Abstract
RAS proteins are commonly mutated in cancerous tumors, but germline RAS mutations are also found in RASopathy syndromes such as Noonan syndrome (NS) and cardiofaciocutaneous (CFC) syndrome. Activating RAS mutations can be subclassified based on their activating mechanisms. Understanding the structural basis for these mechanisms may provide clues for how to manage associated health conditions. We determined high-resolution X-ray structures of the RASopathy mutant KRASP34R seen in NS and CFCS. GTP and GDP-bound KRASP34R crystallized in multiple forms, with each lattice consisting of multiple protein conformations. In all GTP-bound conformations, the switch regions are not compatible with GAP binding, suggesting a structural mechanism for the GAP insensitivity of this RAS mutant. However, GTP-bound conformations are compatible with intrinsic nucleotide hydrolysis, including one that places R34 in a position analogous to the GAP arginine finger or intrinsic arginine finger found in heterotrimeric G proteins, which may support intrinsic GTP hydrolysis. We also note that the affinity between KRASP34R and RAF-RBD is decreased, suggesting another possible mechanism for dampening of RAS signaling. These results may provide a foothold for development of new mutation-specific strategies to address KRASP34R-driven diseases.
Keywords: CFC syndrome, GAP-insensitive, GTPase, Noonan syndrome, RAS
1 ∣. INTRODUCTION
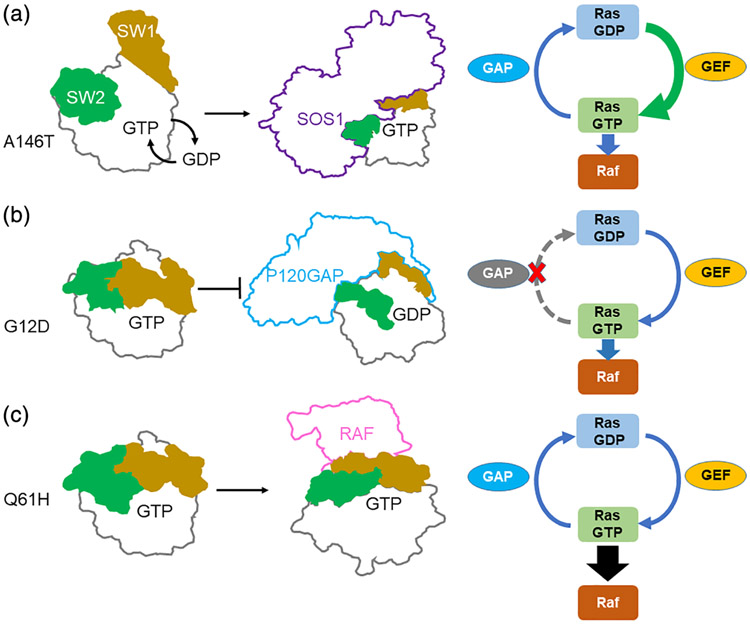
RAS mutations occur both in cancer and developmental disorders. Generally, such mutations lead to overactive signaling downstream of RAS, but the underlying mechanisms of RAS activation can vary by mutation (Hunter et al., 2015). RAS is a molecular switch, cycling between active, GTP-bound and inactive, GDP-bound states (Fernández-Medarde & Santos, 2011). Guanosine nucleotide exchange factors (GEFs) assist in nucleotide exchange, while GTPase activating proteins (GAPs) catalyze GTP hydrolysis (Bos, Rehmann, & Wittinghofer, 2007). Certain RAS mutations prevent GAP-mediated regulatory hydrolysis (Donovan, Shannon, & Bollag, 2002; Vetter & Wittinghofer, 2001), while others increase GEF-dependent nucleotide exchange (Bera et al., 2019; Poulin et al., 2019a; Tartaglia et al., 2007). Still others appear to enhance the ability of RAS to interact with downstream signaling proteins such as those found in the RAF and MEK families (Hunter et al., 2015; Tidyman & Rauen, 2008, 2009; Figure 1). Understanding these mechanistic differences may lead to new approaches for targeting specific forms of mutant RAS, as highlighted by recent breakthroughs in the development of covalent inhibitors of KRASG12C (Canon et al., 2019; Fell et al., 2018; Janes et al., 2018).
FIGURE 1.
Schematic diagram of activation subclasses of RAS mutations. Switch 1 (SW1) in yellow and Switch 2 (SW2) in green. (a) KRASA146T exemplifies rapid nucleotide exchange mutants where RAS activity is elevated by increased efficiency of nucleotide exchange. (b) KRASG12D exemplifies GAP insensitive mutants where GAP-mediated regulatory hydrolysis is impaired. (c) KRASQ61H exemplifies RAS effector sequestration mutants where the affinity of KRAS for RAF kinase is enhanced
RAS mutations generally impact the function of two mobile structural elements in RAS, Switch 1 (SW1) and Switch 2 (SW2), which regulate many of the biological functions of RAS (Figures 1 and 2a). SW1 is involved in the intrinsic GTP hydrolysis mechanism and forms a major interface for interactions with downstream effectors such as RAF and PI3K, while SW2 is primarily involved in GTP hydrolysis (Buhrman, Holzapfel, Fetics, & Mattos, 2010; Scheffzek, Lautwein, Kabsch, Ahmadian, & Wittinghofer, 1996; Vetter & Wittinghofer, 2001). Disease-associated mutations cause changes in the biochemical behavior of RAS by altering SW1 and SW2 dynamics (Hunter et al., 2015; Lu, Bera, Gondi, & Westover, 2018). Understanding mutation-associated structural changes in SW1 and SW2 behavior may aid in understanding the mechanisms that drive biochemical and disease phenotypes.
FIGURE 2.
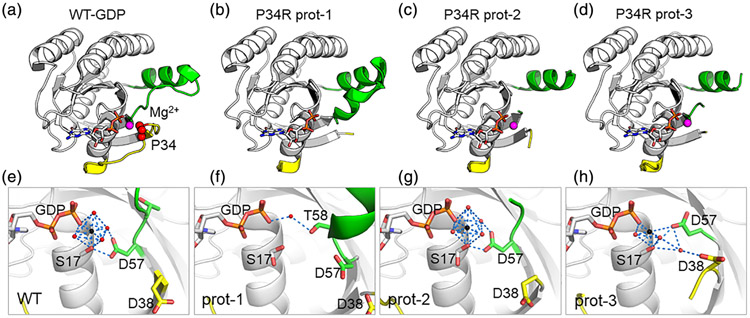
Structures of GDP-KRASP34R. SW1 in yellow, SW2 in green, nucleotide in white sticks. For top row, Mg2+ in magenta; for bottom row, Mg2+ as black spheres and waters as red spheres. Blue dashes show selected interactions. (a) GDP-KRASWT (PDB 4OBE), shown for comparison. Pro34 highlighted by red spheres. (b) GDP-KRASP34R Prot-1 shows disordered SW1 and loss of Mg2+. (c, d) Prot-2 and Prot-3 show disordered SW1 and SW2 but Mg2+ is present. (f) Magnification of Prot-1 shows loss of Mg2+. Asp57 is shifted away from the nucleotide gamma phosphate, and Thr58 rotates toward the gamma phosphate to make secondary interactions with the gamma phosphate through a bridging water. (g) Magnification of Prot-2 shows that Asp57 and Ser17 provide primary or secondary coordination for the octahedral coordination state of Mg2+ (h) Magnification of Prot-3 shows breakdown of the octahedral coordination due to movement of Asp57
Developmental disorders with overactive RAS pathways are collectively known as RASopathy syndromes. KRASP34R occurs in multiple RASopathy syndromes including Noonan Syndrome (NS), an autosomal dominant disorder with relatively high incidence at ~1 in 1000–2,500 (Romano et al., 2010) and Cardiofaciocutaneous Syndrome (CFCS), which is less common (Schubbert et al., 2006). NS patients experience a spectrum of phenotypes that are expressed to varying degrees, including congenital heart defects, lymphatic dysplasia, short stature, coagulation defects, ocular abnormalities, and developmental delay (Romano et al., 2010). NS has also been linked to an increased risk of leukemia and neuroblastoma (Tartaglia, Gelb, & Zenker, 2011). CFC syndrome is characterized by craniofacial dysmorphisms, short stature, and a variable degree of intellectual disability (Kavamura, Peres, Alchorne, & Brunoni, 2002). For both conditions, the standard of care is symptomatic treatment, with no direct therapy available.
KRASP34R, which has an amino acid substitution in the middle of Switch 1 (Figure 2a), was previously shown to exhibit normal rates of intrinsic GTP hydrolysis. However, it is profoundly insensitive to GTPase stimulation by both P120GAP and neurofibromin, two of the common RAS-GAPs (Gremer et al., 2011; Schubbert et al., 2007). Cells expressing KRASP34R under basal growth conditions show elevated levels of GTP-bound RAS and elevated markers of downstream RAS pathways such as pMEK, pERK, pAkt, and pS6 (Schubbert et al., 2007). The structural mechanisms driving these properties have not been reported. We solved crystal structures of KRASP34R both in the inactive (GDP-bound) and active (GTP-bound) states. Both crystallized in lattices consisting of three different protein conformations, illustrating dynamic states of KRASP34R. We use these data to understand the KRASP34R phenotype.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Expression and purification
A KRASP34R expression construct comprising residues 1–169 fused to an N-terminal 6-His tag and TEV cleavage sequence was made by site-directed mutagenesis starting from a human WT V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) bacterial expression construct (Hunter et al., 2015). This was transformed into BL21 (DE3) cells. Cells were grown in Luria broth (LB) to OD600 0.9 and induced with 0.5 M isopropyl β-d-1-thiogalactopyranoside (IPTG) for 16 hr at 16°C. Cells were pelleted and resuspended in lysis buffer (20 mM sodium phosphate [pH 8.0], 500 mM NaCl, 10 mM imidazole, 1 mM 2-mercaptoethanol [BME], 5% [vol/vol] glycerol) containing PMSF, benzamidine, and 1 mg/ml lysozyme. Lysates were flash-frozen and stored at −80° C until use. Protein was purified over an IMAC cartridge (BioRad) and exchanged into 20 mM HEPES (pH 8.0), 150 mM NaCl, 5 mM MgCl2, 0.5 mM DTT buffer using a desalting column. The N-terminal His tag was cleaved by 24 hr digestion with a 1:10 ratio of TEV protease at 4°C, and the TEV and Tag were removed by reverse purification over an IMAC cartridge. Protein was concentrated to 30 mg/ml in a 10 kDa cutoff Amicon filter (Millipore), aliquoted, and then flash-frozen and stored under liquid nitrogen until use. Yields were ~8 mg of purified P34R mutant KRAS per liter of culture.
2.2 ∣. Nucleotide exchange (GNP for GDP)
We used an optimized nucleotide exchange protocol based on methods reported previously wherein EDTA is used to chelate ionic Mg2+, liberating bound nucleotides (Lu et al., 2018). We have been able to obtain KRAS preparations that are >95% GTP-analogue bound as measured by an HPLC.
2.3 ∣. Crystallization and structure determination
Both GDP- and GNP-bound KRASP34R crystals were obtained by hanging drop vapor diffusion using 100 nl KRASP34R and 100 nl crystallization solution by a Mosquito crystallization robot (TTPLabtech) over reservoirs containing 100 μl precipitant solution. The crystals were obtained in 1.8 M Na/K-phosphates at pH 8.2 at 20°C. Crystals were further optimized in 24-well hanging drop vapor diffusion plates and reached a size approximately 100–200 μm within 2 weeks at 20°C. X-ray diffraction data were collected at beamline 19-ID of the Advanced Photon Source. Crystallographic data sets were processed, integrated, and scaled using HKL-3000 packages (HKL Research Inc.; Otwinowski & Minor, 1997). Molecular replacement was performed using Phaser in CCP4 software suite (Winn et al., 2011) with 4OBE (KRASWT:GDP structure) as the initial search model. The atomic model was constructed using Coot (Emsley, Lohkamp, Scott, & Cowtan, 2010) and improved through iterative cycles of refinement using PHENIX (Adams et al., 2010). Model validation was performed with MolProbity (Williams et al., 2018). Final structures were submitted to the Protein Data Bank with ID code 6MS9 (KRASP34R-GDP), 6MTA (KRASP34R-GNP in C2), 6O36 (KRASP34R-GNP in P21), and 6O46 (KRASP34R-GNP in P63). Data collection and refinement statistics of the final structures are listed in Table 1. Molecular graphics figures were prepared using PyMOL (DeLano, 2002), version 1.5.0.4 (Schrödinger, LLC).
TABLE 1.
X-ray crystallographic data collection and refinement statistics
| KRASP34R-GNP (6O36) (6MTA) (6O46) | KRASP34R-GDP (6MS9) | |||
|---|---|---|---|---|
| Space group | P21 | C2 | P63 | C2 |
| Cell parameters, a, b, c (Å) | 95.12, 35.11, 113.42 | 67.32, 83.57, 84.84 | 188.37, 188.37, 34.92 | 67.82, 84.40, 88.17 |
| α, β, γ (°) | 90.00, 111.00, 90.00 | 90.00, 109.00, 90.00 | 90.00, 90.00, 120.00 | 90.00, 110.48, 90.00 |
| Resolution (highest shell) (Å) | 50.00–2.00 (2.03–2.00) | 50.00–2.15 (2.19–2.15) | 50.00–1.90 (1.93–1.90) | 50.00–1.49 (1.53–1.49) |
| No. of unique reflections | 54,159 | 23,697 | 56,142 | 74,438 |
| Redundancy | 4.8 | 6.0 | 16.0 | 6.8 |
| Completeness (last shell) (%) | 93.6 (61.8) | 99.1 (99.0) | 99.0 (86.3.0) | 99.2 (96.9) |
| I/σ(I) (last shell) | 20.9 (3.8) | 13.7 (2.4) | 39.7 (2.0) | 31.0 (1.2) |
| Rmerge (last shell) | 0.044 (0.197) | 0.044 (0.248) | 0.016 (0.307) | 0.056 (0.242) |
| CC1/2 | 0.97 | 0.96 | 0.95 | 0.97 |
| CC* | 0.98 | 0.98 | 0.96 | 0.95 |
| Refinement statistics | ||||
| Resolution range (Å) | 44.40–2.00 | 49.08–2.15 | 40.78–1.90 | 23.63–1.49 |
| No. of reflections used | 42,316 | 22,593 | 54,048 | 74,219 |
| No. of protein atoms | 4,055 | 3,721 | 4,145 | 3,839 |
| No. of water molecules | 203 | 138 | 410 | 464 |
| No. of ligand atoms | 96 | 96 | 96 | 84 |
| No. of metal atoms | 3 | 3 | 3 | 2 |
| Rwork a | 0.20 | 0.19 | 0.16 | 0.14 |
| Rfreeb | 0.22 | 0.23 | 0.19 | 0.19 |
| rmsd bond lengths (Å) | 0.002 | 0.003 | 0.019 | 0.009 |
| rmsd bond angles (°) | 1.113 | 1.037 | 1.518 | 1.246 |
| Ramachandran | ||||
| Favored/allowed | 96.38/3.42 | 96.95/2.83 | 95.81/3.99 | 98.53/1.47 |
| Outlier (%) | 00.20 | 00.22 | 00.20 | 00.00 |
| Average Bfactors (Å2) | ||||
| Protein | 45.35 | 37.53 | 36.01 | 25.68 |
| Ligand | 26.43 | 24.32 | 19.68 | 19.60 |
| Metal | 23.97 | 26.52 | 21.05 | 40.77 |
| Water | 37.47 | 31.04 | 37.71 | 39.67 |
CC* refers to an estimate of the correlation coefficient CC1/2.
Rwork = Σ∥Fobs∣ − ∣Fcal∥/Σ∣Fobs∣.
Rfree = Σ∥Fobs∣ – ∣Fcal∥/Σ∣Fobs∣, where Fobs is from a test set of reflections that are not used in structural refinement.
2.4 ∣. Molecular dynamics simulation
The Schrödinger Maestro package (Schrödinger Release 2016–2: Maestro, version 10.6, Schrödinger, LLC, New York, NY, 2016.) was used to perform molecular dynamics. Systems were prepared from high-resolution crystal structures of KRASWT (PDB code: 4OBE) wherein P34 were computationally mutated to A, G, or R. The Protein Preparation module was used for model construction, including adding missing atoms, H-bond assignments, and restrained minimization. All systems were neutralized by adding charge-neutralizing counter ions with 10 Å buffering distance in SPC solvent model. No ion-excluded region was included. The 50 ns simulations were carried out by Desmond Molecular Dynamics module with constant temperature (300 K) and pressure (1.0 bar) in NPT ensemble.
2.5 ∣. RAS–RAF interaction assay
An AlphaScreen® (Perkin Elmer) assay was used to measure the affinity of KRASP34R:nucleotide complexes for the Ras Binding Domain (RBD) of the Raf protein kinase. Purified RAF kinase RBD was labeled with maleimide PEG biotin (Pierce) following the manufacturer's protocol. Purified flag-tagged KRAS (1 mg/ml) and KRASP34R mutant were loaded with GMPPNP (Sigma-Aldrich) by incubating for 2 hr at 25° C with a 50-fold excess of nucleotide in the presence of alkaline phosphatase (ThermoFisher). RAF–RBD–biotin was diluted to a final concentration of 40 nmol/L and Flag-KRAS to 10 nmol/L in assay buffer (20 mmol/L Tris pH 7.5, 100 mmol/L NaCl, 1 mmol/L MgCl2, 5% glycerol, 0.5% BSA) and added to individual wells of a low volume white 384-well plate (PerkinElmer). Complexes were disrupted by addition of untagged WT KRAS or KRASP34R mutant preloaded with GMPPNP over a range of concentrations. The assay was developed by addition of streptavidin donor and anti-flag acceptor AlphaScreen beads (10 mg/ml). Alpha signal was measured after overnight incubation at 4°C.
3 ∣. RESULTS
3.1 ∣. KRASP34R-GDP shows disorder in the switches and loss of magnesium
We solved crystal structures of the catalytic domain (residues 1–169) of KRASP34R in complex with either GDP or a GTP analogue. The GDP-bound protein crystallized in a C2 space group with three molecules in the asymmetric unit (Table 1, Figure S1a). Reflections extended to 1.5 Å, allowing modeling of water molecules with high confidence. The switch regions were disordered for all but one protomer (Prot-1), which had ordered SW2 residues (Figure 2a-d). RAS GTPase activity requires nucleotide coordination with a divalent metal ion, most commonly Mg2+ (Manne & Kung, 1985). Metals such as Mg2+ typically give strong electron density signal, but prot-1 showed no density for Mg2+ (Figure 2b vs f). Comparison with the other protomers (Prot-2 and Prot-3) revealed a shift in Asp57 for Prot-1 away from the nucleotide gamma phosphate that was not seen in the other promoters (Figure 2f,g). Thr58 is also rotated toward the gamma phosphate to occupy the typical position of Asp57, making secondary interactions with the gamma phosphate through a bridging water molecule (Figure 2f). In Prot-2, Asp57 participates in stabilizing waters and Ser17 to enable an octahedral coordination state of Mg2+ (Figure 2g), similar to the WT structure (Figure 2e). In Prot-3 Asp57 engages the Mg2+ directly, but the octahedral coordination is lost (Figure 2h). These findings support that Mg2+ occupancy in KRAS depends on the position of Asp57 and that the position of Asp57 is influenced by disorder in SW1.
3.2 ∣. KRASP34R-GTP shows switch conformations that modulate GTP hydrolysis
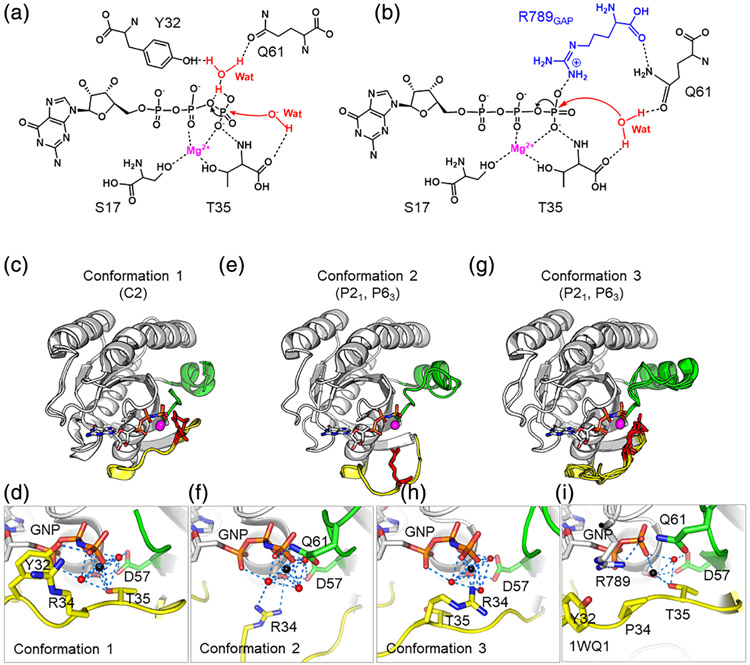
KRASP34R shows preservation of intrinsic GTP hydrolysis but a severe defect in GAP-mediated hydrolysis (Schubbert et al., 2006). The mechanisms of KRAS Intrinsic and GAP-mediated GTP hydrolysis require the receipt of a proton from a catalytic water molecule (Buhrman et al., 2010; Kosloff & Selinger, 2001). In this reaction, a proton is shuttled from the catalytic water molecule via the γ-phosphate of GTP to a nearby bridging water molecule, which can donate hydrogen bonds to both Tyr32 and Gln61 (Figure 3a). In GAP-a stimulated reaction, hydrolysis is enhanced by engagement of an arginine sidechain from GAP which neutralizes developing charges in the transition state, allowing Glu61 to participate in catalysis (Scheffzek et al., 1997) (Figure 3b). To understand the impact of the P34R substitution on these catalytic mechanisms, we solved structures of KRASP34R-GTP.
FIGURE 3.
Structures of GMPPNP-KRASP34R reveal mechanisms of GTP hydrolysis. R34 in red sticks in row 2 and in yellow sticks in row 3. (a) Mechanism for intrinsic GTP hydrolysis in KRAS. Adapted from Buhrman et al. (2010) and Novelli, First, and Webb (2018). (b) Mechanism of GAP-mediated GTP hydrolysis in RAS. Note arginine finger from GAP adapted from Grigorenko, Kots, and Nemukhin (2019) and Scheffzek et al. (1997). (c) Conformation 1, found only in the C2 crystal form. SW1 adopts a closed conformation. (d) Magnification of Conformation 1 shows Tyr32, directly hydrogen-bonded to the γ-phosphate of GNP and Thr35 hydrogen-bonded to the γ-phosphate via coordination with the Mg2+ ion (in black). (e) Open Conformation 2 found in P21 and P63 crystal forms. (f) Magnification of Conformation 2 shows Tyr32 and Thr35 are rotated away from the nucleotide-binding pocket but Arg34 extends toward the gamma phosphate, making interactions with waters that are part of the octahedral coordination with Mg2+. The coordination state is also stabliized by Glu61 and Asp57. (g) Open Conformation 3 found in P21 and P63 space groups. (h) Magnification of Conformation 3 shows that Arg34 directly forms hydrogen bonds to the γ-phosphate of GNP. (i) R34 is similar in principle to the arginine finger found in GAPS and heterotrimeric G-proteins
KRASP34R-GTP crystallized in three different space groups: P21, P63, and C2 (Figure S1b-d). All crystal forms provided high-quality data with complete reflections extending to 1.9–2.1 Å. Each crystal form again showed three protomers in the asymmetric unit (Figure S1b-d). Complete data collection and refinement statistics for the four structures are presented in Table 1. When considering all space groups, three distinct switch conformations were observed, suggesting that KRASP34R is highly dynamic in the switch regions when GTP bound (Figure 3c-h).
All three protomers in the C2 space group had similar switch conformations (Conformation 1, Figure 3c,d). SW1 adopted a closed conformation, with Tyr32 directly hydrogen-bonded to the γ-phosphate of GTP and Thr35 hydrogen-bonded to the γ-phosphate via coordination with the Mg2+ ion, consistent with configuration normally seen for intrinsic GTP hydrolysis. This conformation is also highly similar to the previously described “State 2,” which is relatively closed and compact and able to interact with downstream effectors to promote signaling (compare with PDB ID 4L9W; Ostrem, Peters, Sos, Wells, & Shokat, 2013). In the P63 and P21 space groups, one of the protomers shows a solvent-exposed SW1 associated with rotation of Tyr32 and Thr35 away from the nucleotide-binding pocket (Conformation 2, Figure 3e). However, Arg34 extends toward the gamma phosphate, making interactions with waters that are part of the octahedral coordination of Mg2+. The coordination state is also stabliized by intereactions with Glu61 and Asp57 (Figure 3f). While this interaction likely stabilizes SW1 in the observed conformation, it is not clear that Arg34 in this position would support GTP hydrolysis. Of note, in Conformation 2, we also note a free phosphate ion, which is trapped near the nucleotide binding pocket. We believe this free phosphate ion came from the crystallization buffer (1.8 M phosphate) and could not discern any physiological relevance. In the third conformation, Conformation 3, SW1 was again solvent exposed, but to a lesser degree than Conformation 2 and SW2 was also displaced away from the protein body (Conformation 3, Figure 3g). In this conformation, Arg34 interacted directly with the Mg2+ ion (Figure 3h). This position is conceptually similar to the arginine finger seen in RAS GAPs (Ahmadian, Stege, Scheffzek, & Wittinghofer, 1997) and the intrinsic arginine finger of heterotrimeric G proteins (Mann et al., 2016) and could stimulate GTP hydrolysis. In summary, these models show that Confirmation 2 is inconsistent with the usual mechanism of intrinsic GTP hydrolysis based on displacement of Tyr32 and Gln 61, but that the other conformations may also be capable of supporting intrinsic GTP hydrolysis.
To understand the potential consequences of these conformations on GAP-mediated hydrolysis, we superimposed our structures onto known structures of GAP-RAS complexes (PDB ID 1WQ1 and 6OB2). In all cases, the presence of R34 created a likely steric clash with GAP binding suggesting that GAPS will not efficiently bind to KRAP34R mutated KRAS proteins (Figure S2). This structural incompatibility likely explains the profound lack of GAP-mediated hydrolysis observed in KRASP34R.
3.3 ∣. Effect of P34R on molecular dynamics
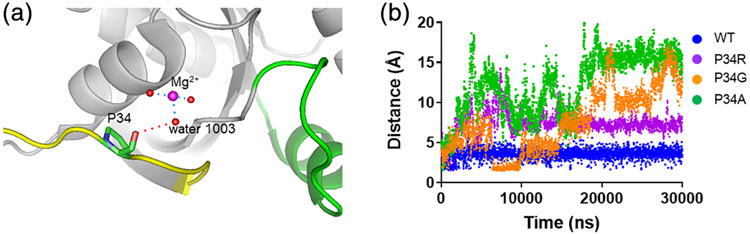
To better understand the mechanisms driving the switch dynamics in KRASP34R structures, we conducted molecular dynamics simulations. Pro34 interacts with the Mg2+ ion indirectly thorough a bridging solvent molecule (Figure 4a). This proline is well-conserved within the RAS family, suggesting that it plays a critical role in stabilizing the conformation of SW1. We performed a molecular dynamics simulation using the crystal structure of GDP-bound WT (4OBE) as the seed model, mutating proline, and monitoring the change in distance between residue 34 and water 1,003 as a surrogate of SW1 stability. The wild-type enzyme maintained this distance over time suggesting relative stability in SW1. However, all other mutations (alanine, glycine, and arginine) resulted in destabilization of this distance, consistent with increased dynamics of SW1 (Figure 4b). This further supports that proline at position 34 is critical for maintaining the conformation of SW1 in RAS subfamily proteins.
FIGURE 4.
Molecular dynamics simulation of KRAS mutated at position 34 show increased dynamics in SW1. (a) In KRASWT, Pro34 interacts with the Mg2+ ion secondarily through hydrogen bonding with a solvent molecule. The distance indicated by the red dashed line is plotted in b. Blue dash lines show nearby polar contacts involving solvent. (b) Distances between side chains of residues 34 and water 1,003 are plotted over time of the simulation
3.4 ∣. Affinity with RAF
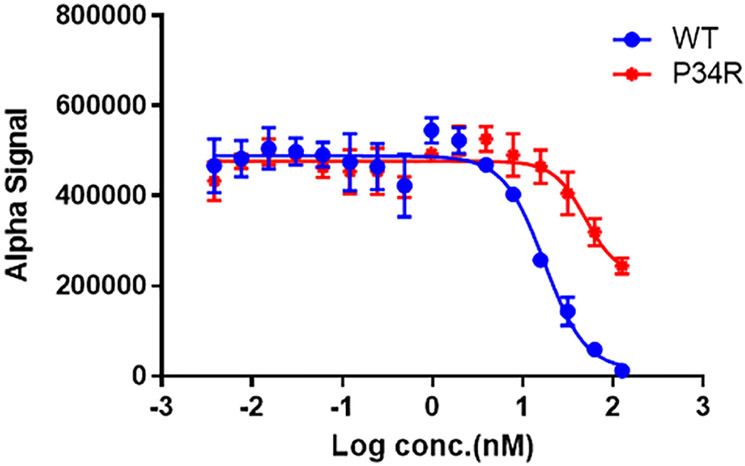
Since the switches serve as the primary interface for interactions with RAS effectors, it is possible that altered switch dynamics might also influence the affinity of RAS for these effectors. RAF kinase is directly downstream of KRAS and is activated through a direct interaction with SW1 of KRAS. Here, we measured the relative affinity of KRASWT and KRASP34R for RAF kinase to detect the effects of P34R substitution using a competitive AlphaScreen bead-based assay (Hunter et al., 2015; Figure 5). Relative to KRAS WT, KRASP34R was impaired for binding, although still within the range of other RAS mutants (Hunter et al., 2015). We speculate that this loss of affinity is attributable to the increased dynamics of SW1 in KRASP34R.
FIGURE 5.
KRASP34R shows decreased affinity of RAF kinase. Untagged KRAS proteins are added to compete apart the preformed complexes of tagged WT KRAS with the RBD of RAF kinase
4 ∣. DISCUSSION
Here, we report four structures of KRASP34R that show variability in the RAS switch regions captured in the crystal lattices. Three of these structures included a nonhydrolyzable GTP analogue that shed light on the previously reported GTPase activities of this mutant. In these structures, the conformations of both switches and the position of Arg34 create steric and electrostatic interactions with GAPs that are predicted to be unfavorable for RAS:GAP binding. This is the likely structural mechanism underlying insensitivity to GAP-stimulated GTP hydrolysis. Nevertheless, we also note that these conformations are compatible with known intrinsic GTP hydrolysis mechanisms, suggesting a possible avenue for preservation of intrinsic GTPase activity (Schubbert, Sovik, et al., 2006). Interestingly one of these has the potential to behave analogously to the intrinsic arginine finger found in heterotrimeric G-proteins, which substantially increase GTP hydrolysis rates in that subfamily of proteins (Mann et al., 2016). While the nature of crystallographic methods limits our ability to know the distribution of these conformations in biological contexts, the fact that all crystal forms obtained during this study showed switch movement is compelling evidence of the dynamic nature of these loops in KRASP34R.
Another consequence of the P34R mutation appears to be a loss of affinity for RAF. This is also compatible with increased dynamics in SW1, because SW1 serves as an interface for the RAS:RAF interaction. This property, together with the preserved intrinsic hydrolysis rate of KRASP34R, may be important for reconciling the seemingly contradictory observations that GAP insensitivity is often seen in cancer-associated RAS mutations, but that KRASP34R mutations are rarely found in cancer. In effect, the loss of affinity for RAF and may serve to dampen overactive RAS signaling, preventing these pathways from crossing signaling thresholds required to induce or support frank malignancy (Haigis, 2017; Poulin et al., 2019b). Indeed, many of the RAS mutations found in RASopathy syndromes appear to demonstrate lower levels of activated RAS signaling when compared to the classical oncogenic mutations (Smith, Neel, & Ikura, 2013).
These structures add to accumulating evidence of biologically meaningful differences between RAS mutations (Hammond et al., 2015; Hobbs et al., 2019; Hunter et al., 2015; Poulin et al., 2019b). In some cases, such as KRASG12C and KRASG12R, these studies have aided in the design of new therapeutic strategies (Haigis, 2017; Montalvo, Li, & Westover, 2017). For KRASG12C, in particular, the opening of a drug pocket beneath SW2 was an important, although serendipitous, aspect of a covalent inhibitor approach that relies on SW2 movement (Ostrem et al., 2013; Zeng et al., 2017). In a similar vein, the increased switch dynamics of KRASP34R may provide therapeutic opportunities. Although highly speculative, one potential strategy would be to discover small molecules that stabilize Conformation 3, the conformation where R34 is directly coordinated with the gamma phosphate. Although our data do not conclusively establish that Conformation 3 is associated with enhanced GTP hydrolysis, the underlying hypothesis would be that such molecules will reduce levels of active GTP-bound KRAS by enhancing the intrinsic GTP hydrolysis activity of KRASP34R by stabilizing the R34 sidechain in a state that facilitates catalysis. Finally, these studies also highlight that despite the large volume of data on RAS, many fundamental discoveries await. This is especially true for understudied RAS mutations, such as D153V, V14I, T58I, and F156L, found in RASopathy syndromes.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the staff at the structural biology laboratory at the University of Texas Southwestern Medical Center and at beamline 19 ID of the Advanced Photon Source for discussions and technical assistance with X-ray data collection and processing. Results shown in this report were derived from work performed at Argonne National Laboratory, Structural Biology Center, at the Advanced Photon Source. Argonne is operated by the University of Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research, under Contract DE-AC02-06CH11357. This work was supported by a career development project through NIH U54 CA196519, Welch Foundation Grant I-1829 and Cancer Prevention and Research Institute of Texas Grant RP170373.
Funding information
Cancer Prevention and Research Institute of Texas, Grant/Award Number: RP170373; National Institutes of Health, Grant/Award Number: U54 CA196519; Welch Foundation, Grant/Award Number: I-1829
Footnotes
CONFLICT OF INTEREST
KDW is a member of the scientific advisory board of Vibliome Therapeutics and is a consultant to Sanofi Pharmaceuticals.
DATA AVAILABILITY STATEMENT
Structural data is available through the RCSB PDB.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, … Zwart PH (2010). PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallographica. Section D, Biological Crystallography, 66(Pt 2), 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian MR, Stege P, Scheffzek K, & Wittinghofer A (1997). Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nature Structural Biology, 4(9), 686–689. [DOI] [PubMed] [Google Scholar]
- Bera AK, Lu J, Wales TE, Gondi S, Gurbani D, Nelson A, … Westover KD (2019). Structural basis of the atypical activation mechanism of KRAS(V14I). The Journal of Biological Chemistry, 294(38), 13964–13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, & Wittinghofer A (2007). GEFs and GAPs: Critical elements in the control of small G proteins. Cell, 129(5), 865–877. [DOI] [PubMed] [Google Scholar]
- Buhrman G, Holzapfel G, Fetics S, & Mattos C (2010). Allosteric modulation of Ras positions Q61 for a direct role in catalysis. Proceedings of the National Academy of Sciences of the United States of America, 107(11), 4931–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, . Koppada N (2019). The clinical KRAS (G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature, 575, 217–223. [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002). The PyMOL molecular graphics system. New York, NY: PyMOL by Schrödinger. [Google Scholar]
- Donovan S, Shannon KM, & Bollag G (2002). GTPase activating proteins: Critical regulators of intracellular signaling. Biochimica et Biophysica Acta, 1602(1), 23–45. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, & Cowtan K (2010). Features and development of coot. Acta Crystallographica. Section D, Biological Crystallography, 66(Pt 4), 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell JB, Fischer JP, Baer BR, Ballard J, Blake JF, Bouhana K, … Burkard MR (2018). Discovery of Tetrahydropyridopyrimidines as irreversible covalent inhibitors of KRAS-G12C with in vivo activity. ACS Medicinal Chemistry Letters, 9(12), 1230–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Medarde A, & Santos E (2011). Ras in cancer and developmental diseases. Genes & Cancer, 2, 344–358. http://dxdoiorg/101177/1947601911411084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremer L, Merbitz-Zahradnik T, Dvorsky R, Cirstea IC, Kratz CP, Zenker M, … Ahmadian MR (2011). Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Human Mutation, 32(1), 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorenko BL, Kots ED, & Nemukhin AV (2019). Diversity of mechanisms in Ras-GAP catalysis of guanosine triphosphate hydrolysis revealed by molecular modeling. Organic & Biomolecular Chemistry, 17(19), 4879–4891. [DOI] [PubMed] [Google Scholar]
- Haigis KM (2017). KRAS alleles: The devil is in the detail. Trends in Cancer, 3(10), 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond DE, Mageean CJ, Rusilowicz EV, Wickenden JA, Clague MJ, & Prior IA (2015). Differential reprogramming of isogenic colorectal cancer cells by distinct activating KRAS mutations. Journal of Proteome Research, 14(3), 1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs GA, Baker NM, Miermont AM, Thurman RD, Pierobon M, Tran TH, … Der CJ (2019). Atypical KRASG12R mutant is impaired in PI3K signaling and macropinocytosis in pancreatic cancer. Cancer Discovery, 10, 104–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, & Westover KD (2015). Biochemical and structural analysis of common cancer-associated KRAS mutations. Molecular Cancer Research, 13(9), 1325–1335. [DOI] [PubMed] [Google Scholar]
- Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, … Ren P (2018). Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell, 172(3), 578–589.e517. [DOI] [PubMed] [Google Scholar]
- Kavamura MI, Peres CA, Alchorne MM, & Brunoni D (2002). CFC index for the diagnosis of cardiofaciocutaneous syndrome. American Journal of Medical Genetics, 112(1), 12–16. [DOI] [PubMed] [Google Scholar]
- Kosloff M, & Selinger Z (2001). Substrate assisted catalysis application to G proteins. Trends in Biochemical Sciences, 26(3), 161–166. [DOI] [PubMed] [Google Scholar]
- Lu J, Bera AK, Gondi S, & Westover KD (2018). KRAS switch mutants D33E and A59G crystallize in the state 1 conformation. Biochemistry, 57(3), 324–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann D, Teuber C, Tennigkeit SA, Schröter G, Gerwert K, & Kötting C (2016). Mechanism of the intrinsic arginine finger in heterotrimeric G proteins. Proceedings of the National Academy of Sciences of the United States of America, 113(50), E8041–E8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manne V, & Kung HF (1985). Effect of divalent metal ions and glycerol on the GTPase activity of H-ras proteins. Biochemical and Biophysical Research Communications, 128(3), 1440–1446. [DOI] [PubMed] [Google Scholar]
- Montalvo SK, Li L, & Westover KD (2017). Rationale for RAS mutation-tailored therapies. Future Oncology, 13(3), 263–271. [DOI] [PubMed] [Google Scholar]
- Novelli ET, First JT, & Webb LJ (2018). Quantitative measurement of intrinsic GTP hydrolysis for carcinogenic glutamine 61 mutants in H-Ras. Biochemistry, 57(44), 6356–6366. [DOI] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, & Shokat KM (2013). K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature, 503(7477), 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, & Minor W (1997). [20] processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology, 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Poulin EJ, Bera AK, Lu J, Lin Y-J, Strasser SD, Paulo JA, … Cook J (2019a). Tissue-specific oncogenic activity of KRASA146T. Cancer Discovery, 9(6), 738–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin EJ, Bera AK, Lu J, Lin Y-J, Strasser SD, Paulo JA, … Haigis KM (2019b). Tissue-specific oncogenic activity of KRAS(A146T). Cancer Discovery, 9(6), 738–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano AA, Allanson JE, Dahlgren J, Gelb BD, Hall B, Pierpont ME, … Noonan JA (2010). Noonan syndrome: Clinical features, diagnosis, and management guidelines. Pediatrics, 126(4), 746–759. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, & Wittinghofer A (1997). The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science, 277(5324), 333–338. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Lautwein A, Kabsch W, Ahmadian MR, & Wittinghofer A (1996). Crystal structure of the GTPase-activating domain of human p120GAP and implications for the interaction with Ras. Nature, 384(6609), 591–596. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Bollag G, Lyubynska N, Nguyen H, Kratz CP, Zenker M, … Shannon K (2007). Biochemical and functional characterization of germ line KRAS mutations. Molecular and Cellular Biology, 27(22), 7765–7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbert S, Sovik O, Bollag G, Nguyen H, Rowe SL, Kratz C, … Shannon K (2006). Biochemical and functional analysis of germline KRAS mutations that cause disorders of the Noonan syndrome spectrum. Blood, 108(11), 1431–1431. [Google Scholar]
- Schubbert S, Zenker M, Rowe SL, Boll S, Klein C, Bollag G, … Kratz CP (2006). Germline KRAS mutations cause Noonan syndrome. Nature Genetics, 38(3), 331–336. [DOI] [PubMed] [Google Scholar]
- Smith MJ, Neel BG, & Ikura M (2013). NMR-based functional profiling of RASopathies and oncogenic RAS mutations. Proceedings of the National Academy of Sciences of the United States of America, 110(12), 4574–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia M, Gelb BD, & Zenker M (2011). Noonan syndrome and clinically related disorders. Best Practice & Research. Clinical Endocrinology & Metabolism, 25(1), 161–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia M, Pennacchio LA, Zhao C, Yadav KK, Fodale V, Sarkozy A, … Gelb BD (2007). Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nature Genetics, 39(1), 75–79. [DOI] [PubMed] [Google Scholar]
- Tidyman WE, & Rauen KA (2008). Noonan, Costello and cardio-facio-cutaneous syndromes: Dysregulation of the Ras-MAPK pathway. Expert Reviews in Molecular Medicine, 10, e37. [DOI] [PubMed] [Google Scholar]
- Tidyman WE, & Rauen KA (2009). The RASopathies: Developmental syndromes of Ras/MAPK pathway dysregulation. Current Opinion in Genetics & Development, 19(3), 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter IR, & Wittinghofer A (2001). The guanine nucleotide-binding switch in three dimensions. Science, 294(5545), 1299–1304. [DOI] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, … Richardson DC (2018). MolProbity: More and better reference data for improved allatom structure validation. Protein Science, 27(1), 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, … Wilson KS (2011). Overview of the CCP4 suite and current developments. Acta Crystallographica. Section D, Biological Crystallography, 67(Pt 4), 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Lu J, Li L, Feru F, Quan C, Gero TW, … Gray NS (2017). Potent and selective covalent Quinazoline inhibitors of KRAS G12C. Cell Chemical Biology, 24(8), 1005–1016.e1003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.