Abstract
Circular RNAs (circRNAs), a new category of noncoding RNA, have emerged in recent years as novel biomolecules with important biological functions. Increasing evidence and reports have revealed that circRNAs play an important role in human carcinogenesis and tumor progression. Gastric cancer (GC) is one of the most prevalent life-threatening malignancies worldwide, and in the present study, a novel circRNA molecule (circRIMS) was shown to be associated GC metastasis using next-generation sequencing. CircRIMS remarkably promoted GC cell metastasis in vitro, functioning as a sponge for hsa-miR-148a-5p and hsa-miR-218-5p. In addition, the results of rescue experiments showed that hsa-miR-148a-5p and hsa-miR-218-5p mimics could reverse the tumor-promoting roles of circRIMS in GC. Thus, circRIMS has potential as an early biomarker for use in predicting invasive metastasis in GC and to guide clinical diagnosis and treatment for precision medicine.
Introduction
Gastric cancer (GC) is one of the most prevalent life-threatening malignancies worldwide, accounting for 1,033,701 new cases and 782,685 deaths worldwide in 2018.1 The 5-year survival rate of GC remains low.2,3 Gastric metastases typically occur in the late stage of malignant disease and frequently indicate short survival.4 Most deaths from GC are caused by metastases. The liver, peritoneum, lung, and bone are most common sites of GC metastasis.5 Therefore, the identification and investigation of novel biomolecules and the mechanisms involved in gastric metastasis remain of great importance.
Circular RNAs (circRNAs) were first identified approximately 30 years ago, and in recent years, these endogenous noncoding RNAs have been recognized as novel biomolecules with important biological functions.6,7 CircRNAs are stable and have the characteristics of a continuous covalently closed loop without a 3′-poly A tail or 5′-cap structure.8 CircRNAs are able to interact with microRNAs,6 proteins9 and so on. In addition, a recent study showed that circRNAs can be translated to generate proteins.10
Increasing evidence has revealed that circRNAs play an important role in human carcinogenesis and tumor progression. For example, the circRNA cSMARCA5, which functions by sponging miR-17-3p and miR-181b-5p to upregulate TIMP3, inhibits growth and metastasis in hepatocellular carcinoma.11 Hsiao et al. observed that the circRNA CCDC66 functions as a microRNA sponge to protect MYC mRNA from attacks by miR-33b and miR-93, thereby promoting colon cancer growth and metastasis.12
In the present study, a novel circRNA molecule (circRIMS) was shown to be related to GC metastasis using next-generation sequencing (NGS). Our data show that circRIMS can act as a tumor promoter in GC metastasis and serve as a valuable marker for GC diagnosis and therapy in the future.
Results and Discussion
CircRNA Expression Profile in Gastric Adenocarcinoma Tissues
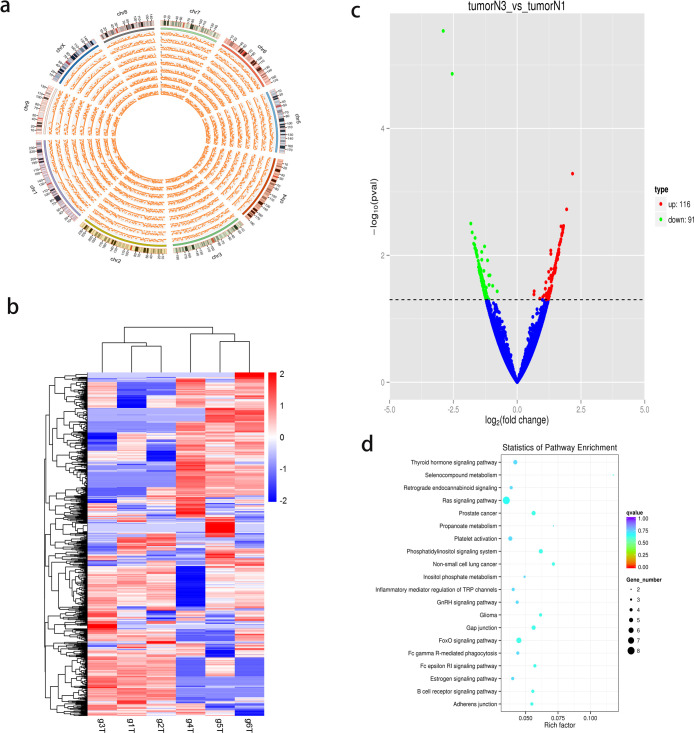
To understand the expression profile of circRNAs in gastric adenocarcinoma tissues, 3 T3N3M0 GC tissues and 3 T3N1M0 GC tissues were used for high-throughput RNA sequencing. According to the globally recognized TNM classification of malignant tumors, T3 means that the tumor has grown into the outer lining of the organ, N1 means 1 or 2 regional lymph nodes has been invaded by the cancer, N3 means more than 7 lymph nodes has been invaded by the cancer, and M0 means the cancer has not spread to other organs. The GC tissues with the TNM status of T3N3M0 and T3N1M0 were collected, and these two groups of cancer tissues only showed the different status in spreading of the regional lymph nodes but not in tumor size or distal spreading. We identified 7048 circRNAs from the intersection of data obtained using the find_circ and CIRI2 tools, 1919 of which were not present in the circBase database (http://www.circbase.org/). All chromosomes in GC cells were observed to encode circRNAs, and the density distribution of circRNAs on chromosomes is shown in Figure 1a. The heat map of the cluster analysis (Figure 1b) shows that all samples from T3N1M0 had the same expression pattern, as well as those from T3N3M0. When comparing the T3N3M0 and T3N1M0 samples, we observed that 116 circRNAs were significantly upregulated and 91 were significantly downregulated (Figure 1c). The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the original genes of all circRNAs is shown in Figure 1d. Some tumor metastasis-related pathways, including the well-known Ras signaling pathway, adherens junction, and FoxO signaling pathway, were enriched. Table 1 shows the 5 circRNAs with the highest differential expression. A novel circRNA molecule, circRIMS, was the most upregulated circRNA between the T3N3M0 and T3N1M0 samples, with P < 0.05.
Figure 1.
Expression profiles of circRNAs in GC. (a) Circos plot showing the density distribution of circRNAs on chromosomes. (b) Cluster heat map displaying the most up and downregulated circRNAs. Each column indicates a sample, while each row indicates an individual circRNA. The red and blue colors represent high and low expression, respectively. All samples from T3N1M0 had the same expression pattern, as well as those from T3N3M0. (c) Volcano plot showing the differences in circRNA expression levels between T3N3M0 and T3N1M0 GC tumor tissues. The red and green dots represent significantly up and downregulated circRNAs, respectively. One hundred 16 circRNAs were significantly upregulated and 91 were significantly downregulated. (d) KEGG pathway enrichment analysis of circRNA-encoding genes. Well-known pathways, including the Ras signaling pathway, adherens junction, and FoxO signaling pathway, were enriched.
Table 1. Five CircRNAs with Highest Rankings for Differential Expression between T3N3M0 and T3N1M0 GC Tumor Tissues.
| IDs | tumorN3_readcount | tumorN1_readcount | log2 fold change | P value |
|---|---|---|---|---|
| circRIMS | 88.68884 | 24.25590323 | 1.3213 | 0.020811 |
| hsa_circ_0002955 | 35.87037 | 11.24297209 | 1.3202 | 0.009505 |
| hsa_circ_0001326 | 84.64352 | 27.56153566 | 1.3167 | 0.00838 |
| hsa_circ_0004365 | 34.14772 | 11.45395604 | 1.2343 | 0.018161 |
| hsa_circ_0079284 | 52.95177 | 13.69650241 | 1.1967 | 0.049898 |
CircRIMS Is a True CircRNA
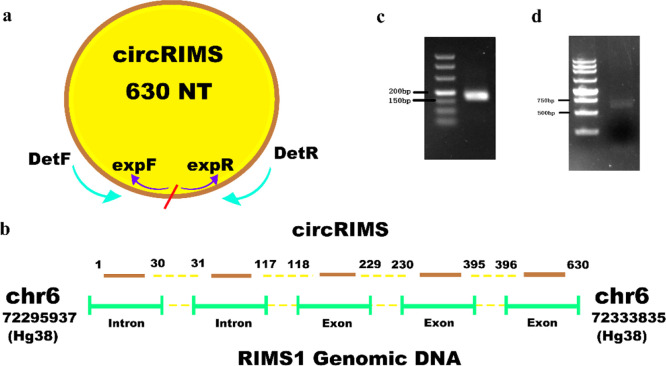
Subsequently, the results of the bioinformatics analysis of the NGS data with respect to the novel circRNA circRIMS were experimentally verified. To this end, two sets of primers were designed to verify the topological structure of circRIMS, as shown in Figure 2a. The convergent primers DetF and DetR face each other across the predicted junction site of circRIMS, while the divergent primers expF and expR face away from each other. Convergent primers only produce a polymerase chain reaction (PCR) product when the RNA has become ligated into a circle. As shown in Figure 2c,d, both sets of primers produced a band of the expected size in the agarose gel. The Sanger sequencing results of these PCR products were consistent with the predicted circRIMS sequence. CircRIMS originates from chr6: 72,295,937–72,333,835 (hg38), which contains 3 exon and 2 intron regions (Figure 2b), where the junction site is located between the first intron and last exon.
Figure 2.
CircRIMS is a true circRNA. (a) Schematic illustration of circRIMS with detection and expression primers. The convergent primers DetF and DetR faced each other across the predicted junction site of circRIMS, while the divergent primers expF and expR faced away from each other. The entire circRIMS sequence comprises 630 nucleotides. (b) CircRIMS transcriptional regions on chromosome 6. The circRIMS originated from chr6: 72,295,937–72,333,835 (hg38), which contains 3 exon and 2 intron regions, and the junction site is located between the first intron and the last exon. (c) Agarose gel of RT-qPCR product using detection primers (DetF and DetR) for circRIMS. The PCR product showed a single band between 150 and 200 bp was visualized on a 3% agarose gel. (d) Agarose gel of RT-qPCR product using expression primers (expF and expR) for circRIMS. The PCR product showed a single band between 500 and 750 bp was visualized on a 1% agarose gel.
CircRIMS Is Upregulated in Metastasis-Prone Clinical Samples
T3N3M0 and T3N1M0 GC samples (30 each) were used to assess the level of circRIMS expression in GC. CircRIMS expression was observed to be significantly correlated with N stage and was upregulated in T3N3M0 samples (Figure 3), consistent with the NGS sequencing results and indicating that it may play an important role in gastric tumor metastasis. An analysis of the correlation between circRIMS expression and patient clinicopathological characteristics (Table 2) demonstrated that circRIMS expression was positively correlated with the presence of lymph node metastasis (P = 0.0008) and clinical stage (P = 0.0008). The remaining pathologic variables, including sex, age, tumor size, lymphovascular invasion, and nerve invasion, exhibited no significant association with circRIMS expression.
Figure 3.
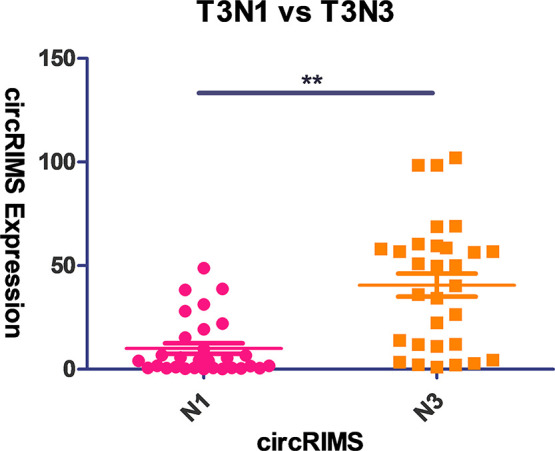
Relative expression of circRIMS in GC tissues as detected by RT-qPCR (n = 60). T3N3M0 and T3N1M0 GC samples (30 each) were used to assess the level of circRIMS expression in GC. CircRIMS expression was observed to be significantly correlated with the N stage and was upregulated in the T3N3M0 samples.
Table 2. Clinicopathological Characteristics of 60 Patients with GC with Respect to the CircRIMS Expression Level.
| circRIMS expression |
||||
|---|---|---|---|---|
| characteristics | total | low | high | P-value |
| Age (Years) | ||||
| <60 | 19 | 10 | 9 | 0.9189 |
| ≥60 | 41 | 21 | 20 | |
| Gender | ||||
| male | 52 | 28 | 24 | 0.6303 |
| female | 8 | 3 | 5 | |
| Tumor Size (cm) | ||||
| <5 | 32 | 17 | 15 | 0.8091 |
| ≥5 | 28 | 14 | 14 | |
| Lymph Node Metastasis | ||||
| N1 | 30 | 22 | 8 | 0.0008 |
| N3 | 30 | 9 | 21 | |
| Clinical Stage | ||||
| I/II | 30 | 22 | 8 | 0.0008 |
| III/IV | 30 | 9 | 21 | |
| Differentiation | ||||
| high/middle | 19 | 12 | 7 | 0.2253 |
| low/undifferentiation | 41 | 19 | 22 | |
| Lauren Classification | ||||
| intestinal | 15 | 7 | 8 | 0.6545 |
| diffuse | 45 | 24 | 21 | |
| Lymphovascular Invasion | ||||
| absent | 33 | 17 | 16 | 0.9793 |
| present | 27 | 14 | 13 | |
| Nerve Invasion | ||||
| absent | 38 | 21 | 17 | 0.4638 |
| present | 22 | 10 | 12 | |
Effect of circRIMS on GC Cell Proliferation
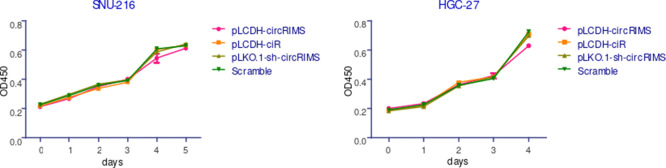
Based on the evidence showing that circRIMS expression is of metastatic relevance in GC, we next investigated the functional role of circRIMS in GC malignant behavior in vitro. We used a lentiviral packaging system to specifically overexpress or knockdown circRIMS in the GC cell lines SNU-216 and HGC-27. Stable circRIMS overexpression or knockdown in these cells was confirmed by realtime quantitative PCR (RT-qPCR) (Figure 4). Interestingly, neither overexpression nor knockdown of circRIMS expression affected the proliferation of HGC-27 or SNU-216 cells, as evaluated by the cell counting kit-8 (CCK-8) assay (Figure 5).
Figure 4.
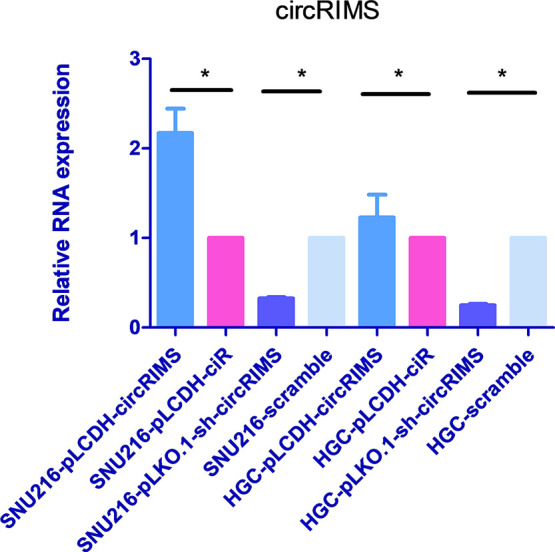
Relative expression of circRIMS in GC cells with stable overexpression or knockdown of circRIMS. Lentiviral packaging systems were used to specifically overexpress or knockdown circRIMS in the GC cell lines SNU-216 and HGC-27. Stable circRIMS overexpression or knockdown in these cells was confirmed by RT-qPCR. The pLCDH-circRIMS sample indicates circRIMS overexpression, with pLCDH-ciR used as a blank control. The pLKO.1-sh-circRIMS sample indicates circRIMS knockdown, with scramble used as a control.
Figure 5.
CircRIMS does not affect GC cell growth in vitro. The CCK-8 assay results showed no significant effect of changes in circRIMS expression on the proliferation rate of HGC-27 or SNU-216 cells. The data are presented as the means ± SD of three independent experiments.
Effect of CircRIMS on GC Cell Migration, Invasion, and Metastatic Potential
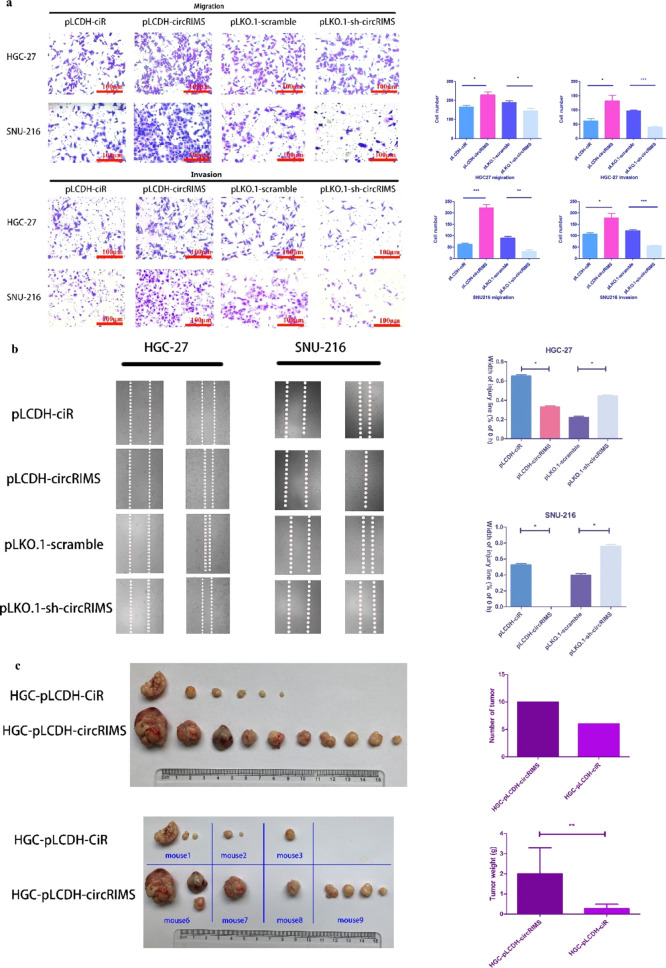
The effect of circRIMS on cell migration was assessed using a Boyden two-chamber assay in which the cells were induced to migrate through a filter by the presence of fetal bovine serum (FBS) on the opposite side. As shown in Figure 6a, circRIMS overexpression enhanced cell migration, whereas circRIMS knockdown suppressed migration. The invasive potential of the GC cells was assessed using a modified Boyden chamber invasion assay, and the results showed that circRIMS overexpression significantly increased the number of cells that invaded through the Matrigel compared to that observed for the control cells. In contrast, circRIMS knockdown in SNU-216 and HGC-27 cells led to a significant decrease in their invasive ability (Figure 6a). A wound-healing/scratch assay was also used to confirm the changes in cell migration, as the extent of wound closure can be used as a direct measure of cell motility. As shown in Figure 6b, the upregulation of circRIMS remarkably accelerated wound closure, whereas the downregulation of circRIMS inhibited wound closure. To determine whether the changes in cell migration and invasion detected in vitro could translate into differences in metastatic potential in vivo, 2 × 106 cells transfected with either pLCDH-ciR-circRIMS-HGC27 or the control pLCDH-ciR-HGC27 were injected into the gastric serosa of NOD/SCID mice. As shown in Figure 6c, 80% of mice in the circRIMS overexpression group exhibited tumor metastasis, while only 60% of mice in the control group exhibited small and few tumor nodules. The numbers and weights of tumor nodules overexpressing circRIMS that formed on the enterocelia of mice were significantly higher than those observed in mice injected with control cells (Figure 6c). These results provide compelling evidence for the important role of circRIMS in gastric carcinoma progression.
Figure 6.
Migration and invasion of stable GC cells. (a) Relative migration and invasion of stable GC cells with circRIMS overexpression or knockdown. CircRIMS overexpression significantly increased the number of cells that invaded through the Transwell pores with or without Matrigel compared with that observed for the control cells. In contrast, circRIMS knockdown in SNU-216 and HGC-27 cells led to a significant decrease in their migratory and invasive abilities. (b) Relative motility as determined by the ability of GC cells with stable circRIMS overexpression or knockdown to close a wound made by creating a scratch through a layer of confluent cells. The upregulation of circRIMS expression remarkably accelerated wound closure, whereas the downregulation of circRIMS expression inhibited wound closure. (c) Enterocelic metastatic burden in mice 8 weeks after the in situ injection of GC cells. The numbers and weights of tumor nodules overexpressing circRIMS that formed on the enterocelia of mice were significantly higher than those observed in mice injected with control cells.
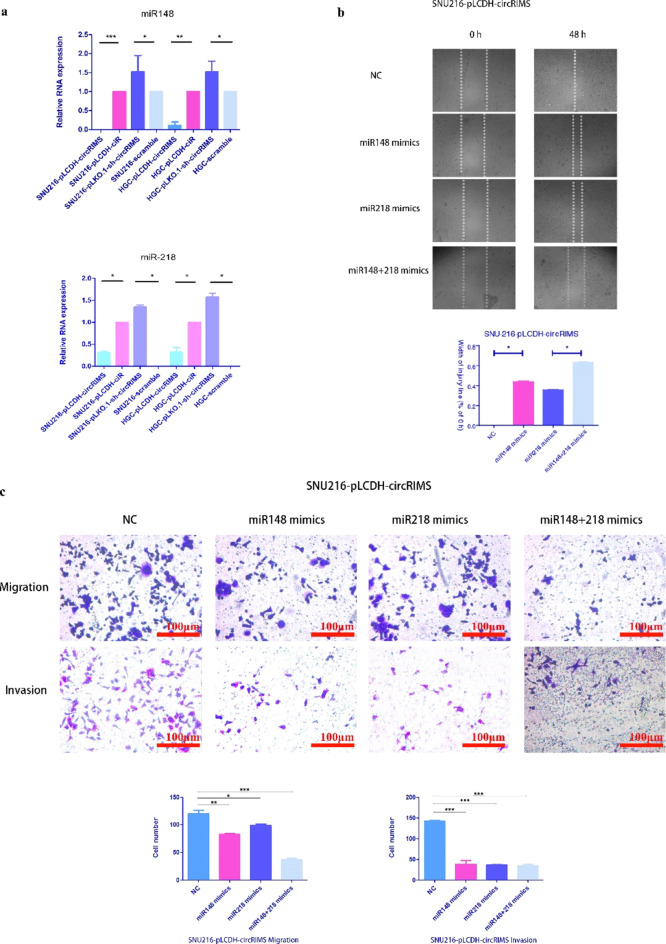
CircRIMS Functions as a Sponge for hsa-miR-148a-5p and hsa-miR-218-5p
To elucidate the molecular mechanism underlying circRIMS activity, we first predicted the potential targets of circRIMS using the microRNA target prediction software miRanda.13,14 A list of the 115 top microRNAs that could be stably absorbed by circRIMS ranked by binding energy is shown in Table S2. As with all GC-related microRNAs that have been reported in the literature,15−19 the intersection of these two data sets was used to obtain hsa-miR-148a-5p and hsa-miR-218-5p, which are well-known tumor-related microRNAs that may serve as downstream molecules and contribute to biological functions. To confirm the relationship between circRIMS and these microRNAs, a microRNA–circRNA capture experiment was performed with total RNA from every stably transduced GC cell line in vitro, and then the remaining RNA was measured using RT-qPCR. As shown in Figure 7a, the residual hsa-miR-148a-5p and hsa-miR-218-5p levels in the cells with high circRIMS expression were less than those observed using the control cells, and vice versa. Subsequently, hsa-miR-148a-5p and hsa-miR-218-5p mimics were transfected into the SNU-216-pLCDH-circRIMS stable cell line to rescue the effect caused by the high expression of circRIMS. As shown in Figure 7c, the results demonstrated that the migration and invasion of cells were inhibited in both the Transwell and wound-healing/scratch assays (Figure 7b). The cotransfection of both mimics resulted in even more pronounced inhibition of tumor cell malignancy compared to that observed with the cotransfection of only one mimic. This result indicates that the effect of circRIMS in GC cells could be reversed by mimics of hsa-miR-148a-5p and hsa-miR-218-5p. Taken together, these results show that hsa-miR-148a-5p and hsa-miR-218-5p are downstream molecules of circRIMS.
Figure 7.
Relative migration and invasion of SNU-216 cells with stable overexpression of circRIMS. (a) Relative expression of miR-148 and miR-218 in each GC cell line with stable circRIMS overexpression or knockdown. The residual hsa-miR-148a-5p and hsa-miR-218-5p levels in the cells with high circRIMS expression were less than those observed in the control cells, and vice versa. (b) Wound closure analysis after creating a scratch through a layer of confluent cells for microRNA rescue experiments. The hsa-miR-148a-5p and hsa-miR-218-5p mimics remarkably inhibited wound closure. (c) Migration ability through a filter toward serum-containing medium in a Boyden chamber assay for microRNA rescue experiments. The hsa-miR-148a-5p and hsa-miR-218-5p mimics were transfected into the SNU-216-pLCDH-circRIMS stable cell line to rescue the effect caused by the high expression of circRIMS successfully.
Discussion
Increasing numbers of circRNAs with multiple biological functions have been discovered in different human tumors in recent years. However, only a few circRNAs have been functionally characterized to date, with the biological functions of most circRNAs remaining unknown. In the present study, through high-throughput RNA sequencing, a circRIMS was identified with significant expression differences between two sets of three GC tissues, the TNM stages of which were T3N3M0 and T3N1M0. CircRIMS was validated by molecular biology experiments to confirm that it is a true circRNA, but the sequence of circRIMS had not yet been reported. We observed that circRIMS expression was markedly upregulated in T3N3M0 GC tissues and significantly correlated with clinical stage in the RNA sequencing data. This result was also confirmed in additional GC tissues using RT-qPCR. Then, in the GC cell model, the functional overexpression of circRIMS in vitro demonstrated that it significantly promoted the metastatic ability of GC cells, while its knockdown using RNA interference showed the opposite effect. These findings confirm that circRIMS is a circRNA molecule that can promote the invasive metastasis of GC cells. Overexpressing or silencing circRIMS did not affect the proliferation of HGC-27 or SNU-216 cells, as evaluated by the CCK-8 assay, emphasizing that the differences in migration and invasion of cells in this study were not caused by differences in cell proliferation. The classic method for in vivo metastasis studies is tail vein injection. However, in the present study, we observed that neither HGC-27 nor SNU-216 cells were able to form tumor nodules by this method, and there are no report in which these two cell lines have been used for tail vein injection. Thus, the alternative in situ injection method was used to perform the in vivo metastasis assay.
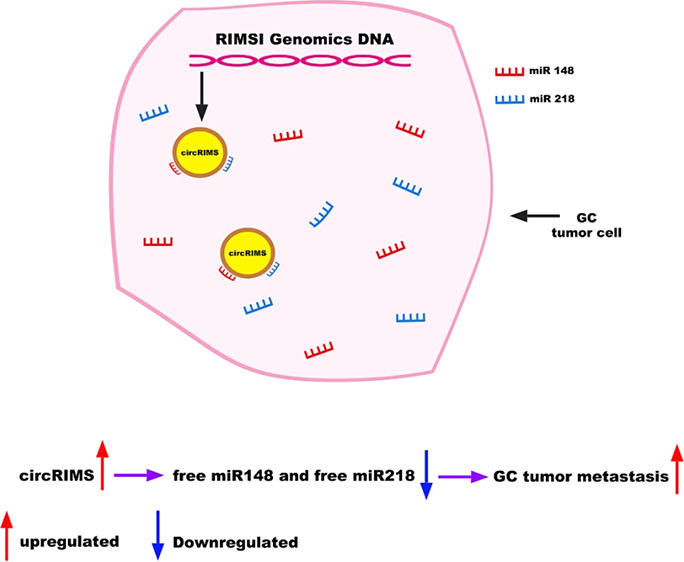
Growing evidence indicates that many circRNAs can serve as microRNA sponges to regulate the expression of downstream microRNA target genes in multiple human cancers. The sponge hypothesis refers to circRNAs with a long half-life that can bind to specific microRNAs through base-pairing and form complexes by hybridization, which causes the free target microRNAs to be deregulated in cells, affecting the expression of downstream genes and proteins regulated by these microRNAs.
In the present study, the circRIMS sequence was assessed through bioinformatic analysis, and the microRNA molecules with the greatest potential to participate in sponge crosstalk with circRIMS were predicted to form the data set α. Then, the data set β was formed by collecting all GC-related microRNA molecules previously reported in the literature. From the intersection of data sets α and β, two microRNAs (hsa-miR-148a-5p and hsa-miR-218-5p) were identified as downstream microRNA targets of circRIMS that were then further studied.
The sponge hypothesis suggests that circRNAs do not have a biological function other than to form a complex with microRNAs. In the process of RNA extraction and microRNA RT-qPCR, the adsorbed microRNA molecules can still be purified and detected. In our study, a reported noncoding RNA capture method was used to remove the circRNA–microRNA complexes in the cell lysate, and the remaining free microRNAs were detected using RT-qPCR.
It is well known that the RNAs with the highest expression levels in human cells are ribosomal RNA (rRNA) and transfer RNA (tRNA), followed by messenger RNA (mRNA). Because the amount of circRNA molecules in human cells is very difficult to detect, circRNAs were scarcely described until 2013. With the development of highly sensitive high-throughput RNA sequencing technology, an increasing number of circRNAs have been identified and have become the focus of investigation.
Both hsa-miR-148a and hsa-miR-218 are key microRNA molecules in oncology research. With respect to the microRNAs hsa-miR-148a and hsa-miR-218 in GC, many previous reports have described the biological functions and mechanisms of these molecules. Both hsa-miR-148a and hsa-miR-218 are anticancer molecules (tumor suppressors) that have a strong inhibitory effect on the development of GC. There are many downstream target genes and proteins of hsa-miR-148a and hsa-miR-218, including PAI-1, VAV2, ITGA5, ITGB8, ROCK1, SMAD2, MMP7, ECOP, and Robo1. These genes and the proteins they produce are regulated by the microRNA molecules hsa-miR-148a and hsa-miR-218.
When circRIMS adsorbs hsa-miR-148a and hsa-miR-218, it leads to the deregulation of these two free microRNAs in human cells. All downstream target genes and proteins regulated by these two microRNAs could also exhibit cascading effects. However, a number of different downstream genes and proteins will be affected by a decrease in the levels of these microRNAs such that the fold change in protein expression relative to each gene itself should exhibit a limited effect. The cell system is very complex, and changes in cellular functions related to circRIMS are certainly the result of the joint action and coregulation of multiple downstream proteins.
The accuracy and sensitivity of western blots for protein quantitation are very limited. In general, western blots require at least twice the amount of protein to obtain bands on film that show differences in brightness and thickness of different samples. Because of the large number of proteins that function downstream hsa-miR-148a and hsa-miR-218, it is difficult to measure the fold change in each protein resulting from the loss of these molecules.
An ideal biomarker for GC should be highly specific, detectable, reliable, and stable. The junction sites of circRNAs are of high specificity that could differ from any other linear mRNA, including mRNA isoforms generated by parental gene, and could be detected by common RT-qPCR, hybridization, or sequencing methods. Compared to linear RNA, circRNAs are resistance to vary types of RNAses and with long half-life period. A reliable biomarker should also be verified in a sufficient number of real specimens. Tumor metastasis involves very complex mechanisms with many biomolecules. Discovery of novel oncogenes is of great significance in deep understand tumor behavior and progress and to improve the puzzle of tumor regulatory network, for farther drug research.
Because of the marked ability of circRIMS to promote the invasion and metastasis of gastric tumor cells, it may be possible to use an inhibitor to knockout circRIMS as a therapy to inhibit the invasion and metastasis of GC. At present, RNA interference technology, as well as CRISPR/Cas13 and other technologies, are rapidly developing and can target RNA molecules in cells to lower the expression of circRIMS. CircRIMS could also be used to predict the possibility of invasive metastasis in GC as an early warning biomarker to guide clinical diagnosis and treatment for precision medicine.
Conclusions
In this study, a novel circRIMS was determined to be involved in GC metastasis to function as a sponge for hsa-miR-148a-5p and hsa-miR-218-5p. This circRNA could be used to predict the possibility of invasive metastasis in GC as an early biomarker to guide clinical diagnosis and treatment for precision medicine.
Methods
Sample Collection
All gastric adenocarcinoma samples were collected during the surgical resection of patients who did not receive preoperative treatment at the Affiliated Hospital of Fujian Medical University. All samples were first pretreated with RNAlater (Invitrogen, USA) according to the manufacturer’s instructions. The use of human tissue samples in this project was approved by the Institutional Review Board and regulatory authorities of Fujian Medical University and written informed consent was obtained from all patients. Clinical and pathological diagnoses were made by the clinical pathology department of the hospital. Clinicopathological classification and staging were determined according to the American Joint Committee on Cancer seventh edition of GC TNM staging.20 The study methodologies conformed to the standards set by the Declaration of Helsinki.
RNA Extraction and Quantitative Reverse Transcription PCR
Total RNA was isolated from cell lines or tissues with TRI Reagent (Ambion, USA) according to the manufacturer’s instructions. RNA was reverse-transcribed using a PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa, Japan) for first-strand complementary DNA synthesis. Quantitative PCR was performed using a SYBR Premix DimerEraser Kit (TaKaRa, Japan). The relative RNA expression level was measured using the classic 2–ΔΔCt method and normalized against endogenous GAPDH levels. Absolute quantification of the circRNA circRIMS in tissues was performed with respect to the standard curve. To generate the standard curve, the PCR product containing the back-splice junction was cloned using the pLB vector system (Tiangen, China). The number of molecules was then calculated by quantifying the PCR product and plotting this number to the serially diluted standard curve.21
CircRNA Sequencing
Five micrograms of total RNA with an RNA integrity number > 8 from each sample was used for library construction. rRNA was removed using a Ribo-Zero Magnetic kit (Epicentre, Madison, WI), and linear RNA was digested with RNase R (Epicentre, Madison, WI) before construction of the RNA-seq libraries. The NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, USA) was used for library construction. High-throughput sequencing was then performed on a HiSeq 4000 system (Illumina, USA) with a 150 bp paired-end run.
Several pipelines have been developed to predict and specifically identify circRNAs, but notable differences between tools and high false-positive rates have been observed.22 In this study, find_circ7 and CIRI223,24 were used to identify circRNAs, the transcripts per million method25 was used for normalization, and the intersection of the results of the two analyses was used for subsequent analyses.
Cell Lines
The human gastric carcinoma cell line HGC-27 was obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), and the SNU-216 cell line26,27 was obtained from the Cobioer Company. The authentication of all cell lines was performed with the standard short tandem repeat method to avoid cross contamination.28,29 The PCR mycoplasma detection set kit (TaKaRa, Japan) was used to monitor and ensure that all cell cultures were free of mycoplasma.
Both cell lines were maintained in RPMI-1640 medium supplemented with 10% FBS, penicillin–streptomycin (HyClone, USA), and mycoplasma removal agent (MPbio, USA) and then incubated under an atmosphere containing 5% CO2 at 37 °C.
Plasmids and Establishment of Stably Transduced GC Cell Lines
To overexpress circRIMS, the full-length cDNA of circRIMS was amplified from the total RNA of tumor tissue and then cloned into the overexpression vector pLCDH-ciR (Geneseed, China), which contains a front and back circular frame, while the control vector contains no circRIMS sequence. To knockdown circRIMS, a short hairpin RNA targeting the back-splice junction site of circRIMS was designed and cloned into the pLKO.1 vector (Addgene, USA) according to the manufacturer’s instructions. The pLKO.1-scramble vector (Addgene, USA) was used as a control.
Every recombinant plasmid was cotransfected with the lentivirus help plasmids pMDL, p-VSV-G, and p-REV into 293T cells in 10 cm culture dishes. The medium was collected after filtration with 0.45 μm filters and added to the culture of GC cells (SNU-216 and HGC-27). After incubation for 48 h, 2 μg·mL–1 puromycin was used to select stably transduced cells for 7–14 days. All oligonucleotides and primers used are listed in Table S1.
Cell Proliferation Assay
Cell proliferation was assessed using the CCK-8 assay (Dojindo, Japan). Cells were seeded at a density of 1000 cells per well into 96-well plates and incubated at 37 °C under an atmosphere with 5% CO2. Ten microliters of CCK-8 solution was added to each well and incubated at 37 °C for 4 h. The absorbance at 450 nm was measured every day using a microplate reader (Tecan, Switzerland).
Wound-Healing Assay
Cells were plated into six-well plates and grown to nearly 90% confluence. The same size scratch was made through the cell monolayer using a 200 μL disposable pipette tip. After washing with PBS, fresh culture medium was added, and the cells were incubated at 37 °C under an atmosphere with 5% CO2. Wound closure was imaged at 0, 24 and 48 h.
Cell Migration and Invasion Assays
For the migration assay, 4 × 104 cells in serum-free media were plated into the upper chamber of a Transwell insert (8 mm pore size; BD Biosciences, San Jose, CA, USA). For the invasion assay, the Transwell insert was coated with Matrigel (BD Biosciences), and 4 × 104 cells were plated onto the top of the coated insert. The medium containing 10% FBS in the lower chamber served as a chemo attractant. After 16 h of incubation at 37 °C in a humidified incubator under an atmosphere with 5% CO2, the cells that did not migrate or invade through the pores in the upper chambers were removed with a cotton swab, and then, the lower surface of the filter was stained with 0.1% crystal violet in 20% methanol for 5 min, imaged, and counted using a Zeiss microscope.
Animal Studies
All work performed with animals was approved by the Institutional Animal Care and Use Committee at Fujian Medical University. For the in situ tumor metastasis study, 10 NOD/SCID mice at the age of 4–5 weeks were randomly divided into two groups. A total of 2 × 106 cells transfected with either pLCDH-ciR-circRIMS-HGC27 or pLCDH-ciR-HGC27 were resuspended in 0.2 mL of buffer composed of PBS and Matrigel at a 1:1 ratio and injected into the gastric serosa of NOD/SCID mice. At 8 weeks post injection, all mice were euthanized, and tumor nodules that formed on the enterocelia were counted and weighed.
In Vitro MicroRNA–CircRNA Capture
Biotin- and spacer 18-labeled circRIMS probe sequences are listed in Table S1. All probes were synthesized by GenScript (Nanjing, China) with high-performance liquid chromatography and mass spectrometry authentication. Probe design and capture were performed according to protocols described by Chu.30 Then, the abundance of circRNA and microRNA was analyzed by RT-qPCR.
Transfection with MicroRNA Mimics
MicroRNA mimics were ordered from GenePharma (Shanghai, China) and dissolved in DEPC-treated water to a final concentration of 20 μM. Lipofectamine 3000 (Invitrogen, USA) was used according to the manufacturer’s instructions to transfect each mimic into the SNU-216-pLCDH-circRIMS stable cell line, which has a high expression level of circRIMS.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5.0 (GraphPad Software, Inc., CA, USA). Student’s t-test was used to determine significant differences in each two-group comparison. All data are presented as the means ± standard deviation (SD) from three independent assays. Differences were considered significant when P < 0.05.
Acknowledgments
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study was approved by the Bioethics Committee of the Fujian Medical University.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c02991.
Author Contributions
J.L. and X.L. conceptualized the study, designed the experiments, and wrote the manuscript. Y.Z. acquired, analyzed, and interpreted the data. X.Z. and C.X. analyzed and interpreted the data. All authors read and approved the final manuscript.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
The authors declare no competing financial interest.
Supplementary Material
References
- Bray F.; Ferlay J.; Soerjomataram I.; Siegel R. L.; Torre L. A.; Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-Cancer J. Clin. 2018, 68, 394–424. 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Karimi P.; Islami F.; Anandasabapathy S.; Freedman N. D.; Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol., Biomarkers Prev. 2014, 23, 700–713. 10.1158/1055-9965.epi-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehdev A.; Catenacci D. V. T. Gastroesophageal cancer: focus on epidemiology, classification, and staging. Discov. Med. 2013, 16, 103–111. [PubMed] [Google Scholar]
- Weigt J.; Malfertheiner P. Metastatic Disease in the Stomach. Gastrointest. Tumors 2015, 2, 61–64. 10.1159/000431304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riihimäki M.; Hemminki A.; Sundquist K.; Sundquist J.; Hemminki K. Metastatic spread in patients with gastric cancer. Oncotarget 2016, 7, 52307–52316. 10.18632/oncotarget.10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T. B.; Jensen T. I.; Clausen B. H.; Bramsen J. B.; Finsen B.; Damgaard C. K.; Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Memczak S.; Jens M.; Elefsinioti A.; Torti F.; Krueger J.; Rybak A.; Maier L.; Mackowiak S. D.; Gregersen L. H.; Munschauer M.; Loewer A.; Ziebold U.; Landthaler M.; Kocks C.; le Noble F.; Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Jeck W. R.; Sharpless N. E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt L. M.; Stahringer A.; Sass K.; Pichler G.; Kulak N. A.; Wilfert W.; Kohlmaier A.; Herbst A.; Northoff B. H.; Nicolaou A.; Gabel G.; Beutner F.; Scholz M.; Thiery J.; Musunuru K.; Krohn K.; Mann M.; Teupser D. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Fan X.; Mao M.; Song X.; Wu P.; Zhang Y.; Jin Y.; Yang Y.; Chen L.-L.; Wang Y.; Wong C. C.; Xiao X.; Wang Z. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.; Xu Q.-g.; Wang Z.-g.; Yang Y.; Zhang L.; Ma J.-z.; Sun S.-h.; Yang F.; Zhou W.-p. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 2018, 68, 1214–1227. 10.1016/j.jhep.2018.01.012. [DOI] [PubMed] [Google Scholar]
- Hsiao K.-Y.; Lin Y.-C.; Gupta S. K.; Chang N.; Yen L.; Sun H. S.; Tsai S.-J. Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer Res. 2017, 77, 2339–2350. 10.1158/0008-5472.can-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D.; Wilson M.; Gabow A.; Marks D. S.; Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2007, 36, D149–D153. 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B.; Enright A. J.; Aravin A.; Tuschl T.; Sander C.; Marks D. S. Human MicroRNA targets. PLoS Biol. 2004, 2, e363 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Li Z.; Li Y.; Zang A. MicroRNA and signaling pathways in gastric cancer. Cancer Gene Ther. 2014, 21, 305–316. 10.1038/cgt.2014.37. [DOI] [PubMed] [Google Scholar]
- Tie J.; Pan Y.; Zhao L.; Wu K.; Liu J.; Sun S.; Guo X.; Wang B.; Gang Y.; Zhang Y.; Li Q.; Qiao T.; Zhao Q.; Nie Y.; Fan D. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet. 2010, 6, e1000879 10.1371/journal.pgen.1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Li Y. miR-148a downregulates the expression of transforming growth factor-β2 and SMAD2 in gastric cancer. Int. J. Oncol. 2016, 48, 1877–1885. 10.3892/ijo.2016.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J.; Guo X.; Yan J.; Deng K. The role of miR-148a in gastric cancer. J. Cancer Res. Clin. Oncol. 2014, 140, 1451–1456. 10.1007/s00432-014-1649-8. [DOI] [PubMed] [Google Scholar]
- Wang S.-H.; Li X.; Zhou L.-S.; Cao Z.-W.; Shi C.; Zhou C.-Z.; Wen Y.-G.; Shen Y.; Li J.-K. microRNA-148a suppresses human gastric cancer cell metastasis by reversing epithelial-to-mesenchymal transition. Tumour Biol. 2013, 34, 3705–3712. 10.1007/s13277-013-0954-1. [DOI] [PubMed] [Google Scholar]
- Wittekind C. TNM-System 2010. Pathologe 2010, 31, 331–332. 10.1007/s00292-010-1349-3. [DOI] [PubMed] [Google Scholar]
- Guarnerio J.; Bezzi M.; Jeong J. C.; Paffenholz S. V.; Berry K.; Naldini M. M.; Lo-Coco F.; Tay Y.; Beck A. H.; Pandolfi P. P. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016, 165, 289–302. 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- Hansen T. B.; Venø M. T.; Damgaard C. K.; Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016, 44, e58 10.1093/nar/gkv1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Zhang J.; Zhao F. Circular RNA identification based on multiple seed matching. Briefings Bioinf. 2018, 19, 803–810. 10.1093/bib/bbx014. [DOI] [PubMed] [Google Scholar]
- Gao Y.; Wang J.; Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L.; Chen J.; Li Z.; Li X.; Hu X.; Huang Y.; Zhao X.; Liang C.; Wang Y.; Sun L.; Shi M.; Xu X.; Shen F.; Chen M.; Han Z.; Peng Z.; Zhai Q.; Chen J.; Zhang Z.; Yang R.; Ye J.; Guan Z.; Yang H.; Gui Y.; Wang J.; Cai Z.; Zhang X. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS One 2010, 5, e15224 10.1371/journal.pone.0015224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku J.-L.; Park J.-G. Biology of SNU cell lines. Cancer Res. Treat. 2005, 37, 1–19. 10.4143/crt.2005.37.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-G.; Yang H.-K.; Kim W. H.; Chung J.-K.; Kang M.-S.; Lee J.-H.; Oh J. H.; Park H.-S.; Yeo K.-S.; Kang S. H.; Song S.-Y.; Kang Y. K.; Bang Y.-J.; Kim Y. I.; Kim J.-P. Establishment and characterization of human gastric carcinoma cell lines. Int. J. Cancer 1997, 70, 443–449. . [DOI] [PubMed] [Google Scholar]
- Yu M.; Selvaraj S. K.; Liang-Chu M. M. Y.; Aghajani S.; Busse M.; Yuan J.; Lee G.; Peale F.; Klijn C.; Bourgon R.; Kaminker J. S.; Neve R. M. A resource for cell line authentication, annotation and quality control. Nature 2015, 520, 307–311. 10.1038/nature14397. [DOI] [PubMed] [Google Scholar]
- Lin J.; Chen L.; Jiang W.; Zhang H.; Shi Y.; Cai W. Rapid detection of low-level HeLa cell contamination in cell culture using nested PCR. J. Cell. Mol. Med. 2019, 23, 227–236. 10.1111/jcmm.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.; Qu K.; Zhong F. L.; Artandi S. E.; Chang H. Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.