoptosis on Sindbis virus infection of Aedes aegypti mosquitoes. J Virol 86:6546–6554. doi: 10.1128/JVI.00125-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelly EM, Moon DC, Bowers DF. 2012. Apoptosis in mosquito salivary glands: Sindbis virus-associated and tissue homeostasis. J Gen Virol 93:2419–2424. doi: 10.1099/vir.0.042846-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Q, Clem RJ. 2011. Defining the core apoptosis pathway in the mosquito disease vector Aedes aegypti: the roles of iap1, ark, dronc, and effector caspases. Apoptosis 16:105–113. doi: 10.1007/s10495-010-0558-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Neill K, Olson BJSC, Huang N, Unis D, Clem RJ. 2015. Rapid selection against arbovirus-induced apoptosis during infection of a mosquito vector. Proc Natl Acad Sci U S A 112:E1152–E1161. doi: 10.1073/pnas.1424469112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khromykh AA, Westaway EG. 1994. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J Virol 68:4580–4588. doi: 10.1128/JVI.68.7.4580-4588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khromykh AA, Sedlak PL, Guyatt KJ, Hall RA, Westaway EG. 1999. Efficient trans-complementation of the flavivirus kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J Virol 73:10272–10280. doi: 10.1128/JVI.73.12.10272-10280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hunter JD. 2007. Matplotlib: a 2D graphics environment. Comput Sci Eng 9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 59.Ciota AT, Chin PA, Kramer LD. 2013. The effect of hybridization of Culex pipiens complex mosquitoes on transmission of West Nile virus. Parasit Vectors 6:305. doi: 10.1186/1756-3305-6-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weger-Lucarelli J, Duggal NK, Bullard-Feibelman K, Veselinovic M, Romo H, Nguyen C, Rückert C, Brault AC, Bowen RA, Stenglein M, Geiss BJ, Ebel GD. 2017. Development and characterization of recombinant virus generated from a New World Zika virus infectious clone. J Virol 91:e01765-16. doi: 10.1128/JVI.01765-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia-Luna SM, Weger-Lucarelli J, Rückert C, Murrieta RA, Young MC, Byas AD, Fauver JR, Perera R, Flores-Suarez AE, Ponce-Garcia G, Rodriguez AD, Ebel GD, Black WC. 2018. Variation in competence for ZIKV transmission by Aedes aegypti and Aedes albopictus in Mexico. PLoS Negl Trop Dis 12:e0006599. doi: 10.1371/journal.pntd.0006599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]