Host factors within the cell are the major mode of restriction of adeno-associated virus (AAV) and keep it from fulfilling its maximum potential as a gene therapy vector. A better understanding of the intricacies of restriction would enable the engineering of better vectors. Via a genome-wide short interfering RNA screen, we identified that proteins of the small ubiquitin-like modifier (SUMO) pathway play an important role in AAV restriction. In this study, we investigate whether this restriction is targeted to the AAV directly or indirectly through host cell factors. The results indicate that both targets act in concert to restrict AAV.
KEYWORDS: AAV, DAXX, SUMO, VP, restriction factor
ABSTRACT
Adeno-associated virus (AAV) has proven to be a promising candidate for gene therapy due to its nonpathogenic nature, ease of production, and broad tissue tropism. However, its transduction capabilities are not optimal due to the interaction with various host factors within the cell. In a previous study, we identified members of the small ubiquitin-like modifier (SUMO) pathway as significant restriction factors in AAV gene transduction. In the present study, we explored the scope of this restriction by focusing on the AAV capsid and host cell proteins as targets. We show that during vector production, the capsid protein VP2 becomes SUMOylated, as indicated by deletion and point mutations of VP2 or the obstruction of its N terminus via the addition of a tag. We observed that SUMOylated AAV capsids display higher stability than non-SUMOylated capsids. Prevention of capsid SUMOylation by VP2 mutations did not abolish transduction restriction by SUMOylation; however, it reduced activation of gene transduction by shutdown of the cellular SUMOylation pathway. This indicates a link between capsid SUMOylation and SUMOylation of cellular proteins in restricting gene transduction. Infection with AAV triggers general SUMOylation of cellular proteins. In particular, the DAXX protein, a putative host cell restriction factor that can become SUMOylated, is able to restrict AAV gene transduction by reducing the intracellular accumulation of AAV vectors. We also observe that the coexpression of a SUMOylation inhibitor with an AAV2 reporter gene vector increased gene transduction significantly.
IMPORTANCE Host factors within the cell are the major mode of restriction of adeno-associated virus (AAV) and keep it from fulfilling its maximum potential as a gene therapy vector. A better understanding of the intricacies of restriction would enable the engineering of better vectors. Via a genome-wide short interfering RNA screen, we identified that proteins of the small ubiquitin-like modifier (SUMO) pathway play an important role in AAV restriction. In this study, we investigate whether this restriction is targeted to the AAV directly or indirectly through host cell factors. The results indicate that both targets act in concert to restrict AAV.
INTRODUCTION
Adeno-associated viruses (AAV) were found originally as a contaminant of purified adenoviruses, and they belong to the genus Dependoparvovirus within the family Parvoviridae (1–3). Currently, 13 primate AAV serotypes that show different cell type and tissue specificity have been isolated (4). AAVs are considered nonpathogenic. This, and the fact that AAV genetic vectors do not require viral sequences on the vector genome besides the inverted terminal repeats (ITRs), has stimulated many efforts to develop AAV as a delivery system for gene therapy (for a review, see reference 5). In fact, with Luxturna, Glybera, and Zolgensma, there are licensed products on the market for clinical use for the treatment of Leber’s congenital disease, lipoprotein lipase deficiency, and spinal muscular atrophy, respectively (6–9).
AAV gene transduction proceeds through receptor-mediated uptake of vector particles into endosomes, followed by endosomal release, nuclear entry, genome release, and conversion of the single-stranded DNA into a double strand (10, 11). Some of these limitations have been overcome, for example, by the use of self-complementary vectors (12). Other steps remain rather inefficient, resulting in high vector doses for efficient gene delivery. While this is not limiting for AAV vector transduction ex vivo, the high vector doses lead to possible immune activation in vivo and also make AAV gene therapy cost intensive (13). The different steps involved in gene transduction are only partially understood; however, the role of the three capsid proteins, VP1, VP2, and VP3, in this process has been analyzed to some extent.
Interactions with the cellular receptor(s) are attributed to VP3. AAV enters host cells by binding to heparin sulfate proteoglycan or other glycans as the primary attachment receptor (for a review, see reference 14). Furthermore, different serotypes interact with various other uptake receptors. Recently, AAVR has been described as a proteinaceous receptor essential for several AAV serotypes (15), although it remains to be elucidated at which stage of AAV entry it plays a role. For entry, AAV can follow different routes; Nonnenmacher and Weber reported that the majority of AAV2 particles are endocytosed via the CLIC/GEEC pathway (16). VP1, an N-terminal extension of VP2 and VP3, is absolutely essential for gene transduction. It plays an important role in escape from the endosomes and nuclear uptake. The unique N terminus of VP1 becomes exposed during endosomal trafficking, thereby presenting a phospholipase A2 domain, putative nuclear localization signals, and further sequence elements involved in intracellular trafficking (17), and it shows proteolytic activity. For VP2, so far no function in the infection process has been found, since VP2 deletion mutants are still infectious (18, 19). Modifications of capsid proteins by phosphorylation and ubiquitination during intracellular trafficking, mainly concerning VP3, are thought to negatively influence transduction efficiency. Zhong and colleagues reported that elimination of tyrosine residues on the capsid surface leads to improvement of AAV vector-mediated gene transduction (3). Insertion of peptide ligands has led to target cell-optimized gene transduction, thereby greatly improving efficacy in vitro and in vivo (for an overview, see reference 20).
Besides capsid-specific properties, host cell restriction factors are believed to limit AAV-mediated gene transduction. Therefore, we and others have performed genome-wide short interfering RNA (siRNA) screens for the identification of such cellular restriction factors (21–23). In two of the studies, the DNA damage response and the U2 snRNA spliceosome were identified to restrict AAV gene transduction. In our screen, the machinery of the small-ubiquitin-like modifier (SUMO) was identified and validated to limit AAV transduction. SUMO belongs to the ubiquitin-like family, as it utilizes a similar mechanism for target conjugation. SUMOylation modulates the activity of target proteins through reversible conjugation of one of the different SUMO paralogs (SUMO1, the nearly identical SUMO2/3, SUMO4, and SUMO5 in mammalian cells) (24). It is carried out by a cascade of different enzymes, namely, the SUMO-activating (SAE1/SAE2), the SUMO-conjugating (UBC9), several SUMO ligases, and SUMO proteases.
Unlike ubiquitination, SUMOylation in most cases does not lead to protein degradation but rather regulates the functional properties of proteins, for example, by determining their protein-protein interactions and subcellular localization (25). The link between AAV and the SUMOylation pathway and the consequence thereof has not been explored so far.
We report here that AAV capsids are directly SUMOylated during vector production, and we identified that the VP2 protein is the target of SUMOylation. Vectors lacking VP2 are not SUMOylated, and N-terminal extensions to VP2 abolish SUMOylation. This explains the lack of SUMO on VP1, although it contains all putative target sequences. Prevention of capsid SUMOylation does not improve AAV-mediated gene transduction per se. We observe that capsid SUMOylation only explains in part the restriction of transduction by SUMOylation. We believe that most of the restrictive effect is mediated through SUMOylation of cellular factors, as suggested by the fact that infection with AAV2 increased SUMOylation of cell proteins in general. In this respect, we could show that DAXX, a highly SUMOylated protein, also restricts AAV gene transduction. AAV particles accumulate to a higher degree in DAXX knockout cells than in wild-type (wt) cells. SUMOylation-dependent restriction of AAV gene transfer can be overcome by coinfection with an adenovirus protein Gam1-expressing AAV vector, leading to transient inactivation of the SUMOylation machinery.
RESULTS
AAV capsids can be SUMOylated.
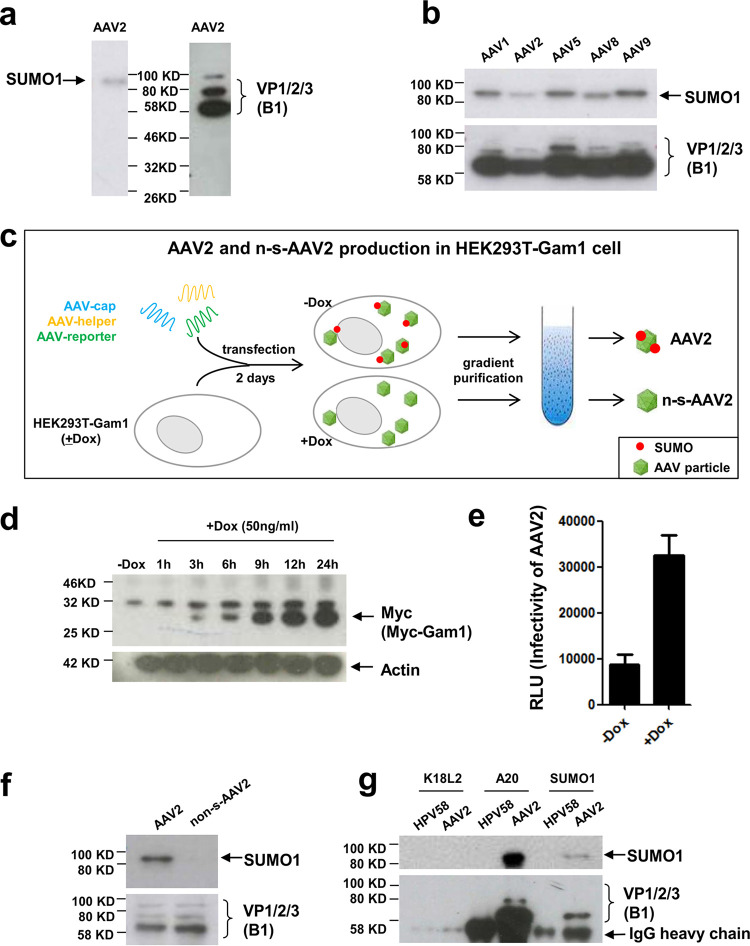
Previously, we reported that SUMOylation restricts transduction of cells by different AAV serotypes (21). To elucidate the mechanism of restriction of AAV transduction by SUMOylation, we were interested in identifying the target of SUMOylation. The restriction of cell transduction could be envisioned to be a direct effect of modification of the capsid proteins by SUMO or indirectly by modification and activation of cellular antiviral factors. To address this question, we first analyzed purified AAV2 vectors for the presence of SUMO tags. Using an anti-SUMO1-specific antibody, a band corresponding to a protein of about 90 kDa became visible in Western blot analysis (Fig. 1a). In addition to AAV2, a SUMO-specific signal also could be detected when analyzing AAV1, -5, -8, and -9 particles (Fig. 1b). To confirm this observation, we produced AAV2 vectors in 293T cells stably transfected with the avian adenovirus CELO Gam1 gene. In these cells, Gam1 expression can be induced by doxycycline (Dox). It has been shown previously that Gam1 effectively inactivates the SUMO E1 enzyme complex formed by Sae1 and Sae2 (26). This should lead to the production of non-SUMOylated AAV vectors in Gam1-expressing HEK293T cells (schematically outlined in Fig. 1c). Analysis of the AAV2 capsids indicates that inactivation of SUMO E1 by Gam1 induction prior to transfection of the AAV2 plasmids completely abolished the SUMO-specific signals on AAV2 capsids (Fig. 1f). Gam1 protein expression increases over time after Dox induction (Fig. 1d), which leads to a strong increase in AAV gene transduction, confirming our previous report about the restrictive role of SUMOylation in AAV gene transduction (21) (Fig. 1e). To demonstrate that the SUMO tag is physically connected to the AAV2 capsids, we performed an immunoprecipitation using either the A20 antibody, recognizing assembled capsids, or using an anti-SUMO1 antibody (Fig. 1g). Both antibodies were able to precipitate AAV2 capsids and revealed a SUMO-specific band at 90 kDa. Taken together, the data indicate that the AAV2 capsid can become SUMOylated.
FIG 1.
AAV2 capsids are SUMOylated. Shown is an analysis of purified AAV2 particles. To determine AAV2 capsid modification by SUMO, we analyzed AAV2 vectors after purification by iodixanol gradient centrifugation. (a and b) AAV2 capsids and AAV1, -5, -8, and -9 capsids produced in 293T cells and analyzed by anti-SUMO1-specific antibodies or an antibody specific for all three AAV capsid proteins (B1 antibody). (c) Strategy to produce non-SUMOylated (n-s-) AAV2. HEK293T-Gam1 cells were transfected with three plasmids for AAV vector production. Induction of the avian adenovirus CELO Gam1 protein by doxycycline (Dox) leads to inactivation of the Sae1/2 E1 complex, leading to inactivation of the SUMOylation machinery. (d) Western blot analysis of Gam1 protein expression over time after Dox induction using Myc-tag antibody. HeLa-Gam1 cells were induced with 50 ng/ml Dox, and then cell lysate was harvested after 1, 3, 6, 9, 12, and 24 h. (e) Induction of Gam1 leads to higher transduction rates by AAV. HEK293T-Gam1 cells were incubated with and without Dox and then transduced with (SUMOylated) AAV-Luc vectors. RLU, relative light units. (f) Western blot analysis of AAV2 particles produced in 293T-Gam1 cells with (n-s-AAV2) or without Dox induction using SUMO1 and VP-specific antibodies. (g) Immunoprecipitation of AAV capsids. Purified AAV2 capsids were precipitated either with the AAV capsid-specific antibody A20 or with an anti-SUMO1 antibody. Precipitated proteins were then detected with either VP-specific antibody B1 or anti-SUMO1 antibody. As a specificity control, purified human papillomavirus type 58 capsids were precipitated using an unrelated antibody (K18L2).
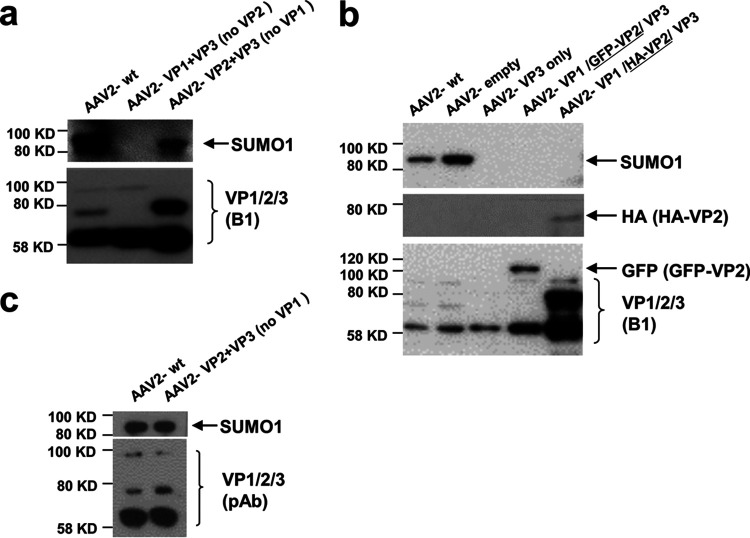
After identifying AAV capsids as a target for SUMOylation, we wanted to determine which of the capsid proteins (VP1, VP2, or VP3) is modified. For this, we analyzed AAV2 vector particles lacking either VP1, VP2, or both. The data show that VP1 is not required for detection of a SUMOylated capsid (Fig. 2a). In contrast, the absence of VP2 in AAV2 capsids abolishes the SUMO-specific signal. Notably, using the B1 antibody in detection of VP proteins for the vectors lacking VP1 did not show a band at 90 kDa but a strong signal for unmodified VP2, indicating that only a fraction of the VP2 molecules becomes SUMOylated in this mutant. B1 also might be less sensitive in detecting SUMOylated VP than the anti-SUMO1 antibody, or the B1 epitope is masked by SUMO. However, a polyclonal serum against VP detects a protein with a size consistent with SUMOylated VP2 (Fig. 2c). When we analyzed AAV2 particles in which either a green fluorescent protein (GFP) or hemagglutinin (HA) tag was fused to the VP2 N terminus, no SUMO-specific signal was detected. Furthermore, capsids composed of VP3 alone also did not possess a SUMOylation signal. We concluded that VP2 is the target for SUMOylation and that a free N terminus of VP2 is required for the modification (Fig. 2b). This conclusion is in line with the migration of the SUMO-specific band at about 90 kDa, assuming a SUMO tag with a molecular weight of 10 kDa added to the VP2 protein, and it is also consistent with the observation that VP1 is not SUMOylated.
FIG 2.
VP2 is a target for SUMOylation. (a) AAV2 vectors lacking either VP2, VP1, or VP1/2 were produced and purified by iodixanol gradient centrifugation. Vectors were analyzed by Western blotting using an anti-SUMO1 or the VP-specific antibody B1. (b) Analysis of AAV2 vectors carrying an N-terminally modified VP2. Vectors were produced as described for panel a. Lane 4 shows AAV vectors carrying a VP2 N-terminally fused GFP, and in lane 5 the HA tag is fused to the N terminus of VP2. (c) Detection of VP using a polyclonal antibody.
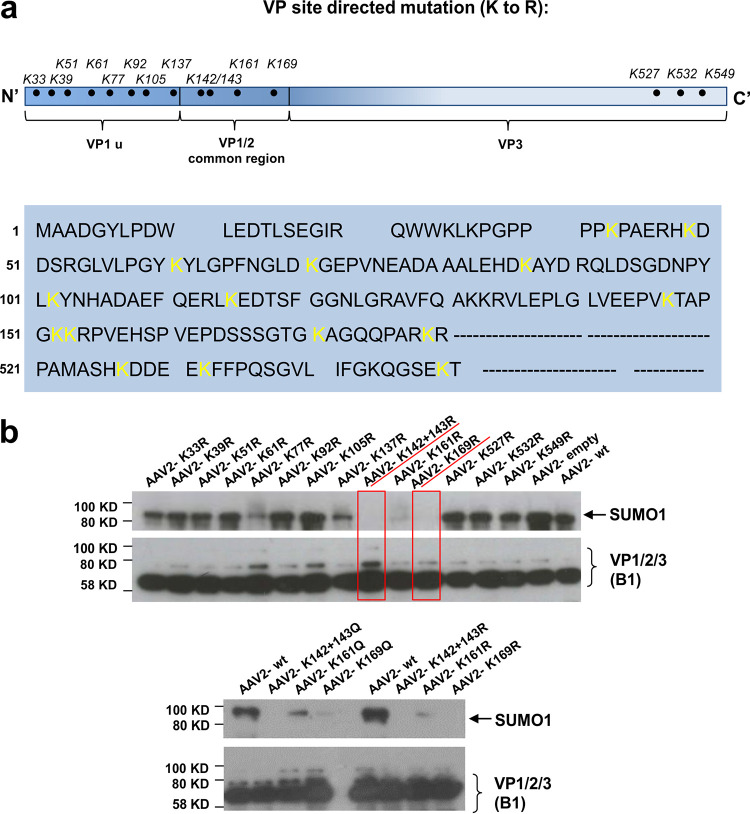
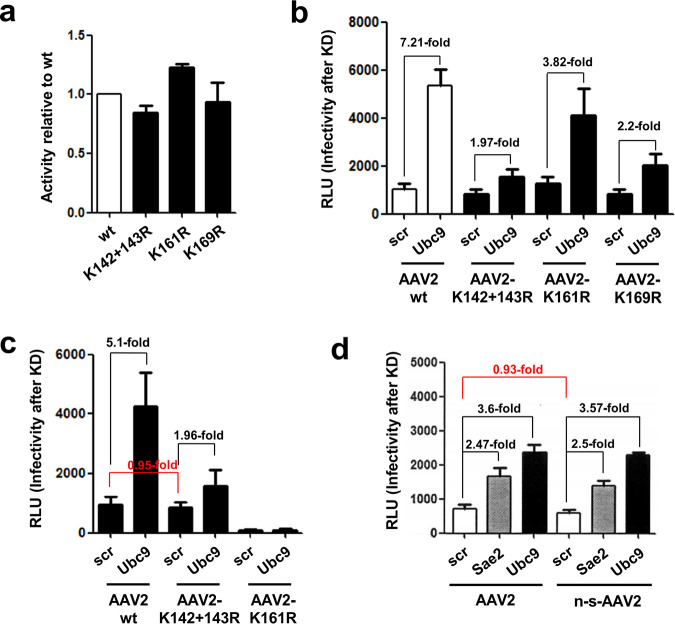
To further determine the site of modification in the VP proteins and to confirm the observations described above, we identified putative lysine residues accessible on assembled capsids. In silico analysis of putative SUMOylation sites by different servers did not reveal a clear pattern of motifs. Therefore, we decided to perform a systematic analysis of VP lysine exchange mutants. To retain the charge and structure of the modified capsids as closely as possible, we changed the lysine residues to arginine residues (K to R). The AAP gene was affected in none of the mutants. A total of 13 mutants were created in which each one carried one exchange. One additional mutant encoded a double exchange of the two adjacent positions K142 and K143 (Fig. 3a). Vectors carrying the lysine exchanges were produced and analyzed for SUMOylation as described above. For most of the modified capsids, the Western blot signal for SUMO was unaltered. However, in particular for two mutants (K142/143R and K169R), the SUMO-specific signal was absent (Fig. 3b). The K161R exchange variant between these two positions showed a much-reduced SUMO signal. These data are consistent with our conclusions above of VP2 being the target for SUMOylation, as the exchanges leading to elimination of SUMOylation are all located at the N terminus of VP2. We also generated K-to-Q exchange variants for the residues K142/143, K161, and K169, and these showed the same pattern for reduced or lack of SUMOylation as seen for the K to R variants (Fig. 3b). As at least two different, albeit closely neighboring, positions both abolished SUMOylation, none of them can be the exclusive site of modification. Rather, structural constraints of the VP2 protein likely contribute to recognition by the SUMOylation machinery, a fact already indicated by the interference of N-terminal fusions to VP2 and the fact that VP1 does not seem to be modified by SUMOylation, although it also contains the respective lysine target residues.
FIG 3.
Lysine residues located in the N terminus of VP2 are essential for SUMOylation. (a) Sequence of VP with the positions of the lysine-arginine exchanges highlighted. (b) Analysis of iodixanol gradient-purified lysine AAV2 capsid exchange mutants by Western blotting using a SUMO1-specific antibody or the anti-VP specific antibody B1. (Top) K to R exchanges. (Bottom) K to Q exchanges for selected lysines.
SUMOylation affects AAV capsid stability.
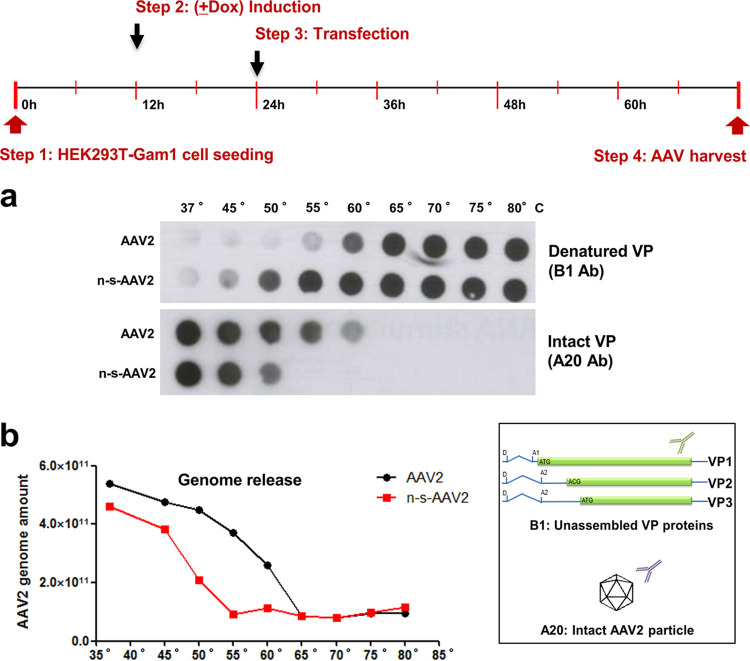
To analyze whether capsid SUMOylation has an influence on capsid integrity, we incubated AAV2 particles with or without (n-s-AAV2, produced in Gam1-expressing HEK293T cells) SUMO tag at different temperatures followed by dot blot and quantitative PCR (qPCR) analysis. Using either an antibody specific for unassembled (B1) or intact (A20) particles (Fig. 4, inset), we could determine the degree of unfolding under the different conditions. Results indicate that, indeed, n-s-AAV2 particles produced in the doxycycline-induced HEK 293T-Gam1 cells are significantly less stable than SUMOylated particles. An increase in signal intensity using the B1 antibody is seen at 50°C for the non-SUMOylated capsids, while only at 60°C or higher temperatures do the SUMOylated capsids become denatured. Labeling with the A20 antibody confirms these results; intact n-s-AAV2 capsids can be detected until 50°C, whereas SUMOylated particles are stable until 60°C (Fig. 4a). Consistently, the vector genomes became sensitive to nuclease at lower temperature in the case of n-s-AAV2 (Fig. 4b). We conclude that SUMOylation significantly increases the stability of the AAV capsids.
FIG 4.
SUMOylated AAV2 capsids display higher thermal stability. (a) Dot blot analysis of AAV2 and n-s-AAV2 particles after incubation at different temperatures. (Inset) The B1 antibody reacts with unassembled VP, while A20 is specific for intact capsids. (b) Quantification of vector genomes of AAV particles shown in panel a. After heat treatment, capsids were incubated with Benzonase to degrade free vector genomes. Packaged vector genomes were quantified by qPCR.
Influence of capsid SUMOylation on AAV gene transduction.
To determine the contribution of AAV capsid SUMOylation on AAV gene transduction, we focused on the mutants that show abolished (K142/143R and K169R) or greatly reduced (K161R) capsid SUMOylation for further analysis. First, it would be conceivable that the abolishment or reduction of capsid SUMOylation by lysine-to-arginine exchanges (K142/143R, K161R, and 169R) would bypass SUMOylation-dependent restriction of transduction activity, but this was not the case (Fig. 5a). All three mutants show infectivity similar to that of the wt AAV vector and also compared to lysine exchange mutants for which SUMOylation was not altered (data not shown). When we determined the effect of inhibition of SUMOylation of capsid proteins by using Ubc9 siRNA (conjugating enzyme E2 of SUMOylation), we observed an approximately 7-fold increase of gene transduction for wt AAV vectors, similar to what we have reported previously (21). However, the capsid mutants showed a reduced but not fully abolished increase (2-fold for the two mutants with abolished SUMOylation) in transduction after knockdown of Ubc9 (Fig. 5b). Accordingly, AAV2 vectors that lacked VP2 and, therefore, cannot be SUMOylated exhibited gene transduction patterns after knockdown of Ubc9 similar to that of the lysine-arginine exchange mutants (Fig. 5c).
FIG 5.
Capsid SUMOylation only contributes partially to host cell restriction of AAV gene transduction. (a) Transduction efficiency of AAV2 carrying lysine-arginine exchanges. HeLa cells were transduced with the different AAV vectors at an MOI of 103, and luciferase activity was measured 48 h postransduction. (b and c) Effect of SUMOylation knockdown (KD) by Ubc9 siRNA on gene transduction of AAV2 lysine-arginine exchange mutants (b) and AAV2 vectors carrying either no VP1 or VP2 protein (c). (d) Gene transduction of non-SUMOylated AAV2 vectors produced in HEK293T cells expressing Gam1. Sae2 siRNA targets the SUMO E1 enzyme complex, and Ubc9 siRNA targets the SUMO E2 enzyme complex. Scr, scrambled siRNA was used as a control.
These results show that prevention of capsid SUMOylation does not rescue restriction of AAV2 gene transduction but influences activation of gene transduction by Ubc9 knockdown, indicating a link between cellular SUMOylation and capsid SUMOylation. In addition to the capsids carrying the lysine-arginine exchanges or VP2 deletion, which cannot be SUMOylated, we also asked whether capsids that lack SUMOylation during vector production, but can possibly become SUMOylated during infection, differ in their susceptibility for restriction by the cellular SUMOylation machinery. To address this question, we transduced HeLa cells with AAV2 and non-SUMOylated AAV2 (n-s-AAV2) produced in HEK293T Gam1-expressing cells (Fig. 1c). No difference in infectivity was observed between AAV2 particles with or without the SUMOylation tag in cells treated with control siRNA (scr) (Fig. 5d). Interestingly, the increase in transduction upon knockdown of the SUMOylation pathway by Sae2 or Ubc9 targeting siRNA also did not differ from that of AAV2 produced in 293T cells lacking Gam1. Taken together, these results show that a preexisting SUMO tag on AAV capsids neither changes infectivity nor influences restriction by the host cell, because capsids might become SUMOylated during infection. This is clearly different from the gene transduction observed with mutant capsids that are not SUMOylated and also cannot become SUMOylated during infection.
AAV activates the host cell SUMOylation machinery.
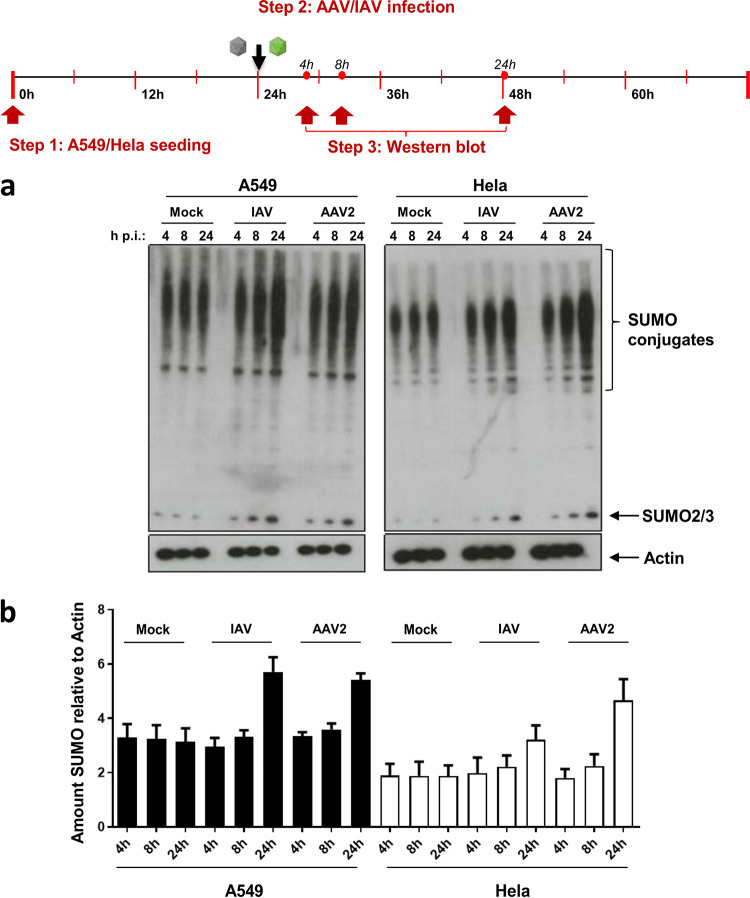
If SUMOylation is considered an antiviral response mechanism, we asked whether the cells are able to sense AAV particles and respond by increasing the total SUMOylation activity, similar to what has been described for influenza A virus (IAV) (27). To address this question, we incubated HeLa and A549 cells with AAV or IAV and analyzed the amount of SUMOylated cellular proteins by Western blotting using an antibody against SUMO2/3 (Fig. 6a and b). As expected, in both cell lines we observed an increase of the SUMO-specific signal for IAV after 8 and 24 h, similar to what has been described by Domingues and colleagues (27). Interestingly, AAV2 also activated total SUMOylation activity in both cell lines. An elevated SUMOylation activity became visible as early as 8 h postransduction, and the effect increased over time. This effect, however, was only visible at a multiplicity of infection (MOI) of 105 or higher.
FIG 6.
AAV activates cellular SUMOylation activity. A549 and HeLa cells were incubated with AAV2 vectors at an MOI of 105. (a) Cell lysates were prepared 4, 8, and 24 h later and analyzed for SUMOylation activity by Western blotting using an antibody specific for SUMO2/3. As a positive control, SUMOylation activity was analyzed after incubating cells with 5 PFU/cell of IAV. (b) Quantification of Western blotting signals shown in panel a, normalized to actin.
Cellular pathways of SUMOylation affecting AAV.
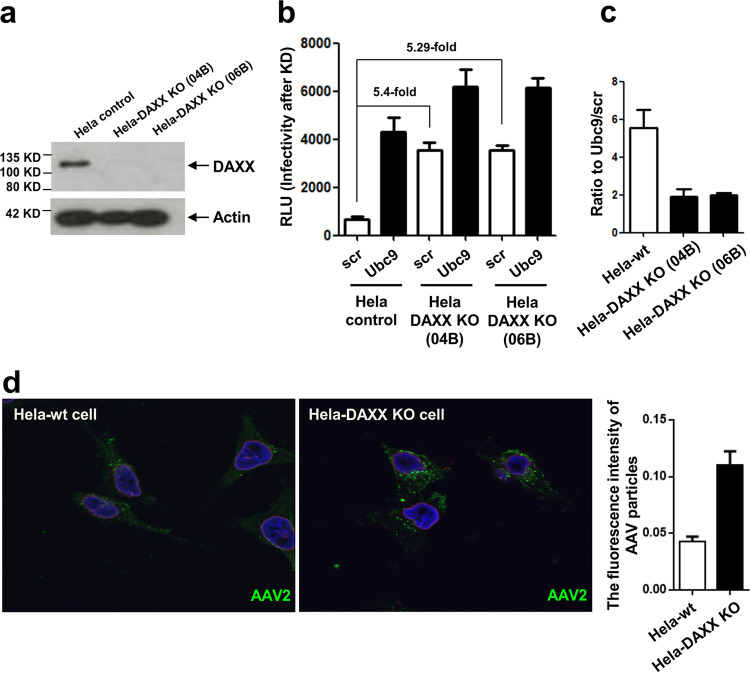
The data presented above show not only that AAV capsids can be SUMOylated but also that this contributes only in part to the restriction in gene transduction, indicating that intracellular factors that are regulated by SUMOylation also play a role in controlling AAV gene transduction. A possible candidate is the cellular DAXX protein, which itself presents a target for SUMOylation, but it can bind to other proteins via their SUMOylation tag (28). Further, in our previous screen, we identified DAXX as a putative AAV restriction factor (21). DAXX is also involved in restriction of other viruses and, therefore, represents a possible cellular factor involved in the observed effects of cellular sumoylation on AAV gene transduction (29). Therefore, we wanted to determine the effect of DAXX on AAV transduction in particular in the context of SUMOylation. We generated DAXX knockout (KO) cells (Fig. 7a) and transduced these with AAV2 vectors. Figure 7b and c show the results for two independent DAXX KO HeLa cell clones. Both cell lines show an approximately 5-fold increase in transduction by AAV2 compared to the parental HeLa cells (Fig. 7b). This difference could also be visualized by detecting transducing AAV2 capsids by immunofluorescence using an anti-capsid antibody (A20). In the DAXX knockout cells, an increased number and intensity of signals were observed in an intracellular localization compared to levels for the parental cells (Fig. 7d). This suggests a role of SUMOylated DAXX, and possibly SUMOylation of other cellular proteins, in intracellular accumulation of AAV vectors. Interestingly, the AAV-mediated gene transduction-enhancing effect of SUMOylation knockdown using Ubc9 siRNA still was present but reduced from about 6-fold to about 2-fold in the knockout cells (Fig. 7c), indicating that DAXX contributes to SUMO-dependent restriction of AAV. Although knockout of DAXX nearly completely rescues restriction of AAV-mediated gene transduction by SUMOylation, there are still other processes involved, as suggested by the 2-fold enhancement of transduction in HeLa DAXX KO cells upon Ubc9 knockdown (Fig. 7b and c).
FIG 7.
DAXX presents a SUMOylation-dependent restriction factor for AAV2. (a) Two HeLa DAXX knockout cell lines (04B and 06B) were generated and verified via Western blot analysis of cell extracts. (b) AAV2 gene transduction was carried out after transfection of Ubc9 or control siRNA (scr). RLU, relative light units. (c) The ratio of AAV transduction after transfection of the cell lines with scrambled or Ubc9 siRNA is shown. (d) Detection of AAV2 particles by indirect immunofluorescence 24 h after transduction using a capsid-specific antibody (A20); shown is a comparison of HeLa-wt cells and HeLa-DAXX KO cells. The graph shows quantification of fluorescence intensity.
Enhancing AAV transduction by coinfection.
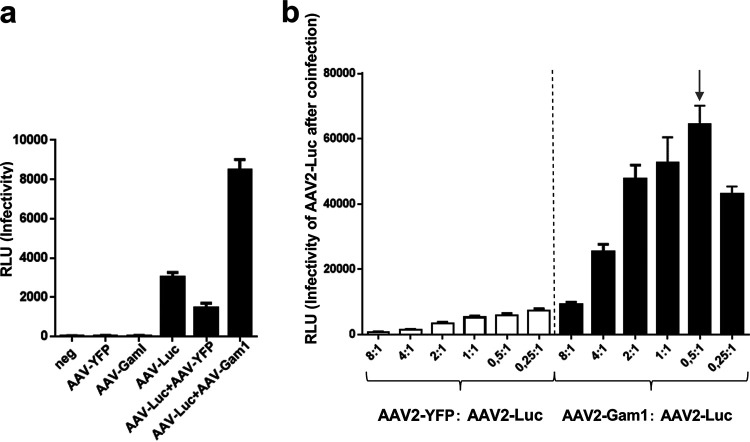
As mentioned above, AAV has been used successfully as a gene transfer vector, and licensed products are already available (for an overview, see reference 5). In gene therapy approaches, lower vector doses would be highly welcome to reduce immune reaction against the vector but also to lower the costs of administration. Knockdown of cellular genes by siRNA is not a feasible strategy. Therefore, we evaluated whether cotransduction with AAV encoding the Gam1 protein could boost transduction by the AAV reporter virus. Figure 8a shows that AAV2-Gam1 indeed enhances transduction by AAV2-Luc vectors. Depending on the ratio of AAV2-Gam1 to AAV-Luc, an up to 10-fold increase was observed (Fig. 8b).
FIG 8.
Inactivation of SUMOylation by cotransduction with AAV2-Gam1 enhances reporter genome delivery. (a) HeLa cells were transduced with AAV-Luc alone or in combination with AAV-Gam1. AAV-YFP vector transduction was performed as a specificity control. (b) Determination of the most effective ratio of AAV2-Gam1 to AAV2-Luc for reporter gene transduction. RLU, relative light units.
DISCUSSION
AAV is a promising gene therapy vector, but AAV vectors do not encode proteins able to counteract cellular restriction factors, and as a result, transduction of cells by AAV is rather inefficient. In vitro and in vivo, large vector doses are applied to compensate for this limitation. Previously, we have identified that the cellular SUMOylation machinery restricts AAV transduction (21). SUMO proteins belong to the family of ubiquitin-like proteins, comprising SUMO, ISG15, Fat10, ATG12, and Nedd8, all of which are covalently linked to target proteins via E3 ligases (for a review, see reference 30). This linkage might influence protein stability, protein-protein interactions, and subcellular localization (see reference 31 and citations therein). In addition, mounting data suggest a cross talk between the ubiquitin and ubiquitin-like modification pathways (32, 33). SUMOylation is a key posttranslational modification that critically regulates a plethora of cellular functions with incredibly diverse consequences, among which are its important roles in gene regulation. In this context, it is often associated with repression of transcription. Restriction of AAV gene transduction by SUMOylation could occur by the SUMOylation of the AAV capsid or by SUMOylation of cellular proteins which then influence the transduction process in an indirect manner. For any in vivo situation, however, it has to be considered that by inhibiting SUMOylation, possible adverse side effects upon gene therapy applications could arise due to improper transcription regulation, general cellular pathway signaling, nuclear import, cell cycle, DNA repair, and chromatin remodeling (24, 34, 35).
In our previous report, we could exclude late events in gene transduction, such as uncoating and DNA single-strand conversion, as well as DNA damage responses leading to restriction by SUMOylation (21). In the present study, we aimed at gaining insight into the mechanisms of AAV restriction by SUMOylation. We show that AAV capsids can be SUMOylated, as AAV vectors assembled in HEK293 cells already carry a SUMO tag. Interestingly, according to our data there are several lines of evidence that only VP2 is SUMOylated. Capsids lacking VP2 do not carry the SUMO tag, and the addition of N-terminal sequences to VP2 abolishes SUMOylation of the capsids. The observation that SUMOylation requires a free VP2 N terminus provides an explanation for the lack of SUMO on VP1, which can, in itself, be considered an N-terminal extension, as it comprises all of VP2. Still, our data do not exclude the possibility that VP1 (or VP3) also becomes SUMOylated during virus entry. It also remains unclear whether non-SUMOylated particles become tagged during entry or whether existing SUMO tags would be a target for poly-SUMOylation. What is the possible significance of VP2 SUMOylation? Notably, AAV vectors with a GFP fused to the N terminus of VP2 remain infectious (18, 19). Prevention of this SUMOylation during vector production, however, has no obvious effect on the infectivity, although SUMOylation impacts capsid stability. With no measurable effect on transduction in vitro, we can only speculate. Until now, the function of VP2 has remained elusive, as AAV vectors lacking VP2 package DNA can transduce cells in vitro (18, 19). On the other hand, VP2 has the same stoichiometry as VP1, and its presence in capsids is conserved among different AAV serotypes, indicating a selective advantage for retaining this protein.
In silico prediction of SUMOylation sites is error prone, and there have been many examples of SUMOylation on nonconsensus sites. Using different servers, such as GPS-SUMO or Jassa, to analyze VP of AAV2, only position K105 was assigned a high score. Therefore, we decided to modify this position and additional lysine residues on VP that are exposed on the mature capsids (36) or become exposed during virus entry (37). To avoid nonspecific effects on assembly, we performed a conservative lysine-to-arginine exchange. When analyzing vector preparations, only capsids with exchanges at positions 142/143 and 169 were lacking SUMOylation. As the sites are located in VP2, it seems plausible, therefore, that one or all positions are required for binding an E3 ligase and then are tagged with SUMO. Given that lysine 142/143 and 169 are highly conserved throughout the different AAV serotypes, it is possible that this is an adaptation of the virus to gain stability during host-to-host transmission. In addition, non-SUMOylated AAV vectors that are not and cannot be SUMOylated do not fully escape the restriction by SUMO, since the enhancing effect of Ubc9 knockdown on transduction was still measurable.
A recent study by Mary and colleagues provides a comprehensive analysis of posttranslational modifications of AAV capsids (38). Using a mass spectrometry approach, the authors report a number of different modifications, such as phosphorylation, ubiquitination, acetylation, glycosylation, and SUMOylation. The latter was found, for example, with AAV2 and AAV9 capsids, consistent with our observations (Fig. 1b), but no SUMOylation was detected for AAV5 and -8 capsids, whereas our data show that SUMO is also present on capsids of these types. Further, according to their mass spectrometry analysis, Mary et al. determined SUMO2/3 was a modifier of the AAV capsids, while we identified SUMO1 modifications. In some of our experiments, we also obtained positive signals using SUMO2/3-specific antibodies, but the signals were faint and the results not consistent. In contrast, SUMO1-specific antibodies reproducibly detected SUMO signals on the AAV capsids.
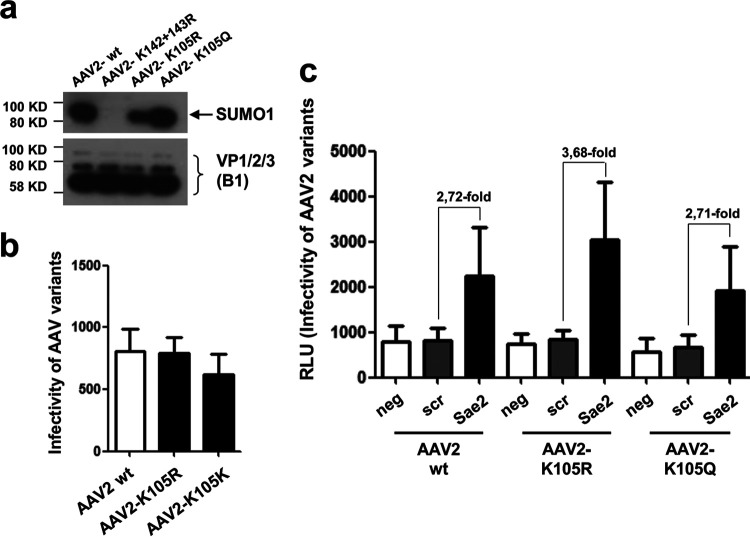
For AAV2, Mary et al. localized the modified site to K258 in a sequence shared by all three capsid proteins. The K258 exchange mutant of AAV2 was not analyzed in this study; however, our data indicate that positions 142/143 and 169 in VP2 are essential for SUMOylation during vector assembly. We cannot exclude that the lysine exchange at these sites also has an effect on distant positions or that, during infection, additional sites become SUMOylated. Of note, Mary et al. report that the K258Q exchange mutant shows reduced, albeit not complete, binding of SUMO1 and also slightly reduced infectivity. The reduced infectivity of a K258E AAV2 mutant, among others, was previously reported in another screen (39), although one would expect higher infectivity if the exchange abolishes SUMOylation of the capsids altogether and if capsid SUMOylation is the prerequisite for the restriction of AAV by SUMO. In a very recent report, Maury et al. report lysine at position 105 as the target for SUMOylation and that a corresponding AAV2 K105Q exchange mutant demonstrates improved transgene expression after ocular gene transfer in mouse retina (40). However, we were not able to confirm this result, as both a K105R and a K105Q AAV2 exchange mutant did not show abolished SUMOylation or improved transduction of cells in vitro. Both mutants also were still susceptible to restriction by the SUMOylation machinery (Fig. 9).
FIG 9.
AAV2 K105R and K105Q capsid variants are SUMOylated. (a) Analysis of wt and mutant AAV capsid by Western blotting using a SUMO1- or VP-specific antibody. (b) Transduction of HeLa cells with wt and mutant AAV vectors. The y axis indicates relative light units. (c) Effect of SUMOylation knockdown on transduction efficiency of wt and mutant vectors.
What leads to SUMOylation of VP2? The SUMOylation pathway parallels that of ubiquitination (41). There are two proteins that form a SUMO activation E1 enzymatic complex, Sae1/Sae2, and one E2 conjugation enzyme, Ubc9. Some substrates can be directly SUMOylated by interacting with Ubc9 if they carry a SUMO interacting motif (SIM), meaning they are able to bind to SUMO tags that are also present on Ubc9. In silico analysis of the VPs did not reveal a conserved SIM for AAV2. Alternatively, SUMOylation of targets is mediated by E3 proteins such as Trim33 (42) or PIAS1 (43), and these two E3 ligases have been identified in our siRNA screen as putative AAV2 restriction factors (21).
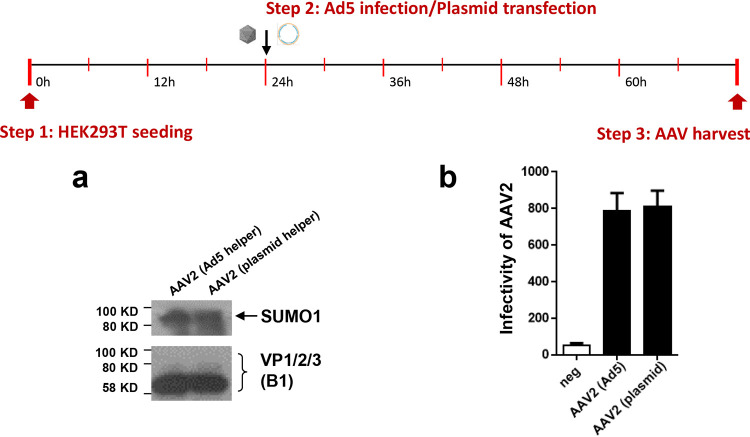
The view that the SUMOylation machinery acts as an antiviral defense against AAV is in accordance with our observation that AAV transduction leads to an upregulation of the cellular SUMOylation activity, as it has been shown for influenza A viruses and herpes simplex viruses (HSV) (27, 44, 45). Cellular stress responses such as the DNA damage response (DDR) were proposed as a trigger (46). As the vectors are not encoding viral proteins, the capsid or the AAV single-stranded vector genomes could act as candidates to induce SUMOylation activity. In particular, the AAV vector genomes have been reported as inducers of DDR (47). It should be noted, however, that in our previous studies we observed that the SUMOylation-dependent restriction on AAV gene transduction is also present in cells lacking the ATR kinase, a key player in the DNA damage response. A number of viruses are able to suppress SUMOylation activity by targeting key components of the SUMO pathway, and among these are AAV helper viruses (for a review, see reference 31). For example, HSV1 encodes a ubiquitin ligase targeting SUMO-tagged proteins of PML bodies for degradation (48). Chicken adenovirus CELO Gam1 protein targets the Sae1/2 SUMO activation complex (26, 49, 50). The elimination of the use of helper viruses in the production and infection of AAV vectors takes these anti-SUMO adaptations out of play and would explain why an increase in total SUMO activity is seen upon AAV transduction. However, when we produced AAV vectors in the presence of adenovirus 5 helper virus, AAV capsids were still found to be SUMOylated, and their infectivity was not increased compared to that of vectors produced by cotransfection of the helper plasmid (Fig. 10). In addition, preventing SUMOylation in the production of AAV vectors by expression of Gam1 only slightly improves yield of vectors (Fig. 11).
FIG 10.
AAV capsids are SUMOylation when produced in the presence of adenovirus 5. Adenovirus helper functions during AAV vector production were provided by helper plasmid cotransfection or by coinfection of cells with adenovirus. (a) Detection of SUMOylated capsid proteins in vector preparations by Western blot analysis. (b) Infectivity of vectors determined by luciferase reporter assay.
FIG 11.
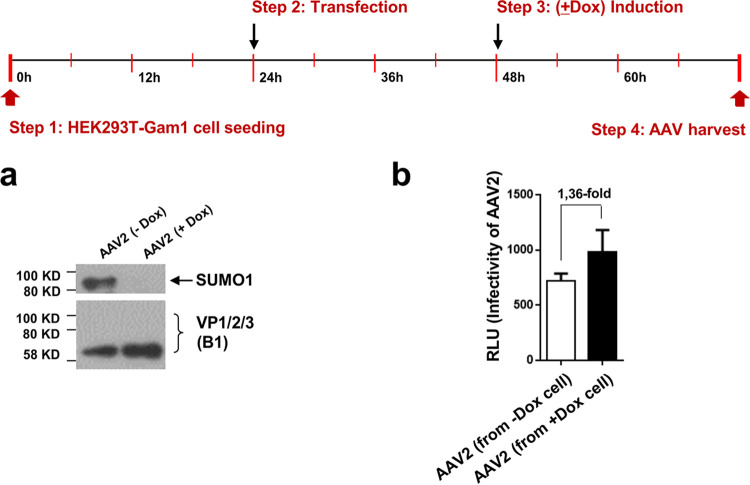
Prevention of SUMOylation during AAV vector production leads to a slight increase in vector yield. AAV vectors were produced in HEK293T-Gam1 cells with (+Dox) or without (−Dox) induction of Gam1. (a) In the presence of Gam1 vector, capsid proteins are not SUMOylated, as shown by Western blot analysis. (b) Transduction efficiency of vector preparation determined by luciferase reporter assay.
According to our findings, SUMOylation represses AAV transduction by directly targeting viral components. However, an even stronger effect of AAV inhibition acts through cellular components activated by SUMOylation. One candidate for the latter is DAXX, a well-known antiviral factor (for a review, see reference 29). DAXX itself is tightly involved in the cellular SUMOylation machinery. It contains C- and N-terminal SUMO-interacting motifs, and through interaction of DAXX with SUMOylated targets, the localization and action of DAXX is regulated (51). We observed strongly increased AAV transduction rates in DAXX knockout cells that are also manifested by increased accumulation of vectors in the cells. This suggests an influence of DAXX and SUMOylation on intracellular trafficking or stability of AAV vectors. At the same time, the activation of AAV transduction through SUMOylation knockdown by Ubc9 siRNA transfection was almost completely reduced in these cells, indicating a functional overlap of DAXX and SUMOylation pathways in the control of AAV transduction.
Until now, the characterization of SUMO as a posttranslational modification that significantly affects AAV has not been explored. Our study relates the restrictive effect of SUMOylation and its activation on AAV gene transduction via two processes: capsid SUMOylation and activation of cellular protein SUMOylation by AAV infection. Here, we were able to validate the interaction and further identify the exact residues of the capsid protein VP2 that are involved. Although the exact functional consequence of this modification remains open, the effect on the stability of the capsid is evident and could be seen as an adaptation of the virus. While capsid SUMOylation has only a minor effect on gene transduction, the strong increase of SUMOylation of cellular proteins induced by AAV indicates an antiviral defense response leading to a decreased accumulation of vectors in the cell, demonstrated here by the effect in the DAXX knockout cells. The increased infectivity of AAV2-luc vectors when used in combination with vectors that prevent cellular SUMOylation offers an interesting possibility in the development of lower-titer gene therapy treatments and presents the SUMOylation process and the interplay with host restriction factors, such as DAXX, as future targets that could improve AAV as a gene therapy vector. We observed that in vitro the transduction efficiency of AAV2 can be increased almost 10-fold by cotransduction with an AAV2-Gam1 vector, while the total vector dose was increased by 50%. It is likely that this effect would be even greater, but Gam1 expression can be carried out only for a limited length of time due to cellular toxicity.
MATERIALS AND METHODS
Cell culture, production, and purification of AAV vectors.
HeLa (ATCC-CCL-2), HEK293T (ATCC ACS-4500), and A549 (ATCC CCL-185) cells were maintained at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Sigma) low glucose supplemented with 10% fetal calf serum (FCS), 100 U penicillin, 100 mg streptomycin, and 2 mM l-glutamine. HEK293T-Gam1 cells (52) were maintained at 37°C and 5% CO2 in DMEM high glucose supplemented with 10% FCS, 100 U penicillin, 100 mg streptomycin, 100 μg/ml Zeocin, 15 μg/ml blasticidin, 2 mM l-glutamine, and 1 μg/ml tetracycline, with 24-h induction when needed. HeLa-Gam1 cell (26) were maintained at 37°C and 5% CO2 in DMEM low glucose supplemented with 10% FCS, 2 mM l-glutamine, 200 μg/ml hygromycin B, 4 μg/ml blasticidin, and 100 ng/ml tetracycline, with 12-h induction when need. The HeLa-DAXX knockout cell line was produced by the CRISPR/Cas system in DMEM, and oligonucleotides (nontargeting control and all Edit-R human DAXX crRNA) were purchased from Dharmacon (Lafayette, CO).
The following oligonucleotides were used: Edit-R crRNA nontargeting control #1, Edit-R CRISPR-Cas9 synthetic tracrRNA, Edit-R human DAXX (1616) crRNA (GATGTTGCAGAACTCCGCCG), Edit-R human DAXX (1616) crRNA (CCTGGGATGCCATTCCACTA), and Edit-R human DAXX (1616) crRNA (GGAAATGTCCGTCTCCACAG).
HPV58 pseudovirions and IAV were produced as described previously (27, 53). For production of different serotypes of rAAV vectors (AAV1, AAV2, AAV5, AAV8, and AAV9) and rAAV2 capsid variants, a three-plasmid transfection system (capsid plus reporter plus helper function vectors) was used as described before (21). Non-SUMOylated AAV2 particles were produced in HEK293T-Gam1 cells after Dox induction.
Generation of plasmid constructs.
All AAV2 capsid protein mutation plasmids, as well as AAV2 VP protein-deficient plasmids VP1+VP3 (no VP2) and VP2+VP3 (no VP1), were constructed in AAV2 wt capsid without ITRs (kindly provided by Dirk Grimm) and then transformed in XL-blue chemically super competent cells after QuikChange mutagenesis (QuikChange II site-directed mutagenesis kit) (see Table S1 in the supplemental material). N-terminal VP2 extension constructs GFP-VP2 and HA-VP2 were prepared via conventional PCR and ligated in N-GFP-pDEST or N-HA-pDEST vector by Gateway cloning (Gateway LR Clonase II enzyme mix) and transformed into electrocompetent MegaX DH10 (Invitrogen) cells, respectively (Table S2). All oligonucleotides were ordered and produced at MWG Eurofins in Ebersberg, Germany.
AAV particle production, purification, buffer exchange, and quantification.
To produce different types of AAV-luc (AAV1, AAV2, AAV5, AAV8, and AAV9) and AAV2-Gam1 particles, 5 × 107 HEK293TT cells were seeded on five 15-cm dishes and transfected using the three-plasmid system with AAV capsid plasmid, helper plasmid, and a firefly reporter plasmid at a molar ratio of 1:1:1. After 48 h of transfection, the cells were harvested. The pellet was resuspended in AAV-lysis buffer (50 mM Tris, pH 8.5, 150 mM NaCl, in H2O, pH 8.5) before undergoing 5 freeze-thaw cycles. Benzonase (50 U/ml), which was used to degrade all forms of DNA and RNA, was added and incubated, followed by centrifugation to harvest the cell crude lysate. Crude lysates containing AAV particles were obtained from the 40% iodixanol phase of a gradient consisting of iodixanol (Sigma) in PBS-MK–NaCl (1 mM MgCl2, 2.5 mM KCl, 1 M NaCl in phosphate-buffered saline), 25% iodixanol in PBS-MK with 3 μl phenol red solution (0.5%), 40% iodixanol in PBS-MK, and 3.8 ml of 60% iodixanol with 5 μl phenol red solution (54). Purified AAV particles were stored at –20°C or –80°C for long-term preservation. For in vivo applications, iodixanol was removed by using a ZEBA desalt spin column (Thermo Scientific) according to the manufacturer’s protocol. AAV particle titer was determined via quantitative real-time PCR (Table S2) (55).
Viral transduction and Luciferase assay.
Viral transduction assays were performed to analyze the infectivity of the viruses containing the firefly reporter gene on different cell lines at different conditions. Cells were seeded at least 1 day before siRNA transduction (Qiagen) or induction with Dox and the viral transduction was done at an MOI of 103. The firefly luciferase signal was analyzed using the Beetle Juice Luciferase assay firefly kit (no. 102511; P.J.K. GmbH) according to the manufacturer’s instructions. The luminescence was analyzed via the Wallac Work Station 1 min after addition of the substrate.
The following siRNAs were used: negative-control siRNA (1022076), AATTCTCCGAACGTGTCACGT; Hs_SAE2_3 (Sea2, SI04234433), CACCGGTTTCTCCCACATCGA; Hs_UBE2I_8 (Ubc9, SI04185937), ACCACCATTATTTCACCCGAA.
Immunoprecipitation.
To investigate the interaction of AAV capsids and SUMO protein, immunoprecipitation was performed using magnetic beads (SureBeads Protein G Magnetic Beads; NEB). Briefly, SureBeads were washed three times with NET-N buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA) containing 1% NP-40. Purified AAV capsid-specific antibody A20 (1:100) or HPV capsid-specific antibody K18L2 (1:100) as a negative control was added to beads in 1% NET-N buffer and incubated on a rotating wheel at 4°C overnight. The beads were washed three times with 1% NET-N buffer on day two and incubated with 5 × 109 purified AAV2 or HPV58 particles (per sample) overnight on a rotating wheel at 4°C. On the third day, the beads containing AAV particles were washed three times with 1% NET-N buffer by resuspension and subsequent magnetization and then moved into a new tube with 40 μl of 1× SDS buffer. The beads were incubated at 95°C for 10 min, followed by moving the eluent to a new tube, which was loaded for analysis via Western blotting.
Immunofluorescence.
To visualize AAV trafficking at different time points after downregulation of SUMOylation, indirect immunofluorescence was performed. HeLa cells (2.5 × 104) were plated on coverslips in a 24-well plate. AAV2 transduction was performed on the cells with an MOI of 105 and incubated at 4°C for 1 h on a shaker. After three washings with PBS, cells were supplemented with fresh medium and transferred to 37°C, 5% CO2. To test AAV infection, slides were collected at different time points after transduction. The slides were washed with 1× PBS and fixed with 2% paraformaldehyde (PFA) for 15 min at room temperature (RT). The cells were quenched in 50 mM ammonium chloride (50 nM NH4Cl) twice for 10 min to avoid artifacts and then permeabilized in 0.2% Triton X-100 for 10 min. After another 3 washings with 1× PBS, the cells were blocked in 1% bovine serum albumin (BSA) for 1 h at 37°C. Thereafter the cells were incubated with primary antibody A20 (1:100) and LamB1 (sc-20011; 1:200; Santa Cruz) diluted in 1% BSA for 1 h at 37°C. The cells were washed three times with 1× PBS and were incubated with secondary antibody AlexaFluor-488/594 (1:100; Life Technologies) as well as 4′,6-diamidino-2-phenylindole (DAPI) diluted in 1% BSA in PBS at 37°C for 1 h. Subsequently, the coverslips were washed three times with 1× PBS and mounted onto slides using mounting medium before being sealed with nail polish. The slides were visualized using the Zeiss Cell Observer or a confocal microscope. The images were processed using Image J, which displayed the cell area and integrated the density to calculate the average optical density.
Dot blotting and Western blotting.
For dot blots, the nitrocellulose (NC) membrane or polyvinylidene difluoride (PVDF) membrane, activated in methanol for 20 min, was assembled with two pieces of filter papers on the filtration plate and fixed on the base of the dot blot chamber. The chamber was sealed with parafilm and connected to a vacuum pump. From the protein sample in lysis buffer (50 mM Tris, pH 8.5, 150 mM NaCl in H2O, pH 8.5), AAV particles were spotted onto the membrane. After completion, the membrane was blocked in 5% milk and then incubated with purified antibody A20 (1:500) or B1 (1:500). The membrane was washed three times with PBS-T (0.3% Tween 20 in PBS) and incubated for 1 h at RT with the secondary antibody (horseradish peroxidase [HRP]-tagged, anti-mouse, or anti-rabbit antibodies). The membrane was washed three times, and proteins were detected using the chemiluminescent kit (A3417-1200); AppliChen according to the manufacturer’s protocol. For Western blots, SDS-PAGE was performed to separate different proteins according to their molecular weights. Each sample (cell lysate or viral particles) was mixed with 3× SDS loading buffer and boiled for 10 min at 95°C and loaded along with a protein ladder. Proteins were separated on the 12.5% SDS gels in 1× TGS running buffer (2.5 mM Tris, 1.45% glycine, 0.1% SDS, in H2O, pH 8.3), and samples were blotted onto activated PVDF or NC membranes. Three soaked blotting pads were placed into the blot module, followed by filter paper. The gel was covered with membrane, followed by more filter paper and another three soaked blotting pads on top, and then fixed into the X Cell II blot module. The transfer was performed at 30 mV for 90 min for proteins with a molecular weight around 80 to 150 kDa, or 30 min for protein smaller than 50 kDa. The membrane was blocked in 5% milk PBS-T at RT and then incubated with the desired primary antibody against SUMO1 (1:500; FL-101; Santa Cruz), SUMO2/3 (PA5-11373; 1:1,000; Sigma), DAXX (25c12, 1:1,000; CST), HA (1:500; sc-805; Santa Cruz), α-actin (1:5,000; MP), and VP (1:500 B1; Progen) in 5% PBS-T milk overnight at 4°C. After incubation, the membrane was washed three times with PBS-T and incubated with the respective HRP-conjugated secondary antibody. The membrane then was washed three times and proteins were detected by a chemiluminescent kit according to the manufacturer’s protocol. Chemiluminescence was detected by the developing machine.
Supplementary Material
ACKNOWLEDGMENT
We are grateful to Macro Binder for providing the A549 cells and the IAV.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Rose JA, Hoggan MD, Shatkin AJ. 1966. Nucleic acid from an adeno-associated virus: chemical and physical studies. Proc Natl Acad Sci U S A 56:86–92. doi: 10.1073/pnas.56.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoggan MD, Blacklow NR, Rowe WP. 1966. Studies of small DNA viruses found in various adenovirus preparations: physical, biological, and immunological characteristics. Proc Natl Acad Sci U S A 55:1467–1474. doi: 10.1073/pnas.55.6.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong L, Li B, Jayandharan G, Mah CS, Govindasamy L, Agbandje-McKenna M, Herzog RW, Weigel-Van Aken KA, Hobbs JA, Zolotukhin S, Muzyczka N, Srivastava A. 2008. Tyrosine-phosphorylation of AAV2 vectors and its consequences on viral intracellular trafficking and transgene expression. Virology 381:194–202. doi: 10.1016/j.virol.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srivastava A. 2016. In vivo tissue-tropism of adeno-associated viral vectors. Curr Opin Virol 21:75–80. doi: 10.1016/j.coviro.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keeler AM, Flotte TR. 2019. Recombinant adeno-associated virus gene therapy in light of luxturna (and zolgensma and glybera): where are we, and how did we get here? Annu Rev Virol 6:601–621. doi: 10.1146/annurev-virology-092818-015530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell S, Bennett J, Wellman JA, Chung DC, Yu ZF, Tillman A, Wittes J, Pappas J, Elci O, McCague S, Cross D, Marshall KA, Walshire J, Kehoe TL, Reichert H, Davis M, Raffini L, George LA, Hudson FP, Dingfield L, Zhu X, Haller JA, Sohn EH, Mahajan VB, Pfeifer W, Weckmann M, Johnson C, Gewaily D, Drack A, Stone E, Wachtel K, Simonelli F, Leroy BP, Wright JF, High KA, Maguire AM. 2017. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet 390:849–860. doi: 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnett JR, Hooper AJ. 2009. Alipogene tiparvovec, an adeno-associated virus encoding the Ser(447)X variant of the human lipoprotein lipase gene for the treatment of patients with lipoprotein lipase deficiency. Curr Opin Mol Ther 11:681–691. [PubMed] [Google Scholar]
- 8.Al-Zaidy SA, Kolb SJ, Lowes L, Alfano LN, Shell R, Church KR, Nagendran S, Sproule DM, Feltner DE, Wells C, Ogrinc F, Menier M, L'Italien J, Arnold WD, Kissel JT, Kaspar BK, Mendell JR. 2019. AVXS-101 (onasemnogene abeparvovec) for SMA1: comparative study with a prospective natural history cohort. J Neuromuscul Dis 6:307–317. doi: 10.3233/JND-190403. [DOI] [PubMed] [Google Scholar]
- 9.Mahajan R. 2019. Onasemnogene abeparvovec for spinal muscular atrophy: the costlier drug ever. Int J Appl Basic Med Res 9:127–128. doi: 10.4103/ijabmr.IJABMR_190_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari FK, Samulski T, Shenk T, Samulski RJ. 1996. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol 70:3227–3234. doi: 10.1128/JVI.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartlett JS, Wilcher R, Samulski RJ. 2000. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J Virol 74:2777–2785. doi: 10.1128/JVI.74.6.2777-2785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarty DM, Monahan PE, Samulski RJ. 2001. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther 8:1248–1254. doi: 10.1038/sj.gt.3301514. [DOI] [PubMed] [Google Scholar]
- 13.Flotte TR, Trapnell BC, Humphries M, Carey B, Calcedo R, Rouhani F, Campbell-Thompson M, Yachnis AT, Sandhaus RA, McElvaney NG, Mueller C, Messina LM, Wilson JM, Brantly M, Knop DR, Ye GJ, Chulay JD. 2011. Phase 2 clinical trial of a recombinant adeno-associated viral vector expressing alpha1-antitrypsin: interim results. Hum Gene Ther 22:1239–1247. doi: 10.1089/hum.2011.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pillay S, Carette JE. 2017. Host determinants of adeno-associated viral vector entry. Curr Opin Virol 24:124–131. doi: 10.1016/j.coviro.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y, Jae LT, Wosen JE, Nagamine CM, Chapman MS, Carette JE. 2016. An essential receptor for adeno-associated virus infection. Nature 530:108–112. doi: 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nonnenmacher M, Weber T. 2011. Adeno-associated virus 2 infection requires endocytosis through the CLIC/GEEC pathway. Cell Host Microbe 10:563–576. doi: 10.1016/j.chom.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popa-Wagner R, Porwal M, Kann M, Reuss M, Weimer M, Florin L, Kleinschmidt JA. 2012. Impact of VP1-specific protein sequence motifs on adeno-associated virus type 2 intracellular trafficking and nuclear entry. J Virol 86:9163–9174. doi: 10.1128/JVI.00282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lux K, Goerlitz N, Schlemminger S, Perabo L, Goldnau D, Endell J, Leike K, Kofler DM, Finke S, Hallek M, Buning H. 2005. Green fluorescent protein-tagged adeno-associated virus particles allow the study of cytosolic and nuclear trafficking. J Virol 79:11776–11787. doi: 10.1128/JVI.79.18.11776-11787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warrington KH Jr, Gorbatyuk OS, Harrison JK, Opie SR, Zolotukhin S, Muzyczka N. 2004. Adeno-associated virus type 2 VP2 capsid protein is nonessential and can tolerate large peptide insertions at its N terminus. J Virol 78:6595–6609. doi: 10.1128/JVI.78.12.6595-6609.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buning H, Srivastava A. 2019. Capsid modifications for targeting and improving the efficacy of AAV vectors. Mol Ther Methods Clin Dev 12:248–265. doi: 10.1016/j.omtm.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holscher C, Sonntag F, Henrich K, Chen Q, Beneke J, Matula P, Rohr K, Kaderali L, Beil N, Erfle H, Kleinschmidt JA, Muller M. 2015. The SUMOylation pathway restricts gene transduction by adeno-associated viruses. PLoS Pathog 11:e1005281. doi: 10.1371/journal.ppat.1005281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mano M, Ippodrino R, Zentilin L, Zacchigna S, Giacca M. 2015. Genome-wide RNAi screening identifies host restriction factors critical for in vivo AAV transduction. Proc Natl Acad Sci U S A 112:11276–11281. doi: 10.1073/pnas.1503607112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schreiber CA, Sakuma T, Izumiya Y, Holditch SJ, Hickey RD, Bressin RK, Basu U, Koide K, Asokan A, Ikeda Y. 2015. An siRNA screen identifies the U2 snRNP spliceosome as a host restriction factor for recombinant adeno-associated viruses. PLoS Pathog 11:e1005082. doi: 10.1371/journal.ppat.1005082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hay RT. 2005. SUMO: a history of modification. Mol Cell 18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Flotho A, Melchior F. 2013. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 26.Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. 2004. A mechanism for inhibiting the SUMO pathway. Mol Cell 16:549–561. doi: 10.1016/j.molcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Domingues P, Golebiowski F, Tatham MH, Lopes AM, Taggart A, Hay RT, Hale BG. 2015. Global reprogramming of host SUMOylation during influenza virus infection. Cell Rep 13:1467–1480. doi: 10.1016/j.celrep.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI, Hung CC, Suen CS, Hwang MJ, Chang KS, Maul GG, Shih HM. 2006. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell 24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 29.Schreiner S, Wodrich H. 2013. Virion factors that target Daxx to overcome intrinsic immunity. J Virol 87:10412–10422. doi: 10.1128/JVI.00425-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cappadocia L, Lima CD. 2018. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem Rev 118:889–918. doi: 10.1021/acs.chemrev.6b00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Everett RD, Boutell C, Hale BG. 2013. Interplay between viruses and host sumoylation pathways. Nat Rev Microbiol 11:400–411. doi: 10.1038/nrmicro3015. [DOI] [PubMed] [Google Scholar]
- 32.Aichem A, Sailer C, Ryu S, Catone N, Stankovic-Valentin N, Schmidtke G, Melchior F, Stengel F, Groettrup M. 2019. The ubiquitin-like modifier FAT10 interferes with SUMO activation. Nat Commun 10:4452. doi: 10.1038/s41467-019-12430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müncheberg S, Hay RT, Ip WH, Meyer T, Weiß C, Brenke J, Masser S, Hadian K, Dobner T, Schreiner S. 2018. E1B-55K-mediated regulation of RNF4 SUMO-targeted ubiquitin ligase promotes human adenovirus gene expression. J Virol 92:e00164-18. doi: 10.1128/JVI.00164-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seeler JS, Dejean A. 2003. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol 4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 35.Muller S, Hoege C, Pyrowolakis G, Jentsch S. 2001. SUMO, ubiquitin's mysterious cousin. Nat Rev Mol Cell Biol 2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- 36.Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, Chapman MS. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci U S A 99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonntag F, Bleker S, Leuchs B, Fischer R, Kleinschmidt JA. 2006. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J Virol 80:11040–11054. doi: 10.1128/JVI.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mary B, Maurya S, Arumugam S, Kumar V, Jayandharan GR. 2019. Post-translational modifications in capsid proteins of recombinant adeno-associated virus (AAV) 1-rh10 serotypes. FEBS J 286:4964–4981. doi: 10.1111/febs.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B, Ma W, Ling C, Van Vliet K, Huang LY, Agbandje-McKenna M, Srivastava A, Aslanidi GV. 2015. Site-directed mutagenesis of surface-exposed lysine residues leads to improved transduction by AAV2, but not AAV8, vectors in murine hepatocytes in vivo. Hum Gene Ther Methods 26:211–220. doi: 10.1089/hgtb.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maurya S, Mary B, Jayandharan GR. 2019. Rational engineering and pre-clinical evaluation of Neddylation and SUMOylation site modified AAV vectors in murine models of hemophilia B and Leber congenital amarousis. Hum Gene Ther 30:1461–1476. doi: 10.1089/hum.2019.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melchior F. 2000. SUMO–nonclassical ubiquitin. Annu Rev Cell Dev Biol 16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 42.Ikeuchi Y, Dadakhujaev S, Chandhoke AS, Huynh MA, Oldenborg A, Ikeuchi M, Deng L, Bennett EJ, Harper JW, Bonni A, Bonni S. 2014. TIF1gamma protein regulates epithelial-mesenchymal transition by operating as a small ubiquitin-like modifier (SUMO) E3 ligase for the transcriptional regulator SnoN1. J Biol Chem 289:25067–25078. doi: 10.1074/jbc.M114.575878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishida T, Yasuda H. 2002. PIAS1 and PIASxalpha function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J Biol Chem 277:41311–41317. doi: 10.1074/jbc.M206741200. [DOI] [PubMed] [Google Scholar]
- 44.Boutell C, Cuchet-Lourenco D, Vanni E, Orr A, Glass M, McFarlane S, Everett RD. 2011. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog 7:e1002245. doi: 10.1371/journal.ppat.1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cuchet-Lourenco D, Boutell C, Lukashchuk V, Grant K, Sykes A, Murray J, Orr A, Everett RD. 2011. SUMO pathway dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog 7:e1002123. doi: 10.1371/journal.ppat.1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saitoh H, Hinchey J. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem 275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 47.Schwartz RA, Carson CT, Schuberth C, Weitzman MD. 2009. Adeno-associated virus replication induces a DNA damage response coordinated by DNA-dependent protein kinase. J Virol 83:6269–6278. doi: 10.1128/JVI.00318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Everett RD, Boutell C, McNair C, Grant L, Orr A. 2010. Comparison of the biological and biochemical activities of several members of the alphaherpesvirus ICP0 family of proteins. J Virol 84:3476–3487. doi: 10.1128/JVI.02544-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boggio R, Chiocca S. 2005. Gam1 and the SUMO pathway. Cell Cycle 4:533–535. doi: 10.4161/cc.4.4.1605. [DOI] [PubMed] [Google Scholar]
- 50.Boggio R, Passafaro A, Chiocca S. 2007. Targeting SUMO E1 to ubiquitin ligases: a viral strategy to counteract sumoylation. J Biol Chem 282:15376–15382. doi: 10.1074/jbc.M700889200. [DOI] [PubMed] [Google Scholar]
- 51.Escobar-Cabrera E, Okon M, Lau DK, Dart CF, Bonvin AM, McIntosh LP. 2011. Characterizing the N- and C-terminal small ubiquitin-like modifier (SUMO)-interacting motifs of the scaffold protein DAXX. J Biol Chem 286:19816–19829. doi: 10.1074/jbc.M111.231647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu F, Chiocca S, Beck WT, Mo YY. 2007. Gam1-associated alterations of drug responsiveness through activation of apoptosis. Mol Cancer Ther 6:1823–1830. doi: 10.1158/1535-7163.MCT-06-0771. [DOI] [PubMed] [Google Scholar]
- 53.Pouyanfard S, Spagnoli G, Bulli L, Balz K, Yang F, Odenwald C, Seitz H, Mariz FC, Bolchi A, Ottonello S, Muller M. 2018. Minor capsid protein L2 polytope induces broad protection against oncogenic and mucosal human papillomaviruses. J Virol 92:e01930-17. doi: 10.1128/JVI.01930-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zolotukhin S, Byrne BJ, Mason E, Zolotukhin I, Potter M, Chesnut K, Summerford C, Samulski RJ, Muzyczka N. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther 6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 55.Veldwijk MR, Topaly J, Laufs S, Hengge UR, Wenz F, Zeller WJ, Fruehauf S. 2002. Development and optimization of a real-time quantitative PCR-based method for the titration of AAV-2 vector stocks. Mol Ther 6:272–278. doi: 10.1006/mthe.2002.0659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.