Merkel cell carcinoma (MCC) is the most aggressive cutaneous tumor without clearly defined treatment. Although MCC has a high propensity for metastasis, little is known about the underlying mechanisms that drive MCC invasion and metastatic progression. MMP-9 has been shown to play a detrimental role in many metastatic human cancers, including melanoma and other nonmelanoma skin cancers. Our study shows that MCV sT-mediated MMP-9 activation is driven through the LSD, a known E3 ligase-targeting domain, in MCC. MMP-9 may serve as the biochemical culprit to target and develop a novel approach for the treatment of metastatic MCC.
KEYWORDS: Merkel cell carcinoma, Merkel cell polyomavirus, small tumor antigen, matrix metallopeptidase-9, FBW7, cell migration, cell invasion
ABSTRACT
Merkel cell polyomavirus (MCV) small T antigen (sT) is the main oncoprotein for the development of Merkel cell carcinoma (MCC). MCC is a rare, clinically aggressive neuroendocrine tumor of the skin with a high propensity for local, regional, and distant spread. The dysregulation of matrix metalloproteinase-9 (MMP-9) has been implicated in multiple essential roles in the development of various malignant tumor cell invasion and metastasis. Previously, MCV sT was shown to induce the migratory and invasive phenotype of MCC cells through the transcriptional activation of the sheddase molecule, ADAM 10 (A disintegrin and metalloprotease domain-containing protein 10). In this study, we show that MCV sT protein stimulates differential expression of epithelial-mesenchymal transition (EMT)-associated genes, including MMP-9 and Snail. This effect is dependent on the presence of the large T stabilization domain (LSD), which is known to be responsible for cell transformation through targeting of promiscuous E3 ligases, including FBW7, a known MMP-9 and Snail regulator. Chemical treatments of MMP-9 markedly inhibited MCV sT-induced cell migration and invasion. These results suggest that MCV sT contributes to the activation of MMP-9 as a result of FBW7 targeting and increases the invasive potential of cells, which can be used for targeted therapeutic intervention.
IMPORTANCE Merkel cell carcinoma (MCC) is the most aggressive cutaneous tumor without clearly defined treatment. Although MCC has a high propensity for metastasis, little is known about the underlying mechanisms that drive MCC invasion and metastatic progression. MMP-9 has been shown to play a detrimental role in many metastatic human cancers, including melanoma and other nonmelanoma skin cancers. Our study shows that MCV sT-mediated MMP-9 activation is driven through the LSD, a known E3 ligase-targeting domain, in MCC. MMP-9 may serve as the biochemical culprit to target and develop a novel approach for the treatment of metastatic MCC.
INTRODUCTION
Merkel cell carcinoma (MCC) is a rare skin cancer of neuroendocrine origin with a high propensity to metastasize (1). Although the incidence rate of MCC is lower than that of melanoma, it is highly aggressive with an estimated mortality rate of 33 to 46%; hence, it is significantly more lethal than malignant melanoma (2). Merkel cell polyomavirus (MCV) is the etiological agent of MCV-positive MCCs and is monoclonally integrated in 80 to 90% of both primary and metastatic MCC tumors (3–5). As a classic polyomavirus, the genomic organization of MCV is similar to that of other known human polyomaviruses. MCV expresses small and large tumor antigens (sT and LT), which are essential for viral replication and pathogenesis. MCC tumor-derived MCV LT sequences integrated into MCC genomes contain mutations prematurely truncating the C-terminal growth inhibitory domain (6), while MCV sT remains intact. The importance of the MCV T antigens has been shown using depletion studies, where it is shown that knockdown of T antigens in MCV-positive MCC cell lines induces growth arrest as well as cell death (7).
MCV sT has been shown to mediate multiple oncogenic mechanisms that contribute to MCC development. Inhibition of SCF (Skp1, Cullin, F-box containing complex) E3 ligases by MCV sT appears to induce activation of several viral and cellular oncoproteins, leading to enhanced MCV replication and cell proliferation (8, 9). Aberrant activation of oncogenic potential in MCV sT-expressing cells also promoted the malignant phenotypes that are involved in genomic instability, such as centrosome amplification, aneuploidy, and micronuclei formation (10). This oncogenic activity of MCV sT requires the LT stabilization domain (LSD), a unique and disordered domain of MCV sT, which is known to interact with SCF E3 ligase complexes (8, 10, 11). Although the exact mechanism by which MCV sT targets E3 ligases is yet to be elucidated, it is clear that the LSD plays a significant role in the distinctive transforming activities induced by MCV sT in vitro and in vivo (8–11).
The E3 ubiquitin ligase F-box and WD repeat domain-containing 7 (FBW7) functions as a putative tumor suppressor and an evolutionarily conserved substrate receptor of SCF ubiquitin ligase complex and plays vital roles in cell proliferation and cell migration (12). In various cancers, including renal cancer (13, 14), gastric cancer (15), and hepatocellular carcinoma (16), FBW7 inhibition promotes metastasis and epithelial-mesenchymal transition (EMT) by upregulating matrix metalloproteinase (MMP) expression, specifically MMP-2, MMP-9, and MMP-13 (13).
Matrix metalloproteinases (MMPs) are a zinc-dependent family of proteolytic enzymes that participate in the degradation of the extracellular matrix (ECM). Dysregulation of these proteases has been observed in multiple cancers where enhanced expression of certain MMP proteins contribute to cell migration, invasion, and angiogenesis (17, 18). Specifically, MMP-9 has been linked to multiple hallmarks of cancer, including but not limited to metastasis, invasion, immunological surveillance, and angiogenesis (19). MMP-9, also known as 92-kDa type IV collagenase (20), plays a vital role in the degradation of elastin and partially hydrolyzed collagen, which are essential for maintaining epithelial structural integrity. Various studies have shown that human tumor virus-associated oncoproteins play a critical role in metastasis and EMT-related mechanisms. Hepatitis B virus (HBV)-encoded X protein (HBx) (21), Kaposi’s sarcoma-associated herpesvirus (KSHV) K1 (22), and Epstein-Barr virus (EBV) latent membrane protein 1 (LMP-1) proteins (23) are known to upregulate MMP-9 expression, thereby contributing to invasiveness and metastasis, key hallmarks of cancer (24).
MCV sT stimulates cell motility by inducing microtubule destabilization (25), actin rearrangement (26), and cell dissociation by disruption of cell junctions (27). Interrogation of previously published quantitative proteomic data sets of MCV sT-expressing cells (25) indicates that MCV sT activated expression of Snail, a transcription factor that enhances mesenchymal genes, and MMP-9. In contrast, MCV sT significantly downregulated genes related to cell adhesion molecules, suggesting the potential function of MCV sT in the regulation of EMT. MMP-9 and Snail activation by MCV sT was strictly dependent on the presence of the LSD, which resulted in the enhancement of cell migration in mouse fibroblast cells and human cancer cell lines. Our findings indicate that the MCV sT targeting of cellular E3 ligases may play a role in MCV sT-induced cell migration and invasion in MCC. Notably, chemical treatment with MMP-9 inhibitors resulted in significant inhibition of MCV sT-induced cell migration and invasion. This suggests that the MMP-9 protein may be a viable target for novel therapeutic intervention for disseminated MCC.
RESULTS
MCV sT induces differential expression of proteins associated with EMT.
Recent studies have highlighted the involvement of MCV sT in the highly migratory and cell dissociation phenotypes of MCC, elucidating its highly multifunctional roles in MCC (25–27). Previously described stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomics data (25) were further interrogated to assess alterations in the host cell proteome upon expression of MCV sT in an HEK293-derived cell line (i293-sT) (Fig. 1A) (25). These results highlighted an alteration in proteins associated with enhancement of cell migration (microtubule-associated cytoskeletal organization) and cell adhesion as previously reported and the basement membrane proteins, a specialized form of the ECM. Specifically, the quantitative proteomic analysis showed an almost 2-fold decrease in the collagen alpha-2(IV) chain (COL4A2) and laminin subunit gamma 1 (LAMC1), two essential components of the basement membrane. The basement membrane is crucial for epithelial structural integrity. It is comprised of a network of glycoproteins and proteoglycans, such as type IV collagen and laminin, and provides a barrier from invasion by tumor cells (28). These results suggest that MCV sT plays a role in basement membrane degradation, an essential process for the metastatic invasion of tumor cells into the circulatory system to occur in MCC. Aligned with these observations, transcriptome analysis has suggested that specific markers associated with EMT are increased upon MCV sT expression (29).
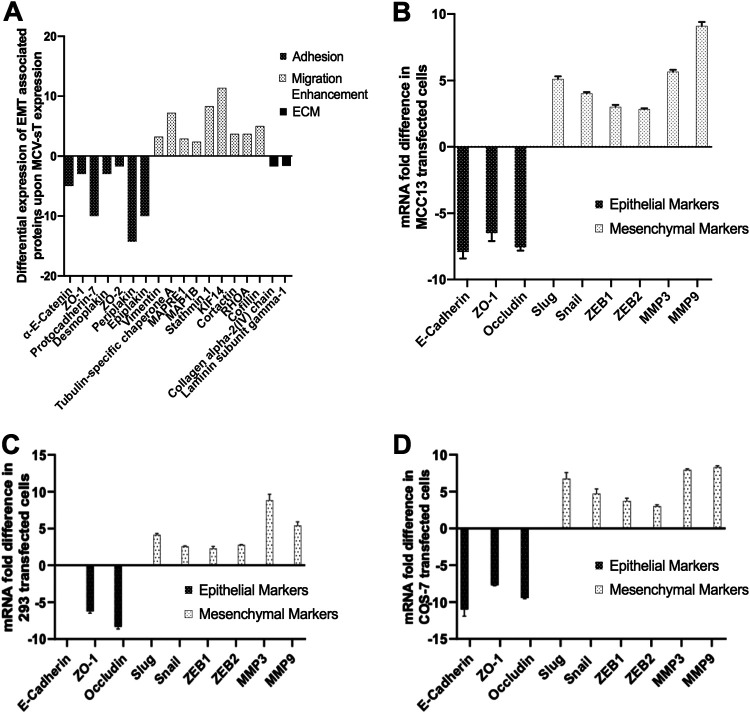
FIG 1.
MCV sT leads to differential expression of proteins associated with epithelial to mesenchymal transition (EMT). (A) Quantitative proteomics analysis illustrating differential expression of EMT-associated proteins upon MCV sT expression. The i293-sT cells were grown in DMEM with unlabeled arginine and lysine amino acids (R0K0) and induced (IN) for 24 h or grown in DMEM with labeled amino acids (R6K4) and remained uninduced (UN) (25). Proteins associated with cell adhesion and structural integrity of the extracellular matrix were downregulated upon MCV sT expression, while expression of proteins involved in cell migration by reorganization of the actin network and microtubule destabilization were upregulated. (B) MCV sT regulates EMT-associated gene expression. MCC13 cells were transfected with control or MCV sT plasmids. While epithelial markers were downregulated, mesenchymal markers were significantly upregulated upon MCV sT expression. Cellular RNA was extracted using a TRIzol reagent, and transcript levels were analyzed by RT-qPCR using the comparative ΔΔCT method (n = 3). HEK293 (C) and COS-7 (D) cells were transfected with control or MCV sT plasmids. While epithelial markers were downregulated, mesenchymal markers were significantly upregulated upon MCV sT expression. E-cadherin expression levels were not assessed in HEK293 cells due to low expression. Cellular RNA was extracted using a TRIzol reagent, and transcript levels were analyzed by RT-qPCR using the comparative ΔΔCT method (n = 3).
To validate the potential regulation of EMT-related markers by MCV sT, reverse transcription-quantitative PCR (RT-qPCR) was performed. Changes in mRNA levels of classic EMT markers were assessed in MCC13 (MCV-negative MCC) cells overexpressing a control or MCV sT construct. Upon MCV sT expression, a significant downregulation in epithelial markers E-cadherin, zonula occludens-1 (ZO-1), and occludin was observed (Fig. 1B). Conversely, mesenchymal markers Slug, Snail, ZEB1, ZEB2, MMP-3, and MMP-9 were upregulated upon MCV sT expression. A similar trend was also observed in HEK293 and COS-7 transfected cells (Fig. 1C and D), inferring the possibility of MCV sT inducing an EMT, which contributes to the metastatic potential of MCV-associated MCC.
MCV sT inhibition of FBW7 contributes to migratory phenotype.
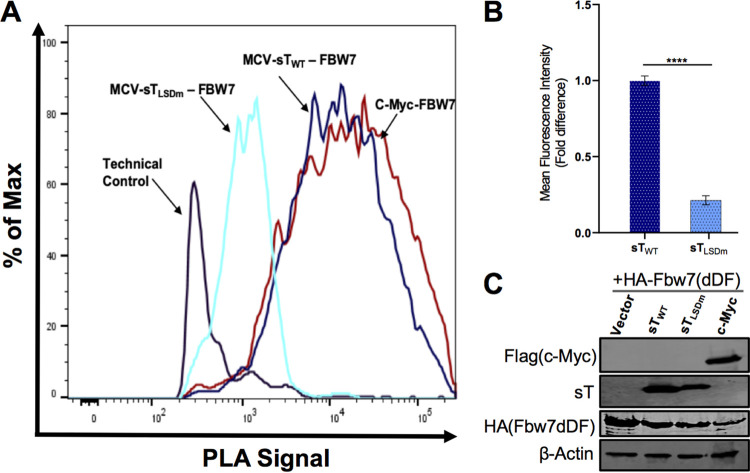
An essential requirement of metastasis involves the dissemination of tumor cells to various organs from the primary tumor (30). Multiple studies have demonstrated that loss of FBW7 promotes cell invasion and migration in numerous cancers through modulation of EMT-related cellular factors such as MMP-9 and Snail (31–33), which are transcriptionally upregulated upon MCV sT expression (Fig. 1). The MCV sT LSD region is known to bind and inhibit FBW7 (8, 10). As a result of this inhibition, FBW7 oncogenic substrates are stabilized in MCV sT-expressing cells, which may contribute to the MCV sT-induced migratory phenotype. Although MCV sT and FBW7 interaction has been characterized in vitro by coimmunoprecipitation (co-IP) in an overexpressing cell, the in vitro techniques do not identify whether this interaction occurs with endogenous proteins and, therefore, may not reflect the native behavior of their endogenous counterparts. In situ proximity ligation assay (PLA) can detect interactions with high specificity and sensitivity due to the coupling of antibody recognition and DNA amplification, which provides a technical advantage over other protein-protein interaction assays often plagued with long preparation times and extensive troubleshooting. For that reason, we utilized PLA combined with flow cytometry to revalidate this interaction in HEK293 cells (34, 35). As shown in Fig. 2, quantification of wild-type MCV sT (sTWT) interaction with FBW7 resulted in high-intensity PLA signal compared to that of our positive control, c-Myc, a well-known FBW7 substrate (36). This interaction was markedly diminished by expression of sTLSDm (Fig. 2A and B), an LSD alanine mutant of MCV sT (Fig. 2B), under comparable protein expression conditions (Fig. 2C), consistent with findings from previous reports (8, 11).
FIG 2.
MCV sT interacts with FBW7. (A) Validation of MCV sT and FBW7 interaction. To confirm the interaction of HA-FBW7 and MCV sT LSD domain, a PLA-flow cytometric analysis was carried out in HEK293 cells. Wild-type MCV sT displayed an interaction with FBW7 comparative to the positive control interaction of c-Myc with FBW7, while mutation of sT LSD greatly ablated sT interaction with FBW7, consistent with the finding from previous reports (8, 11). Primary antibodies were utilized at optimized concentrations with HA-tag (C29F4) (1:500), c-Myc (9E10) (1:500), and 2T2 (1:500). (B) Calculated fold difference of mean fluorescence intensity of PLA analyzed by FlowJo software. (C) Protein expression was evaluated by immunoblot analysis.
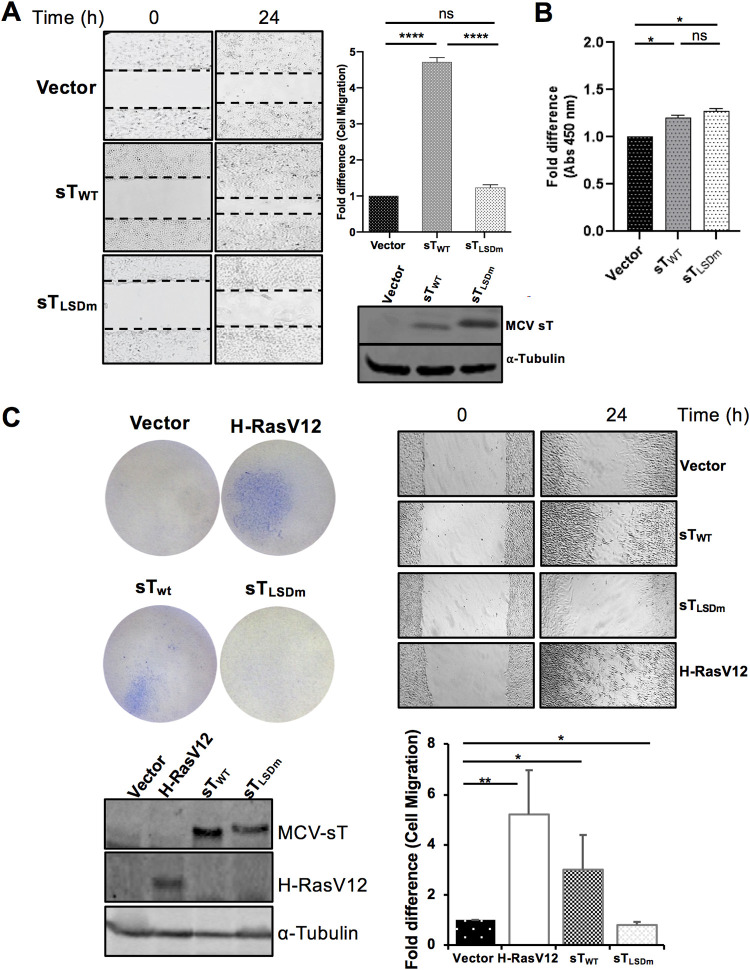
To determine whether MCV sT targeting of FBW7 contributes to the sT-induced migratory phenotype, a scratch assay was performed comparing empty vector control, MCV sTWT, and MCV sTLSDm in U2OS cells. Images of the scratch area were recorded at time point 0 and 24 h postscratch. Compared to empty vector negative control, MCV sT greatly enhanced the motility and migration of U2OS cells, consistent with previous studies (Fig. 3A) (25–27, 37). In contrast, MCV sTLSDm did not show a significant increase in cell migration. Over the 24-h period of the assay, we see no significant positive or negative effect on cell number confirmed by a viability assay, indicating that the resulting phenotype is specific to cell migration (Fig. 3B). Similarly, enhanced cell migration was readily detected with wild-type sT in NIH 3T3 mouse fibroblast (Fig. 3C) and MCC13 cells (4- to 5-fold increase; data not shown), while such a phenotype was not induced by sTLSDm. This suggests that sT targeting of FBW7 may be involved in the MCV sT-induced cell migratory phenotype.
FIG 3.
MCV sT induces cell motility in an LSD-dependent manner. (A) MCV sT-induced cell migration is LSD dependent. Scratch assay. Poly-l-lysine-coated 6-well plates were seeded with U2OS cells and transfected with either a vector control or sTWT or sTLSDm plasmids. Migration of cells toward the scratch was observed over a 24-h period, and images were taken using a Revolve 4 fluorescence microscope (Echo Laboratories). Scratch assays were performed in triplicate and measured using Fiji ImageJ analysis software. Differences between means (P value) were analyzed using a t test with GraphPad Prism software. Protein expression was detected by immunoblot analysis to validate successful transfection using 2T2 antibody for sT antigen and α-tubulin, respectively. (B) No significant differences in cell proliferation were observed between cells expressing MCV sT within 24 h, indicating that cell proliferation does not interfere with the measurement of sT-induced cell migration. (C) MCV sT promotes rodent fibroblast cell migration. NIH 3T3 cells stably expressing an empty vector, H-RasV12, MCV sTWT, and sTLSDm were trypsinized and 2 × 105 cells were used for transwell migration and scratch assay. H-RasV12 was used as a positive control. The experiments were performed two times, and the results were reproducible. The graph indicates the fold difference of migrated cells relative to the vector control sample. Protein expression was determined by immunoblotting.
MCV sT inhibition of FBW7 prevents turnover of MMP-9.
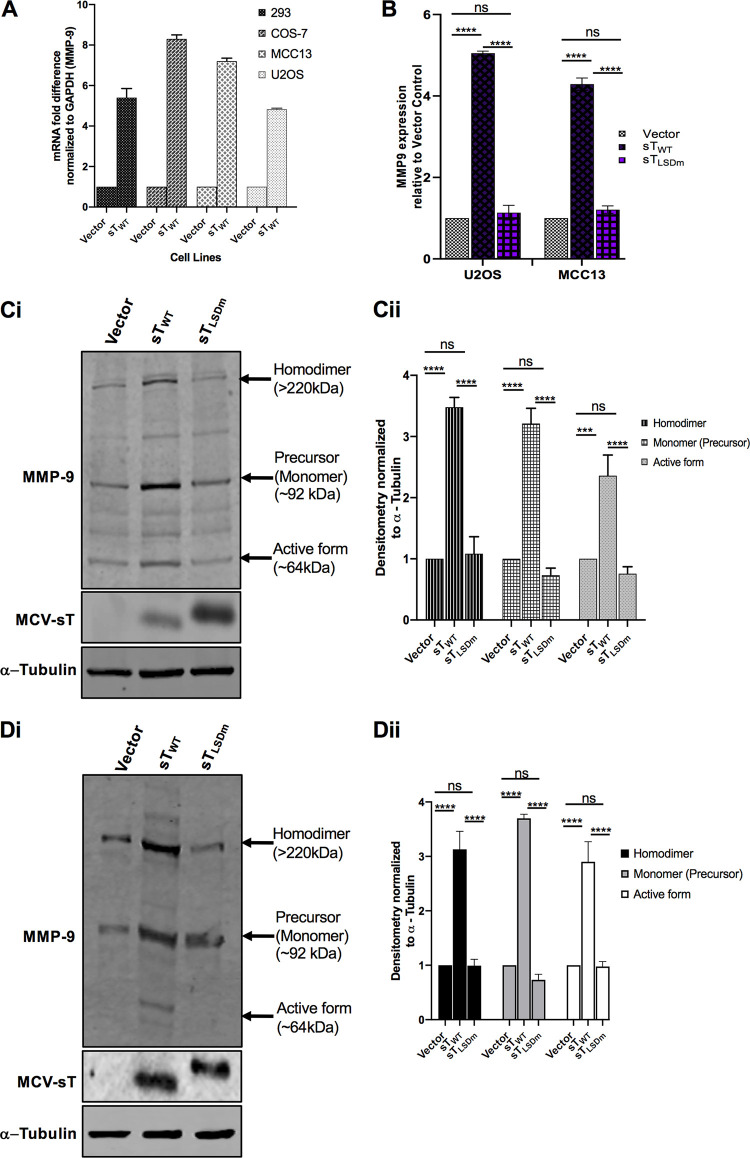
As shown in Fig. 1B, MCV sT induces MMP-9, an essential protein associated with the FBW7-EMT axis in human cancers (38). To confirm MCV sT induction of MMP-9 expression, a variety of cell lines, HEK293, COS-7, MCC13, and U2OS cells, were transfected with a vector control and MCV sTWT plasmids. MMP-9 gene expression is primarily regulated transcriptionally, resulting in low basal levels of these proteases in normal physiology (39). RT-qPCR results showed that MCV sT expression significantly increased MMP-9 transcript levels in all cell lines tested (Fig. 4A).
FIG 4.
MCV sT activates MMP-9. (A) MCV sT expression results in upregulation of MMP-9 mRNA levels. Various cell lines (HEK293, COS-7, MCC13, and U2OS) were transfected with either empty vector or MCV sTWT-expressing plasmids to measure MMP-9 mRNA levels. After 48 h, total RNA was isolated and analyzed by RT-qPCR. (B) MCV sT upregulates MMP-9 transcription through the LSD. U2OS and MCC13 cells transfected with empty vector control, MCV sTWT-, or MCV sTLSDm-expressing plasmids. Transcript levels of MMP-9 were analyzed using the comparative ΔΔCT method. (n = 3). Differences between means (P value) were analyzed using a t test with GraphPad Prism software. (C) MCV sT upregulates MMP-9 protein expression through the LSD. (i) U2OS cells were transfected with empty vector, MCV sTWT, or sTLSDm expression plasmids. After 48 h, immunoblot analysis was performed to analyze expression of MMP-9, sT, and α-tubulin. (ii) Densitometric analysis of immunoblots was carried out using the Image studio software (LI-COR Biosciences) and is shown as a fold change relative to the loading control, α-tubulin. Data analyzed using three biological replicates per experiment (n = 3). (D) MCV sT reproducibly activates MMP-9 expression in MCC13. (i) Quantitative immunoblot analysis was performed to analyze expression of MMP-9, sT, and α-tubulin. (ii) Densitometric analysis of immunoblots (n = 3).
We posited that MCV sT targeting of FBW7 plays a role in promoting the migratory potential of MCC by preventing MMP-9 protein turnover. Both transcriptional and posttranscriptional levels of MMP-9 were assessed in the presence of the MCV sTWT or MCV sTLSDm in U2OS and MCC13 cell lines (Fig. 4B). Results showed that MCV sT significantly induced the upregulation of MMP-9 transcripts when analyzed by RT-qPCR in both cell lines, which was not observed upon mutation of the MCV sT LSD (Fig. 4B). Additionally, we parallelly performed immunoblot analysis to evaluate the effect of sT on MMP-9 protein levels (Fig. 4C and D). Studies have shown that MMP-9 exists in several forms as follows: a monomeric proform (∼92 kDa), a disulfide-bonded homodimeric form (∼220 kDa), and multiple active forms (∼67 and 82 kDa) (40). The active and dimeric forms of MMP-9 play a role in the invasive and migratory phenotypes of cancer cells (41, 42). MCV sTWT expression induced the upregulation of the dimer, monomer, and active forms of MMP-9. However, mutation of the LSD prevented MCV sT-mediated upregulation of MMP-9, as expression levels remained comparable with those of the control, suggesting that MCV sT-mediated upregulation of MMP-9 is LSD dependent (Fig. 4).
MMP-9 inhibition impedes MCV sT-induced cell migration.
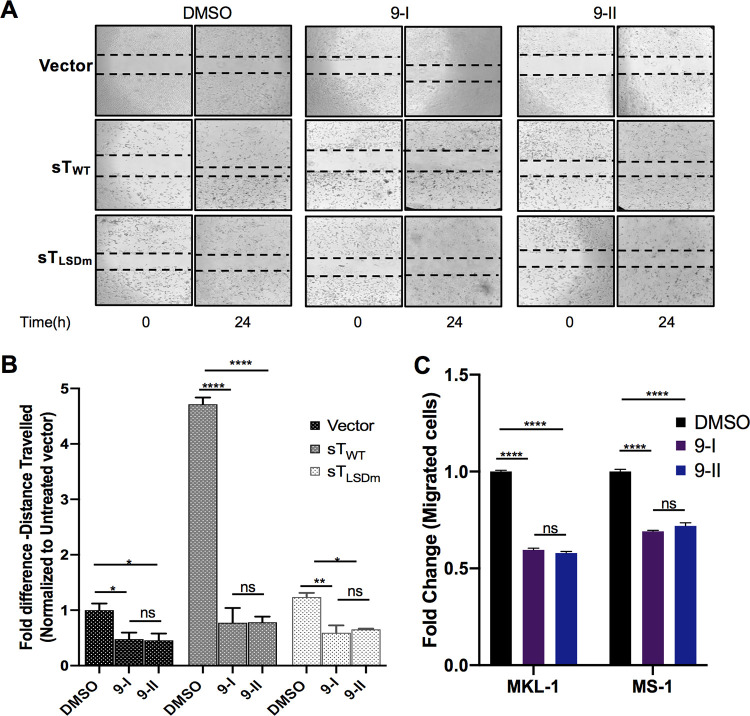
We next sought to determine if MMP-9 inhibition would have an impact on MCV sT-induced motile and migratory potentials. The migratory phenotype of U2OS cells transfected with vector control, sTWT, and sTLSDm was assessed using a scratch assay in the absence or presence of noncytotoxic concentrations of MMP9-I and MMP9-II inhibitors (Fig. 5A). MMP-9 inhibition resulted in a significant decrease in the distance traveled by MCV sT-expressing cells (Fig. 5A and B), confirming that MMP-9 is a critical migratory factor regulated by MCV sT. Incubation of both inhibitors showed a slight decrease in the motility of vector control cells, implying that any changes observed in migratory rates of MCV sT-expressing cells are not due to changes in cell viability or cytotoxicity. Both inhibitors showed a minor impact on the motility of sTLSDm-expressing cells, comparable to that of the vector control cells.
FIG 5.
MMP-9 inhibition impedes MCV sT-induced cell migration. (A) MCV sT promotes MMP-9-induced cell migration. Scratch assay. Poly-l-lysine-coated 6-well plates were seeded with U2OS cells and incubated with specific MMP-9 inhibitors at predetermined concentrations. Cells were transfected with a vector control, sTWT, or sTLSDm plasmids. After 48 h, a scratch was created, and migration of cells toward the scratch was observed over a 24-h period. The size of the wound was measured at 0 and 24 h and presented as the fold change in panel B. Scratch assays were performed in triplicate. (C) MMP-9 is required for MCC migration. MCV-positive MCC cell lines, MKL-1 and MS-1, were incubated with DMSO or the MMP-9 inhibitors as follows: 9-I (0.2 μM) and 9-II (2 μM) for MKL-1 and 9-I (0.1 μM) and 9-II (1 μM) for MS-1. Cells were then transferred into transwell inserts and allowed to migrate for 48 h. Migrated cells were measured using cell counting kit-8 (CCK-8). Data were analyzed using three biological replicates per experiment (n = 3). Differences between means (P value) were analyzed using a t test with GraphPad Prism software.
MMP-9 is essential for cell motility and migration in MCC.
To demonstrate that MMP-9 is vital for cell motility and migration in metastatic MCC, a transwell migration assay was performed using MCV-positive MCC cell lines. This assay quantified the migration ability of MCC cells toward a chemoattractant across a permeable chamber. Previous studies have shown that MCV sT promotes cell migration in MCV-positive MCC cells lines (25), and MCV-infected dermal cells promote MMP induction to support MCV infectivity (43). MCV-positive MCC cell lines, MKL-1 and MS-1 (Fig. 5C), were incubated in the absence or presence of the MMP9-I and MMP9-II inhibitors at nontoxic concentrations. After treatment, cells were allowed to migrate for 48 h, and the total numbers of migrated cells were measured by cell counting kit-8 (CCK-8) assay. Results showed that the migration of MCV-positive MCC cell lines was significantly reduced (∼40% to 50%) upon incubation of both MMP-9 inhibitors in comparison to that of the untreated control, suggesting that MMP-9 expression contributes to the migratory capacity of MCV-positive MCC (Fig. 5C). Together, these results indicate that MMP-9 is required for MCV sT-mediated cell migration enhancement in MCC.
MCV sT invasive phenotype is LSD dependent.
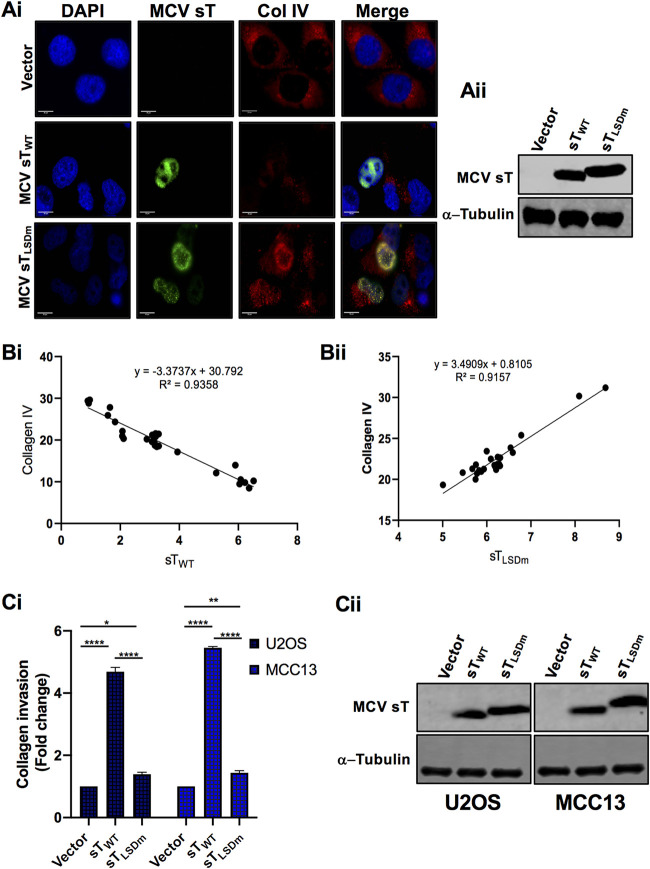
The invasiveness of epithelial cancers is a multistep process, and a key hallmark involves the degradation of the basement membrane. Type IV collagen is a major component in most basement membranes. Multiple studies have correlated overexpression of MMPs with not only an enhancement of cell migration and metastasis but also the invasiveness of cancer cells (17, 19). In particular, MMP-9 is a key protease associated with the degradation of ECM components, including type IV collagen and laminin, which in turn facilitates invasion of tumors into the circulatory system and promotes metastasis. To test if enzymatic activation of MMP-9 is regulated by sT LSD, we evaluated the effect of MCV sT on MMP-9 substrate collagen IV by immunofluorescence staining in U2OS cells. The cells were costained with collagen IV antibody along with T antigen antibody (2T2) to verify sT expression. Our results demonstrate that collagen IV expression in MCV sTWT-expressing cells is significantly reduced in comparison to that of vector control cells, potentially due to MMP-9 activation induced by MCV sT. In contrast, MCV sTLSDm-expressing cells did not show a decrease in collagen IV expression (Fig. 6A). Moreover, our regression analysis revealed that collagen IV expression levels are highly correlated with MCV sTWT or sTLSDm expression levels (Fig. 6B). To further validate the effect of MCV sT LSD on collagen IV degradation, we performed an invasion assay using collagen precoated inserts in U2OS and MCC13 cells. sTWT induced a 4- to 5-fold increase in collagen invasion compared to that of either vector control or sTLSDm (Fig. 6C), inferring that MCV sT induces not only cell migration but also cancer cell invasion through the LSD.
FIG 6.
MCV sT LSD induces collagen degradation. (A) MCV sT decreases collagen IV expression. U2OS cells were transfected with empty vector control, MCV sTWT, or MCV sTLSDm plasmids. (i) Cells were fixed at 48 h posttransfection, and endogenous collagen IV levels were measured by indirect immunofluorescence using a specific antibody. MCV sT expression was detected with 2T2 antigen antibody. Nuclear counterstain (4′,6-diamidino-2-phenylindole [DAPI]; blue), MCV sTWT and MCV sTLSDm (green), and collagen IV (red). (ii) Protein expression levels of wild-type and mutant MCV sT were detected by immunoblot analysis to validate successful transfection. Quantitative infrared fluorescence immunoblotting was performed using a 2T2 antibody for sT antigen and α-tubulin as an equal loading control. (B) MCV sT regulates collagen IV expression through the LSD. Mean fluorescence intensity of collagen IV in wild-type (i) and LSD mutant sT-expressing cells (ii) was analyzed using Fiji ImageJ software. The calculated values were plotted for regression analysis using Prism software. (C) MCV sT induces cell invasion through the LSD. (i) Collagen invasion assay. Serum-starved sT-expressing U2OS and MCC13 cells were seeded on the precoated collagen inserts and incubated for 72 h and then labeled with a cell-staining solution for 20 min. Upon extraction of the cell staining solution, absorbance at optical density at 560 nm (OD560) was measured. Data analyzed using three replicates per experiment; the experiments were performed two times. The results were reproducible, and differences between means (P value) were analyzed using a t test with GraphPad Prism software. (ii) Expression of MCV sT. Protein expression levels of wild-type and mutant MCV sT were detected by immunoblot analysis to validate successful transfection. Quantitative infrared fluorescence immunoblotting was performed using a 2T2 antibody for sT antigens and α-tubulin as an equal loading control.
MCV sT activates expression of EMT regulator, Snail.
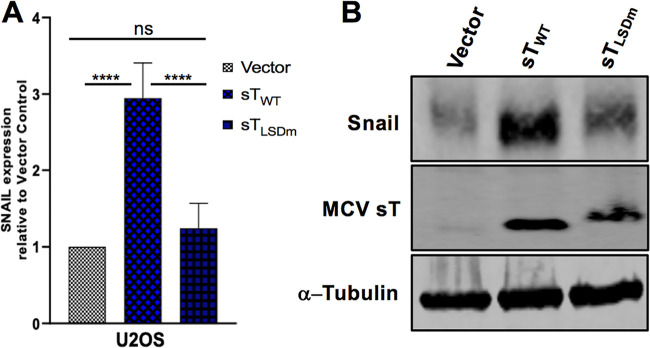
A positive regulatory loop has been identified between MMP-9 and Snail. Small interfering RNA (siRNA)-mediated inhibition of MMP‐9 significantly reduces expression of Snail, and conversely, knockdown of Snail, a transcription factor of MMP-9, suppresses expression of MMP‐9 (44). FBW7 abrogation of Snail protein also inhibits MMP-9 expression (45). Interestingly, MCV sT induced Snail expression in our initial transcriptional analysis (Fig. 1). Since both MMP-9 and Snail are vital mediators of EMT, we assessed the effect of sT LSD on transcriptional and protein levels of Snail in U2OS cells. RT-qPCR results showed that MCV sTWT upregulated mRNA levels of Snail, while this transcriptional change was not observed upon mutation of the LSD (Fig. 7A). Similar to the RT-qPCR data, our results demonstrated a significant increase in Snail protein levels upon MCV sT expression in an LSD-dependent manner (Fig. 7B).
FIG 7.
MCV sT LSD induces Snail expression. (A) MCV sT activates the transcription of Snail in an LSD-dependent manner. U2OS cells were transfected with empty vector control, sTWT-, or sTLSDm-expressing plasmids. Cellular RNA was extracted at 48 h posttransfection, and transcript levels were analyzed using the comparative ΔΔCT method (n = 3). (B) Snail protein expression is induced by MCV sT through the LSD. Immunoblot analysis was performed on the cellular lysates and analyzed using Snail-specific antibody. α-Tubulin was used as a measure of equal loading, and the 2T2 antibody was used to confirm MCV sT wild-type and mutant expression.
DISCUSSION
Metastasis is the endpoint of a series of biological processes by which a tumor cell detaches from the primary tumor and disseminates to a distant site through the circulatory system and establishes a secondary tumor (46). Oncogenic viruses often modulate the EMT axis via regulating E-cadherin repression (47–49), fibroblast growth factor (FGF) ligand modulation (49, 50), cadherin switching (51, 52), induction of transcription factors such as TWIST (53, 54) and Snail (47, 55), and MMP-9 upregulation (21–23). These cellular targets can regulate cancer cell migration and invasion; therefore, they could be exploited for therapeutic strategies in virus-induced metastatic cancers.
Multiple F-box proteins can function as tumor suppressors by negatively regulating oncoproteins, and various studies have focused on elucidating this mechanism in tumorigenesis and EMT progression (38). In this report, we show that inhibition of FBW7 by MCV sT promotes the upregulation of MMP-9 and contributes to MCV sT-mediated cell migration and invasion. Consistent with our data, Verhaegen et al. (9) showed activation of FBW7 substrates by MCV sT using their transgenic mouse model. The mechanism by which FBW7 regulates MMP-9 expression is currently unclear, although many studies have shown that MMP-9 expression is directly and indirectly regulated by FBW7 substrates, such as XBP1, Notch1, and Snail (16, 45, 56). Snail is known to be a substrate of both FBW7 (45) and β-TrCP (57), another major SCF E3 ligase that MCV sT targets through the LSD (10). As previously shown, Snail gene expression induces the loss of epithelial markers and the gain of mesenchymal markers, as well as promoting changes in cell motility and invasive properties (58). Our study initially focused on an MMP-9-specific metastatic progression induced by MCV sT due to limited availability of Snail inhibitors. However, the distressed proteome balance in EMT molecules induced by MCV sT might be triggered by Snail activation through MCV sT targeting multiple E3 ligases, which requires further investigation.
We recently described that the surface charge of the LSD is responsible for MCV sT targeting of the FBW7 WD40 substrate binding domain (11). The discrepancy regarding the interaction between FBW7 and MCV T antigens recently suggested by Dye et al. (59) could be explained by technical difficulties identifying E3 ligase interaction by IP (60, 61). IP for identification of E3 ligase interactions requires abundant bait protein for trapping and stringent washing conditions throughout the process. Kwun et al. (8) treated cells with MG132 proteasomal inhibitor, which allows >250 ng of bait detectable on the membrane by Ponceau staining after transferring proteins. After binding of bait to the resin, the resin needs to be washed several times using stringent lithium chloride (LiCl) or urea wash buffers. These detailed conditions were not reflected in their experimental setting. Moreover, their unconventional IP conditions are reflected by observed inconsistencies of LT interaction with FBW7, which appear to be antibody dependent (59).
To avoid the common technical artifacts observed in IP assays, we established the PLA-flow cytometric analysis. Using this method, we reproducibly revalidated our data on the interaction between FBW7 and T antigens (11). PLA combined with flow cytometry is a more advanced technique than conventional PLA protocol to test E3 ligase interactions. Classical PLA experiments require image processing; therefore, it does not have effective high-throughput advantages, and assessment can be limited to a few hundred cells per experiment. However, PLA combined with flow cytometry has a more simplified data acquisition, and therefore, throughput is significantly improved (35, 62).
It should be noted that our PLA experiments were performed in overexpression systems. Available commercial antibodies do not maintain the required specificity needed to accurately detect FBW7 endogenously. The efficacy of the antibody is important, as PLA requires oligonucleotide complementation of detection probes to the primary antibody, which is followed by a rolling circle polymerization to amplify the detection probes. Antibody specificity is important, as it hinders detection of false positives (63). Additionally, protein-protein interactions should reflect physiological function. FBW7 plays an important role in cell proliferation, and sT LSD has been shown to be responsible for transforming activity of MCV sT in many in vitro and in vivo studies (8–11). If sT LSD mutant still targets FBW7 as shown by Dye et al. (59), an enhancement in cell growth and transformation upon sT LSD mutant expression should be observed. Dye et al. also proposed that sT does not interact with FBW7 because sT does not contain a canonical phosphodegron motif. This is a narrow understanding of the dynamics in FBW7 protein interactions. Within the FBW7 WD40 domain β-propeller structure, multiple modalities of interaction have been observed at the top, the bottom, and the circumference of its surface (64). Therefore, it is likely that there are other binding pockets situated within the FBW7 WD40. sT-FBW7 interaction occurs in an allosteric capacity as an inhibitor, not as a substrate (11).
While the detailed regulatory mechanisms and specificity of sT function in transcription modulation remain unclear, studies have shown that MCV sT mediates cellular transcriptome/chromatin remodeling (29, 65), which may alter transcriptional activity and gene expression. Consistent with our data, Berrios et al. also reported that MCV sT downregulates extracellular matrix organization and cell adhesion molecules in their transcriptome analysis (29). We have demonstrated that MCV sT specifically activates both mRNA and protein levels of the EMT-related cellular proteins MMP-9 and Snail through the LSD; however, our results do not rule out the possibility that this effect is potentially modulated by multiple mechanisms in MCC. Nonetheless, it is clear that MCV sT LSD plays a critical role in regulating metastasis-initiating capacity in MCC and could be a potential target for therapeutic interventions.
MCV T antigens appear to be essential for cell survival among tumors infected with the virus. Because of the rare and aggressive nature of MCC and the lack of standard chemotherapy, it is difficult to prospectively predict outcomes following treatment of distant metastatic MCC. Since recent FDA approvals of avelumab and pembrolizumab represent the only approved treatment options for metastatic MCC (66, 67), it is necessary to evaluate the preclinical anticancer activity of efficient and economical chemotherapeutics through retrospective analysis for both MCV-negative and -positive MCC patients. MCV-negative MCC tumor patients are more likely to present with advanced disease than patients with virus-positive tumors (66.7% versus 48.3%) (68). Therefore, targeting the signaling pathways implicated in regulating tumor invasion could be an effective therapeutic protocol for both types of metastatic MCC treatment.
The underlying mechanism for the high propensity of MCC tumors to metastasize is yet to be elucidated. Our study is the first approach to investigate the therapeutic potential of matrix metalloproteinase in MCC. MCV sT specifically activates the EMT-related cellular proteins MMP-9 through the LSD, which we targeted by commercially available inhibitors, and revealed a potential secondary treatment for distant metastatic MCC.
MATERIALS AND METHODS
Cells.
HEK293, U2OS, and COS-7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) (Seradigm). MCC13, MS-1, and MKL-1 cell lines were maintained in RPMI 1640 medium supplemented with 10% FBS (Seradigm). NIH 3T3 cells were maintained in DMEM with 10% bovine calf serum (Seradigm).
Plasmids, transfection, and transduction.
Plasmids for vector control and codon-optimized cDNA constructs for sTwt, sTLSDm (8), HA-FBW7, and Flag-cMyc (36) plasmids were previously described. FBW7ΔDF(d231-324) was generated by overlapping PCR (11). For sT protein expression, cells were transfected using Lipofectamine 3000 (Invitrogen) or jetOptimus (Polyplus-transfection) according to the manufacturer’s protocol. For lentiviral transduction, codon-optimized cDNAs for MCV sTWT, MCV sTLSDm, (8), and H-RasV12 were inserted into pLVX empty vector using primers (5′-GCA GCG CTA TGA CGG AAT ATA AGC TGG TG-3′ and 5′-CCT GGA TCC TCA GGA GAG CAC ACA CTT GCA-3′). For lentivirus production, 293FT (Invitrogen) cells were used for induction according to the manufacturer’s instructions. Cells were selected with puromycin (3 μg/ml) after infection for 1 week.
Reverse transcription-quantitative PCR.
HEK293, COS-7, U2OS, and MCC13 cell lines are used. RNA was extracted using Monarch total RNA miniprep kit (New England Biolabs) as per the manufacturer’s instruction. A total of 250 ng of RNA was used as a template in each reaction with iTaq Universal one-step RT-qPCR kit (Bio-Rad) or Luna universal one-step RT-qPCR kit (New England Biolabs). Quantitative analysis was performed using the comparative ΔΔCT method. All experiments with SYBR green included a melting curve analysis to confirm the specificity of the amplicons (95°C for 15 s, 60°C for 20 s, and 95°C for 15 s). For comparison of RNA transcription levels between samples, results from the RT-qPCR experiments were normalized to those for two reference genes, the genes for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (5′-CCT CCC GCT TCG CTC TCT-3′, 5′-CTG GCG ACG CAA AAG AAG A-3′) and RNase P (5′-GCG GAG GGA AGC TCA TCA G-3′, 5′-CTG GCC CTA GTC TCA GAC CTT-3′). The following primers are used for the RT-qPCR: MMP-9 (5′-TTT GAG TCC GGT GGA CGA TG-3′, 5′-GCT CCT CAA AGA CCG AGT C-3′) and Snail (5′-GCG AGC TGC AGG ACT CTA AT-3′, 5′-GGA CAG AGT CCC AGA TGA C-3′).
Quantitative immunoblotting and antibodies.
Cells (HEK293, U2OS, MCC13) were lysed in IP buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM benzamidine), and sonicated whole cell lysates were used for direct immunoblotting. Primary antibodies were incubated overnight at 4°C, followed by 1 h secondary antibody incubation at room temperature (RT). All signals were detected using quantitative infrared (IR) secondary antibodies (IRDye 800CW goat anti-mouse, 800CW goat anti-rabbit, 680LT goat anti-rabbit IgG, 680LT goat anti-mouse IgG; LI-COR). Signal intensities were analyzed using a laser-scanning imaging system, Odyssey CLx (LI-COR). Primary antibodies used for this study are Pan Ras (F132), MMP9 (ab26132), Anti-MCPyV T-antigen antibody (2T2), Snail (C15D3), α-tubulin (12G10), collagen IV (ab6586), c-Myc (9E10), hemagglutinin (HA)-tag (C29F4), Anti-Flag (M2), and β-actin (13E5). Protein levels were quantitated and normalized by control, α-tubulin, or β-actin using an Odyssey LI-COR IR imaging system.
SILAC data analysis.
The previously published SILAC-based quantitative proteomic data set analyzed host cell (i293-sT) proteome changes upon MCV sT expression (25) and further interrogated using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (51). For quantitative analysis, a 2.0-fold cutoff was chosen as a basis for investigating potential proteome changes (50).
Proximity ligation assay flow cytometry.
PLA was performed using a Duolink assay kit (Sigma-Aldrich) according to the manufacturer's instructions. To evaluate MCV sT and FBW7 interaction, HA-FBW7ΔDF(d231-324) (11) was coexpressed with sTWT or sTLSDm in HEK293 cells. FBW7ΔDF was also coexpressed with c-Myc, a known FBW7 substrate, as a positive control (36). Primary antibodies were utilized at optimized concentrations with HA-tag (C29F4) (1:500), c-Myc (9E10) (1:500), and Anti-MCPyV T-antigen antibody (2T2) (1:500) (Millipore). Cells were analyzed by flow cytometry on a 16-color BD LSRFortessa. The acquired data were analyzed using FlowJo software (TreeStar, Ashland, OR, USA).
Scratch wound-healing assay.
U2OS cells were seeded into the poly-l-lysine-coated 6-well plates and transfected with either empty vector or sTWT or sTLSDm plasmids. Because MCV sT promotes serum-independent cell growth (69), a serum starvation condition was not considered for our scratch assay to exclude cell proliferation effect by sT. After 48 h, a scratch was created by scraping the monolayer using a p1000 pipette tip. The migration of cells toward the scratch was observed over a 24-h period, and images were taken every 8 h under a Revolve 4 fluorescence microscope (Echo Laboratories). Inhibitor-based scratch assays were incubated for 24 h prior to transfection with 0.1 and 1 μM 9-I and 9-II inhibitors, respectively.
Transwell cell migration assay.
Cells grown in DMEM with 10% FBS were trypsinized and resuspended in DMEM. A total of 1 × 105 cells was gently added to the transwell insert (8 μm; Greiner Bio-One). DMEM with 10% FBS was added to the bottom of the lower chamber (24-well plate). The cells were incubated in the culture incubator at 37°C plus 5% CO2 for the indicated time. The cells migrated from the insert to the well through the filter. The filter was fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min and then stained with 1% crystal violet in 2% ethanol for 20 min for NIH 3T3 cells, and MCC13, MKL-1, and MS-1 cells were counted using a cell counting kit-8 (CCK-8) (Sigma-Aldrich). The stained cells on the lower side were counted under a microscope from 5 different randomly selected views. All conditions were the same for assays performed in triplicate.
Immunofluorescence.
U2OS cells grown on glass coverslips were transfected with empty vector or sT wild-type or LSD mutant expression constructs. After 48 h, cells were fixed in 1:1 methanol/acetone at −20°C, permeabilized, and blocked in PBS with 5% bovine serum albumin (BSA) and 0.3 M glycine for 1 h. Cells were labeled with the appropriate primary antibodies and then incubated with the appropriate conjugated secondary antibodies (anti-mouse Alexa Fluor 488 [A11029], anti-rabbit Alexa Fluor 568 [A11036]). Cells were analyzed with a Revolve 4 fluorescence microscope (Echo Laboratories).
Collagen invasion assay.
U2OS and MCC13 cells were transfected with wild-type and LSD mutant sT constructs for 48 h, followed by overnight serum starvation. A total of 1 × 106 cells resuspending in serum-free medium in each condition was seeded in a 24-well cell invasion plate containing polymerized collagen-coated membrane inserts. The collagen inserts had a pore size of 8 μm (Chemicon QCM collagen cell invasion assay; ECM551). Complete medium was used as a chemoattractant in the lower chamber, and cells were left to incubate for 72 h. Cells/medium were carefully aspirated by pipetting any residual suspension in the transwell insert. Inserts were transferred to a clean well and were carefully stained with 400 μl cell staining solution at room temperature for 20 min, followed by a gentle wash in deionized water. While slightly damp, unattached cells were removed cautiously by cotton swabs from the collagen inserts and allowed to dry at room temperature for 15 min. Dried inserts were transferred to clean wells containing 200 μl of extraction buffer and incubated for 15 min at room temperature. Following the extraction incubation, 100 μl of the extraction solution was pipetted into 96-well plates, and optical density (OD) was measured at 560 nm.
Cell proliferation assay.
U2OS transfected cells (vector control, MCV sTWT and MCV sTLSDm plasmids) were seeded in 96-well plates (1 × 104 cells/well) 48 h posttransfection. Cell proliferation was monitored using a WST-8 based assay cell counting kit-8 (CCK-8) according to the manufacturer’s protocol. OD values were divided by the OD value of day 0 for normalization.
Chemical inhibitors.
MMP-9 inhibitors I and II (EMD Millipore) were used at 0.1 to 0.2 μM and 1 to 2 μM, respectively. Cell toxicity was measured using a cell counting kit-8 (CCK-8) (Sigma-Aldrich) according to the manufacturer’s protocol.
Statistical analysis.
Statistical significance between two groups was determined using one- or two-tailed Student’s t tests in GraphPad Prism (GraphPad Software, Inc., La Jolla, CA, USA). The difference was considered significant when P was <0.05 for multiple testing. The symbols *, **, ***, and **** indicate P values of <0.01, 0.005, 0.001, and 0.0001, respectively.
ACKNOWLEDGMENTS
We thank Patrick S. Moore and Yuan Chang for the kind sharing of MCV-related reagents.
H.J.K. was supported in part by an Institutional Research Grant, IRG-17-175-04, from the American Cancer Society and by the Pennsylvania Department of Health Tobacco CURE Funds. N.N. was supported by training grant T32 CA060395 from the National Cancer Institute, National Institutes of Health.
We declare no competing interests.
REFERENCES
- 1.Becker JC, Stang A, DeCaprio JA, Cerroni L, Lebbé C, Veness M, Nghiem P. 2017. Merkel cell carcinoma. Nat Rev Dis Primers 3:17077. doi: 10.1038/nrdp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller RW, Rabkin CS. 1999. Merkel cell carcinoma and melanoma: etiological similarities and differences. Cancer Epidemiol Biomarkers Prev 8:153–158. [PubMed] [Google Scholar]
- 3.Chang Y, Moore PS. 2012. Merkel cell carcinoma: a virus-induced human cancer. Annu Rev Pathol 7:123–144. doi: 10.1146/annurev-pathol-011110-130227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goedert JJ, Rockville Merkel Cell Carcinoma Group. 2009. Merkel cell carcinoma: recent progress and current priorities on etiology, pathogenesis, and clinical management. J Clin Oncol 27:4021–4026. doi: 10.1200/JCO.2009.22.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shuda M, Feng HC, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. 2008. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci U S A 105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houben R, Shuda M, Weinkam R, Schrama D, Feng HC, Chang YA, Moore PS, Becker JC. 2010. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol 84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwun HJ, Shuda M, Feng H, Camacho CJ, Moore PS, Chang Y. 2013. Merkel cell polyomavirus small T antigen controls viral replication and oncoprotein expression by targeting the cellular ubiquitin ligase SCFFbw7. Cell Host Microbe 14:125–135. doi: 10.1016/j.chom.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verhaegen ME, Mangelberger D, Harms PW, Vozheiko TD, Weick JW, Wilbert DM, Saunders TL, Ermilov AN, Bichakjian CK, Johnson TM, Imperiale MJ, Dlugosz AA. 2015. Merkel cell polyomavirus small T antigen is oncogenic in transgenic mice. J Invest Dermatol 135:1415–1424. doi: 10.1038/jid.2014.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwun HJ, Wendzicki JA, Shuda Y, Moore PS, Chang Y. 2017. Merkel cell polyomavirus small T antigen induces genome instability by E3 ubiquitin ligase targeting. Oncogene 36:6784–6792. doi: 10.1038/onc.2017.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nwogu N, Ortiz LE, Kwun HJ. 2020. Surface charge of Merkel cell polyomavirus small T antigen determines cell transformation through allosteric FBW7 WD40 domain targeting. Oncogenesis 9:53. doi: 10.1038/s41389-020-0235-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ang XL, Wade Harper J. 2005. SCF-mediated protein degradation and cell cycle control. Oncogene 24:2860–2870. doi: 10.1038/sj.onc.1208614. [DOI] [PubMed] [Google Scholar]
- 13.Cai YK, Zhang M, Qiu XF, Wang BW, Fu Y, Zeng J, Bai J, Yang GS. 2017. Upregulation of FBXW7 suppresses renal cancer metastasis and epithelial mesenchymal transition. Dis Markers 2017:8276939. doi: 10.1155/2017/8276939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He H, Dai J, Xu Z, He W, Wang X, Zhu Y, Wang H. 2018. Fbxw7 regulates renal cell carcinoma migration and invasion via suppression of the epithelial-mesenchymal transition. Oncol Lett 15:3694–3702. doi: 10.3892/ol.2018.7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calcagno DQ, Freitas VM, Leal MF, De Souza CRT, Demachki S, Montenegro R, Assumpcao PP, Khayat AS, Smith MDC, dos Santos A, Burbano RR. 2013. MYC, FBXW7 and TP53 copy number variation and expression in gastric cancer. BMC Gastroenterol 13:141. doi: 10.1186/1471-230X-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Zhang J, Zhou L, Sun W, Zheng ZG, Lu P, Gao Y, Yang XS, Zhang ZC, Tao KS, Dou KF. 2015. Fbxw7 regulates hepatocellular carcinoma migration and invasion via Notch1 signaling pathway. Int J Oncol 47:231–243. doi: 10.3892/ijo.2015.2981. [DOI] [PubMed] [Google Scholar]
- 17.Gialeli C, Theocharis AD, Karamanos NK. 2011. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J 278:16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- 18.Nagase H, Woessner JF. 1999. Matrix metalloproteinases. J Biol Chem 274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 19.Huang H. 2018. Matrix metalloproteinase-9 (MMP-9) as a cancer biomarker and MMP-9 biosensors: recent advances. Sensors 18:3249. doi: 10.3390/s18103249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilhelm SM, Collier IE, Marmer BL, Eisen AZ, Grant GA, Goldberg GI. 1989. SV40-transformed human lung fibroblasts secrete a 92-kDa type IV collagenase which is identical to that secreted by normal human macrophages. J Biol Chem 264:17213–17221. [PubMed] [Google Scholar]
- 21.Chung TW, Lee YC, Kim CH. 2004. Hepatitis B viral HBx induces matrix metalloproteinase-9 gene expression through activation of ERK and PI-3K/AKT pathways: involvement of invasive potential. FASEB J 18:1123–1125. doi: 10.1096/fj.03-1429fje. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Wakisaka N, Tomlinson CC, DeWire SM, Krall S, Pagano JS, Damania B. 2004. The Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) K1 protein induces expression of angiogenic and invasion factors. Cancer Res 64:2774–2781. doi: 10.1158/0008-5472.can-03-3653. [DOI] [PubMed] [Google Scholar]
- 23.Yoshizaki T, Sato H, Furukawa M, Pagano JS. 1998. The expression of matrix metalloproteinase 9 is enhanced by Epstein-Barr virus latent membrane protein 1. Proc Natl Acad Sci U S A 95:3621–3626. doi: 10.1073/pnas.95.7.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Knight LM, Stakaityte G, Wood JJ, Abdul-Sada H, Griffiths DA, Howell GJ, Wheat R, Blair GE, Steven NM, Macdonald A, Blackbourn DJ, Whitehouse A. 2015. Merkel cell polyomavirus small T antigen mediates microtubule destabilization to promote cell motility and migration. J Virol 89:35–47. doi: 10.1128/JVI.02317-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stakaitytė G, Nwogu N, Dobson SJ, Knight LM, Wasson CW, Salguero FJ, Blackbourn DJ, Blair GE, Mankouri J, Macdonald A, Whitehouse A. 2018. Merkel cell polyomavirus small T antigen drives cell motility via rho-GTPase-induced filopodium formation. J Virol 92:e00940-17. doi: 10.1128/JVI.00940-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nwogu N, Boyne JR, Dobson SJ, Poterlowicz K, Blair GE, Macdonald A, Mankouri J, Whitehouse A. 2018. Cellular sheddases are induced by Merkel cell polyomavirus small tumour antigen to mediate cell dissociation and invasiveness. PLoS Pathog 14:e1007276. doi: 10.1371/journal.ppat.1007276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horejs CM. 2016. Basement membrane fragments in the context of the epithelial-to-mesenchymal transition. Eur J Cell Biol 95:427–440. doi: 10.1016/j.ejcb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Berrios C, Padi M, Keibler MA, Park DE, Molla V, Cheng JW, Lee SM, Stephanopoulos G, Quackenbush J, DeCaprio JA. 2016. Merkel cell polyomavirus small T antigen promotes pro-glycolytic metabolic perturbations required for transformation. PLoS Pathog 12:e1006020. doi: 10.1371/journal.ppat.1006020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bravo-Cordero JJ, Hodgson L, Condeelis J. 2012. Directed cell invasion and migration during metastasis. Curr Opin Cell Biol 24:277–283. doi: 10.1016/j.ceb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sailo BL, Banik K, Girisa S, Bordoloi D, Fan L, Halim CE, Wang H, Kumar AP, Zheng DL, Mao XL, Sethi G, Kunnumakkara AB. 2019. FBXW7 in cancer: what has been unraveled thus far? Cancers 11:246. doi: 10.3390/cancers11020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Zhang X, Ye M, Jing P, Xiong J, Han Z, Kong J, Li M, Lai X, Chang N, Zhang J, Zhang J. 2018. FBW7 loss promotes epithelial-to-mesenchymal transition in non-small cell lung cancer through the stabilization of Snail protein. Cancer Lett 419:75–83. doi: 10.1016/j.canlet.2018.01.047. [DOI] [PubMed] [Google Scholar]
- 33.Lamouille S, Xu J, Derynck R. 2014. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu XZ, Zelmer A, Wellmann S. 2017. Visualization of protein-protein interaction in nuclear and cytoplasmic fractions by co-immunoprecipitation and in situ proximity ligation assay. J Vis Exp 119:55218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersen SS, Hvid M, Pedersen FS, Deleuran B. 2013. Proximity ligation assay combined with flow cytometry is a powerful tool for the detection of cytokine receptor dimerization. Cytokine 64:54–57. doi: 10.1016/j.cyto.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 36.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. 2004. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J 23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stakaitytė G, Nwogu N, Lippiat JD, Blair GE, Poterlowicz K, Boyne JR, Macdonald A, Mankouri J, Whitehouse A. 2018. The cellular chloride channels CLIC1 and CLIC4 contribute to virus-mediated cell motility. J Biol Chem 293:4582–4590. doi: 10.1074/jbc.RA117.001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song YZ, Lin M, Liu Y, Wang ZW, Zhu XQ. 2019. Emerging role of F-box proteins in the regulation of epithelial-mesenchymal transition and stem cells in human cancers. Stem Cell Res Ther 10:124. doi: 10.1186/s13287-019-1222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fanjul-Fernández M, Folgueras AR, Cabrera S, López-Otín C. 2010. Matrix metalloproteinases: Evolution, gene regulation and functional analysis in mouse models. Biochim Biophys Acta 1803:3–19. doi: 10.1016/j.bbamcr.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Olson MW, Bernardo MM, Pietila M, Gervasi DC, Toth M, Kotra LP, Massova I, Mobashery S, Fridman R. 2000. Characterization of the monomeric and dimeric forms of latent and active matrix metalloproteinase-9: differential rates for activation by stromelysin 1. J Biol Chem 275:2661–2668. doi: 10.1074/jbc.275.4.2661. [DOI] [PubMed] [Google Scholar]
- 41.Dufour A, Zucker S, Sampson NS, Kuscu C, Cao JA. 2010. Role of matrix metalloproteinase-9 dimers in cell migration: design of inhibitory peptides. J Biol Chem 285:35944–35956. doi: 10.1074/jbc.M109.091769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roomi MW, Kalinovsky T, Rath M, Niedzwiecki A. 2014. Effect of a nutrient mixture on matrix metalloproteinase-9 dimers in various human cancer cell lines. Int J Oncol 44:986–992. doi: 10.3892/ijo.2013.2235. [DOI] [PubMed] [Google Scholar]
- 43.Liu W, Yang RF, Payne AS, Schowalter RM, Spurgeon ME, Lambert PF, Xu XW, Buck CB, You JX. 2016. Identifying the target cells and mechanisms of Merkel cell polyomavirus infection. Cell Host Microbe 19:775–787. doi: 10.1016/j.chom.2016.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin CY, Tsai PH, Kandaswami CC, Lee PP, Huang CJ, Hwang JJ, Lee MT. 2011. Matrix metalloproteinase-9 cooperates with transcription factor Snail to induce epithelial-mesenchymal transition. Cancer Sci 102:815–827. doi: 10.1111/j.1349-7006.2011.01861.x. [DOI] [PubMed] [Google Scholar]
- 45.Fu Q, Lu Z, Fu X, Ma S, Lu X. 2019. MicroRNA 27b promotes cardiac fibrosis by targeting the FBW7/Snail pathway. Aging (Albany NY) 11:11865–11879. doi: 10.18632/aging.102465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valastyan S, Weinberg RA. 2011. Tumor metastasis: molecular insights and evolving paradigms. Cell 147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris M, Laverick L, Wei W, Davis A, O’Neill S, Wood L, Wright J, Dawson C, Young L. 2018. The EBV-encoded oncoprotein, LMP1, induces an epithelial-to-mesenchymal transition (EMT) via Its CTAR1 domain through integrin-mediated ERK-MAPK signalling. Cancers 10:130. doi: 10.3390/cancers10050130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Lian Z, Han S, Waye MMY, Wang H, Wu MC, Wu K, Ding J, Arbuthnot P, Kew M, Fan D, Feitelson MA. 2006. Downregulation of E-cadherin by hepatitis B virus X antigen in hepatocellullar carcinoma. Oncogene 25:1008–1017. doi: 10.1038/sj.onc.1209138. [DOI] [PubMed] [Google Scholar]
- 49.Cheng YM, Chou CY, Hsu YC, Chen MJ, Wing L. 2012. The role of human papillomavirus type 16 E6/E7 oncoproteins in cervical epithelial-mesenchymal transition and carcinogenesis. Oncol Lett 3:667–671. doi: 10.3892/ol.2011.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wakisaka N, Murono S, Yoshizaki T, Furukawa M, Pagano JS. 2002. Epstein-barr virus latent membrane protein 1 induces and causes release of fibroblast growth factor-2. Cancer Res 62:6337–6344. [PubMed] [Google Scholar]
- 51.Shair KHY, Schnegg CI, Raab-Traub N. 2009. Epstein-Barr virus latent membrane protein-1 effects on junctional plakoglobin and induction of a cadherin switch. Cancer Res 69:5734–5742. doi: 10.1158/0008-5472.CAN-09-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu DX, Zhou JS, Wang FF, Shi HY, Li Y, Li BH. 2015. HPV-16 E6/E7 promotes cell migration and invasion in cervical cancer via regulating cadherin switch in vitro and in vivo. Arch Gynecol Obstet 292:1345–1354. doi: 10.1007/s00404-015-3787-x. [DOI] [PubMed] [Google Scholar]
- 53.Horikawa T, Yang J, Kondo S, Yoshizaki T, Joab I, Furukawa N, Pagano JS. 2007. Twist and epithelial-mesenchymal transition are induced by the EBV oncoprotein latent membrane protein 1 and are associated with metastatic nasopharyngeal carcinoma. Cancer Res 67:1970–1978. doi: 10.1158/0008-5472.CAN-06-3933. [DOI] [PubMed] [Google Scholar]
- 54.Chen X, Bode AM, Dong ZG, Cao Y. 2016. The epithelial-mesenchymal transition (EMT) is regulated by oncoviruses in cancer. FASEB J 30:3001–3010. doi: 10.1096/fj.201600388R. [DOI] [PubMed] [Google Scholar]
- 55.Horikawa T, Yoshizaki T, Kondo S, Furukawa M, Kaizaki Y, Pagano JS. 2011. Epstein-Barr virus latent membrane protein 1 induces Snail and epithelial-mesenchymal transition in metastatic nasopharyngeal carcinoma. Br J Cancer 104:1160–1167. doi: 10.1038/bjc.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chae U, Lee H, Kim B, Jung H, Kim BM, Lee AH, Lee DS, Min SH. 2019. A negative feedback loop between XBP1 and Fbw7 regulates cancer development. Oncogenesis 8:12. doi: 10.1038/s41389-019-0124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou BHP, Deng J, Xia WY, Xu JH, Li YM, Gunduz M, Hung MC. 2004. Dual regulation of Snail by GSK-3 beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol 6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 58.Przybylo JA, Radisky DC. 2007. Matrix metalloproteinase-induced epithelial-mesenchymal transition: tumor progression at Snail's pace. Int J Biochem Cell Biol 39:1082–1088. doi: 10.1016/j.biocel.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 59.Dye KN, Welcker M, Clurman BE, Roman A, Galloway DA. 2019. Merkel cell polyomavirus tumor antigens expressed in Merkel cell carcinoma function independently of the ubiquitin ligases Fbw7 and β-TrCP. PLoS Pathog 15:e1007543. doi: 10.1371/journal.ppat.1007543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iconomou M, Saunders DN. 2016. Systematic approaches to identify E3 ligase substrates. Biochem J 473:4083–4101. doi: 10.1042/BCJ20160719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Connor HF, Huibregtse JM. 2017. Enzyme-substrate relationships in the ubiquitin system: approaches for identifying substrates of ubiquitin ligases. Cell Mol Life Sci 74:3363–3375. doi: 10.1007/s00018-017-2529-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leuchowius KJ, Weibrecht I, Landegren U, Gedda L, Soderberg O. 2009. Flow cytometric in situ proximity ligation analyses of protein interactions and post-translational modification of the epidermal growth factor receptor family. Cytometry A 75:833–839. doi: 10.1002/cyto.a.20771. [DOI] [PubMed] [Google Scholar]
- 63.Ristic M, Brockly F, Piechaczyk M, Bossis G. 2016. Detection of protein-protein interactions and posttranslational modifications using the proximity ligation assay: application to the study of the SUMO pathway. Methods Mol Biol 1449:279–290. doi: 10.1007/978-1-4939-3756-1_17. [DOI] [PubMed] [Google Scholar]
- 64.Stirnimann CU, Petsalaki E, Russell RB, Müller CW. 2010. WD40 proteins propel cellular networks. Trends Biochem Sci 35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 65.Cheng JW, Park DE, Berrios C, White EA, Arora R, Yoon R, Branigan T, Xiao TF, Westerling T, Federation A, Zeid R, Strober B, Swanson SK, Florens L, Bradner JE, Brown M, Howley PM, Padi M, Washburn MP, DeCaprio JA. 2017. Merkel cell polyomavirus recruits MYCL to the EP400 complex to promote oncogenesis. PLoS Pathog 13:e1006668. doi: 10.1371/journal.ppat.1006668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joseph J, Zobniw C, Davis J, Anderson J, Trinh V. 2018. Avelumab: a review of its application in metastatic Merkel cell carcinoma. Ann Pharmacother 52:928–935. doi: 10.1177/1060028018768809. [DOI] [PubMed] [Google Scholar]
- 67.Shirley M. 2018. Avelumab: a review in metastatic Merkel cell carcinoma. Target Oncol 13:409–416. doi: 10.1007/s11523-018-0571-4. [DOI] [PubMed] [Google Scholar]
- 68.Moshiri AS, Doumani R, Yelistratova L, Blom A, Lachance K, Shinohara MM, Delaney M, Chang O, McArdle S, Thomas H, Asgari MM, Huang ML, Schwartz SM, Nghiem P. 2017. Polyomavirus-negative Merkel cell carcinoma: a more aggressive subtype based on analysis of 282 cases using multimodal tumor virus detection. J Invest Dermatol 137:819–827. doi: 10.1016/j.jid.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shuda M, Kwun HJ, Feng HC, Chang Y, Moore PS. 2011. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest 121:3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]