Understanding the mechanism of HIV-1 resistance to anti-HIV-1 drugs could lead to the development of novel drugs with increased efficiency, resulting in more effective ART. ART composed of more potent and long-acting anti-HIV-1 drugs can greatly improve drug adherence and also provide HIV-1 prevention such as preexposure prophylaxis. NCINIs with a multimodal mode of action exert potent anti-HIV-1 effects through IN overmultimerization during HIV-1 maturation. However, HIV-1 can acquire some mutations that cause IN undermultimerization to alleviate NCINI-induced IN overmultimerization. This undermultimerized IN was efficiently stabilized by HIV-1 RNA and restored to the same level as that of wild-type HIV-1. Our findings revealed that HIV-1 eventually acquires such a conformational escape reaction to overcome the unique NCINI actions. The investigation into drug-resistant mutations associated with HIV-1 protein multimerization may facilitate the elucidation of its molecular mechanism and functional multimerization, allowing us to develop more potent anti-HIV-1 drugs and unique treatment strategies.
KEYWORDS: HIV-1 RNA, HIV-1 integrase, HIV-1 maturation, NCINIs
ABSTRACT
HIV-1 often acquires drug-resistant mutations in spite of the benefits of antiretroviral therapy (ART). HIV-1 integrase (IN) is essential for the concerted integration of HIV-1 DNA into the host genome. IN further contributes to HIV-1 RNA binding, which is required for HIV-1 maturation. Non-catalytic-site integrase inhibitors (NCINIs) have been developed as allosteric IN inhibitors, which perform anti-HIV-1 activity by a multimodal mode of action such as inhibition of the IN-lens epithelium-derived growth factor (LEDGF)/p75 interaction in the early stage and disruption of functional IN multimerization in the late stage of HIV-1 replication. Here, we show that IN undergoes an adaptable conformational change to escape from NCINIs. We observed that NCINI-resistant HIV-1 variants have accumulated 4 amino acid mutations by passage 26 (P26) in the IN-encoding region. We employed high-performance liquid chromatography (HPLC), thermal stability assays, and X-ray crystallographic analysis to show that some amino acid mutations affect the stability and/or dimerization interface of the IN catalytic core domains (CCDs), potentially resulting in the severely decreased multimerization of full-length IN proteins (IN undermultimerization). This undermultimerized IN via NCINI-related mutations was stabilized by HIV-1 RNA and restored to the same level as that of wild-type HIV-1 in viral particles. Recombinant HIV-1 clones with IN undermultimerization propagated similarly to wild-type HIV-1. Our study revealed that HIV-1 can eventually counteract NCINI-induced IN overmultimerization by IN undermultimerization as one of the escape mechanisms. Our findings provide information on the understanding of IN multimerization with or without HIV-1 RNA and may influence the development of anti-HIV-1 strategies.
IMPORTANCE Understanding the mechanism of HIV-1 resistance to anti-HIV-1 drugs could lead to the development of novel drugs with increased efficiency, resulting in more effective ART. ART composed of more potent and long-acting anti-HIV-1 drugs can greatly improve drug adherence and also provide HIV-1 prevention such as preexposure prophylaxis. NCINIs with a multimodal mode of action exert potent anti-HIV-1 effects through IN overmultimerization during HIV-1 maturation. However, HIV-1 can acquire some mutations that cause IN undermultimerization to alleviate NCINI-induced IN overmultimerization. This undermultimerized IN was efficiently stabilized by HIV-1 RNA and restored to the same level as that of wild-type HIV-1. Our findings revealed that HIV-1 eventually acquires such a conformational escape reaction to overcome the unique NCINI actions. The investigation into drug-resistant mutations associated with HIV-1 protein multimerization may facilitate the elucidation of its molecular mechanism and functional multimerization, allowing us to develop more potent anti-HIV-1 drugs and unique treatment strategies.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) infection has been a chronic infectious disease. Patients diagnosed with HIV are treated with antiretroviral therapy (ART), which basically consists of more than two anti-HIV-1 drugs. The life expectancy of HIV-1 patients with ART has been prolonged to almost the same extent as uninfected individuals (1, 2). In spite of the benefits of ART, HIV-1 often acquires drug-resistant mutations (3) resulting in treatment failure. High adherence to ART is required to sustain viral suppression in HIV-1 clinical treatment (4). Recently, the FDA approved certain anti-HIV-1 drugs that can be easily taken once daily in the form of a single tablet, greatly improving ART adherence (5). Long-acting anti-HIV-1 drugs could be also useful for ART adherence and HIV-1 treatment. Moreover, HIV-1 prevention such as preexposure prophylaxis (PrEP) by such long-acting anti-HIV-1 drugs is currently being investigated in clinical trials (6). On the other hand, studying the mechanism of HIV-1 resistance to anti-HIV-1 drugs could lead to the development of novel drugs with increased efficiency and a greater genetic barrier to resistance, resulting in more effective ART for HIV-1 treatment.
Four FDA-approved integrase (IN) strand transfer inhibitors (INSTIs), raltegravir (RAL), elvitegravir (EVG), dolutegravir (DTG), and bictegravir (BTG), which target the active site of IN, are more potent and well tolerated than other classes of anti-HIV-1 drugs due to the lack of homologous human proteins, allowing these INSTIs to be widely used for clinical HIV-1 treatment. The World Health Organization (WHO) recommends INSTIs, especially DTG, for clinical HIV-1 treatment according to December 2018 interim guidelines. A single tablet, Juluca, containing only two anti-HIV-1 drugs (DTG and rilpivirine [RPV]), was approved by the FDA in 2018 as the first maintenance therapy (7), and Dovato (DTG and lamivudine [3TC]) was approved in 2019 for daily first-line HIV-1 treatment in naive patients. Therefore, the development of more potent anti-HIV-1 inhibitor targets for IN would be useful for HIV-1 treatment.
In 2010, Christ et al. identified another new class of integration inhibitors (8) that interfere with the interaction between IN and lens epithelium-derived growth factor (LEDGF)/p75, thereby inhibiting HIV-1 replication. LEDGF/p75 plays an important role in tethering IN to the host genome and stabilizing functional IN multimerization during the integration step (9, 10). They termed such inhibitors LEDGINs. In addition, it has been reported that LEDGINs exert antiviral effects through modulating IN multimerization (IN overmultimerization) in the late stage of HIV-1 replication (11–14). LEDGINs are also called non-catalytic-site integrase inhibitors (NCINIs) (15, 16), allosteric IN inhibitors (ALLINIs) (13, 17), integrase-LEDGF allosteric inhibitors (INLAIs) (18), or multimerization integrase inhibitors (MINIs) (19). We used NCINIs as the inhibitor's name for convenience due to these inhibitors exerting a multimodal mechanism of action as well as because BI224436, which was termed an NCINI (16), advanced to a phase I clinical trial. IN overmultimerization by potent NCINIs leads to the production of noninfectious viruses in which the ribonucleoprotein (RNP) is translocated from the capsid lattice during HIV-1 maturation (15, 17, 20). In 2016, Kessl et al. reported that such noninfectious HIV-1 resulted from the loss of the interaction between overmultimerized IN and HIV-1 RNA during HIV-1 maturation (21).
Many resistance mutations of general HIV-1 strains, including HIV-1NL4-3, against NCINIs have previously been reported (8, 12, 18, 22–25). In the present study, we attempted to examine the mechanism underlying NCINI-related mutations, which could provide details of IN multimerization. The study of drug-resistant mutations concerning such allosteric inhibitors may help to elucidate how HIV-1 proteins undergo functional multimerization, which may aid in the development of more potent anti-HIV-1 drugs and unique treatment strategies.
RESULTS
Emergence of NCINI-related resistance mutations.
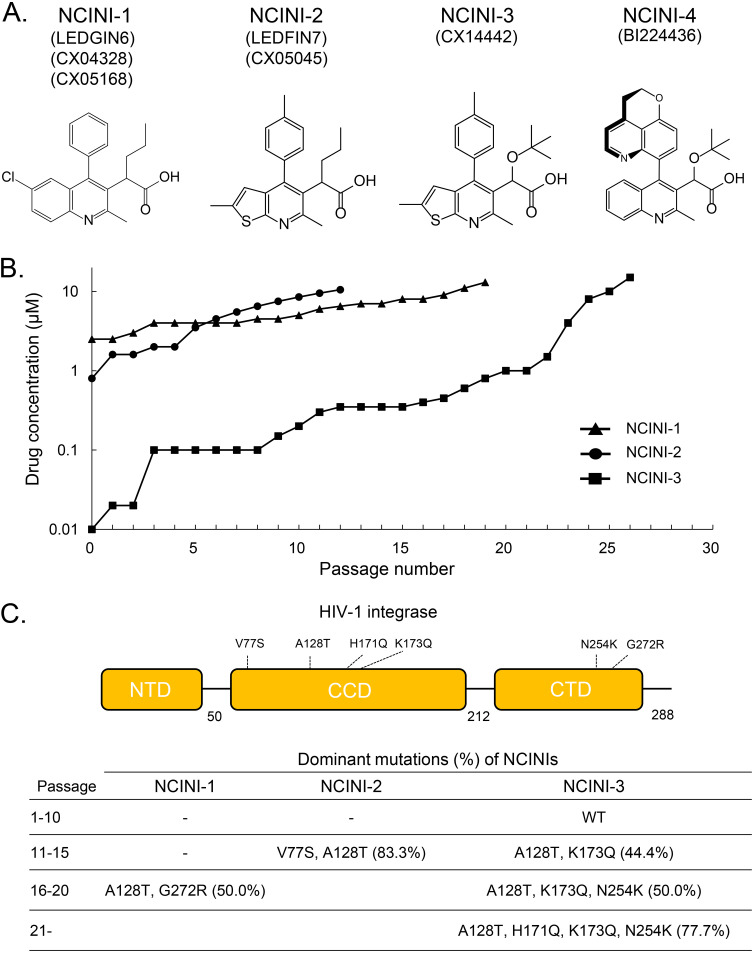
NCINIs with a multimodal mode of action suppress HIV-1 replication by inducing IN overmultimerization (12–14, 17, 20). We selected HIV-1NL4-3 variants resistant to three NCINI compounds known as NCINI-1 (LEDGIN6, CX04328, and CX05168), NCINI-2 (LEDGIN7 and CX05045), and NCINI-3 (CX14442) (Fig. 1A), reported previously (8, 12), by selection experiments (Fig. 1B). As the concentration of NCINIs gradually increased up to 10 μM, HIV-1 acquired several amino acid mutations, such as A128T and K173Q at passage 15 (P15) in the catalytic core domain (CCD) and N254K at P20 in the C-terminal domain (CTD) of IN. Four mutations, A128T, H171Q, K173Q, and N254K, accumulated during P26 in the full-length IN region upon treatment with NCINI-3 at 10 μM. A128T and G272G mutations were acquired in the IN region upon treatment with NCINI-1 at passage 18, and V77S and A128T mutations were acquired upon treatment with NCINI-2 at passage 12 (Fig. 1C). Fifty percent effective concentrations (EC50s) of the drugs NCINI-1, -2, -3, and -4 (BI224436) (16); RAL; and darunavir (DRV) against wild-type HIV-1NL4-3 are shown in Table 1, and EC50s against recombinant HIV-1NL4-3 clones (HIV-1NL4-3 INA128T, INH171Q, INK173Q, INN254K, INP15, INP20, and INP26) and fold changes are shown in Table 2. The accumulation of these mutations gradually gave rise to NCINI-resistant HIV-1. It appeared that each K173Q and N254K mutation did not confer strong resistance to potent NCINIs (NCINI-3 and -4), while A128T alone and the addition of H171Q at P26 conferred substantial resistance (over 10-fold EC50s) to the potent NCINIs; however, none of the clones carrying NCINI-related resistance mutations exhibited cross-resistance to clinically used drugs such as RAL and DRV.
FIG 1.
NCINI information and emergence of NCINI-related resistance mutations in the IN region. (A) Chemical structures and names of NCINIs in this study. (B) Selection experiments for NCINI-1, -2, and -3. HIV-1NL4-3 was propagated in MT-4 cells in the presence of increasing concentrations of NCINI-1, NCINI-2, or NCINI-3, and the selection of virus resistant to the NCINIs continued up to 10 μM. (C) Dominant NCINI-related resistance mutations in the IN region (percent) shown in a schematic of HIV-1 integrase and at passages 1 to 10, 11 to 15, 16 to 20, and over 21. These mutations at each passage were identified from cellular DNA in MT-4 cells infected with NCINI-related resistant HIV-1.
TABLE 1.
| Drug | Mean EC50 (μM) ± SD |
Mean CC50 (μM) ± SD |
||
|---|---|---|---|---|
| MT-2 cells with HIV-1LAI | MT-4 cells with HIV-1NL4-3 | MT-4 cells | HEK293T cells | |
| NCINI-1 | 2.29 ± 1.19 | 7.01 ± 5.41 | 42.23 ± 9.44 | 34.26 ± 4.19 |
| NCINI-2 | 0.77 ± 0.13 | 2.60 ± 0.34 | 66.59 ± 1.70 | >100 |
| NCINI-3 | 0.029 ± 0.003 | 0.19 ± 0.13 | 45.67 ± 6.17 | >100 |
| NCINI-4 | 0.032 ± 0.001 | 0.027 ± 0.004 | 36.09 ± 1.59 | >100 |
| RAL | 0.0024 ± 0.0012 | 0.0019 ± 0.0008 | >100 | >100 |
| EVG | 0.0019 ± 0.0020 | 0.0036 ± 0.0002 | 2.89 ± 0.59 | 4.19 ± 3.89 |
| DTG | 0.0010 ± 0.0010 | 0.0029 ± 0.0010 | 14.33 ± 0.64 | 21.39 ±0.97 |
MT-2 cells were exposed to 100 TCID50s of HIV-1LAI and cultured in the presence of various concentrations of NCINIs and INSTIs, and EC50s were determined by using an MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay.
Cytotoxicities (CC50s) of NCINIs and INSTIs against MT-4 and HEK293T cells were also determined by an MTT assay. All assays were conducted in duplicate, and data shown represent mean values ± SD from three independent experiments.
TABLE 2.
Antiviral activitiesa of NCINIs against NCINI-3-resistant HIV-1NL4-3 clones
| Recombinant HIV-1NL4-3 clone | NCINI-1 |
NCINI-2 |
NCINI-3 |
NCINI-4 |
RAL |
DRV |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean EC50 (μM) ± SD | Fold change | Mean EC50 (μM) ± SD | Fold change | Mean EC50 (μM) ± SD | Fold change | Mean EC50 (μM) ± SD | Fold change | Mean EC50 (μM) ± SD | Fold change | Mean EC50 (μM) ± SD | Fold change | |
| INWT | 7.01 ± 5.41 | 1 | 2.60 ± 0.34 | 1 | 0.19 ± 0.13 | 1 | 0.027 ± 0.004 | 1 | 0.0017 ± 0.0025 | 1 | 0.0037 ± 0.0003 | 1 |
| INA128T | >10 | ND | >10 | ND | 3.12 ± 0.12 | 16.1 | 0.31 ± 0.01 | 11.6 | 0.0028 ± 0.0056 | 1.7 | 0.0094 ± 0.0008 | 2.5 |
| INP15 | >10 | ND | >10 | ND | 3.36 ± 0.11 | 17.3 | 0.32 ± 0.06 | 12.2 | 0.0028 ± 0.0058 | 1.7 | 0.0097 ± 0.0031 | 2.6 |
| INP20 | − | − | − | − | 2.14 ± 0.14 | 11.1 | 0.39 ± 0.04 | 14.7 | 0.0008 ± 0.0041 | 0.5 | 0.0074 ± 0.0018 | 2.0 |
| INP26 | − | − | − | − | 34.83 ± 2.21 | 179.8 | 2.43 ± 0.61 | 91.7 | 0.0017 ± 0.0025 | 1.0 | 0.0034 ± 0.0016 | 0.9 |
| INH171Q | >10 | ND | >10 | ND | 0.42 ± 0.08 | 2.2 | 0.31 ± 0.04 | 11.7 | 0.0027 ± 0.0051 | 1.6 | 0.0085 ± 0.0024 | 2.3 |
| INK173Q | >10 | ND | >10 | ND | 0.14 ± 0.03 | 0.7 | 0.021 ± 0.01 | 0.8 | 0.0022 ± 0.0011 | 1.3 | 0.0032 ± 0.0011 | 0.8 |
| INN254K | − | − | − | − | 0.27 ± 0.04 | 1.4 | 0.092 ± 0.03 | 3.5 | 0.0019 ± 0.0010 | 1.1 | 0.0054 ± 0.0020 | 1.4 |
MT-4 cells (1 × 104 cells/ml) were exposed to 100 TCID50s of each infectious HIV-1 clone, and the inhibition of p24 production by the drug was used as the endpoint on day 7 in culture. The fold changes represent EC50 ratios of each HIV-1 clone to wild-type HIV-1. All assays were performed in duplicate, and the data shown are mean values ± SD derived from the results of three independent experiments. ND, not determined; −, not examined.
Characteristics of recombinant IN proteins carrying NCINI-related resistance mutations.
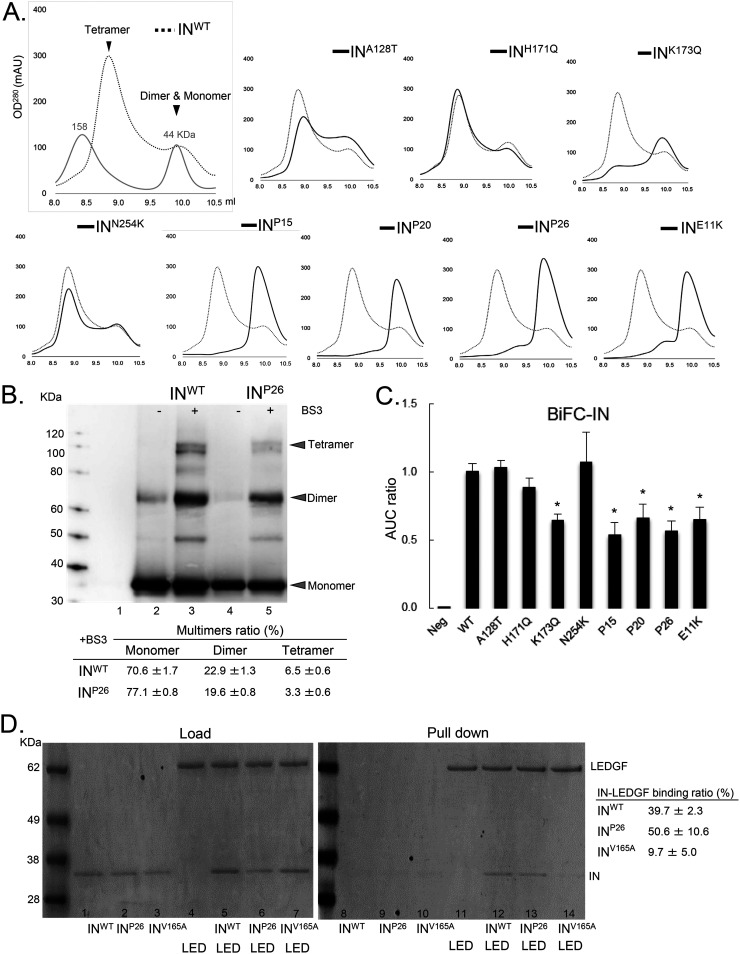
To examine how NCINI-3 resistance mutations acquire resistance against NCINI-3, we purified recombinant His-tagged IN proteins carrying mutations such as A128T, H171Q, K173Q, N254K, P15, P20, P26, and E11K (INA128T, INH171Q, INK173Q, INN254K, INP15, INP20, INP26, and INE11K) expressed in Escherichia coli and analyzed the multimerization of these IN proteins. The E11K substitution was known to impair IN tetramerization (10). Wild-type IN (INWT) multimerization appeared as two peaks of tetramers and a mixture of dimers and monomers using size exclusion chromatography (SEC) (Fig. 2A). INH171Q and INN254K multimerizations were very similar to that of INWT, suggesting that the H171Q and N254K mutations did not affect IN multimerization, whereas INA128T multimerization was seen as a broader shoulder of the two peaks. INK173Q exhibited a moderately decreased tetramer peak. Furthermore, the INP15, INP20, and INP26 proteins carrying the accumulated NCINI-3 resistance mutations completely shifted to one peak consisting of dimers and monomers (Fig. 2A). Next, we examined the proportions of monomers, dimers, and tetramers of INWT and INP26 cross-linking with bis(sulfosuccinimidyl)substrate (BS3) (multimer ratio) using an immunoblot assay. The multimer ratio of INP26 (19.6% dimers and 3.3% tetramers) was slightly decreased compared with that of INWT (22.9% dimers and 6.5% tetramers) (Fig. 2B). In addition, to quantitatively examine IN multimerization, we used a biomolecular fluorescence complementation (BiFC)-IN system reported previously (26). The results of the BiFC-IN system carrying NCINI-3 resistance mutations almost corroborated the SEC data (Fig. 2C).
FIG 2.
Characteristics of recombinant IN proteins carrying NCINI-3 resistance mutations. (A) SEC analysis of IN proteins carrying NCINI-3 resistance mutations. The elution profiles of INWT proteins at a 50 μM concentration are shown together with estimated IN multimers and two molecular weight standards (158 and 44 kDa). Those of the INA128T, INH171Q, INK173Q, INN254K, INP15, INP20, INP26, and INE11K proteins are shown compared with INWT. OD280, optical density at 280 nm; mAU, milli-absorbance unit. (B) Multimer ratios of IN proteins (INWT and INP26) cross-linking with BS3. Each multimer’s intensity of INWT and INP26 measured by ImageJ is shown as a multimer ratio (percent). The molecular weight standard, buffer (lane 1), INWT and INP26 (lanes 2 and 4), and INWT and INP26 cross-linked with BS3 at 0.05 mM (lanes 3 and 5) are shown by immunoblotting with an anti-IN antibody. The results are representative data, and the ratios represent mean values ± standard deviations (SD) from three independent experiments. (C) Results of the BiFC-IN system carrying NCINI-3 resistance mutations and E11K. The fluorescence intensity derived from multimers of BiFC-INA128T, -INH171Q, -INK173Q, -INN254K, -INP15, -INP20, -INP26, and -INE11K in HEK293T cells measured by FACS analysis is shown as a ratio of the area under the curve (AUC) compared to BiFC-INWT. Error bars indicate SD from three independent experiments. The BiFC-IN system was summarized previously (26). Statistical significance was examined using a parametric two-tailed Student t test (*, P < 0.05). (D) LEDGFHis and F-IN pulldown assay. (Left) Loading samples of F-INWT, F-INP26, and F-INV165A (lanes 1 to 3, respectively); LEDGFHis (lane 4); and LEDGFHis together with F-INWT, F-INP26, and F-INV165A (lanes 5 to 7, respectively). (Right) Pulldown samples of F-INWT, F-INP26, and F-INV165A (lanes 8 to 10, respectively); LEDGFHis (lane 11); and LEDGFHis incubated for 30 min with F-INWT, F-INP26, and F-INV165A (lanes 5 to 7, respectively) after His tag column selection. The ability of IN proteins carrying these mutations to bind to LEDGF/p75 is shown as a IN-LEDGF/p75 ratio (percent) by measuring the band intensity using ImageJ. The ratios represent mean values ± SD from two independent experiments.
Next, to understand whether IN undermultimerization affects its interaction with LEDGF/p75, we performed a pulldown assay between His-tagged LEDGF/p75 and a Flag-tagged IN (F-IN) protein carrying P26 or the V165A mutation reported as a class II mutant that fails to interact with LEDGF/p75 (27). As shown in Fig. 2D, the IN-LEDGF/p75 binding ratio of INV165A, which fails to interact with LEDGF/p75, was 9.7%, whereas those of INWT and INP26 with IN undermultimerization were 39.7 and 50.6%, respectively, suggesting that INP26 with IN undermultimerization can interact with LEDGF/p75.
The P26 mutation with accumulated NCINI-3 resistance mutations severely reduced IN multimerization and also did not affect the IN-LEDGF interaction, suggesting that IN undermultimerization may be associated with NCINI-3-related resistance of HIV-1.
Characteristics of recombinant HIV-1NL4-3 clones carrying NCINI-related resistance mutations.
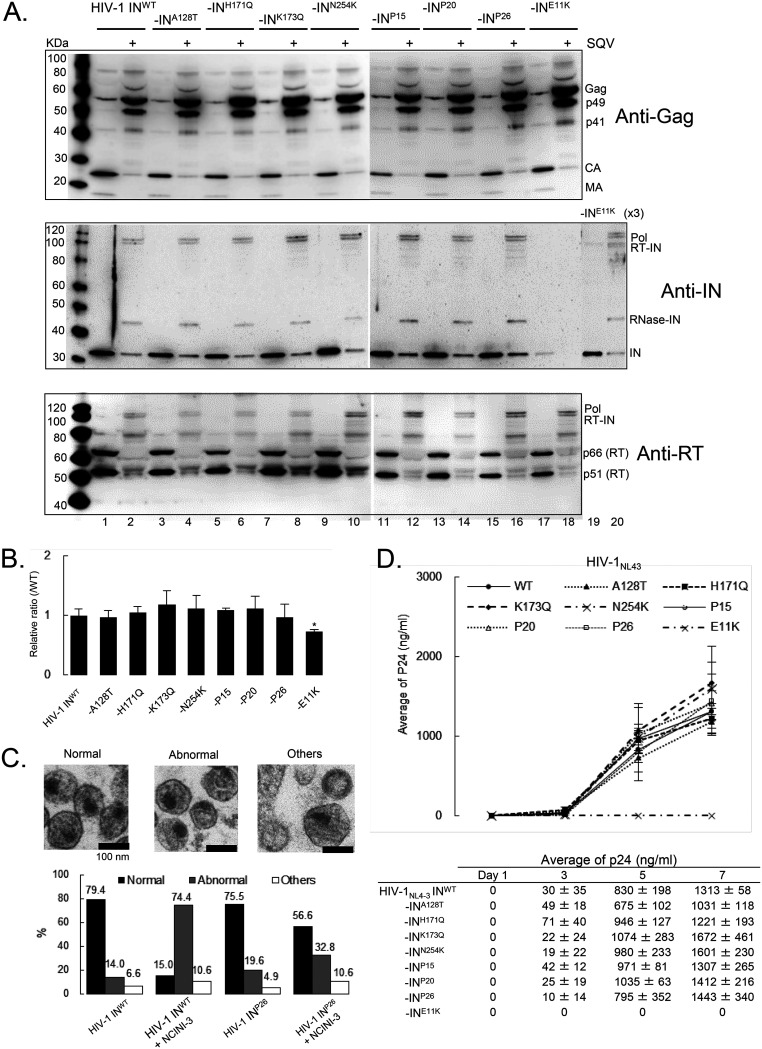
NCINIs induce IN overmultimerization, leading to the production of noninfectious viruses, but are not related to Gag and Gag-Pol proteolytic processing (17, 19, 25). In order to investigate the characteristics of HIV-1 clones carrying the NCINI-3 resistance mutations with IN undermultimerization, we examined the process of viral production, including Gag and Gag-Pol proteolytic processing, and viral maturation of the recombinant HIV-1 INA128T, INH171Q, INK173Q, INN254K, INP15, INP20, INP26, and INE11K clones. As shown in Fig. 2A, the K173Q, P15, P20, and P26 mutations significantly decreased IN multimerization; however, Gag and Gag-Pol proteolytic processing of the recombinant HIV-1 IN clones with or without IN undermultimerization, analyzed by immunoblotting in the presence of saquinavir (SQV) (an HIV-1 protease inhibitor that inhibits Gag-Pol proteolytic processing), was similar to that of HIV-1 INWT (Fig. 3A). Viral production levels (p24 values) of wild-type HIV-1NL4-3 (HIV-1 INWT) and such recombinant HIV-1 IN clones with or without IN undermultimerization, except for HIV-1 INE11K, were also similar (Fig. 3B), indicating that the recombinant HIV-1 clones with IN undermultimerization had normal Gag and Gag-Pol proteolytic processing and viral production.
FIG 3.
Characteristics of recombinant HIV-1 IN clones carrying NCINI-3 resistance mutations with IN undermultimerization. (A) Gag and Gag-Pol proteolytic processing products of recombinant HIV-1NL4-3 carrying NCINI-related mutations or INE11K clones, respectively. The HIV-1 IN clones were produced in the presence (even lanes) or absence (odd lanes) of 2 μM SQV. Gag and Gag-Pol proteolytic processing products normalized to p24 levels of each clone were visualized by immunoblotting with anti-HIV-1 Gag, IN, and reverse transcriptase (RT) antibodies. Three times the amount of INE11K (lanes 19 and 20) was examined due to the epitope of the anti-IN antibody including the E11 residue. (B) Viral production and p24 levels of HIV-1 INWT and HIV-1 clones carrying NCINI-3 resistance mutations or the E11K mutation. Data indicate relative ratios normalized to HIV-1 INWT and mean values ± SD from three independent experiments. Statistical significance was examined using two-tailed Student’s t test (*, P < 0.05). (C) Morphologies of HIV-1 INWT and the HIV-1 INP26 clone in the presence or absence of 20 μM NCINI-3 using TEM. (Top) Representative images of normal, abnormal, and other HIV-1 particles (magnification, ×168,000). (Bottom) Over 100 HIV-1 particles were examined from several images of each HIV-1 sample, and the percentages of HIV-1 morphology classified as normal, abnormal, and others are shown in a graph. (D) Replication kinetics of HIV-1 carrying the NCINI-3 resistance mutation or INE11K clones. MT-4 cells were exposed to an HIV-1 preparation normalized to p24 levels, and the production of each HIV-1 clone from MT-4 cells was monitored at days 1, 3, 5, and 7 by a p24 ELISA. The assays were performed in triplicate, and error bars indicate SD from two independent experiments.
Also, the morphologies of HIV-1 INWT and the HIV-1 INP26 clone in the presence or absence of NCINI-3 were observed using a transmission electron microscope (TEM). HIV-1 particles were classified by viral structures under three phenotypes, normal mature conical core, abnormal (or empty) core, and other core, including immature and ambiguous (Fig. 3C), as previously reported (17, 20, 28). The morphology of HIV-1 INWT included 79.4% normal, 14.0% abnormal, and 6.6% other structures. By the addition of NCINI-3, an abnormal morphology of HIV-1 INWT was identified in 74.4%, which indicated RNP translocation from the capsid lattice. On the other hand, the morphology of the HIV-1 INP26 clone with IN undermultimerization was similar to that of HIV-1 INWT and exhibited resistance against NCINI-3 (56.6% normal, 32.8% abnormal, and 10.6% other structures). Finally, we examined the replication fitness of HIV-1 INWT and the recombinant HIV-1 IN clones. The HIV-1 IN clones with IN undermultimerization in infected MT-4 cells showed robust replication fitness similar to that of HIV-1 INWT up to day 7, except for the HIV-1 INE11K clone (p24, from 1,031 to 1,672 ng/ml) (Fig. 3D). Thus, IN undermultimerization induced by the NCINI-3 resistance mutations in this study did not significantly affect viral production, including Gag and Gag-Pol processing, and viral maturation in the HIV-1 life cycle.
Effect of certain mutations on the conformation and stability of the IN catalytic core domain.
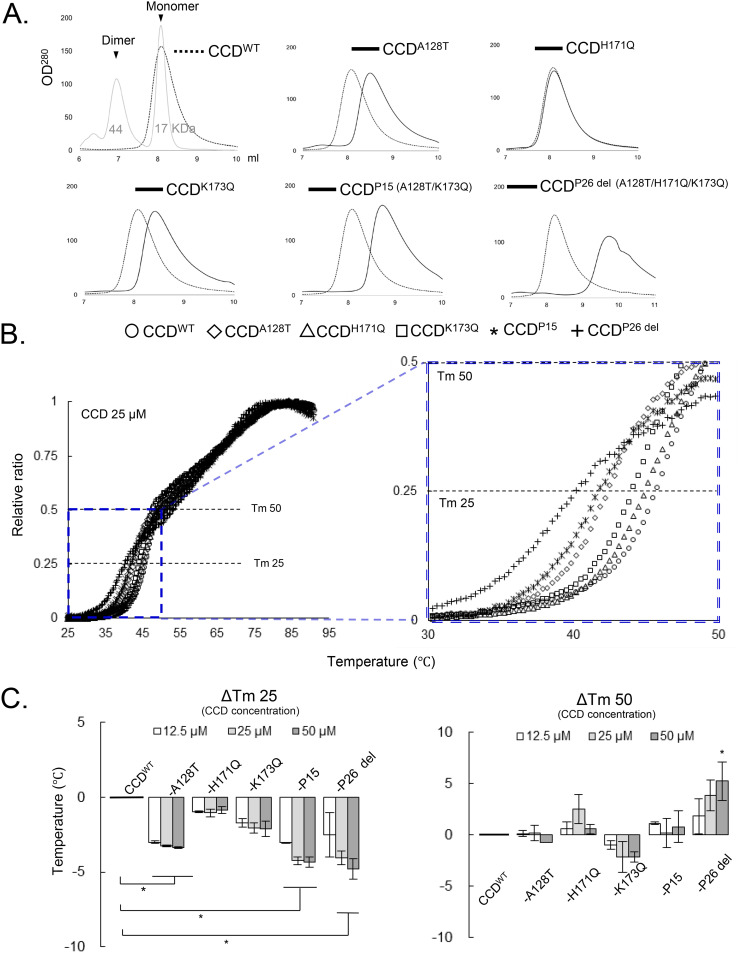
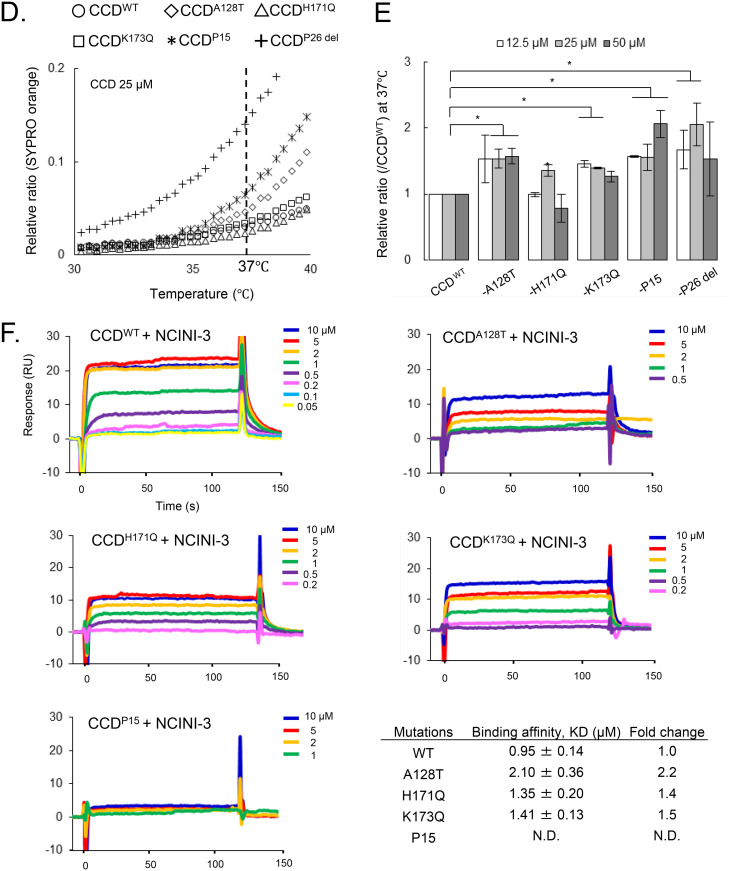
It has been reported that NCINIs bind to two pockets formed in CCD proteins (8) carrying a solubilizing amino acid substitution, F185K (CCDF185K) (29). To examine whether the NCINI-3 resistance mutations affect the structure of the CCD, we produced and purified CCDF185K proteins carrying NCINI-3 resistance mutations (CCDA128T, CCDH171Q, CCDK173Q, CCDP15, and CCDP26 del) expressed in E. coli. P26 del indicates P26 without the N254K mutation in the CTD. It appeared that a CCDWT peak eluted at roughly the same monomer size as CCD: 17 kDa by SEC under our method conditions (Fig. 4A). CCDH171Q, which did not reduce full-length IN multimerization, eluted at the same size fraction as CCDWT. Notably, the CCDA128T and CCDK173Q proteins, which changed full-length IN multimerization, eluted at a decreased size compared to CCDWT. CCDP15 and CCDP26 del eluted at further decreased sizes. These results suggest that the A128T, K173Q, P15, and P26 del mutations affect the form of the CCD monomer (Fig. 4A).
FIG 4.
Effect of NCINI-3 resistance mutations on the CCD. (A) SEC analysis of CCD proteins carrying NCINI-3 resistance mutations together with F185K. The elution profile of CCDWT is shown with two molecular weight standards (44 and 17 kDa) and estimating the size positions of CCD dimers and monomers. Those of CCDA128T, CCDH171Q, CCDK173Q, CCDP15, and CCDP26 del at a 50 μM concentration was are shown compared with CCDWT. (B) DSF analysis of CCD proteins carrying NCINI-3 resistance mutations. (Left) The fluorescence of CCDWT, CCDA128T, CCDH171Q, CCDK173Q, CCDP15, and CCDP26 del was detected and is plotted as a relative ratio from 25°C to 95°C. (Right) Expanded plots from 30°C to 50°C. (C) ΔTm 25 or 50 values indicating the Tm 25 or 50 differences between CCDWT and CCD proteins carrying the mutations at 12.5, 25, and 50 μM CCD concentrations. The assays were performed in triplicate, and error bars indicate SD from two independent experiments. The statistical significance of ΔTm 25 or 50 was examined using two-tailed Student’s t test (*, P < 0.05). (D) Relative ratios of SYPRO orange fluorescence of CCDWT and CCD proteins carrying the mutations from 30°C to 40°C at a 25 μM concentration. (E) Proportions of surface hydrophobic sites of CCD proteins at 37°C. The relative ratios were obtained from SYPRO orange binding to the surface hydrophobic sites of CCD proteins carrying the mutations compared to CCDWT at 12.5, 25, and 50 μM concentrations. The assays were performed in triplicate, and error bars indicate SD from two independent experiments. Statistical significance was examined using Student’s t test (*, P < 0.05). (F) SPR analysis of NCINI-3 interactions with CCDWT, CCDA128T, CCDH171Q, CCDK173Q, and CCDP15. Data shown represent results derived from two independent experiments. Shown are KD values of NCINI-3 for CCDWT and CCD proteins carrying the mutations, and fold changes indicate KD ratios of CCD proteins carrying the mutations to CCDWT. Error bars indicate SD from two independent experiments. N.D., not determined.
In addition, to examine the thermal stability of CCDA128T, CCDH171Q, CCDK173Q, CCDP15, and CCDP26 del, we performed differential scanning fluorimetry (DSF) (30). The melting temperature (Tm) value for proteins generally represents the temperature at which the protein is 50% folded (Tm 50). As shown in Fig. 4B and C, the Tm 50 values of all CCD proteins did not indicate significant differences. On the other hand, we especially noticed the Tm 25 value, indicating the temperature at which CCD proteins are 25% folded upon heating. The Tm 25 values of the CCDA128T, CCDK173Q, CCDP15, and CCDP26 del proteins, but not CCDH171Q, decreased compared with that of CCDWT (Fig. 4C), suggesting that CCD proteins carrying these mutations were unstable at lower temperatures from 35°C to 45°C compared with CCDWT. Additionally, the surface hydrophobic sites of CCDA128T, CCDK173Q, CCDP15, and CCDP26 del at 37°C increased, as shown in Fig. 4D and E. It seems that the A128T, K173Q, P15, and P26 del mutations affect the CCD conformation in the region of body temperature. These results suggest that the A128T, K173Q, P15, and P26 del mutations but not H171Q probably change the CCD conformation, possibly affecting full-length IN multimerization.
Furthermore, to investigate the binding affinity of NCINI-3 for each CCDA128T, CCDH171Q, CCDK173Q, and CCDP15 protein, we used surface plasmon resonance (SPR) (Fig. 4F). The binding affinity (equilibrium dissociation constant [KD] value) of NCINI-3 for CCDWT was 0.95 μM, while those for CCDA128T, CCDH171Q, and CCDK173Q decreased by 2.10 μM (2.2-fold), 1.35 μM (1.4-fold), and 1.41 μM (1.5-fold), respectively. Furthermore, CCDP15 did not interact at 10 μM NCINI-3 (Fig. 4F). The interactions of NCINI-3 with CCD proteins carrying the A128T, H171Q, or K173Q mutation decreased, while K173Q did not confer significant resistance against NCINI-3 (Table 2). These results suggest that K173Q causing mild IN undermultimerization may have a different resistant profile from those of A128T and H171Q.
The P15 mutation changes NCINI binding pockets formed in the CCD.
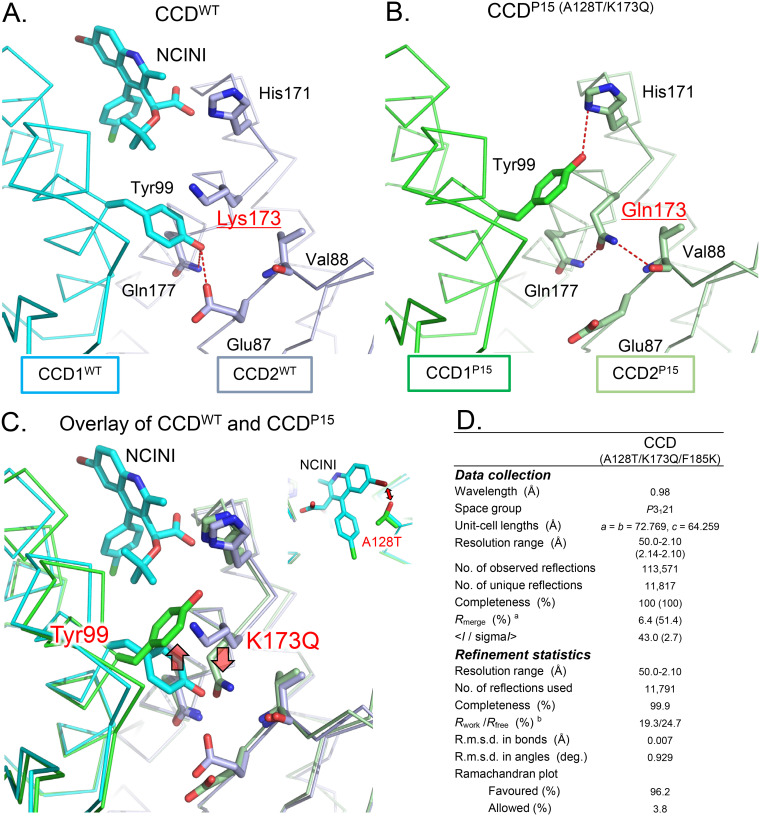
It has been reported that NCINI-related compounds bind to CCD proteins containing the F185K substitution by crystallographic analysis (8, 14, 31). To confirm the CCD conformation carrying P15 (A128T/K173Q), which mostly lost the tetramer peak of full-length IN (Fig. 2A), we analyzed a crystal structure of CCDP15 carrying F185K using X-ray crystallography.
As shown in Fig. 5A, the NCINI-related compound binds to the binding pocket in CCDWT (Protein Data Bank [PDB] accession number 4GW6) (22), while a phenolic side chain (Y99 position) in CCDP15 (PDB accession number 6L0C) shifts closer to the NCINI binding pocket than that in CCDWT (Fig. 5B and C), potentially interfering with the binding of the NCINI to CCDP15. Additionally, the A128T mutation also affects the positioning of the NCINI, as previously reported (22) (Fig. 5C). Interestingly, the phenolic side chain of the Y99 residue in a CCDWT monomer (CCD1WT) participates in intermolecular interactions with two hydrogen bonds (H-bonds) to the side chains of E87 and Q177, respectively, in the other CCDWT monomer (CCD2WT) (Fig. 5A). However, the interaction with Y99 in CCD1P15 reduces one H-bond with the side chain of H171 in CCD2P15; specifically, Q173 forms new intramolecular interactions with the V88 and Q177 residues in CCD2P15 (Fig. 5B). Such H-bond alterations at the dimerization interface in CCDP15 might cause an unstable CCDP15 dimer compared with CCDWT, affecting NCINI binding pockets in CCDP15.
FIG 5.
The P15 mutation changes the NCINI binding space formed in the CCD. (A) X-ray crystal structure of the CCDWT dimer (PDB accession number 4GW6). The NCINI-related compound interacts with the binding pocket formed from CCD1WT and CCD2WT monomers shown in cyan and gray, respectively. Intermolecular hydrogen bonds between CCD1WT and CCD2WT are shown by red dashed lines. Locations of side chains for the Y99 and K173 residues in the monomer-monomer interface of CCDWT are indicated. (B) X-ray crystal structure of the CCDP15 dimer (PDB accession number 6L0C). CCD1P15 and CCD2P15 monomers are shown in green and light green, respectively. Intermolecular and intramolecular hydrogen bonds in CCDP15 dimers are shown by red dashed lines. Locations of side chains for the Y99 and Q173 residues in CCDP15 are indicated. (C) Overlay of the crystal structures of CCDWT in complex with the NCINI-related compound (PDB accession number 4GW6) and CCDP15 (PDB accession number 6L0C). (D) X-ray data collection and refinement statistics. The numbers in parentheses indicate the highest-resolution shell. aRmerge = Σhkl Σi |Ii (hkl) − ⟨I(hkl)⟩|/Σhkl Σi Ii (hkl), where Ii (hkl) is the observed intensity and ⟨I(hkl)⟩ is the mean value of Ii (hkl). bRwork = ΣǁFo| − |Fcǁ/Σ|Fo|. Rfree was calculated from the test set (5% of the total data). R.m.s.d., root mean square deviation.
The P15 mutation reduces the NCINI binding space formed in CCDP15 dimers (Fig. 5C) and also might disrupt the binding pocket by producing unstable CCDP15 dimers, shifting to CCDP15 monomers, potentially resulting in reduced full-length IN multimerization (IN undermultimerization).
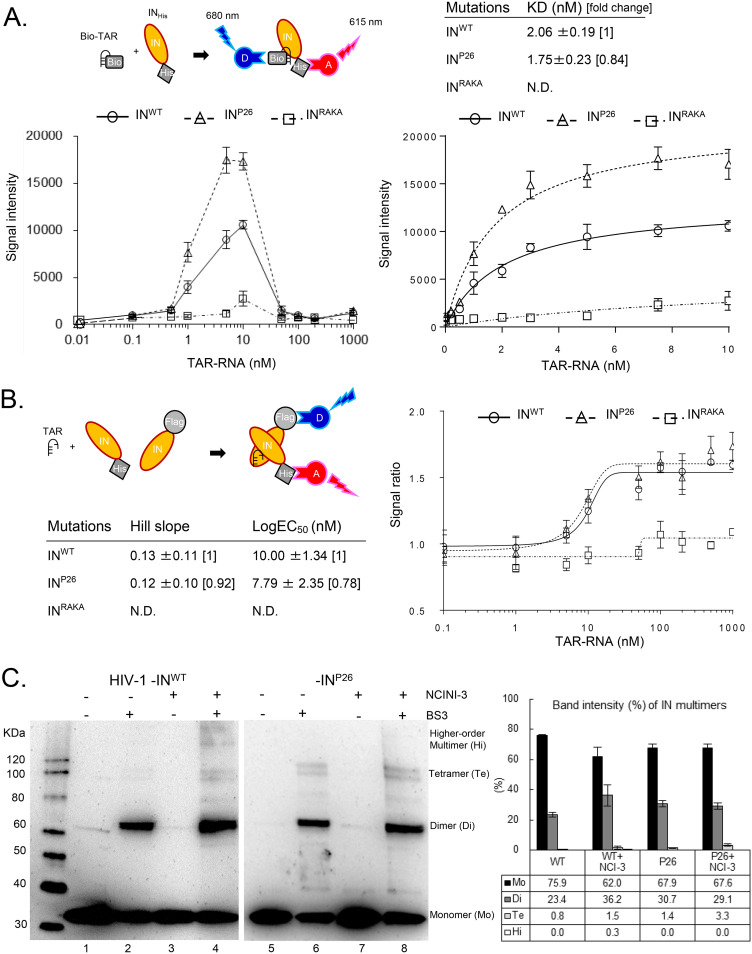
INP26 undermultimerization is stabilized by HIV-1 RNA and restored in viral particles.
Overmultimerized IN, directed by NCINIs, fails to interact with HIV-1 RNA, leading to the production of noninfectious HIV-1 particles with an abnormal core (Fig. 3C) (21). To examine whether IN undermultimerization affects direct interactions with HIV-1 RNA, we analyzed the interaction between purified 6×His-tagged INP26 proteins and a biotinylated transactivation response (TAR) element, of 54 RNA nucleotides, using Alpha, which is a bead-based nonradioactive amplified luminescent proximity homogeneous assay (Alpha 1) (21, 32, 33). In this assay, we employed recombinant INRAKA proteins carrying the R269A/K273A mutation, with reported normal IN multimerization but a loss of the interaction with HIV-1 RNA (21). The Alpha signals of INWT and INP26 at 10 nM were each sufficiently increased by the addition of TAR RNA at concentrations up to 10 nM, fitting to saturation curves, and then were decreased because of hooking effects (Fig. 6A). Interestingly, the binding affinity (KD value) of INRAKA in Alpha 1 was ambiguous, whereas that of INP26 was 1.76 nM (95% confidence interval [CI], 1.22 to 2.30 nM) (0.84-fold), which was slightly lower than that of INWT, at 2.06 nM (95% CI, 1.42 to 2.70 nM) (Fig. 6A), suggesting that the ability of INP26 to bind to TAR RNA (HIV-1 RNA) may be slightly reinforced.
FIG 6.
IN undermultimerization is stabilized by HIV-1 RNA and restored in viral particles. (A) Alpha 1 for direct binding between biotinylated TAR RNA (Bio-TAR) and INHis. The schematic illustration at the top left indicates that streptavidin-coated donor (D) and anti-His acceptor (A) beads bind to the Bio-TAR–INHis complex. The Alpha signal intensity between INHis (INWT, INP26, and INRAKA at 10 nM) and Bio-TAR (from 0 to 1,000 nM) is shown at the bottom left. The intensities of INWT, INP26, and INRAKA from 0 to 10 nM Bio-TAR are fitted to saturated curves at the bottom right. The curves and mean KD values ± SD were determined from representative fitting data from three independent experiments. (B) Alpha 2 for indirect IN multimerization between F-IN and INHis. The schematic illustration indicates that anti-Flag-coated donor and anti-His coated acceptor beads bind to a dimer of F-IN and INHis. Alpha signal ratios between F-IN and INHis proteins (INWT, INP26, and INRAKA, respectively) by the addition of 0.1 to 1,000 nM concentrations of TAR RNA were fitted to sigmoid curves. Mean Hill slopes and log EC50s ± SD were calculated by representative sigmoid curves from three independent experiments. (C) IN multimerization of HIV-1NL4-3 INWT and INP26 viral particles generated in the presence or absence of 20 μM NCINI-3. The HIV-1 INWT and INP26 clones were purified by ultracentrifugation. Highly concentrated HIV-1 INWT and INP26 were cross-linked with (lanes 2, 4, 6, and 8) or without (lanes 1, 3, 5, and 7) BS3 and visualized by SDS-PAGE with anti-HIV-1 IN antibody. Monomers, dimers, tetramers, and high-order multimers of INWT and INP26 are shown as multimer ratios (percent) determined by measuring each band intensity using ImageJ. The ratios represent mean values ± SD from two independent experiments.
Next, Alpha 2 was performed to confirm the effect of HIV-1 RNA on IN multimerization (Fig. 6B). The Alpha 2 signal ratios of INWT and INP26 were determined at concentrations of up to 1,000 nM TAR RNA. The Alpha 2 signal ratios of INWT and INP26, but not INRAKA, increased according to the concentration of TAR RNA until it reached 100 nM, and then showed a plateau without a hooking effect, fitting to sigmoid curves (Fig. 6B). Hill slopes of INWT and INP26 but not INRAKA were almost similar, while the log EC50 of INP26 was 7.79 nM (95% CI, 2.94 to 12.64 nM) (0.78-fold), which was slightly lower than that of INWT, at 10.00 nM (95% CI, 7.23 to 12.80 nM) (Fig. 6B). These results suggest that INP26 multimerization may be supported efficiently by HIV-1 RNA compared to INWT.
Finally, in order to investigate the effect of NCINIs on INWT and INP26 multimerization during HIV-1 maturation, we analyzed IN multimerization of HIV-1 INWT and INP26 clones produced in the absence or presence of 20 μM NCINI-3 using cross-linking with a BS3 immunoblot assay. IN dimers and tetramers obviously increased, and high-order multimers clearly appeared in HIV-1 INWT, which were produced in the presence of NCINI-3, while IN dimers and tetramers of HIV-1 INP26 produced in the absence and presence of NCINI-3 were shown to be similar (Fig. 6C), indicating that the P26 mutation suppressed NCINI-3-induced IN overmultimerization during HIV-1 maturation and also that INP26 undermultimerization was restored to the same level as INWT multimerization in viral particles.
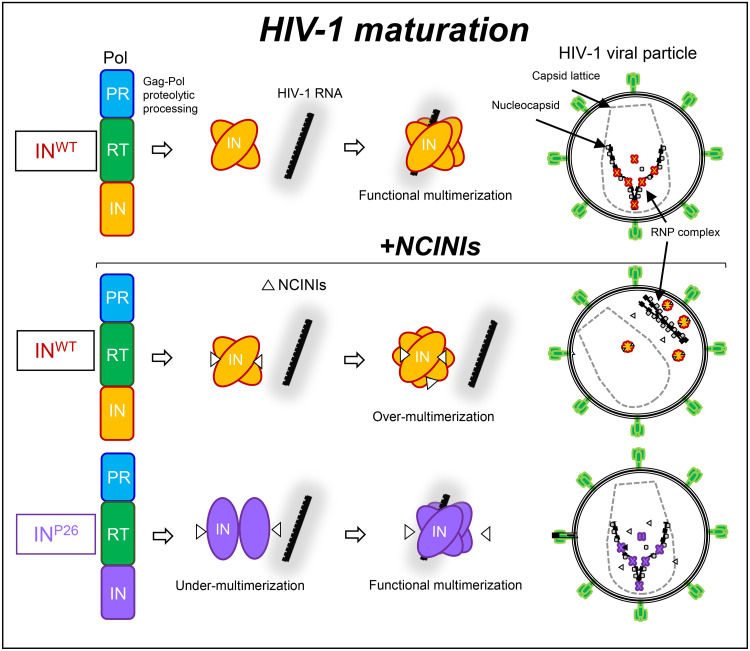
Taken together, IN undermultimerized proteins via the accumulated NCINI-related resistance mutations were stabilized by HIV-1 RNA and eventually restored in HIV-1 particles during HIV-1 maturation. These results reveal that IN undermultimerization via NCINI-resistant mutations may be one of the HIV-1 escape mechanisms allowing NCINI-resistant HIV-1 to propagate within cells (Fig. 7).
FIG 7.
Graphical representation of the HIV-1 escape reaction only during HIV-1 maturation. The schematic illustration focuses on the interaction of IN multimerization with HIV-1 RNA and the location of RNP in viral particles during HIV-1 maturation from Gag-Pol proteolytic processing in this study. The top illustration indicates that the normal interaction of functional IN multimerization with HIV-1 RNA results in mature HIV-1. The middle illustration shows that overmultimerized IN proteins induced by NCINIs fail to interact with HIV-1 RNA, producing immature HIV-1 in which the RNP complex is translocated from the capsid lattice. The bottom illustration indicates that undermultimerized INP26 proteins can escape from NCINI binding and are restored to the same level as INWT multimerization in the viral particles, producing mature HIV-1 in the presence of NCINIs as one of the HIV-1 escape mechanisms.
DISCUSSION
In the present study, we found that IN undergoes an adaptable conformational alteration to escape NCINI activity. It has been reported that CCD dimers formed at the central core are material components of full-length IN tetramers with HIV-1 nucleotides (10, 34). However, without HIV-1 nucleotides, the P15 (A128T/K173Q) mutation changed the conformation of CCD monomers, which might cause unstable CCD dimers and also affect the form of NCINI binding sites. Such conformational changes could induce severe reductions of full-length IN multimerization before IN binding to HIV-1 RNA. INP26 undermultimerized proteins carrying A128T/K173Q were efficiently recovered by HIV-1 TAR RNA and restored to the same levels as INWT multimerization in the viral particles. The morphology and replication fitness of the HIV-1 INP26 clone were similar to those of wild-type HIV-1. These results suggest that IN undermultimerized proteins carrying A128T/K173Q still retained intrinsic IN function.
On the other hand, NCINIs exert a multimodal mode of action, including an anti-IN-LEDGF interaction in the early stage of HIV-1 replication. The A128T mutation was first identified in HIV-1 mutants that were selected using cell lines that stably expressed a C-terminal fragment of LEDGF/p75 (35), indicating that A128T is related to the IN-LEDGF interaction in the early stage of HIV-1 replication. In this study, we did not investigate the roles of IN undermultimerization by the A128T/K173Q mutation in the early stage, which requires further experimental data to fully understand the NCINI-related resistance mutations of HIV-1 against the multimodal mode of action. Additionally, the A128T mutation has been reported to be a typical and critical NCINI-related resistance mutation (8, 11, 12, 17–19, 22). We observed that A128T emerged early at P15 together with K173Q in the presence of NCINI-3 and later at P17 and P12 in the presence of NCINI-1 and -2, respectively. As shown in Table 2, HIV-1 INA128T exhibited strong resistance to NCINI-3 and -4, suggesting that A128T represents a critical mutation against NCINI action.
The HIV-1 INK173Q clone carrying K173Q, which moderately reduces IN multimerization, showed resistance (over 10 μM) against weak NCINIs (NCINI-1 and -2) but no observed resistance against potent NCINIs (NCINI-3 and -4). By the addition of A128T, HIV-1 INP15 clones acquired severe resistance to the potent NCINIs (Table 2). Collectively, K173Q moderately decreased IN multimerization (Fig. 2A) and reduced the direct binding of NCINI-3 (Fig. 4F), but the EC50 of the HIV-1 INK173Q clone did not indicate critical resistance compared with that of the HIV-1 INA128T clone, suggesting that K173Q plays a secondary role among the NCINI-related resistance mutations. The H171Q mutation has been reported to be an NCINI-related resistance mutation (11, 18). In this study, the H171Q mutation, which did not affect IN multimerization, was acquired at P26, together with A128T, K173Q, and N254K, and conferred sufficient resistance against the potent NCINIs (Table 2). The characteristics of H171Q were similar to those of H171T reported previously (23) at the same amino acid position. Moreover, we constructed and examined an HIV-1 INA128T/H171Q clone carrying a double and potent NCINI resistance mutation. The replication fitness of the HIV-1 INA128T/H171Q clone was severely lower than that of wild-type HIV-1 (data not shown), suggesting that the HIV-1 INA128T/H171Q clone may require secondary mutations such as K173Q and N254K to propagate within cells.
The Y99H mutation was also reported (12, 18) to be an important resistance mutation among NCINI-related mutations. Y99H was not acquired by our selection assay in this study. Interestingly, according to the crystal structure of CCDP15 (Fig. 5), the side chain of Q173 in one CCDP15 protein pushed that of Y99 in the other CCDP15 protein to the NCINI binding pocket, suggesting that K173Q might be a substitute mutation for Y99H. The T124N/T174I resistance mutation against a pyridine-based inhibitor, KF116 (MINI), was also reported to disrupt IN multimerization (24). T124N/T174I decreased the Gag-Pol proteolytic processing, viral maturation, and viral infectivity of the HIV-1 INT124N/T174I variant compared with those in wild-type HIV-1. It seems that the T124N/T174I mutation, which induced abnormal IN multimerization, confers a phenotype similar to that of class II IN mutant HIV-1 (36); however, A128T/K173Q showed a clearly different phenotype from that of the class II IN mutant. Moreover, it was reported that the CTD of IN plays a critical role in IN overmultimerization (37), and NCINIs can interact with both the CCD and CTD, which are contributed by two different dimers so that IN forms an open polymer mediated by inhibitor-bridged contacts in the crystal lattice (38). Although N254K did not confer sufficient resistance against potent NCINIs, N254K is possibly associated with the inhibition of such an IN overmultimerization mechanism.
In viral particles, HIV-1 nucleocapsid (NC) binds to HIV-1 RNA, while IN also binds to HIV-1 RNA at different positions from NC (21, 28). It has yet to be shown how IN binding to HIV-1 RNA maintains RNP in the capsid lattice during HIV-1 maturation. NCINIs are not yet approved by the FDA, and NCINI-related resistance mutations in vivo (clinical use) are unknown, which presents the major limitations of our study.
Our findings reveal that HIV-1 can counteract NCINI-inducing IN overmultimerization by IN undermultimerization as one of the escape mechanisms possibly allowing NCINI-resistant HIV-1 to propagate within cells. Investigation into the drug-resistant mutations associated with HIV-1 protein multimerization may facilitate the elucidation of its molecular mechanisms, allowing the development of more potent anti-HIV-1 drugs and unique treatment strategies.
MATERIALS AND METHODS
Cells.
293T (JCRB Cell Bank, Japan) and HEK293T (ATCC CRL-11268) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Wako, Japan) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich), penicillin (P+) (100 IU/ml), and kanamycin (K+) (100 mg/ml) (Meiji, Japan) at 37°C with 5% CO2. MT-2 and MT-4 cells (JCRB) were cultured in RPMI 1640 medium (Wako, Japan) supplemented with 10% FBS, P+, and K+ at 37°C with 5% CO2.
Plasmid constructs.
Full-length IN and CCD sequences derived from pNL4-3 were introduced into pET30a vectors (Novagen) with a 6×His tag at the C terminus or N terminus, respectively, producing pET30a IN-6×His (INHis) or pET30a 6×His-CCD (CCDHis) by using an In-Fusion HD cloning kit (TaKaRa Bio Inc., Japan). Flag-tagged IN (F-IN) was also introduced into the pET30a vector at the N terminus by the In-Fusion method, producing pET30a Flag-IN. The LEDGF/p75 sequence derived from 293T cells was introduced into pET30a vectors (Novagen) with a 6×His tag at the N terminus, producing pET30a His-LEDGF/p75 (LEDGF/p75His). Site-directed mutagenesis of single mutations was performed using PrimeSTAR Max (TaKaRa) to introduce the A128T, H171Q, K173Q, N254K, E11K, V165A, and R269A/K273A mutations into pET30a INHis, producing the pET30a INA128T, INH171Q, INK173Q, INN254K, INE11K, INV165A, and INRAKA vectors, respectively. The NCINI-3 resistance mutations in the CCD were introduced into pET30a CCDHis with F185K, producing the pET30a CCDA128T, CCDH171Q, and CCDK173Q vectors. Plural-resistance mutations such as P15 (A128T/K173Q), P20 (A128T/K173Q/N254K), and P26 (A128T/H171Q/K173Q/N254K) were introduced into the pET30a INHis or CCDHis vector by the In-Fusion method, producing pET30a INP15, pET30a INP20, pET30a INP26, and pET30a CCDP15 with F185K. These NCINI-3 resistance mutations were also introduced into the pNL4-3 and pBiFC-IN vectors (26) by using the above-described method.
Protein expression and purification.
IN and CCD proteins were produced from pET30a INHis and CCDHis, respectively, in E. coli Rosetta(DE3)/pLysS competent cells (Novagen) grown in LB medium supplemented with K+ and chloramphenicol (Cam+) at 37°C and induced with 1.0 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) for 4 h at 30°C. Bacterial cells were harvested and stored at −80°C. Pellets of INHis were resuspended and sonicated in lysis buffer (50 mM HEPES-NaOH [pH 7.4], 100 mM NaCl). The lysates were cleared by centrifugation for 15 min at 3,500 rpm at 4°C. The supernatants were removed, and the pellets of IN were resuspended in IN reservoir buffer {50 mM HEPES (pH 7.4), 1 M NaCl, 7.5 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 10 mM MgSO4, 10% glycerol} supplemented with 5 mM β-mercaptoethanol (BME), 10 mM imidazole, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF). The pellets of CCD proteins were resuspended and sonicated in CCD reservoir buffer (50 mM HEPES [pH 7.4], 500 mM NaCl, 7.5 mM CHAPS, 10 mM MgSO4, 5% glycerol) supplemented with 5 mM BME, 10 mM imidazole, and 0.5 mM PMSF. The lysates were precleared for 30 min at 15,000 rpm at 4°C and filtered through a 0.45-μm filter. IN and CCD proteins were loaded onto a His-Talon column (TaKaRa), and the columns were washed with IN wash buffer (20 mM HEPES [pH 7.4], 1 M NaCl, 3.75 mM CHAPS) or CCD wash buffer (20 mM HEPES [pH 7.4], 500 mM NaCl, 3.75 mM CHAPS). IN or CCD proteins were eluted with IN or CCD reservoir buffer, respectively, supplemented with 300 mM imidazole using AKTAprime plus (GE Healthcare). IN or CCD fractions were concentrated in each reservoir buffer using an Amicon Ultra-30K or -10K device (Merck Millipore). LEDGF/p75 was produced in E. coli Rosetta(DE3)/pLysS competent cells grown in LB medium supplemented with 2% ethanol, K+, and Cam+ at 37°C and induced with 0.5 mM IPTG for 5 h at 30°C. Bacterial cells were harvested and sonicated in LEDGF reservoir buffer (50 mM Tris-HCl [pH 7.4], 500 mM NaCl, 5 mM BME) supplemented with 10 mM imidazole and 0.5 mM PMSF. The lysates were precleared for 30 min at 15,000 rpm at 4°C and filtered. LEDGF/p75 was loaded onto a His-Talon column, and the column was washed with LEDGF wash buffer (20 mM Tris-HCl [pH 7.4], 500 mM NaCl). LEDGF/p75 was eluted with LEDGF reservoir buffer supplemented with 300 mM imidazole. LEDGF/p75 was concentrated in LEDGF reservoir buffer using an Amicon Ultra-50K device. F-IN was expressed according to the above-described method and purified by using methods from a previous report (13). The protein concentration was determined using a bicinchoninic acid (BCA) protein assay reagent kit (Thermo Fisher Scientific).
Recombinant HIV-1 clones.
293T cells (1.5 × 105 cells/ml) were seeded onto 6-well tissue culture (TC) plates (Greiner Bio-One) and incubated for 24 h at 37°C. Cells were transfected with 5 μg of pNL4-3 encoding NCINI-3 resistance mutations (A128T, H171Q, K173Q, N254K, P15, P20, and P26) or E11K in the IN region using Attractene transfection reagent (Qiagen). Cells were washed 24 h later, and cell supernatants containing viruses were collected after 48 h of incubation. The HIV-1 clones were measured by using an HIV-1 p24 enzyme-linked immunosorbent assay (ELISA) (Lumipulse G1200; Fujirebio Inc., Japan), normalized to determine the viral concentration, and stored at −80°C.
Western blotting.
293T cells were plated onto 6-well TC plates (1.5 × 105 cells/ml) and incubated for 24 h at 37°C in 5% CO2. Cells were cotransfected with BiFC-IN vectors carrying NCINI-3 resistance mutations using Attractene and incubated for 48 h. Subsequently, the cells were lysed in lysis buffer [20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 7.0), 400 mM NaCl, 10% (wt/vol) sucrose, 0.5 mM dithiothreitol (DTT), 0.5% NP-40, Halt protease inhibitor cocktail (Thermo Fisher Scientific)] (9). After centrifugation for 30 min at 15,000 rpm at 4°C, the samples were titrated using a BCA protein assay kit and stored at −80°C. Wild-type and recombinant HIV-1 clones were filtered, purified by ultracentrifugation (at 35,000 rpm for 30 min) in 15% sucrose–phosphate-buffered saline (PBS), normalized by the p24 levels, and stored in PBS supplemented with the Halt protease inhibitor cocktail at −80°C. The samples were prepared in NuPAGE lithium dodecyl sulfate (LDS) and reducing sample buffer (Thermo Fisher Scientific), separated by SDS-PAGE (4 to 12% Bis-Tris gel; Thermo Fisher Scientific), and transferred to a nitrocellulose membrane. The samples were detected with anti-HIV-1 IN antibody (catalog number ab66645; Abcam), anti-HIV-1 Gag (p55 + p24 + p17) antibody (catalog number ab63917; Abcam), anti-HIV1 p24 (39/5.4A) (catalog number ab9071; Abcam), and secondary mouse or rabbit antibody (MBL Co. Ltd.) and anti-beta-actin antibody (horseradish peroxidase [HRP] conjugated) (Abcam) and then visualized using SuperSignal WestPico chemiluminescent substrates (Thermo Fisher Scientific).
Cross-linking IN proteins with BS3.
Purified IN proteins were diluted at a 300 nM final concentration in a buffer (20 mM HEPES [pH 7.4], 100 mM NaCl, 2 mM DTT, 5 mM MgCl2, and 15 μM ZnCl2). Samples were mixed with BS3 (Thermo Scientific Pierce) at a 0.05 mM final concentration and incubated for 20 min at room temperature (RT), and the reactions were quenched by the addition of mixed LDS and reducing sample buffer. The cross-linked proteins with BS3 were visualized by immunoblotting. In the case of IN in viral particles (15), the highly concentrated wild-type and recombinant HIV-1 clones were titrated, treated with 0.5% Triton X-100 in PBS, and cross-linked with 0.05 mM BS3. The samples employed at 100 ng (p24 level) per well were visualized by immunoblotting with anti-IN antibody and SDS-PAGE (4 to 12% Bis-Tris gel).
Generation of drug-resistant HIV-1 variants.
Selection experiments for resistant HIV-1 variants were performed as previously described (39). In brief, MT-4 cells (1.0 × 105 cells/ml) were exposed to HIV-1NL4-3 (100 50% tissue culture infective doses [TCID50s]) and cultured in the presence of NCINIs, each at an initial concentration of each EC50. Viral replication in MT-4 cells was monitored by p24 levels in the culture medium at intervals of 1 week. The selection procedure was continued until the compound concentration was over 10 μM. Proviral DNA sequences of resistant HIV-1 variants were extracted from infected MT-4 cells using a NucleoSpin tissue kit (TaKaRa). DNA sequences of the IN regions were amplified with 1st primers (5′-AAA TTT AAA TTA CCC ATA CAA AAG GAA ACA TGG GAA GC-3′ and 5′-GGT CTG CTA GGT CAG GGT CTA CTT GTG TGC TAT ATC-3′) followed by 2nd nested primers (5′-AAA AGG AAA AAG TCT ACC TGG CAT GGG TAC CAG CAC AC-3′ and 5′-AGT CCT TAG CTT TCC TTG AAA TAT ACA TAT GG-3′) and then analyzed using IN sequence primers (5′-AAA AGT TAT CTT GGT AGC AGT TCA TGT AGC CAG TGG-3′ and 5′-TT TAG TTT GTA TGT CTG TTG CTA TTA TGT CTA CTA TTC-3′).
Biomolecular fluorescence complementation analysis.
The BiFC-IN system was used as previously described (26). This assay was modified as follows. In brief, HEK293T cells were seeded (2 × 104 cells/well) on collagen I-coated 96-well microplates (BD BioCoat) and incubated for 24 h at 37°C in 5% CO2. The cells were cotransfected with pBiFC-IN vectors (Venus N terminus [VN]-IN and Venus C terminus [VC]-IN) carrying the NCINI-3 resistance mutations using Attractene and incubated for 48 h at 37°C. Next, Venus fluorescence in the cells was measured using a fluorescence-activated cell sorter (FACS) (FACSVerse flow cytometer; BD Bioscience). The area under the curve (AUC) of the histogram emitting from BiFC-IN was measured using ImageJ. Data were compared as AUC ratios to BiFC-INWT.
IN and LEDGF/p75 binding assay.
A pulldown assay was performed to confirm the interaction of F-IN with LEDGF/p75His (20, 24). Briefly, IN (2 μM) and LEDGF/p75 (2 μM) in binding buffer (50 mM HEPES [pH 7.0], 300 mM NaCl, 2 mM MgCl2, 2 mM BME, 5% glycerol, and 0.2% [vol/vol] Nonidet P-40) supplemented with 20 mM imidazole were incubated for 30 min at RT and loaded onto His 60 Ni resin (TaKaRa) after equilibration. The reaction mixture was incubated for 30 min at RT, followed by repeated washing with wash buffer (50 mM HEPES [pH 7.0], 1 M NaCl, 5 mM BME, and 5% glycerol). The samples were eluted with binding buffer supplemented with 300 mM imidazole and visualized by Coomassie brilliant blue staining.
High-performance liquid chromatography (HPLC).
Recombinant INHis proteins were examined using a TSKgel-SWXLG3000 column (Tosoh Corp., Japan) at 1.0 ml/min in running buffer (20 mM HEPES [pH 7.4], 750 mM NaCl, 3.75 mM CHAPS) (10). IN proteins (50 μM) in sample buffer (50 mM HEPES [pH 7.4], 1 M NaCl, 7.5 mM CHAPS, 10 mM MgSO4, 50 μM ZnCl2, 2 mM DTT, 10% glycerol) were subjected to an Agilent Technologies 1220 Infinity liquid chromatography (LC) system (Tosoh) for SEC and detected by measuring the optical density (OD) at 280 nm. Recombinant CCDHis proteins were examined with a Cosmosil 5Diol-120-II column (Nacalai Tesque, Japan) at 1.0 ml/min in running buffer (20 mM HEPES [pH 7.4], 500 mM NaCl, 3.75 mM CHAPS). CCD proteins (50 μM) in sample buffer (50 mM HEPES [pH 7.4], 500 mM NaCl, 7.5 mM CHAPS, 10 mM MgSO4, 5 mM BME, 5% glycerol) were subjected to SEC. The column was calibrated with standard proteins containing bovine gamma globulin (158 kDa), chicken ovalbumin (44 kDa), and horse myoglobin (17 kDa). All the procedures were controlled at 4°C when possible.
Crystallization, X-ray data collection, and structure determination.
The crystallization procedure was performed according to methods in a previous report (14). Briefly, crystallization was performed by the hanging-drop vapor diffusion method using EasyXtal 15-well tools (Qiagen). CCDP15 proteins carrying the A128T, K173Q, and F185K mutations were expressed and purified as described above. The solution buffer was changed to one containing 50 mM morpholineethanesulfonic acid (MES)-NaOH (pH 5.5), 50 mM NaCl, and 5 mM DTT using an Amicon Ultra-10K device (Millipore). The protein concentration was adjusted to 2 mg/ml. The reservoir solution consists of 50 mM sodium cacodylate-HCl (pH 6.5) and 1.2 M ammonium sulfate. The crystals reached 0.2 to 0.4 mm within 1 week at 16°C. The crystals were transferred to a reservoir solution supplemented with 25% glycerol and flash-frozen at 100 K. The X-ray diffraction experiments were carried out on beamlines of Photon Factory (Tsukuba, Japan) and of SPring-8 (Hyogo, Japan). The diffraction data used for the following structure determination were collected on BL17A of Photon Factory and processed with HKL2000 (40). The phases were determined by molecular replacement with MOLREP (41) using the coordinates of CCDWT in complex with the NCINI-related compound (PDB accession number 4GW6) as a search model (22). Structural refinements were carried out using PHENIX (42) and COOT (43). Molecular graphics were prepared using PyMOL version 2.0 (Schrödinger LLC). Data collection and refinement statistics of CCDP15 are listed in Fig. 5D.
DSF.
Recombinant CCDWT, CCDA128T, CCDH171Q, CCDK173Q, CCDP15, and CCDΔP26 proteins, together with F185K (at 12.5, 25, and 50 μM), were prepared in sample buffer (50 mM HEPES [pH 7.4], 500 mM NaCl, 7.5 mM CHAPS, 10 mM MgSO4, 5 mM BME, 5% glycerol). SYPRO orange (Life Technologies) was added to the samples (final concentration of SYPRO orange, 5×) (30). The samples were successively heated from 25°C to 95°C, and increasing fluorescence intensity was detected by using the 7500 fast real-time PCR system (Applied Biosystems). Data are indicated as relative ratios between minimum and maximum intensities of SYPRO orange from 25°C to 95°C detected for each sample.
SPR.
SPR was examined using a Biacore X100 instrument (GE Healthcare) at RT. Recombinant CCD proteins carrying the A128T, H171Q, K173Q, and P15 (A128T/K173Q) mutations together with F185K at 20 μg/ml diluted in HBS-EP buffer (GE Healthcare) were immobilized on two flow cells of a CM5 sensor chip by direct amine coupling to 3,000 response units (RU). NCINI-3 diluted in HBS-EP buffer was flowed with a 2-min injection at 30 μl/min followed by a 2-min dissociation step. KD values of NCINI-3 for the CCD proteins were calculated from the sensorgrams using Biacore X100 control software.
TEM.
Viruses were produced by transfection with pNL4-3 INWT or INP26 into 293T cells. The cells were washed 12 h later, and fresh medium with or without 20 μM NCINI-3 was added. After 48 h of incubation, the samples were fixed in a solution of 4% formaldehyde and 4% glutaraldehyde in 0.1 M phosphate (pH 7.4) buffer for 1 h at 4°C. After centrifugation, the supernatants were filtered through a 0.45-μm filter, and ultracentrifugation was then performed using a model 7780 centrifuge (Kubota, Japan) at 22,000 rpm for 5 h at 4°C. Virus pellets were resolved in a minuscule quantity of 2% glutaraldehyde in 0.1 M phosphate (pH 7.4) buffer and shipped to Tokai Electron Microscopy Inc., Japan. Digital images (3,296 by 2,472 pixels) were taken with a charge-coupled-device camera (EM-14830RUBY2; JEOL Ltd., Tokyo, Japan). Over 100 virus images for each sample were classified as three types by the locations of RNP and the capsid lattice in the viral particles according to previous reports (17, 20).
Alpha.
Alpha 1 (21, 32) was used for the recombinant INHis proteins (10 nM final concentration), which were incubated with various concentrations of synthetic biotinylated TAR RNA (biotin-5′-dT-GGU CUC UCU GGU UAG ACC AGA UCU GAG CCU GGG AGC UCU CUG GCU AAC UAG GGA ACC-3′-biotin-dT) (Integrated DNA Technologies) at 4°C for 3 h in AlphaLISA buffer (PerkinElmer). Anti-His donor and streptavidin acceptor AlphaLISA beads (PerkinElmer) were added at a final concentration of 10 mg/ml to each well and incubated at RT for 3 h. The Alpha signal was detected by using an EnSpire plate reader (PerkinElmer) with an AlphaScreen module setting. KD values were determined by saturation binding curves using one-site-specific binding analysis in GraphPad Prism (44). Alpha 2 was used for the recombinant INHis and F-IN proteins, which were incubated with various concentrations of synthetic TAR RNA (Integrated DNA Technologies) at 4°C for 3 h in AlphaLISA buffer. Anti-His donor and anti-Flag acceptor AlphaLISA beads were added at a final concentration of 10 mg/ml to each well and incubated at RT for 3 h, and the Alpha signal was then detected by using the same method as the one described above. Hill slope and EC50 values were determined using sigmoidal x is log analysis with GraphPad Prism.
HIV-1 replication fitness.
Wild-type or recombinant HIV-1NL4-3 clones carrying E11K or NCINI-3 resistance mutations at 30 ng/ml of p24 were exposed to MT-4 cells (2.4 × 105 cells/ml) in 6-well TC plates for 3 h, the cells were washed and divided into three fractions in fresh medium, and p24 levels in each culture were measured by using an HIV-1 p24 ELISA at 1, 3, 5, and 7 days (39).
Drug susceptibility assay.
Evaluation of EC50 and 50% cytotoxic concentration (CC50) values of drugs and test compounds was performed as previously reported (39).
Compounds.
NCINI-1, -2, and -3 and DRV were synthesized using previously reported synthetic methods (8, 12, 45). RAL, EVG, and DTG were purchased from Selleck Chemicals, and NCINI-4 (BI224436) was obtained from MedChemExpress LLC.
Data availability.
Atomic coordinates and structure factors of CCDP15 were deposited in the Protein Data Bank under accession number 6L0C.
ACKNOWLEDGMENTS
We thank Haruo Aikawa and Hirokazu Tamamura for NCINI synthesis, Sachiko Otsu for technical assistance with the experiments, and Tokai Electron Microscopy Inc. staff for technical advice on TEM sample preparation and data collection. We also thank the beamline staff at Photon Factory and SPring-8 for their help with the data collection.
This work was supported by Japan Society for the Promotion of Science grants JP16K15521, JP18K16180, and JP18K08436 and Development of Novel Drugs for Treating HIV-1 Infection and AIDS.
Tomofumi Nakamura designed and performed almost all the experiments. Teruya Nakamura and Yuriko Yamagata performed X-ray crystallography. Toshikazu Miyakawa and Masao Matsuoka discussed the data and supported the preparation of the manuscript. Hirotomo Nakata supervised, managed the project, and acquired the necessary funding. Tomofumi Nakamura and Hirotomo Nakata wrote and edited the manuscript. All authors read, commented on, and approved the final manuscript.
We declare that we have no conflicts of interest.
REFERENCES
- 1.May MT, Gompels M, Delpech V, Porter K, Orkin C, Kegg S, Hay P, Johnson M, Palfreeman A, Gilson R, Chadwick D, Martin F, Hill T, Walsh J, Post F, Fisher M, Ainsworth J, Jose S, Leen C, Nelson M, Anderson J, Sabin C, UK Collaborative HIV Cohort Study. 2014. Impact on life expectancy of HIV-1 positive individuals of CD4+ cell count and viral load response to antiretroviral therapy. AIDS 28:1193–1202. doi: 10.1097/QAD.0000000000000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antiretroviral Therapy Cohort Collaboration. 2017. Survival of HIV-positive patients starting antiretroviral therapy between 1996 and 2013: a collaborative analysis of cohort studies. Lancet HIV 4:e349–e356. doi: 10.1016/S2352-3018(17)30066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Günthard HF, Calvez V, Paredes R, Pillay D, Shafer RW, Wensing AM, Jacobsen DM, Richman DD. 2019. Human immunodeficiency virus drug resistance: 2018 recommendations of the International Antiviral Society-USA Panel. Clin Infect Dis 68:177–187. doi: 10.1093/cid/ciy463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanters S, Park JJ, Chan K, Socias ME, Ford N, Forrest JI, Thorlund K, Nachega JB, Mills EJ. 2017. Interventions to improve adherence to antiretroviral therapy: a systematic review and network meta-analysis. Lancet HIV 4:e31–e40. doi: 10.1016/S2352-3018(16)30206-5. [DOI] [PubMed] [Google Scholar]
- 5.Flexner C, Tierney C, Gross R, Andrade A, Lalama C, Eshleman S, Aberg J, Sanne I, Parsons T, Kashuba A, Rosenkranz S, Kmack A, Ferguson E, Dehlinger M, Mildvan D, ACTG A5073 Study Team. 2010. Comparison of once-daily versus twice-daily combination antiretroviral therapy in treatment-naive patients: results of AIDS Clinical Trials Group (ACTG) A5073, a 48-week randomized controlled trial. Clin Infect Dis 50:1041–1052. doi: 10.1086/651118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gulick RM, Flexner C. 2019. Long-acting HIV drugs for treatment and prevention. Annu Rev Med 70:137–150. doi: 10.1146/annurev-med-041217-013717. [DOI] [PubMed] [Google Scholar]
- 7.Dowers E, Zamora F, Barakat LA, Ogbuagu O. 2018. Dolutegravir/rilpivirine for the treatment of HIV-1 infection. HIV AIDS (Auckl) 10:215–224. doi: 10.2147/HIV.S157855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, Marchand D, Bardiot D, der Veken N, Remoortel B, Strelkov SV, Maeyer M, Chaltin P, Debyser Z. 2010. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol 6:442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem 278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 10.Hare S, Nunzio DF, Labeja A, Wang J, Engelman A, Cherepanov P. 2009. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog 5:e1000515. doi: 10.1371/journal.ppat.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsiang M, Jones GS, Niedziela-Majka A, Kan E, Lansdon EB, Huang W, Hung M, Samuel D, Novikov N, Xu Y, Mitchell M, Guo H, Babaoglu K, Liu X, Geleziunas R, Sakowicz R. 2012. New class of HIV-1 integrase (IN) inhibitors with a dual mode of action. J Biol Chem 287:21189–21203. doi: 10.1074/jbc.M112.347534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christ F, Shaw S, Demeulemeester J, Desimmie BA, Marchand A, Butler S, Smets W, Chaltin P, Westby M, Debyser Z, Pickford C. 2012. Small-molecule inhibitors of the LEDGF/p75 binding site of integrase block HIV replication and modulate integrase multimerization. Antimicrob Agents Chemother 56:4365–4374. doi: 10.1128/AAC.00717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kessl JJ, Jena N, Koh Y, Taskent-Sezgin H, Slaughter A, Feng L, de Silva S, Wu L, Le Grice SF, Engelman A, Fuchs JR, Kvaratskhelia M. 2012. Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J Biol Chem 287:16801–16811. doi: 10.1074/jbc.M112.354373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouzic LE, Bonnard D, Chasset S, Bruneau JM, Chevreuil F, Strat LF, Nguyen J, Beauvoir R, Amadori C, Brias J, Vomscheid S, Eiler S, Levy N, Delelis O, Deprez E, Saib A, Zamborlini A, Emiliani S, Ruff M, Ledoussal B, Moreau F, Benarous R. 2013. Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage. Retrovirology 10:144. doi: 10.1186/1742-4690-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balakrishnan M, Yant SR, Tsai L, O’Sullivan C, Bam RA, Tsai A, Niedziela-Majka A, Stray KM, Sakowicz R, Cihlar T. 2013. Non-catalytic site HIV-1 integrase inhibitors disrupt core maturation and induce a reverse transcription block in target cells. PLoS One 8:e74163. doi: 10.1371/journal.pone.0074163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenwick C, Amad M, Bailey MD, Bethell R, Bös M, Bonneau P, Cordingley M, Coulombe R, Duan J, Edwards P, Fader LD, Faucher A-M, Garneau M, Jakalian A, Kawai S, Lamorte L, LaPlante S, Luo L, Mason S, Poupart M-A, Rioux N, Schroeder P, Simoneau B, Tremblay S, Tsantrizos Y, Witvrouw M, Yoakim C. 2014. Preclinical profile of BI 224436, a novel HIV-1 non-catalytic-site integrase inhibitor. Antimicrob Agents Chemother 58:3233–3244. doi: 10.1128/AAC.02719-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurado KA, Wang H, Slaughter A, Feng L, Kessl JJ, Koh Y, Wang W, Ballandras-Colas A, Patel PA, Fuchs JR, Kvaratskhelia M, Engelman A. 2013. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc Natl Acad Sci U S A 110:8690–8695. doi: 10.1073/pnas.1300703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amadori C, Velden Y, Bonnard D, Orlov I, Bel N, Rouzic E, Miralles L, Brias J, Chevreuil F, Spehner D, Chasset S, Ledoussal B, Mayr L, Moreau F, García F, Gatell J, Zamborlini A, Emiliani S, Ruff M, Klaholz B, Moog C, Berkhout B, Plana M, Benarous R. 2017. The HIV-1 integrase-LEDGF allosteric inhibitor MUT-A: resistance profile, impairment of virus maturation and infectivity but without influence on RNA packaging or virus immunoreactivity. Retrovirology 14:50. doi: 10.1186/s12977-017-0373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma A, Slaughter A, Jena N, Feng L, Kessl JJ, Fadel HJ, Malani N, Male F, Wu L, Poeschla E, Bushman FD, Fuchs JR, Kvaratskhelia M. 2014. A new class of multimerization selective inhibitors of HIV-1 integrase. PLoS Pathog 10:e1004171. doi: 10.1371/journal.ppat.1004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desimmie BA, Schrijvers R, Demeulemeester J, Borrenberghs D, Weydert C, Thys W, Vets S, Remoortel VB, Hofkens J, Rijck DJ, Hendrix J, Bannert N, Gijsbers R, Christ F, Debyser Z. 2013. LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology 10:57. doi: 10.1186/1742-4690-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessl JJ, Kutluay SB, Townsend D, Rebensburg S, Slaughter A, Larue RC, Shkriabai N, Bakouche N, Fuchs JR, Bieniasz PD, Kvaratskhelia M. 2016. HIV-1 integrase binds the viral RNA genome and is essential during virion morphogenesis. Cell 166:1257–1268.e12. doi: 10.1016/j.cell.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng L, Sharma A, Slaughter A, Jena N, Koh Y, Shkriabai N, Larue RC, Patel PA, Mitsuya H, Kessl JJ, Engelman A, Fuchs JR, Kvaratskhelia M. 2013. The A128T resistance mutation reveals aberrant protein multimerization as the primary mechanism of action of allosteric HIV-1 integrase inhibitors. J Biol Chem 288:15813–15820. doi: 10.1074/jbc.M112.443390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slaughter A, Jurado K, Deng N, Feng L, Kessl J, Shkriabai N, Larue R, Fadel H, Patel P, Jena N, Fuchs J, Poeschla E, Levy R, Engelman A, Kvaratskhelia M. 2014. The mechanism of H171T resistance reveals the importance of Nδ-protonated His171 for the binding of allosteric inhibitor BI-D to HIV-1 integrase. Retrovirology 11:100. doi: 10.1186/s12977-014-0100-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoyte AC, Jamin AV, Koneru PC, Kobe MJ, Larue RC, Fuchs JR, Engelman AN, Kvaratskhelia M. 2017. Resistance to pyridine-based inhibitor KF116 reveals an unexpected role of integrase in HIV-1 Gag-Pol polyprotein proteolytic processing. J Biol Chem 292:19814–19825. doi: 10.1074/jbc.M117.816645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desimmie BA, Weydert C, Schrijvers R, Vets S, Demeulemeester J, Proost P, Paron I, De Rijck J, Mast J, Bannert N, Gijsbers R, Christ F, Debyser Z. 2015. HIV-1 IN/Pol recruits LEDGF/p75 into viral particles. Retrovirology 12:16. doi: 10.1186/s12977-014-0134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura T, Campbell JR, Moore AR, Otsu S, Aikawa H, Tamamura H, Mitsuya H. 2016. Development and validation of a cell-based assay system to assess human immunodeficiency virus type 1 integrase multimerization. J Virol Methods 236:196–206. doi: 10.1016/j.jviromet.2016.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu R, Limón A, Devroe E, Silver PA, Cherepanov P, Engelman A. 2004. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J Virol 78:12735–12746. doi: 10.1128/JVI.78.23.12735-12746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontana J, Jurado KA, Cheng N, Ly NL, Fuchs JR, Gorelick RJ, Engelman AN, Steven AC. 2015. Distribution and redistribution of HIV-1 nucleocapsid protein in immature, mature, and integrase-inhibited virions: a role for integrase in maturation. J Virol 89:9765–9780. doi: 10.1128/JVI.01522-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenkins TM, Hickman AB, Dyda F, Ghirlando R, Davies DR, Craigie R. 1995. Catalytic domain of human immunodeficiency virus type 1 integrase: identification of a soluble mutant by systematic replacement of hydrophobic residues. Proc Natl Acad Sci U S A 92:6057–6061. doi: 10.1073/pnas.92.13.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niesen FH, Berglund H, Vedadi M. 2007. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 31.Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. 1994. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 32.Demeulemeester J, Tintori C, Botta M, Debyser Z, Christ F. 2012. Development of an AlphaScreen-based HIV-1 integrase dimerization assay for discovery of novel allosteric inhibitors. J Biomol Screen 17:618–628. doi: 10.1177/1087057111436343. [DOI] [PubMed] [Google Scholar]
- 33.Ullman EF, Kirakossian H, Singh S, Wu ZP, Irvin BR, Pease JS, Switchenko AC, Irvine JD, Dafforn A, Skold CN. 1994. Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence. Proc Natl Acad Sci U S A 91:5426–5430. doi: 10.1073/pnas.91.12.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Passos DO, Li M, Yang R, Rebensburg SV, Ghirlando R, Jeon Y, Shkriabai N, Kvaratskhelia M, Craigie R, Lyumkis D. 2017. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science 355:89–92. doi: 10.1126/science.aah5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hombrouck A, De Rijck J, Hendrix J, Vandekerckhove L, Voet A, De Maeyer M, Witvrouw M, Engelborghs Y, Christ F, Gijsbers R, Debyser Z. 2007. Virus evolution reveals an exclusive role for LEDGF/p75 in chromosomal tethering of HIV. PLoS Pathog 3:e47. doi: 10.1371/journal.ppat.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engelman A. 1999. In vivo analysis of retroviral integrase structure and function. Adv Virus Res 52:411–426. doi: 10.1016/s0065-3527(08)60309-7. [DOI] [PubMed] [Google Scholar]
- 37.Shkriabai N, Dharmarajan V, Slaughter A, Kessl JJ, Larue RC, Feng L, Fuchs JR, Griffin PR, Kvaratskhelia M. 2014. A critical role of the C-terminal segment for allosteric inhibitor-induced aberrant multimerization of HIV-1 integrase. J Biol Chem 289:26430–26440. doi: 10.1074/jbc.M114.589572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta K, Turkki V, Sherrill-Mix S, Hwang Y, Eilers G, Taylor L, McDanal C, Wang P, Temelkoff D, Nolte RT, Velthuisen E, Jeffrey J, Van Duyne GD, Bushman FD. 2016. Structural basis for inhibitor-induced aggregation of HIV integrase. PLoS Biol 14:e1002584. doi: 10.1371/journal.pbio.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amano M, Koh Y, Das D, Li J, Leschenko S, Wang YF, Boross PI, Weber IT, Ghosh AK, Mitsuya H. 2007. A novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI), GRL-98065, is potent against multiple-PI-resistant human immunodeficiency virus in vitro. Antimicrob Agents Chemother 51:2143–2155. doi: 10.1128/AAC.01413-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.Vagin A, Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr 66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Cassel JA, Blass BE, Reitz AB, Pawlyk AC. 2010. Development of a novel nonradiometric assay for nucleic acid binding to TDP-43 suitable for high-throughput screening using AlphaScreen technology. J Biomol Screen 15:1099–1106. doi: 10.1177/1087057110382778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosh AK, Leshchenko S, Noetzel M. 2004. Stereoselective photochemical 1,3-dioxolane addition to 5-alkoxymethyl-2(5H)-furanone: synthesis of bis-tetrahydrofuranyl ligand for HIV protease inhibitor UIC-94017 (TMC-114). J Org Chem 69:7822–7829. doi: 10.1021/jo049156y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Atomic coordinates and structure factors of CCDP15 were deposited in the Protein Data Bank under accession number 6L0C.