The current pandemic of respiratory illness, COVID-19, is caused by a recently emerged coronavirus named SARS-CoV-2. This virus infects airway and lung cells causing fever, dry cough, and shortness of breath. Severe cases of COVID-19 can result in lung damage, low blood oxygen levels, and even death. As there are currently no vaccines approved for use in humans, studies of the mechanisms of SARS-CoV-2 infection are urgently needed. Our research identifies an excellent system to model SARS-CoV-2 infection of the human airways that can be used to test various treatments. Analysis of infection in this model system found that human airway epithelial cell cultures induce a strong proinflammatory cytokine response yet block the production of type I and III IFNs to SARS-CoV-2. However, treatment of airway cultures with the immune molecules type I or type III interferon (IFN) was able to inhibit SARS-CoV-2 infection. Thus, our model system identified type I or type III IFN as potential antiviral treatments for COVID-19 patients.
KEYWORDS: COVID-19, lung, SARS-CoV-2, cytokines, innate immunity, type I interferon
ABSTRACT
The newly emerged human coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has caused a pandemic of respiratory illness. Current evidence suggests that severe cases of SARS-CoV-2 are associated with a dysregulated immune response. However, little is known about how the innate immune system responds to SARS-CoV-2. In this study, we modeled SARS-CoV-2 infection using primary human airway epithelial (pHAE) cultures, which are maintained in an air-liquid interface. We found that SARS-CoV-2 infects and replicates in pHAE cultures and is directionally released on the apical, but not basolateral, surface. Transcriptional profiling studies found that infected pHAE cultures had a molecular signature dominated by proinflammatory cytokines and chemokine induction, including interleukin 6 (IL-6), tumor necrosis factor alpha (TNF-α), and CXCL8, and identified NF-κB and ATF-4 as key drivers of this proinflammatory cytokine response. Surprisingly, we observed a complete lack of a type I or III interferon (IFN) response to SARS-CoV-2 infection. However, pretreatment and posttreatment with type I and III IFNs significantly reduced virus replication in pHAE cultures that correlated with upregulation of antiviral effector genes. Combined, our findings demonstrate that SARS-CoV-2 does not trigger an IFN response but is sensitive to the effects of type I and III IFNs. Our studies demonstrate the utility of pHAE cultures to model SARS-CoV-2 infection and that both type I and III IFNs can serve as therapeutic options to treat COVID-19 patients.
IMPORTANCE The current pandemic of respiratory illness, COVID-19, is caused by a recently emerged coronavirus named SARS-CoV-2. This virus infects airway and lung cells causing fever, dry cough, and shortness of breath. Severe cases of COVID-19 can result in lung damage, low blood oxygen levels, and even death. As there are currently no vaccines approved for use in humans, studies of the mechanisms of SARS-CoV-2 infection are urgently needed. Our research identifies an excellent system to model SARS-CoV-2 infection of the human airways that can be used to test various treatments. Analysis of infection in this model system found that human airway epithelial cell cultures induce a strong proinflammatory cytokine response yet block the production of type I and III IFNs to SARS-CoV-2. However, treatment of airway cultures with the immune molecules type I or type III interferon (IFN) was able to inhibit SARS-CoV-2 infection. Thus, our model system identified type I or type III IFN as potential antiviral treatments for COVID-19 patients.
INTRODUCTION
In December 2019, a novel human coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged in Wuhan, China, causing an outbreak of severe respiratory disease (1, 2). In the span of several months, SARS-CoV-2 has rapidly escalated to a pandemic, with over 12 million infections and 550,000 deaths worldwide (3). There are currently no vaccines or antivirals approved for use in humans that can prevent or treat infection. SARS-CoV-2 infection manifests as an upper and lower respiratory disease (named COVID-19 by the World Health Organization) characterized by fever, dry cough, and shortness of breath. SARS-CoV-2 targets lower respiratory tract cells, with one study finding 93% of their patients’ bronchial lavages were positive for SARS-CoV-2 by quantitative reverse transcription-PCR (qRT-PCR) (4). Correspondingly, lung abnormalities have been observed in several patients with COVID-19, and severe infection can lead to respiratory failure and lung tissue destruction (5). Severe disease has also been associated with a low level of lymphocytes in the blood and high levels of proinflammatory cytokines, such as interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-α) (6). However, the pathways through which inflammation occurs are largely unknown.
The SARS-CoV-2 genome is 29.8 kb in length and predicted to contain 12 open reading frames. This includes 15 putative nonstructural proteins, envelope and capsid proteins, an RNA-dependent RNA polymerase (RDRP), and a spike protein (7, 8). SARS-CoV-2 is closely related to the β-coronavirus SARS-CoV, which caused an outbreak of acute respiratory distress syndrome in China in 2003 (9). β-Coronaviruses use the spike protein receptor binding domain to gain entry to target cells, and recent studies have found that SARS-CoV-2 and SARS-CoV utilize the same receptor, ACE-2 (10). SARS-CoV-2 entry was also found to require the expression of the cellular protease TMPRSS2 (10). ACE-2 and TMPRSS2 are expressed in epithelial tissue from the lung and gut, with highest expression in ciliated cells from the nasal cavity (11). Accordingly, in vitro systems have confirmed that SARS-CoV-2 productively infects primary human airway epithelial (pHAE) cultures, with cells isolated from the nasal cavity sustaining the highest level of SARS-CoV-2 replication (12). pHAE cultures isolated from the bronchial region are also highly susceptible to SARS-CoV-2, and similarly to SARS-CoV, this replication occurs primarily in ciliated cells, where ACE-2 expression is localized (12, 13).
Type I interferon (IFN) is the first line of defense and is critical for blocking early virus replication, spread, and tropism as well as promoting the adaptive immune response. Type I IFN induces a systemic response that impacts nearly every cell in the host, while type III IFNs are restricted to anatomic barriers and selected immune cells (14). This selectivity is due to the receptor expression patterns: type I IFN binds IFNAR1 and IFNAR2, which are ubiquitously expressed, while type III IFN binds IFNLR1 and IL-10-Rβ, which are expressed preferentially on epithelial cells (14). Despite using different receptors, type I and III IFNs use the same downstream signaling complex (ISGF3) and induce similar gene expression profiles. Type III IFN induces lower levels of ISG (interferon-stimulated gene) expression, and at a lower rate than type I IFN (15, 16). Thus, type III IFN produces a less inflammatory, localized response compared to type I IFN. Recent research found that treatment of immortalized lung epithelial cells with type I IFN was effective at reducing SARS-CoV-2 viral burden (17). However, the role of type I and type III IFNs in restricting SARS-CoV-2 infection of primary airway epithelial cells has not been studied.
In this study, we sought to address some of these unanswered questions about the innate immune response to SARS-CoV-2. We found that SARS-CoV-2 infects and replicates in pHAE cells and is released from the apical surface. We performed transcriptional profiling and found that infection triggers a robust inflammatory cytokine response characterized by the induction of IL-6, CXCL8, and IL-1 family cytokines. In contrast, we observed a lack of induction of type I or type III IFN and IFN-stimulated genes even though other viral infections are able to induce IFN in pHAE cultures. We found that both pre- and posttreatment of pHAE cultures with type I or III IFNs reduced SARS-CoV-2 replication, and this was correlated with increased expression of IFN-stimulated antiviral effector genes. Combined, our studies demonstrate the utility of pHAE cultures to model SARS-CoV-2 infection and identify type I and III IFNs as potential therapeutics to restrict infection in the airways of COVID-19 patients.
RESULTS
Human bronchial airway epithelial cells are permissive to SARS-CoV-2 infection.
To understand the response of airway epithelial cells to SARS-CoV-2 infection, we utilized primary human airway epithelial (pHAE) cells isolated from the bronchial or tracheal region. These cells were cultured using an air-liquid interface model to create a polarized, pseudostratified epithelial layer. This culture system excellently recapitulates the unique features of the human respiratory tract, including mucus production and coordinated cilium movement (18). To determine if these cultures are permissive to SARS-CoV-2, pHAE cultures were infected on the apical side with SARS-CoV-2 at multiplicities of infection (MOI) of 0.1 and 0.25 (as determined on VeroE6 cells). In our study, we used a low-cell-culture-passaged and sequence-verified SARS-CoV-2 strain, 2019-nCoV/WA1, which was isolated in January 2020 from nasopharyngeal and oropharyngeal swab specimens collected 3 days post-symptom onset (19). Using two different measurements, we demonstrated that SARS-CoV-2 infects and replicates in pHAE cultures. On the apical surface, we detected infectious SARS-CoV-2 beginning 24 h postinfection (p.i.) and increasing through 48 h p.i. as measured by plaque assay (Fig. 1A). In contrast, we were unable to detect infectious SARS-CoV-2 virus on the basolateral side at any time point or MOI, suggesting directional release of the virus from pHAE cultures. We next confirmed the presence of viral RNA in the cells by qRT-PCR with primer/probes that anneal to the SARS-CoV-2 RNA-dependent RNA polymerase (RDRP). We observed an increase in viral RNA between MOI 0.1 and 0.25 at 48 h p.i. (Fig. 1B). To determine what percentage of cells in the pHAE cultures were infected, we utilized an infectious clone SARS-CoV-2 with a green fluorescent protein (GFP) reporter (icSARS-CoV-2-mNG) (20). We confirmed that icSARS-CoV-2-mNG had a viral burden at 72 h p.i. (MOI = 0.5) similar to that of 2019-nCoV/WA1 using a focus-forming assay (FFA) (Fig. 1C). Confocal microscopy of icSARS-CoV-2-mNG-infected samples revealed widespread distribution of GFP at 72 h p.i., with an average of 40% of cells expressing detectable GFP. Additionally, z-stack reconstruction determined that GFP expression was preferentially localized to the apical side (Fig. 1D). Combined, these findings demonstrate that pHAE cultures are permissive for SARS-CoV-2 infection.
FIG 1.
pHAE cultures are permissive to SARS-CoV-2 infection. (A) Differentiated pHAE cultures were infected by adsorption to the apical side at the indicated MOI. Supernatant was collected from the apical or basolateral side of the epithelial layer and the virus was measured by plaque assay. (B) Viral RNA was measured by probing for the SARS-CoV-2 RDRP RNA at 48 h p.i. by qRT-PCR. CT values are represented as relative fold change over mock (log10). (C) pHAE cultures were infected with 2019-nCoV/WA isolate or icSARS-CoV-2-mNG at an MOI of 0.5. At 72 h p.i., viral burden was measured via FFA for the apical side. (D) Representative images of mock or icSARS-CoV-2-mNG infected (MOI = 0.5, 72 h p.i.) cultures stained for nuclei and phalloidin and imaged at ×60 on a confocal microscope. Percentage of GFP+ cells was determined using ImageJ software; z-stack reconstruction was created using Imaris software. All experiments were repeated twice with biological triplicates. Statistical analysis was performed using Prism software; a t test was performed to determine significance of qRT-PCR data and percent GFP+. *, P < 0.05; ****=, P < 0.0001.
SARS-CoV-2 infection prompts a proinflammatory response in pHAE cultures.
We next evaluated the innate immune response to SARS-CoV-2 infection. To this end, we performed bulk mRNA-sequencing analysis on differentiated pHAE cultures infected with SARS-CoV-2 (MOI = 0.25) at 48 h p.i. Following infection, we observed 1,039 differentially expressed genes (DEGs (P < 0.01; 1.5-fold change cutoff), with 458 upregulated (Fig. 2A, in red) and 581 downregulated (Fig. 2A, in blue) DEGs. We detected viral transcripts representing coverage over the viral genome, although there was minor variation between the three replicates (Fig. 2B).
FIG 2.
Bulk RNA-Seq analysis of SARS-CoV-2-infected pHAE cultures. pHAE cultures were infected apically with SARS-CoV-2 (MOI = 0.25) for 48 h, at which point mock- and SARS-CoV-2-infected (n = 3) samples were harvested for bulk RNA-Seq analysis. (A) Volcano plot demonstrating DEGs. Lines indicate cutoffs; P value < 0.01; fold change less than −1.5 or greater than 1.5. Highlighted in red are the most highly upregulated genes (P value < 0.001; fold change > 1.5), in blue are most highly downregulated genes (P value < 0.001; fold change less than −1.5). (B) Normalized read counts (log2) of SARS-CoV-2 RNA products, using the MT246667.1 reference sequence.
We next investigated genes associated with proinflammatory cytokine/chemokine production and signaling. Several genes showed increased expression in SARS-CoV-2-infected cells compared to mock-infected cells (Fig. 3A, purple highlighted genes). We then performed gene set enrichment analysis (GSEA) using the Hallmarks data set from MSigDB. Compared to mock-infected cells, SARS-CoV-2-infected cells had significant enrichment for TNF-α signaling, with a normalized enrichment score (NES) of 1.53 and P value of <0.0001, and IL-6-STAT3-related signaling (NES = 1.34; P < 0.04) (Fig. 3B). Together, these findings indicate that SARS-CoV-2 infection induces a proinflammatory phenotype in pHAE cultures. We also analyzed genes associated with barrier immunity, which include genes related to the production of mucus and antimicrobial/antiviral peptides (AMPs) (Fig. 3C, highlighted in orange). While these genes were expressed in mock-infected and infected cells, we observed little to no change in gene expression and only a few genes approached threshold levels. This suggests that SARS-CoV-2 does not alter mucus and AMP production in pHAE cultures.
FIG 3.
SARS-CoV-2 infection promotes a proinflammatory and ER stress response in pHAE cultures. pHAE cultures were infected apically with SARS-CoV-2 (MOI = 0.25) for 48 h, at which point mock- and SARS-CoV-2-infected (n = 3) samples were harvested for bulk RNA-Seq analysis. For global DEG analysis, see Fig. 2. (A) Volcano plot with all DEGs in gray and the indicated gene set highlighted (purple, proinflammatory signaling). (B) GSEA plots of the enrichment score plotted against gene rank. Individual gene hits are indicated by the solid black line below the enrichment score curve. NES and P value are indicated on the plot. Gene sets are from the Hallmarks gene set from MSigDB. (C) Volcano plot illustrating barrier immunity-associated genes in orange. (D) Network map illustrating regulatory nodes for our DEGs. (E) GSEA plots for the indicated gene sets.
To determine the transcriptional regulatory network induced in SARS-CoV-2-infected cells, we performed cis-regulatory sequence analysis using iRegulon to computationally predict regulatory nodes. iRegulon identifies enrichment of transcription factor binding motifs within a DEG list (21). Consistent with pathway enrichment for proinflammatory cytokines, our analysis identified NF-κB and ATF-4 as top predicted transcriptional regulators following SARS-CoV-2 infection (Fig. 3D; see also Table S1 in the supplemental material). NF-κB regulates a substantial portion of the DEGs, with a network of 312 DEGs. This network includes some of the top upregulated genes, such as the IL-6 and CXCL8 genes. ATF-4 has a slightly smaller enriched network of 219 DEGs, which includes CHAC1, which is a key marker of the unfolded protein response (Fig. 3D) (22). The identification of NF-κB as a key transcriptional node is consistent with our observations that SARS-CoV-2 triggers induction of a proinflammatory response. ATF-4 is associated with the promotion of a cellular stress response. Thus, we used GSEA to investigate if ATF4 corresponds to an enrichment of cellular stress pathways in SARS-CoV-2-infected cells (23). Compared to mock-infected cells, SARS-CoV-2-infected pHAE cultures showed an enrichment of genes related to the unfolded protein response, such as the ASNS, CHAC1, and STC2 genes (NES = 1.39; P value < 0.0001) (Fig. 3E). CEBP plays a critical role in maintaining epithelial barriers by regulating the epithelial-to-mesenchymal transition (24). Accordingly, GSEA also revealed enrichment of genes associated with the transition from epithelial to mesenchymal tissue (Fig. 3E). Overall, transcriptional analysis revealed that in response to SARS-CoV-2 infection, the normal homeostatic functions of pHAE cultures are disrupted and they induce a proinflammatory phenotype characterized by NF-κB signaling, endoplasmic reticulum (ER) stress response and IL-6 production.
Correspondingly, SARS-CoV-2-infected cells had increased transcript expression of IL-6, TNF-α, and other cytokine genes, including the IL-17 family (IL17C and IL-23A) and the IL-1 family (IL-18 and IL1B) (Fig. 4A). In addition, several chemokines were upregulated in SARS-CoV-2-infected cells, including molecules that promote monocyte migration (CCL4 and CCL5) and neutrophil migration (CXCL8 and CXCL6) (Fig. 4A). To confirm our transcriptional findings, cytokine levels were analyzed for both apical and basolateral supernatants from SARS-CoV-2-infected or mock-infected pHAE cultures at 72 h p.i. (MOI = 0.5) using a Luminex assay. In agreement with our transcriptome sequencing (RNA-Seq) data, SARS-CoV-2 infection upregulated the production of IL-6, IL-8, and IL-18 from both the apical and basolateral surfaces (Fig. 4B). Other cytokines identified in the transcriptional data (IL-1α, IP-10, and MIP-1b) were induced by SARS-CoV-2 infection, but only from the basolateral side. Cytokine and chemokine levels were higher on the basolateral surface in both mock- and SARS-CoV-2-infected cultures, suggesting that cytokines/chemokines are preferentially released to the basolateral side. Luminex analysis identified several additional proinflammatory cytokines not included in the transcriptional analysis, such as IL-9 and SDF-1a, which were induced up to 1,000-fold in infected pHAE cells. A few cytokines were notably not detected in SARS-CoV-2-infected samples, including IFN-α1 and TNF-α (Fig. 4B). Together, these data show that SARS-CoV-2 infection of pHAE cultures induces a robust proinflammatory cytokine response.
FIG 4.
pHAE cultures produce proinflammatory cytokines and chemokines in response to SARS-CoV-2 infection. RNA-Seq analysis of the cytokine and chemokine response is shown. For global DEG analysis, see Fig. 2. (A) Heat map illustrating z-scores for the indicated genes in mock- and SARS-CoV-2-infected samples. (B) Supernatants were harvested from the apical and basolateral sides of SARS-CoV-2-infected pHAE cultures (MOI = 0.5) at 72 h p.i. Cytokine protein levels were determined by Luminex assay. Values below the limit of detection are designated nd. All statistical analysis was performed using Prism software. All data were analyzed using one-way or two-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
SARS-CoV-2 does not induce IFN production or signaling in pHAE cultures.
Viral sensing by pathogen recognition receptors (PRRs) is a key initial step in responding to viral infections. Activation of PRR pathways triggers a signaling cascade that induces expression of type I IFN, interferon-regulatory factor (IRF)-dependent and IFN-dependent stimulated genes. First, we determined that SARS-CoV-2 induces minimal type I or III IFNs at the transcript level (Fig. 5A). In both mock- and SARS-CoV-2-infected samples, there was no detectable IFN-α of any subtype, and low induction of IFN-β1 and IFN-λ1, with normalized read counts less than 10 (Fig. 5A). We next determined whether this was due to the lack of expression of molecules required for the induction of IFN transcription, such as the RIG-I-like receptor signaling pathway. We found that several genes within this pathway, including the RIG-I, MDA5, TBK1, TRAF6, IRF-3, and IRF-7 genes, are expressed at baseline but show little to no induction in response to SARS-CoV-2 (Fig. 5B, highlighted in green).
FIG 5.
pHAE cultures do not upregulate type I or III IFNs in response to SARS-CoV-2 infection. RNA-Seq analysis of the IFN response is shown; for global DEG analysis, see Fig. 2. (A) Normalized read counts in mock-infected (white) and SARS-CoV-2-infected (gray) samples of type I and type III IFNs. (B and C) Volcano plots illustrating all genes in gray and genes associated with IFN production in green (B) and genes associated with IFN signaling in pink (C). (D and E) Bar graphs indicating the normalized read count for interferon-stimulated genes (D) and type I and type III IFN receptors (E) for mock- and SARS-CoV-2-infected samples. (F and G) pHAE cultures were infected at an MOI of 0.5 with SARS-CoV-2 or H1N1 (California 2009) for 24 or 48 h, and qRT-PCR was used to quantify viral RNA for SARS-CoV-2 RdRp or H1N1 nucleoprotein (NP) (F) and type I or III IFNs and selected ISGs (G). All qRT-PCR data are represented as fold change over mock, untreated pHAE samples. Data are representative of those from two independent experiments, performed in biological triplicate. All statistical analysis was performed using Prism software. All data were analyzed using one-way or two-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We observed little to no change in expression for several genes related to type I IFN signaling, including transcription factors and antiviral effector genes, such as the IFIT2, IFIT3, IFITM1, OAS1, and MX1 genes (Fig. 5C and D, highlighted in pink). We next evaluated expression of both type I IFN (IFNAR1 and IFNAR2) and type III IFN (IFNLR1 and IL10Rβ) receptors. At the transcript level, the type I and III IFN receptors were present in pHAE cultures, but the expression of these receptors did not change with infection (Fig. 5E). Combined, these data demonstrate that SARS-CoV-2 infection does not induce a type I or III IFN response in pHAE cultures, but these cells express the signaling components to respond to type I or III IFN signaling.
To determine if the lack of IFN production was a unique feature of SARS-CoV-2 infection, we compared the responses of pHAE cultures to SARS-CoV-2 and influenza virus (H1N1 California 2009). qRT-PCR analysis confirmed the presence of SARS-CoV-2 or H1N1 viral RNA, with increasing levels from 24 to 48 h p.i. (Fig. 5F). In agreement with the RNA-Seq analysis, SARS-CoV-2-infected pHAE cultures did not significantly upregulate type I and III IFN or selected ISGs compared to mock-infected samples. In contrast, H1N1-infected pHAE cultures had substantial upregulation compared to mock-infected samples of IFN-β1, IFN-λ1, the innate immune sensor DDX58, and antiviral effectors such as MX1 and IFIT3, with expression increasing between 24 and 48 h p.i. (Fig. 5G). Thus, pHAE cultures are capable of mounting an IFN response but there is a lack of induction during SARS-CoV-2 infection.
Pretreatment with type I and type III IFN restricts SARS-CoV-2 replication.
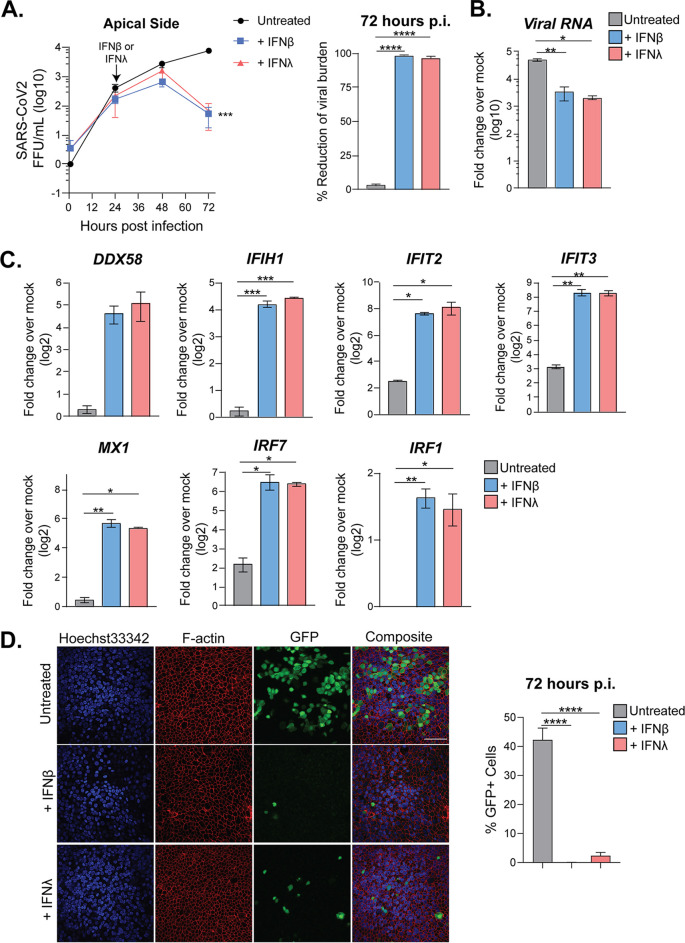
To determine whether SARS-CoV-2 is sensitive to type I and III IFNs, we pretreated the basolateral side of pHAE cultures with IFN-β1 or IFN-λ1 (100 IU/ml) for 24 h prior to infection. The next day, pHAE cultures were infected on the apical side with SARS-CoV-2 (Fig. 6A). Compared to untreated cells, we observed significantly reduced viral RNA in type I (3-fold less) and III (3-fold less) IFN-treated cells by 24 h p.i. (Fig. 6B). We next evaluated infectious virus release and found that pHAE cultures pretreated with type I or III IFNs significantly reduced (14-fold and 12-fold, respectively) SARS-CoV-2 burden by 24 h p.i., resulting in greater than 90% reduction in virus replication compared to that of untreated SARS-CoV-2-infected cells (Fig. 6C).
FIG 6.
Pretreatment with type I or III IFNs restricts SARS-CoV-2 replication in pHAE cultures. pHAE cultures were pretreated from the basolateral side with IFN-β1 or IFN-λ1 (100 IU/ml) for 24 h, after which cultures were infected apically (MOI = 0.25) and harvested at 24 h p.i. (A) Experimental schematic. (B) SARS-CoV-2 burden in untreated, IFN-β1-treated, and IFN-λ1-treated cultures as assessed via focus-forming assay. Percent reduction was calculated as the percentage of the untreated sample at 24 h p.i. (C and D) qRT-PCR analysis was performed at 24 h p.i. for viral RNA (C) or ISGs (D). qRT-PCR data are represented as fold change over mock, untreated pHAE samples. Data are representative of those from two independent experiments, performed in biological triplicate. All statistical analysis was performed using Prism software. All data were analyzed using one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.00001.
To better understand how type I and III IFN signaling promotes restriction of SARS-CoV-2 replication, we performed qRT-PCR analysis of SARS-CoV-2-infected treated and untreated cells. Both type I and III IFN treatment upregulated ISGs in uninfected and infected pHAE cells. This included innate immune sensors RIG-I and MDA5 and the IFIT family of antiviral effector genes (Fig. 6D). Changes in transcription factors were also observed with increases in IRF-7 and, to a lesser extent, IRF-1. Treatment with type I and III IFNs upregulated ISGs regardless of infection status, but SARS-CoV-2-infected samples had larger fold changes than mock-infected cells. In SARS-CoV-2-infected samples, treatment with type I IFN induced higher expression of certain ISGs (IFIH1 and IFIT2) than type III IFN (Fig. 6D). Taken together, our data demonstrate that pretreatment with type I or III IFN increases antiviral effector expression and restricts SARS-CoV-2 replication in pHAE cultures.
Treatment of SARS-CoV-2-infected pHAE cultures with type I and III IFN reduces viral burden.
We next evaluated the antiviral potential of type I and III IFNs in a therapeutic model of infection. In this case, pHAE cultures were infected with SARS-CoV-2 (MOI = 0.5) and, at 24 h p.i., the basolateral side was treated with IFN-β1 or IFN-λ1 (100 IU/ml). Infectious virus release was measured before and after treatment (Fig. 7A). Twenty-four hours after treatment (48 h p.i.), there was no significant difference in viral burden between treatments. However, by 72 h p.i., treatment with both type I and III IFNs had reduced SARS-CoV-2 levels 50-fold compared to those in untreated samples, resulting in a 98% reduction in viral burden at 72 h p.i. (Fig. 7A). Analysis of RNA at 72 h p.i. confirmed that both IFN-β1 and IFN-λ1 treatment reduced viral RNA compared to that in untreated cells, 12-fold and 20-fold, respectively (Fig. 7B). Similar to the case with our pretreated samples (Fig. 6), treatment after infection with type I or III IFNs upregulated ISGs, such as RIG-I, MDA5, and IFIT family members, compared to those in untreated samples (Fig. 7C). Type I and III IFNs increased expression of IRF transcription factors IRF-7 and IRF-1 compared to that in mock samples. Treatment with type I and type III IFNs also significantly reduced the percentage of infected cells, as determined by confocal microscopy. icSARS-CoV-2-mNG-infected cells treated with IFN-λ1 or IFN-β1 had less than 5% or 1% GFP-positive cells, respectively, compared to 40% GFP positive in untreated samples (Fig. 7D). Combined, therapeutic treatment with type I or III IFNs was effective at reducing viral burden and upregulating ISGs in SARS-CoV-2-infected pHAE cultures.
FIG 7.
Posttreatment with type I or III IFNs decreases viral burden in pHAE cultures. pHAE cultures were infected with SARS-CoV-2 (MOI = 0.5) apically. At 24 h p.i., cultures were treated from the basolateral side with IFN-β1 or IFN-λ1 (100 IU/ml). At 72 h p.i. (48 h posttreatment), cultures were harvested for qRT-PCR analysis. (A) SARS-CoV-2 burden was assessed via FFA for the apical side of untreated, IFN-β1-treated, and IFN-λ1-treated cultures. Percent reduction was calculated for the 72-h time point. (B and C) qRT-PCR analysis at 72 h p.i. compared to mock, untreated samples, measuring viral RNA (B) or ISGs (C). qRT-PCR data are represented as fold change over mock. (D) Representative images of icSARS-CoV-2-mNG-infected pHAE cultures (MOI = 0.5), untreated or treated with IFN-β1 or IFN-λ1 at 72 h p.i. Percentage of GFP+ cells was counted using ImageJ software. Results are representative of those from two independent experiments performed in triplicate. All statistical analysis was performed using Prism 8 software. Growth curves were analyzed using two-way ANOVA. qRT-PCR data and microscopy data were analyzed using one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
DISCUSSION
In this study, we found that human pHAE cultures, which model the air-liquid interface of the lung, are permissive to SARS-CoV-2 infection and the virus is unilaterally released from the apical surface. Transcriptional profiling revealed that SARS-CoV-2-infected pHAE cultures trigger a proinflammatory response driven by ATF-4 and NF-κB. This analysis also suggested that the normal cellular functions of pHAE cultures were disrupted during SARS-CoV-2 infection, as evidenced by the enrichment of ER stress pathways. Despite having a baseline expression of viral sensing pathway components, pHAE cultures did not produce type I or III IFN in response to SARS-CoV-2 infection, and there was little to no induction of antiviral effector genes. However, SARS-CoV-2 is sensitive to the effects of type I and III IFN, as pretreatment with exogenous IFN was able to significantly reduce viral burden in pHAE cultures. Therapeutic administration of type I and III IFNs was also effective at restricting viral replication and induced upregulation of ISGs in pHAE cultures, thus demonstrating that SARS-CoV-2 is susceptible to both type I and type III IFNs and identifying them as potential antiviral treatments.
The baseline expression of molecules such as RIG-I, MDA5, IRF-3, and IRF-7 in pHAE cultures suggests that these cells are capable of producing type I and III IFNs. In other models of viral infection, pHAE cultures produce a robust IFN response, and we confirmed this in our system using influenza virus (15, 25) (Fig. 5G). Recent work comparing the transcriptional profiles of pHAE cultures infected with SARS-CoV-2 or influenza also confirmed our findings (17). We performed transcriptional analysis and similarly found that influenza virus-infected or type I IFN-treated pHAE cultures upregulated IFN and ISGs, but SARS-CoV-2-infected cultures lacked a detectable IFN response. Previous studies with SARS-CoV showed that the virus is able to block the production of type I IFN by inhibiting the phosphorylation and nuclear translocation of IRF-3 and interfering with STAT signaling (26–31). However, studies of both in vitro and in vivo SARS-CoV infection found that delayed production of type I IFN did occur eventually (32, 33). Follow-up studies assessing whether SARS-CoV-2 uses similar mechanisms to block type I and III IFN production will be essential for understanding viral antagonism of innate immune signaling pathways during infection.
pHAE cultures did not produce IFN in response to SARS-CoV-2 infection; however, these cells were sensitive to the effects of IFN. Both type I and III IFNs were able to reduce SARS-CoV-2 burden and upregulate ISGs compared to those in untreated cells when pretreated or treated therapeutically. Conducting airway epithelial cells are highly polarized and often have differential localization of receptors, such as ACE-2, on the apical versus basolateral side (34). Accordingly, we noted a distinct directionality in the release of both infectious virus and cytokines from our pHAE cultures (Fig. 1 and 4) In this study, IFN was added to the basolateral side to better mimic the actions of infiltrating immune cells or an intravenous therapeutic. However, an interesting follow-up study would be to compare the differential effects of cytokine application to the apical side. A recent clinical study suggests that type I IFN may be an effective COVID-19 treatment when applied apically, as the aerosolized administration of IFN-α2b reduced the time from symptom onset to viral clearance (35). However, the impact of type III IFN on SARS-CoV-2 infection has not yet been assessed in a clinical setting.
In our study, the reductions in viral load were similar between both IFNs, but type I IFN was able to induce higher expression than type III IFN (∼2-fold) of most ISGs in pretreated pHAE cultures. However, in our posttreated samples, this difference had disappeared. Studies of the differential effects of type I and III IFNs in other viral infection models have shown differences in both magnitude and timing of ISG and IRF transcription factor expression. Type I IFN responses often peak early and then decline sharply, while type III IFN responses take longer to initiate and are more sustained (15, 16). Detailed analysis of the transcriptional response to type I and III IFNs noted that these differences are driven by IRF-1 (36). Correspondingly, we did see higher induction of IRF-1 in type I IFN-treated pHAE cultures than with type III IFN during SARS-CoV-2 infection. Thus, while type I and III IFNs have similar effects on SARS-CoV-2 replication, type III IFN might have a more localized, less inflammatory effect on the immune response. Dissecting the signaling pathways of type I and type III IFN could provide a clearer understanding of the innate immune response in airway epithelial cells and inform the application of these cytokines as therapeutics.
In response to SARS-CoV-2 infection, pHAE cultures primarily produced proinflammatory cytokines, which promote localized edema, fever, and the recruitment of immune cells into the respiratory tract. Many of the cytokines induced by SARS-CoV-2 infection, such as CXCL6, CXCL8, and CXCL5, are chemotactic factors that primarily recruit neutrophils (37). However, the recruitment of neutrophils may be of limited use in controlling SARS-CoV-2, as neutrophils are not particularly effective against intracellular pathogens and have been associated with poor prognosis in other models of respiratory disease (38). Several cytokines primarily associated with a TH17 response (IL-23 and IL-17) were also highly upregulated in SARS-CoV-2-infected pHAE cultures. While TH17 cells are important for barrier immunity, they are most potent against extracellular pathogens and have the potential to cause chronic inflammation (39). IL-6, neutrophil infiltration, and TH17 immunity have all been previously implicated in the development of fibrotic tissue during respiratory disease (40). Additionally, SARS-CoV-2-infected pHAE cultures were highly enriched in genes associated with the unfolded protein response and the epithelial-to-mesenchymal transition, which are both pathways directly linked to the formation of fibrotic tissue (41–43). Taken together, our RNA-Seq data suggest that SARS-CoV-2 may produce a suboptimal immune response in pHAE cultures, as evidenced by a preference for the production of cytokines whose primary function is in the defense against extracellular pathogens and the complete lack of type I and III IFN signaling. Furthermore, this dysregulated immune response combined with an increase in ER stress may promote the formation of fibrotic epithelial tissue during SARS-CoV-2 infection. Additional studies are needed to explore the specific role these cytokines play during SARS-CoV-2 infection of the respiratory system and their implications in the formation of fibrotic tissue.
In this study, we characterized the innate immune response to SARS-CoV-2 infection using an in vitro model of the airway epithelial barrier. We found that pHAE cultures are permissive to SARS-CoV-2 but mount a weak antiviral innate response that notably lacks type I and III IFN production, signaling, and induction of ISGs. Instead, the innate immune response signature of infected pHAEs is dominated by proinflammatory cytokines and chemokines that recruit neutrophils and monocytes. Most importantly, therapeutic treatments of pHAE cultures with type I and III IFNs were able to reduce SARS-CoV-2 burden. Overall, these data suggest that pHAE cultures mount a misdirected innate immune response to SARS-CoV-2 infection, but the early administration of type I or III IFN could potentially decrease virus replication and disease.
MATERIALS AND METHODS
Viruses and cells.
SARS-CoV-2 (2019-nCoV/USA_WA1/2020) was isolated from the first reported case in the United States (19). A plaque-purified passage 4 stock was kindly provided by Natalie Thornburg (CDC, Atlanta, GA). The infectious clone GFP-tagged SARS-CoV-2 (icSARS-CoV-2-mNG) was kindly provided to us by Vineet Menachery (UTMB) (20). Viral titers were determined by plaque assay on VeroE6 cells (ATCC). Vero cells were cultured in complete Dulbecco modified Eagle medium (DMEM) consisting of 1× DMEM (Corning Cellgro), 10% fetal bovine serum (FBS), 25 mM HEPES buffer (Corning Cellgro), 2 mM l-glutamine, 1 mM sodium pyruvate, 1× nonessential amino acids, and 1× antibiotics. Influenza virus (A/California/04/2009 (H1N1)) was propagated in Madin-Darby canine kidney (MDCK) cells cultured in complete DMEM. Viral stock titer was determined on MDCK cells and stored at −80°C until use.
Quantification of infectious virus.
To perform plaque assays, 10-fold dilutions of viral supernatant in serum-free DMEM (VWR; no. 45000-304) were overlaid on VeroE6 cell monolayers and adsorbed for 1 h at 37°C. After adsorption, 0.8% Oxoid agarose in 2× DMEM supplemented with 10% FBS (Atlanta Biologics) and 5% sodium bicarbonate was overlaid, and cultures were incubated for 72 h at 37°C. Plaques were visualized using crystal violet staining (70% methanol in double-distilled water [ddH2O]). For focus-forming assays, 10-fold dilutions of viral supernatant on VeroE6 cells were incubated with a methylcellulose overlay (1.0% methylcellulose in 2× DMEM) for 24 h at 37°C. Methylcellulose was then removed, and cells were fixed with 2% paraformaldehyde (PFA) and permeabilized with 0.1% bovine serum albumin (BSA)-saponin in phosphate-buffered saline (PBS). Cells were incubated with an anti-SARS-CoV-2 spike protein primary antibody conjugated to biotin (generously provided by Jens Wrammert, Emory University) for 2 h at room temperature and then with avidin-horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. Foci were visualized using True Blue HRP substrate and imaged on an enzyme-linked immunosorbent spot assay (ELISPOT) reader (CTL Analyzers).
Generation of pHAE cultures.
Bronchial primary human airway epithelial (pHAE) cultures were kindly provided by C. U. Cotton (Case Western Reserve University) and cultured as described previously (18). Briefly, after initial expansion in F medium supplemented with ROCK inhibitor (Selleck Chemical LLC), bronchial lung specimens were seeded on Transwell permeable support inserts (Costar-Corning), cultured until confluent, and then transferred to an air-liquid interface. Cultures were differentiated for 3 weeks and maintained in DMEM/Ham’s F-12 differentiation medium supplemented with 2% Ultroser G (Pall Corp., France), until they had transepithelial electrical resistance (TEER) measurements greater than 1,000 Ω and were deemed ready for use. In some studies, modifications of the above-described protocol were utilized with primary cells obtained from a commercial source and methods provide by vendor (STEMCELL, Vancouver, BC, Canada) as described by Zhu et al. (2).
SARS-CoV-2 infection of pHAE cultures.
Prior to infection, the apical side of the pHAE culture was washed 3 times with PBS. Virus was diluted to the specified MOI (cell number based on a representative count at the time of infection) in PBS and allowed to adsorb for 1 h at 37°C. After adsorption, the apical side was washed 3 times with PBS to remove excess virus, and the basolateral medium was changed. To collect viral supernatant, PBS was added to the apical side and incubated for 30 min at 37°C. Treatment of pHAE cultures with type I or type III IFN was performed by adding 100 IU/ml of human IFN-β or human IFN-λ1 (PBL Assay Science). Cytokine levels were measured using the cytokine and chemokine 34-plex human Pro-Carta Plex Panel 1 kit (Thermo Fisher).
RNA sequencing and bioinformatics.
pHAE cultures were infected at an MOI of 0.5 for 48 h. RNA was harvested from mock-infected and infected pHAE cultures (n = 3) by treating with RNA lysis buffer for >5 min and gently pipetting to recover cells. Total RNA was extracted using the Zymo Quick-RNA miniprep kit (VWR; R1055) according to the manufacturer’s protocol. mRNA sequencing libraries were prepared by the Yerkes Genomics Core (http://www.yerkes.emory.edu/nhp_genomics_core/). Libraries were generated using the Clontech SMART-Seq v4 kit; barcoding and sequencing primers were added using the NexteraXT library prep kit. Libraries were validated my microelectrophoresis, quantified, pooled, and sequenced on an Illumina NovaSeq 6000 in 100-base single-read reactions. Illumina bcl2fastq v2.17.1.14 was used for demultiplexing, and reads were mapped to a merged reference of the hg38 human reference genome. Viral genes were mapped to the FDAARGOS_983 strain of the 2019-nCoV/USA-WA1/2020 SARS-CoV2 isolate (GenBank accession no. MT246667.1), using STAR v2.7.3a (44). Reads were normalized and differentially expressed genes were analyzed using DESeq2. Gene set enrichment analysis was performed using the software provide by the Broad Institute and the MSigDB database. Pathway analysis was performed using Cytoscape software.
Quantitative reverse transcription-PCR (qRT-PCR).
RNA was extracted from the pHAE cultures using the method described above. Purified RNA was reverse transcribed into cDNA using the high-capacity cDNA reverse transcription kit (Thermo Fisher, 43-688-13). RNA levels were quantified using the IDT Prime Time gene expression master mix and TaqMan gene expression Primer/Probe sets (IDT). All qPCR was performed in 384-well plates and run on a QuantStudio5 qPCR system. To quantify viral RNA, SARS-CoV-2 RDRP-specific primers (forward [F], GTGARATGGTCATGTGTGGCGG; reverse [R], CARATGTTAAASACACTATTAGCATA) and probe (56-6-carboxyfluorescein [FAM]/CAGGTGGAA/ZEN/CCTCATCAGGAGATGC/3IABkFQ) were used. To quantify influenza viral RNA, H1N1 nucleoprotein-specific primers (F, CCCTAAGAAAACAGGAGGACCC; R, TTGGCGCCAAACTCTCCTTA) and probe (FAM-AGAC(+G)(+G)(+A)AA(+G)T(+G)GATGA-IBFQ) were used. The following TaqMan primer/probe sets were used in this analysis: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Hs02758991_g1), IFIT2 (Hs01922738_s1), IFIT3 (Hs01922752_s1), DDX58 (Hs01061436_m1), IFIH1 (Hs00223420_m1), OAS1 (Hs00973637_m1), IRF1 (Hs00971960_m1), IRF7 (Hs01014809_g1), and MX1 (Hs00895608_m1). Threshold cycle (CT) values were normalized to the reference gene GAPDH and represented as fold change over mock samples.
Confocal imaging.
pHAE cultures were fixed in 2% PFA in Dulbecco's phosphate-buffered saline (DPBS) for 20 min. PFA was quenched with 0.1 M glycine, and then cultures were washed twice with DPBS. Cells were permeabilized using 1% Triton X-100 for 5 min and then blocked with PBS-BGT (PBS, 0.5% BSA, 0.1% glycine, 0.05% Tween 20) for 10 min. Phalloidin conjugated to AF647 (Thermo Fisher) was diluted 1:100 in PBS-BGT and incubated on the cells for 1 h at room temperature. Samples were washed 3 times with PBS and counterstained with Hoechst 33342 (0.5% [vol/vol] in PBS-BGT) for 5 min at 37°C. Cells were washed 3 times with PBS, removed from the insert, and mounted on a glass slide using ProLong glass mounting media (Thermo Fisher). Cultures were imaged using a Olympus IX81 confocal microscope, and the images were analyzed using ImageJ and Imaris software.
Statistical analysis.
Statistical analyses were performed using GraphPad Prism 8, ggplot2 R package, and GSEA software. Statistical significance was determined as P value of <0.05 using Student’s t test or a one-way analysis of variance (ANOVA). All comparisons were made between treatment or infection conditions with time point-matched, uninfected and untreated controls.
Data availability.
The raw data of all RNA sequencing in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO accession number GSE153970.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. U. Cotton (Case Western Reserve University) for generously providing our pHAE specimens, Natalie Thornburg (CDC, Atlanta, GA) for providing our SARS-CoV-2 viral stock, Jacob Kohlmeier (Emory University, Atlanta, GA) for providing our H1N1 California 2009 viral stock, and Jens Wrammert (Emory University, Atlanta, GA) for making the anti-SARS-CoV-2 spike protein (biotin-conjugated CR3022) antibody. We also thank the Yerkes Genomics Core (Emory University, Atlanta, GA) for their help in obtaining RNA-Seq data.
Research reported in this publication was supported in part by the Emory University Integrated Cellular Imaging Microscopy Core.
The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
The research reported in this publication was supported in part by an Emory EVPHA Synergy Fund award (M.S.S.), COVID-Catalyst-I3 Funds from the Woodruff Health Sciences Center and Emory School of Medicine (M.S.S.), by Emory University and the MP3 Initiative (M.S.S. and A.C.L.), Center for Childhood Infections and Vaccines, Children’s Healthcare of Atlanta, and National Institutes of Health ORIP/OD P51OD011132 (M.S.S.), HHSN272201400004C (NIAID Centers of Excellence for Influenza Research and Surveillance, CEIRS; M.S.S., J.E.K.), R00 AG049092 (V.D.M.), and World Reference Center for Emerging Viruses and Arboviruses R24 AI120942 (V.D.M.). The Yerkes NHP Genomics Core is supported in part by NIH P51 OD011132 and an equipment grant, NIH S10 OD026799 (S.E.B.).
The funders had no role in study design, data collection and analysis, manuscript preparation, or the decision to publish.
A.V., P.R., and M.S. contributed to the acquisition, analysis, and interpretation of the data. T.C., M.Z., and A.G. contributed to the acquisition and interpretation of the data. A.U., K.P., and S.E.B. contributed to the acquisition and analysis of the data. C.M., E.S., B.M., J.L.L., J.E.K., S.D., A.L., and L.A. contributed to the acquisition of the data. T.C., A.L., L.A., V.D.M., P.S., G.M.T., H.A., and S.E.B. contributed to the interpretation of the data, as well as the conception and design of the work. A.V., P.R., A.G., and M.S. contributed to the acquisition, analysis, and interpretation of the data, as well as the conception and design of the work and writing of the manuscript.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. 2020. COVID-19 dashboard. https://covid19.who.int/.
- 4.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. 11 March 2020. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan W-J, Ni Z-Y, Hu Y, Liang W-H, Ou C-Q, He J-X, Liu L, Shan H, Lei C-L, Hui DSC, Du B, Li L-J, Zeng G, Yuen K-Y, Chen R-C, Tang C-L, Wang T, Chen P-Y, Xiang J, Li S-Y, Wang J-L, Liang Z-J, Peng Y-X, Wei L, Liu Y, Hu Y-H, Peng P, Wang J-M, Liu J-Y, Chen Z, Li G, Zheng Z-J, Qiu S-Q, Luo J, Ye C-J, Zhu S-Y, Zhong N-S. 2020. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, Zhang X, Zhang M, Wu S, Song J, Chen T, Han M, Li S, Luo X, Zhao J, Ning Q. 2020. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, Yuen KY. 2020. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect 9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JM, Chung YS, Jo HJ, Lee NJ, Kim MS, Woo SH, Park S, Kim JW, Kim HM, Han MG. 2020. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health Res Perspect 11:3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsang KW, Ho PL, Ooi GC, Yee WK, Wang T, Chan-Yeung M, Lam WK, Seto WH, Yam LY, Cheung TM, Wong PC, Lam B, Ip MS, Chan J, Yuen KY, Lai KN. 2003. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med 348:1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, Talavera-López C, Maatz H, Reichart D, Sampaziotis F, Worlock KB, Yoshida M, Barnes JL, Banovich NE, Barbry P, Brazma A, Collin J, Desai TJ, Duong TE, Eickelberg O, Falk C, Farzan M, Glass I, Gupta RK, Haniffa M, Horvath P, Hubner N, Hung D, Kaminski N, Krasnow M, Kropski JA, Kuhnemund M, Lako M, Lee H, Leroy S, Linnarson S, Lundeberg J, Meyer KB, Miao Z, Misharin AV, Nawijn MC, Nikolic MZ, Noseda M, Ordovas-Montanes J, Oudit GY, Pe’er D, Powell J, Quake S, Rajagopal J, Tata PR, HCA Lung Biological Network. 2020. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH, Kato T, Lee RE, Yount BL, Mascenik TM, Chen G, Olivier KN, Ghio A, Tse LV, Leist SR, Gralinski LE, Schäfer A, Dang H, Gilmore R, Nakano S, Sun L, Fulcher ML, Livraghi-Butrico A, Nicely NI, Cameron M, Cameron C, Kelvin DJ, de Silva A, Margolis DM, Markmann A, Bartelt L, Zumwalt R, Martinez FJ, Salvatore SP, Borczuk A, Tata PR, Sontake V, Kimple A, Jaspers I, O’Neal WK, Randell SH, Boucher RC, Baric RS. 27 May 2020. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sims AC, Baric RS, Yount B, Burkett SE, Collins PL, Pickles RJ. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J Virol 79:15511–15524. doi: 10.1128/JVI.79.24.15511-15524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazear HM, Schoggins JW, Diamond MS. 2019. Shared and distinct functions of type I and Type III interferons. Immunity 50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML, Staeheli P, Wack A. 2013. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog 9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jilg N, Lin W, Hong J, Schaefer EA, Wolski D, Meixong J, Goto K, Brisac C, Chusri P, Fusco DN, Chevaliez S, Luther J, Kumthip K, Urban TJ, Peng LF, Lauer GM, Chung RT. 2014. Kinetic differences in the induction of interferon stimulated genes by interferon-alpha and interleukin 28B are altered by infection with hepatitis C virus. Hepatology 59:1250–1261. doi: 10.1002/hep.26653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanco-Melo D, Nilsson-Payant BE, Liu W-C, Uhl S, Hoagland D, Møller R, Jordan TX, Oishi K, Panis M, Sachs D, Wang TT, Schwartz RE, Lim JK, Albrecht RA, tenOever BR. 2020. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chirkova T, Lin S, Oomens AGP, Gaston KA, Boyoglu-Barnum S, Meng J, Stobart CC, Cotton CU, Hartert TV, Moore ML, Ziady AG, Anderson LJ. 2015. CX3CR1 is an important surface molecule for respiratory syncytial virus infection in human airway epithelial cells. J Gen Virol 96:2543–2556. doi: 10.1099/vir.0.000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, Queen K, Tao Y, Paden CR, Zhang J, Li Y, Uehara A, Wang H, Goldsmith C, Bullock HA, Wang L, Whitaker B, Lynch B, Gautam R, Schindewolf C, Lokugamage KG, Scharton D, Plante JA, Mirchandani D, Widen SG, Narayanan K, Makino S, Ksiazek TG, Plante KS, Weaver SC, Lindstrom S, Tong S, Menachery VD, Thornburg NJ. 2020. Severe acute respiratory syndrome coronavirus 2 from patient with 2019 novel coronavirus disease, United States. Emerg Infect Dis 26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie X, Muruato A, Lokugamage KG, Narayanan K, Zhang X, Zou J, Liu J, Schindewolf C, Bopp NE, Aguilar PV, Plante KS, Weaver SC, Makino S, LeDuc JW, Menachery VD, Shi P-Y. 2020. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmerman MG, Bowen JR, McDonald CE, Young E, Baric RS, Pulendran B, Suthar MS. 2019. STAT5: a target of antagonism by neurotropic flaviviruses. J Virol 93:e00665-19. doi: 10.1128/JVI.00665-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bettigole SE, Glimcher LH. 2015. Endoplasmic reticulum stress in immunity. Annu Rev Immunol 33:107–138. doi: 10.1146/annurev-immunol-032414-112116. [DOI] [PubMed] [Google Scholar]
- 23.Minakshi R, Padhan K, Rani M, Khan N, Ahmad F, Jameel S. 2009. The SARS coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One 4:e8342. doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lourenço AR, Roukens MG, Seinstra D, Frederiks CL, Pals CE, Vervoort SJ, Margarido AS, van Rheenen J, Coffer PJ. 2020. C/EBPɑ is crucial determinant of epithelial maintenance by preventing epithelial-to-mesenchymal transition. Nat Commun 11:785. doi: 10.1038/s41467-020-14556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klinkhammer J, Schnepf D, Ye L, Schwaderlapp M, Gad HH, Hartmann R, Garcin D, Mahlakoiv T, Staeheli P. 2018. IFN-lambda prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. Elife 7:e33354. doi: 10.7554/eLife.33354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Subbarao K. 2007. The immunobiology of SARS. Annu Rev Immunol 25:443–472. doi: 10.1146/annurev.immunol.25.022106.141706. [DOI] [PubMed] [Google Scholar]
- 27.Hu Y, Li W, Gao T, Cui Y, Jin Y, Li P, Ma Q, Liu X, Cao C. 2017. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J Virol 91:e02143-16. doi: 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jauregui AR, Savalia D, Lowry VK, Farrell CM, Wathelet MG. 2013. Identification of residues of SARS-CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PLoS One 8:e62416. doi: 10.1371/journal.pone.0062416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spiegel M, Pichlmair A, Martínez-Sobrido L, Cros J, García-Sastre A, Haller O, Weber F. 2005. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J Virol 79:2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong HH, Fung TS, Fang S, Huang M, Le MT, Liu DX. 2018. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology 515:165–175. doi: 10.1016/j.virol.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang SH, Lee TY, Lin YJ, Wan L, Lai CH, Lin CW. 2017. Phage display technique identifies the interaction of severe acute respiratory syndrome coronavirus open reading frame 6 protein with nuclear pore complex interacting protein NPIPB3 in modulating type I interferon antagonism. J Microbiol Immunol Infect 50:277–285. doi: 10.1016/j.jmii.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menachery VD, Eisfeld AJ, Schafer A, Josset L, Sims AC, Proll S, Fan S, Li C, Neumann G, Tilton SC, Chang J, Gralinski LE, Long C, Green R, Williams CM, Weiss J, Matzke MM, Webb-Robertson BJ, Schepmoes AA, Shukla AK, Metz TO, Smith RD, Waters KM, Katze MG, Kawaoka Y, Baric RS. 2014. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio 5:e01174-14. doi: 10.1128/mBio.01174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Channappanavar R, Fehr Anthony R, Vijay R, Mack M, Zhao J, Meyerholz David K, Perlman S. 2016. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J, Farzan M, Wohlford-Lenane C, Perlman S, McCray PB Jr. 2005. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol 79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Q, Chen V, Shannon CP, Wei X-S, Xiang X, Wang X, Wang Z-H, Tebbutt SJ, Kollmann TR, Fish EN. 2020. Interferon-α2b treatment for COVID-19. Front Immunol 11:1061. . doi: 10.3389/fimmu.2020.01061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forero A, Ozarkar S, Li H, Lee CH, Hemann EA, Nadjsombati MS, Hendricks MR, So L, Green R, Roy CN, Sarkar SN, von Moltke J, Anderson SK, Gale M Jr, Savan R. 2019. Differential activation of the transcription factor IRF1 underlies the distinct immune responses elicited by type I and type III interferons. Immunity 51:451–464.e6. doi: 10.1016/j.immuni.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sokol CL, Luster AD. 2015. The chemokine system in innate immunity. Cold Spring Harb Perspect Biol 7:a016303. doi: 10.1101/cshperspect.a016303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang BM, Shojaei M, Teoh S, Meyers A, Ho J, Ball TB, Keynan Y, Pisipati A, Kumar A, Eisen DP, Lai K, Gillett M, Santram R, Geffers R, Schreiber J, Mozhui K, Huang S, Parnell GP, Nalos M, Holubova M, Chew T, Booth D, Kumar A, McLean A, Schughart K. 2019. Neutrophils-related host factors associated with severe disease and fatality in patients with influenza infection. Nat Commun 10:3422. doi: 10.1038/s41467-019-11249-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weaver CT, Elson CO, Fouser LA, Kolls JK. 2013. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol 8:477–512. doi: 10.1146/annurev-pathol-011110-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le TT, Karmouty-Quintana H, Melicoff E, Le TT, Weng T, Chen NY, Pedroza M, Zhou Y, Davies J, Philip K, Molina J, Luo F, George AT, Garcia-Morales LJ, Bunge RR, Bruckner BA, Loebe M, Seethamraju H, Agarwal SK, Blackburn MR. 2014. Blockade of IL-6 trans signaling attenuates pulmonary fibrosis. J Immunol 193:3755–3768. doi: 10.4049/jimmunol.1302470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalluri R, Neilson EG. 2003. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest 112:1776–1784. doi: 10.1172/JCI200320530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim KK, Kugler MC, Wolters PJ, Robillard L, Galvez MG, Brumwell AN, Sheppard D, Chapman HA. 2006. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci U S A 103:13180–13185. doi: 10.1073/pnas.0605669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin L, Han Q, Xiong Y, Li T, Liu Z, Xu H, Wu Y, Wang N, Liu X. 2017. Krüpple-like-factor 4 attenuates lung fibrosis via inhibiting epithelial-mesenchymal transition. Sci Rep 7:15847. doi: 10.1038/s41598-017-14602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of all RNA sequencing in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO accession number GSE153970.