Abstract
Background/Aim:
Esophageal cancer (EC) is a common malignancy with significant morbidity and mortality. As individual cancers exhibit unique mutation patterns, identifying and characterizing gene mutations in EC that may serve as biomarkers might help predict patient outcome and guide treatment. Traditionally, personalized cancer DNA sequencing was impractical and expensive. Recent technological advancements have made targeted DNA sequencing more cost- and time-effective with reliable results. This technology may be useful for clinicians to direct patient treatment.
Materials and Methods:
The Ion PGM and AmpliSeq Cancer Panel was used to identify mutations at 737 hotspot loci of 45 cancer-related genes in 64 EC samples from Chinese patients.
Results:
Frequent mutations were found in TP53 and less frequent mutations in PIK3CA, FBXW7 and KRAS.
Conclusion:
These results demonstrate that targeted sequencing can reliably identify mutations in individual tumors that make this technology a possibility for clinical use.
Keywords: Esophageal cancer, next-generation sequencing, Ion PGM, genetic mutations, targeted therapy, personalized medicine
Esophageal cancer (EC) is the eighth most common cancer and the sixth most common cause of death from cancer worldwide. In 2012, an estimated 456,000 new cases of esophageal cancer and roughly 400,000 esophageal cancer-related deaths were reported globally (1). Of those, China alone accounted for nearly 250,000 cases and close to 200,000 deaths (1). There are two major subtypes of esophageal cancer: adenocarcinoma (EAC) and squamous cell carcinoma (ESCC). EAC is much more common in Western countries where poor diet and obesity are major risk factors, whereas ESCC is more prevalent in Eastern countries and is strongly associated with alcohol consumption and smoking (2, 3). Similar to other cancer types, survival of patients with EC largely depends on disease stage and progression. More than 50% of EC patients have advanced, unresectable disease or present with distant metastases upon diagnosis, with an average of 8 to 10 months overall survival and a dismal 5%-17% 5-year survival rate (4, 5).
While treatment for EC varies with disease stage and subtype (ESCC or EAC), general treatment regimens for resectable tumors utilize broad-acting chemotherapeutic agents like cisplatin and fluorouracil; however, these drugs can have toxic effects, particularly in older patients who might be afflicted by comorbid conditions (6). Targeted therapies, based on DNA sequencing of cancer-associated gene mutations, have become the focus of current research. These targets include specific gene mutations in disrupted signaling pathways, such as those associated with vascular endothelial growth factor (VEGF), epidermal growth factor receptor (EGFR), receptor tyrosine-protein kinase erbB-2 (ERBB2) and others (5). Many of the drugs targeting those mutations have shown promising results with minimal side effects in patients of other cancer types and are currently in clinical trials in EC patients (7–9).
Individual cancer DNA sequencing is also useful to identify gene mutations that may interfere with drug effectiveness. For example, KRAS mutations, which are found in a small percentage of ECs, have been found to confer resistance to EGFR inhibitors, including tyrosine kinase inhibitors and monoclonal antibodies that slow or halt uncontrolled cell growth (10, 11). Hence, the identification of KRAS mutations may spare patients from unnecessary drug toxicity from an EGFR inhibitor rendered ineffective by the mutation. In addition to predicting drug resistance, personalized cancer sequencing may also reveal gene mutations with prognostic value. For example, TP53 mutations, which are found in more than 40% of ECs, have been shown to correspond to poorer patient responses to the neoadjuvant chemotherapeutic agents fluorouracil and cisplatin, and patients with these mutations have reduced overall survival compared to those with wild-type TP53 (12). Effective methods to identify such mutations may help clinicians guide treatment for EC patients.
As new targeted drug therapies are developed, and expanded clinical trials show promising results, the need to easily and reliably detect these mutations and identify new targets is heightened. Traditional Sanger sequencing, next-generation sequencing (NGS) platforms and whole exome sequencing have been used to identify mutations in ECs (13, 14), but these platforms are generally impractical for clinical use due to the high cost and lengthy run times. However, recent technological advancements have brought NGS to the benchtop, making affordable and time-efficient individual genome sequencing possible (15). Specifically, sequencing with the semiconductor-based Ion Personal Genome Machine (PGM) is able to circumvent many of the issues associated with other sequencing methods (16). In the current study, we used the Ion PGM and Ion AmpliSeq Cancer Panel to analyze 737 mutational hotspots from 45 known tumor-suppressor genes and oncogenes to identify genetic mutations in 64 esophageal cancer samples from Chinese patients.
Materials and Methods
Ethics statement.
The study has been approved by the Human Research Ethics Committee of the First Hospital of Qiqihar City, China. The institutional ethics committee waived the need for consent for formalin-fixed, paraffin embedded (FFPE) tumor samples from the tumor tissue bank at the Department of Pathology of the hospital. All samples and medical data used in this study have been irreversibly anonymized.
Patient information.
A total of 64 FFPE tumor samples were collected from the First Hospital of Qiqihar City, China, from esophageal cancer patients. Patients’ characteristics can be found in Table I. For analyses, the patients were further categorized based on gender and tumors categorized by sub-types of esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC).
Table I.
Clinical features of 64 esophageal cancer patients.
| Characteristic | n (%) |
|---|---|
| Age (years) | |
| Median=64.1 | |
| Range=45-85 | |
| Gender | |
| Male | 37 (57.8%) |
| Female | 27 (42.2%) |
| Pathological diagnosis | |
| EAC | 28 (43.8%) |
| ESCC | 35 (54.7%) |
| ND | 1 (1.6%) |
| Differentiation | |
| Low | 3 (4.7%) |
| Middle | 20 (31.3%) |
| High | 0 (0.0%) |
| ND | 41 (64.1%) |
EAC: Esophageal adenocarcinoma; ESCC: esophageal squamous cell carcinoma; ND: not determined.
DNA preparation, Ion Torrent PGM library preparation and sequencing.
Sections of FFPE tissue samples (3-5 μm thick) were deparaffinized in xylene and DNA was then isolated using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA) following manufacturer’s instructions. The Ion AmpliSeq Library Kit 2.0 (Life Technologies, Carlsbad, CA, USA; Part #4475345 Revs. A) was used to construct an adapter-ligated library as per the manufacturer’s protocol, while the Ion PGM 200 Sequencing Kit (Part # 4474004 Revs. B) was used for sequencing reactions according to the recommended protocol and as in our previous publications (17, 18). The AmpliSeq Cancer Panel used for this study is designed to target 737 mutational hotspot loci in the following 45 key cancer-related genes: ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNAS, HNF1A, HRAS, IDH1, JAK3, KDR, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53 and VHL. Detailed methods of Ion Ampliseq Cancer Panel sequencing have been described previously (17).
Variant calling.
The Ion Torrent platform-specific pipeline software Torrent Suite used data from the initial PGM runs to generate sequence reads, trim adapter sequences and filter and remove poor signal profile reads. Then, Torrent Suite Software v3.2, with a plugin “variant caller v3.2” program, was used to generate variant calling from the initial Ion AmpliSeq sequencing data. In order to eliminate base calling errors, several filtering steps were employed to generate final variant calling: the first filter was fixed at an average total coverage depth >100, each variant coverage >20, a variant frequency of each sample >5% and p-value <0.01; the second filter was visually inspecting the mutations using Integrative Genomics Viewer (IGV) software (http//www.broadinstitute.org/igv) or SAMtools software (http://samtools.sourceforge.net), along with eliminating possible DNA strand-specific errors; the third filter was set as variants within 737 hotspots, as per manufacturer’s instructions; and the final filtering step eliminated variants in amplicon AMPL339432 (PIK3CA, exon13, chr3:178938822-178938906), which is not uniquely matched in the human genome.
Bioinformatical and experimental validation.
We used the COSMIC database (19) and MyCancerGenome database (http://www.mycancergenome.org/) to assess reappearing esophageal cancer mutations. Additionally, the accuracy of the Ion PGM was compared to the Sanger sequencing method when sufficient sample DNA was available.
Statistical analysis.
The Fisher’s exact test was used to calculate p-values in the detected mutated genes and total variants using GraphPad QuickCalcs Online Calculator for Scientists (http://www.graphpad.com/quickcalcs). All p-values are two-sided and statistical significance was defined as p<0.05.
Results
Sequence coverage.
For the 64 samples analyzed, the mean read length of each sequence was 78 bp and the average sequence was approximately 24.4 Mb per sample. With reads normalized to 329,000 per specimen, there was an average of 1,788 reads per amplicon (range=44-4,574) (Figure 1A), 181/189 (95.8%) amplicons averaged at least 100 reads and 172/189 (91.0%) amplicons averaged at least 300 reads (Figure 1B).
Figure 1.
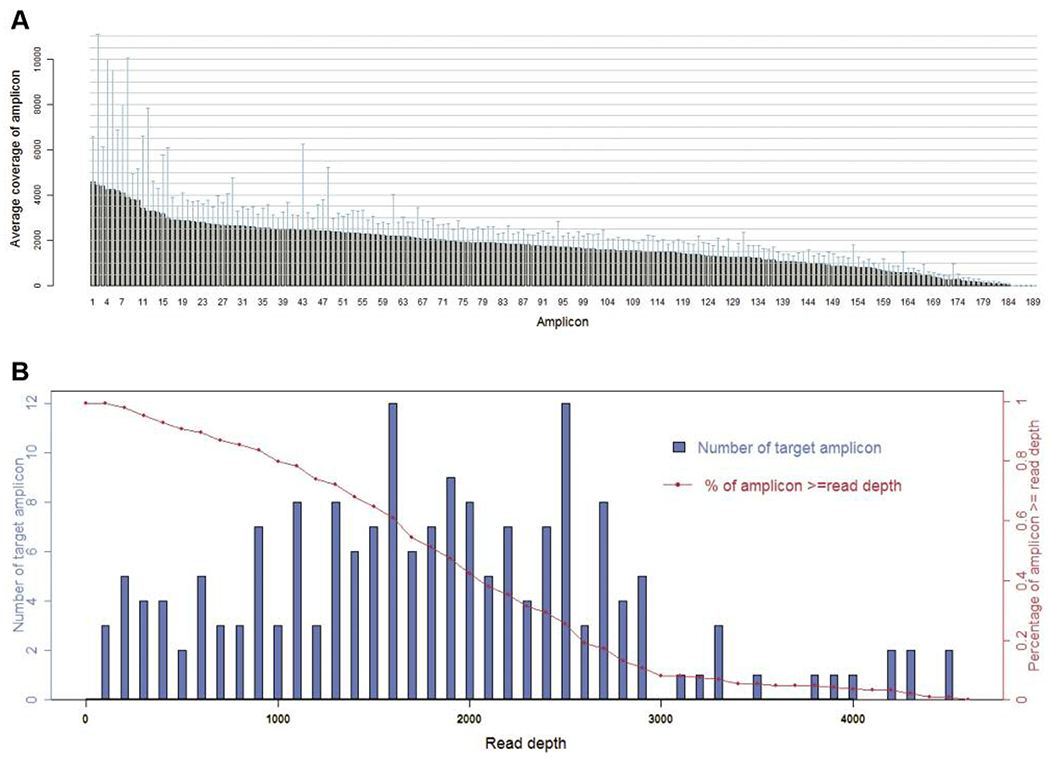
Sequence read distribution across 189 amplicons generated from 64 specimens, normalized to 300,000 reads per sample. A. Average number of reads observed for each amplicon. B. Number of targets with a given read depth, sorted in bins of 100 reads.
TP53, PIK3CA, FBXW7 and KRAS gene mutations.
Sequencing with the Ion PGM revealed that 18 of the 64 (28.1%) esophageal cancers in our sample set had one mutation in various genes and one of these samples contained a combination of three missense mutations. Of the 45 genes sequenced, we detected the highest mutation frequency in TP53 (20.3%) and lower frequencies of mutations were found in PIK3CA (4.7%), FBXW7 (3.1%) and KRAS (1.6%). A detailed list of individual point mutations can be found in Table II. There was no statistical difference between mutation rates in males vs. females (32.4% vs. 22.2%, respectively; p=0.370). A higher mutation rate was found in ESCC samples compared to EAC samples (31.4% vs. 21.4%, respectively), but again this difference was not significant (p=0.374). Figure 2 summarizes the detected mutations based on pathological type of EC and patient sex and age ranges in years.
Table II.
Specific point-mutations detected among 64 esophageal cancer samples.
| Gene | Exon | Mutation | Gender | Age (years) | Pathological diagnosis |
|---|---|---|---|---|---|
| FBXW7 | 8 | p.R465C# | F | 55 | EAC |
| FBXW7 | 9 | p.R505L | M | 58 | ESCC |
| KRAS | 2 | p.G13D# | F | 55 | EAC |
| KRAS | 3 | p.A59T# | F | 55 | EAC |
| PIK3CA | 9 | p.E542K | M | 69 | EAC |
| PIK3CA | 9 | p.E545K | M | 71 | ESCC |
| PIK3CA | 9 | p.E545K | M | 79 | ESCC |
| TP53 | 5 | p.A159V | M | 72 | EAC |
| TP53 | 5 | p.R175H | F | 64 | ESCC |
| TP53 | 5 | p.R175H | F | 70 | ESCC |
| TP53 | 5 | p.C176F | M | 75 | EAC |
| TP53 | 5 | p.C176Y | M | 66 | EAC |
| TP53 | 5 | p.H179R | M | 61 | ESCC |
| TP53 | 7 | p.S241F | M | 49 | ND |
| TP53 | 8 | p.C275Y | M | ND | ESCC |
| TP53 | 8 | p.C275Y | F | 70 | ESCC |
| TP53 | 8 | p.P278S | F | 62 | EAC |
| TP53 | 8 | p.E298* | M | 67 | ESCC |
| TP53 | 10 | p.R342* | M | 65 | ESCC |
| TP53 | 10 | p.R342* | F | 68 | ESCC |
Mutations found within the same sample.
Nonsense mutations resulting in STOP codon; EAC: esophageal adenocarcinoma; ESCC: esophageal squamous cell carcinoma; ND: not determined.
Figure 2.
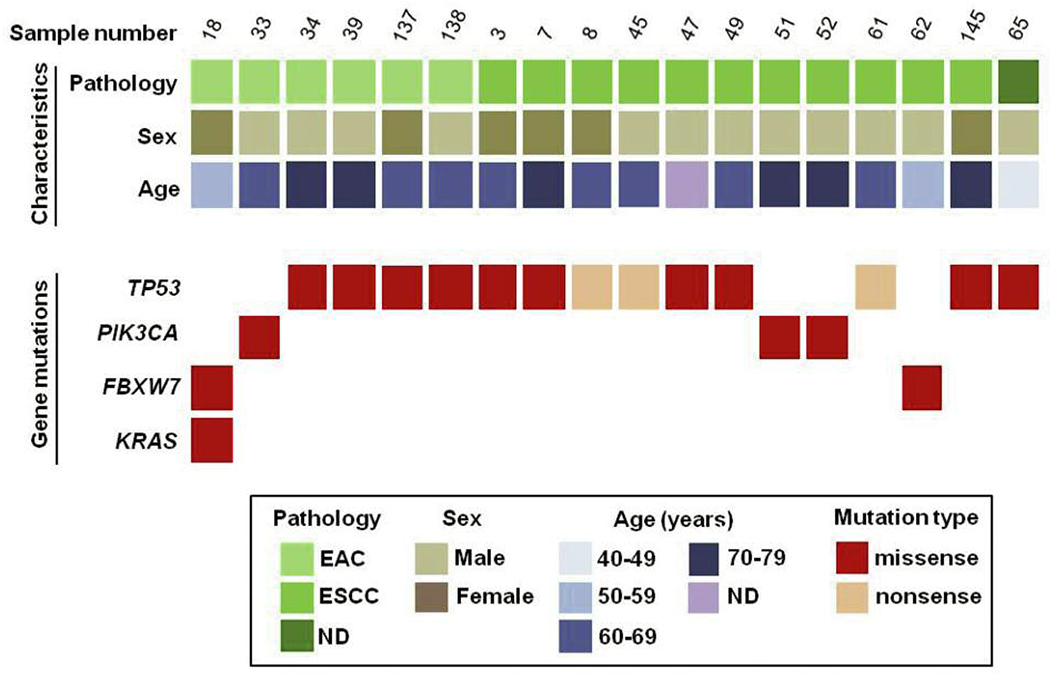
Summary of mutated genes detected in 64 esophageal cancer samples. Eighteen (18) samples harbor mutations in TP53, PIK3CA, FBXW7 and KRAS. Samples are classified by three methods: (i) Pathological type (EAC or ESCC); (ii) sex (M or F); (iii) age (years).
EAC: esophageal adenocarcinoma; ESCC: esophageal squamous cell carcinoma; ND: not determined.
We identified TP53 mutations in 20.3% (13/64) of samples at known hotspot locations in exon 5 (p.A159V, p.R175H, p.C176F, p.C275Y and p.H179R), exon 7 (p.S241F), exon 8 (p.C275Y, p.P278S and p. E298*) and exon 10 (p.R342*). While more TP53 mutations were found in ESCCs vs. EACs (22.9% vs. 14.3%, respectively), this difference was not significant (p=0.523). Additionally, TP53 mutations occurred at roughly equal proportions between males and females (18.5% vs. 21.6%, respectively).
PIK3CA mutations were identified in 3/64 samples (4.7%): one EAC and two ESCC, all from male patients. These were all missense mutations located in exon 9 at the known hotspot residues p.E542K and p.E545K.
Two samples (3.1%) contained a mutation in the FBXW7 gene: one in exon 8 (p.R465C) and one in exon 9 (p.R505L). Interestingly, the EAC sample with the FBXW7 p.A465C mutation also contained two KRAS mutations, one in exon 2 (p.G13D) and the other in exon 3 (p.A59T).
Bioinformatical and experimental validation.
Our detected mutations were compared to those in esophageal cancer from the COSMIC database (19) and MyCancerGenome database; we found that two of our mutations (FBXW7 p.R505L and KRAS p.A59T) have not been previously reported in esophageal cancers. Additionally, the accuracy of the Ion PGM was compared to the Sanger sequencing method for nine samples with sufficient DNA available. All nine samples gave consistent results between the Ion PGM and Sanger sequencing (Table III).
Table III.
Ion PGM versus Sanger sequencing results for 9 esophageal cancer samples.
| # | Sample ID | Pathological diagnosis | Gene | Mutation AA | Ion PGM Variant Frequency (%) | Sanger result Consistent? |
|---|---|---|---|---|---|---|
| 1 | 7 | ESCC | TP53 | p.R175H | 47.01 | YES |
| 2 | 39 | EAC | TP53 | p.A159V | 30.53 | YES |
| 3 | 34 | EAC | TP53 | p.C176F | 39.57 | YES |
| 4 | 3 | ESCC | TP53 | p.R175H | 35.16 | YES |
| 5 | 49 | ESCC | TP53 | p.H179R | 20.20 | YES |
| 6 | 33 | EAC | PIK3CA | p.E542K | 11.14 | YES |
| 7 | 45 | ESCC | TP53 | p.R342* | 66.92 | YES |
| 8 | 47 | ESCC | TP53 | p.C275Y | 12.92 | YES |
| 9 | 8 | ESCC | TP53 | p.R342* | 14.4 | YES |
Nonsense mutations resulting in STOP codon; EAC: esophageal adenocarcinoma; ESCC: esophageal squamous cell carcinoma.
Discussion
In the current study we used the high-throughput Ion PGM and AmpliSeq Cancer Panel to sequence 64 esophageal cancers from Chinese patients by which we identified mutations in TP53, PIK3CA, FBXW7 and KRAS in the sample population. While we did not have access to patient information regarding disease stage, treatments or patient outcome, many of these mutations have already been identified as biomarkers in EC patients (14, 20). Esophageal cancer DNA sequencing has previously been performed with Sanger sequencing and on a variety of NGS platforms (13, 14). We used Sanger sequencing to confirm our mutations when sufficient sample DNA was available, and all of these samples had consistent results between both methods. While the AmpliSeq Cancer Panel may only provide information on a pre-defined set of genes, it is useful for identifying known point mutations associated with disease. Additionally, the Ion PGM has been demonstrated to have greater sensitivity than the Sanger method: the Ion PGM can detect an allele variant frequency of 5%, whereas Sanger sequencing has been shown to miss mutations where the allele variant frequency is less than 10% (21, 22). Of further clinical relevance, the Ion PGM is considerably more cost- and time-effective than other NGS platforms (23).
Of the mutations identified in our study, TP53 was most commonly mutated with 20.3% of samples containing a mutation in this gene. TP53 plays many roles as a tumor suppressor gene and its protein product p53 works in cell cycle regulation, DNA repair, maintaining genomic stability and apoptosis (24–26). Additionally, TP53 mutations significantly impair the regulatory tumor suppressor activity of p53. Although an estimated 80% of TP53 mutations are missense resulting in a stable full-length protein (27), most mutant p53 proteins lose their DNA-binding activity, leading to faulty growth inhibition and apoptotic properties (28). TP53 mutations have been widely studied, as these are some of the most common gene mutations present in greater than 50% of all patients with various types of cancer, and TP53 mutations are specifically present in 36-80% of esophageal cancers (29–31). The TP53 mutation rate found in our sample set was lower than previous reports, which may reflect our relatively small sample size and the tendency for mutation rates to vary greatly depending on the population and geographic location.
TP53 mutations have previously been used as prognostic markers for patient survival in various cancers. In one clinical study, EC patients without TP53 mutations who underwent curative resection survived nearly twice as long as those who had TP53 mutations (30). While this study did not find any correlation between treatment response or patient survival and specific TP53 mutations, other research suggests that different TP53 point mutations may indeed influence the patient outcome or response to treatment. One such clinical study found that patients with TP53 mutations in the zinc-binding domains (L2 and L3, amino acids 163195 and 236-251, respectively (32)) were more resistant to chemotherapy or radiation and had significantly poorer prognoses compared to patients without TP53 mutations or with TP53 mutations outside L2 or L3 (33). Additional studies indicate that L2-L3 mutations are correlated with decreased survival time in patients with breast and colorectal cancer (34, 35). Five of the TP53 mutations identified in our study (p.R175H, p.C176F, p.C176Y, H179R, p.S241F) were found within the L2-L3 zinc binding domain. Knowledge of such mutations may help to better predict a patient’s response to treatment or outcome, thus highlighting the importance of genetic sequencing for each patient.
The PIK3CA gene encodes for the catalytic subunit p110a of class IA phosphatidylinositol 3-kinases (PI3Ks) (36) and mutations in this gene, while common in many cancers, including breast and colon, are only found in roughly 5% of ECs (19, 37). The two PIK3CA mutations we identified at p.E542K and p.E545K are known hotspot mutations in the PIK3CA helical domain that have previously been identified in various cancers (20, 37). These mutations alter interactions with other regulatory proteins like p85 and RAS and elevate lipid kinase activity that leads to an activation of downstream Akt signaling (38), which in turn regulates several signaling pathways controlling, among others, cell survival, proliferation and apoptosis (39, 40). Mutations in PIK3CA may offer valuable prognostic information as recent clinical studies indicate that these mutations are associated with a better prognosis in certain ESCC patients (20, 41). PIK3CA mutations have been found to interfere with anti-EGFR therapy (42) and, as some of these drugs are currently being tested in some EC patients (43, 44), identifying these mutations prior to drug administration may save a number of patients from unnecessary toxicities from treatments rendered ineffective by the mutations.
In addition to the common mutations found in this study, other less frequent mutations in ECs, such as FBXW7 and KRAS, may serve as prognostic markers or have clinical implications in directing patient treatment. FBXW7 is a TP53-dependent tumor suppressor gene that encodes for a subunit of a ubiquitin protein ligase that regulates levels of Cyclin E, Notch and other proteins. Mutations in FBXW7 impair Cyclin E degradation and are associated with decreased genetic stability and impaired growth regulation (45, 46). A recent clinical study found EC patients with low FBXW7 expression to have a significantly poorer overall survival than those with higher expression levels (47). RAS proteins are critical components of signaling pathways that help regulate cell proliferation, differentiation, cell cycle regulation and angiogenesis (48), while mutations in KRAS lead to constitutive activation and impaired regulatory functions (49). While KRAS mutations are uncommon events in ECs that are found in only 2-3% of samples (19, 50), they are nonetheless clinically relevant as KRAS mutations cause resistance to currently used anti-EGFR therapies in various cancers, such as colorectal and lung (51,52).
In conclusion, individualized cancer sequencing may be the next critical step in improving patient treatments and outcomes by guiding therapy for those with disease. Our current study supports the applicability of the Ion PGM and AmpliSeq Cancer Panel to sequence esophageal cancer samples in a clinical setting to potentially provide patient-specific information that could help make personalized medicine a feasible option for cancer patients.
Acknowledgements
The Authors would like to thank Rong Shi at the Wu Jieping Foundation, Haibo Wang, Ying Li and other members of San Valley Biotechnology Inc. Beijing, for their assistance in sample and data collection. We would also like to thank the staff at the First Hospital of Qiqihar and Beijing Military Hospital for their generous support for DNA sequencing and data collection. This research was supported by an institutional grant and grants from the Wu Jieping Medical Foundation and the National Institutes of Health (R01 CA90427 & R01 AI084811 to SYC).
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: GLOBOCAN 2012 v1.0, Cancer incidence and mortality worldwide: IARC CancerBase No. 11 [Internet]. IARC Publ, 2013. [Google Scholar]
- 2.Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y, Ueda J, Wei W, Inoue M and Tanaka H: Epidemiology of esophageal cancer in Japan and China. J Epidemiol 23: 233–242, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McManus DT, Olaru A and Meltzer SJ: Biomarkers of esophageal adenocarcinoma and Barrett’s esophagus. Cancer Res 64: 1561–1569, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Wiedmann MW and Mossner J: New and emerging combination therapies for esophageal cancer. Cancer Manag Res 5: 133–146, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tew WP, Kelsen DP and Ilson DH: Targeted therapies for esophageal cancer. Oncologist 10: 590–601, 2005. [DOI] [PubMed] [Google Scholar]
- 6.Enzinger PC and Mayer RJ: Esophageal cancer. N Engl J Med 349: 2241–2252, 2003. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, Chau I and Van Cutsem E: Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 351: 337–345, 2004. [DOI] [PubMed] [Google Scholar]
- 8.Sugawara S, Oizumi S, Minato K, Harada T, Inoue A, Fujita Y, Maemondo M, Yoshizawa H, Ito K, Gemma A, Nishitsuji M, Harada M, Isobe H, Kinoshita I, Morita S, Kobayashi K, Hagiwara K, Kurihara M and Nukiwa T: Randomized phase II study of concurrent versus sequential alternating gefitinib and chemotherapy in previously untreated non-small cell lung cancer with sensitive EGFR mutations: NEJ005/TCOG0902. Ann Oncol 00: 1–7, 2015. [DOI] [PubMed] [Google Scholar]
- 9.Gefitinib Extends Survival in Some Esophageal Cancers. Cancer Discov 5: OF4–OF4, 2015. [DOI] [PubMed] [Google Scholar]
- 10.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, Bencardino K, Cercek A, Chen C-T, Veronese S, Zanon C, Sartore-Bianchi A, Gambacorta M, Gallicchio M, Vakiani E, Boscaro V, Medico E, Weiser M, Siena S, Di Nicolantonio F, Solit D and Bardelli A: Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486: 532–536, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD and Chang DD: Wild-Type KRAS Is Required for Panitumumab Efficacy in Patients With Metastatic Colorectal Cancer J Clin Oncol 26: 1626–1634, 2008. [DOI] [PubMed] [Google Scholar]
- 12.Kandioler D, Schoppmann SF, Zwrtek R, Kappel S, Wolf B, Mittlböck M, Kührer I, Hejna M, Pluschnig U, Ba-Ssalamah A, Wrba F and Zacherl J: The biomarker TP53 divides patients with neoadjuvantly treated esophageal cancer into 2 subgroups with markedly different outcomes. A p53 Research Group study. J Thorac Cardiovasc Surg 148: 2280–2286, 2014. [DOI] [PubMed] [Google Scholar]
- 13.Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, McKenna A, Carter SL, Cibulskis K, Sivachenko A, Saksena G, Voet D, Ramos AH, Auclair D, Thompson K, Sougnez C, Onofrio RC, Guiducci C, Beroukhim R, Zhou Z, Lin L, Lin J, Reddy R, Chang A, Landrenau R, Pennathur A, Ogino S, Luketich JD, Golub TR, Gabriel SB, Lander ES, Beer DG, Godfrey TE, Getz G and Bass AJ: Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet 45: 478–486, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miura JT, Xiu J, Thomas J, George B, Carron BR, Tsai S, Johnston FM, Turaga KK and Gamblin TC: Tumor profiling of gastric and esophageal carcinoma reveal different treatment options. Cancer Biol Ther 16: 764–769, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J and Pallen MJ: Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30: 434–439, 2012. [DOI] [PubMed] [Google Scholar]
- 16.Hadd AG, Houghton J, Choudhary A, Sah S, Chen L, Marko AC, Sanford T, Buddavarapu K, Krosting J, Garmire L, Wylie D, Shinde R, Beaudenon S, Alexander EK, Mambo E, Adai AT and Latham GJ: Targeted, high-depth, next-generation sequencing of cancer genes in formalin-fixed, paraffin-embedded and fine-needle aspiration tumor specimens. J Mol Diagn 15: 234–247, 2013. [DOI] [PubMed] [Google Scholar]
- 17.Bai X, Zhang E, Ye H, Nandakumar V, Wang Z, Chen L, Tang C, Li J, Li H, Zhang W, Han W, Lou F, Zhang D, Sun H, Dong H, Zhang G, Liu Z, Dong Z, Guo B, Yan H, Yan C, Wang L, Su Z, Li Y, Jones L, Huang XF, Chen SY and Gao J: PIK3CA and TP53 gene mutations in human breast cancer tumors frequently detected by Ion Torrent DNA sequencing. PLoS One 9: e99306, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai X, Sheng J, Tang C, Nandakumar V, Ye H, Ji H, Tang H, Qin Y, Guan H, Lou F, Zhang D, Sun H, Dong H, Zhang G, Liu Z, Dong Z, Guo B, Yan H, Yan C, Wang L, Su Z, Li Y, Jones L, Huang XF, Chen SY, Wu T and Lin H: Frequent mutations in EGFR, KRAS and TP53 genes in human lung cancer tumors detected by Ion Torrent DNA sequencing. PLoS One 9: e95228, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, Kok CY, Jia M, De T, Teague JW, Stratton MR, McDermott U and Campbell PJ: COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43: D805–D811, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou J, Jiang D, Zhang J, Gavine PR, Xu S, Liu Y, Xu C, Huang J, Tan Y, Wang H, Lu Y, Zheng L, Hou Y and Tan L: Frequency, characterization, and prognostic analysis of PIK3CA gene mutations in Chinese esophageal squamous cell carcinoma. Hum Pathol 45: 352–358, 2014. [DOI] [PubMed] [Google Scholar]
- 21.Meldrum C, Doyle MA and Tothill RW: Next-generation sequencing for cancer diagnostics: a practical perspective. Clin Biochem Rev 32: 177–195, 2011. [PMC free article] [PubMed] [Google Scholar]
- 22.Malapelle U, Vigliar E, Sgariglia R, Bellevicine C, Colarossi L, Vitale D, Pallante P and Troncone G: Ion Torrent next-generation sequencing for routine identification of clinically relevant mutations in colorectal cancer patients. J Clin Pathol 68: 64–68, 2015. [DOI] [PubMed] [Google Scholar]
- 23.Glenn TC: Field guide to next-generation DNA sequencers. Mol Ecol Resour 11: 759–769, 2011. [DOI] [PubMed] [Google Scholar]
- 24.Kastan MB, Onyekwere O, Sidransky D, Vogelstein B and Craig RW: Participation of p53 protein in the cellular response to DNA damage. Cancer Res 51: 6304–6311, 1991. [PubMed] [Google Scholar]
- 25.Lane DP: Cancer. p53, guardian of the genome. Nature 358: 15–16, 1992. [DOI] [PubMed] [Google Scholar]
- 26.Levine AJ and Oren M: The first 30 years of p53: growing ever more complex. Nat Rev Cancer 9: 749–758, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edlund K, Larsson O, Ameur A, Bunikis I, Gyllensten U, Leroy B, Sundström M, Micke P, Botling J and Soussi T: Data-driven unbiased curation of the TP53 tumor suppressor gene mutation database and validation by ultradeep sequencing of human tumors. Proc Natl Acad Sci USA 109: 9551–9556, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soussi T and Beroud C: Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer 1: 233–240, 2001. [DOI] [PubMed] [Google Scholar]
- 29.Wang DY, Xiang YY, Tanaka M, Li XR, Li JL, Shen Q, Sugimura H and Kino I: High prevalence of p53 protein overexpression in patients with esophageal cancer in Linxian, China and its relationship to progression and prognosis. Cancer 74: 3089–3096, 1994. [DOI] [PubMed] [Google Scholar]
- 30.Ribeiro U Jr., Finkelstein SD, Safatle-Ribeiro AV, Landreneau RJ, Clarke MR, Bakker, Swalsky PA, Gooding WE and Posner MC: p53 sequence analysis predicts treatment response and outcome of patients with esophageal carcinoma. Cancer 83: 7–18, 1998. [PubMed] [Google Scholar]
- 31.Agrawal N, Jiao Y, Bettegowda C, Hutfless SM, Wang Y, David S, Cheng Y, Twaddell WS, Latt NL, Shin EJ, Wang L-D, Wang L, Yang W, Velculescu VE, Vogelstein B, Papadopoulos N, Kinzler KW and Meltzer SJ: Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov 2: 899–905, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho Y, Gorina S, Jeffrey PD and Pavletich NP: Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 265: 346–355, 1994. [DOI] [PubMed] [Google Scholar]
- 33.Kihara C, Seki T, Furukawa Y, Yamana H, Kimura Y, van Schaardenburgh P, Hirata K and Nakamura Y: Mutations in zinc-binding domains of p53 as a prognostic marker of esophageal-cancer patients. Jpn J Cancer Res 91: 190–198, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Børresen A-L, Andersen TI, Eyfjörd JE, Cornelis RS, Thorlacius S, Borg Å, Johansson U, Theillet C, Scherneck S, Hartman S, Cornelisse CJ, Hovig E and Devilee P: TP53 mutations and breast cancer prognosis: particularly poor survival rates for cases with mutations in the zinc-binding domains. Genes Chromosomes Cancer 14: 71–75, 1995. [DOI] [PubMed] [Google Scholar]
- 35.Børresen-Dale AL, Lothe RA, Meling GI, Hainaut P, Rognum TO and Skovlund E: TP53 and long-term prognosis in colorectal cancer: mutations in the L3 zinc-binding domain predict poor survival. Clin Cancer Res 4: 203–210, 1998. [PubMed] [Google Scholar]
- 36.Fruman DA, Meyers RE and Cantley LC: Phosphoinositide kinases. Annu Rev Biochem 67: 481–507, 1998. [DOI] [PubMed] [Google Scholar]
- 37.Bader AG, Kang S and Vogt PK: Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci USA 103: 1475–1479, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao L and Vogt PK: Class I PI3K in oncogenic cellular transformation. Oncogene 27: 5486–5496, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, Incio J, Digumarthy SR, Pollack SF, Song Y, Muzikansky A, Lifshits E, Roberge S, Coffman EJ, Benes CH, Gómez HL, Baselga J, Arteaga CL, Rivera MN, Dias-Santagata D, Jain RK and Engelman JA: BIM expression in treatment-naïve cancers predicts responsiveness to kinase inhibitors. Cancer Discov 1: 352–365, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang S, Bader AG, Zhao L and Vogt PK: Mutated PI 3-kinases: cancer targets on a silver platter. Cell Cycle 4: 578–581, 2005. [DOI] [PubMed] [Google Scholar]
- 41.Shigaki H, Baba Y, Watanabe M, Murata A, Ishimoto T, Iwatsuki M, Iwagami S, Nosho K and Baba H: PIK3CA mutation is associated with a favorable prognosis among patients with curatively resected esophageal squamous cell carcinoma. Clin Cancer Res 19: 2451–2459, 2013. [DOI] [PubMed] [Google Scholar]
- 42.Sood A, McClain D, Maitra R, Basu-Mallick A, Seetharam R, Kaubisch A, Rajdev L, Mariadason JM, Tanaka K and Goel S: PTEN gene expression and mutations in the PIK3CA gene as predictors of clinical benefit to anti-epidermal growth factor receptor antibody therapy in patients with KRAS wild-type metastatic colorectal cancer. Clin Colorectal Cancer 11: 143–150, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhai Y, Hui Z, Wang J, Zou S, Liang J, Wang X, Lv J, Chen B, Zhu H and Wang L: Concurrent erlotinib and radiotherapy for chemoradiotherapy-intolerant esophageal squamous cell carcinoma patients: results of a pilot study. Dis Esophagus 26: 503–509, 2013. [DOI] [PubMed] [Google Scholar]
- 44.Ayyappan S, Prabhakar D and Sharma N: Epidermal growth factor receptor (EGFR)-targeted therapies in esophagogastric cancer. Anticancer Res 33: 4139–4155, 2013. [PubMed] [Google Scholar]
- 45.Welcker M and Clurman BE: FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 8: 83–93, 2008. [DOI] [PubMed] [Google Scholar]
- 46.Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dofou D, Marth C, Mueller-Holzner E, Corcoran M, Dagnell M, Nejad SZ, Nayer BN, Zali MR, Hansson J, Egyhazi S, Petersson F, Sangfelt P, Nordgren H, Grander D, Reed SI, Widschwendter M, Sangfelt O and Spruck C: FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res 67: 9006–9012, 2007. [DOI] [PubMed] [Google Scholar]
- 47.Kurashige J, Watanabe M, Iwatsuki M, Kinoshita K, Saito S, Hiyoshi Y, Kamohara H, Baba Y, Mimori K and Baba H: Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer 106: 182–188, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adjei AA: Blocking oncogenic Ras signaling for cancer therapy. J Natl Cancer Inst 93: 1062–1074, 2001. [DOI] [PubMed] [Google Scholar]
- 49.Downward J: Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 3: 11–22, 2003. [DOI] [PubMed] [Google Scholar]
- 50.Bettstetter M, Berezowska S, Keller G, Walch A, Feuchtinger A, Slotta-Huspenina J, Feith M, Drecoll E, Höfler H and Langer R: Epidermal growth factor receptor, phosphatidylinositol-3-kinase catalytic subunit/PTEN, and KRAS/NRAS/BRAF in primary resected esophageal adenocarcinomas: loss of PTEN is associated with worse clinical outcome. Human Pathol 44: 829–836. [DOI] [PubMed] [Google Scholar]
- 51.Tao S, Wang S, Moghaddam SJ, Ooi A, Chapman E, Wong PK and Zhang DD: Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer Res 74: 7430–7441, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong NA, Gonzalez D, Salto-Tellez M, Butler R, Diaz-Cano SJ, Ilyas M, Newman W, Shaw E, Taniere P and Walsh SV: RAS testing of colorectal carcinoma – a guidance document from the Association of Clinical Pathologists Molecular Pathology and Diagnostics Group. J Clin Pathol 67: 751–757, 2014. [DOI] [PubMed] [Google Scholar]


