Abstract
Objective
Epidemiological studies show that shift workers are at increased risk for type 2 diabetes. As modern societies increasingly require shift work, it seems crucial to determine whether there are long‐lasting health effects of rotational shifts.
Methods
This study examined the after‐effects of 4 weeks of time‐restricted feeding (TRF) during the light period (= light‐fed) in rats, an animal model for shift work. This study also included a TRF‐dark (= dark‐fed) control group. The aligned and misaligned feeding times of light and dark feeding are associated with poor and good health outcomes, respectively. Several physiological measures were monitored continuously; blood, liver, brown adipose tissue, and soleus and gastrocnemius muscle were collected following 11 days of ad libitum (AL) feeding after ending the TRF.
Results
In the dark‐fed animals, the day/night differences in food intake, activity, and respiratory exchange ratio were still enhanced at the end of the experiment. Light‐fed animals displayed the smallest day/night differences for these measures, as well as for body temperature. In both the light‐ and dark‐fed animals, rhythms in plasma glucose, nonesterified fatty acids, and gene expression had not fully recovered after 11 days of AL feeding. Importantly, the effects on gene expression were both tissue and gene dependent.
Conclusions
Our data indicate that rotational shift workers may have an increased risk of long‐lasting disturbed rhythms in several physiological measures after a period of shift work. Clearly, such disturbances may harm their health.
Study Importance.
What is already known?
-
►
Time‐restricted feeding (TRF) is an often‐used animal model for shift work, but studies on potential after‐effects of shift work using this feeding paradigm are scarce.
What does this study add?
-
►
We show that following a 4‐week TRF protocol, several physiological measures, including feeding behavior, metabolic substrate preference (respiratory exchange ratio), and blood metabolites show after‐effects for more than 1 week.
-
►
In four metabolically important tissues, circadian clock–related and glucose/lipid‐related metabolic gene expression had not fully recovered; the soleus and gastrocnemius muscles were most affected, and liver and brown adipose tissue were affected to a lesser extent.
Introduction
Virtually all living species have evolved an internal timing system to anticipate predictable daily changes in the environment, such as the light/dark (L/D) cycle and food availability. This internal timing system optimizes many physiological processes through the timed orchestration of cellular processes important for metabolism and other cellular functions.
On a molecular level, this circadian clock consists of a transcriptional translational feedback loop with both a positive and a negative arm to ensure molecular oscillations with a period of ~24 hours. At its core, the molecular clock consists of various clock genes, including brain and muscle aryl hydrocarbon receptor nuclear translocator–like factor 1 (Bmal1); period 1 (Per1), period 2 (Per2), and period 3 (Per3); cryptochrome 1 (Cry1) and cryptochrome 2 (Cry2); nuclear receptor subfamily 1, group D α (Reverbα) and nuclear receptor subfamily 1, group D β (Reverbβ); retinoic acid receptor–related orphan receptor α (RORα) and retinoic acid receptor–related orphan receptor β (RORβ); and several other auxiliary genes that facilitate fine‐tuning the period, phase, and flexibility of the clock (1, 2, 3, 4, 5, 6).
As the biological clock plays a major role in energy metabolism, including lipid and glucose metabolism, disturbance of this clock could have detrimental effects on energy balance. Indeed, an increasing amount of evidence from both human and animal studies indicates that disturbed biological rhythms, (i.e., circadian disruption resulting from, e.g., shift work and [social] jet lag) can cause imbalances in energy metabolism and possibly even lead to metabolic disorders such as type 2 diabetes mellitus (T2DM). Epidemiological studies, for example, indicate that shift workers are at an increased risk of developing T2DM (7, 8, 9, 10). Although the mechanisms that link shift work and/or circadian disruption to T2DM remain largely unknown, one suspected contributing factor is the erratic eating patterns of shift workers, who regularly eat during nighttime, the natural inactive and fasting period of humans (11).
Time‐restricted feeding (TRF) during the light period is an animal model for shift work, particularly for the erratic eating patterns (12). In TRF, access to food is restricted to a specific time window during the day/night cycle. Food access is usually restricted to several hours during the light period (= inactive phase of nocturnal rodents such as rats and mice; TRF thus results in a misalignment between the feeding activity of animals and the natural timing of their sleep/wake behavior) or during the dark period (= active phase of nocturnal rodents; TRF thus strengthens the alignment between feeding behavior and the natural timing of their sleep/wake behavior). TRF in general is often associated with metabolic health improvement and longevity in nematodes, fruit flies, and mammals, including rodents (13). However, TRF during the inactive period is associated with unhealthiness, including adiposity, T2DM, and cardiovascular disease (13, 14). Indeed, a wide variety of metabolic (whole‐body) measures, as well as peripheral clock‐gene expression, is deregulated when TRF is timed during the regular sleep period (15, 16, 17, 18, 19). Moreover, the specific effects of TRF appear to be tissue dependent (18, 20). However, very little is known about possible after‐effects of TRF (i.e., how animals recover from the TRF regimen when being reverted to ad libitum [AL] feeding conditions again). An important part of our modern society participates in shift work, but usually not in a continuous fashion, which could result in continuously disturbed circadian clocks and disease development. Unfortunately, studies on the long‐lasting effects of shift work either in humans or using animal models such as TRF are lacking. Therefore, the aim of this study was to determine whether TRF (either aligned or misaligned with the natural sleep/wake cycle) has long‐lasting effects on physiology. To characterize any possible after‐effects of shift work, we subjected rats to TRF for 4 weeks, during either the light or dark period, after which animals could recover from this shift‐work condition during 11 days of AL feeding conditions. Several physiological measures (including food intake, body temperature, respiratory exchange ratio [RER], and locomotor activity) were measured throughout the entire duration of the experiment. Eleven days after returning to the AL feeding condition, animals were sacrificed, and blood metabolites and clock‐gene and metabolic‐gene expression in liver, brown adipose tissue (BAT), and soleus and gastrocnemius muscles were measured. We hypothesized that light‐fed animals, (i.e., TRF misaligned with the regular sleep/wake cycle) would show long‐lasting negative effects on health outcomes, whereas dark‐fed animals (i.e., TRF aligned with the regular sleep/wake cycle) would show long‐lasting positive effects on health outcomes.
Methods
Animals and housing
A total of 72 ~8‐week‐old male Wistar rats weighing 240 to 280 g on arrival (Wistar WU rats; Charles River, Den Bosch, The Netherlands) were used for the experiments. All animals were individually housed under a controlled 12‐hour/12‐hour L/D cycle (lights on at Zeitgeber Time [ZT]0 and lights off at ZT12) and ate only pelleted chow (Teklad Global Diets; Envigo, Horst, The Netherlands). All experiments were approved by the Dutch government and performed in accordance with the guidelines on animal experimentation from the Netherlands Institute for Neuroscience.
Experimental design
After acclimatization and 1 to 2 weeks of AL baseline feeding conditions, the TRF conditions started, during which animals were randomly distributed over three groups: AL‐fed, dark‐fed, and light‐fed (Figure 1A). Dark‐fed animals could only eat between ZT13 and ZT23, and light‐fed animals could only eat between ZT1 and ZT11. Tap water was provided AL for all groups. After 4 weeks (28 days) of TRF, the final experimental condition started, the Revert condition, during which all animals could eat AL again. After 11 (10.5‐11.5) days of the Revert condition, animals were sacrificed at ZT0, ZT6, ZT12, or ZT18, and liver, BAT, and soleus and gastrocnemius muscle tissues were snap‐frozen in liquid nitrogen and stored at −80 °C. Trunk blood was also collected and centrifuged at 4,000 rpm, and plasma was stored at −20 °C.
Figure 1.
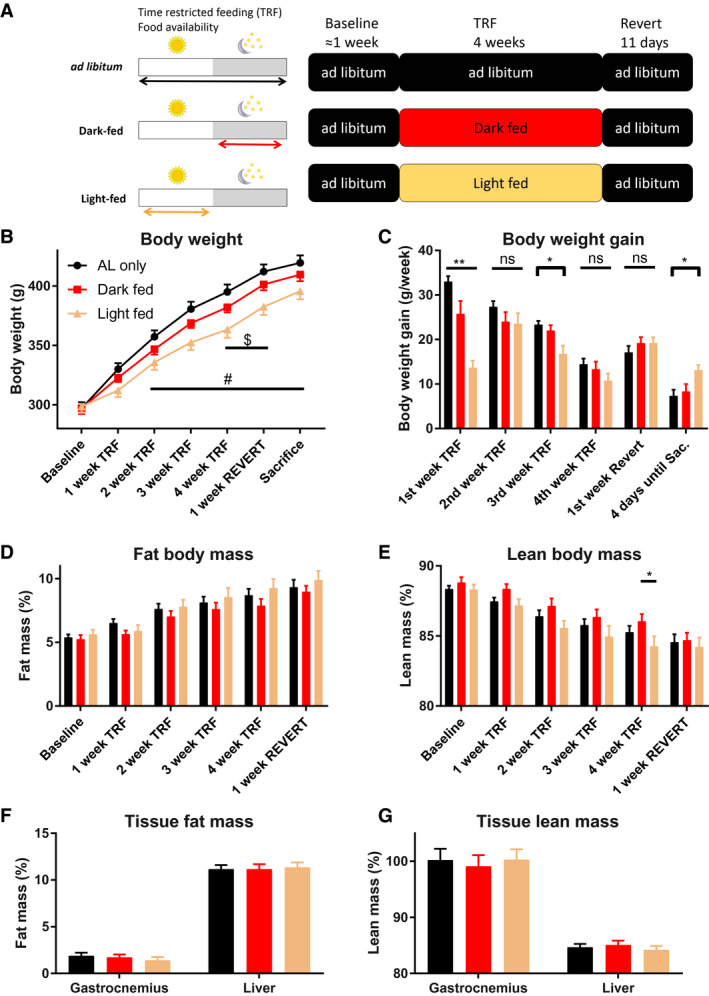
Experimental design and basic physiological measures of the rats. (A) Experimental design. During the experiment, there were three different phases: (1) the baseline phase, during which all animals had ad libitum (AL) access to food; (2) the time‐restricted feeding (TRF) phase, during which the dark‐ and light‐fed groups had limited access to food; and (3) the Revert phase, during which all animals had AL access to food again and thus could recover from the TRF phase. During the TRF phase, the light‐ and dark‐fed animals had access to chow pellets for 10 hours during either the light period (Zeitgeber Time [ZT]1‐ZT11) or the dark period (ZT13‐ZT23), respectively. (B) Body weight during the experiment. Animals were weighed weekly, as well as on the day of sacrifice. (C) Body weight gain during the experiment. (D,E) Body composition of the animals during the experiments. Relative (D) fat and (E) lean body mass steadily increased and decreased, respectively, during the course of the experiments. Relative (F) fat and (G) lean tissue mass of the liver and gastrocnemius muscle of the rats after sacrifice. $Significant post hoc difference between dark‐fed and light‐fed animals. #Significant post hoc difference between AL‐fed and light‐fed animals. *P value < 0.05 after Tukey post hoc test. **P value < 0.01 after Tukey post hoc test. (B‐E) n = 9 per experimental group. (F,G) For the liver composition, n = 19, n = 15, and n = 15 for the AL, dark‐fed, and light‐fed animals, respectively. For the gastrocnemius muscle measures, n = 15, n = 11, and n = 11 for the AL, dark‐fed, and light‐fed animals, respectively. ns, nonsignificant.
Body temperature
A subset of 20 animals underwent surgery, during which a temperature logger (DST nano‐T; STAR:ODDI, Gardabaer, Iceland) was implanted to sample body temperature every 15 minutes for the entire duration of the experiments. Thus, temperature data were collected during all three experimental conditions.
Metabolic cages
Another subset of 16 animals was placed in metabolic cages (TSE Systems, Inc., Midland, Michigan) for the entire duration of the experiments to measure physiological parameters, including locomotor activity, RER, and food consumption. Food and water were replaced twice a week for these animals.
EchoMRI analysis
Twenty‐seven animals were weighed weekly and had their relative fat and lean mass percentages measured in the EchoMRI (EchoMRI LLC, Houston, Texas), a body composition–analyzing device suitable for tissue measurements, throughout the experiment. At the end of the experiment, the remaining left lateral liver lobe and the entire gastrocnemius muscle of the left leg were taken out entirely in a subset of animals, weighed, and scanned in the EchoMRI.
Blood metabolites
Blood glucose was measured in plasma using blood‐glucose test strips (FreeStyle‐lite; Abbott Diabetes Care, Witney, United Kingdom). Plasma insulin and corticosterone were measured using a radioimmunoassay (Millipore, Burlington, Massachusetts). Plasma nonesterified fatty acid (NEFA) levels were measured as described previously (21).
RNA isolation, complementary DNA synthesis, and quantitative polymerase chain reaction
Methods to perform RNA isolation, complementary DNA synthesis, and quantitative polymerase chain reaction were as described previously (18, 22). Primer sequences used are listed in Supporting Information Table S3.
Statistics
Rhythmicity of the physiological parameters RER and body temperature was determined using SigmaPlot version 14.0 (Systat Software, San Jose, California), as this software package can account for within‐individual (repeated) measurements (RMs). Data were fitted to the following cosine regression: y = a + b × cos(2π(x − c) / 24), in which a is the mean level, b is the amplitude, and c is the acrophase of the rhythm.
Rhythmicity of gene expression and plasma metabolites was determined using the JTK_Cycle version 3.0 script (Hughes Lab, St. Louis, Missouri) for nonparametric cosine regression, as this software package is best suited for independent sampling. Graphs were plotted using GraphPad Prism version 7 (GraphPad Software, San Diego, California). All other statistics (i.e., the ANOVAs, including their respective post hoc tests) were executed by using GraphPad Prism version 7.
Results
Body weight
Body weight continuously increased for all three experimental groups throughout the experiment (Figure 1B; P < 0.0001, P < 0.0001, and P = 0.0112 for the main effects of Time, Group, and the interaction Time × Group, respectively; two‐way ANOVA). However, the animals grew at different rates, especially in the first week of TRF, during which the AL group grew the most, and the light‐fed group grew the least (Figure 1C; P < 0.0001, P = 0.0088, and P < 0.0001 for the main effects of Time, Group, and the interaction Time × Group, respectively; two‐way ANOVA).
Body composition
Throughout the experiment, the relative fat mass continuously increased, whereas relative lean mass decreased (Figures 1D‐1E; main effect of Time [P < 0.0001] for both lean and fat mass). No significant effects of group or Time × Group were found for either whole‐body lean or whole‐body fat mass measures.
Tissue composition
After sacrifice, a liver lobe and the entire gastrocnemius muscle were taken out, weighed, and measured to determine relative fat and lean mass (Figures 1F‐G). Muscle lean and fat mass did not differ among the three experimental groups, nor did liver fat or lean mass (P > 0.5; one‐way ANOVA).
Food intake
In the baseline (i.e., AL) experimental phase, the three experimental groups displayed a highly similar pattern of daily feeding behavior, with most food intake during the dark period and two distinct peaks: one at the beginning and one at the end of the dark period (Figure 2A).
Figure 2.

Metabolic parameters (food intake, activity, respiratory exchange ratio [RER], and body temperature) during the three different experimental phases. Analysis of the (A‐C) daily food intake, (D‐F) locomotor activity, (G‐I) RER, and (J‐L) subcutaneous body temperature of the animals during each of the three experimental phases. Graphs at the left, middle, and right are the 24‐hour traces of the baseline, time‐restricted feeding (TRF), and Revert phases, respectively, averaged per experimental group. (M‐P) Average 24‐hour values of the metabolic parameters for the three experimental groups. (Q‐T) Difference within the metabolic parameters between light and dark period (L/D) for the ad libitum (AL), dark‐fed, and light‐fed animals in the Revert phase. All food intake, activity, and RER values are averages of the last 2 days of each experimental phase; body temperature values are averages of the last 3 days of each experimental phase. Locomotor activity is presented as arbitrary units (AU). ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05 after Tukey post hoc test. Shaded areas represent the dark period. (A‐I,M‐O,Q‐S) For the metabolic cage measures, n = 4, n = 6, and n = 6 for the AL, dark‐fed, and light‐fed animals, respectively. (J‐L,P,T) For the body temperature measures, n = 6, n = 7, and n = 7 for the AL, dark‐fed, and light‐fed animals, respectively. L/D, light/dark; ns, nonsignificant; ZT, Zeitgeber Time.
During the TRF phase, this bimodal eating behavior persisted for the light‐ and dark‐fed groups, with the obvious difference that light‐ and dark‐fed animals could only eat during the light and dark periods, respectively (Figure 2B).
During the Revert phase, the three experimental groups still displayed different feeding patterns, with the dark‐fed animals having the largest L/D difference and the light‐fed animals having the smallest L/D difference (Figures 2C and 2Q).
Locomotor activity
During the baseline phase, all animals were more active during the dark period as compared with the light period (Figure 2D; P < 0.0001 for all three groups; RM two‐way ANOVA with Sidak post hoc tests). During the TRF phase, this L/D difference in activity was lost for the light‐fed animals (Figure 2E and Supporting Information Figure S1; P = 0.5790). During the Revert phase, all animals were again most active during the dark period as compared with the light period (Figure 2F; P ≤ 0.0006 for all three groups), but dark‐fed animals had the most distinct L/D difference in activity, and the light‐fed animals had the smallest L/D difference in activity (Figure 2R).
RER findings
During the baseline phase, the experimental groups showed a highly similar daily rhythm in RER, with the peak in RER during the dark period at around ZT19 (Figure 2G; P < 0.005 for all groups; cosine regression and RM two‐way ANOVA with Sidak post hoc tests).
In the TRF phase, both the dark‐fed and light‐fed group showed an increased amplitude in RER as compared with the AL group, together with shifted acrophases to ZT21 and ZT9, respectively (Figure 2H). Moreover, 24‐hour mean RER was lower in the dark‐fed and light‐fed groups (Figure 2O).
During the Revert phase, the acrophases of the dark‐ and light‐fed animals quickly returned to values similar to that of the AL‐fed animals (Figure 2I and Supporting Information Figure S2C). However, the light‐fed animals persisted in having the peak in RER significantly earlier during the dark period as compared with the AL‐fed and dark‐fed groups (P = 0.0228 and P = 0.0275, respectively; Figure 2I and Supporting Information Figure S2C). Additionally, a significant main effect of Group and a significant ZT × Group interaction was found for the Revert phase (P = 0.0491 and P = 0.0015, respectively; Figure 2I), with dark‐fed animals having an overall lower RER as compared with AL‐fed animals. When comparing the RER in the light period among the three different groups, dark‐fed animals showed significantly lower RER compared with AL‐fed animals and showed a trend toward lowered RER as compared with light‐fed animals (P = 0.0139 and P = 0.0664, respectively; Figure 2S). Although the 24‐hour RER also showed a significant difference among the groups (P = 0.046) during the Revert phase, post hoc tests only showed a trend for lowered RER in the dark‐fed group as compared with both AL‐fed and light‐fed animals (P = 0.071 and P = 0.087 respectively; Figure 2O).
Body temperature
During the baseline phase, all animals had higher subcutaneous body temperature during the dark period as compared with the light period (Figure 2J; P < 0.0001 for all three groups; RM two‐way ANOVA with Sidak post hoc tests).
During the TRF phase, this L/D difference in body temperature was lost for the light‐fed animals (Figure 2K and Supporting Information Figure S3; P = 0.3285). However, cosinor analysis indicated that during the TRF phase, the body temperature was still significantly rhythmic for light‐fed animals, but the amplitude of the rhythm was drastically reduced, and the acrophase was phase‐advanced by ~6 hours to ZT12 (Supporting Information Figures S2E‐S2F).
During the Revert phase, light‐fed animals persisted in having higher body temperature during the light period when compared with AL‐fed animals (Figure 2T; P = 0.0247; two‐way ANOVA using Tukey post hoc tests). In line with this finding, cosinor analysis indicated that 11 days after the reversion to AL feeding, the peak in body temperature was still phase‐advanced for the light‐fed group, compared with both AL‐fed and dark‐fed animals (Supporting Information Figure S2F).
Supporting Information Figures [Link], [Link] depict the same metabolic measures (food intake, locomotor activity, RER, and body temperature) as Figures 2A‐2T but are organized in a within‐group manner instead of in a between‐group manner (used in Figures 2A‐2T).
Plasma metabolites
Although two‐way ANOVA showed that plasma levels of glucose, insulin, NEFAs, and corticosterone all showed an effect of ZT, significant rhythms were only found for glucose (AL‐fed), NEFAs (dark‐fed and light‐fed), and corticosterone (dark‐fed) (Figure 3, Table 1), and trends for rhythmicity for the AL‐fed group were observed for NEFAs and corticosterone.
Figure 3.
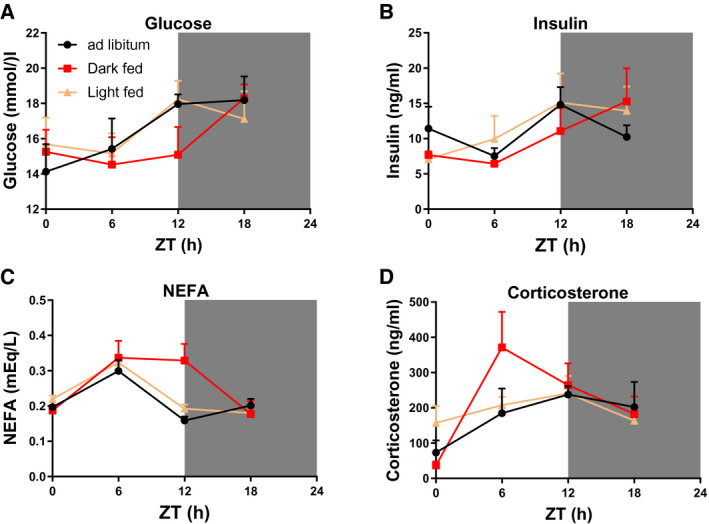
After‐effects of 4 weeks of time‐restricted feeding (TRF) on daily profiles of plasma metabolite levels. Plasma levels of (A) glucose, (B) insulin, (C) nonesterified fatty acids (NEFA), and (D) corticosterone in the rats 11 days after the reversion to AL feeding conditions. Shaded areas represent the dark period. N = 4 × 6 per time point and experimental group. ZT, Zeitgeber Time.
TABLE 1.
After‐effects of 4 weeks of TRF on plasma metabolites 11 days after return to AL feeding
| Plasma metabolite | Two‐way ANOVA (P value) | JTK_Cycle (acrophase in ZT) | JTK_Cycle (P value) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ZT | TRF | ZT × TRF | AL‐fed | Dark‐fed | Light‐fed | AL‐fed | Dark‐fed | Light‐fed | |
| Glucose | 0.022 | 0.697 | 0.673 | 15 | NR | NR | 0.036 | 0.321 | 0.769 |
| Insulin | 0.026 | 0.774 | 0.580 | NR | NR | NR | 0.379 | 0.176 | 0.423 |
| NEFA | 0.000 | 0.0826 | 0.010 | NR | 9 | 6 | 0.083 | 0.002 | 0.008 |
| Corticosterone | 0.001 | 0.576 | 0.196 | NR | 12 | NR | 0.056 | 0.007 | 0.718 |
Rhythmic analysis of the data presented in Figure 3 was performed by JTK_Cycle. The acrophases (in ZT) are only given for metabolites that are significantly rhythmic (P < 0.05). 0.000 = P < 0.0001. Significant P values (P < 0.05) are formatted in bold.
AL, ad libitum; NEFA, nonesterified fatty acid; NR, not significantly rhythmic; TRF, time‐restricted feeding; ZT, Zeitgeber Time.
Gene expression profiles
Liver
Several clock and metabolic genes in the liver of light‐fed animals had not yet fully recovered their normal daily rhythm, (i.e., they differed significantly from the AL control group, after 11 days of AL feeding in the Revert phase) (Figures 4, 5, Supporting Information Figure S6, Table 2, and Supporting Information Table S1). Compared with the AL feeding condition, light‐fed animals showed a 3‐hour shift in the acrophase for Cry2, Per1, Per2, and albumin D‐site‐binding protein (Dbp). Gene expression of the dark‐fed group showed a gain of rhythm for circadian locomotor output cycles kaput (Clock) and a 3‐hour shift in the Per2 acrophase as compared with the AL‐fed control group. Compared with AL feeding, fatty acid synthase, lipin, and pyruvate dehydrogenase kinase 4 (Pdk4) showed a loss of rhythm, whereas peroxisome proliferator–activated receptor γ coactivator 1 α (Pgc1a) showed a gain of rhythm in the light‐fed group. Nocturnin (Noct), glucose transporter 2, and carnitine palmitoyltransferase 1a (Cpt1a) showed 3‐hour shifts for the light‐fed group, whereas for the dark‐fed groups, 3‐hour shifts were observed for fatty acid synthase, glucokinase, phosphoglycerate kinase, and Cpt1a.
Figure 4.
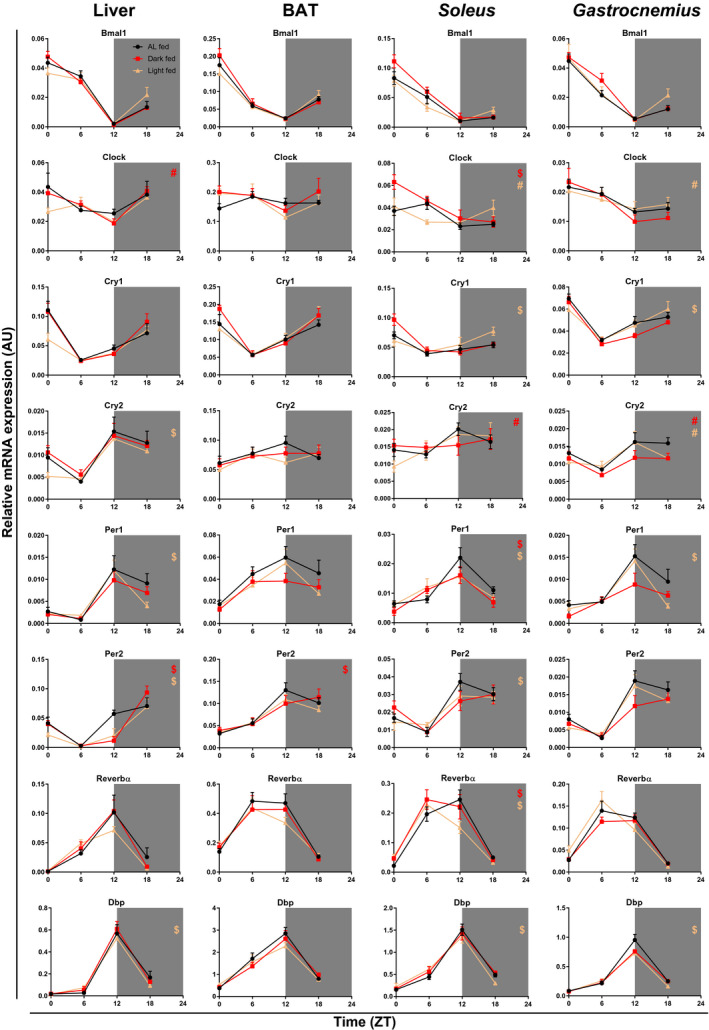
After‐effects of 4 weeks of time‐restricted feeding (TRF) on expression profiles of clock genes and the clock‐controlled gene albumin D‐site‐binding protein (Dbp) in liver, brown adipose tissue (BAT), and soleus and gastrocnemius tissues in rats 11 days after the reversion to ad libitum (AL) feeding conditions. Shaded areas represent the dark period. N = 4 × 6 per time point and experimental group. #Difference in rhythmicity compared with AL feeding conditions (gain/loss of rhythm). $Difference in the acrophase compared with AL feeding conditions. Bmal1, brain and muscle aryl hydrocarbon receptor nuclear translocator–like factor 1; Clock, circadian locomotor output cycles kaput; Cry, cryptochrome; Dbp, albumin D‐site‐binding protein; Reverbα, nuclear receptor subfamily 1, group D α; ZT, Zeitgeber Time.
Figure 5.
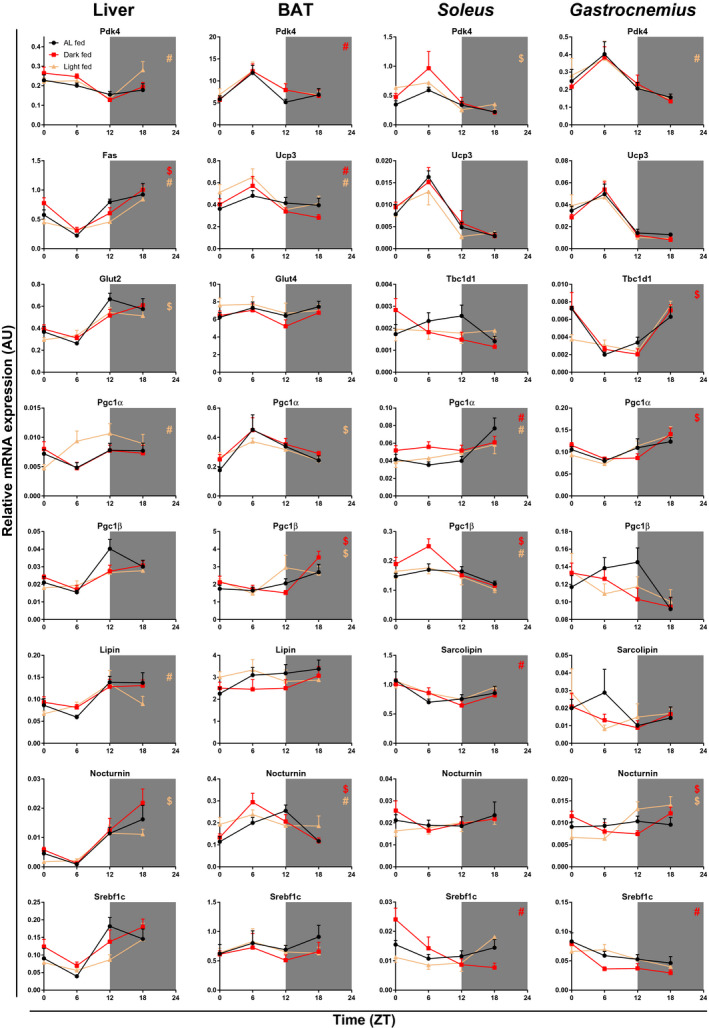
After‐effects of 4 weeks of time‐restricted feeding (TRF) on expression profiles of metabolic genes in liver, brown adipose tissue (BAT), and soleus and gastrocnemius tissues in rats 11 days after the reversion to ad libitum (AL) feeding conditions. Shaded areas represent the dark period. N = 4 × 6 per time point and experimental group. #Difference in rhythmicity compared with AL feeding conditions (gain/loss of rhythm). $Difference in the acrophase compared with AL feeding conditions. Fas, fatty acid synthase; Glut, glucose transporter; Pgc, peroxisome proliferator–activated receptor γ coactivator; Pdk, pyruvate dehydrogenase kinase; Srebf, sterol regulatory element–binding protein; Tbc1d1, trehalase 2/Bub2/cell division cycle 16 domain family member 1; Ucp, uncoupling protein; ZT, Zeitgeber Time.
TABLE 2.
After‐effects of 4 weeks of TRF on daily rhythms in molecular clock and metabolic gene expression in the liver, BAT, and soleus and gastrocnemius muscle 11 days after return to AL feeding
| Liver | BAT | Soleus | Gastrocnemius | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AL‐fed | Dark‐fed | Light‐fed | AL‐fed | Dark‐fed | Light‐fed | AL‐fed | Dark‐fed | Light‐fed | AL‐fed | Dark‐fed | Light‐fed | |
| Clock genes | ||||||||||||
| Bmal1 | 3 | 3 | 3 | 0 | 0 | 0 | 3 | 3 | 3 | 3 | 3 | 3 |
| Clock | NR | 0 | NR | NR | NR | NR | 6 | 3 | NR | 3 | 3 | NR |
| Cry1 | 21 | 21 | 21 | 21 | 21 | 21 | 0 | 0 | 21 | 0 | 0 | 21 |
| Cry2 | 18 | 18 | 15 | NR | NR | NR | 15 | NR | 15 | 18 | NR | NR |
| Per1 | 18 | 18 | 15 | 12 | 12 | 12 | 15 | 12 | 12 | 15 | 15 | 12 |
| Per2 | 18 | 21 | 21 | 15 | 18 | 15 | 18 | 18 | 15 | 18 | 18 | 18 |
| Reverbα | 12 | 12 | 12 | 9 | 9 | 9 | 12 | 9 | 9 | 9 | 9 | 9 |
| Dbp | 15 | 15 | 12 | 12 | 12 | 12 | 15 | 15 | 12 | 15 | 15 | 12 |
| Metabolic genes | ||||||||||||
| Pdk4 | 3 | 3 | NR | NR | 9 | NR | 6 | 6 | 3 | 9 | 9 | NR |
| Fas | 18 | 21 | NR | NR | NR | NR | ||||||
| Ucp3 | NR | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | |||
| Glut2 | 18 | 18 | 15 | |||||||||
| Glut4 | NR | NR | NR | |||||||||
| Tbc1d1 | NR | NR | NR | 21 | 0 | 21 | ||||||
| Pgc1a | NR | NR | 12 | 12 | 12 | 9 | 21 | NR | NR | 18 | 21 | 18 |
| Pgc1b | 18 | 18 | 18 | NR | 21 | 18 | 9 | 6 | NR | NR | NR | NR |
| Lipin | 18 | 18 | NR | NR | NR | NR | ||||||
| Sarcolipin | NR | 3 | NR | NR | NR | NR | ||||||
| Noct | 18 | 18 | 15 | 12 | 9 | NR | NR | NR | NR | NR | 21 | 18 |
| Srebf1c | 18 | 18 | 18 | NR | NR | NR | NR | 3 | NR | NR | 3 | NR |
| Glucokinase | 21 | 0 | 21 | |||||||||
| Pgk | 21 | 0 | 21 | |||||||||
| Cpt1a | 15 | 12 | 12 | 12 | NR | 6 | ||||||
| Cpt1b | 15 | NR | NR | NR | NR | NR | ||||||
| Ppara | 12 | NR | NR | |||||||||
| Hexokinase II | NR | 3 | NR | NR | NR | NR | ||||||
| MyoD | 21 | 0 | 21 | |||||||||
Rhythmic analysis of the data presented in Figures 4, 5 was performed by JTK_Cycle. The acrophases (in ZT) are only given for genes that are rhythmically expressed (P < 0.05). P values are given in Supporting Table S1.
AL, ad libitum; BAT, brown adipose tissue; Bmal1, brain and muscle aryl hydrocarbon receptor nuclear translocator–like factor 1; Clock, circadian locomotor output cycles kaput; Cpt1, carnitine palmitoyltransferase 1; Cry, cryptochrome; Dbp, albumin D‐site‐binding protein; Fas, fatty acid synthase; Glut, glucose transporter; MyoD, myogenic differentiation; Noct, nocturnin; NR, not significantly rhythmic; Pdk4, pyruvate dehydrogenase kinase 4; Per, period; Pgc1, peroxisome proliferator–activated receptor γ coactivator 1; Pgk, phosphoglycerate kinase; Ppara, peroxisome proliferator–activated receptor α; Reverbα, nuclear receptor subfamily 1, group D α; Srebf1c, sterol regulatory element–binding transcription factor 1c; Tbc1d1, trehalase 2/Bub2/cell division cycle 16 domain family member 1; TRF, time‐restricted feeding; Ucp3, uncoupling protein 3; ZT, Zeitgeber Time.
BAT findings
Gene expression profiles of the molecular‐clock components in BAT of dark‐fed and light‐fed animals had largely recovered toward the normal rhythm of the AL‐fed animals. Compared with the AL group, dark‐fed animals showed a 3‐hour shift in the acrophase for Per2 only (Figure 4 and Table 2). In the light‐fed group, no differences as compared with the AL group were found in the clock genes. Of the metabolic genes tested, a loss of rhythm was observed for carnitine palmitoyltransferase 1b (Cpt1b) (dark‐fed & light‐fed), Noct (light‐fed), and peroxisome proliferator–activated receptor α (Pgc1a) (dark‐fed & light‐fed), whereas a gain of rhythm was observed for Pdk4 (dark‐fed), peroxisome proliferator–activated receptor γ coactivator 1 β (Pgc1b) and uncoupling protein 3 (Ucp3) (dark‐fed & light‐fed). Compared with AL feeding, 3‐hour shifts in the acrophase were found for Noct (dark‐fed) and Pgc1a (light‐fed).
Soleus
Daily clock‐gene expression rhythms in the soleus muscle were not fully recovered yet in either TRF group. In the light‐fed group, only Bmal1 and Cry2 had an expression pattern similar to AL conditions. Expression of Clock was not rhythmic anymore and 3‐hour shifts in the acrophase were found for Cry1, Per1, Per2, Reverbα, and Dbp (Figure 4 and Table 2). In the dark‐fed group, Cry2 showed a loss of its rhythmic expression, and 3‐hour shifts were observed in the acrophase for Clock, Per2, and Reverbα. Additionally, in the dark‐fed animals, the overall daily mean expression level of Clock was higher as compared with the AL group, and the overall daily mean expression level of Bmal1 was higher as compared with the light‐fed animals (two‐way ANOVA with Tukey post hoc tests; Supporting Information Table S2). Of the metabolic genes tested, a loss of rhythm was observed for Cpt1a (dark‐fed), Pgc1a (dark‐fed & light‐fed), and Pgc1b (light‐fed), whereas a gain of rhythm was observed for Hexokinase II, Sarcolipin, and sterol regulatory element–binding transcription factor 1c (Srebf1c) (dark‐fed). Compared with AL feeding, 3‐hour shifts were found for Pdk4 (light‐fed) and Pgc1b (dark‐fed), and a 6‐hour shift was found for Cpt1a (light‐fed).
Gastrocnemius
In addition, in the gastrocnemius muscle, several of the clock genes in the TRF groups had not yet completely recovered their daily rhythm to that of AL feeding (Figure 4 and Table 2). In the dark‐fed group, Cry2 had lost its rhythmic expression. In the light‐fed group both Clock and Cry2 had lost their rhythmic expression. Additionally, compared with the AL feeding conditions, the light‐fed group displayed 3‐hour shifts in the acrophase for Cry1, Per1, and Dbp. Of the metabolic genes tested, a gain of rhythm was observed for Noct (light‐fed & dark‐fed) and Srebf1c (dark‐fed), whereas rhythmic expression of Pdk4 was lost in the light‐fed group. Compared with AL feeding, 3‐hour shifts were observed for trehalase 2/Bub2/cell division cycle 16 domain family member 1 (Tbc1d1), Pgc1a, and the myogenic differentiation gene (MyoD) in the dark‐fed group.
Discussion
We characterized in rats the after‐effects of TRF on a molecular as well as systemic level in four different metabolically important peripheral tissues (liver, BAT, and soleus and gastrocnemius muscle). Eleven days after returning to AL feeding, TRF animals still differed from AL animals in several metabolic parameters. RER levels were lower during a part of the light period in the dark‐fed group as compared with both the light‐ and AL‐fed animals. Furthermore, in both the dark‐ and light‐fed groups, the daily rhythm in plasma glucose levels was abolished, whereas plasma levels of NEFAs became rhythmic. In addition, at the molecular level, the effects of TRF persisted until 11 days after returning to the AL feeding condition. Importantly, these after‐effects of TRF on the molecular clock and metabolic gene expression were both tissue and gene dependent.
Physiological measures and their relation and meaning
At the end of the Revert phase, feeding behavior, activity, body temperature, and RER had not yet fully recovered to AL feeding conditions (Figures 2C, 2F, 2I, and 2L), indicating persisting changes in the daily rhythms of these measures. RER, for example, was lower in dark‐fed animals during the light period (Figures 2O and 2S), whereas in the light‐fed animals, a phase advance in body temperature and RER persisted (Supporting Information Figures S2C and S2F). Most of these persisting effects were found in the dark‐fed group and confirmed an increased day/night difference in these measures. Although body composition was mostly not significantly different among the three groups, dark‐fed animals displayed the highest fat mass and lowest lean body mass throughout the TRF phase (Figures 1D‐1E). It was only during the fourth week of TRF that lean body mass significantly differed between the dark‐fed and light‐fed animals, but this effect was abolished after 1 week of the Revert phase. Tissue fat/lean composition of the liver and gastrocnemius did not differ after 11 days of Revert feeding (Figures 1F‐1G). However, because we only performed a single measurement on these measures at the very end of the experiment, we were unable to determine if the changes in whole‐body composition observed during the TRF phase were caused by the liver and gastrocnemius muscle or by changes in other tissues, such as the adipose depots.
Taken together, our results imply that the after‐effects of the TRF regimen on these metabolic measures were most pronounced in the dark‐fed animals and can be thought of as beneficial for health, as decreased RER (indicative of increased lipid substrate usage), increased lean body mass, and strengthened biological rhythms are associated with health benefits (20, 23). Although mostly not significant, in the light‐fed animals, the day/night amplitude during the Revert phase was reduced for food intake, activity, and body temperature (Figures 2Q‐2T). As reduced biological rhythms are associated with disease, the after‐effects of light‐period feeding could indicate metabolic health complications or at least increased vulnerability (13, 24).
Furthermore, our continuous metabolic cage and body temperature measurements showed that the changes in RER amplitude were established more quickly than the amplitude changes in body temperature in the light‐fed animals (Supporting Information Figures S1B and S1E). Additionally, the change in the RER acrophase in the light‐fed animals was established more quickly after the transition from AL feeding to TRF compared with the transition from TRF back to AL feeding. In fact, 11 days after returning to AL feeding, this acrophase still had not fully recovered (Supporting Information Figure S2C). Taken together, our results indicate that alignment of the TRF protocol with the natural sleep/wake cycle has long‐lasting effects on health that can be considered as beneficial, whereas misalignment of the TRF protocol with the natural sleep/wake cycle has long‐lasting effects that eventually could lead to metabolic health complications. Moreover, the alterations from AL feeding to the TRF phase were established more quickly than those back from the TRF phase to AL feeding in the Revert phase, which could mean that constantly changing work shifts, as frequently occurs in, for example, nurses, does not allow for a full recovery of physiological rhythms during free days.
Last, it should be noted that locomotor activity, RER, and body temperature all strongly depend on food intake; therefore, the persisting effects on these measures of whole‐body metabolism could be the result of the incomplete recovery of the feeding behavior.
Clock‐gene expression
Only Bmal1 expression had fully recovered in all four tissues tested, whereas the rhythmic expression of Clock, Cry2, Per1, Per2 and Dbp had still not fully recovered in 3 out of the 4 tissues investigated. The tissue‐dependent effects are clearly demonstrated by the finding that in soleus muscle, 10 out of the 16 expression rhythms investigated were still changed, whereas in liver, gastrocnemius muscle, and BAT respectively, six, five, and one expression pattern(s) had not yet recovered completely. Thus, of the four peripheral tissues tested here, BAT appears to have recovered most quickly from the TRF phase, as only Per2 still showed a 3‐hour shift in the dark‐fed groups. Previously, we showed that after 5 weeks of light‐period TRF, clock‐gene expression rhythms in BAT had shifted by several hours (6‐9 hours) for most of the clock genes tested, whereas the liver showed completely antiphasic rhythms. In the soleus and gastrocnemius muscles, the daily rhythms in clock‐gene expression were completely abolished, with the exception of Reverbα in gastrocnemius muscle (25, 26, 27, 28). Therefore, a possible explanation for the quick recovery of BAT rhythms is that during TRF conditions, the BAT clock is least affected, and recovery of the BAT clock thus takes less time. Another explanation could be that the different tissues respond differently to the TRF‐induced changes of feeding behavior and locomotor activity. It has been suggested that feeding affects mostly the liver clock and locomotor activity, especially the muscle clock (22, 29, 30, 31, 32, 33, 34). Therefore, experiments that can specifically manipulate activity behavior, without affecting feeding rhythms, should be performed to better understand the separate influences of feeding and activity on the peripheral clocks. Examples of such experiments could be time‐restricted access to a running wheel, with or without manipulating feeding behavior at the same time.
Metabolic gene expression
An important output of peripheral clocks is the expressional (mRNA/protein/posttranslational) regulation of (glucose/lipid) metabolic processes to anticipate the time‐of‐day–dependent alterations in substrate preferences. Our finding of after‐effects in metabolic gene expression in the four tested tissues are thus in line with the after‐effects both in the peripheral clocks and in the physiological measures. Similar to the clock genes, the metabolic genes are affected in both a tissue‐ and gene‐dependent manner, likely depending on both the alterations of the peripheral clock and the specific needs regarding substrate use within the tissues. For example, Pdk4, which acts like a switch between glucose and lipid metabolism, showed after‐effects in all four tissues, whereas transcription factor Srebf1c, important for both glucose and lipid metabolism, was only altered in the two skeletal muscles. In addition, differences were found between the two muscle types, as, for example, metabolic regulator Noct became rhythmic only in the gastrocnemius muscle, whereas Tbc1d1 (important for glucose transporter 4 translocation) was shifted by 3 hours for dark‐fed animals in gastrocnemius muscle, but was not significantly rhythmic in any group in soleus muscle. A consequence of the differential effects on metabolic gene expression among these four tissues is a disturbed balance between substrate demand (mainly muscles, but also BAT for thermogenesis) and substrate supply (liver, white adipose tissue). Our results on liver and gastrocnemius fat and lean composition at the end of the experiment do not reflect such changes of metabolic substrates (Figures 1F‐1G), but the results on whole‐body fat and lean mass composition do indicate such changes. As lipid and glucose substrate demand is highly dependent on the time of day, long‐lasting alterations in both the molecular clock (which is an important regulator of these time‐dependent changes in metabolism) and metabolic genes themselves could lead to inefficient metabolism and ultimately metabolic diseases.
Other studies on the long‐term effects of shift work
Although many groups have studied the effects of shift work on metabolism in both humans and rodents, the after‐effects of shift work have been studied only rarely. We could find only one study on the molecular clock, in which after several weeks of shift work, animals were allowed to recover from the shift‐work protocol. In this study, rats were subjected to forced activity in the sleep period for 5 days a week, instead of food restriction (i.e., food was available AL) (35). After 4 weeks, the shift‐work paradigm was ended, and animals could recover for 3, 5, or 7 days, after which they were sacrificed at four different time points along the L/D cycle to measure liver gene expression. The authors reported that several clock and metabolic genes did not fully recover to control conditions within 7 days of the recovery period. Similar to our results, they found that hepatic expression of Bmal1 was restored at the end of the 7‐day recovery period, whereas Per2 was not. Unfortunately, this study did not provide data on other tissues or on metabolic phenotypes other than food intake and activity (which quickly and fully returned to control conditions). In another study, using a slightly different experimental setup, mice could eat a diet rich in fat and were placed on a 5‐day dark‐period TRF regimen followed by 2 days of AL feeding (i.e., similar to some human intermittent fasting protocols), which protected them from the weight gain seen in animals that could eat AL (36). Although this study did not characterize possible alterations in the molecular clock, the results are in line with the current ones, as they imply that it is not strictly necessary to adhere to TRF on a daily basis to reap its beneficial effects.
Clinical relevance
Epidemiological studies have found that rotating night‐shift work increased the risk of T2DM (7, 9). Moreover, experimental studies have found that simulation of shift work as well as circadian misalignment by itself can impair glucose metabolism and insulin sensitivity (37, 38, 39, 40, 41). It is therefore essential to characterize whether and (if yes) how quickly disturbed rhythms that result from shift work can recover. Our results indicate that (partial) recovery from long‐lasting TRF conditions is possible at the whole‐body, systemic, and molecular‐clock levels. Although not all measures reported here had fully recovered after 11 days, most of them recovered within a few days. It should be stressed that these persisting deviations from AL conditions are not unhealthy per se. TRF during the active period is often associated with metabolic health improvements, whereas TRF during the inactive phase is associated with negative effects on health outcomes (13, 14). It is therefore expected that the after‐effects in the dark‐fed group are beneficial, whereas the after‐effects in the light‐fed group are mainly harmful for metabolic health. Although studies in humans have indicated that shift work alters rhythms in both core body temperature and gene expression in blood cells, no studies have been performed yet on the recovery of such alterations (42, 43). Therefore, studies in humans are necessary to determine how quickly disturbed rhythms in physiology and clock‐gene expression recover after shift work.
Conclusion
Our data show that following 4 weeks of simulated shift work by daytime TRF, persisting metabolic changes (food intake, activity, RER, plasma glucose, and NEFAs) can still be found, even 11 days after return to AL feeding. Additionally, the molecular clocks in the liver, BAT, and soleus and gastrocnemius muscles had not yet completely recovered from the TRF conditions. Importantly, these persisting effects of the TRF protocol were both tissue and clock gene dependent, with BAT displaying the most and soleus muscle displaying the least recovery. As the specific outputs of the molecular clock govern a wide variety of metabolic processes, including many lipid and glucose metabolic pathways, these differences in recovery could have implications for the synchrony between the metabolic supply and demands of the different tissues. This persisting desynchrony between different metabolic tissues is probably an important risk factor for the development of metabolic disorders, including (pre)diabetes.
Therefore, optimized protocols for shift workers should be designed in which the amount of shift days and nonworking/resting days are balanced in such a way that the detrimental effects of shift work on metabolic health are minimized.
Contrasting, persisting effects in the dark‐fed TRF group (food intake, activity, RER, and both clock‐gene and metabolic‐gene expression in the four different tissues) likely reflect positive effects on health, as they often included strengthened rhythms or increased lipid oxidation. The persistence of these positive effects could mean that when partaking in TRF as a lifestyle program to improve health, TRF should be timed in the active period, but it is not strictly necessary to adhere to the TRF regimen every single day, which probably would make it easier for participants to successfully comply with such a lifestyle program.
Funding agencies
PdG was funded by a ZonMW TOP grant (91214047).
Disclosure
The authors declared no conflict of interest.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Table S1
Table S2
Table S3
References
- 1. Gekakis N, Staknis D, Nguyen HB, et al. Role of the CLOCK Protein in the Mammalian Circadian Mechanism. Science 1998;280:1564‐1569. [DOI] [PubMed] [Google Scholar]
- 2. Hogenesch JB, Gu Y‐Z, Jain S, Bradfield CA. The basic‐helix–loop–helix‐PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A 1998;95:5474‐5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature 2002;418:935‐941. [DOI] [PubMed] [Google Scholar]
- 4. Ohno T, Onishi Y, Ishida N. A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res 2006;35:648‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoo S‐H, Ko CH, Lowrey PL, et al. A noncanonical E‐box enhancer drives mouse Period2 circadian oscillations in vivo. Proc Natl Acad Sci U S A 2005;102:2608‐2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moynihan Ramsey K, Marcheva B, Kohsaka A, Bass J. The clockwork of metabolism. Annu Rev Nutr 2007;27:219‐240. [DOI] [PubMed] [Google Scholar]
- 7. Pan A, Schernhammer ES, Sun Q, Hu FB. Rotating night shift work and risk of type 2 diabetes: two prospective cohort studies in women. PLoS Med 2011;8:e1001141. doi: 10.1371/journal.pmed.1001141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vetter C, Dashti HS, Lane JM, et al. Night shift work, genetic risk, and type 2 diabetes in the UK biobank. Diabetes Care 2018;41:762‐769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shan Z, Li Y, Zong G, et al. Rotating night shift work and adherence to unhealthy lifestyle in predicting risk of type 2 diabetes: results from two large US cohorts of female nurses. BMJ 2018;363:k464. doi: 10.1136/bmj.k4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hansen AB, Stayner L, Hansen J, Andersen ZJ. Night shift work and incidence of diabetes in the Danish Nurse Cohort. Occup Environ Med 2016;73:262‐268. [DOI] [PubMed] [Google Scholar]
- 11. Bonham MP, Leung GKW, Davis R, et al. Does modifying the timing of meal intake improve cardiovascular risk factors? Protocol of an Australian pilot intervention in night shift workers with abdominal obesity. BMJ Open 2018;8:e020396. doi: 10.1136/bmjopen-2017-020396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Opperhuizen A‐L, van Kerkhof LW, Proper KI, Rodenburg W, Kalsbeek A. Rodent models to study the metabolic effects of shiftwork in humans. Front Pharmacol 2015;6:50. doi: 10.3389/fphar.2015.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Manoogian ENC, Panda S. Circadian rhythms, time‐restricted feeding, and healthy aging. Ageing Res Rev 2017;39:59‐67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Longo VD, Panda S. Fasting, circadian rhythms, and time‐restricted feeding in healthy lifespan. Cell Metab 2016;23:1048‐1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bray MS, Ratcliffe WF, Grenett MH, Brewer RA, Gamble KL, Young ME. Quantitative analysis of light‐phase restricted feeding reveals metabolic dyssynchrony in mice. Int J Obes (Lond) 2013;37:843‐852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dyar KA, Ciciliot S, Tagliazucchi GM, et al. The calcineurin‐NFAT pathway controls activity‐dependent circadian gene expression in slow skeletal muscle. Mol Metab 2015;4:823‐833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oosterman JE, Foppen E, van der Spek R, Fliers E, Kalsbeek A, la Fleur SE. Timing of fat and liquid sugar intake alters substrate oxidation and food efficiency in male Wistar rats. Chronobiol Int 2015;32:289‐298. [DOI] [PubMed] [Google Scholar]
- 18. de Goede P, Sen S, Oosterman JE, et al. Differential effects of diet composition and timing of feeding behavior on rat brown adipose tissue and skeletal muscle peripheral clocks. Neurobiol Sleep Circadian Rhythms 2017;4:24‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A 2009;106:21453‐21458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reznick J, Preston E, Wilks DL, Beale SM, Turner N, Cooney GJ. Altered feeding differentially regulates circadian rhythms and energy metabolism in liver and muscle of rats. Biochim Biophys Acta 2013;1832:228‐238. [DOI] [PubMed] [Google Scholar]
- 21. Milanova IV, Kalsbeek MJT, Wang X‐L, et al. Diet‐induced obesity disturbs microglial immunometabolism in a time‐of‐day manner. Front Endocrinol (Lausanne) 2019;10:424. doi: 10.3389/fendo.2019.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Goede P, Sen S, Su Y, et al. An ultradian feeding schedule in rats affects metabolic gene expression in liver, brown adipose tissue and skeletal muscle with only mild effects on circadian clocks. Int J Mol Sci 2018;19:E3171. doi: 10.3390/ijms19103171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hatori M, Vollmers C, Zarrinpar A, et al. Time‐restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high‐fat diet. Cell Metab 2012;15:848‐860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reutrakul S, Knutson KL. Consequences of circadian disruption on cardiometabolic health. Sleep Med Clin 2015;10:455‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oosterman JE, Koekkoek LL, Foppen, E , et al. Synergistic effect of feeding time and diet on hepatic steatosis and gene expression in male Wistar rats. Obesity 2020. doi: 10.1002/OBYSUP.22832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Goede P. Daily Rhythms in Muscle Mitochondria Effects of Time‐Restricted Feeding and Exercise. Dissertation. University of Amsterdam; 2019. Published October 2019. Accessed April 10, 2019. https://hdl.handle.net/11245.1/eb869988‐9e82‐4ea6‐9246‐734b217951e1 [Google Scholar]
- 27. de Goede P, Sen S, Oosterman JE, et al. Differential effects of diet composition and timing of feeding behavior on rat brown adipose tissue and skeletal muscle peripheral clocks. Neurobiol Sleep Circadian Rhythms 2018;4:24‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eggink HM, Oosterman JE, de Goede P, et al. Complex interaction between circadian rhythm and diet on bile acid homeostasis in male rats. Chronobiol Int 2017;34:1339‐1353. [DOI] [PubMed] [Google Scholar]
- 29. Aoyama S, Shibata S. The role of circadian rhythms in muscular and osseous physiology and their regulation by nutrition and exercise. Front Neurosci 2017;11:63. doi: 10.3389/fnins.2017.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Froy O. Metabolism and circadian rhythms—implications for obesity. Endocr Rev 2010;31:1‐24. [DOI] [PubMed] [Google Scholar]
- 31. Hamaguchi Y, Tahara Y, Hitosugi M, Shibata S. Impairment of circadian rhythms in peripheral clocks by constant light is partially reversed by scheduled feeding or exercise. J Biol Rhythms 2015;30:533‐542. [DOI] [PubMed] [Google Scholar]
- 32. Sasaki H, Hattori Y, Ikeda Y, et al. Forced rather than voluntary exercise entrains peripheral clocks via a corticosterone/noradrenaline increase in PER2:LUC mice. Sci Rep 2016;6:27607. doi: 10.1038/srep27607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schibler U, Ripperger J, Brown SA. Peripheral circadian oscillators in mammals: time and food. J Biol Rhythms 2003;18:250‐260. [DOI] [PubMed] [Google Scholar]
- 34. Stephan FK. The “other” circadian system: food as a Zeitgeber. J Biol Rhythms 2002;17:284‐292. [DOI] [PubMed] [Google Scholar]
- 35. Saderi N, Báez‐Ruiz A, Azuara‐Álvarez LE, Escobar C, Salgado‐Delgado RC. Differential recovery speed of activity and metabolic rhythms in rats after an experimental protocol of shift‐work. J Biol Rhythms 2019;34:154‐166. [DOI] [PubMed] [Google Scholar]
- 36. Chaix A, Zarrinpar A, Miu P, Panda S. Time‐restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab 2014;20:991‐1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharma A, Laurenti MC, Dalla Man C, et al. Glucose metabolism during rotational shift‐work in healthcare workers. Diabetologia 2017;60:1483‐1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scheer FAJL, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A 2009;106:4453‐4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morris CJ, Purvis TE, Mistretta J, Scheer FAJL. Effects of the internal circadian system and circadian misalignment on glucose tolerance in chronic shift workers. J Clin Endocrinol Metab 2016;101:1066‐1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qian J, Dalla Man C, Morris CJ, Cobelli C, Scheer FAJL. Differential effects of the circadian system and circadian misalignment on insulin sensitivity and insulin secretion in humans. Diabetes Obes Metab 2018;20:2481‐2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wefers J, van Moorsel D, Hansen J, et al. Circadian misalignment induces fatty acid metabolism gene profiles and compromises insulin sensitivity in human skeletal muscle. Proc Natl Acad Sci U S A 2018;115:7789‐7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kervezee L, Cuesta M, Cermakian N, Boivin DB. Simulated night shift work induces circadian misalignment of the human peripheral blood mononuclear cell transcriptome. Proc Natl Acad Sci U S A 2018;115:5540‐5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Resuehr D, Wu G, Johnson RL, Young ME, Hogenesch JB, Gamble KL. Shift work disrupts circadian regulation of the transcriptome in hospital nurses. J Biol Rhythms 2019;34:167‐177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Table S1
Table S2
Table S3


