Abstract
Less than 20% of the protein coding genome is thought to be targetable using small molecules. mRNA therapies are not limited in the same way since in theory, they can silence or edit any gene by encoding CRISPR nucleases, or alternatively, produce any missing protein. Yet not all mRNA therapies are equally likely to succeed. Over the past several years, an increasing number of clinical trials with siRNA- and antisense oligonucleotide-based drugs have revealed three key concepts that will likely extend to mRNA therapies delivered by nonviral systems. First, scientists have come to understand that some genes make better targets for RNA therapies than others. Second, scientists have learned that the type and position of chemical modifications made to an RNA drug can alter its therapeutic window, toxicity, and bioavailability. Third, scientists have found that safe and targeted drug delivery vehicles are required to ferry mRNA therapies into diseased cells. In this study, we apply these learnings to cystic fibrosis (CF). We also describe lessons learned from a subset of CF gene therapies that have already been tested in patients. Finally, we highlight the scientific advances that are still required for nonviral mRNA- or CRISPR-based drugs to treat CF successfully in patients.
Keywords: CRISPR, cystic fibrosis, mRNA, LNP, nanoparticle, barcoding
Simple Genetic Diseases are more Amenable to RNA Therapies than Complex Diseases
As the number of DNA and RNA reads from next-generation sequencing has grown exponentially,1 biologists have increasingly been able to identify genes that drive disease.2 An ideal way to address any genetic disease is by targeting the protein that the gene encodes using small molecules. However, less than 20% of encoded proteins are thought to be addressable using these compounds, since many diseases are caused by the absence of a wild-type protein or a protein that lacks an appropriate binding site.3
RNA therapies are drugs partially or fully comprising RNA nucleotides that can either downregulate or overexpress any desired gene. For example, siRNAs and antisense oligonucleotides (ASOs) can bind complementary mRNA and cleave it using Argonaute or RNase H1, respectively.4,5 In contrast, mRNA therapies seek to express a protein that is mutated in the patient, nonexistent or underexpressed by utilizing the cell's ribosomes.6 Alternatively, DNA-based therapies delivered with adeno-associated virus (AAV) enable long-term expression of a particular gene.7 As a result, DNA-based therapies lead to long-term gene expression, whereas mRNA therapies lead to transient gene expression.
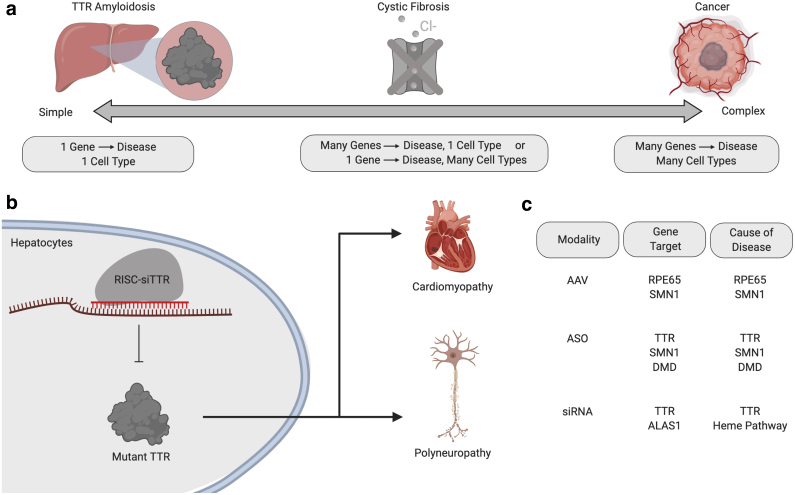
It is important to note that not all genes are equally likely to be effectively manipulated by an RNA drug; the likelihood an RNA therapy will work is influenced by the gene itself (Fig. 1a). One key trait that differentiates genes is the simplicity of the cell signaling that causes disease. Monogenic diseases, that is, diseases that are caused by mutations in a single gene, are preferable to diseases driven by mutations in several genes. One such example is the first FDA-approved siRNA drug, which consists of an ionizable lipid nanoparticle (LNP) that delivers siRNA targeting the transthyretin (TTR) gene to hepatocytes.8
Figure 1.
The complexity of a disease influences how likely it is a gene therapy will work. (a) An ideal target for gene therapy is a gene that is solely responsible for disease progression and is expressed primarily in one cell type. However, CF is a disease caused by CFTR affecting multiple cell types and, thus, lies in the middle of this complexity spectrum. (b) An example of an ideal target for gene therapy is TTR amyloidosis, a disease caused by mutant TTR expression in hepatocytes. Thus, siTTR delivered to hepatocytes slows disease progression. (c) Of the seven FDA-approved AAV, ASO, or siRNA gene therapies, only one does not meet the criteria for an ideal gene target: ALAS1 siRNA for acute hepatic porphyria. The other six diseases and the underlying gene targets are RPE65 DNA for Leber's congenital amaurosis, SMN1 DNA, and ASO for spinal muscular atrophy, TTR siRNA and ASO for TTR amyloidosis, and DMD ASO for Duchenne's muscular dystrophy. AAV, adeno-associated virus; ALAS1, aminolevulinic acid synthase 1; ASO, antisense oligonucleotide; CF, cystic fibrosis; CFTR, cystic fibrosis transmembrane conductance regulator; TTR, transthyretin.
The cell signaling diagram of TTR amyloidosis is ideal for an RNA therapy (Fig. 1b); mutant TTR protein drives disease, therefore, reducing the production of mutant TTR protein with siTTR halts disease. Notably, an antisense oligonucleotide that reduces mutant TTR expression is also approved.9 While different from RNA therapies, simple cell signaling diagrams can also be drawn with early successful AAV therapies, which use DNA to express a protein for long durations. For example, clinical data have been generated in patients treated with AAVs carrying RPE65 or SMN1, the absence of which drives Leber's congenital amaurosis and spinal muscular atrophy, respectively.10
In fact, six of the first seven FDA-approved AAV, siRNA, and ASO therapies, silence, modify splicing, or replace a gene that is singly responsible for disease progression (Fig. 1c). The seventh FDA-approved drug, termed Givosiran, is more genetically complicated. Patients are born with mutations that lead to buildup of toxic heme intermediates. Rather than targeting mutated genes directly, Givosiran silences aminolevulinic acid synthase 1 (ALAS1), an enzyme upstream of the heme synthesis pathway.11 Finally, genetically simple diseases benefit from a second advantage: sequencing enables the selection of patients with a specific mutation for clinical trials.
A second trait that makes a gene amenable to RNA therapies is cell type-specific expression. TTR is again an ideal case; mutant TTR does not play a significant role in the normal function of other cell types that could be targeted by the siRNA therapy. In the case of systemic mRNA therapies, off-target cells are likely to include macrophages and Kupffer cells,12 and, as a result, an mRNA therapy targeting a given gene may lead to significant off-target effects if that gene alters macrophage or Kupffer cell function.
Within the context of the lung, two analogous off-target cell types are pulmonary macrophages and endothelial cells; for example, developing a siRNA or ASO gene therapy for a disease in pulmonary epithelial cells would be challenging if that gene is critical for the function of either of the off-target cell types. In the context of cystic fibrosis (CF), there is evidence to suggest that cystic fibrosis transmembrane conductance regulator (CFTR) expression in cells of hematopoietic lineage may be physiologically significant,13,14 whereas CFTR expression is more limited in pulmonary endothelial cells. A related challenge is when the proposed gene target is a transcription factor (e.g., NF-κB) or other “master regulator” (e.g., Kras) that influences the function of many cell types.
In addition to genes with important functions in off-target cells and master regulatory genes, it may be difficult to develop mRNA therapies for diseases that are driven by a combination of genes (Fig. 1a). For example, RNA therapies have been designed to treat a single gene in cancers, even when the cancers are characterized by several driving mutations as well as genomic instability. One such case is treatment of KrasG12D in pancreatic ductal adenocarcinoma, where the cancer is characterized by somatic mutations in other genes.15,16 It remains unclear whether these RNA therapies will succeed in late-stage clinical trials.
One way to address cancers and other complex phenotypes is to develop combination therapies, wherein two to five distinct siRNAs are packaged into a single delivery system.17,18 Although promising, successful clinical data using combination RNA therapies have not been generated to date. Ongoing efforts involving large single-cell RNA-seq datasets (e.g., the Cell Atlas) may help deal with the challenge of picking ideal gene targets by uncovering how gene expression varies in different cell subtypes.19,20
Some CFTR Mutations can be Treated without RNA Therapies
Most gene targets lie somewhere on a spectrum between TTR, which is ideal, and a genetically unstable cancer, which is not ideal. One such example is CF, a disease caused by autosomal recessive mutations in the CFTR, located on chromosome 7 (Fig. 1a).21 In the United States, about 1 in 2,500 Caucasian children are diagnosed with the disease; considerably lower incidences are observed in both African American and Asian populations.22 Prognoses have significantly improved since the discovery of the disease; while still far lower than a person without CF, the average life expectancy now surpasses 40 years, largely due to development of treatments that manage bacterial infections, replace pancreatic enzymes, enhance nutrient absorption, and facilitate chest clearance of thick mucus23,24
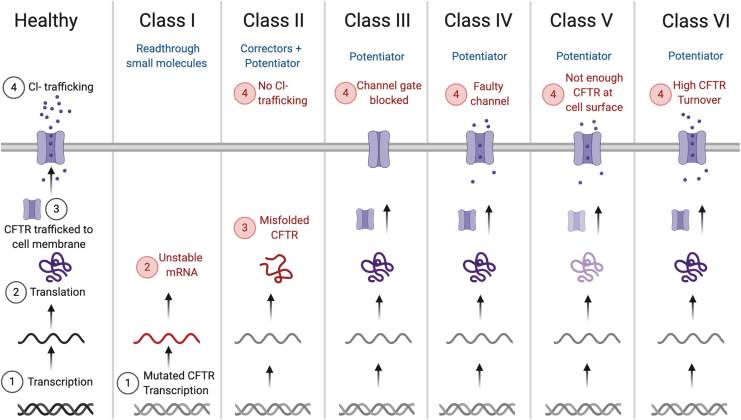
There are many CFTR mutant genotypes; recently, scientists estimated that over 1,000 genetic variations in the CFTR gene are represented in less than five patients each worldwide.25 Yet this suite of mutations can be grouped into just six classes, distinguished by the effect on the CFTR protein and its downstream phenotype within the cell.26 Some mutations within these classes can already be treated with FDA-approved small molecules.25,26 Other mutations cannot and may therefore be more dependent on RNA therapies in the future (Fig. 2).
Figure 2.
CF mutations fall under six different classes. These classes range from the generation of mutant CFTR mRNA transcripts that do not get translated to the generation of faulty CFTR protein, which does not allow for enough ion trafficking through the cell membrane. Different small molecules have been used to treat the six classes, including readthrough small molecules, corrector, and potentiator-based therapeutics. CFTR mutations that are unlikely to be addressed by existing small molecules may be better candidates for RNA gene therapies.
Class I mutations such as G542X, W1282X, or R553X result in a premature stop codon, which prevents functional protein from being created.27,28 To treat mutations of this kind, scientists have developed readthrough small molecules. These small molecules bypass premature stop codons to encourage transcription.26 Readthrough small molecules have been tested in several studies and clinical trials; however, it is unknown whether they will be approved by the FDA.29 This potential lack of small-molecule drugs to treat Class I mutations may alternatively be addressed by mRNA therapies designed to replace the missing CFTR gene.
Class II mutations create misfolded proteins that are unable to reach the cell membrane.26 Notable among these mutations is the ΔF508 variant: a mutation caused by the deletion of three base pairs in exon 10 that leads to the absence of a phenylalanine residue from the CFTR protein.30 More than 70% of defective CFTR alleles include this mutation, making it the leading cause of CF in the United States.23,27 As a result, a tremendous amount of research has been invested in finding treatments for patients with the ΔF508 mutation, resulting in approved small-molecule therapies.
As an example, the combination lumacaftor/ivacaftor therapeutic acts to partially restore CFTR function. Specifically, lumacaftor is a CFTR corrector that helps during the protein folding process, allowing CFTR to be trafficked to the cell surface.31 Once CFTR reaches the cell surface, ivacaftor acts as a potentiator, ensuring that CFTR allows chloride ions to pass through.32 Notably, for the case of ΔF508, ivacaftor is not effective on its own, since it requires properly folded CFTR protein to act upon.32,33
Next-generation correctors have also been developed to deal with different processing abnormalities, including tezacaftor and elexacaftor.25 Specifically, elexacaftor binds to a different pocket in the CFTR protein and results in the improvement of lung function and other respiratory-related factors when administered in combination with tezacaftor and ivacaftor.34 In fact, the FDA recently approved Trikafta, a triple combination therapy that combines elexacaftor, ivacaftor, and tezacaftor. Trikafta is approved for use in patients with at least one ΔF508 mutation, whereas previous single or combination therapies only treated patients homozygous for the ΔF508 mutation.35,36 These small-molecule treatments mark a huge success for CF scientists and patients. Future CF therapies, including mRNA therapies, will need to demonstrate extraordinarily high safety and efficacy to supplant these existing drugs.
Other observed CFTR mutations fall within classes III–VI. A Class III mutation lacks a functional ion channel gate, a Class IV has defective ion conductance across the CFTR channel, a Class V results in insufficient CFTR protein reaching the cell surface, and a Class VI has high CFTR turnover as a result of low protein stability at the cell membrane.27 Ivacaftor, the CFTR potentiator, has been approved by the FDA to treat 38 variants of CFTR mutations, mostly within Class III, but interestingly, shows differing degrees of improvement depending on the patient genotype.25,37
Relatedly, there are many CFTR variants that are extremely rare (sometimes as rare as a single person).25 As a result, it can be difficult to obtain sufficiently powered clinical evidence to support an approval for small-molecule drugs, leaving these patients in limbo. Even if there is clinical evidence that a rare or yet-to-be studied mutation may be treated by an existing small molecule, accessing these “off-label” drugs can be expensive, blocking patient access.25 There is a concerted effort to further understand which mutations are treatable with existing small-molecule therapies, and how they differentiate from untreatable mutation variants.25,27 Understanding these differences and the challenges they pose to CF patients is important for identifying mutations that may need to be addressed using RNA therapies.
RNA Therapies can be used to replace or edit CFTR
Given the complexities and potential genotype-specific efficacy of small molecules, there is an interest in developing gene therapies (including mRNA) to treat CF. In this study, we focus on gene therapies that require nonviral delivery vehicles. Gene therapies that utilize viral delivery vehicles have been reviewed.38 One such approach, which is agnostic to genotype, is to use mRNA to replace CFTR protein. A second approach is to use mRNA encoding for CRISPR-based gene editors to edit genomic DNA in target cells. The CRISPR approach is not agnostic to genotype, since each CRISPR drug would need to be targeted to the diseased locus (i.e., the mutation site). However, it is theoretically modular, since only the sequence of the sgRNA and the potential DNA template would need to be changed to address different mutations.
Treating CF by delivering mRNA that encodes CFTR has the potential to work in any CF patient, independent of the underlying mutation. It has been estimated that restoring 5% of wild-type CFTR mRNA in the cytosol is enough to ameliorate the symptoms of CF, but a somewhat higher threshold is necessary to avoid complications later in life.39 To this end, groups have shown that delivering exogenous CFTR mRNA to mice lacking wild-type CFTR results in production of functional ion channels.40,41 In both examples, scientists delivered CFTR mRNA using nanoparticles; these treatments led to improvements in lung functional parameters, including improved forced expiratory volume (FEV) values, and improved ion conductivity in the nasal epithelia in a manner resembling FDA-approved CF drugs.
These results suggest a CFTR mRNA therapy is feasible, but to be clinically relevant, it will require long-lived CFTR protein expression after mRNA delivery. If the CFTR protein is only produced for a short period of time, a patient will be required to re-administer the drug often, which could lead to toxicities from high doses of the drug delivery vehicle carrying the CFTR mRNA. Notably, the CFTR gene is transcribed at low levels and the mature protein product can be stable for extended periods of time, with a half-life of >15 h after reaching the plasma membrane.42–44
Furthermore, mRNAs (∼1–10 kb) are much larger than siRNA (∼20 bases) and sgRNAs (∼150 bases). It is feasible that the size and structure of the formulated RNA influence how it interacts with the LNP. For example, it was demonstrated that LNPs containing the same components, but using different molar ratios, have different efficiencies when delivering siRNA or mRNA. More specifically, a vehicle optimized for siRNA delivery to liver hepatocytes performed poorly for mRNA delivery, but orthogonal experimental design allowed for optimization of an mRNA vehicle that performed considerably better using different mole ratios.45 In the case of formulating Cas9 mRNA, sgRNA, and a donor strand, a high mass ratio of LNP: nucleic acid may be required to account for the fact that three distinct nucleic acids must be stably formulated together.
Another potential treatment is utilizing mRNA encoding nucleases such as CRISPR-Cas9 accompanied by gRNA and using them to edit DNA in target cells. Alternative options to facilitate CRISPR-based editing include the use of a Cas-gRNA complex—known as ribonucleoprotein (RNP)—plasmids encoding Cas9 and gRNA, or viral vectors.46 While all these options are viable and have been shown to work, there are drawbacks to using each. RNPs are ideal for timing—the editor and guide RNA complex are active immediately upon delivery to a target cell type. While formulation of RNPs into lipid-based delivery vehicles is often difficult due to their size, there are other approaches for RNP delivery such as conjugation with cell-penetrating peptides or other ligands, formulation with amphiphilic peptides, the use of polymeric or metal nanoparticles, and the use of techniques such as electroporation.47,48
While the use of plasmids to co-express an editor and a gRNA has seen wide success in simple cell models, it has also been shown to cause higher off-target editing relative to RNPs,49 and unintended genome integration and cell toxicity.50 Viral vectors, which can consist of either DNA genomes (e.g., adenovirus and AAV) or RNA genomes (e.g., lentivirus), are ideal for long-term expression and can induce potent editing, but target cell transduction is long lasting, which is undesirable for many Cas9 applications. To address this, researchers have developed methods to inactivate the expression cassette, or its Cas9 product, allowing for the reduction and modulation of cell exposure to the nuclease over time in vivo.51,52
The co-delivery of Cas9 mRNA and guide RNA to facilitate editing can bypass some of the drawbacks of delivering protein, using a plasmid, or using a viral vector. For example, mRNA is transient, allowing for editing to take place during a specific timeframe instead of being long lived. In addition, mRNA and guide RNA can be co-delivered within an LNP, which can be optimized to induce delivery in a specific on-target cell type or tissue. This approach has been used to facilitate potent on-target editing.53,54
In the context of CF, which requires a therapy to either increase expression of functional CFTR or make nonfunctional CFTR protein functional, these gene-editing therapies (i) require the nuclease to make a cut at a target site and then (ii) edit that cut site by “pasting” a DNA template encoding the desired sequence. Critically, Cas nucleases can efficiently make targeted cuts in DNA, but in most cell types, the pasting efficiency remains low. More specifically, most double-stranded DNA breaks (DSBs) are repaired using the error-prone nonhomologous end-joining (NHEJ) mechanism instead of homology-directed repair (HDR), which is necessary to repair the CFTR gene with a wild-type copy.55,56
To improve the efficiency of HDR, several approaches have been used. In one example, scientists use Scr7 to inhibit DNA ligase IV, a protein required for NHEJ, in mouse embryos harvested at E10 (10th day of pregnancy). Specifically, mouse embryos were treated with Cas9, two sgRNAs targeting different genes, a template strand, and the inhibitor. The presence of Scr7 compared to the control favored HDR in two ways, by (i) completely eliminating NHEJ-mediated deletions at the two loci and (ii) increasing the rate of HDR up to fourfold.57 In another example, scientists used a reporter screen to identify genes that enhance or inhibit HDR but using a dsDNA template to simulate homologous recombination (HR). Interestingly, most of the genes necessary for HR belonged to the Fanconi Anemia pathway, a set of genes that was central to execute the HDR mechanism when the DNA template was single stranded.
In addition, small-molecule inhibition of genes counterproductive to HDR (as identified in the screen) yielded a candidate gene target, CDC7; its inhibition upregulated HDR compared to the control with both ssDNA and dsDNA templates across a multitude of gene loci.58 It will be critical to understand whether these approaches can be used in a cell type-agnostic manner, including in human pulmonary epithelial cells. Improving the efficiency of HDR-mediated gene editing is paramount if CRISPR therapies are used to treat CF. More specifically, in every cell that is cut, but precise gene editing does not occur, there is a risk of creating an insertion or deletion that effectively converts a Class II–VI mutation into a Class I mutation within that cell.
Nonetheless, there have been several advances in applying gene-editing approaches to CF. One of the first applications of the technology to the treatment of this disease was demonstrated in epithelial organoids grown from patient-derived intestinal stem cells with a homozygous ΔF508 mutation. Using HR with plasmid DNA as the donor, scientists demonstrated restored organoid swelling after forskolin exposure in treated samples versus the control.59 Similar approaches corrected the same mutation in induced pluripotent stem cells (iPSCs) derived from patients, which were later differentiated into lung epithelial cells with normal CFTR expression and function.60
In a different approach, researchers used CRISPR to target three different mutations in the intronic regions of CFTR mRNA that cause alternative splicing and produce a nonfunctional protein.61 Instead of using a DNA template to repair the cut site, the authors targeted two adjacent sites in the intronic regions spanning the mutation, which resulted in the deletion of that region and the DSB being repaired by NHEJ. More recently, a group delivered Cas9 RNP containing chemically modified sgRNA within AAV6 to upper airway basal stem cells and bronchial epithelial cells containing the ΔF508 mutation. They achieved 30–50% editing efficiencies that restored CFTR function equivalent to 20–50% of wild-type controls.62
Finally, scientists used helper-dependent adenoviruses to deliver Cas9, guide and template DNA to iPSCs, significantly improving gene editing efficiencies. While the majority of recombinants were able to integrate the donor DNA into one of the mutated alleles, the second allele had a high rate of indels at the Cas9 cleavage site, sometimes up to 8 kb deletions.63 In the context of CF, heterozygous wild-type CFTR expression is enough to avoid any pathology, but large deletions could have malignant effects.
One alternative is to use modified CRISPR systems with inactivated nuclease domains and proteins that promote base editing. These “base editors” allow scientists to edit SNPs in genes of interest without creating a DSB in the DNA, thereby minimizing the risk of potential NHEJ-mediated indels. Specifically, if the protein is a cytidine deaminase, a C-G pair can be changed to T-A (CBE), whereas an adenosine deaminase converts an A-T pair to a G-C (ABE).64 Recent studies have shown that these next-generation base editors have 1.5% indel rates for cytidine base editors, but <0.1% for adenine base editors.65,66
Adenine base editors have been used to correct nonsense mutations in a CF patient-derived organoid biobank that overrepresents rare CF mutations.67 In this example, authors used spCas9- and xCas9-derived adenine base editors to correct the R553X and R785X mutations. In another example, researchers used adenosine deaminases to correct the W496X CF mutation, which leads to a premature stop codon.68 Even though these base editors may provide a safer platform from where to perform gene editing and have proven efficient at correcting some CFTR mutations,67,68 they are not capable of addressing all mutant phenotypes. For example, for the ΔF508 mutation, base editors are unable to correct the mutant CFTR gene since an entire codon is missing from the sequence. However, a novel approach known as prime editing may be able to correct the ΔF508 mutation through base pair insertion.69
In addition, prime editing can mediate all 12 base-to-base conversions, rendering it useful for additional CF mutations, such as those discussed above. This form of editing uses an RNA-programmable nickase that is fused to a reverse transcriptase and a prime editing guide RNA (pegRNA). The pegRNA facilitates recognition of the target site and can be reverse transcribed to replace the target nucleotides with a desired template. Thus, prime editing may enable targeted editing for specific CF mutations.
Chemically Modifying MRNA Drugs improves their Safety and Efficacy
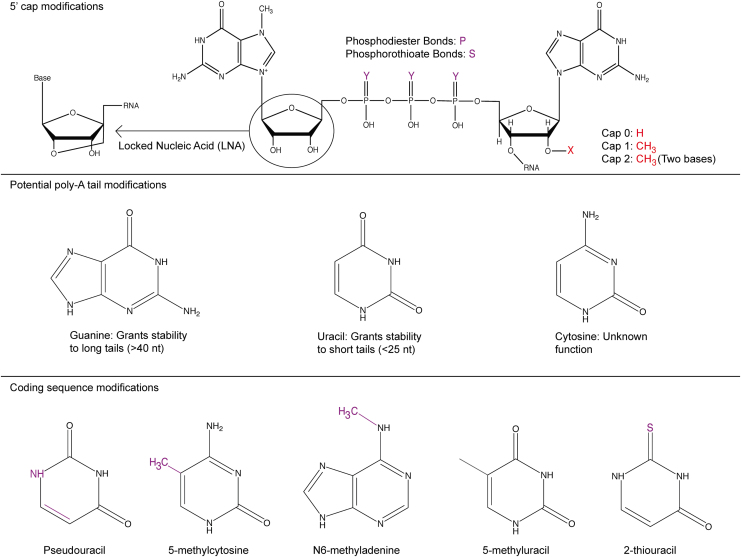
In addition to the selection of the gene itself, scientists have learned that chemical modifications made to RNA drugs can impact their safety and efficacy (Fig. 3). Chemical modifications improve the efficacy of RNA drugs through three potential mechanisms: (i) decreasing immune activation; (ii) increasing the stability of the drug, and therefore the duration of its effect; and (iii) influencing the affinity with which the RNA drugs bind other nucleic acids and proteins.70
Figure 3.
Chemical modifications made to mRNA can improve its half-life by occluding nucleases that degrade it or decreasing binding to receptors that initiate immune responses.
Chemical modifications made to mRNA drugs are often made at positions in the mRNA that are naturally modified in endogenous eukaryotic mRNAs.71 For example, endogenous mRNAs are modified in human cells to include a 5′ cap; mRNA drugs are also modified at the 5′ end. These 5′ cap modifications have been used to decrease activation of innate immunity,72 and can often be divided into three types: cap 0, 1, and 2, respectively.
Cap 0 is typically referred to as a m7G cap and is made up of an N7-methyl guanosine connected to the first transcribed nucleotide through a 5′–5′ triphosphate linkage. The presence of guanosine on the 5′ cap can reduce exonuclease activity, thereby stabilizing the mRNA and increasing the amount of protein produced per unit mRNA delivered into the cell.73
Cap 1 describes methylation of the 2′ sugar group on the initiating nucleotide, whereas cap 2 describes methylation of the 2′ hydroxy of the sugar on the first two nucleotides.74 Additional modifications include the use of a locked nucleic acid on the N7-methyl guanosine and phosphorothioate modifications to the triphosphate backbone, among others.75 In all cases, these modifications have been shown to improve mRNA, relative to unmodified mRNA. Yet future work is needed to study whether the ideal 5′ modification is universal for all mRNAs, or whether it changes based on the (i) mRNA sequence or (ii) cell signaling within the desired cell.
Endogenous mRNAs are also modified with a poly-A tail, which decreases exonuclease-mediated degradation of the mRNA. Scientists developed a method, known as TAIL-Seq, to study how poly-A tails influence mRNA stability. In TAIL-Seq, authors used RNA sequencing to understand the distribution of poly-A tail lengths in common cell lines (e.g., HeLa and NIH 3T3). They then correlated the poly-A tail lengths to mRNA half-life and determined that longer tail lengths were correlated with an increased half-life, but not improved translation. Using this approach, the authors identified that 50–100 nt poly-A tail lengths were most common in the cell lines tested. In addition, they noted the presence of uridylation on shorter poly-A tails and guanylation on longer poly-A tails, suggesting that both modifications at the 3′ end could improve mRNA stability.76 Techniques such as TAIL-Seq could be used to identify characteristics of long-lived mRNAs, helping scientists tailor mRNA expression profiles to fit the therapeutic needs of a particular disease.
In a separate example, researchers developed a technique known as FLAM-seq, which used RNA sequencing to understand co-dependencies between poly-A tail length and the 3′ untranslated region (UTR) and transcription start site. They also identified the presence of cystine residues in the poly-A tail of human cells.77 Others have used similar approaches based on sequencing to understand how poly-A tails impact translation and stability.78,79
Scientists have also optimized the 3′ UTR of the mRNA to increase its stability and augment protein expression.80 Specifically, researchers created a library of naturally occurring mRNA sequences from human dendritic cells that were attached to the end of an enhanced green fluorescent protein sequence, serving as the 3′ UTR. After six rounds of consecutive sequence selections, the optimized sequences were six times more stable in vitro than the initial mRNAs and had two to three times more protein expression in vivo than the commonly used 2hBg 3′ UTR sequence.
Similar approaches have been carried out by other research groups to optimize UTR sequences yielding encouraging results.81 Scientists selected 10 different 5′ UTR and 3′ UTR sequences from liver proteins and performed a combinatorial screen measuring ARG1 protein expression with these UTR variants. Interestingly, for the genes selected, 5′ UTR played a more important role in driving higher protein expression than 3′ UTR, suggesting the effect was driven by improved translation efficiency instead of greater mRNA stability.
mRNA drugs are made using in vitro transcription. As a result, they can also be rationally engineered to include chemically modified bases such as pseudouridine, methylated adenosine, 2-thiouridine, or 5-methyl-cytidine.71,82 In most cases, these modified bases are used to reduce signaling by pattern recognition receptors, most notably toll-like receptors (TLRs) and RIG-I, which activate the innate immune system upon sensing exogenous mRNA.83–85
In one example, by modifying an mRNA sequence so that 25% of uridine and cytidine bases were replaced with 2-thiouridine and 5-methyl-cytidine, authors reduced mRNA binding to TLRs and RIG-I; this led to reductions in production of IL-12 and IFN-γ, key mediators of innate immune response. When compared to mRNA containing other naturally found modified nucleosides, the incorporation of pseudouridine improved translation and decreased immune activation. In this case, uridine 5′-triphosphate (UTP) was replaced with a pseudouridine before in vitro transcription to make mRNA.86 Pseudouridine modifications are now commonplace and have been used instead of UTP to make mRNAs for vaccination against Zika virus and influenza.87,88
More recently, it has been found that chemically modifying mRNA can influence the activation of protein kinase R (PKR) in cells. Incorporation of pseudouridine into mRNA reduced activation of PKR, which was not previously known to be activated by mRNA.89 Using a firefly luciferase reporter system, authors demonstrated that unmodified mRNA bound and activated PKR, leading to downstream reductions in firefly luciferase translation. By contrast, pseudouridine-containing mRNAs were unable to bind PKR and resulted in a fourfold to fivefold increase in mRNA translation at similar doses. Interestingly, the concurrent delivery of unmodified and modified mRNAs reduced translation of the modified mRNA transcript, suggesting that PKR phosphorylation led to transcription-wide reductions of protein production.
Complementing this work, we found that that low doses of a TLR4 agonist, which is immediately upstream of PKR, dramatically reduced the amount of LNP-mediated mRNA delivery in vivo, in all tested cell types. Importantly, mRNA delivery was rescued by inhibiting PKR and TLR4 with small molecules, illustrating the value of understanding cellular pathways that block the translation of therapeutic mRNA.90
Modified mRNAs have been used in mRNA drugs treating CF patients. Specifically, Translate Bio, an mRNA company, is currently evaluating a treatment for CF known as MRT5005. MRT5005 is an inhaled mRNA therapy that contains a 5′ cap and a poly (A) tail to increase translation efficacy and prevent mRNA degradation. To mimic endogenous mRNA, the bases were left unmodified; it is unclear whether additional modifications could further improve the efficacy of this drug.
Preclinical studies of MRT5005 conducted in vitro using Fischer rat thyroid and human bronchial epithelial91 cells showed improved CFTR channel activity. In vivo delivery of CFTR mRNA to healthy rats and nonhuman primates led to increased CFTR mRNA in lung tissue after a single dose, and no adverse effect was detected at all dose levels. Furthermore, a study on CFTR−/− rats exhibited significantly improved chloride transport as indicated by nasal potential difference. Finally, Translate Bio conducted a placebo-controlled interim study in 12 subjects using MRT5005 that showed the drug was well tolerated at low- and medium-dose levels. Notably, subjects in the medium-dose group saw a significant improvement in ppFEV1 (percent predicted forced expiratory volume in one second) after 8 days of treatment.
These studies demonstrate that mRNA can be modified to increase its stability and reduce immune recognition, but mRNA half-lives are still considered to be on the order of hours, not days. As a result, repeat administration of CFTR mRNA will be necessary to achieve considerable protein production in diseased patients. This transient activity may benefit Cas9 therapies, since off-target gene editing may increase with the duration of protein expression.
Delivering RNA Therapies to the CF Lung is Challenging
Given their large size and anionic charge, RNA drugs do not enter cells on their own. As a result, they require drug delivery vehicles to enter diseased cells. To achieve lung delivery from the airway within the context of CF, these delivery vehicles must (i) migrate across viscous mucus, (ii) avoid clearance and uptake by phagocytes within this mucus and the airways, (iii) reach pulmonary epithelial cells, (iv) be endocytosed, (v) escape the endosome, and (vi) release their contents into the cytoplasm. While the last three requirements are common to all RNA therapies, the first three requirements represent specialized criteria that are unique to CF. To this end, when evaluating drug delivery vehicles, it is important to choose a CF animal model characterized by lung mucus phenotype, and ideally, bacterial infections, since these barriers will influence nanoparticle delivery.
This suggests that early-on CFTR−/− mice are not ideal for characterizing CF delivery vehicles since they fail to exhibit characteristic lung, pancreatic, intestinal, and liver disease phenotypes observed in humans.92,93 However, a plethora of animal models have been developed in the past decade to overcome the limitations of the original mouse model; no animal model exactly recapitulates all aspects of the disease observed in humans, but each comes with advantages that enhance our understanding of the pathophysiology and potential treatments of CF.
One such model is the CFTR−/− pig, which exhibits lung mucus accumulation, decreased bacterial clearance, airway inflammation, and remodeling, in addition to other phenotypes, including meconium ileus (MI), exocrine pancreatic destruction, and focal biliary cirrhosis.93,94 However, 100% of newborn pigs develop MI in contrast with 15% of human newborns93,95; severe MI frequently results in mortality and renders the pigs difficult to study. Recently, one group created a pig model expressing CFTR under the intestine-specific FABP promoter, which corrects MI complications.95 Subsequent studies in cloned gut-corrected pigs found elevated neonatal mortalities, prompted by systemic edema, hepatic abnormalities, and chronic hypoproteinemia immediately after birth.96
Scientists have developed ferret CFTR−/− knockout models that also express CFTR under FABP promoter, reducing the rates of MI from 75% to nonexistent. Their size and faster gestation period provide some advantages compared to pigs, and they develop vas deferens deficiencies like humans and unlike pigs.97 However, researchers have found two important traits may be considered limitations: first, the tendency to develop inflammatory and structural lung disease with mucus obstruction, despite potent antibiotic cocktails that kill bacteria and fungi, and second, the poor nutritional status, which is a consequence of a shorter intestine and the lack of a cecum.97,98 More recently, scientists were able to improve prognosis for ferrets with a G551D mutation on both alleles by treating pregnant ferrets with ivacaftor followed by neonatal administration. This animal model does not develop MI, has reduced mucus accumulation and bacterial infection, and improved pancreatic function.99 However, it is important to note that G551D is a hypomorphic mutation, where CFTR function is not completely abrogated.
A third model, which was created using CRISPR, is a sheep CFTR knockout. While CFTR−/− sheep still exhibit MI that has not been corrected to date, the models have traits that may be useful for some studies. First, they exhibit longer gestation periods that allow observation of CF-related complications that begin in utero. Second, newborn sheep only develop lung complications after birth, which is similar to humans. Third, the observed pancreatic fibrosis is similar to human pathophysiology with well-advanced cases at birth.100
Using CRISPR, scientists also developed a ΔF508 rat that recapitulated some of the disease phenotypes observed in humans such as MI and poor growth. However, the rats do not recapitulate damage in other organs at birth.101 Second, mice harboring the G542X mutation, a nonsense mutation belonging to Class I, were also created with CRISPR Cas9. Given the lack of treatment options for patients harboring this mutation, this animal model proves timely to evaluate therapies such as readthrough molecules.102 As described, there have been significant advances made in the development of CF animal models, which have been reviewed in detail.93–102
There have been advances in the last decade that address the first three steps in the drug delivery process of RNA therapies for CF. First, there have been reports that densely coating nanoparticles with polyethylene glycol (PEG) seems to improve transport past different mucosal surfaces in humans. In one example, scientists measured transport across human endotracheal mucus of PEG-coated polystyrene nanoparticles and found up to 35-fold greater diffusion rates compared to their uncoated counterparts.103
In another example, scientists used PEG-coated poly(lactic-co-glycolic acid) nanoparticles to understand transport across human cervical mucus, finding that the presence of PEG increases the diffusion coefficient eightfold.104 However, for LNP applications, a dense PEG coating might be counterproductive to successful delivery; studies have demonstrated that large PEG amounts may not facilitate efficient cellular uptake of LNPs.105 Studies often utilize mucosal layers from nondiseased humans; delivery of nanoparticles through CF sputum becomes increasingly challenging due to its increased viscosity and thickness, as well as a chronic inflammation state in the surrounding tissue.
Second, administration of nanoparticles through nebulization has considerably improved in the recent past. In one example, scientists were able to uniformly transfect epithelial cells across the entire mouse lung using polymeric particles containing luciferase mRNA.106 A single dose of 1 mg of mRNA produced protein concentrations over 100 ng/g 24 h after administration without notable toxic effects. Finally, some groups have reported limited success in CF treatment with mRNA therapies. For example, one group achieved intranasal delivery of 0.1 mg/kg of CFTR mRNA to CFTR knockout mice for two consecutive days, leading to mice having a third of WT CFTR function for 2 weeks in nasal epithelial cells.40
Another group compared intravenous and intratracheal delivery41 of CFTR mRNA using polymeric nanoparticles. These results show improvements in lung function parameters after a 2 mg/kg injection intravenously and 4 mg/kg intratracheally; it will be interesting to quantify how the mRNA was delivered to other organs. One potential next step will be to evaluate this delivery system in animal models with lung mucus phenotypes.
Third, extracellular vesicles (EVs), including microvesicles (MVs) and exosomes, are another delivery vehicle that has been frequently employed to correct CFTR mutations. In one study,107 researchers used Calu-3 cells to produce EVs containing high quantities of CFTR proteins and CFTR mRNA, with MVs containing considerably higher amounts of both compared to exosomes. They then transfected CF15 cells containing the ΔF508 mutation and found that function was significantly restored, although CFTR channel activity in CF15s was still much lower than in naturally functional Calu-3 cells. They also showed that viral transduction of A549 cells using HAdV5 containing GFP-CFTR fusion proteins and GFP-CFTR mRNA (for tracking the uptake of the EVs) could create an “EV-donor cell” capable of producing EVs containing the CFTR glycoprotein and CFTR mRNA at much higher quantities than Calu-3 cells; however, the EVs also contained viral fragments and viral DNA. Transducing CF15 cells with GFP-CFTR EVs resulted in dose-dependent restoration of function, quantified by iodide efflux, which was similar to the native CFTR function of Calu-3 cells. Surprisingly, both MVs and exosomes restored function equally, although MVs contained a significantly higher amount of CFTR protein and mRNA than exosomes, which was explained by MVs containing metabolically inactive CFTR mRNA and following a different pathway of cellular uptake than exosomes.
A separate study delivered siRNA to human airway epithelial cells (HAEs) using exosomes and corrected the Cl− channel defect in HAEs from CF donors by delivering the CFTR protein using MVs.108 Another study found that sgRNA and Cas9 (although not Cas9 mRNA) can be packaged into exosomes and form a functional sgRNA:Cas9 RNP complex, which can be delivered to target cells.109 The researchers also engineered a modified exosome by fusing CD63, a membrane protein often found on exosomes, with GFP and used GFP nanobody-fused Cas9 since it readily binds with the CD63-GFP for more efficient loading of sgRNA:Cas9 RNPs into the exosome. The modified exosome delivered larger quantities of sgRNA and Cas9 proteins without altering the exosome morphology or the function of the CRISPR-Cas9 system and successfully removed the stop sequence in A549 cells containing stop-DsRed sequences, resulting in greater red fluorescent signals than their unmodified counterparts. One key limitation is that these studies were all done in vitro; it will be interesting to evaluate the performance of exosomes as a delivery platform in some of the animal models aforementioned.
Finally, researchers have shown that the defects associated with loss of CFTR, such as hyperabsorption of sodium and upregulation of the epithelial Na+ channel (ENaC), can be alleviated through downregulation of ENaC using siRNA.110,111 In the first example, studies used targeted nanocomplexes made up of a liposome and an epithelial targeting peptide to deliver ENaC siRNA to primary CF airway epithelial cells in an air-liquid interface culture, as well as to the lungs of C57BL6 mice using oropharyngeal administration.110 In the second example, researchers compared the translocation kinetics of receptor-targeted nanocomplexes carrying labeled siRNA across pig gastric mucus, healthy human airway mucus, and CF human airway mucus, showed that they could transfect bronchial epithelial cells transduced with CF and growing in an air-liquid interface, and used oropharyngeal administration to successfully transfect the lungs of healthy mice with ENaC siRNA.111
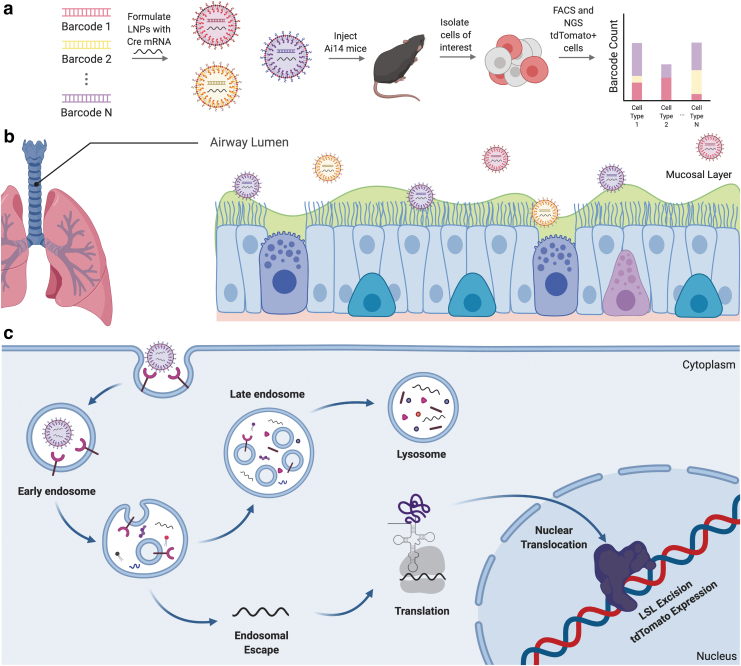
Both the mucus and cellular barriers described above make it likely that most nanoparticles will fail to efficiently and safely deliver RNA to pulmonary epithelial cells in CF. As a result, one key challenge for the field will be identifying new systems to test hundreds or thousands of chemically distinct nanoparticles. Notably, traditional nanoparticle discovery pipelines rely on one-by-one testing, which is usually performed in vitro. One promising alternative that has not yet been tested in a relevant CF animal model is the use of high-throughput DNA barcoding to test more than 100 different nanoparticles in a single animal (Fig. 4a).112,113 These DNA barcoding systems can either quantify nanoparticle biodistribution114–116 or functional delivery (i.e., mRNA translated into functional protein) (Fig. 4a).12,117
Figure 4.
High-throughput methods to identify lung tropic nanomaterials can be used to treat CF. (a) High-throughput barcoding is a novel method of identifying LNPs that efficiently deliver functional nucleic acids in animal models to different tissues. For example, (b) intratracheally delivered LNPs that target epithelial cells in the airway lumen can be identified for treatment of CF. (c) The FIND system requires LNPs to functionally deliver their payload to a target cell, escape the endosome, and translate Cre protein that translocates to the nucleus to facilitate tdTomato expression. LNP, lipid nanoparticle.
Testing for functional delivery is particularly relevant because it allows for identification of LNPs that deliver their payload to the cytoplasm of a cell, while disregarding LNPs that cannot escape endosomes. In one such system, LNPs are formulated to each carry a distinct DNA barcode that is chemically modified to avoid degradation by exonucleases and designed to bypass polymerase chain reaction bias. LNPs are also formulated to contain Cre mRNA; for functional delivery to occur, an LNP must reach a target cell type, escape the endosome, and reach the cytoplasm. Upon successful cytoplasmic delivery into cells in Ai14 mice, which contain a Lox-Stop-Lox-tdTomato construct, Cre mRNA is translated to Cre protein, which translocates to the nucleus and excises the stop, thereby leading to tdTomato expression. Cells in which functional delivery occurred accumulate tdTomato protein and are identified and sorted using fluorescence-activated cell sorting; the barcodes are then sequenced from these cells using next-generation sequencing.
These techniques have been used to identify LNPs that deliver functional RNA to splenic T cells,113 splenic endothelial cells,117 liver endothelial cells,115 liver Kupffer cells,12 and bone marrow endothelial cells.112 Alternative barcoding systems have also been designed. In one, researchers formulated nanoparticles with a chemotherapeutic drug and a barcode, then injected LNPs into a mouse, and looked at accumulation in tumors. The researchers then correlated the number of barcodes found in live and dead cells with the therapeutic potency of four different drugs and a placebo at once.118 In another, researchers barcoded the mRNA contained in LNPs such that each nanoparticle was formulated to carry mRNA with a unique barcode in the 3′ UTR. They then isolated tissues and determined LNP biodistribution using luminescence. Tissues that were functionally delivered to were then sequenced to find the best performing LNPs.1
One alternative approach to treating CF is to avoid the mucus barrier encountered during nebulization (Fig. 4b) by intravenously injecting nanoparticles. However, in order for an intravenously injected LNP to reach the lung epithelium, it must get past the endothelial tight junctions that create a continuous barrier between the blood flow and the lung. Upon bypassing this barrier, the delivery vehicle would need to traverse the extracellular matrix and reach the epithelial layer beyond. The delivery vehicle must then escape the endosome and reach the cytoplasm of a cell (Fig. 4c).
To date, several efforts have been made to target the lung, with a focus on endothelial cells. Improved high-throughput in vivo screening methods have been able to identify LNPs that are capable of bypassing the liver to deliver their cargo to lung endothelial cells. One such LNP formulation contains a lipomer known as 7C1,119 which can effectively deliver siRNA to lung endothelial cells with minimal alteration of gene expression in off-target cell types. Another notable lung tropic formulation is made with cKK-E12.120 LNP shifts in tropism have mainly been facilitated by either changing the cationic lipid or the phospholipid that is included in the LNP.
A recent method known as selective organ targeting was developed to shift tropism of hepatic-targeting LNPs to organs like the spleen and lung.53 Researchers showed that the inclusion of differentially charged phospholipids could shift LNP delivery; specifically, cationic phospholipids (e.g., DOTAP) shifted delivery to the lungs, anionic phospholipids (e.g., 18PA) shifted delivery to the spleen, and neutral phospholipids (e.g., DODAP) maintained hepatic delivery. However, LNPs will have to efficiently target the correct cell type to prove viable. Recent scRNA-seq studies uncovered a cell type lining the lung epithelium called ionocytes. These cells may serve as an important source for CFTR transcripts in the lung and play a key role in maintaining airway surface physiology, including mucus viscosity.121,122 Ionocytes express CFTR mRNA at much higher levels than surrounding airway cell types, suggesting that gene therapies should target basal progenitor cells for long-lasting lung function improvements in patients with CF.
A second barrier to systemic delivery is off-target delivery to the liver. Delivery to organs outside of the liver has been traditionally difficult to do. In particular, the liver has been the primary organ targeted by genetic therapies as a result of its discontinuous endothelium and slowed blood flow through hepatic sinusoids. These factors facilitate clearance of nanomaterials to cell types in the extracellular matrix—notably hepatocytes—ensuring that most systemically delivered therapeutics are cleared before making it to on-target tissues.
Learning from CF Gene Therapy Trials
The lessons described above are applicable to most RNA therapies, independent of the disease. However, there are also many CF-specific lessons that can be learned from CF gene therapy trials that are ongoing or have been completed (Supplementary Table S1). Before establishing drug efficacy, it is necessary to determine if a CF treatment is safe in single or multiple doses, as well as over time, as the disease already poses a daily risk to the patient and a significantly shortened life span. In 2001, the first aerosolized AAV encoding for CFTR (tgAAVCF) successfully demonstrated lung delivery without severe side effects.123 Later, Moss et al. conducted a phase IIa study to test the repeated use of tgAAVCF, concluding that nebulization was an effective and safe route of administration for AAVs.124 Both of these trials contrasted AAVs that were sprayed or instilled into the bronchi, which resulted in side effects that potentially outweighed their benefits. However, a phase IIb study seeking to establish tgAAVCF's efficacy was inconclusive125; FEV was not significantly improved 30 days after repeat administrations.
Nonviral therapies were developed as an alternative for treating CF, since viral therapies can result in antibody production that precludes repeat dosing. Alton et al. performed a trial delivering DNA encoding CFTR with a nonviral vehicle.126,127 Their GL67A liposome, made up of the cationic lipid GL67, was able to functionally deliver CFTR cDNA through a plasmid vector and restore partial function as demonstrated by a more positive potential difference across the lung epithelium, thereby indicating the presence of functional CFTR to pump out chloride ions.
Before this clinical trial, most trials only looked at the nasal epithelium; it was assumed to be analogous to the lung epithelium and was easier to study. However, Alton et al. decided to validate this assumption by studying delivery to both tissues. In phase I, nasal results differed from lung results in both immune and isoprenaline response, leading them to conclude that both tissues must be analyzed for future CF treatments.126 Further clinical trials conducted in 2015 showed that their therapy could be re-administered monthly for over a 1-year period with no significant adverse effects.127 Unfortunately, the authors observed only modest improvements in FEV, and further studies were dropped.
Complementing these efforts have been recent attempts to use nonviral vehicles to deliver mRNA encoding CFTR. Translate Bio's MRT5005 is the only mRNA treatment currently being investigated for CF in active clinical trials.128 As previously described, delivering a functional mRNA (or cDNA) bypasses the need to identify what exact mutation in a gene causes a disease state; one can simply deliver the correct mRNA (or cDNA) to produce a functional protein. Furthermore, MRT5005 is encapsulated in an LNP, which differs from many current treatments at the clinical trial level because it is not cDNA encoded by a plasmid vector inside of a viral vehicle. Their single ascending dose trial showed promising results; there were no serious adverse effects and FEV significantly improved at low and mid doses.
Future Perspectives
These CF clinical trials provide evidence that a genotype-agnostic gene therapy for CF is possible. However, to achieve this goal, several challenges will need to be addressed. First, it will be important to identify drug delivery vehicles that can reach pulmonary epithelial cells at low doses. Given the fact that drug delivery vehicles will likely struggle to penetrate the viscous mucus in CF patients, it will be important that all delivery vehicles are tested in models that recapitulate the mucus phenotype; this is true whether delivery vehicles are tested using traditional one-by-one methodologies or with new high-throughput barcoding approaches. The CF field has done an excellent job developing and characterizing models; one very useful advance would be a published consensus (e.g., by the CF Foundation) describing ideal animal models for drug delivery scientists to use as models for delivery. This would be particularly helpful for the many chemistry laboratories that are interested in CF drug delivery, but do not have expertise in CF biology.
When considering the approaches that can be used to design drug delivery systems for CF, we are optimistic about directed evolution approaches,112 which use large drug delivery datasets to design and predict nanoparticles that exhibit specific traits. If such approaches are applied to animal models that have mucus phenotypes, it may be feasible to identify nanoparticle traits that promote mucus penetration. If these studies occur, it will be interesting to see which nanoparticle structures are most effective. Specifically, it is feasible that nanoparticles similar to those that were previously optimized for other epithelial cells or intranasal/intratracheal administrations may be best at delivering RNA in the context of CF. Alternatively, entirely new nanoparticle design rules may be required.
One interesting alternative to designing a nanoparticle that penetrates CF mucus is to use other clinical advances in concert with RNA delivery. The past decade has seen the advancement of agents to the clinic that help clear out excessive mucus and restore airway surface liquid volume in CF patients129,130; pretreatment with hypertonic saline or mannitol followed by administration of chemically modified CFTR mRNA might result in increased delivery efficiency. These types of studies may work nicely with small molecules that work on some genotypes more than others. For example, it is conceivable that for some genotypes, a small-molecule drug resolves enough lung disease for a nanoparticle to target pulmonary epithelial cells, but not enough lung disease to treat the patient with the small molecule only. In all cases, these studies will need to be performed in preclinical models that closely resemble the CF disease phenotype in the lung.
The second required improvement is the duration of CFTR protein following the administration of CFTR mRNA. First-generation siRNA therapies have been safely administered to patients every few weeks, for several years. Second-generation siRNA therapies are likely to be administered once every 3 months, and scientists are working on third-generation therapies that may be administered only once per year. While siRNA therapeutics necessitate long-term stability for continuous target-gene silencing, mRNA therapeutics need long-term protein expression to continuously modulate a disease state. To facilitate long-term mRNA expression, a combination of RNA modifications will need to be optimized for CFTR mRNA. One alternative is to use recently reported computational methods to design a new CFTR protein variant with a longer half-life.131 Although these advances may seem daunting, the clinical progress made with RNA therapies within the last 10 years provides a rational source of hope. Coupled with the potential to treat CF patients independent of their genotype, there is a clear reason to pursue these approaches.
Supplementary Material
Acknowledgments
J.E.D. thanks Taylor E. Shaw.
Author Disclosure
J.E.D. is a co-founder of Guide Therapeutics.
Funding Information
This work was funded by the National Institutes of Health (R01-GM132985 and UG3-TR002855, awarded to James E. Dahlman).
Supplementary Material
References
- 1. Guimaraes PPG, Zhang R, Spektor R, et al. Ionizable lipid nanoparticles encapsulating barcoded mRNA for accelerated in vivo delivery screening. J Control Release 2019;316:404–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lander ES. Initial impact of the sequencing of the human genome. Nature 2011;470:187–197 [DOI] [PubMed] [Google Scholar]
- 3. Dixon SJ, Stockwell BR. Identifying druggable disease-modifying gene products. Curr Opin Chem Biol 2009;13:549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Setten RL, Rossi JJ, Han SP. The current state and future directions of RNAi-based therapeutics. Nat Rev Drug Discov 2019;18:421–446 [DOI] [PubMed] [Google Scholar]
- 5. Levin AA. Treating disease at the RNA level with oligonucleotides. N Engl J Med 2019;380:57–70 [DOI] [PubMed] [Google Scholar]
- 6. Sahin U, Kariko K, Tureci O. mRNA-based therapeutics—developing a new class of drugs. Nat Rev Drug Discov 2014;13:759–780 [DOI] [PubMed] [Google Scholar]
- 7. Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet 2014;15:445–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adams D, Gonzalez-Duarte A, O'Riordan WD, et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med 2018;379:11–21 [DOI] [PubMed] [Google Scholar]
- 9. Benson MD, Waddington-Cruz M, Berk JL, et al. Inotersen treatment for patients with hereditary transthyretin amyloidosis. N Engl J Med 2018;379:22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. High KA, Roncarolo MG. Gene therapy. N Engl J Med 2019;381:455–464 [DOI] [PubMed] [Google Scholar]
- 11. Sardh E, Harper P, Balwani M, et al. Phase 1 trial of an RNA interference therapy for acute intermittent porphyria. N Engl J Med 2019;380:549–558 [DOI] [PubMed] [Google Scholar]
- 12. Paunovska K, Da Silva Sanchez AJ, Sago CD, et al. Nanoparticles containing oxidized cholesterol deliver mRNA to the liver microenvironment at clinically relevant doses. Adv Mater 2019;31:e1807748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bruscia EM, Bonfield TL. Cystic fibrosis lung immunity: the role of the macrophage. J Innate Immun 2016;8:550–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Di A, Brown ME, Deriy LV, et al. CFTR regulates phagosome acidification in macrophages and alters bactericidal activity. Nat Cell Biol 2006;8:933–944 [DOI] [PubMed] [Google Scholar]
- 15. Cancer Genome Atlas Research Network. Electronic address aadhe, Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 2017;32:185–203.e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Golan T, Khvalevsky EZ, Hubert A, et al. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget 2015;6:24560–24570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xue W, Dahlman JE, Tammela T, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci U S A 2014;111:E3553–E3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y, Chen Y, Li J, et al. Co-delivery of microRNA-21 antisense oligonucleotides and gemcitabine using nanomedicine for pancreatic cancer therapy. Cancer Sci 2017;108:1493–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han X, Wang R, Zhou Y, et al. Mapping the mouse cell atlas by Microwell-Seq. Cell 2018;172:1091–1107.e1017. [DOI] [PubMed] [Google Scholar]
- 20. Aizarani N, Saviano A, Sagar, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 2019;572:199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zielenski J, Rozmahel R, Bozon D, et al. Genomic DNA sequence of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Genomics 1991;10:214–228 [DOI] [PubMed] [Google Scholar]
- 22. Hamosh A, FitzSimmons SC, Macek M Jr., et al. Comparison of the clinical manifestations of cystic fibrosis in black and white patients. J Pediatr 1998;132:255–259 [DOI] [PubMed] [Google Scholar]
- 23. Cystic Fibrosis Foundation. 2018 Patient Registry Annual Report. www.cff.org/Research/Researcher-Resources/Patient-Registry/2018-Patient-Registry-Annual-Data-Report.pdf (last accessed September2, 2020)
- 24. Cohen-Cymberknoh M, Shoseyov D, Kerem E. Managing cystic fibrosis: strategies that increase life expectancy and improve quality of life. Am J Respir Crit Care Med 2011;183:1463–1471 [DOI] [PubMed] [Google Scholar]
- 25. Joshi D, Ehrhardt A, Hong JS, et al. Cystic fibrosis precision therapeutics: emerging considerations. Pediatr Pulmonol 2019;54(Suppl 3):S13–S17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De Boeck K, Amaral MD. Progress in therapies for cystic fibrosis. Lancet Respir Med 2016;4:662–674 [DOI] [PubMed] [Google Scholar]
- 27. Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. N Engl J Med 2005;352:1992–2001 [DOI] [PubMed] [Google Scholar]
- 28. Cystic Fibrosis Foundation. CFTR Mutation Classes. www.cff.org/Care/Clinician-Resources/Network-News/August-2017/Know-Your-CFTR-Mutations.pdf (last accessed September2, 2020)
- 29. Dabrowski M, Bukowy-Bieryllo Z, Zietkiewicz E. Advances in therapeutic use of a drug-stimulated translational readthrough of premature termination codons. Mol Med 2018;24:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ghanem N, Fanen P, Martin J, et al. Exhaustive screening of exon 10 CFTR gene mutations and polymorphisms by denaturing gradient gel electrophoresis: applications to genetic counselling in cystic fibrosis. Mol Cell Probes 1992;6:27–31 [DOI] [PubMed] [Google Scholar]
- 31. Wainwright CE, Elborn JS, Ramsey BW, et al. Lumacaftor-ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N Engl J Med 2015;373:220–231 [DOI] [PubMed] [Google Scholar]
- 32. Edmondson C, Davies JC. Current and future treatment options for cystic fibrosis lung disease: latest evidence and clinical implications. Ther Adv Chronic Dis 2016;7:170–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Flume PA, Liou TG, Borowitz DS, et al. Ivacaftor in subjects with cystic fibrosis who are homozygous for the F508del-CFTR mutation. Chest 2012;142:718–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoy SM. Elexacaftor/ivacaftor/tezacaftor: first approval. Drugs 2019;79:2001–2007 [DOI] [PubMed] [Google Scholar]
- 35. Middleton PG, Mall MA, Drevinek P, et al. Elexacaftor-tezacaftor-ivacaftor for cystic fibrosis with a single Phe508del allele. N Engl J Med 2019;381:1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heijerman HGM, McKone EF, Downey DG, et al. Efficacy and safety of the elexacaftor plus tezacaftor plus ivacaftor combination regimen in people with cystic fibrosis homozygous for the F508del mutation: a double-blind, randomised, phase 3 trial. Lancet 2019;394:1940–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brodlie M, Haq IJ, Roberts K, et al. Targeted therapies to improve CFTR function in cystic fibrosis. Genome Med 2015;7:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cooney AL, McCray PB Jr., Sinn PL. Cystic fibrosis gene therapy: looking back, looking forward. Genes (Basel) 2018;9:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ramalho AS, Beck S, Meyer M, et al. Five percent of normal cystic fibrosis transmembrane conductance regulator mRNA ameliorates the severity of pulmonary disease in cystic fibrosis. Am J Respir Cell Mol Biol 2002;27:619–627 [DOI] [PubMed] [Google Scholar]
- 40. Robinson E, MacDonald KD, Slaughter K, et al. Lipid nanoparticle-delivered chemically modified mRNA restores chloride secretion in cystic fibrosis. Mol Ther 2018;26:2034–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Haque A, Dewerth A, Antony JS, et al. Chemically modified hCFTR mRNAs recuperate lung function in a mouse model of cystic fibrosis. Sci Rep 2018;8:16776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McCarthy VA, Harris A. The CFTR gene and regulation of its expression. Pediatr Pulmonol 2005;40:1–8 [DOI] [PubMed] [Google Scholar]
- 43. Wei X, Eisman R, Xu J, et al. Turnover of the cystic fibrosis transmembrane conductance regulator (CFTR): slow degradation of wild-type and delta F508 CFTR in surface membrane preparations of immortalized airway epithelial cells. J Cell Physiol 1996;168:373–384 [DOI] [PubMed] [Google Scholar]
- 44. Ward CL, Kopito RR. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J Biol Chem 1994;269:25710–25718 [PubMed] [Google Scholar]
- 45. Cheng Q, Wei T, Jia Y, et al. Dendrimer-based lipid nanoparticles deliver therapeutic FAH mRNA to normalize liver function and extend survival in a mouse model of hepatorenal tyrosinemia type I. Adv Mater 2018;30:e1805308. [DOI] [PubMed] [Google Scholar]
- 46. Lino CA, Harper JC, Carney JP, et al. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv 2018;25:1234–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu C, Zhang L, Liu H, et al. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release 2017;266:17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee K, Conboy M, Park HM, et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat Biomed Eng 2017;1:889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liang X, Potter J, Kumar S, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol 2015;208:44–53 [DOI] [PubMed] [Google Scholar]
- 50. Kim S, Kim D, Cho SW, et al. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 2014;24:1012–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li A, Lee CM, Hurley AE, et al. A self-deleting AAV-CRISPR system for in vivo genome editing. Mol Ther Methods Clin Dev 2019;12:111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kelkar A, Zhu Y, Groth T, et al. Doxycycline-dependent self-inactivation of CRISPR-Cas9 to temporally regulate on- and off-target editing. Mol Ther 2020;28:29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cheng Q, Wei T, Farbiak L, et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat Nanotechnol 2020;15:313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Finn JD, Smith AR, Patel MC, et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep 2018;22:2227–2235 [DOI] [PubMed] [Google Scholar]
- 55. Cho SW, Kim S, Kim Y, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 2014;24:132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fu Y, Foden JA, Khayter C, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 2013;31:822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Maruyama T, Dougan SK, Truttmann MC, et al. Corrigendum: increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 2016;34:210. [DOI] [PubMed] [Google Scholar]
- 58. Wienert B, Nguyen DN, Guenther A, et al. Timed inhibition of CDC7 increases CRISPR-Cas9 mediated templated repair. Nat Commun 2020;11:2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schwank G, Koo BK, Sasselli V, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013;13:653–658 [DOI] [PubMed] [Google Scholar]
- 60. Firth AL, Menon T, Parker GS, et al. Functional gene correction for cystic fibrosis in lung epithelial cells generated from patient iPSCs. Cell Rep 2015;12:1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sanz DJ, Hollywood JA, Scallan MF, et al. Cas9/gRNA targeted excision of cystic fibrosis-causing deep-intronic splicing mutations restores normal splicing of CFTR mRNA. PLoS One 2017;12:e0184009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vaidyanathan S, Salahudeen AA, Sellers ZM, et al. High-efficiency, selection-free gene repair in airway stem cells from cystic fibrosis patients rescues CFTR function in differentiated epithelia. Cell Stem Cell 2020;26:161–171.e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Palmer DJ, Turner DL, Ng P. A single “All-in-One” helper-dependent adenovirus to deliver donor DNA and CRISPR/Cas9 for efficient homology-directed repair. Mol Ther Methods Clin Dev 2020;17:441–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017;551:464–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Komor AC, Zhao KT, Packer MS, et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv 2017;3:eaao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zuo E, Sun Y, Wei W, et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 2019;364:289–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Geurts MH, de Poel E, Amatngalim GD, et al. CRISPR-based adenine editors correct nonsense mutations in a cystic fibrosis Organoid Biobank. Cell Stem Cell 2020;26:503–510.e507. [DOI] [PubMed] [Google Scholar]
- 68. Montiel-Gonzalez MF, Vallecillo-Viejo I, Yudowski GA, et al. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc Natl Acad Sci U S A 2013;110:18285–18290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019;576:149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gaj T, Gersbach CA, Barbas CF, 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 2013;31:397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA modifications in gene expression regulation. Cell 2017;169:1187–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ramanathan A, Robb GB, Chan SH. mRNA capping: biological functions and applications. Nucleic Acids Res 2016;44:7511–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Virtanen A, Henriksson N, Nilsson P, et al. Poly(A)-specific ribonuclease (PARN): an allosterically regulated, processive and mRNA cap-interacting deadenylase. Crit Rev Biochem Mol Biol 2013;48:192–209 [DOI] [PubMed] [Google Scholar]
- 74. Decroly E, Ferron F, Lescar J, et al. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol 2011;10:51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kore AR, Shanmugasundaram M, Charles I, et al. Locked nucleic acid (LNA)-modified dinucleotide mRNA cap analogue: synthesis, enzymatic incorporation, and utilization. J Am Chem Soc 2009;131:6364–6365 [DOI] [PubMed] [Google Scholar]
- 76. Chang H, Lim J, Ha M, et al. TAIL-seq: genome-wide determination of poly(A) tail length and 3’ end modifications. Mol Cell 2014;53:1044–1052 [DOI] [PubMed] [Google Scholar]
- 77. Legnini I, Alles J, Karaiskos N, et al. FLAM-seq: full-length mRNA sequencing reveals principles of poly(A) tail length control. Nat Methods 2019;16:879–886 [DOI] [PubMed] [Google Scholar]
- 78. Subtelny AO, Eichhorn SW, Chen GR, et al. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 2014;508:66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Workman RE, Tang AD, Tang PS, et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat Methods 2019;16:1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Orlandini von Niessen AG, Poleganov MA, Rechner C, et al. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3’ UTRs identified by cellular library screening. Mol Ther 2019;27:824–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Asrani KH, Farelli JD, Stahley MR, et al. Optimization of mRNA untranslated regions for improved expression of therapeutic mRNA. RNA Biol 2018;15:756–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kormann MS, Hasenpusch G, Aneja MK, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol 2011;29:154–157 [DOI] [PubMed] [Google Scholar]
- 83. Hornung V, Ellegast J, Kim S, et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science 2006;314:994–997 [DOI] [PubMed] [Google Scholar]
- 84. Fimognari C, Nusse M, Iori R, et al. The new isothiocyanate 4-(methylthio)butylisothiocyanate selectively affects cell-cycle progression and apoptosis induction of human leukemia cells. Invest New Drugs 2004;22:119–129 [DOI] [PubMed] [Google Scholar]
- 85. Heil F, Hemmi H, Hochrein H, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004;303:1526–1529 [DOI] [PubMed] [Google Scholar]
- 86. Kariko K, Muramatsu H, Welsh FA, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther 2008;16:1833–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Richner JM, Himansu S, Dowd KA, et al. Modified mRNA vaccines protect against Zika virus infection. Cell 2017;168:1114–1125.e1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Freyn AW, Ramos da Silva J, Rosado VC, et al. A multi-targeting, nucleoside-modified mRNA influenza virus vaccine provides broad protection in mice. Mol Ther 2020;28:1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Anderson BR, Muramatsu H, Nallagatla SR, et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res 2010;38:5884–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lokugamage MP, Gan Z, Zurla C, et al. Mild innate immune activation overrides efficient nanoparticle-mediated RNA delivery. Adv Mater 2020;32:e1904905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Colledge WH, Abella BS, Southern KW, et al. Generation and characterization of a delta F508 cystic fibrosis mouse model. Nat Genet 1995;10:445–452 [DOI] [PubMed] [Google Scholar]
- 92. Shah VS, Meyerholz DK, Tang XX, et al. Airway acidification initiates host defense abnormalities in cystic fibrosis mice. Science 2016;351:503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rogers CS, Stoltz DA, Meyerholz DK, et al. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science 2008;321:1837–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Stoltz DA, Meyerholz DK, Pezzulo AA, et al. Cystic fibrosis pigs develop lung disease and exhibit defective bacterial eradication at birth. Sci Transl Med 2010;2:29ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Stoltz DA, Rokhlina T, Ernst SE, et al. Intestinal CFTR expression alleviates meconium ileus in cystic fibrosis pigs. J Clin Invest 2013;123:2685–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ballard ST, Evans JW, Drag HS, et al. Pathophysiologic evaluation of the transgenic Cftr “Gut-Corrected” porcine model of cystic fibrosis. Am J Physiol Lung Cell Mol Physiol 2016;311:L779–L787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sun X, Sui H, Fisher JT, et al. Disease phenotype of a ferret CFTR-knockout model of cystic fibrosis. J Clin Invest 2010;120:3149–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Rosen BH, Evans TIA, Moll SR, et al. Infection is not required for mucoinflammatory lung disease in CFTR-knockout ferrets. Am J Respir Crit Care Med 2018;197:1308–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sun X, Yi Y, Yan Z, et al. In utero and postnatal VX-770 administration rescues multiorgan disease in a ferret model of cystic fibrosis. Sci Transl Med 2019;11:eaau7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Fan Z, Perisse IV, Cotton CU, et al. A sheep model of cystic fibrosis generated by CRISPR/Cas9 disruption of the CFTR gene. JCI Insight 2018;3:e123529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Dreano E, Bacchetta M, Simonin J, et al. Characterization of two rat models of cystic fibrosis-KO and F508del CFTR-Generated by Crispr-Cas9. Animal Model Exp Med 2019;2:297–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. McHugh DR, Steele MS, Valerio DM, et al. A G542X cystic fibrosis mouse model for examining nonsense mutation directed therapies. PLoS One 2018;13:e0199573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Schuster BS, Suk JS, Woodworth GF, et al. Nanoparticle diffusion in respiratory mucus from humans without lung disease. Biomaterials 2013;34:3439–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Cu Y, Saltzman WM. Controlled surface modification with poly(ethylene)glycol enhances diffusion of PLGA nanoparticles in human cervical mucus. Mol Pharm 2009;6:173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mui BL, Tam YK, Jayaraman M, et al. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol Ther Nucleic Acids 2013;2:e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Patel AK, Kaczmarek JC, Bose S, et al. Inhaled nanoformulated mRNA polyplexes for protein production in lung epithelium. Adv Mater 2019;31:e1805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Vituret C, Gallay K, Confort MP, et al. Transfer of the cystic fibrosis transmembrane conductance regulator to human cystic fibrosis cells mediated by extracellular vesicles. Hum Gene Ther 2016;27:166–183 [DOI] [PubMed] [Google Scholar]
- 108. Singh BK, Cooney AL, Krishnamurthy S, et al. Extracellular vesicle-mediated siRNA delivery, protein delivery, and CFTR complementation in well-differentiated human airway epithelial cells. Genes (Basel) 2020;11:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ye Y, Zhang X, Xie F, et al. An engineered exosome for delivering sgRNA:Cas9 ribonucleoprotein complex and genome editing in recipient cells. Biomater Sci 2020;8:2966–2976 [DOI] [PubMed] [Google Scholar]
- 110. Manunta MDI, Tagalakis AD, Attwood M, et al. Delivery of ENaC siRNA to epithelial cells mediated by a targeted nanocomplex: a therapeutic strategy for cystic fibrosis. Sci Rep 2017;7:700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Tagalakis AD, Munye MM, Ivanova R, et al. Effective silencing of ENaC by siRNA delivered with epithelial-targeted nanocomplexes in human cystic fibrosis cells and in mouse lung. Thorax 2018;73:847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sago CD, Lokugamage MP, Islam FZ, et al. Nanoparticles that deliver RNA to bone marrow identified by in vivo directed evolution. J Am Chem Soc 2018;140:17095–17105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lokugamage MP, Sago CD, Gan Z, et al. Constrained nanoparticles deliver siRNA and sgRNA to T cells in vivo without targeting ligands. Adv Mater 2019;31:e1902251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Paunovska K, Sago CD, Monaco CM, et al. A direct comparison of in vitro and in vivo nucleic acid delivery mediated by hundreds of nanoparticles reveals a weak correlation. Nano Lett 2018;18:2148–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Paunovska K, Gil CJ, Lokugamage MP, et al. Analyzing 2000 in vivo drug delivery data points reveals cholesterol structure impacts nanoparticle delivery. ACS Nano 2018;12:8341–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sago CD, Lokugamage MP, Lando GN, et al. Modifying a commonly expressed endocytic receptor retargets nanoparticles in vivo. Nano Lett 2018;18:7590–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Sago CD, Lokugamage MP, Paunovska K, et al. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc Natl Acad Sci U S A 2018;115:E9944–E9952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Yaari Z, da Silva D, Zinger A, et al. Theranostic barcoded nanoparticles for personalized cancer medicine. Nat Commun 2016;7:13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Dahlman JE, Barnes C, Khan O, et al. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat Nanotechnol 2014;9:648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Dong Y, Love KT, Dorkin JR, et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc Natl Acad Sci U S A 2014;111:3955–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Montoro DT, Haber AL, Biton M, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 2018;560:319–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Plasschaert LW, Zilionis R, Choo-Wing R, et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 2018;560:377–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Aitken ML, Moss RB, Waltz DA, et al. A phase I study of aerosolized administration of tgAAVCF to cystic fibrosis subjects with mild lung disease. Hum Gene Ther 2001;12:1907–1916 [DOI] [PubMed] [Google Scholar]
- 124. Moss RB, Rodman D, Spencer LT, et al. Repeated adeno-associated virus serotype 2 aerosol-mediated cystic fibrosis transmembrane regulator gene transfer to the lungs of patients with cystic fibrosis: a multicenter, double-blind, placebo-controlled trial. Chest 2004;125:509–521 [DOI] [PubMed] [Google Scholar]
- 125. Moss RB, Milla C, Colombo J, et al. Repeated aerosolized AAV-CFTR for treatment of cystic fibrosis: a randomized placebo-controlled phase 2B trial. Hum Gene Ther 2007;18:726–732 [DOI] [PubMed] [Google Scholar]
- 126. Alton EW, Boyd AC, Porteous DJ, et al. A phase I/IIa safety and efficacy study of nebulized liposome-mediated gene therapy for cystic fibrosis supports a multidose trial. Am J Respir Crit Care Med 2015;192:1389–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Alton E, Armstrong DK, Ashby D, et al. Repeated nebulisation of non-viral CFTR gene therapy in patients with cystic fibrosis: a randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Respir Med 2015;3:684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Translate Bio I. Translate Bio Announces Interim Results from Phase 1/2 Clinical Trial of MRT5005 in Patients with Cystic Fibrosis. www.globenewswire.com/news-release/2019/07/31/1894563/0/en/Translate-Bio-Announces-Interim-Results-from-Phase-1-2-Clinical-Trial-of-MRT5005-in-Patients-with-Cystic-Fibrosis.html (last accessed September2, 2020)
- 129. Jaques A, Daviskas E, Turton JA, et al. Inhaled mannitol improves lung function in cystic fibrosis. Chest 2008;133:1388–1396 [DOI] [PubMed] [Google Scholar]
- 130. Donaldson SH, Bennett WD, Zeman KL, et al. Mucus clearance and lung function in cystic fibrosis with hypertonic saline. N Engl J Med 2006;354:241–250 [DOI] [PubMed] [Google Scholar]
- 131. Huang PS, Boyken SE, Baker D. The coming of age of de novo protein design. Nature 2016;537:320–327 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.