Summary
The antifungal echinocandin lipopeptide, acrophiarin, was circumscribed in a patent in 1979. We confirmed that the producing strain NRRL 8095 is Penicillium arenicola and other strains of P. arenicola produced acrophiarin and acrophiarin analogues. Genome sequencing of NRRL 8095 identified the acrophiarin gene cluster. Penicillium arenicola and echinocandin-producing Aspergillus species belong to the family Aspergillaceae of the Eurotiomycetes, but several features of acrophiarin and its gene cluster suggest a closer relationship with echinocandins from Leotiomycete fungi. These features include hydroxy-glutamine in the peptide core instead of a serine or threonine residue, the inclusion of a non-heme iron, α-ketoglutarate-dependent oxygenase for hydroxylation of the C3 of the glutamine, and a thioesterase. In addition, P. arenicola bears similarity to Leotiomycete echinocandin-producing species because it exhibits self-resistance to exogenous echinocandins. Phylogenetic analysis of the genes of the echinocandin biosynthetic family indicated that most of the predicted proteins of acrophiarin gene cluster exhibited higher similarity to the predicted proteins of the pneumocandin gene cluster of the Leotiomycete Glarea lozoyensis than to those of the echinocandin B gene cluster from A. pachycristatus. The fellutamide gene cluster and related gene clusters are recognized as relatives of the echinocandins. Inclusion of the acrophiarin gene cluster into a comprehensive phylogenetic analysis of echinocandin gene clusters indicated the divergent evolutionary lineages of echinocandin gene clusters are descendants from a common ancestral progenitor. The minimal 10-gene cluster may have undergone multiple gene acquisitions or losses and possibly horizontal gene transfer after the ancestral separation of the two lineages.
Introduction
The echinocandins are a family of lipohexapeptide fungal metabolites that non-competitively bind to the catalytic unit of β−1,3-glucan synthase leading to osmotic instability and fungal cell wall lysis. The antifungal drug caspofungin (CANCIDAS™) was developed from the echinocandin variant, pneumocandin B0 (Balkovec et al., 2013). Two more echinocandin-type antifungals, micafungin (MYCAMINE™) derived from FR901370 (WF11899A), and anidulafungin (ERAXIS™) derived from echinocandin B have been brought to market (Vazquez and Sobel, 2006; Balkovec et al., 2013). A new long-acting echinocandin, rezafungin (CD101 IV), is based on the echinocandin B natural product (Zhao et al., 2016). Because of the importance of echinocandin lipopeptides in antifungal therapy, understanding the full range of natural echinocandin variants and their underlying biosynthesis will provide options to generate and optimize new echinocandins. Furthermore, comprehensive mapping of these gene clusters across the Ascomycetes will contribute to understanding the natural functions of echinocandins in their respective producing organisms.
Our group artificially made the echinocandin acrophiarin (antibiotic S31794/F-1, Fig. 1, Table 1) (Dreyfuss and Tscherter, 1979; Dreyfuss, 1986) during mutasynthesis experiments (Chen et al., 2016b). The polyketide synthase (GLPKS4) of Glarea lozoyensis that is responsible for the 10,12-methyl myristate side-chain of pneumocandins was inactivated by disruption of glpks4 (Chen et al., 2016b). By exploiting the relaxed substrate specificity of the pathway’s acetyl-CoA ligase (GLligase), feeding this mutant with straight-chain fatty acid precursors, e.g. myristic acid (C14) resulted in acrophiarin and other pneumocandin variants with substituted side-chains (Chen et al., 2016b).
Fig. 1.
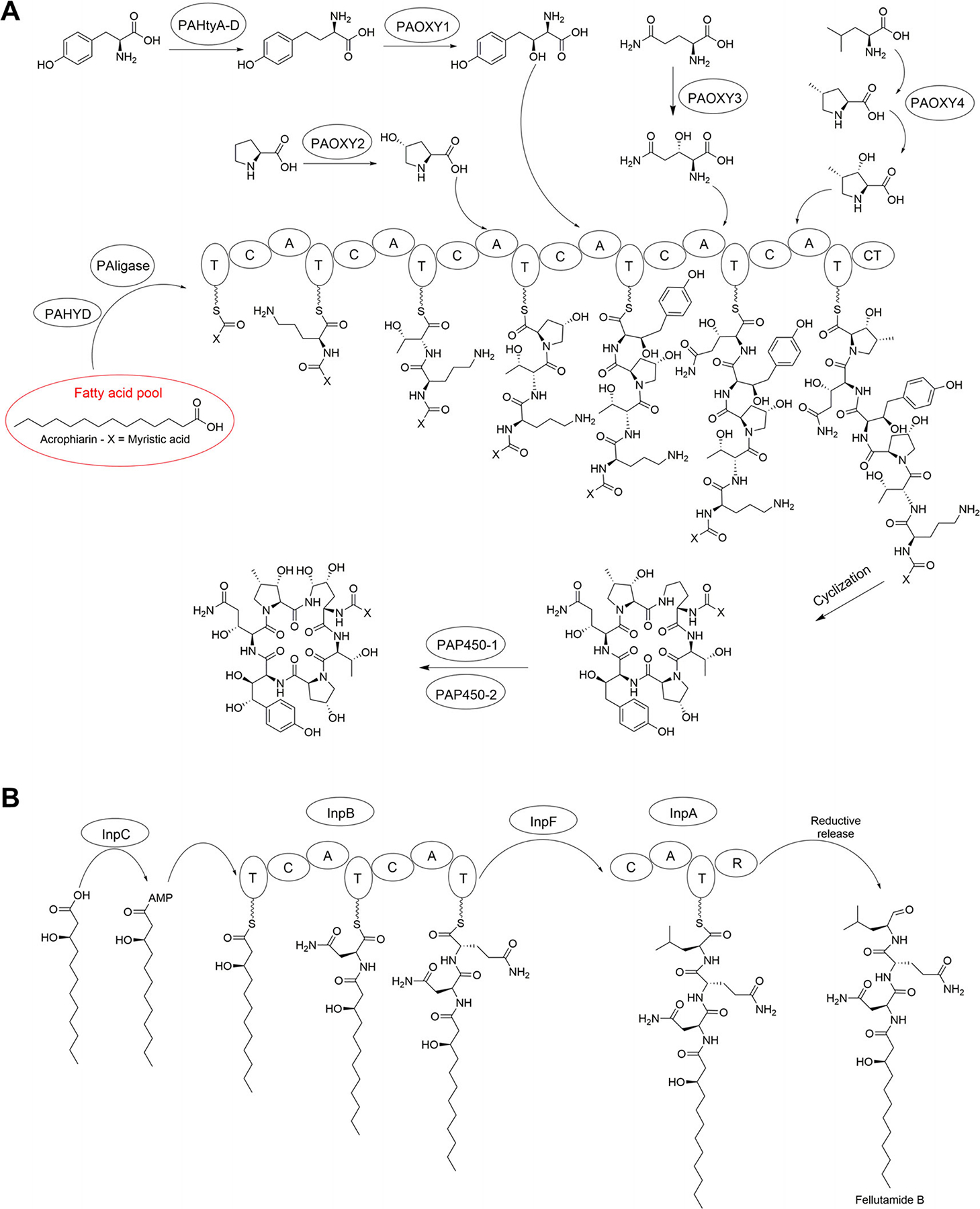
Biosynthetic pathways of echinocandin and fellutamide lipopeptides mentioned in this report. A. Acrophiarins. B. Fellutamides. The fellutamide pathway is redrawn from Yeh et al. (2016).
Table 1.
Principal natural echinocandins, representative strains that produce them, acyl side-chains, amino acids (AA) in positions 1–6 of peptide core and accession numbers for genomes or gene clusters.
| Naturally occurring echinocandins | Representative strain | Class, family classification | Acyl side-chain | AA1 | AA2 | AA3 | AA4 | AA5 | AA6 | Genome or gene cluster accession |
|---|---|---|---|---|---|---|---|---|---|---|
| Acrophiarin (antibiotic S31794/ | Penicillium arenicola | Eurotiomycetes, Aspergillaceae | Myristic acid | 4R,5R-dihydroxy-L-Orn | L-Thr | 4R-hydroxy-L-Pro | 3S,4S-dihydroxy-L-homoTyr | 3R-hydroxy-L-GIn | 3S-hydoxy-4S-methyl-L-Pro | MN518690 |
| Echinocandin B | Aspergillus pachycristatus NRRL 11440 (ATCC 58397) | Eurotiomycetes, Aspergillaceae | Linoleic acid | 4R,5R-dihydroxy-L-Orn | L-Thr | 4R-hydroxy-L-Pro | 3S,4S-dihydroxy-L-homoTyr | L-Thr | 3S-hydoxy-4S-methyl-L-Pro | JX421684 |
| Mulundocandin | Aspergillus mulundensis DSMZ5745 | Eurotiomycetes, Aspergillaceae | 12-Methylmyristic acid | 4R,5R-dihydroxy-L-Orn | L-Thr | 4R-hydroxy-L-Pro | 3S,4S-dihydroxy-L-homoTyr | L-Ser | 3S-hydoxy-4S-methyl-L-Pro | KP742486, PVWQ00000000 |
| Aculeacin A | Aspergillus aculeatus ATCC 16872 (NRRL 5094) | Eurotiomycetes, Aspergillaceae | Palmitic acid | 4R,5R-dihydroxy-L-Orn | L-Thr | 4R-hydroxy-L-Pro | 3S,4S-dihydroxy-L-homoTyr | L-Thr | 3S-hydoxy-4S-methyl-L-Pro | JGI ATCC16872 v1.1 |
| Pneumocandin A0 | Glarea lozoyensis ATCC 20868 | Leotiomycetes, Helotiaceae | 10,12-Dimethylmyristic | 4R,5R-dihydroxy-L-Orn | L-Thr | 4R-hydroxy-L-Pro | 3S,4S-dihydroxy-L-homoTyr | 3R-hydroxy-L-Gln | 3S-hydoxy-4S-methyl-L-Pro | ALVE00000000 |
| Sporiofungin | Pezicula radicicola NRRL 12192 | Leotiomycetes, Dermateaceae | 10,12-dimethyl myristic acid | 4R,5R-dihydroxy-L-Orn | L-Ser | 4R-hydroxy-L-Pro | 3S-hydroxy-L-homoTyr | 3R-hydroxy-L-Gln | 3S-hydoxy-4S-methyl-L-Pro | PDUO00000000 |
| FR901379 (WF11899A) | Coleophoma cylindrospora FERM BP 6252 | Leotiomycetes, Dermateaceae | Palmitic acid | 4R,5R-dihydroxy-L-Orn | L-Thr | 4R-hydroxy-L-Pro | 3S, 7-dihydroxyl-L-homoTyr-7-O-sulfate | 3R-hydroxy-L-Gln | 3S-hydoxy-4S-methyl-L-Pro | AB723722, AB720725, PDLN00000000 |
| FR209602 | Coleophoma crateriformis FERM BP 5796 | Leotiomycetes, Dermateaceae | Palmitic acid | 4R,5R-dihydroxy-L-Orn | L-Ser | 4R-hydroxy-L-Pro | 3S,4S,7-trihydroxyl-L-homoTyr-7-O-sulfate | 3R-hydroxy-L-Gln | 3S-hydoxy-4S-methyl-L-Pro | AB720076, PDLN00000000 |
| FR190293 | Venustampulla echinocandica FERM BP 5553 | Leotiomycetes, Pleuroascaceae | 10,12-dimethyl myristic acid | 4R,5R-dihydroxy-L-Orn | L-Thr | 4R-hydroxy-L-Pro | 3S, 7-dihydroxyl-L-homoTyr-7-O-sulfate | 3R-hydroxy-L-Gln | 3S-hydoxy-4S-methyl-L-Pro | AB720726, NPIC00000000 |
| FR227673a | Chalara sp. | Leotiomycetes, family unknown | 12,14-dimethylpalmitic acid | 4R,5R-dihydroxy-L-Orn | L-Thr | 4R-hydroxy-L-Pro | 3S, 7-dihydroxyl-L-homoTyr-7-O-sulfate | 3R-hydroxy-L-Gln | 3S-hydoxy-4S-methyl-L-Pro | None available |
Not studied. Included here for comparative purposes. See reference.
All echinocandins produced by Aspergillus species (Eurotiomycetes, Aspergillaceae) bear serine or threonine in the fifth position (Table 1), and have been referred to Aspergillus- or Eurotiomycete-type echinocandins (Yue et al., 2015). In contrast, Leotiomycete-type echinocandins invariably bear hydroxy-l-glutamate in the peptide’s fifth position (Table 1). Based on this classification of echinocandins (Yue et al., 2015), acrophiarin from P. arenicola appears to be a Leotiomycete-type echinocandin from a fungus in the Aspergillaceae, albeit in a genus distinct from Aspergillus. Therefore, acrophiarin is unique among the echinocandins because it combines a feature of Leotiomycete-type echinocandins, hydroxy-glutamate in the cyclic peptide’s fifth position with a straight-chain myristate side-chain (Table 1; Fig. 1), which presumably originates from the cellular fatty acid pool.
A patent from the Swiss pharmaceutical company Sandoz (now Novartis) first disclosed acrophiarin as antibiotic S31794/F-1 (Dreyfuss and Tscherter, 1979). In the patent, infrared, UV, 1H, and 13C NMR spectra, and basic elemental composition data circumscribed a purified fermentation product and antifungal molecule effective against Candida species, however, a complete chemical structure was not proposed. The new antifungal metabolite was produced from strain NRRL 8095 from soil from British Columbia (Dreyfuss and Tscherter, 1979). In 1981, a patent from workers at Eli Lilly (Abbott and Fukuda, 1981) disclosed a chemical structure for antibiotic S31794/F-1 and proposed that it was a cyclic peptide consisting of dihydroxy-l-ornithine, l-threonine, hydroxy-l-proline, dihydroxy-l-homo-tyrosine, hydroxy-l-glutamine, hydroxy-4-methyl-l-proline that was N-acylated at the ornithine residue to myristoyl. In other words, acrophiarin was an echinocandin with a peptide core like that of the pneumocandins but with a straight myristoyl side-chain rather than the dimethyl-myristoyl side-chain characteristic of the pneumocandins (Table 1; Fig. 1). The Lilly patent repeated the NMR spectra and physiochemical data from the Sandoz patent but offered no new spectral data to verify the structural assignments (Abbott and Fukuda, 1981). Antibiotic S31794/F-1 was later renamed ‘acrophiarin’, and the producing strain was identified as P. arenicola (Dreyfuss, 1986). Penicillium arenicola has been recognized as morphologically and phylogenetically distinct from the narrow phylogenetic definition of Penicillium and is closely related to Phialomyces macrosporus of the Aspergillaceae (Pitt, 1980; Houbraken and Samson, 2011; Frisvad et al., 2013). At the time of writing, P. arenicola has yet to be formally reclassified as a species of Phialomyces, therefore, we are obliged to continue to use the name Penicillium arenicola. Other metabolites reported from P. arenicola are the γ-butyrolactone canadensolide (McCorkindale et al., 1968) and the C-glycosylated depsides arenicolins A and B (Perlatti et al., 2020).
The biosynthetic gene clusters (BGCs) of most of the known echinocandins have been mapped and consist of 10–16 contiguous and co-regulated genes (Yue et al., 2015; Hüttel, 2017). Based on phylogenetic analysis of the protein sequences of echinocandin enzymes and comparisons structural variations among different echinocandins, they were classified into two types from divergent lineages of fungi (Yue et al., 2015). Aspergillus-types incorporate either serine or threonine in the core peptide’s fifth position and lack a polyketide synthase (PKS) dedicated to the synthesis of the side-chain. Leotiomycete-type echinocandins incorporating hydroxy-glutamate in the fifth position have either a fatty acid- or highly reducing PKS-derived side-chain. Some Leotiomycete-type echinocandins undergo O-sulfation of the homotyrosine residue (e.g. FR901370, Table 1). As noted above, acrophiarin incorporates features of both echinocandin types.
To better comprehend the evolutionary processes leading to diversification of the echinocandin lipopeptides, we sequenced the genome of P. arenicola NRRL 8095 to identify the acrophiarin gene cluster. This newly characterized BGC is integrated into a revised evolutionary framework of echinocandins and the phylogenetically related fellutamide BCGs. The analysis indicates the acrophiarin biosynthetic and transport genes bridge the deep phylogenetic hiatus between the two types of echinocandins. These new data associate a non-ribosomal peptide synthetase (NRPS) adenylation domain (A domain) for the incorporation of hydroxy-l-glutamine in the echinocandin core with a species in the Aspergillaceae of the Eurotiomycetes. Based on these new findings, we offer a revised hypothesis on the origin of ancestral echinocandins and their relationship to the fellutamide gene clusters. Phylogenetic and synteny analyses of echinocandin gene clusters indicate divergent evolutionary lineages of echinocandin and fellutamide gene clusters are descendants from common ancestral progenitors. Subsequent to the ancestral separation, echinocandin structural diversity appears to have undergone additional elaboration among the Leotiomycete fungi. Horizontal gene transfer (HGT) of a Leotiomycete-type gene cluster could plausibly explain the re-introduction of the acrophiarin gene cluster into the Aspergillaceae lineage. Furthermore, we demonstrate that acrophiarin and a series of natural acrophiarin analogues are a chemotaxonomic feature for strains of P. arenicola. Finally, the structural characterization of naturally occurring acrophiarin is updated with new data on its spectral properties, and its antifungal activity towards the producing strains is evaluated.
Results
Confirmation of NRRL 8095 as Penicillium arenicola
The patent describing antibiotic S31794/F-1 identified the producing strain as Acrophialophora limonispora (Dreyfuss and Tscherter, 1979). This invalid name was later corrected, and the strain was correctly identified as P. arenicola (Dreyfuss, 1986). By microscopy, we observed the typical conidial state described in the literature (Fig. S1) (Pitt, 1980). Database searches with internal transcribed spacer and large subunit ribosomal DNA from the genome sequence of NRRL 8095 (GenBank MN512717) sequences from strains of P. arenicola, including the ex-type strain NRRL 3392 (Table 2) as the top BLAST hits. Furthermore, as previously noted (Pitt, 1980), strains in Table 2 were highly similar in their colony characteristics and microscopic features. We, therefore, concluded that NRRL 8095 is conspecific with authentic strains of P. arenicola.
Table 2.
Strains of Penicillium arenicola and Phialomyces macrosporus examined in this work and their geographic origin.
| Speciesa | Strain number | Habitat | Geographic origin |
|---|---|---|---|
| P. arenicola | NRRL 8095 | Soil | British Columbia, Canada |
| P. arenicola | NRRL 3392a | Soil, pine forest | Near Kyiv, Ukraine |
| P. arenicola | NRRL 31507 | Mineral soil, under Pinus resinosa | Ontario, Canada |
| P. arenicola | NRRL 31509 | Oil soaked soil | Norman Wells, Northwest Territories, Canada |
| Ph. macrosporus | IBT 31128b | Thermally heated soil | Rotorua, New Zealand |
| Ph. macrosporus | IBT 31129 | Decaying needles of Pinus luchuensis | Iriomoto-jima Island, Okinawa, Japan |
Ex-lectotype strain.
Ex-holotype strain.
Acrophiarin and related analogues detection and chemical structure determination
Bioassay of extracts of all four P. arenicola strains (Table 2, Fig. S2) in the five media tested resulted in strong inhibition of growth of C. albicans, but extracts were only weakly or not active towards Cryptococcus neoformans (Fig. 2). This pattern of antifungal activity is suggestive of echinocandins because they do not affect C. neoformans, possibly due to their failure to reach the site of β-glucan assembly (Thompson et al., 1999). All extracts from the two strains of the closely related Ph. macrosporus were inactive towards C. albicans indicating echinocandin-type metabolites were not produced (Fig. 2).
Fig. 2.
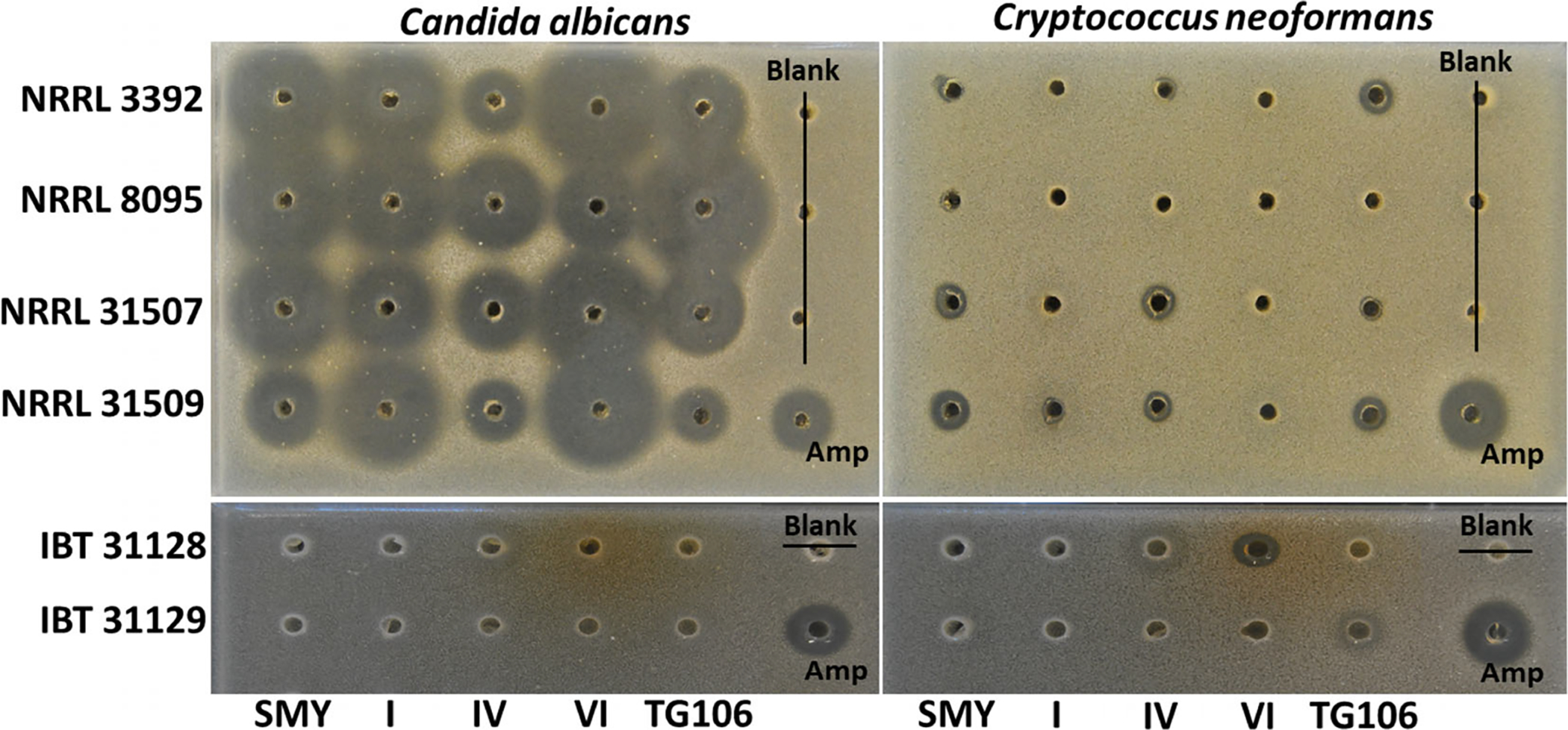
Agar diffusion assay of extracts of four strains of P. arenicola (NRRL 3392, 8095, 31507 and 31509) and two strains of Ph. macrosporus (IBT 31128, 31129) grown in five different fermentation media against C. albicans ATCC 10231 (left) and C. neoformans H99 (right). The extracts of each strain on each medium were arrayed in agar wells from left to right. Amphotericin B was the positive control (lower right corner). Assay wells are 4 mm in diameter.
HPLC–MS analysis confirmed that strains NRRL 8095, 3392, 31507, and 31509 produced acrophiarin and highly related analogues (Figs 3 and 4) indicating that the acrophiarin BGC may be a consistent feature of P. arenicola (see results below). Additionally, we could infer that like other echinocandins, the production of acrophiarin and its analogues is facile. They could be detected after a few days when significant growth accumulated (data not shown) indicating that the BGC is transcribed during exponential growth of the culture.
Fig. 3.
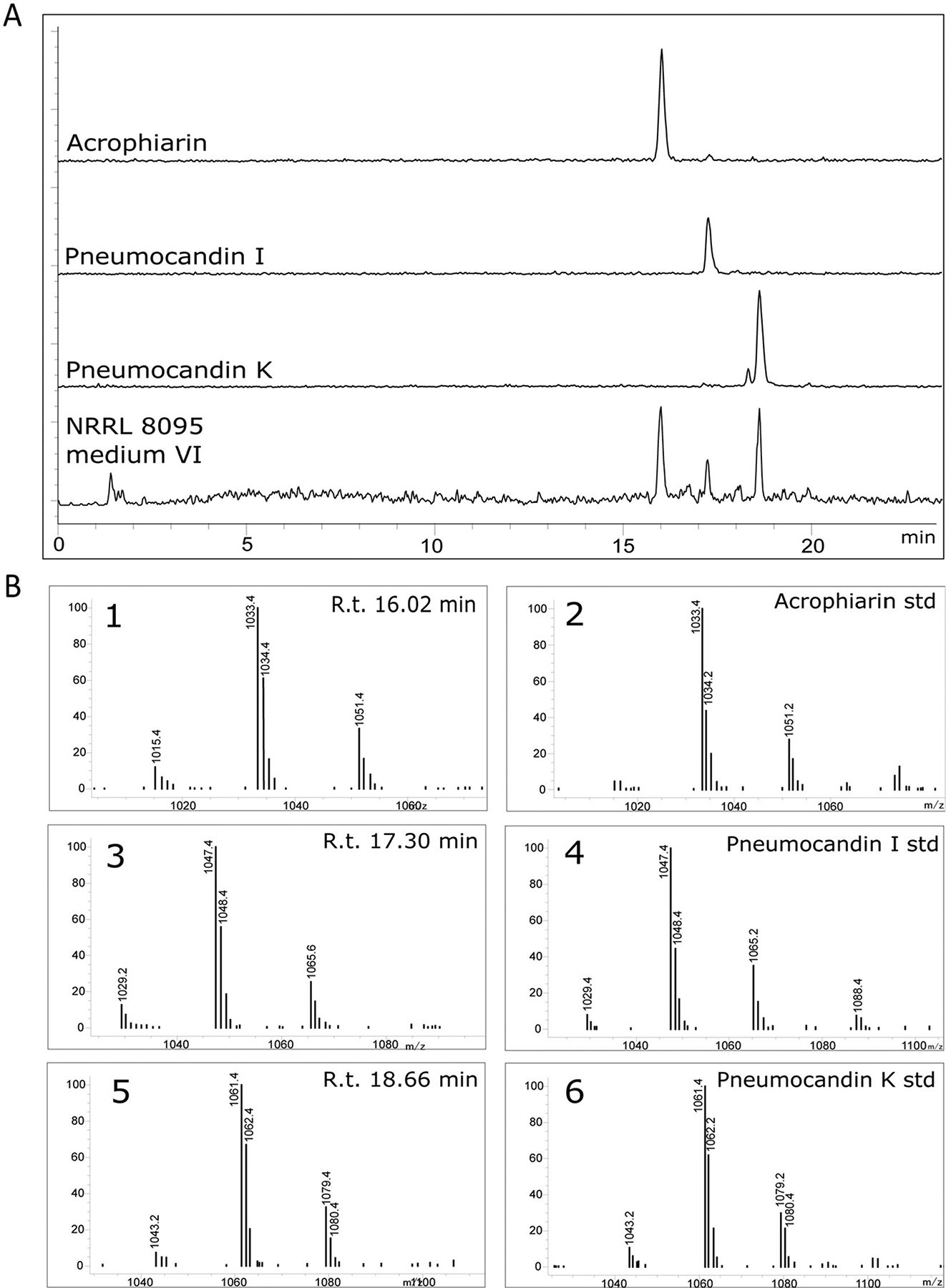
A. EIC chromatograms of purified standards of acrophiarin, pneumocandin I and pneumocandin K obtained from G. lozoyensis (+ESI, m/z 900–1250). The chromatogram obtained for P. arenicola NRRL 3392 in medium VI was selected among crude extract chromatograms as an example of acrophiarin and related pneumocandins in crude extracts. B. MS fragments observed for: 1. Peak with retention time 16.02 min; 2. Purified acrophiarin from G. lozoyensis; 3. Peak with retention time 17.30 min; 4. Purified pneumocandin I from G. lozoyensis; 5. Peak with retention time 18.66 min; 6. Purified pneumocandin K from G. lozoyensis. See Chen and colleagues (2016b) for the production of standards.
Fig. 4.
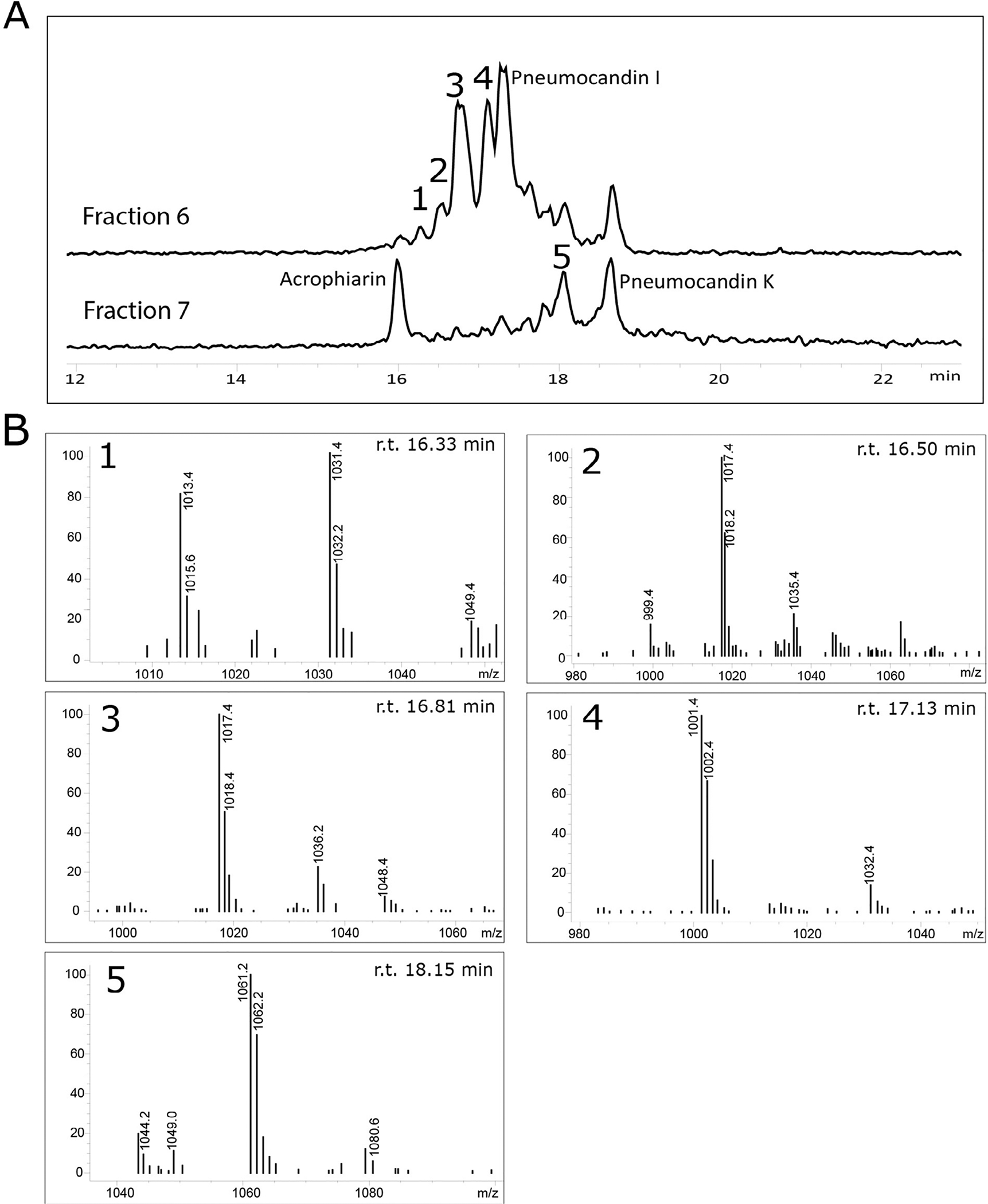
A. LC-MS analysis of fractions 6 and 7 showing the presence of several acrophiarin and pneumocandin analogues. B. MS fragmentation observed for unidentified acrophiarin analogues 1–5.
Acrophiarin was characterized by the comparison of its mass spectrometer (MS) and 1H NMR spectroscopic data (Fig. S3) with those described in the literature and co-injection of purified standards made during G. lozoyensis mutasynthesis experiments (Dreyfuss and Tscherter, 1979; Chen et al., 2016b). As the data agreed well with the published values, the principal echinocandin from P. arenicola was confirmed as acrophiarin. Additional peaks were observed by HPLC–MS, with m/z corresponding to the 15C and 16C side-chain variants of pneumocandins I and K, and their presence was confirmed by co-injection with analytical standards (Chen et al., 2016b) (Fig. 3). Additional minor peaks were also observed, and analysis by MS fragmentation indicated the existence of analogues lacking some of the core peptide hydroxyls (Fig. 4), consistent with minor products observed in other pneumocandin mixtures resulting from incomplete oxidation of the core’s amino acids (Masurekar et al., 1992; Li et al., 2015).
Identification of the acrophiarin BGC
Only one BGC that comprised an acetyl-CoA ligase, an NRPS with six adenylation domains, along with the hty genes encoding the biosynthesis of homotyrosine from tyrosine was found in the draft genome assembly for NRRL 8095. Genes in this BGC exhibited high sequence similarity to other BGCs encoding echinocandins and had a gene cluster organization and order that was intermediate between Aspergillus- and Leotiomycete-types (Fig. 5; Table 3). The high level of identity and synteny with the pneumocandin BGC and other echinocandin clusters indicated with a high degree of certainty it encoded the biosynthesis of acrophiarin. Surprisingly, most of the predicted proteins of acrophiarin BGC exhibited higher similarity to the predicted proteins of the pneumocandin BGC from G. lozoyensis than to those of the echinocandin B BGC (Table 3). Consistent with the chemical structure of acrophiarin, the BGC lacks a PKS orthologue of GLPKS4 that assembles the pneumocandin side-chain (Fig. 5; Fig. S4, Table S1). To rule out the possibility that a side-chain encoding PKS gene might lie outside the acrophiarin gene cluster, we used the predicted amino acid sequence of GLPKS4 to search a database of NRRL 8095 protein models. Two distantly related highly reducing PKS (identities, 43% and 40% respectively) were found at other loci on other contigs, and both contained a C-methyltransferase domain, thus it seems unlikely either would be responsible for the myristoyl side-chain (Fig. S5). Thus, the hypothesis that the acrophiarin side-chain originates from the fatty acid pool rather from a highly reducing PKS seems parsimonious with the available data and previous mutasynthesis experiments with pneumocandins (Chen et al., 2016b).
Fig. 5.
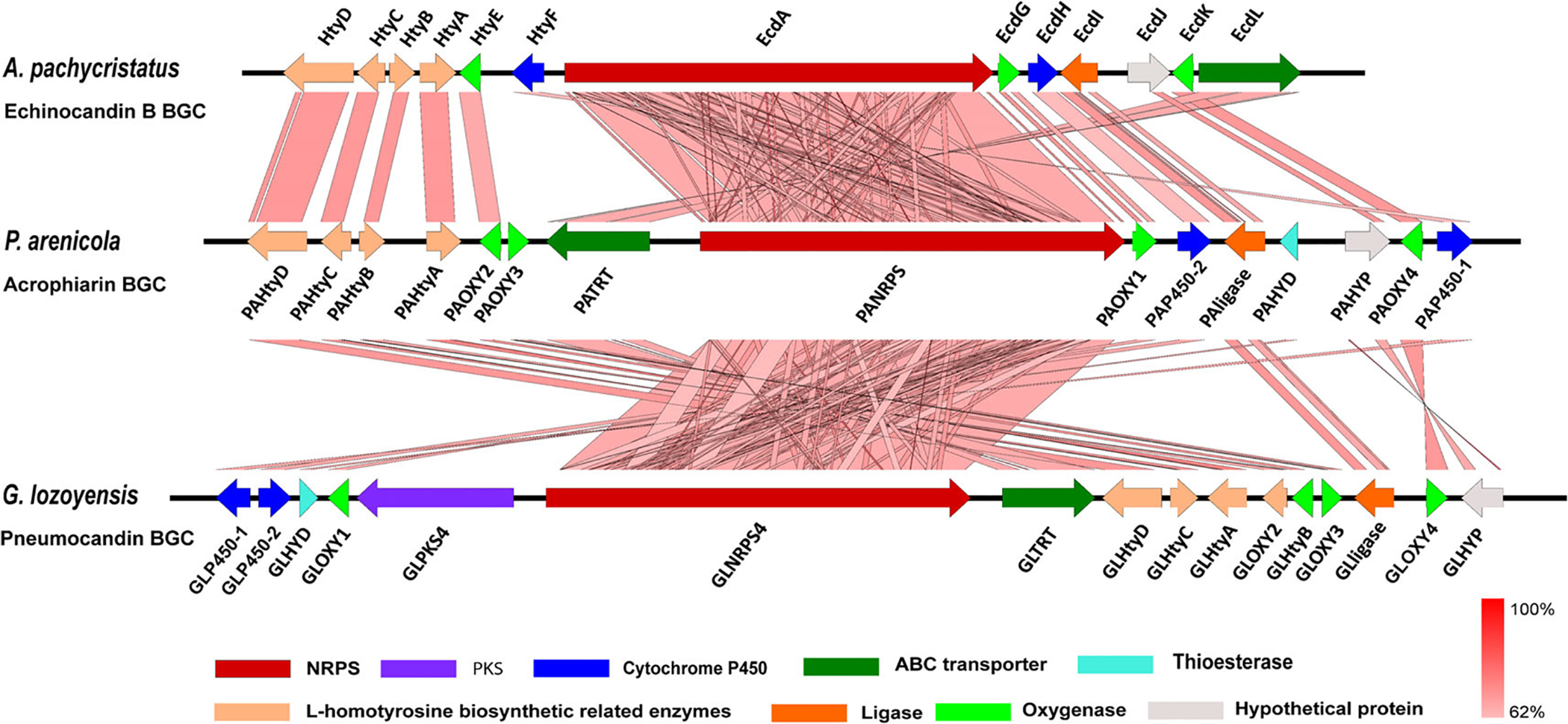
Graphic representation of the echinocandin-type gene clusters and their microsynteny. Acrophiarin gene cluster from Penicillium arenicola NRRL 8095, echinocandin B gene cluster from Aspergillus pachycristatus NRRL 11440, and pneumocandin gene cluster from Glarea lozoyensis. Gene functions are colour coded. The intensity of the red scale bar indicates the degree of nucleotide similarity.
Table 3.
Sequence comparisons between proteins in the pneumocandin, echinocandin B and acrophiarin gene clusters.
| Pneumocandin biosynthetic proteins | Predicted function | Acrophiarin biosynthetic proteins | Coverage %; Identity % | Echinocandin B biosynthetic proteins | Coverage %; Identity % |
|---|---|---|---|---|---|
| GLNRPS4 | Non-ribosomal peptide synthetase | PANRPS4 | 98; 65 | EcdA | 98; 55 |
| GLPKS4 | Polyketide synthase | Absent | No | Absent | No |
| GLligase | AMP-dependent ligase | PALigase | 100; 59 | EcdI | 98; 57 |
| GLTRT | ABC transporter | PATRT | 99; 60 | EcdL | 99; 51 |
| GLHYD | Thioesterase | PAHYD | 100; 57 | Absent | no |
| GLP450–1 | Cytochrome P450 | PAP450–1 | 98; 57 | HtyF | 93; 52 |
| GLP450–2 | Cytochrome P450 | PAP450–2 | 99; 55 | EcdH | 98; 49 |
| GLOXY-1 | Oxygenase | PAOXY-1 | 99; 67 | EcdG | 99; 56 |
| GLOXY-2 | Oxygenase | PAOXY-2 | 100; 69 | HtyE | 100; 64 |
| GLOXY-3 | Oxygenase | PAOXY-3 | 93; 66 | Absent | No |
| GLOXY-4 | Oxygenase | PAOXY-4 | 100; 71 | EcdK | 100; 62 |
| GLHtyA | 2-Isopropylmalate synthase | PAHtyA | 82; 67 | HtyA | 93; 64 |
| GLHtyB | d-Amino acid aminotransferase | PAHtyB | 98; 74 | HtyB | 100; 64 |
| GLHtyC | Isopropylmalate dehydrogenase | PAHtyC | 98; 73 | HtyC | 99; 71 |
| GLHtyD | 3-Isopropylmalate dehydratase | PAHtyD | 100; 65 | HtyD | 95; 62 |
| GLHYP | Hypothetical protein | PAHYP | 99; 38 | EcdJ | 56; 51 |
See Cacho and colleagues (2012) and Li and colleagues (2015) for gene nomenclature.
Another unexpected feature of the acrophiarin gene cluster is the presence of an orthologue of GLHYD (Figs 1 and 5; Table 3), a gene encoding a thioesterase that apparently assists in offloading the side-chain from the highly reducing PKS (Chen et al., 2016b). However, this enzyme could also conceivably hydrolyse myristoyl CoA/myristoryl-ACP to myristate, thus making it available to the first adenylation domain of the NRPS. Current data indicate that thioesterase-encoding genes are absent in all other Aspergillus-type echinocandin BGCs. Furthermore, unlike other Aspergillus-type, echinocandin BGCs described to date, the acrophiarin gene cluster contains an orthologue of GLOXY3 that encodes a non-heme iron, α-ketoglutarate-dependent oxygenase responsible for hydroxylation of the C3 of the glutamine (Yue et al., 2015; Hüttel, 2017). These latter two features indicate a strong affinity of the acrophiarin BGC to those of the Leotiomycete-type echinocandins.
Evolution of the acrophiarin NRPS and other pathway enzymes
In order to test whether acrophiarin NRPS modules were more similar to Eurotiomycete- or Leotiomycete-type NRPS modules, the predicted A domains were extracted from the deduced amino acid sequences for each of the echinocandin NRPSs and were aligned with our previously published data set of echinocandin-type NRPS A domains (Yue et al., 2015). The analysis also included the A domains of InpB, the first NRPS of the fellutamide BGC (Fig. 1) (Yeh et al., 2016), because in our previous work, the InpB NRPS and the InpC acetyl-CoA ligase were found to be close phylogenetic relatives of the echinocandin NRPSs and acetyl-CoA ligases respectively. BLAST searches of public databases and bioinformatic analysis of genes identified additional fellutamide gene clusters in species of Aspergillus sect. Nidulantes (A. sydowii, A. versicolor, A. mulundensis and A. pachycristatus) and fellutamide-like gene clusters in other unrelated ascomycetes, including Spathularia flavida (Leotiomycetes, Rhytismatales), Bisporella sp. (Leotiomycetes, Helotiales) and Lobaria pulmonaria (Lecanoromycetes, Peltigerales) indicating that the fellutamide family of lipopeptides may more widespread than previously recognized (Shigemori et al., 1991; Lee and Hong, 2011; Xu et al., 2011; Wu et al., 2014; Kjærbølling et al., 2019). Because the products of these BGCs in these latter fungi are unknown, we refer to them as fellutamide-like BGCs. These InpB and InpC orthologues were added to the analysis to explore possible relationships between the fellutamides and echinocandins, and they fell within their respective fellutamide subclades of the echinocandin lineage with significant statistical support (Figs 6 and 7).
Fig. 6.
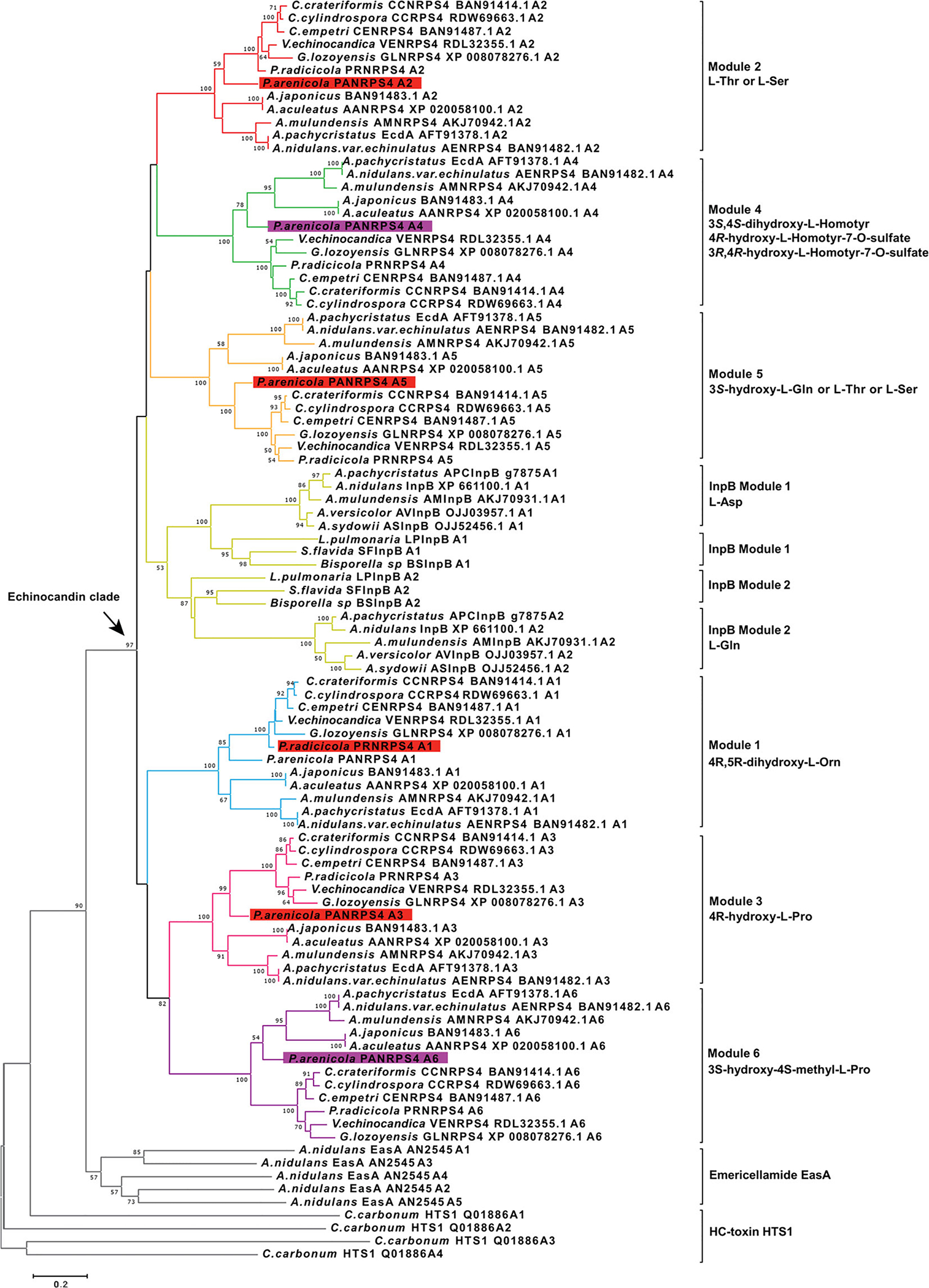
Phylogenetic analysis of the echinocandin and fellutamide NRPS A domains. Evolutionary analyses were conducted in MEGA 7.0 by using the maximum likelihood method based on the JTT matrix-based model. The tree is rooted with A domains of emericellamide synthetase (EasA). The corresponding amino acids that are activated by each subclade of A domains are indicated to the right. Numbers at nodes are likelihood bootstrap support values.
Fig. 7.
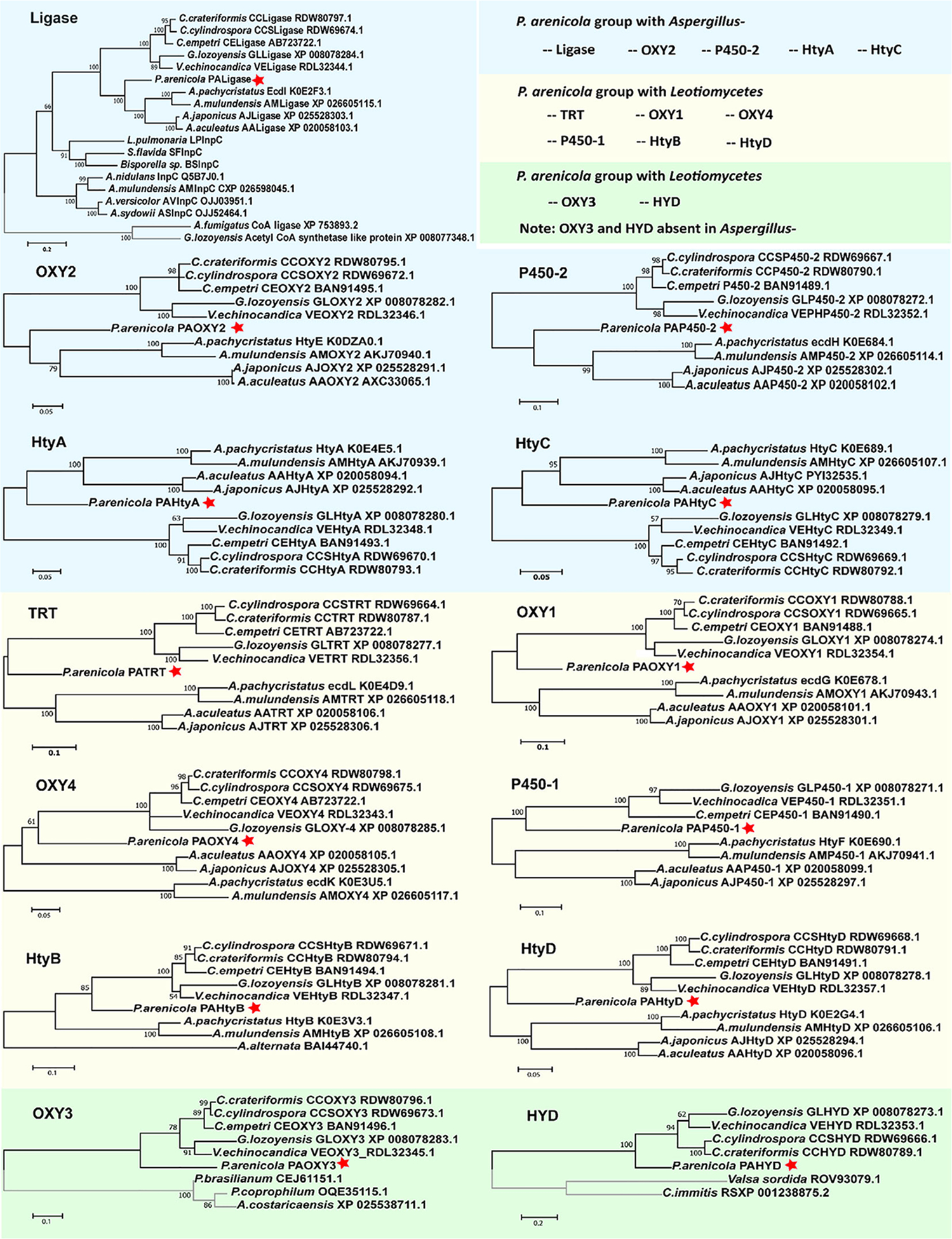
Abbreviated trees for maximum likelihood phylogenies inferred from genes in the echinocandin biosynthetic gene clusters. Maximum likelihood phylogeny was inferred from each of the echinocandin biosynthetic-related genes. Each of the echinocandin biosynthetic-related genes forms a monophyletic lineage. Eight of 13 P. arenicola acrophiarin biosynthetic genes group with genes Leotiomycete-type echinocandins. A. Phylogeny of acyl-AMP and AMP-dependent ligases. B, G, H and L. Phylogeny of non-heme iron, α-ketoglutarate-dependent oxygenases. C and I. Phylogeny of cytochrome P450s. D. Phylogeny of 2-isopropylmalate synthases. E. Phylogeny of isopropylmalate dehydrogenases. F. Phylogeny of ABC transporters. J. Phylogeny of d-amino acid aminotransferases. K. Phylogeny of isopropylmalate dehydratases. M. Phylogeny of thioesterases. Numbers at nodes are likelihood support values.
A maximum likelihood (ML) tree of these A domains (Fig. 6) reproduced similar topological features observed in a prior analysis (Yue et al., 2015). The ML tree indicated that all the echinocandin A domains and both A domains of the InpB NRPS (ANID_03496) of the fellutamide tripeptides from A. nidulans (Yeh et al., 2016), and its orthologues ASInpB from A. sydowii, AVInpB from A. versicolor, AMInpB from A. mulundensis, and APCInpB from A. pachycristatus NRRL 1140, and newly found fellutamide-like BGCs formed a distinct clade (97% support value). Therefore, the echinocandin NRPSs along with the InpB module 2 and its orthologues formed a well-supported monophyletic lineage within the euascomycete clade synthetase subfamily of fungal NRPSs (Bushley and Turgeon, 2010) (Fig. 6). It should be noted that the InpB module 2 activates l-glutamine while the InpB module 1 activates L-asparagine. It is unclear whether L-glutamine activation by the InpB module 2 and the Leotiomycete-type fifth position A domains or is indicative of a past evolutionary connection between the fellutamide and echinocandin peptide sequences, or whether incorporation of a common amino acid is coincidence.
Within the echinocandin A domain clade, the six individual A domains exhibited highly resolved intra-clade relationships (Fig. 6). The echinocandin clade was resolved into seven well-supported subclades, each corresponding to one of the six amino acid positions in the echinocandin nucleus plus a clade for modules 1 and 2 of InpB and similar fellutamide-like NRPSs (Fig. 6). As is the case with other NRPS orthologues from distantly related fungi (Bushley and Turgeon, 2010), this topology indicates that the A domains for each amino acid position from each distantly related fungus are more similar to each other than the individual A domains from a given echinocandin-producing strain. As expected based on its function, the acrophiarin NRPS module 5, which activates glutamine, formed a cluster (100% bootstrap value) with its corresponding Leotiomycete A domains. Although, the Aspergillus-type NRPS A domains were phylogenetically distinct from those of the Leotiomycete-types, the phylogenetic placement of acrophiarin A domains were in nearly all cases (except for module 4 and 6) more closely related to those of the Leotiomycetes although the fungus belongs to the Eurotiomycetes. Within each of these echinocandin A-domain clades, the branching of terminal leaves was congruent with the expected phylogenetic species trees except for the A domains from the acrophiarin NRPS. These analyses strongly supported the hypothesis that the extant echinocandin and fellutamide NRPSs were descendants from a common ancestral peptide synthetase (Fig. 6). However, the phylogenetic affinity of the acrophiarin NRPS A domains deviated from those of the Aspergillus species, and this phylogenetic incongruency could be interpreted as evidence for a closer relationship to echinocandin BGCs of Leotiomycete fungi.
To further explore the relationships of the acrophiarin cluster genes and the variations in their echinocandin gene orthologues, we built phylogenetic trees for individual echinocandin and related fellutamide pathway genes (Fig. 7). In trees inferred from orthologues of individual echinocandin genes, relationships among more closely related genes of the Leotiomycete- and Aspergillus-types were generally well resolved (bootstrap values >80%). In most single-gene trees (except for OXY4), the aculeacin genes (A. japonicus/aculeatus) formed a distinct branch from the echinocandin/mulundocandin genes (A. pachycristatus/mulundensis). Also, in the acetyl-CoA ligase tree, the fellutamide acetyl-CoA ligases (InpC) and orthologues from fellutamide-like BGCs appeared to be basal to the echinocandin ligases. InpC from A. nidulans fellutamide BGC exhibited about 51% identity to EcdI from NRRL 1440 echinocandin B gene cluster. This observation, along with the fact that in some Nidulantes species (A. pachycristatus/mulundensis) the fellutamide and echinocandin BGCs coexist in the same genome, prompts further speculation that the fellutamide gene cluster may share common ancestry with progenitors of the echinocandin gene clusters. It should also be pointed out that fellutamides are also antifungal, albeit with a mode of action distinct from that of echinocandins (Xu et al., 2011).
The relationships among the acrophiarin cluster genes and the corresponding genes of the two lineages of echinocandins were generally unresolved (Fig. 7). In some cases, acrophiarin gene sequences fell on a distinct branch intermediate between the two echinocandin types. A few branch conflicts were observed in some gene trees, but the conflicts were statistically unsupported by bootstrap analysis. Only in the case of the acetyl-CoA ligase, did an acrophiarin gene clearly fall into the Aspergillus-type echinocandin clade (100% bootstrap value). On the one hand, the P450–1, HtyB and OXY4 gene were strongly associated with the Leotiomycete-type clade with support values of 100%, 85%, and 61% respectively. Nevertheless, the overall high similarities of the acrophiarin and the Leotiomycete-type echinocandin cluster including the presence of GLHYD in the acrophiarin BCG, which is thought to assist the acetyl-CoA ligase in off-loading 10,12-methyl myristate in G. lozoyensis (Chen et al., 2016b), suggested a close relationship between acrophiarin and Leotiomycete-type echinocandins, e.g. pneumocandins. On the other hand, the absence of an orthologue of GLPKS4, which provides 10,12-methyl myristate in some Leotiomycetes, suggests a hypothetical scenario where the Aspergillus-type echinocandins clusters were lost in most lineages of the Aspergillaceae, including the Phialomyces lineage. Subsequently, an acrophiarin BGC could have been reintroduced into Phialomyces lineage via HGT soon after the ancestral divergence of the Leotiomycete and Aspergillus lineages.
To compare phylogenetic relationships of echinocandin cluster orthologues to the phylogeny of the corresponding fungi in which they occur, we inferred a species tree from the concatenated sequences of six housekeeping genes (18S rRNA, 28S rRNA, ITS RNA region, β-tubulin, translation elongation factor 1-α and RNA polymerase II subunit 2) to a tree based on the 10 shared enzymes of the echinocandin pathways (NRPS, acetyl-CoA ligase, TRT, P450–2, OXY1, OXY2, OXY4, HtyA, HtyC and HtyD) (Fig. 8). We analysed trees inferred from each housekeeping gene individually, which indicated that the individual housekeeping-gene trees were congruent and consistent with current models of ascomycete species phylogeny (data not shown). The high bootstrap values for almost all branches in the species tree provided evidence for the hierarchical relationships of most of the genera examined (Fig. 8A). Even though P. arenicola clearly grouped with the Eurotiomycetes in the housekeeping gene tree, it fell outside the Eurotiomycetes in the echinocandin gene tree. Thus, the topologies of the species tree and the concatenated echinocandin-gene tree were conflicted, and the incongruent branch with P. arenicola was the cause of the conflict (Shimodaira–Hasegawa test, P = 0; weighted Shimodaira–Hasegawa test, P = 0) (Fig. 8B; Fig. S6). Such phylogenetically incongruent gene sets are classic evidence of a past HGT event. This conflict suggested a scenario where the acrophiarin BGC may have been horizontally transferred from a Leotiomycete ancestor, and this putative HGT event occurred subsequent to the divergence of Leotiomycete- and Aspergillus-type echinocandins.
Fig. 8.
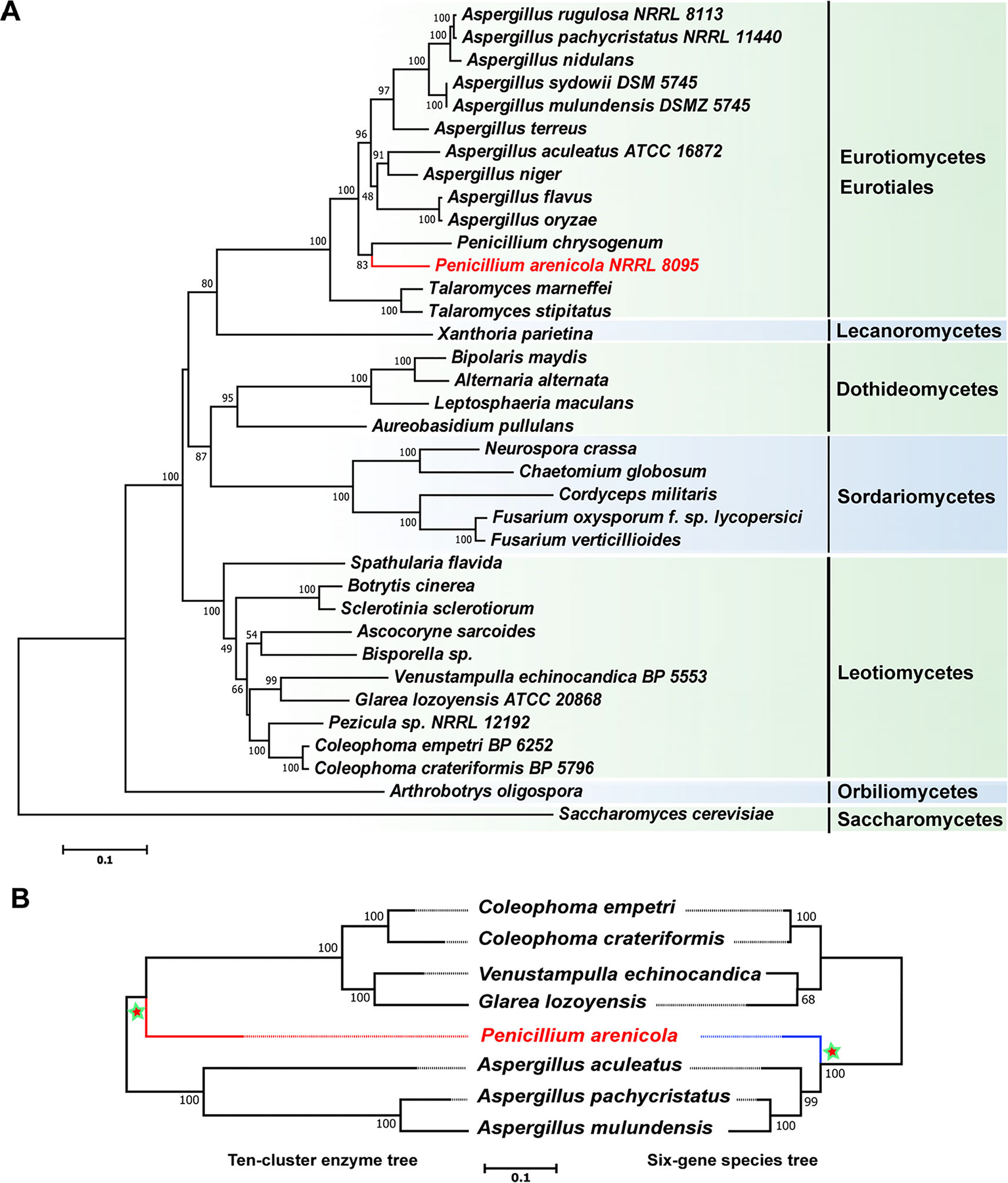
Maximum likelihood phylogenies of fungal species and the enzymes of the echinocandin pathway. A. Phylogenetic reconstruction of the echinocandin-producing fungi and other Ascomycetes using maximum likelihood analysis of a six-gene dataset consisting of DNA fragments of the 18S rRNA gene, the 28S rRNA gene, the ITS RNA region, the β-tubulin gene, the translation elongation factor 1-α gene and the RNA polymerase II subunit 2 gene. B. Consensus phylogenetic pattern for gene tree for 10 shared enzymes of all echinocandin pathways (left) and the established phylogenetic tree extracted from the data in panel A. Trees are rooted at the midpoint. Numbers at nodes are likelihood support values. The 10-cluster enzyme tree significantly conflicted with the species phylogeny tree (Shimodaira–Hasegawa test, P = 0; weighted Shimodaira–Hasegawa test, P = 0). Dotted lines are aids to connect branch tips to strain name.
Penicillium arenicola is self-resistant to acrophiarin and other echinocandins.
Previous studies demonstrated that the growth of echinocandin-producing strains of Aspergillus is sensitive to exogenously applied echinocandins, while the Leotiomycete-type echinocandin-producing strains are generally insensitive or have reduced sensitivity to echinocandins (Tóth et al., 2012; Yue et al., 2018). To test whether the acrophiarin-producing P. arenicola are susceptible to echinocandins as in Aspergilli or have innate elevated resistance to echinocandins like Leotiomycetes that produce echinocandins, we carried out a zone of inhibition (ZOI) assay with three P. arenicola strains, the pneumocandin-producing strain G. lozoyensis, the echinocandin B producing-strain A. pachycristatus and C. albicans against pure echinocandins. Natural echinocandins (pneumocandin B0, echinocandin B and acrophiarin) caused large ZOIs with A. pachycristatus and C. albicans, while the same compound set had little effect on the three P. arenicola strains (Fig. 9 and Fig. S7). Glarea lozoyensis exhibited minimal sensitivity to pneumocandin B0 and was unaffected by acrophiarin and echinocandin B. Bioinformatic analysis of the P. arenicola genome sequence revealed only a single copy of FKS1. These results indicated that the acrophiarin-producing strains of P. arenicola are intrinsically resistant to echinocandins as is the case for echinocandin-producing species of the Leotiomycete lineage, e.g. G. lozoyensis. To date, echinocandin resistance mechanisms have been attributed to amino acid mutations in the FKS1 protein (a plasma membrane-embedded enzyme), a compensatory increase in chitin synthase, or a second auxiliary copy of FKS1 associated with an echinocandin BGC (Johnson et al., 2011; Tóth et al., 2012; Walker et al., 2015; Slot, 2017; Yue et al., 2018). Therefore, the mechanism underlying the intrinsic resistance to echinocandins in G. lozoyensis and P. arenicola remains unclear.
Fig. 9.
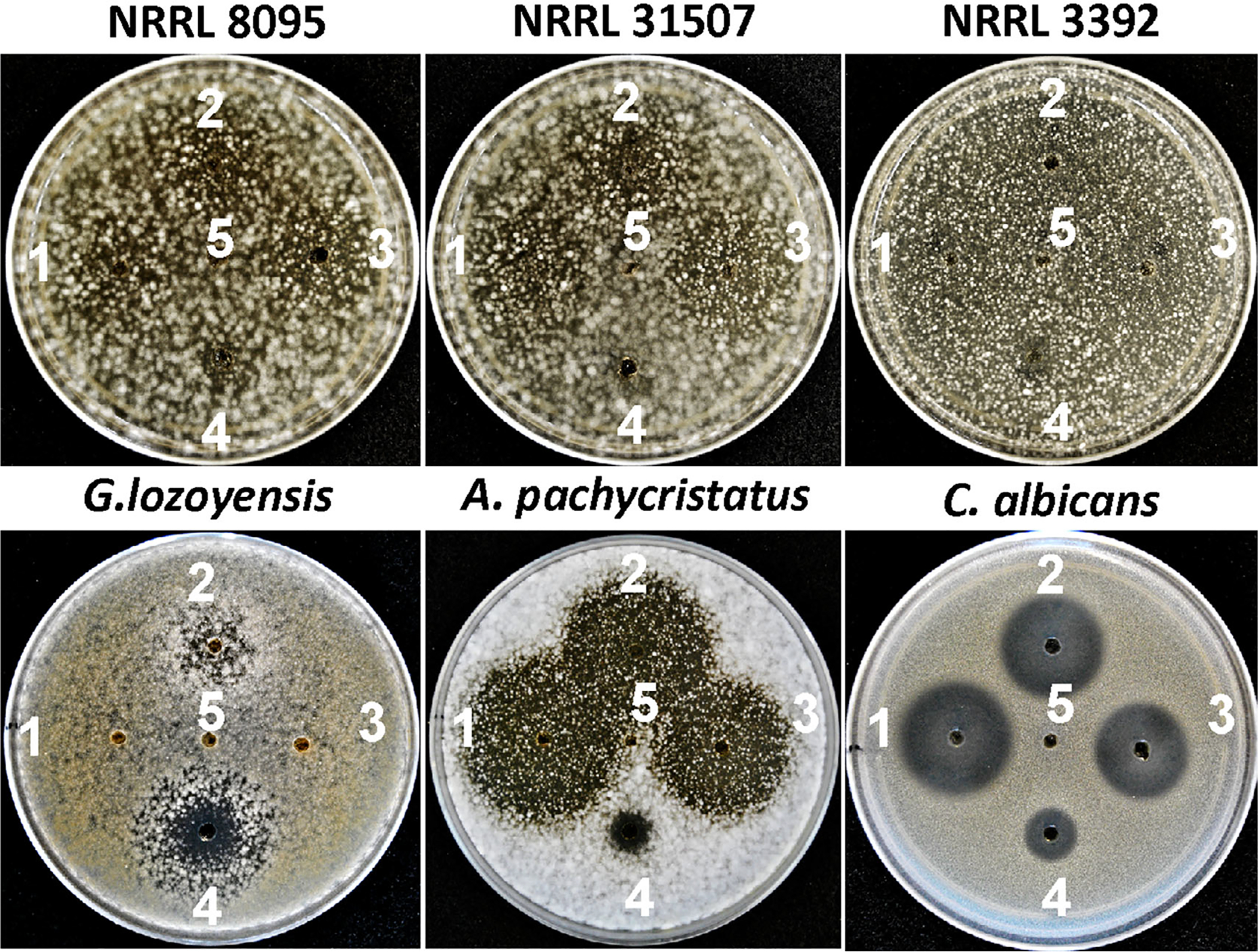
Zone of inhibition assays for evaluation of echinocandin susceptibility of P. arenicola (NRRL 8095, NRRL 31507 and NRRL 3392), G. lozoyensis (ATCC 20868), A. pachycristatus (NRRL 11440) and C. albicans (ATCC 10231). 1. Acrophiarin (250 μg ml−1), 2. Pneumocandin B0 (250 μg ml−1), 3. Echinocandin B (250 μg ml−1), 4. Amphotericin B (250 μg ml−1) and 5. DMSO.
Discussion
The genetic mechanisms leading to the formation, persistence, and loss of complex secondary metabolite BGCs remain poorly understood. A combination of genomic, phylogenetic, functional and biochemical analyses has provided insights into the evolutionary history of echinocandin biosynthesis in species of diverse genera of Pezizomycotina (Yue et al., 2015; Hüttel, 2017; Yue et al., 2018). The above results build upon previous analyses and indicated that structural diversity of echinocandins produced by these fungi has arisen largely from gains and losses of genes and changes in specificities of adenylation sites of the core NRPS gene during the evolutionary histories of the echinocandin BGCs. A variety of mechanisms, including gene duplication, neofunctionalization, introgression, HGT and horizontal chromosome transfer, have the potential to contribute to gain and diversification of secondary metabolic biosynthetic genes in fungi (Wisecaver et al., 2014; Koczyk et al., 2015; Florea et al., 2017; Slot, 2017; Feurtey and Stukenbrock, 2018; Thynne et al., 2019). One of the most important requisites for inferring relationships and prediction of HGT events is that all extant forms of the genes be included in the analysis (Feurtey and Stukenbrock, 2018). The inclusion of the acrophiarin BGC along with the discovery of the fellutamide BGC (Yeh et al., 2016) have filled important gaps in previous analyses of the echinocandin family. The addition of the acrophiarin BGC to our phylogenetic analyses indicates that the evolutionary histories of echinocandin-encoding genes do not always parallel the phylogenetic relationships of echinocandin-producing fungi, in contrast to previous conclusions (Yue et al., 2015). Therefore, the revised evolutionary pattern among echinocandin BGCs more resembles reticulate patterns of pathway evolution observed for other families of fungal secondary metabolites, and where conflicted gene phylogenies of some of the pathways genes have been offered as classic evidence of past HGT events (Wisecaver et al., 2014; Lind et al., 2017). One scenario supported by our phylogenetic analyses and comparative mapping of gene clusters suggests that the origin of acrophiarin BGC in P. arenicola of the Phialomyces lineage was derived from an ancestral Leotiomycete after the canonical Aspergillus-type echinocandin BGC had been lost during lineage sorting (Fig. 10).
Fig. 10.
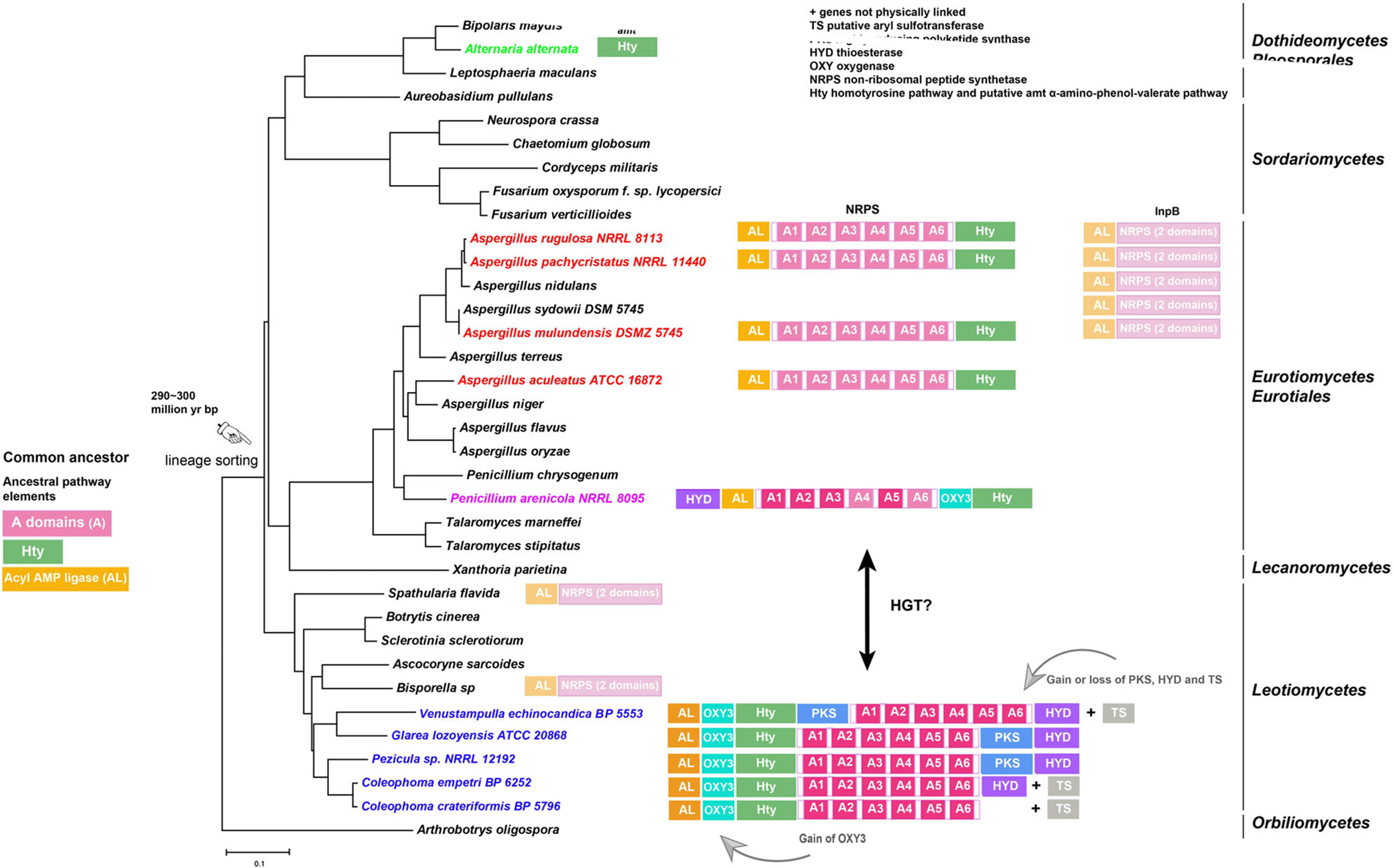
Evolutionary hypothesis for the family of echinocandin and fellutamide biosynthetic gene clusters. Possible ancestral elements observed in the Aspergillaceae include adenylation domains on the NRPSs, an acyl-CoA ligase of the fellutamide and echinocandin pathways and their orthologues, and the Hty pathway. The origin of the Hty pathway is unknown, and it is illustrated separately from the core NRPS of the echinocandins. In the Leotiomycete lineage, a mutation in A domain 5 leads to incorporation of glutamine in position 5, and an oxygenase for glutamine hydroxylation (OXY3) and a thioesterase (HYD) were recruited. A highly reducing PKS and a presumed aryl sulfotransferase (TS) were recruited in some species of the Leotiomycetes. An arrow indicates the possible origin of the acrophiarin gene cluster in Penicillium arenicola by horizontal transfer (HGT) from Leotiomycetes. The date of the estimated divergence of Leotiomycete–Eurotiomycete lineages is from Lutzoni and colleagues (2018).
A more complete picture of the extant echinocandin biosynthetic gene family allows for the inference of ancestral states of the echinocandin cluster and biosynthetic boundaries of the pathway genes. Genome sequences (including draft genomes) for at least 12 strains representing eight species have enabled comparisons of the echinocandin BGC locus across representatives of nearly all types of producing strains with varying capabilities to produce echinocandins (Fig. 10). Chemotype differences among these diverse species can be attributed to the presence or absence of genes encoding key pathway steps and variations in NRPS A domains (Yue et al., 2015; Hüttel, 2017). However, some anomalous reactions remain to be discovered, e.g. the mechanism of homotyrosine O-sulfation in some Leotiomycete-type echinocandins (Table. 1). Interestingly, to date, we have not detected evidence of widespread degenerate echinocandin gene clusters and single echinocandin genes among genomes of the Pezizomycotina suggesting that these genes clusters are quickly lost from the genome when the gene cluster degenerates. Thus, the distribution of echinocandin BGCs remains relatively narrow and is based almost entirely on detection of the corresponding echinocandins in antifungal assays from a limited number of species (Dreyfuss, 1986; Peláez et al., 2011; de la Cruz et al., 2012; Yue et al., 2015; Hüttel, 2017). However, the discovery of additional echinocandin and related NRPS clusters remains a possibility with more widespread genome sequencing of the Pezizomycotina and continued screening for cell-wall-active antifungal metabolites.
Hybridization and introgression may be important forces for interspecific dispersal of biosynthetic genes in fungi and in diversifying and generating novel metabolites (Olarte et al., 2015; Moore et al., 2017; Hubka et al., 2018). Although the number of available genomes of echinocandin-producing fungi precludes a detailed analysis, interspecific hybridization and introgression could have played a role in interspecific dispersion of the echinocandin BGCs, at least in species complexes where closely related species produce echinocandins. The greatest concentration of echinocandin-producing species recognized to date is in Aspergillus section Nidulantes. Species of this section reported to produce echinocandins include A. pachycristatus, A. spinulosporus, A. rugulosus, A. quad-rilineatus and A. navahoensis which produce echinocandin B and its variants (de la Cruz et al., 2012; Chen et al., 2016a; Hüttel, 2017), and A. mulundensis which produces mulundocandin (Bills et al., 2016). Because of the large number of species in section Nidulantes (65 species, Chen et al., 2016a), additional producers of echinocandin B and its variants are likely to be found with more targeted genome surveys or focused screening for cell-wall-active metabolites. Section Nidulantes encompasses a large complex of rapidly evolving species, with both homothallic and heterothallic species, consequently, interspecific dispersal of BGCs and their chemical evolution in part may be driven by hybridization and introgression as has been observed in the A. flavus complex (Olarte et al., 2015; Moore et al., 2017), and section Fumigati (Hubka et al., 2018).
Phylogenetic analysis of ascomycete NRPS A domains also provided evidence that the echinocandin clade of NRPSs also incorporates the InpB NRPS of the fellutamide pathway and similar orthologous NRPSs from diverse fungi (Fig. 6). Phylogenetic analyses suggested a degree of common ancestry between the two metabolite pathways (Figs 6 and 7), but because the relationship appears to be ancient and predating the divergence of major ascomycete lineages, it is difficult to speculate on the relationships between the echinocandin and fellutamide gene clusters. During the first step of fellutamide B biosynthesis (Fig. 1) (Yeh et al., 2016), InpC activates 3-hydroxydodecanoic acid to form 3-hydroxydodecanoyl-AMP that is then loaded onto the T0 domain of InpB. The 3-hydroxydodecanoyl-S-phosphopantetheinyl-T0 is extended stepwise with l-asparagine and l-glutamine by the two condensation- adenylation-thiolation modules of InpB. The linear lipodipeptide is then transferred from InpB onto InpA for the addition of the last amino acid, l-leucine. The lipotripeptide undergoes reductive release by the thioesterase domain of InpA resulting in (2S)-fellutamide B (Yeh et al., 2016). InpF might be involved in the release and transfer of the lipodipeptide from inpB to inpA. The relatively high protein sequence similarity between echinocandin acetyl-CoA ligases and InpC orthologues also suggests both these ligases might have shared a common ancestor and their divergence in function is a result of neofunctionalization, thus resulting in paralogues.
Additionally, most echinocandin genes bear some significant similarity to non-echinocandin genes. This suggests that the neofunctionalization of closely related non-echinocandin genes has contributed to the evolution of the gene clusters. For example, the four genes comprising the homo-L-tyrosine subcluster of all echinocandins BGCs and the four-gene cassette from the Alternaria alternata AM toxin pathway (amt gene cluster, Fig. 10) that is predicted to encode synthesis of the α-amino-4-phenyl-valeric acid monomer of AM toxin are examples of neofunctionalization. The genes of the Hty subpathway would need to be inherited with other pathway genes since the incorporation of homotyrosine has been shown to be essential for product formation (Cacho et al., 2012).
Ten orthologues are shared in all extant echinocandin BGCs across both lineages of the echinocandin-type BGC clusters (Fig. 8B) and are necessary for the conserved core lipopeptide skeleton structure, the peripheral genes that build the non-proteinogenic amino acids, the initiation and activation of side-chain variants, and ligation of the side-chain with the ornithine residue (Fig. 1). Thus, the ancestral echinocandin cluster and its products could be the same as simple echinocandins produced by extant species of Aspergilli, e.g. aculeacin and echinocandin B (Fig. 10, Table 1). It seems plausible the ancient echinocandin progenitor descended into both the Leotiomycete and Eurotiomyctes lineages. The above phylogenetic and synteny analyses point to a Leotiomycete ancestor as the likely donor pathway for the acrophiarin BGC because of the glutamine-specific A-domain 5 of the core NRPS and the presence of the thioesterase gene (GLHYD orthologues) and GLOXY3 orthologues. During the evolution of the Leotiomycete lineage, the addition of a thioesterase may have facilitated the recruitment of highly reducing PKS responsible for the branched PKS side-chain of the pneumocandins, sporiofungins, and FR190239. The different species of echinocandin-producing fungi have varied habitats (soil, endophytes and litter). The dispersal by HGT to a heavily sporulating species with a broad geographic distribution, e.g. the circumboreal P. arenicola, would provide ample opportunities for intimate contact needed for HGT from a rare, host-specific or geographically restricted species of the Leotiomycetes, e.g. G. lozoyensis, Venustampulla echinocandica, or Coleophoma cylindrospra. Finally, Penicillium arenicola also appears to be more similar to the Leotiomycete echinocandin-producing species with regard to self-resistance to exogenous echinocandins. Whether this is a coincidence or there is a mechanistic relationship remains to be determined.
In conclusion, the identification of the acrophiarin BGC in P. arenicola and the inclusion of the fellutamide gene cluster in the echinocandin biosynthetic family expands the known boundaries of echinocandin biosynthetic capabilities. The echinocandins exhibit a highly disjunct distribution among ascomycetous fungi from very different ecologies (e.g. soil, plant litter and endophytes). Therefore, a common ecological function for these metabolites remains unclear except for the fact that the discovery of auxiliary copies of FKS1 in some species clearly indicates their selection for interaction with fungal cell wall biosynthesis (Yue et al., 2018). Moreover, the diversity of fungi that produce echinocandins suggests their ecological roles may vary among plant-, litter- and soil-associated species. The strong phylogenetic relationships and coexistence of genes from the fellutamide BGC cluster in some Aspergillus genomes suggests an origin of both pathways prior to the divergence on the major lineages of filamentous ascomycete more than 400 million years ago (Lutzoni et al., 2018) leading to independent evolutionary courses for echinocandin pathways in the Eurotiomycete and Leotiomycete fungi. The origin of the Leotiomycete-like acrophiarin BGC in P. arenicola could be explained by a subsequent dispersal of a Leotiomycete echinocandin gene cluster into P. arenicola during an HGT event. This is a significant finding for the field of fungal lipopeptide biosynthesis and expands the possibilities to identify additional antifungal compounds and biosynthetic reactions encoded from the echinocandin-fellutamide family.
Experimental procedures
Strains and culture conditions.
The acrophiarin-producing strain P. arenicola NRRL 8095 (Dreyfuss and Tscherter, 1979) and three additional strains of P. arenicola were obtained from the U.S.D.A National Regional Research Laboratories (NRRL) (Table 2). Two strains of the closely related Phialomyces macrosporus were obtained from the IBT Culture Collection of Fungi (Table 2). The agar medium for growth and sporulation was YM agar (glucose 10 g, malt extract 3 g, yeast extract 3 g, peptone 5, agar 20 g per 1000 ml of deionized H2O). Five different fermentation media were tested for the production of acrophiarin based on their previous history for producing echinocandins. These media were: Medium I (glucose 20 g, casein peptone 5 g, NaNO3 3 g, of KHPO4 1 g, KCl 0.5 g, MgSO4·7H2O 0.5 g, FeSO4·7H2O 10 mg per 1000 ml of deionized H2O) (Dreyfuss and Tscherter, 1979); Medium IV (Debono, 1980) (ZnSO4·7H2O 4.5 mg, meat peptone 30.5 g, soybean meal 15.5 g, dextrin 2.0 g, blackstrap molasses (Brer Rabbit) 10.5 g, Na2HPO4 4.5 g, MgSO4·7H2O 5.5 g, FeSO4·7H2O 0.10 g, cottonseed oil 40 ml per 1000 ml of deionized H2O); Medium VI (Boeck and Kastner, 1981); Medium TG 106 (Tkacz et al., 1993) (D-mannitol 100 g, NZ amine 33 g, yeast extract 10 g, (NH4)2SO4 5 g, KH2PO4 9 g per 1000 ml of deionized H2O) and medium SMY (Bacto neopeptone 10 g, maltose 40 g, yeast extract 10 g per 1000 ml of deionized H2O).
For the seed cultures stage, six agar discs from 3-week old YM agar culture were inoculated into Medium SMY (maltose 40 g, neopeptone 10 g, yeast extract 10 g, 1000 ml deionized H2O) with 0.4% agar in 50-ml aliquots in 250-ml flasks. Seed cultures were grown at 24 °C, 220 rpm for 4 days. For the production cultures, 1-ml aliquots of the seed growth were transferred to flasks with 50 ml of Medium I, Medium IV, Medium IV, Medium TG 106 and Medium SYM respectively. For quantitative measurements of titers, five replicates of each fermentation were grown at 24 °C, 220 rpm for 12 days.
Genome sequencing and annotation.
NRRL 8095 was grown in a static culture of 100 ml SMY for 14 days at 23 °C. Mycelium was filtered, pressed dry, frozen at −80 °C and lyophilized. Genomic DNA was purified from ground mycelial powder with a Zymo Research Corporation Quick-DNA™ Fungal/Bacterial Miniprep Kit. For preparation of sequencing libraries, 500 ng of total genomic DNA was used as the template and processed using the KAPA HyperPlus Kit for PCR-free workflows (Roche, Switzerland) according to the manufacturer’s instructions. Sequencing libraries were size selected for 600–800 bp fragments using a LightBench (Coastal Genomics, Canada). Whole-genome sequencing was run on a HiSeq 4000 Sequencing System (Illumina, USA). The genome was assembled by SPAdes using standard parameters (Bankevich et al., 2012). Ab initio gene predictions from the genome assembly were made with Augustus (Stanke et al., 2004) using A. fumigatus as the reference genome.
The acrophiarin BGC in NRRL 8095 (GenBank MN518690) was identified by submitting the unannotated contig sequences for antiSMASH analysis (https://fungismash.secondarymetabolites.org/) and by reciprocal BLAST searches with sequences of known echinocandin biosynthetic genes from A. pachycristatus NRRL 11440 (JX421684, JX421685) and G. lozoyensis ATCC 20868 (PRJNA246203). The acrophiarin gene cluster occupied a single continuous locus on contig 7. Additional fellutamide BGCs from Spathularia flavida, Bisporella sp. and Lobaria pulmonaria were identified by BLAST searches and from annotated draft genomes available at the Department of Energy Joint Genome Institute’s Mycocosm portal (https://mycocosm.jgi.doe.gov/mycocosm/home).
Screening strains for echinocandin activity.
To detect echinocandin-type activity, strains (Table 2) were fermented using Medium SMY, Medium I, Medium IV, Medium VI and TG106. Each fermentation was extracted by the addition of an equal volume of methanol (MeOH) followed by shaking for 2 h. The H2O-MeOH mixture was filtered and evaporated under vacuum (Fig. S2). Residues were dissolved in DMSO at 10× of the original culture volume, and 20 μl of each DMSO extract was applied to a 4-mm well aspirated from a plate of YM agar seeded with an overnight culture of Candida albicans (ATCC 10231) or Cryptococcus neoformans H99. Plates were incubated at 25 °C and examined after 24–48 h for zones of inhibition.
Antifungal assay for determining the sensitivity of P. arenicola to echinocandins.
For the ZOI assays, fresh conidia suspensions of P. arenicola strains, pneumocandin-producing strain G. lozoyensis and echinocandin B-producing strain A. pachycristatus were added to melted YM agar at 45 °C and adjusted to a final conidial concentration to 2 × 103 conidia/ml. Twenty-five milliliter aliquots of the seeded agar media were poured into 9-cm Petri dishes. When the seed-agar plates are cooled and solidified, wells were made by aspirating agar with a 4-mm diameter syringe tip. Acrophiarin, pneumocandin B0 and echinocandin B were dissolved in DMSO to 250 μg/ml, and amphotericin B (250 μg/ml) was used as the control antimicrobial. Ten microliter aliquots of these compounds were added to each well. The plates of P. arenicola, G. lozoyensis and A. pachycristatus were incubated at 24 °C for 3, 6 and 2 days respectively, then, ZOIs were measured and photographed.
Extraction and analysis for HPLC-MS.
For each fermentation sample described in the previous step, a 5-ml aliquot of the aqueous-MeOH filtrate was evaporated to dryness, resuspended in 0.5 ml of MeOH, filtered through 0.2 μm cellulose membrane, and a 10-μL aliquot was analysed by HPLC-MS on an Agilent 1260 HPLC equipped with a diode array detector and coupled to an Agilent 6120 single quadrupole MS. Samples were eluted on a C18 reverse-phase column (Ace Equivalence 5C18, 4.6 × 150 mm, 5 μm) with a solvent gradient of 10%–100% B for 28 min [solvent A, 0.1% formic acid in H2O; solvent B, 0.1% formic acid in acetonitrile (ACN)], with a flow rate of 1.0 ml min−1. The chromatographic profiles were monitored by wavelength scanning from 190 to 400 nm and acquisition set at 210 nm and by positive and negative ESI-MS from m/z 160–1500.
Isolation and identification of acrophiarin and pneumocandin-enriched fractions.
To obtain sufficient mass for purification and biological tests, the crude extracts from different fermentations were pooled, evaporated to dryness and dissolved in 300 ml of H2O:MeOH (1:1 v/v). The extract was evaporated under vacuum until most of the MeOH was removed. The remaining aqueous sample was extracted with an equal volume of methyl ethyl ketone. After 2 h, the organic phase was separated and evaporated under vacuum. The crude extract was dissolved in a mixture of ACN and H2O (1:1 v/v) and fractionated with a Grace Reveleris X2 flash chromatography system using a Reveleris C18 RP 12 g cartridge (10%–100% ACN over 16 min, flow rate 30 ml min−1), using UV and ELSD detection, resulting in 16 fractions.
Fractions 2–5 were pooled, dried, dissolved in ACN: H2O (1:1 v/v) and further purified by semipreparative HPLC (Agilent Zorbax SB-C18 column; 5 μm; 9.4 × 250 mm; gradient of 30%–60% B for 20 min (solvent A, 0.1% formic acid in H2O; solvent B, 0.1% formic acid in ACN), 4.0 ml min−1), yielding 2.1 mg of acrophiarin. Fractions 6 and 7 were analysed by HPLC-MS with the same method described for crude extracts and showed the presence of other pneumocandin analogues, which were not further purified. NMR data were collected on a Bruker 500 MHz NMRs equipped with a 5-mm triple resonance cryoprobe at 298 K, with tetramethylsilane used as internal standard (TMS δH 0).
Prediction of adenylation domain specificity.
The adenylation domain (A-domain) structure determines the specific amino acids incorporated during peptide elongation and is a critical step in predicting the essential binding-pocket residues that correlate with the amino acid sequence of the NRPS product. Key positions of the A-domain binding pockets were determined at the PKS/NRPS Analysis website (http://nrps.igs.umaryland.edu) (Bachmann and Ravel, 2009).
Phylogenetic analysis and hypothesis testing.
Phylogenetic trees for all the enzymes in the echinocandin biosynthetic pathway were explored in the context of orthologous functional enzymes from across the Ascomycota. Proteins encoded by G. lozoyensis and A. pachycristatus echinocandin clusters and the A. pachycristatus fellutmide cluster were used as queries in BLAST searches against NCBI databases. Each set of proteins was aligned by using ClustalW implemented in MEGA 7.0 (Kumar et al., 2016), and the resulting alignment was manually adjusted. Phylogenies were inferred by the ML method implemented in MEGA 7.0 under a JTT + G model. Bootstrap supports were calculated using the default options in MEGA 7.0 with 100 replicates per run.
To estimate the phylogenetic affinities of NRRL 8095, a combined six-gene dataset, including the DNA fragments of the small-subunit (18S) rRNA gene, the large-subunit (28S) rRNA gene, the ITS RNA gene region, the β-tubulin gene, the EF1 gene and the RPB2 gene were resampled from previous phylogenetic studies of the echinocandin-producing fungi and other Ascomycota (Yue et al., 2015) and aligned with ClustalW implemented in MEGA 7.0. The best-fit nucleotide substitution models were determined for the alignment based on the lowest Bayesian information criterion scores. Positions containing gaps and missing data were eliminated. A GTR + G + I model was applied the alignment to construct phylogenies using the ML method with MEGA 7.0.
To test whether the phylogenies of the echinocandin pathway genes were congruent with the current classification of the Ascomycota, the concatenated phylogenetic marker gene sequences for the eight echinocandin-producing fungi were aligned with ClustalW and analysed with MEGA 7.0 by ML method using a GTR + G + I model. Ten pathway enzymes were found to be common among the eight echinocandin-producing fungi. The amino acid sequences of the 10 enzymes (NRPS, acyl AMP-dependent ligase, ABC transporter, oxygenase 1, oxygenase 2, oxygenase 4, P450–2, isopropylmalate dehydrogenase, 2-isopropylmalate synthase and aconitase) were combined to construct a phylogenetic tree. The concatenated amino acid sequences of these enzymes were aligned with ClustalW and analysed with MEGA 7.0 by the ML method using a JTT + G + I + F model. Alternative hypotheses based on the tree topologies assessed under the null hypothesis that all topologies were equally good explanations of the data were tested with the Shimodaira–Hasegawa test (Shimodaira and Hasegawa, 1999) and with a weighted Shimodaira–Hasegawa test as implemented in TREEFINDER (Jobb et al., 2004).
Supplementary Material
ACKNOWLEDGEMENTS
Parts of this work were supported by the Ben and Kay Fortson Endowment (to GFB) and a grant from the National Institutes of Health (R01 GM121458). We thank Michael Dreyfuss for historical information regarding the discovery of acrophiarin.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site:
Appendix S1: Supporting Information
REFERENCES
- Abbott BJ, and Fukuda DS 1981. S 31794/F-1 Nucleus. US Patent 4,304,716. [Google Scholar]
- Bachmann BO, and Ravel J (2009) Chapter 8. Methods for in silico prediction of microbial polyketide and non-ribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol 458: 181–217. [DOI] [PubMed] [Google Scholar]
- Balkovec JM, Hughes DL, Masurekar P, Sable CA, Schwartz RA, and Singh SB (2013) Discovery and develpment of first in class antifungal caspofungin (Cancidas). A case study. Nat Prod Rep 31: 15–34. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bills GF, Yue Q, Chen L, Li Y, An Z, and Frisvad JC (2016) Aspergillus mulundensis sp. nov., a new species for the fungus producing the antifungal echinocandin lipopeptides, mulundocandins. J Antibiot 69: 141–148. [DOI] [PubMed] [Google Scholar]
- Boeck LD, and Kastner RE 1981. Method of producing the A-30912 antibiotics. US Patent 4,288,549. [Google Scholar]
- Bushley KE, and Turgeon BG (2010) Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol Biol 10: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacho RA, Jiang W, Chooi YH, Walsh CT, and Tang Y (2012) Identification and characterization of the echinocandin B biosynthetic gene cluster from Emericella rugulosa NRRL 11440. J Am Chem Soc 134: 16781–16790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AJ, Frisvad JC, Sun BD, Varga J, Kocsubé S, Dijksterhuis J, et al. (2016a) Aspergillus section Nidulantes (formerly Emericella): polyphasic taxonomy, chemistry and biology. Stud Mycol 84: 1–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li Y, Yue Q, Loksztejn A, Yokoyama K, Felix EA, et al. (2016b) Engineering of new pneumocandin side-chain analogues from Glarea lozoyensis by mutasynthesis and evaluation of their antifungal activity. ACS Chem Biol 11: 2724–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz M, Martín J, González-Menéndez V, Pérez-Victoria I, Moreno C, Tormo JR, et al. (2012) Chemical and physical modulation of antibiotic activity in Emericella species. Chem Biodivers 9: 1095–1113. [DOI] [PubMed] [Google Scholar]
- Debono M 1980. Derivatives of cyclic peptide nuclei. European Patent Application 0031220 A1. [Google Scholar]
- Dreyfuss MM (1986) Neue Erkenntnisse aus einem pharmakologischen Pilz-screening. Sydowia 39: 22–36. [Google Scholar]
- Dreyfuss MM, and Tscherter H 1979. Antibiotic S31794/F-1. US Patent 4,173,629. [Google Scholar]
- Feurtey A, and Stukenbrock EH (2018) Interspecific gene exchange as a driver of adaptive evolution in fungi. Ann Rev Micobiol 72: 377–398. [DOI] [PubMed] [Google Scholar]
- Florea S, Panaccione DG, and Schardl CL (2017) Ergot alkaloids of the family Clavicipitaceae. Phytopathology 107: 504–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad JC, Houbraken J, Popma S, and Samson RA (2013) Two new Penicillium species Penicillium buchwaldii and Penicillium spathulatum, producing the anticancer compound asperphenamate. FEMS Microbiol Lett 339: 77–92. [DOI] [PubMed] [Google Scholar]
- Houbraken J, and Samson RA (2011) Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol 70: 1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V, Barrs V, Dudová Z, Sklenář F, Kubátová A, Matsuzawa T, et al. (2018) Unravelling species boundaries in the Aspergillus viridinutans complex (section Fumigati): opportunistic human and animal pathogens capable of interspecific hybridization. Persoonia 41: 142–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttel W (2017) Structural diversity in echinocandin biosynthesis: the impact of oxidation steps and approaches toward an evolutionary explanation. Z Naturforsch C 72: 1–20. [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, and Strimmer K (2004) TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol 4: 18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Johnson ME, Katiyar SK, and Edlind TD (2011) New FKS hot spot for acquired echinocandin resistance in Saccharomyces cerevisiae and its contribution to intrinsic resistance of Scedosporium species. Antimicrob Agents Chemother 55: 3774–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjærbølling I, Vesth T, and Andersen MR (2019) Resistance gene-directed genome mining of 50 Aspergillus species. mSystems 4: e00085–00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koczyk G, Dawidziuk A, and Popiel D (2015) The distant siblings—a phylogenomic roadmap illuminates the origins of extant diversity in fungal aromatic polyketide biosynthesis. Genome Biol Evol 7: 3132–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, and Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-M, and Hong J (2011) A cytotoxic fellutamide analogue from the sponge-derived fungus Aspergillus versicolor. Bull Korean Chem Soc 32: 3817–3820. [Google Scholar]
- Li Y, Chen L, Yue Q, Liu X, An Z, and Bills GF (2015) Genetic manipulation of the pneumocandin biosynthetic pathway for generation of analogues and evaluation of their antifungal activity. ACS Chem Biol 10: 1702–1710. [DOI] [PubMed] [Google Scholar]
- Lind AL, Wisecaver JH, Lameiras C, Wiemann P, Palmer JM, Keller NP, et al. (2017) Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLoS Biol 15: e2003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzoni F, Nowak MD, Alfaro ME, Reeb V, Miadlikowska J, Krug M, et al. (2018) Contemporaneous radiations of fungi and plants linked to symbiosis. Nat Commun 9: 5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masurekar PS, Fountoulakis JM, Hallada TC, Sosa MS, and Kaplan L (1992) Pneumocandins from Zalerion arboricola 2. Modification of product spectrum by mutations and medium manipulation. J Antibiot 45: 1867–1874. [DOI] [PubMed] [Google Scholar]
- McCorkindale NJ, Wright JLC, Brian PW, Clarke SM, and Hutchinson SA (1968) Canadensolide - an antifungal metabolite of Penicillium canadense. Tetra Lett 9: 727–730. [DOI] [PubMed] [Google Scholar]
- Moore GG, Olarte RA, Horn BW, Elliott JL, Singh R, O’Neal CJ, and Carbone I (2017) Global population structure and adaptive evolution of aflatoxin-producing fungi. Ecol Evol 7: 9179–9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarte RA, Worthington CJ, Horn BW, Moore GG, Singh R, Monacell JT, et al. (2015) Enhanced diversity and aflatoxigenicity in interspecific hybrids of Aspergillus flavus and Aspergillus parasiticus. Mol Ecol 24: 1889–1909. [DOI] [PubMed] [Google Scholar]
- Peláez F, Collado J, Platas G, Overy DP, Martín J, Vicente F, et al. (2011) Phylogeny and intercontinental distribution of the pneumocandin-producing anamorphic fungus Glarea lozoyensis. Mycology 2: 1–17. [Google Scholar]
- Perlatti B, Lan N, Earp CE, AghaAmiri S, Vargas SH, Azhdarinia A, et al. (2020) Arenicolins: C-glycosylated depsides from Penicillium arenicola. J Nat Prod 83: 668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt JI (1980) The Genus Penicillium and Its Teleomorphic States Eupenicillium and Talaromyces. London, UK: Academic Press. [Google Scholar]
- Shigemori H, Wakuri S, Yazawa K, Nakamura T, Sasaki T, and Kobayashi J.i. (1991) Fellutamides A and B, cytotoxic peptides from a marine fish-possessing fungus Penicillium fellutanum. Tetrahedron 47: 8529–8534. [Google Scholar]
- Shimodaira H, and Hasegawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol 16: 1114–1116. [Google Scholar]
- Slot JC (2017) Fungal gene cluster diversity and evolution. Adv Genet 100: 141–178. [DOI] [PubMed] [Google Scholar]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, and Morgenstern B (2006) AUGUSTUS: ab initio prediction of alternative transcripts. Nuc Acids Res 34: W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Douglas CM, Li W, Jue CK, Pramanik B, Yuan X, Rude TH, Toffaletti DL, Perfect JR, and Kurtz M (1999) A glucan synthase FKS1 homolog in Cryptococcus neoformans is single copy and encodes an essential function. J Bacteriol 181: 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thynne E, Mead OL, Chooi Y-H, McDonald MC, and Solomon PS (2019) Acquisition and loss of secondary metabolites shaped the evolutionary path of three emerging phytopathogens of wheat. Genome Biol Evol 11: 890–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacz JS, Giacobbe RA, and Monaghan RL (1993) Improvement in the titer of echinocandin-type antibiotics: a magnesium-limited medium supporting the biphasic production of pneumocandins A0 and B0. J Ind Microbiol 11: 95–103. [DOI] [PubMed] [Google Scholar]
- Tóth V, Nagy CT, Pócsi I, and Emri T (2012) The echinocandin B producer fungus Aspergillus nidulans var. roseus ATCC 58397 does not possess innate resistance against its lipopeptide antimycotic. Appl Microbiol Biotechnol 95: 113–122. [DOI] [PubMed] [Google Scholar]
- Vazquez JA, and Sobel JD (2006) Anidulafungin: a novel echinocandin. Clin Infect Dis 43: 215–222. [DOI] [PubMed] [Google Scholar]
- Walker LA, Lee KK, Munro CA, and Gow NAR (2015) Caspofungin treatment of Aspergillus fumigatus results in ChsG-dependent upregulation of chitin synthesis and the formation of chitin-rich microcolonies. Antimicrob Agents Chemother 59: 5932–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisecaver JH, Slot JC, and Rokas A (2014) The evolution of fungal metabolic pathways. PLoS Genet 10: e1004816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-J, Li C-W, and Cui C-B (2014) Seven new and two known lipopeptides as well as five known polyketides: the activated production of silent metabolites in a marine-derived fungus by chemical mutagenesis strategy using diethyl sulphate. Mar Drugs 12: 1815–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Ondeyka J, Harris GH, Zink D, Kahn JN, Wang H, et al. (2011) Isolation, structure, and biological activities of fellutamides C and D from an undescribed Metulocladosporiella (Chaetothyriales) using the genome-wide Candida albicans fitness test. J Nat Prod 74: 1721–1730. [DOI] [PubMed] [Google Scholar]
- Yeh HH, Ahuja M, Chiang YM, Oakley CE, Moore S, Yoon O, et al. (2016) Resistance gene-guided genome mining: serial promoter exchanges in Aspergillus nidulans reveal the biosynthetic pathway for fellutamide B, a proteasome inhibitor. ACS Chem Biol 11: 2275–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Q, Chen L, Zhang X, Li K, Sun J, Liu X, et al. (2015) Evolution of chemical diversity in echinocandin lipopeptide antifungal metabolites. Eukaryotic Cell 14: 698–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Q, Li Y, Chen L, Zhang X, Liu X, An Z, and Bills GF (2018) Genomics-driven discovery of a novel self-resistance mechanism in the echinocandin-producing fungus Pezicula radicicola. Environ Microbiol 20: 3154–3167. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Perez WB, Jiménez-Ortigosa C, Hough G, Locke JB, Ong V, et al. (2016) CD101: a novel long-acting echinocandin. Cell Microbiol 18: 1308–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


