Abstract
Background & Aims
Recently, the variant rs72613567:TA in the 17‐beta‐hydroxysteroid dehydrogenase 13 (HSD17B13) has been associated with reduced levels of ALT and AST and a reduced risk of alcohol‐related liver disease (ALD) in the European population. Therefore, the aim of this study was to investigate the association between the polymorphisms of HSD17B13 and ALD, liver serum markers and patatin‐like phospholipase domain‐containing protein 3 (PNPLA3) p.I148M in the Chinese Han population.
Methods
A case‐control study was performed from five centres and included 769 ALD patients and 767 healthy controls. Two SNPs (rs72613567 and rs6834314) in HSD17B13 were genotyped using the Sequenom MassArray system and allele association analysis was performed using PLINK 1.90 software.
Results
HSD17B13 rs72613567:TA allele was associated with a reduced risk of ALD by 19% (95% confidence interval [CI]: 0.05‐0.31, P = .01), uniformly, the G allele in the rs6834314 reduced the risk of ALD by 19% (95% CI: 0.05‐0.31, P = 8.28 × 10−3). And the genotypes of two SNPs were associated with reducing the risk of ALD in three genetic model analysis. In addition, we found that TA allele was associated with lower levels of serum ALT, AST and GGT (P = .005, .007 and .02, respectively), higher level of serum ALB (P = .02), but not associated with ALP. In this cohort, the interaction between HSD17B13 rs72613567 and the steatogenic allele PNPLA3 rs738409 was not validated.
Conclusion
The present study revealed that HSD17B13 rs72613567 was significantly associated with a reduced risk of ALD in Chinese Han population.
Keywords: alcohol‐related liver disease, HSD17B13, single‐nucleotide polymorphism
Abbreviations
- ALD
alcohol‐related liver disease
- ASH
alcoholic steatohepatitis
- HCC
hepatocellular carcinoma
- DALY
disability‐adjusted life years
- GWAS
genome‐wide association study
- SNP
single nucleotide polymorphism
- GWAS
genome‐wide association study
- PNPLA3
patatin‐like phospholipase domain‐containing 3
- TM6SF2
transmembrane 6 superfamily 2
- MBOAT7
membrane bound O‐acetyltransferase domain containing 7
- NAFLD
nonalcoholic fatty liver disease
- HSD17B13
hydroxysteroid 17‐beta dehydrogenase 13
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- GGT
gamma‐glutamyl transpeptidase
- ALP
alkaline phosphatase
- ALB
albumin
- HWE
Hardy‐Weinberg equilibrium
- RDH
retinol dehydrogenase
Key points.
In this large‐sample study, the association between the polymorphism of the susceptibility gene HSD17B13 and alcohol‐related liver disease was verified in the Chinese Han population.
Two variants of HSD17B13, rs72613567 and rs6834314 (high linkage with each other) were found to reduce the risk of alcohol‐related liver disease.
1. INTRODUCTION
Alcohol‐related liver disease (ALD) is a common liver disease as a consequence of long‐term alcohol consumption, and includes the onset of steatosis up to alcohol‐related steatohepatitis (ASH), alcohol‐related liver fibrosis, alcohol‐related cirrhosis and even hepatocellular carcinoma (HCC). 1 These pathological processes overlap and are not independent of each other in ALD progression. According to the 2018 Global status report on alcohol and health by the World Health Organization (WHO), 2 the harmful use of alcohol caused approximately 3 million deaths worldwide in 2016 (5.3% of all deaths) and 132.6 million disability‐adjusted life years (DALYs), representing the 5.1% of all the DALYs in that year. Among these results, alcohol‐induced cirrhosis caused 607,000 deaths and 22.2 million DALY in the same year. However, not all people who abuse alcohol develop ASH and alcohol‐related cirrhosis. More than 90% of long‐term heavy drinkers may develop fatty liver, and 10%‐35% of these people may further develop inflammation and progressive fibrosis, whereas 10%‐20% may suffer from alcoholic cirrhosis. 1 , 3 With the constant improvement of living standards, ALD is emerging in many Asian countries, such as China and India, while it is decreasing in Western Europe. 4 , 5 The susceptibility of ALD is determined by genetic and environmental factors, such as gender, ethnicity, obesity, hepatitis virus infection, eating habits and metabolic diseases. 6 , 7 , 8
In recent years, a number of single nucleotide polymorphisms (SNPs) associated with ALD have been identified by the genome‐wide association studies (GWAS) 9 , 10 with the intent of exploring the genetic mechanisms of ALD to provide new ideas in preventing ALD. Several SNPs are reported and verified as associated with the development of ALD in different populations, including rs738409 in patatin‐like phospholipase domain‐containing 3 (PNPLA3), 9 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 rs58542926 in transmembrane 6 superfamily 2 (TM6SF2) 9 , 17 and rs671738 in membrane bound O‐acetyltransferase domain containing 7 (MBOAT7). 9 , 19 In addition, these SNPs are associated with the susceptibility of nonalcoholic liver disease (NAFLD). 20 , 21 , 22 , 23 , 24 Recently, Abul‐Husn and colleagues 25 demonstrated that the splice variant rs72613567 in hydroxysteroid 17‐beta dehydrogenase 13 (HSD17B13) is associated with a reduced risk of progression from steatosis to steatohepatitis, and the variant is associated with a decreased level of alanine aminotransferase (ALT) and aspartate aminotransferase (AST). HSD17B13 rs72613567:TA mitigates liver injury associated with a reduced risk of ALD and NAFLD development. 25 A more recent study by Yang et al 26 showed that the loss‐function variant rs72613567 of HSD17B13 has a protective effect on the development of hepatocellular carcinoma in patients with alcoholic liver disease. Ma et al studied another SNP in the downstream of the HSD17B13 gene to identify the associations with the histological features of NAFLD and their results showed that the minor allele of the rs6834314 was significantly associated with cirrhosis in the general population and with increased steatosis but decreased inflammation, ballooning, Mallory‐Denk bodies and liver enzyme levels in NAFLD patients. 27
It is of utmost importance to verify whether these variants exert the observed influential effect on the susceptibility of ALD in other populations. Therefore, the aims of the present study were to determine whether rs72613567 and rs6834134 in HSD17B13 were associated with ALD in the Chinese Han population; to explore the association between the polymorphisms in HSD17B13 and serum biomarkers of liver; and to explore potential interaction between the polymorphisms in HSD17B13 and other steatogenic genes.
2. MATERIALS AND METHODS
2.1. Study population
A case‐control study was designed, which included 769 male ALD patients and 767 male healthy controls from five medical centres. All participants were from the Chinese Han population and unrelated to each other. This study was approved by the Ethics Committee of the first Hospital of Jilin University. The patients of the case group (n = 769 males) were outpatients or inpatients diagnosed with ALD in five centres such as the first Hospital of Jilin University (Changchun, China), Peking Union Medical College Hospital (Beijing, China), the Fifth Medical Center of the General Hospital of the People's Liberation Army (Beijing, China), Shengjing Hospital affiliated with China Medical University (Shenyang, China) and Hepatobiliary Hospital of Jilin (Changchun, China) from August 2016 to November 2018. The 767 male control individuals were selected from those who received a routine physical examination at the Peking Union Medical College Hospital from April 2016 to November 2018. All patients with ALD had a history of alcohol abuse for more than 5 years, which was equivalent to an alcohol consumption of ≥40 g/d, or a large amount of alcohol consumption in the last 2 weeks averaging more than 80 g/d. Imaging examinations, such as abdominal ultrasound, CT or MRI, revealed the presence of fatty liver or cirrhosis. Patients with other liver diseases, such as viral hepatitis, drug‐induced liver disease, autoimmune liver disease and hereditary metabolic liver disease, were excluded from this cohort of ALD patients. In addition, hepatocellular carcinoma was not a parameter considered in this study. The healthy control group matched the ALD group regarding the gender and age, and liver diseases, tumour or other chronic diseases were excluded by ultrasonography and laboratory examination in the physical examination (including blood routine, urine routine, liver function test and renal function test) to ensure the absence of any abnormality. The men of the control group did not have any drinking habit or they had an average alcohol consumption <40 g/d. All basic personal information (gender, age), medical history (hypertension, type 2 diabetes) and clinical laboratory indicators were collected at baseline.
2.2. Sample preparation and genotyping
Samples were collected in EDTA tubes and stored at −80°C immediately after centrifugation until the extracts from all samples could be analysed at the same time point. The genomic DNA of each subject was extracted from 2 mL of peripheral blood sample using the Tiangen DNA extraction kit (Beijing, China), according to the manufacturer's instructions. The quality measurement was performed on all DNA samples.
In this study, two SNPs (rs72613567 and rs6834314 in HSD17B13) whose minor allele frequency was more than 0.05 in the Chinese Han population were selected. The genotyping of the two SNPs of the HSD17B13 was performed by the Sequenom MassArray genotyping system, which was combined with multiplex polymerase chain reaction (PCR), MassArray iPLEX single‐base extension technique and matrix‐assisted laser desorption ionization time‐of‐flight mass spectrometry (MALDI‐TOFMS). In brief, after performing multiplex PCR and locus‐specific single‐base extension reactions, the DNA was genotyped by the Sequenom MassArray system (iPLEX assay). Eventually, allele analysis was performed by MALDI‐TOFMS and the data were analysed using the MassArray Typer 4.0 software. The variant PNPLA3 rs738409 was previously genotyped in our previous study, and these previous genotyping data partially overlapped the genotyping data in this cohort. 16
2.3. Laboratory analysis
Clinical laboratory indicators, including red blood cell (RBC) count, mean corpuscular volume (MCV), platelet (PLT) count, serum ALT, AST, gamma‐glutamyl transpeptidase (GGT), alkaline phosphatase (ALP) and albumin (ALB), were measured using standard hospital assays (Beckman, HITACHI, and SYSMEX).
2.4. Statistical analysis
Statistical analysis was performed using PLINK v1.90 software, SPSS Statistics V.25 (IBM) and Prism V.8 (GraphPad). Continuous traits were expressed as median (and interquartile ranges) and categorical traits were expressed as numbers (and percentages) to describe the study population. The Hardy‐Weinberg equilibrium (HWE) test was performed on the genotype frequency distribution in the ALD patients and controls. If P ≥ .05, the genotype frequency of the population did not deviate from the Hardy‐Weinberg equilibrium. Differences in baseline characteristics and variant genotype frequency among ALD patients (n = 769) and controls (n = 767) were calculated by Student's t test (Mann‐Whitney U test) and chi‐square test respectively. The association between variant alleles and ALD was tested with logistic regression. Three model analyses were performed by logistic regression analysis, such as additive model, dominant model and recessive model to explore the association between the HSD17B13 genotypes and ALD. For statistical tests, HSD17B13 rs72613567 genotypes were coded 0, 1 and 2 for common homozygotes (T/T), heterozygotes (TA/T) and rare homozygotes (TA/TA) respectively. The association between HSD17B13 genotype and serum markers of liver was tested with linear regression, and all regressions were subjected to age adjustment. All serum marker measurements were transformed to natural logarithms before performing the regression because of the non‐normal distribution of the values. To test the interaction between HSD17B13 rs72613567 and PNPLA3 rs738409, the linear regression model was used. Linkage disequilibrium was carried out using Haploview software v4.2. P < .05 were considered statistically significant.
3. RESULTS
3.1. Baseline characteristics of patients and controls
The baseline characteristics of the 769 male ALD patients and 767 male healthy controls are shown in Table 1. The baseline characteristics between the two groups were significantly different (all P < .05) except age.
TABLE 1.
Baseline characteristics of study subjects
| Characteristics | ALD patients | Healthy controls | P value |
|---|---|---|---|
| Number | 769 men | 767 men | — |
| Alcoholic cirrhosis | 708 (92%) | — | — |
| Age (ys) | 52 (47‐58) | 52 (46‐58) | .48 |
| ALT (U/L) | 24 (17‐33) | 23 (18‐29) | .04 |
| AST (U/L) | 41 (30‐60) | 20 (17‐24) | <.0001 |
| GGT (U/L) | 66 (32‐145) | 26 (19‐39) | <.0001 |
| ALP (U/L) | 122 (96‐165) | 62 (53‐74) | <.0001 |
| ALB (g/L) | 32 (28‐35) | 46 (45‐47) | <.0001 |
| PLT (109/L) | 77 (50‐121) | 224 (194‐261) | <.0001 |
| RBC (1012/L) | 3.5 (2.8‐4.1) | 4.9 (4.7‐5.2) | <.0001 |
| MCV (fL) | 96 (89‐104) | 90 (88‐93) | <.0001 |
Values are numbers (and percentages) for categorical traits, or medians (and interquartile ranges) for continuous traits. P values were performed using Student's t test or Mann‐Whitney U test. Bold indicates P < .05.
Abbreviations: ALB, albumin; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, gamma‐glutamyl transpeptidase; MCV, mean corpuscular volume; PLT, platelet; RBC, red blood cell count.
3.2. Allele and genotype analysis
The details of the two variants are shown in Table 2. Rs72613567 and rs6834314 followed the Hardy‐Weinberg equilibrium (both not significant). Figure 1 shows the allele frequency of the two SNPs in ALD patients and controls. The frequency of both the rs72613567 TA allele and rs6834314 G allele was lower in ALD patients (24.3% and 25.0%, respectively) compared to the control cohort (28.3% and 29.2%, respectively) with a protective effect against ALD (OR = 0.81 [0.69‐0.95], P = .01; OR = 0.81 [0.69‐0.95], P = 8.78 × 10−3, respectively).
TABLE 2.
Details of candidate SNPs
| SNP | Region | SNP category | Allele | P value a (case/control) | LD test b |
|---|---|---|---|---|---|
| rs72613567 | chr4 | Indel (insertion of A) | T > TA | >.05/>.05 | D' = 0.98, r 2 = .93 |
| rs6834314 | chr4 | Downstream variant | A > G | >.05/>.05 |
P value was analysed by Hardy‐Weinberg equilibrium test on the genotype frequency distribution in ALD patients and controls. If P > .05, then the population is in accordance with HWE.
LD test*: Linkage disequilibrium test performed by Haploview V4.2.
FIGURE 1.
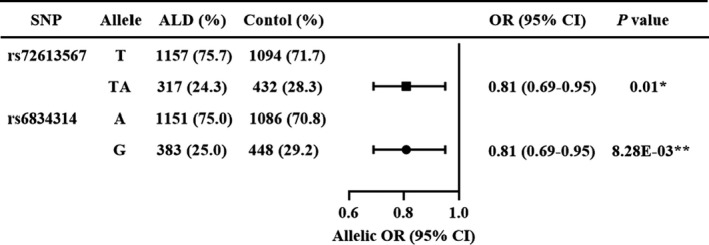
Allele frequency distribution in ALD Patients and Healthy Controls. ORs and P values by logistic regression were adjusted for age
The genotype frequency analysis (Figure 2) revealed that the genotype distribution of the two SNPs was significantly different between ALD patients and controls. As regard HSD17B13 rs72613567, the frequency of TA/TA, TA/T and T/T genotypes in ALD patients was 6.0% (46), 36.5% (279) and 57.5% (439), respectively, while in the controls was 8.5% (65), 39.6% (302) and 51.9% (396) respectively. The heterozygote genotype TA/T and homozygote genotype TA/TA were lower in ALD patients than in the controls (P = .04). As regard HSD17B13 rs6834314, the frequency of G/G, G/A and A/A genotype in ALD patients was 5.7% (44), 38.5% (295) and 55.8% (428), respectively, while in the controls was 8.9% (68), 40.7% (312) and 50.4% (387) respectively. Similarly, the heterozygote genotype G/A and the homozygote genotype G/G were significantly lower in ALD patients than in the controls (P = .02).
FIGURE 2.
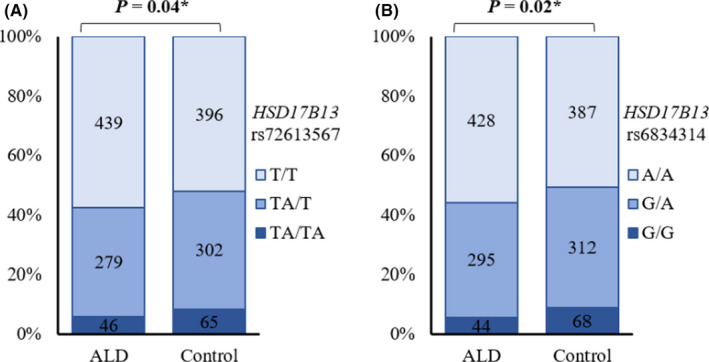
Genotype distribution in ALD patients and healthy controls. The genotype distribution of two SNPs (A) HSD17B13 rs72613567 and (B) HSD17B13 rs6834314 in ALD patients and controls analysed by the Chi‐square test
3.3. Genetic model analysis
The analysis in three genetic models was performed to explore the effect of different genotypes on ALD (Figure 3). In the additive model, each allele TA was identified to significantly reduce the risk of ALD by 19%, (95% CI: 0.69‐0.95, P = .01). The combination of TA/T and TA/TA genotypes (defining the TA allele carriers) also reduced the risk of ALD (OR = 0.79 [0.64‐0.97], P = .03) in the dominant model. However, the protective effect of TA/TA homozygote was lost in the recessive model compared with T/T and TA/T genotypes (OR = 0.68 [0.46‐1.02], P = .06). As regard HSD17B13 rs6834314, each allele‐G was also confirmed to reduce the risk of ALD by 20% in the additive model (95% CI: 0.68‐0.94, P = .007). Uniformly, the combination of G/A and G/G genotypes confirmed the association with a reduced risk of ALD in the dominant model (OR = 0.81 [0.66‐0.99], P = .04, respectively). However, the G/G homozygote was identified as the one reducing the risk of ALD in the recessive model (OR = 0.62 [0.42‐0.92], P = .02). Carrying a TA or G allele conferred a potential protective effect against ALD, thus, TA/TA or G/G homozygote resulted in a significant reduction in ALD risk.
FIGURE 3.
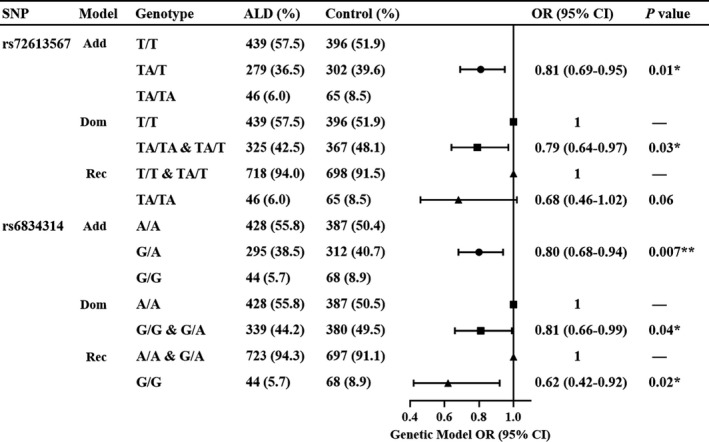
Risk of ALD according to the Genotype in the Genetic Model. ORs and P values by logistic regression are adjusted for age. Add, additive model; Dom, dominant model; Rec, recessive model
The above data revealed that rs72613567 might be consistent with rs6834314. The linkage disequilibrium was evaluated to determine whether rs72613567 was associated with rs6834314. The two tag SNPs were in strong linkage disequilibrium (D’ = 0.98, r 2 = .93) with each other (Table 2), similar with populations of European ancestors. 27 This result suggested that these two variants could be considered as the same.
3.4. HSD17B13 variant and biochemical liver markers
Owing to the high linkage disequilibrium of the two tag SNPs, the association between HSD17B13 rs72613567 and biochemical liver markers was evaluated. The study by Abul‐Husn et al discovered that HSD17B13 rs72613567:TA was associated with a reduced plasma ALT level. 25 To confirm whether rs72613567:TA was associated with the ALT level or other biochemical liver markers in the serum of Chinese Han cohorts, the effect of HSD17B13 rs72613567 on ALT, AST, ALP, GGT and ALB was evaluated (Figure 4). The allele TA reduced the level of serum ALT (P = .005). The TA/TA homozygote was 5.0 U/L (relative change: 17.4%) lower than the T/T homozygote and 3.8 U/L (relative change: 14.0%) lower than the TA/T heterozygote. The TA allele was also associated with lower serum AST level (P = .007) and GGT level (P = .02), while the serum ALB level was higher (P = .02), but this allele was not associated with ALP. The serum levels of AST and GGT in TA/TA homozygote was 6.6 U/L (relative change was 17.3%), 10.4 U/L (11.7%) lower than T/T homozygote and the serum level of ALB in TA/TA homozygote was 1.8 U/L (4.5%) higher than the T/T homozygote.
FIGURE 4.
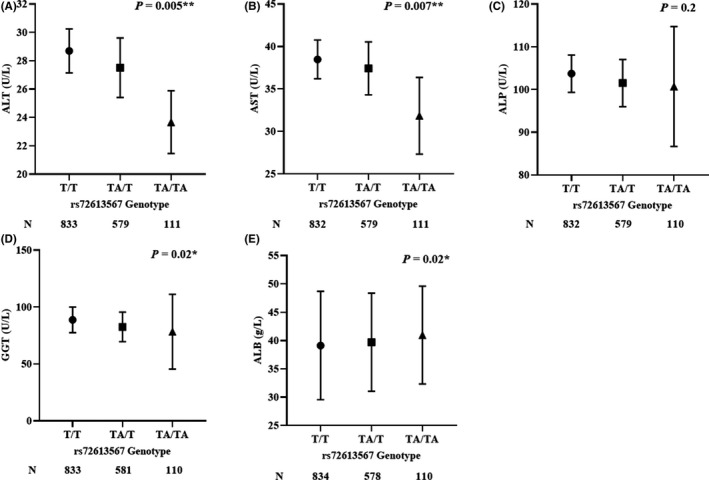
Effect of HSD17B13 rs72613567 Genotype on Liver Biomarkers. Estimates depict mean liver biomarker levels, and error bars are 95% CIs. The P value by linear regression was adjusted for age. For visual clarity, the y‐axis was truncated. The association of biochemical liver markers (A) ALT, (B) AST, (C) ALP, (D) GGT, (E) ALB with genotypes of rs72613567. ALT, alanine aminotransferase; AST, aspartate amino‐transferase; ALP, alkaline phosphatase; ALB, albumin; GGT, gamma‐glutamyl transpeptidase
3.5. Interaction between HSD17B13 rs72613567 and PNPLA3 rs738409
Our previous study was performed to confirm the association between steatogenic genes (PNPLA3 rs738409, MBOAT7 rs626283 and rs641738, SUGP1 rs10401969 and TM6SF2 rs58542926) and ALD, 16 and these previous genotyping data partially overlapped the genotyping data in this cohort (685 individuals in the overlap). Only PNPLA3 rs738409 was associated with an increased risk of ALD in the Chinese Han population (OR = 1.93 [1.63‐2.28], P = 1.25 × 10−14) and with a higher serum ALT level [OR = 1.67 [1.20‐2.32], P = 2.12 × 10−3]. The ALT‐lowering effect of HSD17B13 was explored in the overlapping 685 individuals by the interaction between HSD17B13 rs72613567 and PNPLA3 rs738409 (Figure 5B). Finally, the ALT‐lowering effect of HSD17B13 rs72613567 was lost in this cohort after stratifying by PNPLA3 rs738409 genotype, which was not consistent with the study of Gellert‐Kristensen in a Danish cohort. 28 To confirm the effect of rs738409 and rs72613567 on serum ALT in this cohort (685 individuals), the association between two SNPs and serum ALT level was evaluated in a linear regression (Figure 5A). In this cohort, the genotypes of PNPLA3 rs738409 and HSD17B13 rs72613567 were not associated with serum ALT (P = .64, P = .51, respectively), which explained the inconsistent result with the study of Gellert‐Kristensen.
FIGURE 5.
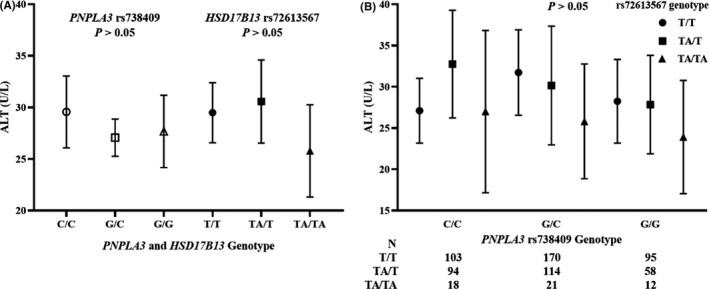
Effect of HSD17B13 rs72613567 on ALT, stratified by PNPLA3 rs738409 genotype. A, Effect of PNPLA3 rs738409 and HSD17B13 rs72613567 genotype on ALT. B, Effect of the interaction between PNPLA3 rs738409 and HSD17B13 rs72613567 on ALT. Estimates depict mean liver biomarker levels, and error bars are 95% CIs. The P values in A and B were found with the linear and logistic regression, respectively, and adjusted for age. For visual clarity, the y‐axis was truncated. ALT, alanine aminotransferase
4. DISCUSSION
This study demonstrated that the polymorphisms of HSD17B13 gene were associated with ALD in the Chinese Han population and identified the effect of HSD17B13 variant on liver biomarkers.
Alcohol‐related liver disease is a common chronic liver disease caused by long‐term heavy alcohol consumption. Genetic factors are involved in the pathogenesis of ALD, and the prevalence of these factors differs among different ethnicity and genetic background. Recently, Abul‐Husn et al reported that rs72613567 in HSD17B13 was associated with a reduced risk of chronic liver disease and progression from steatosis to steatohepatitis. 25 Abul‐Husn and colleagues found that HSD17B13 rs72613567:TA was associated with a reduced risk of ALD and liver damage caused by the genetic susceptibility of PNPLA3 P.I148M variant of steatosis. 25 Our findings revealing that the variant in HSD17B13 reduced the risk of ALD were consistent with the study by Abul‐Husn. Several studies confirmed that PNPLA3 rs738409 was associated with an increased risk of alcoholic cirrhosis. 9 , 11 , 29 More recently, Gellert‐Kristensen et al combined PNPLA3 p.I148M and TM6SF2 p.E167K variants to validate the interaction with HSD17B13 rs72613567, and they discovered that an increasing genetic risk score amplified the ALT‐lowering effect of HSD17B13. 28 Our previous study confirmed that PNPLA3 rs738409:G allele was associated with an increased risk of ALD in the Chinese Han population. 16 However, in this study, we did not find a role of rs72613567:TA variant in mitigating ALT‐increasing effect of PNPLA3 rs738409. Although there was a tendency of ALT‐lowing effect by HSD17B13:TA in G/C and G/G genotypes of PNPLA3. This result could be caused by the small sample size considered in this cohort.
Abul‐Husn discovered that the rs72613567 TA allele was associated with lower plasma ALT and AST levels. 25 Recently, Gellert‐Kristensen et al demonstrated that rs72613567:TA was associated with stepwise lower levels of plasma ALT, AST, GGT and bilirubin in the Danish general population, but not with ALP and ALB. 28 Our work confirmed the association between rs72613567:TA and lower serum ALT, AST and GGT levels, and higher serum ALB level, but no association was found with ALP. A higher level of albumin indicates better liver function. Our findings showed that rs72613567:TA was associated not only with a reduced liver injury by alcohol consumption but was also associated with a better liver function.
The HSD17B13 gene is located on the long arm of human chromosome 4, and it encodes the HSD17B13 protein that belongs to the hydroxysteroid 17‐beta dehydrogenase family, which includes enzymes involved in the steroid and fatty acid metabolism. 30 HSD17B13 rs72613567 is a loss‐of‐function variant owing to the insertion of adenine (A) that results in the production of shortened proteins and reduced enzyme activity. 25 , 31 Molecular and proteomic studies revealed that the HSD17B13 protein is mainly expressed in the liver and located on the surface of lipid droplets. 32 , 33 The study of Horiguchi et al 32 showed that HSD17B13 has an N‐terminal sequence similar to that of the known HSD17B11. However, HSD17B13 is mainly expressed in the liver and is not enhanced by the peroxisome proliferator‐activated receptor alpha (PPARα) and its ligands. Moreover, HSD17B13 overexpressed in the liver of normal C57BL/6 mice through an adenovirus‐based approach showed an increased hepatic amount of triglyceride, but exerting a small effect on plasma triglyceride and cholesterol. 33 Therefore, HSD17B13 may be a lipid droplet‐related protein that plays a specific role in lipid metabolism. 32 , 34 However, the specific biological function of HSD17B13 is still unclear. 33 , 34 Ma et al recently found that HSD17B13 had a retinol dehydrogenase (RDH) activity in vitro, 27 playing an important role in retinol metabolism associated with the activation of hepatic stellate cells. Of note, the activation of hepatic stellate cells promotes the development of hepatic fibrosis. This may indicate that HSD17B13 participates in the development of liver disease through the hepatic stellate cell activity. In order to clarify the biological role of HSD17B13, substantial studies are needed in the future to identify the substrate and metabolites of HSD17B13, and to clarify the mechanism of HSD17B13 in liver diseases.
This study has certain limitations, for example, no females are included in the case group and control group. A reasonable explanation for this choice is offered by the traditional Chinese custom, which requires that women do not drink as much as men, resulting in a lower incidence of ALD in women than that of men. The female ALD samples collected in this study were <1% of the case group, thus, women were excluded from this study. Moreover, the ALT‐lowering effect of rs72613567:TA was not found among carriers of PNPLA3 rs738409 because of the small useful sample size. Of note, the findings of the interaction between PNPLA3 rs738409 and HSD17B13 rs72613567 from the conclusion of other studies are very precious. 25 , 28 The effect of HSD17B13 rs72613567 in reducing the risk of liver injury from PNPLA3 rs738409 variant may provide a new perspective to prevent the progression of ALD or other fatty liver disease.
With the improvement of the living standards, alcohol consumption and the prevalence of ALD gradually increased. The incidence of ALD is higher in some developing countries, such as China and India, thus, its prevention is essential. The HSD17B13 gene is a recently discovered candidate gene associated with ALD and may serve as a therapeutic target to combat ALD. More and more studies recently confirmed that the polymorphisms of HSD17B13 are associated with ALD or NAFLD. 25 , 26 , 27 , 28 , 31 , 35 , 36 , 37 However, more attention should be paid in further studies to the role of HSD17B13 protein in ALD.
In conclusion, our results showed that the two protective SNPs of HSD17B13, such as rs72613567 and rs6834314, reduced the risk of ALD in the Han Chinese men and were associated with serum liver biomarkers level.
CONFLICT OF INTEREST
All authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
All authors contributed to the study conception and design. Material preparation was performed by TSG, YLM, FNY, YFZ, LBL, CXL, HDG and YTJ, data collection and analysis by YFZ and HZC. The first draft of the manuscript was written by YFZ and HZC. Experiment supervision and paper review were performed by YZL and JH.
ETHICAL STATEMENTS
The study was approved by the ethical committees of The First Hospital of Jilin University.
ACKNOWLEDGEMENTS
We thank Chunwei Cao of the Beijing Institute of Genomics for help and suggestions in genotyping and data analysis.
Chen H, Zhang Y, Guo T, et al. Genetic variant rs72613567 of HSD17B13 gene reduces alcohol‐related liver disease risk in Chinese Han population. Liver Int. 2020;40:2194–2202. 10.1111/liv.14616
Haizhen Chen, Yanfang Zhang and Tongsheng Guo have contributed equally to this work.
Handling Editor: Stefano Romeo
Contributor Information
Yongzhe Li, Email: yongzhelipumch@126.com.
Jing Huang, Email: jluhuangjing1@126.com.
REFERENCES
- 1. Seitz HK, Bataller R, Cortez‐Pinto H, et al. Alcoholic liver disease. Nat Rev Dis Primers. 2018;4:16. [DOI] [PubMed] [Google Scholar]
- 2. WHO . Global status report on alcohol and health 2018. Geneva: World Health Organization; 2018. https://apps.who.int/iris/bitstream/handle/10665/274603/9789241565639‐eng.pdf. [Google Scholar]
- 3. Teli MR, Day CP, Burt AD, Bennett MK, James OF. Determinants of progression to cirrhosis or fibrosis in pure alcoholic fatty liver. The Lancet. 1995;346:987‐990. [DOI] [PubMed] [Google Scholar]
- 4. Liangpunsakul S, Haber P, McCaughan GW. Alcoholic liver disease in Asia, Europe, and North America. Gastroenterology. 2016;150:1786‐1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Levy R, Catana AM, Durbin‐Johnson B, Halsted CH, Medici V. Ethnic differences in presentation and severity of alcoholic liver disease. Alcohol Clin Exp Res. 2015;39:566‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Louvet A, Mathurin P. Alcoholic liver disease: mechanisms of injury and targeted treatment. Nat Rev Gastroenterol Hepatol. 2015;12:231‐242. [DOI] [PubMed] [Google Scholar]
- 7. European Association for the Study of the Liver , Thursz M, Gual A, et al. EASL Clinical Practice Guidelines: management of alcohol‐related liver disease. J Hepatol. 2018;69(1):154‐181. [DOI] [PubMed] [Google Scholar]
- 8. Gao B, Zakhari S. Epidemiology and pathogenesis of alcoholic liver disease In: Sanyal AJ, Boyer TD, Terrault N, Lindor KD, eds. Zakim and Boyer's Hepatology, Vol. 22. Philadelphia PA: Elsevier; 2018:334‐344. [Google Scholar]
- 9. Buch S, Stickel F, Trépo E, et al. A genome‐wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol‐related cirrhosis. Nat Genet. 2015;47:1443‐1448. [DOI] [PubMed] [Google Scholar]
- 10. Agrawal A, Bierut LJ. Identifying genetic variation for alcohol dependence. Alcohol Res. 2012;34:274‐281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burza MA, Molinaro A, Attilia ML, et al. PNPLA3 I148M (rs738409) genetic variant and age at onset of at‐risk alcohol consumption are independent risk factors for alcoholic cirrhosis. Liver Int. 2014;34:514‐520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Salameh H, Raff E, Erwin A, et al. PNPLA3 gene polymorphism is associated with predisposition to and severity of alcoholic liver disease. Am J Gastroenterol. 2015;110:846‐856. [DOI] [PubMed] [Google Scholar]
- 13. Falleti E, Cussigh A, Cmet S, Fabris C, Toniutto P. PNPLA3 rs738409 and TM6SF2 rs58542926 variants increase the risk of hepatocellular carcinoma in alcoholic cirrhosis. Dig Liver Dis. 2016;48:69‐75. [DOI] [PubMed] [Google Scholar]
- 14. Atkinson SR, Way MJ, McQuillin A, Morgan MY, Thursz MR. Homozygosity for rs738409: G in PNPLA3 is associated with increased mortality following an episode of severe alcoholic hepatitis. J Hepatol. 2017;67:120‐127. [DOI] [PubMed] [Google Scholar]
- 15. Beaudoin JJ, Long N, Liangpunsakul S, et al. An exploratory genome‐wide analysis of genetic risk for alcoholic hepatitis. Scand J Gastroenterol. 2017;52:1263‐1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Y, Guo T, Yang F, et al. Single‐nucleotide rs738409 polymorphisms in the PNPLA3 gene are strongly associated with alcoholic liver disease in Han Chinese males. Hepatol Int. 2018;12:429‐437. [DOI] [PubMed] [Google Scholar]
- 17. Stickel F, Buch S, Nischalke HD, et al. Genetic variants in PNPLA3 and TM6SF2 predispose to the development of hepatocellular carcinoma in individuals with alcohol‐related cirrhosis. Am J Gastroenterol. 2018;113:1475‐1483. [DOI] [PubMed] [Google Scholar]
- 18. Yang J, Trépo E, Nahon P, et al. PNPLA3 and TM6SF2 variants as risk factors of hepatocellular carcinoma across various etiologies and severity of underlying liver diseases. Int J Cancer. 2019;144:533‐544. [DOI] [PubMed] [Google Scholar]
- 19. Basyte‐Bacevice V, Skieceviciene J, Valantiene I, et al. TM6SF2 and MBOAT7 gene variants in liver fibrosis and cirrhosis. Int J Mol Sci. 2019;20:1277‐1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rotman Y, Koh C, Zmuda JM, Kleiner DE, Liang TJ. The association of genetic variability in patatin‐like phospholipase domain‐containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 2010;52:894‐903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461‐1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kozlitina J, Smagris E, Stender S, et al. Exome‐wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352‐356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Y‐L, Reeves HL, Burt AD, et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non‐alcoholic fatty liver disease. Nat Commun. 2014;5:4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mancina RM, Dongiovanni P, Petta S, et al. The MBOAT7‐TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology. 2016;150(5):1219‐1230.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abul‐Husn NS, Cheng X, Li AH, et al. A protein‐truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med. 2018;378:1096‐1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang J, Trépo E, Nahon P, et al. A 17‐beta‐hydroxysteroid dehydrogenase 13 variant protects from hepatocellular carcinoma development in alcoholic liver disease. Hepatology. 2019;70:234‐240. [DOI] [PubMed] [Google Scholar]
- 27. Ma Y, Belyaeva OV, Brown PM, et al. 17‐beta hydroxysteroid dehydrogenase 13 is a hepatic retinol dehydrogenase associated with histological features of nonalcoholic fatty liver disease. Hepatology. 2019;69:1504‐1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gellert‐Kristensen H, Nordestgaard BG, Tybjaerg‐Hansen A, Stender S. High risk of fatty liver disease amplifies the alanine transaminase‐lowering effect of a HSD17B13 variant. Hepatology. 2020;71:56‐66. [DOI] [PubMed] [Google Scholar]
- 29. Kolla BP, Schneekloth TD, Biernacka J, et al. PNPLA3 association with alcoholic liver disease in a cohort of heavy drinkers. Alcohol Alcohol. 2018;53:357‐360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Su W, Mao Z, Liu Y, et al. Role of HSD17B13 in the liver physiology and pathophysiology. Mol Cell Endocrinol. 2019;489:119‐125. [DOI] [PubMed] [Google Scholar]
- 31. Pirola CJ, Garaycoechea M, Flichman D, et al. Splice variant rs72613567 prevents worst histologic outcomes in patients with nonalcoholic fatty liver disease. J Lipid Res. 2019;60:176‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Horiguchi Y, Araki M, Motojima K. 17beta‐Hydroxysteroid dehydrogenase type 13 is a liver‐specific lipid droplet‐associated protein. Biochem Biophys Res Commun. 2008;370:235‐238. [DOI] [PubMed] [Google Scholar]
- 33. Su W, Wang Y, Jia X, et al. Comparative proteomic study reveals 17beta‐HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc Natl Acad Sci USA. 2014;111:11437‐11442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu S, Huang C, Li D, et al. Molecular cloning and expression analysis of a new gene for short‐chain dehydrogenase/reductase 9. Acta Biochim Pol. 2007;54:213‐218. [PubMed] [Google Scholar]
- 35. Stickel F, Lutz P, Buch S, et al. Genetic variation in HSD17B13 reduces the risk of developing cirrhosis and hepatocellular carcinoma in alcohol misusers. Hepatology. 2020;72:88‐102. [DOI] [PubMed] [Google Scholar]
- 36. Kallwitz E, Tayo BO, Kuniholm MH, et al. Association of HSD17B13 rs72613567:TA with non‐alcoholic fatty liver disease in Hispanics/Latinos. Liver Int. 2020;40:889‐893. [DOI] [PubMed] [Google Scholar]
- 37. Seko Y, Yamaguchi K, Tochiki N, et al. Attenuated effect of PNPLA3 on hepatic fibrosis by HSD17B13 in Japanese patients with non‐alcoholic fatty liver disease. Liver Int. 2020;40:1686‐1692. [DOI] [PubMed] [Google Scholar]


