Abstract
PURPOSE
Women with breast cancer have a 4%-16% lifetime risk of a second primary cancer. Whether mutations in genes other than BRCA1/2 are enriched in patients with breast and another primary cancer over those with a single breast cancer (S-BC) is unknown.
PATIENTS AND METHODS
We identified pathogenic germline mutations in 17 cancer susceptibility genes in patients with BRCA1/2-negative breast cancer in 2 different cohorts: cohort 1, high-risk breast cancer program (multiple primary breast cancer [MP-BC], n = 551; S-BC, n = 449) and cohort 2, familial breast cancer research study (MP-BC, n = 340; S-BC, n = 1,464). Mutation rates in these 2 cohorts were compared with a control data set (Exome Aggregation Consortium [ExAC]).
RESULTS
Overall, pathogenic mutation rates for autosomal, dominantly inherited genes were higher in patients with MP-BC versus S-BC in both cohorts (8.5% v 4.9% [P = .02] and 7.1% v 4.2% [P = .03]). There were differences in individual gene mutation rates between cohorts. In both cohorts, younger age at first breast cancer was associated with higher mutation rates; the age of non–breast cancers was unrelated to mutation rate. TP53 and MSH6 mutations were significantly enriched in patients with MP-BC but not S-BC, whereas ATM and PALB2 mutations were significantly enriched in both groups compared with ExAC.
CONCLUSION
Mutation rates are at least 7% in all patients with BRCA1/2 mutation–negative MP-BC, regardless of age at diagnosis of breast cancer, with mutation rates up to 25% in patients with a first breast cancer diagnosed at age < 30 years. Our results suggest that all patients with breast cancer with a second primary cancer, regardless of age of onset, should undergo multigene panel testing.
INTRODUCTION
Patients with breast cancer (BC) have a 4%-16% risk of developing a second primary cancer, including contralateral BC (CBC).1,2 The increased risk of a second cancer may be due to genetic factors, treatment effects, environmental exposures, or a combination. Although multiple primary (MP) cancers may indicate genetic susceptibility, there is a paucity of data examining mutation rates in patients with MP-BC other than in women with breast and ovarian cancer. In one small series, 20% of patients with a primary BC and a nonovarian primary cancer had mutations in BRCA1/2.3
CONTEXT
Key Objective
This study aimed to determine the frequency of mutations in genes commonly found on multiplex genetic testing panels in patients with breast and another primary cancer compared with patients with a single breast cancer.
Knowledge Generated
Mutation rates in BRCA1/2-negative patients with multiple primary breast cancer were approximately 7%-9% compared with approximately 5% in BRCA1/2-negative patients with a single breast cancer. Mutation rates for breast cancer susceptibility genes negatively correlate with increasing age of onset of breast cancer but do not correlate with age of onset of the non–breast cancer.
Relevance
These results confirm added diagnostic yield of multiplex panel testing in patients with multiple primary breast cancer.
As survival from BC improves, the development of a second primary cancer has become a significant issue in BC survivorship.2 Patients with MP cancers have worsened overall survival4 and increased rates of depression.5 It is therefore critical to understand the frequency of mutations in genes other than BRCA1/2 in patients with MP-BC such that second primaries might be prevented or detected earlier. Under-referral for genetic testing is well documented in the United States.6,7 Thus, it is important to clearly delineate which patients need to be referred for genetic testing and to identify subgroups of those who had undergone prior BRCA1/2 testing and may benefit from updated panel testing. Finally, understanding the cancer risk spectrum associated with various genes may help to determine appropriate screening protocols.
A number of large, multiplex panel testing studies of patients with BRCA1/2-negative BC have shown rates of mutations in germline cancer susceptibility genes to be between 5% and 15%,8 with enrichment in certain subgroups of patients, including triple-negative BC,9 early-onset BC,10 and family history of BC.11 In this study, we investigated whether mutations in cancer susceptibility genes were enriched in patients with MP-BC compared with a single BC (S-BC).
PATIENTS AND METHODS
Patient Cohorts
For both cohorts, eligibility criteria for patients with S-BC were diagnosis of BC, high-risk criteria (age of BC onset < 40 years or family history of BC as defined by the presence of ≥ 3 first- to third-degree relatives with BC), and negative BRCA1/2 sequencing in a Clinical Laboratory Improvement Amendment (CLIA)–approved laboratory. Eligibility criteria for patients with MP-BC were diagnosis of BC and another primary cancer, including BC and excluding nonmelanoma skin cancers as per SEER Multiple Primary Rules,12 and negative BRCA1/2 sequencing in a CLIA-approved laboratory. One patient was analyzed per family; if DNA samples from > 1 family member were available, the patient with the youngest age at BC diagnosis was chosen. Cohort 1 included 1,000 patients ascertained from Penn Medicine–affiliated clinical practices seen in a high-risk cancer genetics or BC clinic within a single institution (MP-BC, n = 551; S-BC, n = 449).10 Cohort 2 included 1,804 patients enrolled in a familial BC research registry with either MP-BC (n = 340) or S-BC (n = 1,464), as previously described,11 excluding patients from cohort 1 (Table 1).
TABLE 1.
Phenotypic and Pathologic Features of Patients With BC With and Without Additional Primary Cancers
DNA Sequencing by Targeted Enrichment
DNA samples from cohorts 1 and 2 underwent targeted, massively parallel research-based sequencing. Acquisition of the patient samples was approved by the institutional review boards (IRBs) of the corresponding institutions, and informed consent was obtained from each participant for use of their samples in genetic studies (Data Supplement).
Sequencing by targeted enrichment for cohorts 1 and 2 was done as previously described.10,11 Briefly, DNA libraries of sufficient quality were pooled precapture and hybridized to an Agilent SureSelect (Agilent Technologies, Santa Clara, CA) custom target library. The captured fragments underwent paired-end sequencing on an Illumina HiSeq system (Illumina, San Diego, CA). Samples underwent quality control as previously described.10,11
Sequencing Data Analysis
Sequencing data were analyzed with a custom bioinformatics pipeline10,11 to identify all presumed heterozygous (allele frequency [AF], 30%-70%) single nucleotide variants, small- and medium-sized insertion/deletions, and large genomic rearrangements. Variants were called into a 5-tiered classification scheme per guidelines of the American College of Medical Genetics,13 as described in Maxwell et al,14 and patients were considered to be mutation carriers if a pathogenic or likely pathogenic mutation was identified (Data Supplement). Genes analyzed for heterozygous mutations included high-penetrance BC susceptibility genes (high-breast; CDH1 [lobular breast cancer only], PALB2, PTEN, STK11, TP53), moderate-penetrance BC susceptibility genes (mod-breast; ATM, CHEK2, NBN [c.657_661del5 mutation only]),15 genes known to cause a high risk of other non-BCs (high-other; CDKN2A, MLH1, MSH2, MSH6, PMS2, MUTYH [biallelic mutations only]), and genes known to cause moderate risk of other non-BCs (mod-other; BRIP1, RAD51C, RAD51D). In addition, we performed an exploratory analysis of heterozygous mutations in proposed BC susceptibility genes (BARD1, BLM, MRE11A, RAD50, XRCC2). High penetrance was defined as a greater than fivefold increased risk of cancer compared with the general population, and moderate penetrance was defined as a two- to fivefold increased risk. Low penetrance (less than twofold) alleles were excluded.
Case-Control Analysis
Mutations in the study genes in the Exome Aggregation Consortium (ExAC) version 0.3.1, excluding samples from The Cancer Genome Atlas (accessed December 29, 2014) were identified and classified in Slavin et al.11 ExAC is a set of reference genomes that were made publicly available as a set of population data.15a Allele counts for mutations were summed for each gene in European patients with MP-BC and S-BC; non-Finnish European ExAC AFs were deter-mined previously.9 Associations with BC for each gene were generated using a 2-sided Fisher exact test using the minimum likelihood method.
Clinical and Statistical Analysis
Clinical data were obtained for the patients in both cohorts by IRB-approved chart review. Comparisons of rates in different groups were determined using a 2-sided Fisher exact test of significance. Correlation of age and mutation rate in 10-year age-of-onset groups was performed by a linear regression model.
RESULTS
Cancer Susceptibility Gene Mutations in Patients With BRCA1/2-Negative MP-BC
To determine whether mutations in 17 cancer susceptibility genes are enriched in patients with MP-BC versus those with S-BC, 2 different BRCA1/2 mutation–negative cohorts were studied (Data Supplement). As described previously, cohort 1 included 1,000 patients with either MP-BC (n = 551) or S-BC (n = 449), and cohort 2 included 1,804 patients with either MP-BC (n = 340) or S-BC (n = 1,464; Table 1). Compared with cohort 2, cohort 1 had significantly more patients with early-onset BC (35.4% v 21.2%; P < .0001) and significantly fewer patients with familial BC (49.8% v 100%; P < .0001; Table 1; Data Supplement). The average age of onset of BC was significantly lower in cohort 1 than in cohort 2 (46.6 v 48.1; P = .0003). The 2 cohorts also differed in the spectrum of second cancers. Cohort 1 had significantly fewer patients with a second BC (44.1% v 65.0%; P < .0001) and significantly more patients with another second primary cancer than cohort 2 (78.8% v 37.6%; P < .0001; Data Supplement).
Mutation rates overall were higher in patients with MP-BC versus those with S-BC in both cohorts (cohort 1, 8.5% v 4.9% [P = .02]; cohort 2, 7.1% v 4.2% [P = .03]; Table 2). However, the individual genes responsible for the differences in mutation rates between patients with MP-BC and S-BC differed in the 2 cohorts (Table 2; Data Supplement). In cohort 1, with an early median age of diagnosis, the mutation rates in high-breast and high-other genes were significantly higher in patients with MP-BC than in those with S-BC (2.4% v 0.5% [P = .007] and 1.8% v 0.2% [P = .03], respectively). However, mutation rates in mod-breast and mod-other genes were nearly identical in patients with MP-BC and S-BC in cohort 1 (3.6% v 3.6% [P = 1.000] and 0.7% v 0.7% [P = 1.000], respectively). In cohort 2, mutation rates in high-breast and high-other genes were higher but not statistically significant (2.4% v 1.3% [P = .21] and 0.9% v 0.3% [P = .18], respectively). In cohort 2, mutation rates in mod-breast genes were also similar in patients with MP-BC compared with those with S-BC (3.8% v 2.3%; P = .13).
TABLE 2.
Mutation Rates for Known and Cancer Susceptibility Genes in Patients With MP-BC v S-BC
A bilateral mastectomy at the time of initial BC diagnosis would prevent the development of a CBC. Surgical data were available for 78% of patients with S-BC in cohort 1 (Data Supplement). Mutation rates in patients with MP-CBC (9.1%) were higher than that of patients with S-BC both before (5.1%) and after (3.2%) exclusion of patients who underwent bilateral mastectomy (Data Supplement). Mutation rates for individual genes were similar in patients with S-BC before and after exclusion of patients who underwent bilateral mastectomy (Data Supplement).
MP Cancer Spectrum in Mutation Carriers
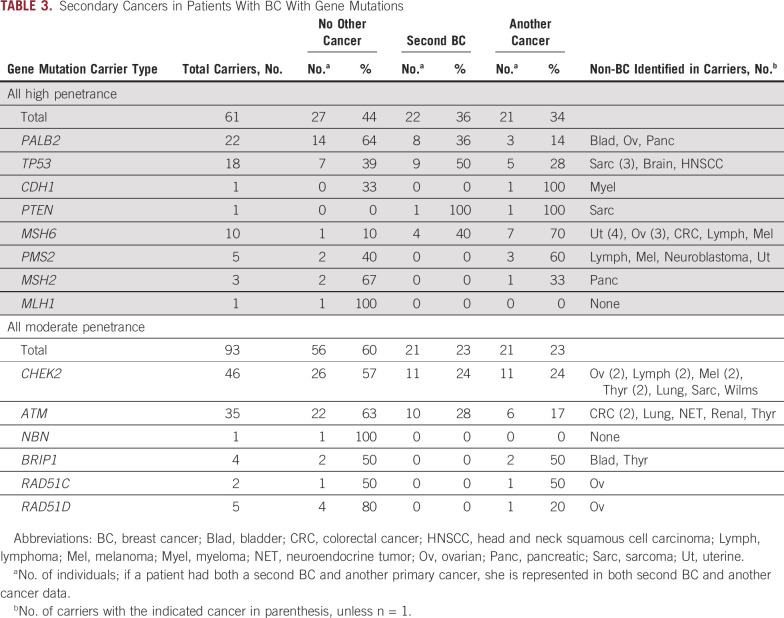
The percentage of MP-BC mutation carriers with a second BC and a primary non-BC varied among gene groups (Table 3). Among 61 v 93 carriers of high- versus moderate-penetrance gene mutations, 36% v 23% (P = .10) had a second primary BC, 34% v 23% (P = .14) had at least 1 primary non-BC, and 44% v 60% (P = .07) had no other cancer. Second primary BCs were seen in 43% and 26% of high- and mod-breast gene mutation carriers but no carriers of mutations in other genes (except MSH6; Table 3). Of 26 other primary cancers in mutation carriers of genes with a known non-BC association, 62% were part of the associated cancer spectrum for that gene (Data Supplement).
TABLE 3.
Secondary Cancers in Patients With BC With Gene Mutations
Of the 11 carriers of BRIP1, RAD51C, and RAD51D mutations, 6 (55%) had a personal or family history of ovarian cancer, which was significantly higher than individuals with no mutations in both cohorts (18% [P = .008] v 15% [P = .003] in cohorts 1 and 2, respectively; Data Supplement). Of the 19 carriers of a Lynch syndrome gene mutation, 12 (63%) had a personal or family history of a Lynch syndrome cancer compared with 56% (P = .645) and 36% (P = .028) of cohorts 1 and 2, respectively. Five Lynch mutation–positive families fulfilled Bethesda criteria (26%) compared with only 9% of cohort 1 (P = .028). Among TP53 mutation carriers, 50% of the 18 carriers either had BC at age ≤ 31 years or met modified Chompret criteria (Data Supplement). Hormone receptor (HR) status was available for 99 of the BC mutation carriers. The majority of BC (72%) in carriers of ATM, CHEK2, and Lynch syndrome gene mutations were estrogen receptor positive and human epidermal growth factor receptor 2 (HER2) negative (Data Supplement). HR distribution was significantly altered in PALB2 and TP53 carriers compared with individuals with no mutations, with triple-negative BC enriched in PALB2 carriers (40% v 15%; P = .004) and HER2-positive BC enriched in TP53 carriers (58% v 24%; P = .01; Data Supplement).
Mutation Rates Stratified by Age at Cancer Diagnosis
To investigate whether age of either BC or non-BC in patients with MP-BC was related to mutation status, patients were stratified by age-group (Data Supplement). Mutation rates were highest in patients with S-BC with BC diagnosed at age < 30 at 8.6% (n = 92), and rates progressively decreased with older age of onset of BC (Fig 1A). Mutation rates for high-breast and mod-breast genes were negatively correlated with age-group in patients with S-BC up to age 60 years (r2 = 0.68 and 0.81, respectively), whereas mutation rates in high-other and mod-other genes showed no relationship with age (Data Supplement). A similar result was seen for age at BC diagnosis but not for age at diagnosis of non-BC in patients with MP-BC (Figs 1B and 1C). Mutation rates were 25.0% in patients with MP-BC with the first BC at age < 30 years (n = 24), 11.9% at age 31-40 years (n = 143), 8.8% at age 41-50 years (n = 143), and 4.5% at age 51-60 years (n = 319; Fig 1B). In contrast, mutation rates were nearly identical regardless of the age at diagnosis of non-BC (9%-15%; Fig 1C). In patients with MP-BC, mutation rates in high-breast and mod-breast genes were negatively correlated with age-group of first BC up to age 60 years (r2 = 0.72 and 0.97, respectively), and there was no relationship of age with mutation rates in high-other or mod-other genes (Data Supplement). Mutation rates were not correlated with age of non-BC diagnosis (Data Supplement).
FIG 1.

Relationship of age of first breast cancer (BC) or other cancer diagnosis in 5 age-groups (< 30 years, 31-40 years, 41-50 years, 51-60 years, and > 60 years) with mutation rates in genes other than BRCA1/2. (A) Patients with a single BC (S-BC) from sequencing cohorts 1 and 2 in whom mutations were analyzed in high-penetrance BC susceptibility genes (high-breast), moderate-penetrance BC susceptibility genes (mod-breast), high-penetrance other cancer susceptibility genes (high-other), and moderate-penetrance other cancer susceptibility genes (mod-other). (B) Patients with multiple primary BC (MP-BC) from sequencing cohorts 1 and 2 in whom mutations were analyzed in high-breast, mod-breast, high-other, and mod-other genes. (C) Patients with MP-BC from sequencing cohorts 1 and 2 analyzed on the basis of age at diagnosis of their first non-BC; mutations were analyzed in high-breast, mod-breast, high-other, and mod-other genes.
Case-Control Study With ExAC
Mutation rates in European individuals in the MP-BC and S-BC combined cohort were compared with mutation rates in the non-Finnish European ExAC population11 for genes with > 5 mutation carriers (Table 4). Mutation rates in TP53 and MSH6 were significantly enriched in patients with MP-BC but not in those with S-BC compared with ExAC. Mutations in ATM and PALB2 were significantly enriched in patients with MP-BC and those with S-BC relative to ExAC, although in both cases, the association was stronger in patients with MP-BC. Associations did not reach significance in either cohort for CHEK2.
TABLE 4.
Case-Control in MP-BC and S-BC Cohorts Compared With ExAC Non-Finnish European Individuals
Other Genetic Variations in MP-BC
To explore other possible genetic associations with MP-BC, mutations in a set of genes were analyzed in cohort 1 (BARD1, BLM, MRE11A, RAD50, XRCC2). Heterozygous BLM mutations were found at modestly higher rates in patients with MP-BC compared with those with S-BC (Data Supplement).
DISCUSSION
In this study, mutation rates in genes commonly found on multigene panels for cancer susceptibility were studied in patients with MP-BC or S-BC from 2 different cohorts. Our results indicate that mutation rates are higher in patients with BRCA1/2-negative MP-BC versus S-BC. However, the genes responsible for the differences in rates varied between the 2 cohorts. High-penetrance cancer genes were responsible for higher mutation rates in the cohort enriched for early-onset BC and non-BC second primaries, whereas moderate-penetrance cancer genes were responsible for the higher mutation rates in the cohort enriched for familial BC and second BC primaries. However, our data suggest that mutations in genes other than BRCA1/2 do not explain the majority of MP-BC cases.
We found that 7%-9% of patients with BRCA1/2-negative MP-BC carried a pathogenic mutation in 1 of 17 genes. Studies from the United Kingdom and Singapore found pathogenic mutations in the genes in our study in 5.2% (n = 440) and 13% (n = 110) of patients with multiple primary cancers of any type, respectively.16,17 Patients with MP-BC with 1 of 6 other cancer types from a genetic testing company cohort found mutations in these genes in 7.4%-11% of patients.18
More than half of the non-BCs in mutation carriers in this study were cancers known to be associated with inherited mutations in the identified gene. In moderate-risk ovarian cancer gene carriers, Lynch syndrome gene mutation carriers, and TP53 carriers, the cancers associated with these genes were enriched in personal and family histories compared with the cohort overall. Approximately half of patients with BC who are found to have mutations in non-BC–related genes fit the genetic testing criteria for the identified gene. In the case that BC is the presenting cancer, the finding of a genetic mutation could lead to implementation of cancer prevention and screening strategies to identify future cancers, hopefully at an early stage to reduce morbidity and mortality.2
Mutation rates in BC susceptibility genes, but not genes associated with other cancers, were correlated with age at diagnosis of BC. Of note, although not the aim of the current study, we found that in patients with S-BC diagnosed at age > 61 years, 0.6% had pathogenic variants in high-penetrance and 1.9% in moderate-penetrance genes. The age of the other cancer was not correlated with the presence of a mutation in a BC susceptibility gene. Particularly for DNA repair germline mutation carriers where poly (ADP-ribose) polymerase inhibitors may be effective targeted therapy,19 it will be important to determine whether the non-BC in these patients is related to the germline mutation or is a sporadic cancer. Several lines of evidence suggest that noncanonical cancers (eg, other than breast, ovarian, prostate, and pancreatic for BRCA1/2 mutations) are less likely to have arisen as a result of the inherited mutation.20,21
The limitations of this study include small numbers of patients in both cohorts, although, to our knowledge this is the largest study of patients with MP-BC to date. The patient cohorts were from academic cancer genetics clinics and research studies; therefore, they may not reflect mutation rates in the general population. In addition, other known or proposed cancer susceptibility genes were not evaluated, and there may be additional genes associated with MP-BC. Conversely, although there are studies supportive22,23 of the groupings used for individual genes in this study, these categories may change with additional data. Among MP-BC cohorts, we noted a slightly increased rate of mutations in women with BC diagnosed at age ≥ 61 years compared with those age 51-60 years, which may reflect a particularly strong family history for women diagnosed at late ages to meet our eligibility criteria; therefore, studies in unselected patients will be helpful to generalize our results. Our results could be influenced by the possibility that some patients with S-BC may truly be patients with MP-BC if they had not had a bilateral mastectomy because those patients were excluded from cohort 2; however, we did not see a difference in mutation rates in patients with S-BC before and after excluding patients with bilateral mastectomy in cohort 1. Finally, mutation rates were compared with the ExAC population, which could inflate odds ratios; a well-matched case-control study is needed in patients with MP-BC.
The majority of patients with cancer who present for genetic testing are now undergoing multigene panel testing compared with gene- or syndrome-specific testing.8,24 One potential benefit from this practice is the identification of unexpected cancer risk in a family.25,26 However, mutation rates in genes other than BRCA1/2 are low, which leads to concerns about overtesting of patients who present for cancer risk evaluation.27,28 Therefore, studies that define cohorts with both lower and higher mutation rates are critical. Our finding of a low rate of mutations in women age > 61 years with solitary breast cancer supports that if genetic testing is done, a limited panel, including BRCA1, BRCA2, and PALB2, may be appropriate, although the prevalence of PALB2 mutations in our study was 0.2%. Given that resources are often constrained and many individuals with a much higher risk of having pathogenic variants often do not undergo testing, the low yield must be recognized. Our study shows that regardless of age, mutation rates in genes other than BRCA1/2 are found in at least 5% of patients with BC and another primary cancer, with up to 25% in patients with their first BC at age < 30 years. These data support the current practice of multigene panel testing in all patients with BC and another primary cancer, regardless of age of onset or family history, and substantiate that all such patients should be referred for genetic counseling.
Presented at the American Society of Clinical Oncology 2018 Annual Meeting, Chicago, IL, June 1-5, 2018.
SUPPORT
Supported by Department of Defense Breast Cancer Research Program Postdoctoral Fellowship W81XWH-13-1-0338 (K.N.M.); ASCO/Breast Cancer Research Foundation (K.N.M.); National Institutes of Health grants K08-CA215312 (K.N.M.), P30-CA016520 (Abramson Cancer Center), and R01-CA176785 (K.O., F.J.C., K.L.N.); Burroughs Wellcome Foundation (K.N.M.); Breast Cancer Research Foundation (S.M.D., K.L.N.); Rooney Family Foundation (A.R.B., S.M.D., K.L.N.); Basser Center for BRCA at the University of Pennsylvania (S.M.D., K.L.N.); MacDonald Cancer Risk Evaluation Program (A.R.B., S.M.D., K.L.N.); Susan G. Komen Foundation (S.M.D.); and Commonwealth Universal Research Enhancement Program (K.L.N.).
The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions. Views and opinions of and endorsements by the authors do not reflect those of the US Army or the Department of Defense.
AUTHOR CONTRIBUTIONS
Conception and design: Kara N. Maxwell, Abha Kulkarni, Mark E. Robson, Angela M. DeMichele, Kenneth Offit, Susan M. Domchek, Katherine L. Nathanson
Financial support: Kara N. Maxwell, Katherine L. Nathanson
Administrative support: Benita Weathers, Judy E. Garber, Katherine L. Nathanson
Provision of study material or patients: Paolo Peterlongo, Bernardo Bonanni, James M. Ford, Judy E. Garber, Susan L. Neuhausen, Angela M. DeMichele, Kenneth Offit, Fergus J. Couch, Susan M. Domchek, Katherine L. Nathanson
Collection and assembly of data: Kara N. Maxwell, Brandon M. Wenz, Abha Kulkarni, Kurt D’Andrea, Benita Weathers, Noah Goodman, Joseph Vijai, Jenna Lilyquist, Steven N. Hart, Thomas P. Slavin, Kasmintan A. Schrader, Vignesh Ravichandran, Tinu Thomas, Chunling Hu, Paolo Peterlongo, Bernardo Bonanni, James M. Ford, Judy E. Garber, Susan L. Neuhausen, Angela M. DeMichele, Jeffrey N. Weitzel, Fergus J. Couch, Susan M. Domchek, Katherine L. Nathanson
Data analysis and interpretation: Kara N. Maxwell, Brandon M. Wenz, Bradley Wubbenhorst, Steven N. Hart, Kasmintan A. Schrader, Vignesh Ravichandran, Tinu Thomas, Chunling Hu, Mark E. Robson, Payal D. Shah, Angela R. Bradbury, Susan M. Domchek, Katherine L. Nathanson
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Joseph Vijai
Patents, Royalties, Other Intellectual Property: Title: Diagnosis & Treatment of ERCC3-Mutant Cancer; inventors: Joseph Vijai, Sabine Topka, Kenneth Offit; US National Stage Patent Application No.: 16/493,214; filing date: September 11, 2019 (Inst)
Kasmintan A. Schrader
Consulting or Advisory Role: Pfizer, AstraZeneca
Research Funding: AstraZeneca (Inst)
Mark E. Robson
Consulting or Advisory Role: Change Health Care
Research Funding: AstraZeneca (Inst), InVitae (Inst), AbbVie (Inst), Pfizer (Inst), Merck (Inst)
Travel, Accommodations, Expenses: AstraZeneca, Pfizer, Merck
Other Relationship: Research to Practice, Clinical Care Options, Physician Education Resource
Uncompensated Relationships: Merck, Pfizer, Daiichi Sankyo, Epic Sciences
Open Payments Link: https://openpaymentsdata.cms.gov/physician/612669/summary
James M. Ford
Research Funding: Genentech (Inst), AstraZeneca (Inst), Puma Biotechnology (Inst), Pfizer (Inst)
Judy E. Garber
Consulting or Advisory Role: Novartis (I), GTx (I), Helix BioPharma, Konica Minolta, Aleta BioTherapeutics (I), H3 Biomedicine (I), Kronos Bio (I)
Research Funding: Novartis (I), Ambry Genetics, Invitae Genetics, Myriad Genetics
Other Relationship: Susan G. Komen for the Cure (I), American Association for Cancer Research, Diane Helis Henry Medical Foundation (I), James P. Wilmot Foundation (I), Adrienne Helis Malvin Medical Research Foundation (I), Breast Cancer Research Foundation, Facing our Risk of Cancer Empowered
Payal D. Shah
Stock and Other Ownership Interests: Johnson & Johnson (I), Novartis (I), Novo Nordisk (I), Pfizer (I), Merck (I), Amgen, Johnson & Johnson, Novo Nordisk
Consulting or Advisory Role: Tmunity Therapeutics
Angela R. Bradbury
Consulting or Advisory Role: AstraZeneca, Merck
Angela M. DeMichele
Consulting or Advisory Role: Context Therapeutics, Pfizer (I)
Research Funding: Pfizer (Inst), Genentech (Inst), Calithera Biosciences (Inst), Menarini (Inst), Silicon Biosystems (Inst)
Jeffrey N. Weitzel
Speakers’ Bureau: AstraZeneca
Fergus J. Couch
Consulting or Advisory Role: AstraZeneca
Speakers’ Bureau: Ambry Genetics, QIAGEN
Research Funding: Grail
Travel, Accommodations, Expenses: Grail, QIAGEN
Other Relationship: Ambry Genetics
Susan M. Domchek
Honoraria: AstraZeneca, Clovis Oncology, Bristol Myers Squibb
Research Funding: AstraZeneca (Inst), Clovis Oncology (Inst)
Open Payments Link: https://openpaymentsdata.cms.gov/physician/917904
No other potential conflicts of interest were reported.
REFERENCES
- 1.Murphy CC, Gerber DE, Pruitt SL. Prevalence of prior cancer among persons newly diagnosed with cancer: An initial report from the Surveillance, Epidemiology, and End Results Program. JAMA Oncol. 2018;4:832–836. doi: 10.1001/jamaoncol.2017.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogt A, Schmid S, Heinimann K, et al. Multiple primary tumours: Challenges and approaches, a review. ESMO Open. 2017;2:e000172. doi: 10.1136/esmoopen-2017-000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shih HA, Nathanson KL, Seal S, et al. BRCA1 and BRCA2 mutations in breast cancer families with multiple primary cancers. Clin Cancer Res. 2000;6:4259–4264. [PubMed] [Google Scholar]
- 4.Raymond JS, Hogue CJ. Multiple primary tumours in women following breast cancer, 1973-2000. Br J Cancer. 2006;94:1745–1750. doi: 10.1038/sj.bjc.6603172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gotay CC, Ransom S, Pagano IS. Quality of life in survivors of multiple primary cancers compared with cancer survivor controls. Cancer. 2007;110:2101–2109. doi: 10.1002/cncr.23005. [DOI] [PubMed] [Google Scholar]
- 6.Kurian AW, Ward KC, Howlader N, et al. Genetic testing and results in a population-based cohort of breast cancer patients and ovarian cancer patients. J Clin Oncol. 2019;37:1305–1315. doi: 10.1200/JCO.18.01854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia C, Harrison K, Ring KL, et al. Genetic counseling referral for ovarian cancer patients: A call to action. Fam Cancer. 2019;18:303–309. doi: 10.1007/s10689-019-00129-5. [DOI] [PubMed] [Google Scholar]
- 8.Afghahi A, Kurian AW. The changing landscape of genetic testing for inherited breast cancer predisposition. Curr Treat Options Oncol. 2017;18:27. doi: 10.1007/s11864-017-0468-y. [DOI] [PubMed] [Google Scholar]
- 9.Couch FJ, Hart SN, Sharma P, et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol. 2015;33:304–311. doi: 10.1200/JCO.2014.57.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maxwell KN, Wubbenhorst B, D’Andrea K, et al. Prevalence of mutations in a panel of breast cancer susceptibility genes in BRCA1/2-negative patients with early-onset breast cancer. Genet Med. 2015;17:630–638. doi: 10.1038/gim.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. doi: 10.1038/s41523-017-0024-8. Slavin TP, Maxwell KN, Lilyquist J, et al: The contribution of pathogenic variants in breast cancer susceptibility genes to familial breast cancer risk. NPJ Breast Cancer 3:22, 2017 [Erratum: NPJ Breast Cancer 3:44, 2017] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson CH, Peace S, Adamo P, et al: Multiple Primary and Histology Coding Rules, 2007. https://seer.cancer.gov/tools/mphrules/2007_mphrules_manual_08242012.pdf.
- 13.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maxwell KN, Hart SN, Vijai J, et al. Evaluation of ACMG-guideline-based variant classification of cancer susceptibility and non-cancer-associated genes in families affected by breast cancer. Am J Hum Genet. 2016;98:801–817. doi: 10.1016/j.ajhg.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang G, Zeng Y, Liu Z, et al. Significant association between Nijmegen breakage syndrome 1 657del5 polymorphism and breast cancer risk. Tumour Biol. 2013;34:2753–2757. doi: 10.1007/s13277-013-0830-z. [DOI] [PubMed] [Google Scholar]
- 15a.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan GHJ, Ong PY, Low JJH, et al. Clinical genetic testing outcome with multi-gene panel in Asian patients with multiple primary cancers. Oncotarget. 2018;9:30649–30660. doi: 10.18632/oncotarget.25769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitworth J, Smith PS, Martin JE, et al. Comprehensive cancer-predisposition gene testing in an adult multiple primary tumor series shows a broad range of deleterious variants and atypical tumor phenotypes. Am J Hum Genet. 2018;103:3–18. doi: 10.1016/j.ajhg.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. doi: 10.1038/s41436-019-0633-8. LaDuca H, Polley EC, Yussuf A, et al: A clinical guide to hereditary cancer panel testing: Evaluation of gene-specific cancer associations and sensitivity of genetic testing criteria in a cohort of 165,000 high-risk patients. Genet Med 22:407-415, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20:764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. doi: 10.1016/j.cell.2018.03.039. Huang KL, Mashl RJ, Wu Y, et al: Pathogenic germline variants in 10,389 adult cancers. Cell 173:355-370.e14, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsons DW, Roy A, Yang Y, et al. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol. 2016;2:616–624. doi: 10.1001/jamaoncol.2015.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Couch FJ, Shimelis H, Hu C, et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 2017;3:1190–1196. doi: 10.1001/jamaoncol.2017.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. doi: 10.1038/s41436-018-0361-5. Lee K, Seifert BA, Shimelis H, et al: Clinical validity assessment of genes frequently tested on hereditary breast and ovarian cancer susceptibility sequencing panels. Genet Med 21:1497-1506, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hooker GW, Clemens KR, Quillin J, et al. Cancer genetic counseling and testing in an era of rapid change. J Genet Couns. 2017;26:1244–1253. doi: 10.1007/s10897-017-0099-2. [DOI] [PubMed] [Google Scholar]
- 25.O’Leary E, Iacoboni D, Holle J, et al. Expanded gene panel use for women with breast cancer: Identification and intervention beyond breast cancer risk. Ann Surg Oncol. 2017;24:3060–3066. doi: 10.1245/s10434-017-5963-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenthal ET, Evans B, Kidd J, et al. Increased identification of candidates for high-risk breast cancer screening through expanded genetic testing. J Am Coll Radiol. 2017;14:561–568. doi: 10.1016/j.jacr.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Doherty J, Bonadies DC, Matloff ET. Testing for hereditary breast cancer: Panel or targeted testing? Experience from a clinical cancer genetics practice. J Genet Couns. 2015;24:683–687. doi: 10.1007/s10897-014-9796-2. [DOI] [PubMed] [Google Scholar]
- 28.Obeid EI, Hall MJ, Daly MB. Multigene panel testing and breast cancer risk: Is it time to scale down? JAMA Oncol. 2017;3:1176–1177. doi: 10.1001/jamaoncol.2017.0342. [DOI] [PubMed] [Google Scholar]