Abstract
Purpose of Review
The goal of this review is to summarize the state of big data analyses in the study of heart failure (HF). We discuss the use of big data in the HF space, focusing on “omics” and clinical data. We address some limitations of this data, as well as their future potential.
Recent Findings
Omics are providing insight into plasmal and myocardial molecular profiles in HF patients. The introduction of single cell and spatial technologies is a major advance that will reshape our understanding of cell heterogeneity and function as well as tissue architecture. Clinical data analysis focuses on HF phenotyping and prognostic modeling.
Summary
Big data approaches are increasingly common in HF research. The use of methods designed for big data, such as machine learning, may help elucidate the biology underlying HF. However, important challenges remain in the translation of this knowledge into improvements in clinical care.
Keywords: Heart failure, Big data, Omics, Single cell, Machine learning
Introduction
In the past 5–10 years, big data has become an integral part of the study of cardiovascular disease. There are many definitions of big data; however, one definition is data large or complex enough that they cannot be analyzed or interpreted by traditional methods. As a result, computational methods, primarily statistics and machine learning (ML), are used to analyze this data. Several big data technologies are starting to be applied in the clinic: for example, genomics and transcriptomics are used for patient stratification in breast cancer diagnosis and treatment [1, 2] and can be used to determine acute cardiac allograft rejection [3, 4]. However, due to challenges in clinical implementation and questions about the benefits of these methods [5], most big data approaches are implemented in preclinical research.
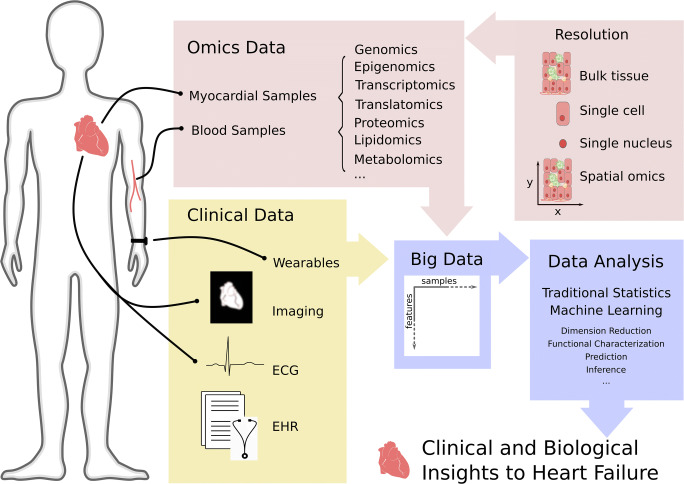
Chronic heart failure (HF) is a prime target for big data research due to the complex etiology of the syndrome, the large number of risk factors, the high degree of comorbidity in patients, and the prolonged and progressive course of disease. Big data used for the study of HF are derived from a variety of sources (Fig. 1). Some of these sources are dependent on tissue such as blood or myocardial samples, while others are ascertained through clinical care or wearable devices.
Fig. 1.
Types of big data in heart failure and the body location from which samples are taken for that data type. Omics and clinical data are the two common big data types to study HF. Clinical data can be gathered via wearables, imaging techniques, echocardiography (ECG), and electronic health records (EHRs). Different omics technologies primarily analyze cardiac tissue or blood and include genomics, transcriptomics, translatomics, proteomics, metabolomics, and lipidomics. Specimen can be studied at different resolutions, including bulk, single cell, single nucleus, and spatial level. To date the different tissue resolutions are not yet available for every omic. Data analysis is challenged by accuracy, structure, and volume of omics and clinical data. Traditional statistical as well as machine learning methods are employed to extract essential information to improve biological understanding and clinical care in HF.
In this review, we discuss the biological and clinical impact of the application of common big data types and computational approaches across the spectrum of human HF etiologies and subtypes, including inherited and acquired HF as well as HF with preserved and reduced ejection fraction (HFpEF and HFrEF, respectively). While many types of big data are used for the study of HF (Table 1), this review will focus on several areas of omics, including genomics, epigenomics, transcriptomics, and proteomics, as well as big clinical data. We also address several current issues with big data collection and analysis, and reflect on the future of these methods in HF.
Table 1.
Big data of types in heart failure. Many types of big data used in the study of HF are listed below along with a brief description. Data types specifically addressed in this review are in italics
| Types of big data | Description | Examples in HF |
|---|---|---|
| Genomics | ||
| Genome-wide association study (GWAS) | Observational study testing the association of genome-wide common genetic variation with a trait in a population of individuals. | Reviewed in [6, 7] |
| Whole-genome sequencing (WGS) | Sequencing of the whole genome. Usually applied in the study of inherited disorders resulting in HF. | [8, 9] |
| Whole-exome sequencing (WES) | Sequencing of the exome (protein-coding portion) of the genome. Usually used to study forms of HF with known genetic etiologies. | Reviewed in [6] |
| Transcriptomics | ||
| Microarray | Quantification of RNA by fluorescence measurement of cDNA using chips. Limited to genes targeted by array chip. | [10, 11] |
| Bulk RNAseq | Quantification of RNA though sequencing of cDNA, alignment to reference genome, and counting. | [12, 13] |
| Single-cell RNAseq | Single cell or nucleus isolation prior to RNAseq | [14••] |
| Spatial transcriptomics | RNAseq performed on patches of tissue on slides | [15] |
| Proteomics | The study of proteins or peptides in a targeted or agnostic manner. | Reviewed in [16–18] |
| Metabolomics | The agnostic or targeted study of metabolites. | Reviewed in [19] |
| Lipidomics | The study of the complete or targeted lipid profile in an individual or population | [20, 21] |
| Wearables | An item worn externally that provides continuous data on parameters like heart rate, blood pressure, or fitness activity. | Reviewed in [22] |
| Clinical data | ||
| Electronic health records | Electronic data representing patients or patient groups produced for the purpose of managing clinical care | [23] |
| Imaging data | The process of creating visual representation of physiology. Examples include CT, MRI, echocardiography, EKG, X-ray. | Reviewed in [24] |
Big Data Computational Methods
In order to analyze big data, methods that account for both the size and the complexity of the dataset are required. Data noise, spurious correlations, and limitations in computational power are a few of the challenges that these types of analysis methods are designed to overcome [25]. Big data analysis protocols vary based on the nature of the data collected as well as the specific research question. Nonetheless, there are shared concepts that are common in these analyses that we will briefly discuss: dimension reduction, ML, and a popular branch of ML called deep learning (DL). For more detailed methodological overview, see [26–28].
When a large number of features are measured, data visualization and interpretation is difficult. Thus, feature or dimension reduction is a common aim of computational methods [29]. Data-driven approaches like principal component analysis transform features to a lower dimensional space by finding combinations of features that capture most of the variability in the data. These combined features, however, are often hard to interpret as they are a mathematical combination of arbitrary molecules. Thus, an alternative is to perform dimension reduction based on interpretable features. For this purpose, molecules are grouped into processes such as cellular pathways, based on prior knowledge that is available in databases. There are many methodological approaches that then try to estimate which processes are more or less active, based on the levels of the molecules that belong to them (e.g., gene set enrichment analysis [30]).
After preprocessing and potentially dimension reduction, statistical learning and modeling is often applied to make estimations about populations (inference) or predictions of new experiments. These analyses often rely on ML, a variety of algorithms to carry out computational tasks without detailed instructions provided by the user. ML algorithms can be classified as unsupervised or supervised. Unsupervised algorithms learn underlying patterns from data while supervised algorithms learn from labeled data to perform tasks like classification or prediction. For this, a portion of available data, referred to as the training data, is used to fit a model. The model performance is then assessed on test data, data that has not been used for model training.
A common supervised ML algorithm is a neural network. A neural network is characterized by nodes that are organized in layers, inspired by biological neuronal circuits. Each node can be regarded as a function that processes inputs to generate outputs that become the inputs of the next layer of nodes. The information processing of each node is learned from data by an optimization method that adjusts the node’s function parameters to minimize the error in performing a task, learning from each sample in a data set. Deep learning (DL) is a specific type of machine learning that uses neural networks with many layers. Deep learning is difficult to interpret since information processing via different nodes and layers becomes incomprehensible. However, it is very powerful in performing without human input highly difficult tasks such as interpretation of biomedical imaging or clinical health records that otherwise would require high domain knowledge.
Omics in Heart Failure
High-throughput methods enable researchers to study molecular profiles of tissues at high resolutions (Table 1). This field is generally referred to with the suffix -omics. Omics technologies can be described as non-targeted—those that aim to measure complete molecular profiles in an unbiased manner—and targeted—those that have predefined molecules of interest. While non-targeted omics are often treated as a complete representation of a molecular profile, the biases that underlie these representations need to be considered as possible sources of error. Technical bias often results from favoring abundant or easy to read molecules and can be only partially corrected during normalization procedures before data analysis. As technologies develop, technical biases are addressed (e.g., long-read RNAseq can overcome negative bias towards long transcripts and improves isoform detection [31]). Further biases are introduced at the analysis level, when prior biological knowledge is used to reduce the dimensions of omics data (see “Big Data Computational Methods”). The consultation of prior knowledge can forestall new discoveries by disregarding valuable information. Furthermore, bioinformatic databases are a main source of biological knowledge and their inherent biases and inaccuracies are integrated into analyses that use them.
In HF, the specimen for omics analysis usually is cardiac tissue or blood (e.g., peripheral blood mononuclear cells (PBMCs)). While myocardial omic analyses can help elucidate disease mechanisms and identify biomarkers and therapeutic targets, the tissue availability for human samples is limited. Blood samples are easier to access and can help survey HF patients at a higher temporal resolution. They are used for biomarker detection and to study genomics as well as the role of circulating cells while the origin and pathophysiology of circulating molecules can be difficult to define.
Different omics technologies pose similar challenges on data analysis and evaluation, including problems concerning accuracy, imputation, integration, replication, and interpretation. We will discuss recent advances and challenges in important omics, in particular genomics, epigenomics, transcriptomics, and proteomics.
Genomics
Genomics is one of the classic areas of big data, studying the role of the genome in disease. Within genomics, analyses are split by technique: genome-wide association studies (GWAS) investigate specific predetermined single nucleotide polymorphisms (SNPs), while whole-exome sequencing (WES) and whole-genome sequencing (WGS) use next generation sequencing to identify all variations in the coding regions or the complete genome, respectively. GWAS is typically used to study common HF, while WES and WGS are more frequently used to study subtypes of HF with known genetic etiologies such as familial dilated cardiomyopathy (DCM) [32]. In both GWAS and whole-genome association studies (WGAS) difficulties include limited information at each single nucleotide polymorphism (SNP), difficulty understanding the biological mechanism that may drive the association for SNPs not in genes, and the need for large populations to gain sufficient statistical power [33].
As a result of years of HF GWAS with limited associations that provided little insight into the biology of the disease [34], recent GWAS have focused on gathering sufficiently large samples, looking at HF subgroups [35], and investigating biomarker and intermediate quantitative traits relevant to HF [7, 36, 37]. While GWAS of HF have been reviewed elsewhere [6] [7], we want to highlight the recent publication of the largest GWAS of HF to date: Shah et al. found 12 independent signals at 11 loci associated with HF risk factors or structural parameters of the left ventricle (LV) [38••]. After statistical analysis to determine causality, several loci that remained as risk factors mapped to genes involved in cellular senescence, cardiac development, and protein homeostasis. These results indicate that while much of the genetics of all cause HF is due to risk factors, other innate biological pathways relevant to cardiac function also play a role in genetic predisposition to disease. However, like much of the GWAS literature, these results are also limited in scope as the population studied only included European ancestry individuals.
Epigenomics
Epigenomics is the study of alterations across the genome that regulate genome expression and function without altering the DNA sequence. Some of the epigenomic alterations that are known to be relevant in HF include DNA methylation [39], chromatin conformation mapping [40], and histone modifications [41, 42]. Many studies that apply epigenomics also use data from other omics techniques, often either genomics or transcriptomics into a multi-omics approach [39, 43, 44].
Despite these examples, there are many challenges in both the analysis and interpretation of epigenomic data. Different groups use different analysis workflows and there is limited consensus on the best way to analyze data. The epigenome is highly dynamic, meaning that with only a single sample at a single time-point, it can be difficult to determine which changes are causes and which are consequences of the cell state [45]. In data interpretation, the cell and tissue specificity of the epigenomic landscape means that it can be difficult to be certain which changes are relevant to disease state [45, 46]. Despite these challenges, epigenomics provide a natural bridge between knowledge of the genetic state in HF and potential biological consequences.
Transcriptomics
The transfer from genetic code to cellular function is mediated by the transcription of ribonucleic acid (RNA). RNA can be translated to proteins (coding RNA or messenger RNA), or execute structural (e.g., ribosomal RNA) or gene regulatory functions (e.g., micro RNA, long non-coding RNA). The quantification of the set of RNA molecules (transcripts) produced by the genome is generally referred to as transcriptomics and provides important understanding of disease mechanisms [47]. As transcriptome profiles can cover up to ~ 20.000 coding and ~ 15.000 non-coding genes [48], analysis and functional interpretation is challenging. To extract relevant information from transcriptome data, dimension reduction methods, enrichment based analysis, linear modeling, clustering algorithms and other ML techniques are routinely applied. The combination of these methods with prior biological knowledge constitutes a key concept of functional interpretation of large-scale gene expression data [49]. Among the disadvantages of using bulk transcriptomics is their susceptibility to fluctuations in cellular composition, which can lower sample comparability. One solution to this is computational cell deconvolution methods [50], which calculate cell fractions from bulk measurements and can estimate cell-specific expression profiles [51]. Cell deconvolution of human heart tissue has been performed [52] and might serve as a first example to enhance future HF transcriptome analyses.
The first high-throughput transcriptomic study on myocardial human HF was published in 2000 [53]. In the subsequent decades, technological and bioinformatic advances in transcriptomics have improved our comprehension of cardiac hypertrophy [54], reverse remodeling [55], cardiac metabolism [10, 56], cardiac fibrosis [57], and immune dysregulation [58] in HF. Several studies made their data sets and protocols publicly available on platforms like NCBI’s gene expression omnibus. However, few attempts have been made to compare transcriptomic HF studies [59–61]. The continuing development of sophisticated data analysis methods invites the retrospective re-analysis and integration of published HF studies, although data integration from different platforms, centers, and technologies presents many challenges [62].
Transcriptome study of myocardial remodeling in HF is complicated by tissue accessibility. Thus, for patient safety, most studies analyzed tissue from HF patients undergoing heart transplantation or LVAD treatment, leaving a knowledge gap of gene expression profiling in HFpEF patients. A recent study compared myocardial transcriptomes in patients with clinical profiles suggesting HFpEF with those not displaying signs of HF [63•] gene dysregulations similar to those observed in HFrEF (TNNT2, LUM and p53). Future research is required to specify differences between cardiac remodeling in HFpEF and HFrEF patients to enable development of disease specific therapy, which is currently lacking.
The profiling of non-coding RNA has provided targets for diagnostic and therapeutic purposes in HF [64, 65]. As HF specific non-coding RNAs can be detected in bloodstream, developing miRNA panels to stratify HF patients by prognostic or diagnostic aspects has been a major focus. However, the clinical utility still has to be demonstrated and the (patho-)physiologic role of circulating RNAs remains unclear. Here, a functional microRNA screening approach could help to prioritize candidates [66].
Translatomics can be described as the quantification of translating mRNAs and ribosomes providing important information of subsequent RNA regulation [67]. To address how these layers of gene regulation connect in the failing heart, a study by van Heesch et al. combined genotypes, transcriptomes, and translatomes in 80 hearts (control vs. dilated cardiomyopathy) [13••]. Deciphering regulation in protein biosynthesis, the balance between transcriptional and translational gene regulation was elucidated, for example mitochondrial processes are initiated during transcription and significantly enhanced on the translational level. Furthermore, protein-truncating variants of DCM causing genes were reported to inefficiently terminate translation, providing insights in the pathogenicity of genetic variants. Excitingly, the authors report that circular RNAs (circRNAs) and long non-coding RNAs (lncRNAs) were found to be also translated to novel microproteins.
Single-Cell RNAseq
With single-cell RNAseq, the transcriptome of individual cells can be measured, providing tissue profiling at unprecedented granularity. Bulk RNAseq fails to account for a functional diversity of cell types that might be crucial in understanding the orchestration of myocardial syncytium in health and disease. Single-cell expression profiles can inform about cell lineage heterogeneity [68], inter cell communications [69], individual transcription factor and pathway activity levels [70], or can be integrated within multi-omic approaches [71].
The main challenges in the application of this technology included separation of single, viable cells and subsequent amplification of a minute amount of RNA. Different approaches to overcome these hurdles vary in gene coverage and multiplexing ability (i.e., the capacity to process in parallel) [72, 73]. The cardiac tissue poses additional challenges. Since cardiomyocytes (CM) are too large for many cell sorting approaches, single nucleus RNAseq can be applied, which involves isolating the nucleus rather than the whole cell prior to sequencing. The transcriptional profile of single cell and single nucleus RNAseq has been reported to be comparable during CM differentiation [74]. As the transcriptional profiles of mono- and polynucleated CM were reported to be similar [75], application of single nucleus RNAseq on cardiac tissue is encouraged. The plethora of information gathered by single cell RNAseq poses new challenges to big data analysis that have only partially been met. These include the need to quantify uncertainty in measurements and efficiently handle gene dropout rates; the limited benchmarking possibilities; the need to scale to higher dimensional data, as more cells and more genes can be measured; and the integration of multiple levels of single-cell omics [76].
Single-cell RNAseq has already been applied to study the cardiovascular system (reviewed in [77–80]). To date, studies have focused on the description of cardiac cell lineage heterogeneity and trajectory in mice [79, 81–83], as well as on human cardiogenesis [84–88].
HF was studied in murine models, investigating cell cycle arrest [89] and adaptive remodeling of CM [90]. Human HF description on single cell level was recently reported [14••]. A total of 21,422 single cells from 14 control and 8 failing hearts were compared. The reported cellular heterogeneity was suggested to reflect functional specializations. Macrophages fulfill very heterogeneous tasks in the human heart [91], which was mirrored by different macrophage clusters, some of which presumably specialize in regulating the conduction system, whereas other clusters displayed traits that suggested involvement in immune response regulation. A cluster of endothelial cells (EC) was characterized to highly express ACKR1. These ECs decreased in HF and might exert cardioprotective functions, as injection of ACKR1+ ECs into a murine HF model improved cardiac function compared to control. This study highlighted that cell heterogeneity within lineages relates to functional specialization and pathological condition, aspects that can only be studied with single-cell resolution. The knowledge of such functional cellular subgroups could enable the targeting of such clusters to boost their cardioprotective ability.
Spatially Resolved Transcriptomics
As single-cell analysis is exhibiting a soaring development, spatially resolved transcriptomics add yet another layer of information and complexity. Study of the single cell or bulk transcriptome does not regard the complex tissue architecture of the heart. Further, many pathological processes like fibrotic scarring or ischemia can be local aberrations that require to be studied within tissue context. Spatially resolved transcriptomics could overcome these hurdles by gathering gene expression data while retaining respective spatial information. Many approaches are limited in gene coverage and/or in spatial resolution [92]. Stahl et al. developed spatial transcriptomics, a technique that compartmentalizes tissue into patches which are subsequently profiled via RNAseq [93] and set the path for further development [94]. Other technologies are under active development to improve the spatial resolution and coverage of genes of these methods [95, 96]. The data generated by these technologies still require a sound computational framework to integrate both layers of information.
In a pilot study, Asp et al. analyzed cardiac fibrotic samples from three HFpEF patients and demonstrated the feasibility of performing spatial transcriptomics on adult human heart tissue [15]. A hallmark study combined spatial transcriptomics, single cell technology and in situ sequencing to create a spatiotemporal map of gene expression patterns during embryonic heart development at an hitherto unseen resolution [85•]. This combinatorial approach is a highly promising field for HF research and is likely to fundamentally improve our understanding of cardiac remodeling in the near future.
Proteomics
Proteomics is the quantification of proteins, as transcriptomics do not consider the subsequent and frequent regulations of RNA translation or protein modification. Targeted and non targeted proteomic technologies can be applied, differing in protein coverage, protocol complexity and analytical throughput [97]. Data generated by proteomics provides similar challenges to the other omics analysis as discussed above. In HF, proteomics have been applied to unravel protein posttranslational regulation and temporal dynamics [16]. Due to tissue availability, animal models of HF have been more frequently analyzed [18]. In 2017, the first human proteomic heart atlas described 10,700 proteins in 16 anatomical regions, providing a rich resource of known and unknown protein distributions [98].
The plasma proteome of HF patients has been analyzed extensively to identify biomarkers for HF and their genetic risk-association as well as to understand organ crosstalk via blood stream [99–101]. Egerstedt et al. studied the plasma proteome of HF patients in different clinical stages (early HF development, manifest advanced HF, and reversal of HF after heart transplantation) [102••]. They identified 33 candidate proteins that were associated with HF development. The origin of those circulating proteins was investigated by querying public transcriptome and proteome datasets as well as applying spatial transcriptomics on two failing hearts. GWAS was then used to determine genetic loci that were associated with protein candidates. This study demonstrates that the arsenal of omics technologies can be successfully applied to complete biological characterization of candidate biomarkers.
Clinical Data in Heart Failure
While omics data provides information about the cellular state during disease, how the disease state is viewed and treated in real patients provides additional insight. Clinical data can be described as information about a patient’s health status that is gathered mainly for the purpose of clinical care. These include imaging data, electronic health records (EHRs), and data captured by wearables (Fig. 1). Clinical and omic data types can be analyzed with similar methods; however, they differ regarding their data structure. While omic data are structured measurements, clinical data is often a combination of unstructured, semi-structured, and structured data with the added complication that free text can be subjective or spurious. Thus, clinical data often requires significant pre-processing prior to analysis, a major hurdle for clinical data analysis on a large scale. Highly promising approaches to this challenge of extracting relevant information from unstructured clinical data include natural language processing [103–105], but even structured clinical data is subject to noise resulting from entry errors. Clinical data is frequently sparse, subject to care utilization and documentation habits, and biased, in that health states outside of clinical encounters are rarely reported. Once preprocessing challenges are overcome, clinical data analysis is often subjected to similar statistical and mathematical modeling as omics data for predictive or inference purposes. In HF, patient outcomes have been associated with the presence of a wide variety of comorbid conditions and ejection fraction sub-group. Despite this, mortality and risk of rehospitalization in HF patients remains high. As a result, three major trends have emerged in the use of clinical data for the study of HF: sub-phenotyping, deep phenotyping, and imaging data.
The emergence of sub-phenotyping has caused a shift from the tendency to view HF patients as a single population (or as two clearly defined populations) towards the tendency to view them as a large heterogeneous supergroup composed of many smaller and potentially unknown subgroups [106, 107]. Predicting the outcomes of HF patients, especially within subgroups, is a major area within big data studies using EHR data or other data relevant to clinical care [108]. Adler et al. were able to divide HF patients into those at high and low risk of death based on clinical variables, and their classifier had a better predictive power than any of the individual classifier components, and better than other comparison markers including NT-proBNP [109]. Ahmad et al. divided a group of HF patients into four clusters which differed in age, sex, clinical measures, and comorbid conditions, before building a classifier to predict survival. They found that cluster membership had a modest predictive ability, but performed better than left ventricular ejection fraction alone as the gold standard measure of cardiac function [110]. Other studies have tested multiple types of algorithms for predicting outcomes including HF hospitalization and mortality among HFpEF patients [111], and phenogrouped HF patients who had been randomized to cardiac resynchronization therapy with a regular or implantable cardiac defibrillator prior to evaluation of the effect on HF events and death [112].
Deep phenotyping—the characterization of a phenotype through the comprehensive evaluation of components and intermediate manifestations—has resulted in the use of many diverse types of data. Data including echocardiography [113], electrocardiography (ECG) [114], cardiac magnetic resonance imaging (MRI) [115], tissue imaging [116], implantable monitors [112], and other wearable and non-invasive cardiac monitors [117, 118] are used in combination with ML and DL methods for the prediction and monitoring of HF patients. Laboratory values and intermediate phenotypes are also widely analyzed. The diversity of data types used for the study of HF is rapidly expanding. Analyzing populations that have multiple data in the same individuals can provide detailed information about the progression of disease as well as insights into clinical characteristics that may indicate negative outcomes.
Imaging data constitutes a major branch of big data analysis, facilitating automated assessment of echocardiography, computed tomography, magnetic resonance imaging, and nuclear imaging results. The rise of imaging data from clinical care has happened partially due to improvements in the ability of ML analysis methods for this data. As DL approaches are especially useful to consider the vast amount of features in raw images and integrate those with other clinical variables. For a detailed discussion of these topics, we point the reader to dedicated reviews [24, 119]). However, as with other big data types, important limitations remain. Most importantly, the lack of interpretability of DL models based on image data is a major obstacle for relevance to clinical care.
As a whole, big data from clinical populations has provided great insight into the true phenotypic diversity of HF and has begun to provide links between that diversity and patient outcomes. However, despite the increased understanding of phenotypic heterogeneity, there is still a significant amount to be learned about the relationship between sub-phenotypes and outcomes. While this is a rapidly expanding field, questions about the necessary manual curation of certain data types, inconsistencies in imaging between clinical sites, and privacy concerns remain. The promise of the interface between large scale clinical imaging and electronic health records holds great promise.
Conclusions
“Since we can never know all the factors that a problem entails, we can never solve it. [..] To arrive at the truth we would need more data along with the intellectual resources for exhaustively interpreting the data.” - Fernando Pessoa, from The Book of Disquiet (translated by Richard Zenith)
Despite advances in the use of big data in HF, we are still learning how to use this information to understand the complexities of HF. To date, many challenges remain, as reflected by high mortality and morbidity rates and limited treatment options. However, the direction that HF research has taken towards big data science promises to advance our knowledge substantially. Relevant data is being collected and analyzed in larger numbers with emerging data types forthcoming. Those include image data, wearable data, environmental data and data generated by cardiac monitors such as CardioMEMs [120]. And, the combination of multiple data types, either clinical and molecular data or multi-omic integration is becoming more common. However, despite the innovation in big data and HF, unresolved questions remain.
Data storage and sharing are key aspects of big data research and security breaches on patient data can lead to serious person rights infringements. While research data is often anonymized to protect participants, data re-identification is a serious threat. To minimize the risks of infringing on participant privacy, different data sharing strategies including open consent, controlled access and registered access are used [121].
Among omics data, genomic information has highest re-identification risk and thus elaborate sharing regulations are needed [122]. Approaches include sharing only the subset of data that is not sensitive, sharing only more common genetic variants that will be less specific to a single individual, and requiring strict protocols for data access. Another approach are search engines for genomic mutations where allelic information can be queried with no reference to a patient [123]. Other omic data in general are less specific to an individual, and data is made public on servers like the gene expression omnibus. However, even transcriptome data can be used to infer genetic structural variants and thus facilitate re-identification [124].
Clinical data is highly sensitive and its use in big data research is subjected to strict regulations concerning data privacy and security. Databases like UK Biobank or dbGAP provide clinical and molecular profiles of participants at great depth. Here, controlled access has to be requested by scientists with a research proposal and an agreement to a data handling framework. Most databases store data and regulate access in a centralized fashion, which constitutes a weak point for security breaches as frequently reported in the US [125]. Methods that rely on decentralized networks, such as blockchain, have been suggested to provide additional security and data ownership for individuals [126–128]. However, as with all data security measures, there remains a trade-off between affordable protection and making data sufficiently accessible to researchers. Beyond critical aspects of data safety, costs of storage, energy consumption, and environmental consequences need to be considered.
One of the major issues across big data domains are biases in the collection and analysis of that data. As we rush to collect ever larger sample sizes, we should pause to carefully consider whether we are merely enthralled by ever increasing data samples (so-called data chauvinism [129]) or whether the biological question is best answered by data of the type and quality available. For many omics technologies, the number of features considered requires large samples, or the noise introduced will result in inferior model fitting. In other cases, a large sample size can be less informative if the sampling is of lower quality, for instance if non-probabilistic sampling was applied [130]. Thus, many omic studies, especially those analyzing sparse myocardial tissue, suffer from small patient cohorts that cannot compensate for the biological and clinical variability. A large-scale effort to acquire and comprehensively characterize relevant tissue samples with a variety of omics techniques would ameliorate this issue and potentially provide great insight into the biology of HF. Such efforts have proven valuable in other areas, most notably in oncology. In clinical data analysis we must balance the desire to find subsets of patients that share characteristics, with the goal of making sure that all patients benefit from the potential of precision medicine. Concerns about sampling bias, data missingness, and measurement error in big data, and especially big clinical data, are all relevant to research in HF [131–133]. These data quality concerns are also important because they will directly affect the output of machine learning analyses [134].
Lastly, despite the excitement about big data, the ultimate goal in medicine must always be to improve human health. Physicians should receive additional training allowing them to appropriately evaluate the potential of big data in clinical care [135]. To successfully implement precision medicine approaches based on big data technologies, clinicians will need to understand the strengths and weaknesses of methodologies and have confidence in their relevance to disease. The role of big data in HF prevention and treatment necessitates a multi-disciplinary discussion where physicians are needed to take a leading role.
A significant amount of big data has been generated and analyzed for the study of HF to promote a digitalization of medicine [136] and are adapted to deal with the particular problems posed by HF biology on the various levels that have been discussed. However, challenges still lie ahead. Some are data governance issues, such as patient privacy as well as data access and sharing, while others are more biological or technical, such as integration of multiple data types to describe HF from different perspectives. As the amount of big data generated by different methods continues to accrue, we must piece the biology together, and harness that knowledge to benefit patients. While a unified theory explaining complete clinic and biology of HF might still be unattainable, the era of big data analysis helps us to consider more and more factors and thus brings us much closer to the goal of treating the right patient with the right treatment at the right time.
Acknowledgments
JDL and RTL are supported by Informatics for Life funded by the Klaus Tschira Foundation.
Funding Information
Open Access funding provided by Projekt DEAL.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
Rebecca T. Levinson and Julio Saez-Rodriguez - co-advised the review
This article is part of the Topical Collection on Translational Research in Heart Failure
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer CE, Jr, Dees EC, Goetz MP, Olson JA, Jr, Lively T, Badve SS, Saphner TJ, Wagner LI, Whelan TJ, Ellis MJ, Paik S, Wood WC, Ravdin PM, Keane MM, Gomez Moreno HL, Reddy PS, Goggins TF, Mayer IA, Brufsky AM, Toppmeyer DL, Kaklamani VG, Berenberg JL, Abrams J, Sledge GW., Jr Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379:111–121. doi: 10.1056/NEJMoa1804710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jang Y, Choi T, Kim J, Park J, Seo J, Kim S, Kwon Y, Lee S, Lee S. An integrated clinical and genomic information system for cancer precision medicine. BMC Med Genet. 2018;11:34. doi: 10.1186/s12920-018-0347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pham MX, Teuteberg JJ, Kfoury AG, Starling RC, Deng MC, Cappola TP, Kao A, Anderson AS, Cotts WG, Ewald GA, Baran DA, Bogaev RC, Elashoff B, Baron H, Yee J, Valantine HA, IMAGE Study Group Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med. 2010;362:1890–1900. doi: 10.1056/NEJMoa0912965. [DOI] [PubMed] [Google Scholar]
- 4.Kobashigawa J, Patel J, Azarbal B, Kittleson M, Chang D, Czer L, Daun T, Luu M, Trento A, Cheng R, Esmailian F. Randomized pilot trial of gene expression profiling versus heart biopsy in the first year after heart transplant: early invasive monitoring attenuation through gene expression trial. Circ Heart Fail. 2015;8:557–564. doi: 10.1161/CIRCHEARTFAILURE.114.001658. [DOI] [PubMed] [Google Scholar]
- 5.Tebani A, Afonso C, Marret S, Bekri S. Omics-based strategies in precision medicine: toward a paradigm shift in inborn errors of metabolism investigations. Int J Mol Sci. 2016;17. 10.3390/ijms17091555. [DOI] [PMC free article] [PubMed]
- 6.Tayal U, Prasad S, Cook SA. Genetics and genomics of dilated cardiomyopathy and systolic heart failure. Genome Med. 2017;9:20. doi: 10.1186/s13073-017-0410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Ende MY, Said MA, van Veldhuisen DJ, Verweij N, van der Harst P. Genome-wide studies of heart failure and endophenotypes: lessons learned and future directions. Cardiovasc Res. 2018;114:1209–1225. doi: 10.1093/cvr/cvy083. [DOI] [PubMed] [Google Scholar]
- 8.Zar G, Smith JG, Smith ML, Andersson B, Nilsson J. Whole-genome sequencing based on formalin-fixed paraffin-embedded endomyocardial biopsies for genetic studies on outcomes after heart transplantation. PLoS One. 2019;14:e0217747. doi: 10.1371/journal.pone.0217747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bagnall RD, Ingles J, Dinger ME, Cowley MJ, Ross SB, Minoche AE, Lal S, Turner C, Colley A, Rajagopalan S, Berman Y, Ronan A, Fatkin D, Semsarian C. Whole genome sequencing improves outcomes of genetic testing in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2018;72:419–429. doi: 10.1016/j.jacc.2018.04.078. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Morley M, Brandimarto J, Hannenhalli S, Hu Y, Ashley EA, Tang WH, Moravec CS, Margulies KB, Cappola TP, Li M, MAGNet consortium RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics. 2015;105:83–89. doi: 10.1016/j.ygeno.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kittleson MM, Minhas KM, Irizarry RA, Ye SQ, Edness G, Breton E, Conte JV, Tomaselli G, Garcia JGN, Hare JM. Gene expression analysis of ischemic and nonischemic cardiomyopathy: shared and distinct genes in the development of heart failure. Physiol Genomics. 2005;21:299–307. doi: 10.1152/physiolgenomics.00255.2004. [DOI] [PubMed] [Google Scholar]
- 12.Sweet ME, Cocciolo A, Slavov D, Jones KL, Sweet JR, Graw SL, Reece TB, Ambardekar AV, Bristow MR, Mestroni L, Taylor MRG. Transcriptome analysis of human heart failure reveals dysregulated cell adhesion in dilated cardiomyopathy and activated immune pathways in ischemic heart failure. BMC Genomics. 2018;19:812. doi: 10.1186/s12864-018-5213-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Heesch S, Witte F, Schneider-Lunitz V, et al. The translational landscape of the human heart. Cell. 2019;178:242–260.e29. doi: 10.1016/j.cell.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Yu P, Zhou B, et al. Single-cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nat Cell Biol. 2020;22:108–119. doi: 10.1038/s41556-019-0446-7. [DOI] [PubMed] [Google Scholar]
- 15.Asp M, Salmén F, Ståhl PL, Vickovic S, Felldin U, Löfling M, Fernandez Navarro J, Maaskola J, Eriksson MJ, Persson B, Corbascio M, Persson H, Linde C, Lundeberg J. Spatial detection of fetal marker genes expressed at low level in adult human heart tissue. Sci Rep. 2017;7:12941. doi: 10.1038/s41598-017-13462-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam MPY, Ping P, Murphy E. Proteomics research in cardiovascular medicine and biomarker discovery. J Am Coll Cardiol. 2016;68:2819–2830. doi: 10.1016/j.jacc.2016.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giudice G, Petsalaki E. Proteomics and phosphoproteomics in precision medicine: applications and challenges. Brief Bioinformatics. 2019;20:767–777. doi: 10.1093/bib/bbx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kooij V, Venkatraman V, Tra J, Kirk JA, Rowell J, Blice-Baum A, Cammarato A, Van Eyk JE. Sizing up models of heart failure: proteomics from flies to humans. Proteomics Clin Appl. 2014;8:653–664. doi: 10.1002/prca.201300123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGarrah RW, Crown SB, Zhang G-F, Shah SH, Newgard CB. Cardiovascular metabolomics. Circ Res. 2018;122:1238–1258. doi: 10.1161/CIRCRESAHA.117.311002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tham YK, Bernardo BC, Huynh K, Ooi JYY, Gao XM, Kiriazis H, Giles C, Meikle PJ, McMullen JR. Lipidomic profiles of the heart and circulation in response to exercise versus cardiac pathology: a resource of potential biomarkers and drug targets. Cell Rep. 2018;24:2757–2772. doi: 10.1016/j.celrep.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Halade GV, Dorbane A, Ingle KA, Kain V, Schmitter J-M, Rhourri-Frih B. Comprehensive targeted and non-targeted lipidomics analyses in failing and non-failing heart. Anal Bioanal Chem. 2018;410:1965–1976. doi: 10.1007/s00216-018-0863-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeVore AD, Wosik J, Hernandez AF. The future of wearables in heart failure patients. JACC Heart Fail. 2019;7:922–932. doi: 10.1016/j.jchf.2019.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Blecker S, Katz SD, Horwitz LI, Kuperman G, Park H, Gold A, Sontag D. Comparison of approaches for heart failure case identification from electronic health record data. JAMA Cardiol. 2016;1:1014–1020. doi: 10.1001/jamacardio.2016.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin-Isla C, Campello VM, Izquierdo C, Raisi-Estabragh Z, Baeßler B, Petersen SE, Lekadir K. Image-based cardiac diagnosis with machine learning: a review. Front Cardiovasc Med. 2020;7:1. doi: 10.3389/fcvm.2020.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan J, Han F, Liu H. Challenges of big data analysis. Natl Sci Rev. 2014;1:293–314. doi: 10.1093/nsr/nwt032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson KW, Torres Soto J, Glicksberg BS, Shameer K, Miotto R, Ali M, Ashley E, Dudley JT. Artificial intelligence in cardiology. J Am Coll Cardiol. 2018;71:2668–2679. doi: 10.1016/j.jacc.2018.03.521. [DOI] [PubMed] [Google Scholar]
- 27.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 28.Gandomi A, Haider M. Beyond the hype: big data concepts, methods, and analytics. Int J Inf Manag. 2015;35:137–144. [Google Scholar]
- 29.Meng C, Zeleznik OA, Thallinger GG, Kuster B, Gholami AM, Culhane AC. Dimension reduction techniques for the integrative analysis of multi-omics data. Brief Bioinformatics. 2016;17:628–641. doi: 10.1093/bib/bbv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019;20:631–656. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- 32.Fu Y, Eisen HJ. Genetics of dilated cardiomyopathy. Curr Cardiol Rep. 2018;20:121. doi: 10.1007/s11886-018-1061-0. [DOI] [PubMed] [Google Scholar]
- 33.King CR, Nicolae DL. GWAS to sequencing: divergence in study design and analysis. Genes (Basel) 2014;5:460–476. doi: 10.3390/genes5020460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rau CD, Lusis AJ, Wang Y. Genetics of common forms of heart failure: challenges and potential solutions. Curr Opin Cardiol. 2015;30:222–227. doi: 10.1097/HCO.0000000000000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aragam KG, Chaffin M, Levinson RT, McDermott G, Choi SH, Shoemaker MB, Haas ME, Weng LC, Lindsay ME, Smith JG, Newton-Cheh C, Roden DM, London B, For the GRADE Investigators. Wells QS, Ellinor PT, Kathiresan S, Lubitz SA, Bloom HL, Dudley SC, Shalaby AA, Weiss R, Gutmann R, Saba S. Phenotypic refinement of heart failure in a national biobank facilitates genetic discovery. Circulation. 2018;139:489–501. doi: 10.1161/CIRCULATIONAHA.118.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wild PS, Felix JF, Schillert A, Teumer A, Chen MH, Leening MJG, Völker U, Großmann V, Brody JA, Irvin MR, Shah SJ, Pramana S, Lieb W, Schmidt R, Stanton AV, Malzahn D, Smith AV, Sundström J, Minelli C, Ruggiero D, Lyytikäinen LP, Tiller D, Smith JG, Monnereau C, di Tullio MR, Musani SK, Morrison AC, Pers TH, Morley M, Kleber ME, Aragam J, Benjamin EJ, Bis JC, Bisping E, Broeckel U, Cheng S, Deckers JW, del Greco M F, Edelmann F, Fornage M, Franke L, Friedrich N, Harris TB, Hofer E, Hofman A, Huang J, Hughes AD, Kähönen M, investigators K, Kruppa J, Lackner KJ, Lannfelt L, Laskowski R, Launer LJ, Leosdottir M, Lin H, Lindgren CM, Loley C, MacRae CA, Mascalzoni D, Mayet J, Medenwald D, Morris AP, Müller C, Müller-Nurasyid M, Nappo S, Nilsson PM, Nuding S, Nutile T, Peters A, Pfeufer A, Pietzner D, Pramstaller PP, Raitakari OT, Rice KM, Rivadeneira F, Rotter JI, Ruohonen ST, Sacco RL, Samdarshi TE, Schmidt H, Sharp ASP, Shields DC, Sorice R, Sotoodehnia N, Stricker BH, Surendran P, Thom S, Töglhofer AM, Uitterlinden AG, Wachter R, Völzke H, Ziegler A, Münzel T, März W, Cappola TP, Hirschhorn JN, Mitchell GF, Smith NL, Fox ER, Dueker ND, Jaddoe VWV, Melander O, Russ M, Lehtimäki T, Ciullo M, Hicks AA, Lind L, Gudnason V, Pieske B, Barron AJ, Zweiker R, Schunkert H, Ingelsson E, Liu K, Arnett DK, Psaty BM, Blankenberg S, Larson MG, Felix SB, Franco OH, Zeller T, Vasan RS, Dörr M. Large-scale genome-wide analysis identifies genetic variants associated with cardiac structure and function. J Clin Invest. 2017;127:1798–1812. doi: 10.1172/JCI84840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aung N, Vargas JD, Yang C, Cabrera CP, Warren HR, Fung K, Tzanis E, Barnes MR, Rotter JI, Taylor KD, Manichaikul AW, Lima JAC, Bluemke DA, Piechnik SK, Neubauer S, Munroe PB, Petersen SE. Genome-wide analysis of left ventricular image-derived phenotypes identifies fourteen loci associated with cardiac morphogenesis and heart failure development. Circulation. 2019;140:1318–1330. doi: 10.1161/CIRCULATIONAHA.119.041161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shah S, Henry A, Roselli C, et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun. 2020;11:163. doi: 10.1038/s41467-019-13690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meder B, Haas J, Sedaghat-Hamedani F, Kayvanpour E, Frese K, Lai A, Nietsch R, Scheiner C, Mester S, Bordalo DM, Amr A, Dietrich C, Pils D, Siede D, Hund H, Bauer A, Holzer DB, Ruhparwar A, Mueller-Hennessen M, Weichenhan D, Plass C, Weis T, Backs J, Wuerstle M, Keller A, Katus HA, Posch AE. Epigenome-wide association study identifies cardiac gene patterning and a novel class of biomarkers for heart failure. Circulation. 2017;136:1528–1544. doi: 10.1161/CIRCULATIONAHA.117.027355. [DOI] [PubMed] [Google Scholar]
- 40.Rosa-Garrido M, Chapski DJ, Schmitt AD, Kimball TH, Karbassi E, Monte E, Balderas E, Pellegrini M, Shih TT, Soehalim E, Liem D, Ping P, Galjart NJ, Ren S, Wang Y, Ren B, Vondriska TM. High-resolution mapping of chromatin conformation in cardiac Myocytes reveals structural remodeling of the Epigenome in heart failure. Circulation. 2017;136:1613–1625. doi: 10.1161/CIRCULATIONAHA.117.029430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKinsey TA, Vondriska TM, Wang Y. Epigenomic regulation of heart failure: integrating histone marks, long noncoding RNAs, and chromatin architecture. [version 1; peer review: 2 approved]. F1000Res. 2018. 10.12688/f1000research.15797.1. [DOI] [PMC free article] [PubMed]
- 42.Marín-García J, Akhmedov AT. Epigenetics of the failing heart. Heart Fail Rev. 2015;20:435–459. doi: 10.1007/s10741-015-9483-x. [DOI] [PubMed] [Google Scholar]
- 43.Andersson C, Lin H, Liu C, Levy D, Mitchell GF, Larson MG, Vasan RS. Integrated multiomics approach to identify genetic underpinnings of heart failure and its echocardiographic precursors: Framingham heart study. Circ Genom Precis Med. 2019;12:e002489. doi: 10.1161/CIRCGEN.118.002489. [DOI] [PubMed] [Google Scholar]
- 44.Arvanitis M, Tampakakis E, Zhang Y, et al. Genome-wide association and multi-omic analyses reveal ACTN2 as a gene linked to heart failure. Nat Commun. 2020;11:1122. doi: 10.1038/s41467-020-14843-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter AC, Chang HY, Church G, Dombkowski A, Ecker JR, Gil E, Giresi PG, Greely H, Greenleaf WJ, Hacohen N, He C, Hill D, Ko J, Kohane I, Kundaje A, Palmer M, Snyder MP, Tung J, Urban A, Vidal M, Wong W. Challenges and recommendations for epigenomics in precision health. Nat Biotechnol. 2017;35:1128–1132. doi: 10.1038/nbt.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu C-F, Tang WHW. Epigenetics in cardiac hypertrophy and heart failure. JACC Basic Transl Sci. 2019;4:976–993. doi: 10.1016/j.jacbts.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willyard C. New human gene tally reignites debate. Nature. 2018;558:354–355. doi: 10.1038/d41586-018-05462-w. [DOI] [PubMed] [Google Scholar]
- 49.Dugourd A, Saez-Rodriguez J. Footprint-based functional analysis of multi-omic data. Current Opinion in Systems Biology. 2019;15:82–90. doi: 10.1016/j.coisb.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Avila Cobos F, Vandesompele J, Mestdagh P, De Preter K. Computational deconvolution of transcriptomics data from mixed cell populations. Bioinformatics. 2018;34:1969–1979. doi: 10.1093/bioinformatics/bty019. [DOI] [PubMed] [Google Scholar]
- 51.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, Khodadoust MS, Esfahani MS, Luca BA, Steiner D, Diehn M, Alizadeh AA. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donovan MKR, D’Antonio-Chronowska A, D’Antonio M, Frazer KA. Cellular deconvolution of GTEx tissues powers discovery of disease and cell-type associated regulatory variants. Nat Commun. 2020;11:955. doi: 10.1038/s41467-020-14561-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Moravec CS, Sussman MA, DiPaola NR, Fu D, Hawthorn L, Mitchell CA, Young JB, Francis GS, McCarthy PM, Bond M. Decreased SLIM1 expression and increased gelsolin expression in failing human hearts measured by high-density oligonucleotide arrays. Circulation. 2000;102:3046–3052. doi: 10.1161/01.cir.102.25.3046. [DOI] [PubMed] [Google Scholar]
- 54.Raghow R. An “omics” perspective on cardiomyopathies and heart failure. Trends Mol Med. 2016;22:813–827. doi: 10.1016/j.molmed.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 55.Kim GH, Uriel N, Burkhoff D. Reverse remodelling and myocardial recovery in heart failure. Nat Rev Cardiol. 2018;15:83–96. doi: 10.1038/nrcardio.2017.139. [DOI] [PubMed] [Google Scholar]
- 56.Peterzan MA, Lygate CA, Neubauer S, Rider OJ. Metabolic remodeling in hypertrophied and failing myocardium: a review. Am J Physiol Heart Circ Physiol. 2017;313:H597–H616. doi: 10.1152/ajpheart.00731.2016. [DOI] [PubMed] [Google Scholar]
- 57.Louzao-Martinez L, Vink A, Harakalova M, Asselbergs FW, Verhaar MC, Cheng C. Characteristic adaptations of the extracellular matrix in dilated cardiomyopathy. Int J Cardiol. 2016;220:634–646. doi: 10.1016/j.ijcard.2016.06.253. [DOI] [PubMed] [Google Scholar]
- 58.Mann DL, Topkara VK, Evans S, Barger PM. Innate immunity in the adult mammalian heart: for whom the cell tolls. Trans Am Clin Climatol Assoc. 2010;121:34–50. [PMC free article] [PubMed] [Google Scholar]
- 59.Alimadadi A, Munroe PB, Joe B, Cheng X. Meta-analysis of dilated cardiomyopathy using cardiac RNA-Seq Transcriptomic datasets. Genes (Basel). 2020;11. 10.3390/genes11010060. [DOI] [PMC free article] [PubMed]
- 60.Sharma UC, Pokharel S, Evelo CTA, Maessen JG. A systematic review of large scale and heterogeneous gene array data in heart failure. J Mol Cell Cardiol. 2005;38:425–432. doi: 10.1016/j.yjmcc.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 61.Barth AS, Kumordzie A, Frangakis C, Margulies KB, Cappola TP, Tomaselli GF. Reciprocal transcriptional regulation of metabolic and signaling pathways correlates with disease severity in heart failure. Circ Cardiovasc Genet. 2011;4:475–483. doi: 10.1161/CIRCGENETICS.110.957571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toro-Domínguez D, Villatoro-García JA, Martorell-Marugán J, Román-Montoya Y, Alarcón-Riquelme ME, Carmona-Sáez P. A survey of gene expression meta-analysis: methods and applications. Brief Bioinformatics. 2020. 10.1093/bib/bbaa019. [DOI] [PubMed]
- 63.Das S, Frisk C, Eriksson MJ, et al. Transcriptomics of cardiac biopsies reveals differences in patients with or without diagnostic parameters for heart failure with preserved ejection fraction. Sci Rep. 2019;9:3179. doi: 10.1038/s41598-019-39445-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gomes CPC, Schroen B, Kuster GM, et al. Regulatory rnas in heart failure. Circulation. 2020;141:313–328. doi: 10.1161/CIRCULATIONAHA.119.042474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vegter EL, van der Meer P, de Windt LJ, Pinto YM, Voors AA. MicroRNAs in heart failure: from biomarker to target for therapy. Eur J Heart Fail. 2016;18:457–468. doi: 10.1002/ejhf.495. [DOI] [PubMed] [Google Scholar]
- 66.Verjans R, Derks WJA, Korn K, Sönnichsen B, van Leeuwen REW, Schroen B, van Bilsen M, Heymans S. Functional screening identifies MicroRNAs as multi-cellular regulators of heart failure. Sci Rep. 2019;9:6055. doi: 10.1038/s41598-019-41491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao J, Qin B, Nikolay R, Spahn CMT, Zhang G. Translatomics: the global view of translation. Int J Mol Sci. 2019;20. 10.3390/ijms20010212. [DOI] [PMC free article] [PubMed]
- 68.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 70.Holland CH, Tanevski J, Perales-Patón J, Gleixner J, Kumar MP, Mereu E, Joughin BA, Stegle O, Lauffenburger DA, Heyn H, Szalai B, Saez-Rodriguez J. Robustness and applicability of transcription factor and pathway analysis tools on single-cell RNA-seq data. Genome Biol. 2020;21:36. doi: 10.1186/s13059-020-1949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Argelaguet R, Velten B, Arnol D, Dietrich S, Zenz T, Marioni JC, Buettner F, Huber W, Stegle O. Multi-Omics factor analysis-a framework for unsupervised integration of multi-omics data sets. Mol Syst Biol. 2018;14:e8124. doi: 10.15252/msb.20178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Svensson V, Vento-Tormo R, Teichmann SA. Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc. 2018;13:599–604. doi: 10.1038/nprot.2017.149. [DOI] [PubMed] [Google Scholar]
- 73.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 74.Selewa A, Dohn R, Eckart H, Lozano S, Xie B, Gauchat E, Elorbany R, Rhodes K, Burnett J, Gilad Y, Pott S, Basu A. Systematic comparison of high-throughput single-cell and single-nucleus Transcriptomes during Cardiomyocyte differentiation. Sci Rep. 2020;10:1535. doi: 10.1038/s41598-020-58327-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yekelchyk M, Guenther S, Preussner J, Braun T. Mono- and multi-nucleated ventricular cardiomyocytes constitute a transcriptionally homogenous cell population. Basic Res Cardiol. 2019;114:36. doi: 10.1007/s00395-019-0744-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Laehnemann D, Köster J, Szczurek E, et al (2019) 12 grand challenges in single-cell data science. doi: 10.7287/peerj.preprints.27885v3. [DOI] [PMC free article] [PubMed]
- 77.Chaudhry F, Isherwood J, Bawa T, Patel D, Gurdziel K, Lanfear DE, Ruden DM, Levy PD. Single-cell RNA sequencing of the cardiovascular system: new looks for old diseases. Front Cardiovasc Med. 2019;6:173. doi: 10.3389/fcvm.2019.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ackers-Johnson M, Tan WLW, Foo RS-Y. Following hearts, one cell at a time: recent applications of single-cell RNA sequencing to the understanding of heart disease. Nat Commun. 2018;9:4434. doi: 10.1038/s41467-018-06894-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Molenaar B, van Rooij E. Single-cell sequencing of the mammalian heart. Circ Res. 2018;123:1033–1035. doi: 10.1161/CIRCRESAHA.118.313531. [DOI] [PubMed] [Google Scholar]
- 80.Meilhac SM, Buckingham ME. The deployment of cell lineages that form the mammalian heart. Nat Rev Cardiol. 2018;15:705–724. doi: 10.1038/s41569-018-0086-9. [DOI] [PubMed] [Google Scholar]
- 81.Skelly DA, Squiers GT, McLellan MA, Bolisetty MT, Robson P, Rosenthal NA, Pinto AR. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 2018;22:600–610. doi: 10.1016/j.celrep.2017.12.072. [DOI] [PubMed] [Google Scholar]
- 82.Wolfien M, Galow A-M, Müller P, Bartsch M, Brunner RM, Goldammer T, et al. Single-nucleus sequencing of an entire mammalian heart: cell type composition and velocity. Cells. 2020;9. 10.3390/cells9020318. [DOI] [PMC free article] [PubMed]
- 83.Tabula Muris Consortium, Overall coordination, Logistical coordination, Organ collection and processing, Library preparation and sequencing, Computational data analysis, Cell type annotation, Writing group, Supplemental text writing group, Principal investigators Single-cell transcriptomics of 20 mouse organs creates a tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiong H, Luo Y, Yue Y, Zhang J, Ai S, Li X, Wang X, Zhang YL, Wei Y, Li HH, Hu X, Li C, He A. Single-cell Transcriptomics reveals Chemotaxis-mediated Intraorgan crosstalk during Cardiogenesis. Circ Res. 2019;125:398–410. doi: 10.1161/CIRCRESAHA.119.315243. [DOI] [PubMed] [Google Scholar]
- 85.Asp M, Giacomello S, Larsson L, et al. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell. 2019;179:1647–1660.e19. doi: 10.1016/j.cell.2019.11.025. [DOI] [PubMed] [Google Scholar]
- 86.Li G, Tian L, Goodyer W, Kort EJ, Buikema JW, Xu A, Wu JC, Jovinge S, Wu SM. Single cell expression analysis reveals anatomical and cell cycle-dependent transcriptional shifts during heart development. Development. 2019;146:dev173476. doi: 10.1242/dev.173476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Phansalkar R, Red-Horse K. Techniques converge to map the developing human heart at single-cell level. Nature. 2020;577:629–630. doi: 10.1038/d41586-020-00151-z. [DOI] [PubMed] [Google Scholar]
- 88.Cui Y, Zheng Y, Liu X, et al. Single-cell Transcriptome analysis maps the developmental track of the human heart. Cell Rep. 2019;26:1934–1950.e5. doi: 10.1016/j.celrep.2019.01.079. [DOI] [PubMed] [Google Scholar]
- 89.See K, Tan WLW, Lim EH, Tiang Z, Lee LT, Li PYQ, Luu TDA, Ackers-Johnson M, Foo RS. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nat Commun. 2017;8:225. doi: 10.1038/s41467-017-00319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nomura S, Satoh M, Fujita T, Higo T, Sumida T, Ko T, Yamaguchi T, Tobita T, Naito AT, Ito M, Fujita K, Harada M, Toko H, Kobayashi Y, Ito K, Takimoto E, Akazawa H, Morita H, Aburatani H, Komuro I. Cardiomyocyte gene programs encoding morphological and functional signatures in cardiac hypertrophy and failure. Nat Commun. 2018;9:4435. doi: 10.1038/s41467-018-06639-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leuschner F, Nahrendorf M. Novel functions of macrophages in the heart: insights into electrical conduction, stress, and diastolic dysfunction. Eur Heart J. 2019. 10.1093/eurheartj/ehz159. [DOI] [PMC free article] [PubMed]
- 92.Moor AE, Itzkovitz S. Spatial transcriptomics: paving the way for tissue-level systems biology. Curr Opin Biotechnol. 2017;46:126–133. doi: 10.1016/j.copbio.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 93.Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, Mollbrink A, Linnarsson S, Codeluppi S, Borg Å, Pontén F, Costea PI, Sahlén P, Mulder J, Bergmann O, Lundeberg J, Frisén J. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 94.Vickovic S, Eraslan G, Salmén F, Klughammer J, Stenbeck L, Schapiro D, Äijö T, Bonneau R, Bergenstråhle L, Navarro JF, Gould J, Griffin GK, Borg Å, Ronaghi M, Frisén J, Lundeberg J, Regev A, Ståhl PL. High-definition spatial transcriptomics for in situ tissue profiling. Nat Methods. 2019;16:987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eng C-HL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, Cronin C, Karp C, Yuan GC, Cai L. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature. 2019;568:235–239. doi: 10.1038/s41586-019-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F, Macosko EZ. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schubert OT, Röst HL, Collins BC, Rosenberger G, Aebersold R. Quantitative proteomics: challenges and opportunities in basic and applied research. Nat Protoc. 2017;12:1289–1294. doi: 10.1038/nprot.2017.040. [DOI] [PubMed] [Google Scholar]
- 98.Doll S, Dreßen M, Geyer PE, Itzhak DN, Braun C, Doppler SA, Meier F, Deutsch MA, Lahm H, Lange R, Krane M, Mann M. Region and cell-type resolved quantitative proteomic map of the human heart. Nat Commun. 2017;8:1469. doi: 10.1038/s41467-017-01747-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Benson MD, Yang Q, Ngo D, Zhu Y, Shen D, Farrell LA, Sinha S, Keyes MJ, Vasan RS, Larson MG, Smith JG, Wang TJ, Gerszten RE. Genetic architecture of the cardiovascular risk proteome. Circulation. 2018;137:1158–1172. doi: 10.1161/CIRCULATIONAHA.117.029536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lygirou V, Latosinska A, Makridakis M, Mullen W, Delles C, Schanstra JP, Zoidakis J, Pieske B, Mischak H, Vlahou A. Plasma proteomic analysis reveals altered protein abundances in cardiovascular disease. J Transl Med. 2018;16:104. doi: 10.1186/s12967-018-1476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cao TH, Jones DJL, Voors AA, Quinn PA, Sandhu JK, Chan DCS, Parry HM, Mohan M, Mordi IR, Sama IE, Anker SD, Cleland JG, Dickstein K, Filippatos G, Hillege HL, Metra M, Ponikowski P, Samani NJ, van Veldhuisen DJ, Zannad F, Lang CC, Ng LL. Plasma proteomic approach in patients with heart failure: insights into pathogenesis of disease progression and potential novel treatment targets. Eur J Heart Fail. 2020;22:70–80. doi: 10.1002/ejhf.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Egerstedt A, Berntsson J, Smith ML, et al. Profiling of the plasma proteome across different stages of human heart failure. Nat Commun. 2019;10:5830. doi: 10.1038/s41467-019-13306-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Datta S, Bernstam EV, Roberts K. A frame semantic overview of NLP-based information extraction for cancer-related EHR notes. J Biomed Inform. 2019;100:103301. doi: 10.1016/j.jbi.2019.103301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zeng Z, Deng Y, Li X, Naumann T, Luo Y. Natural language processing for EHR-based computational Phenotyping. IEEE/ACM Trans Comput Biol Bioinform. 2019;16:139–153. doi: 10.1109/TCBB.2018.2849968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sheikhalishahi S, Miotto R, Dudley JT, Lavelli A, Rinaldi F, Osmani V. Natural language processing of clinical notes on chronic diseases: systematic review. JMIR Med Inform. 2019;7:e12239. doi: 10.2196/12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Iorio A, Pozzi A, Senni M. Addressing the heterogeneity of heart failure in future randomized trials. Curr Heart Fail Rep. 2017;14:197–202. doi: 10.1007/s11897-017-0332-1. [DOI] [PubMed] [Google Scholar]
- 107.Altman RB, Ashley EA. Using “big data” to dissect clinical heterogeneity. Circulation. 2015;131:232–233. doi: 10.1161/CIRCULATIONAHA.114.014106. [DOI] [PubMed] [Google Scholar]
- 108.Desai RJ, Wang SV, Vaduganathan M, Evers T, Schneeweiss S. Comparison of machine learning methods with traditional models for use of administrative claims with electronic medical records to predict heart failure outcomes. JAMA Netw Open. 2020;3:e1918962. doi: 10.1001/jamanetworkopen.2019.18962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Adler ED, Voors AA, Klein L, Macheret F, Braun OO, Urey MA, Zhu W, Sama I, Tadel M, Campagnari C, Greenberg B, Yagil A. Improving risk prediction in heart failure using machine learning. Eur J Heart Fail. 2020;22:139–147. doi: 10.1002/ejhf.1628. [DOI] [PubMed] [Google Scholar]
- 110.Ahmad T, Lund LH, Rao P, Ghosh R, Warier P, Vaccaro B, et al. Machine learning methods improve prognostication, identify clinically distinct phenotypes, and detect heterogeneity in response to therapy in a large cohort of heart failure patients. J Am Heart Assoc. 2018. 10.1161/JAHA.117.008081. [DOI] [PMC free article] [PubMed]
- 111.Angraal S, Mortazavi BJ, Gupta A, Khera R, Ahmad T, Desai NR, Jacoby DL, Masoudi FA, Spertus JA, Krumholz HM. Machine learning prediction of mortality and hospitalization in heart failure with preserved ejection fraction. JACC Heart Fail. 2020;8:12–21. doi: 10.1016/j.jchf.2019.06.013. [DOI] [PubMed] [Google Scholar]
- 112.Cikes M, Sanchez-Martinez S, Claggett B, Duchateau N, Piella G, Butakoff C, Pouleur AC, Knappe D, Biering-Sørensen T, Kutyifa V, Moss A, Stein K, Solomon SD, Bijnens B. Machine learning-based phenogrouping in heart failure to identify responders to cardiac resynchronization therapy. Eur J Heart Fail. 2019;21:74–85. doi: 10.1002/ejhf.1333. [DOI] [PubMed] [Google Scholar]
- 113.Tabassian M, Sunderji I, Erdei T, Sanchez-Martinez S, Degiovanni A, Marino P, Fraser AG, D’hooge J. Diagnosis of heart failure with preserved ejection fraction: machine learning of spatiotemporal variations in left ventricular deformation. J Am Soc Echocardiogr. 2018;31:1272–1284.e9. doi: 10.1016/j.echo.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 114.Acharya UR, Fujita H, Oh SL, Hagiwara Y, Tan JH, Adam M, Tan RS (2018) Deep convolutional neural network for the automated diagnosis of congestive heart failure using ECG signals. Appl Intell 1–12.
- 115.Ambale-Venkatesh B, Yang X, Wu CO, Liu K, Hundley WG, McClelland R, Gomes AS, Folsom AR, Shea S, Guallar E, Bluemke DA, Lima JAC. Cardiovascular event prediction by machine learning: the multi-ethnic study of atherosclerosis. Circ Res. 2017;121:1092–1101. doi: 10.1161/CIRCRESAHA.117.311312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nirschl JJ, Janowczyk A, Peyster EG, Frank R, Margulies KB, Feldman MD, Madabhushi A. A deep-learning classifier identifies patients with clinical heart failure using whole-slide images of H&E tissue. PLoS One. 2018;13:e0192726. doi: 10.1371/journal.pone.0192726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Inan OT, Baran Pouyan M, Javaid AQ, et al. Novel wearable seismocardiography and machine learning algorithms can assess clinical status of heart failure patients. Circ Heart Fail. 2018;11:e004313. doi: 10.1161/CIRCHEARTFAILURE.117.004313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stehlik J, Schmalfuss C, Bozkurt B, et al. Continuous wearable monitoring analytics predict heart failure hospitalization: the LINK-HF multicenter study. Circ Heart Fail. 2020;13:e006513. doi: 10.1161/CIRCHEARTFAILURE.119.006513. [DOI] [PubMed] [Google Scholar]
- 119.Al’Aref SJ, Anchouche K, Singh G, et al. Clinical applications of machine learning in cardiovascular disease and its relevance to cardiac imaging. Eur Heart J. 2019;40:1975–1986. doi: 10.1093/eurheartj/ehy404. [DOI] [PubMed] [Google Scholar]
- 120.Pour-Ghaz I, Hana D, Raja J, Ibebuogu UN, Khouzam RN. CardioMEMS: where we are and where can we go? Ann Transl Med. 2019;7:418. doi: 10.21037/atm.2019.07.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Joly Y, Dyke SOM, Knoppers BM, Pastinen T. Are data sharing and privacy protection mutually exclusive? Cell. 2016;167:1150–1154. doi: 10.1016/j.cell.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 122.Berger B, Cho H. Emerging technologies towards enhancing privacy in genomic data sharing. Genome Biol. 2019;20:128. doi: 10.1186/s13059-019-1741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fiume M, Cupak M, Keenan S, Rambla J, de la Torre S, Dyke SOM, Brookes AJ, Carey K, Lloyd D, Goodhand P, Haeussler M, Baudis M, Stockinger H, Dolman L, Lappalainen I, Törnroos J, Linden M, Spalding JD, Ur-Rehman S, Page A, Flicek P, Sherry S, Haussler D, Varma S, Saunders G, Scollen S. Federated discovery and sharing of genomic data using beacons. Nat Biotechnol. 2019;37:220–224. doi: 10.1038/s41587-019-0046-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Harmanci A, Gerstein M. Analysis of sensitive information leakage in functional genomics signal profiles through genomic deletions. Nat Commun. 2018;9:2453. doi: 10.1038/s41467-018-04875-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.U.S. Department of Health & Human Services - Office for Civil Rights Breach Portal: Notice to the Secretary of HHS Breach of Unsecured Protected Health Information. In: Breach Portal: Notice to the Secretary of HHS Breach of Unsecured Protected Health Information. https://ocrportal.hhs.gov/ocr/breach/breach_report.jsf. Accessed 16 Jun 2020.
- 126.Kassab M, DeFranco J, Malas T, Graciano Neto VV, Destefanis G (2019) Blockchain: a panacea for electronic health records? 2019 IEEE/ACM 1st international workshop on software engineering for healthcare (SEH). IEEE, pp 21–24.
- 127.Angraal S, Krumholz HM, Schulz WL. Blockchain technology: applications in health care. Circ Cardiovasc Qual Outcomes. 2017. 10.1161/CIRCOUTCOMES.117.003800. [DOI] [PubMed]
- 128.Karafiloski E, Mishev A (2017) Blockchain solutions for big data challenges: a literature review. IEEE EUROCON 2017 -17th international conference on smart technologies. IEEE, pp 763–768.
- 129.Lee EWJ, Viswanath K. Big data in context: addressing the twin perils of data absenteeism and chauvinism in the context of health disparities research. J Med Internet Res. 2020;22:e16377. doi: 10.2196/16377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Meng X-L. Statistical paradises and paradoxes in big data (I): law of large populations, big data paradox, and the 2016 US presidential election. Ann Appl Stat. 2018;12:685–726. [Google Scholar]
- 131.Kaplan RM, Chambers DA, Glasgow RE. Big data and large sample size: a cautionary note on the potential for bias. Clin Transl Sci. 2014;7:342–346. doi: 10.1111/cts.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wells BJ, Chagin KM, Nowacki AS, Kattan MW. Strategies for handling missing data in electronic health record derived data. EGEMS (Wash DC) 2013;1:1035. doi: 10.13063/2327-9214.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Beaulieu-Jones BK, Lavage DR, Snyder JW, Moore JH, Pendergrass SA, Bauer CR. Characterizing and managing missing structured data in electronic health records: data analysis. JMIR Med Inform. 2018;6:e11. doi: 10.2196/medinform.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gianfrancesco MA, Tamang S, Yazdany J, Schmajuk G. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern Med. 2018;178:1544–1547. doi: 10.1001/jamainternmed.2018.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Moskowitz A, McSparron J, Stone DJ, Celi LA. Preparing a new generation of clinicians for the era of big data. Harv Med Stud Rev. 2015;2:24–27. [PMC free article] [PubMed] [Google Scholar]
- 136.Topol EJ. A decade of digital medicine innovation. Sci Transl Med. 2019;11:eaaw7610. doi: 10.1126/scitranslmed.aaw7610. [DOI] [PubMed] [Google Scholar]