Abstract
Alcohol dehydrogenase 5 (ADH5) is a member of medium‐chain dehydrogenase/reductase family and takes part in cellular formaldehyde and S‐nitrosoglutathione metabolic network. 2‐tridecanone (2‐TD) is a toxic compound in many Solanaceae crops to defend against a variety of herbivory insects. In the broader context of insect development and pest control strategies, this study investigates how a new ADH5 from Helicoverpa armigera (HaADH5) regulates the expression of CYP6B6, a gene involved in molting and metamorphosis, in response to 2‐TD treatment. Cloning of the HaADH5 complementary DNA sequence revealed that its 1002 bp open reading frame encodes 334 amino acids with a predicted molecular weight of 36.5 kD. HaADH5 protein was purified in the Escherichia coli Transetta (pET32a‐HaADH5) strain using a prokaryotic expression system. The ability of HaADH5 protein to interact with the 2‐TD responsive region within the promoter of CYP6B6 was confirmed by an in vitro electrophoretic mobility shift assay and transcription activity validation in yeast. Finally, the expression levels of both HaADH5 and CYP6B6 were found to be significantly decreased in the midgut of 6th instar larvae after 48 h of treatment with 10 mg/g 2‐TD artificial diet. These results indicate that upon 2‐TD treatment of cotton bollworm, HaADH5 regulates the expression of CYP6B6 by interacting with its promoter. As HaADH5 regulation of CYP6B6 expression may contribute to the larval xenobiotic detoxification, molting and metamorphosis, HaADH5 is a candidate target for controlling the growth and development of cotton bollworm.
Keywords: alcohol dehydrogenase, DNA‐protein interaction, Helicoverpa armigera, regulatory factor, 2‐tridecanone
Introduction
Alcohol dehydrogenases (ADHs) of medium‐chain dehydrogenase/reductase (MDR) superfamily play important roles in many physiological processes, including the metabolism of ethanol, aldehydes, steroids, retinoids, lipid peroxidation products, hydroxy fatty acids, and xenobiotic alcohols (Ramchandani et al., 2001; Oscar & Marinkovic, 2003; Luo et al., 2008; Chase et al., 2009; Anand et al., 2014).
ADH5 belongs to the MDR family and functions as a homodimer that localizes to the nucleus and cytoplasm (Yang et al., 1997; Fernández et al., 2003). Each subunit of the homodimer has two tightly bound zinc atoms, the one catalytic zinc at the active site and the other structural zinc in a lobe of the catalytic domain (Kaiser et al., 1988; Östberg et al., 2016). ADH5 does not normally metabolize ethanol, aldehydes or retinoids, and it was identical to formaldehyde dehydrogenase and S‐nitrosoglutathione reductase based on its amino acid sequence and its structural and kinetic properties (Koivusalo et al., 1989; Jensen et al., 1998; Deltour et al., 1999).
The characterization of ADH5 has focused on its functions in the formaldehyde and nitrogen oxide metabolism pathways. Formaldehyde can rapidly attack electron‐rich thiol and amino groups to form covalent adducts, such as DNA interstrand crosslinks and DNA‐protein crosslinks; the resulting blockage of transcription and replication can lead to mutagenesis and cell death (Kawanishi et al., 2014). ADH5 plays a central role in formaldehyde metabolism to prevent cytotoxicity and provides one carbon unit for nucleotide biosynthesis (Tibbetts & Appling, 2010; Pontel et al., 2015; Burgos et al., 2017). ADH5 can also convert S‐nitrosoglutathione, a stable nitrogen oxide reserve, to S‐nitrosothiol, resulting in the post‐translational S‐nitrosylation of a protein at the same time (Wei et al., 2010; Smith & Marletta, 2012; Corti et al., 2014). The S‐nitrosylation protein modification is a potential signal in physiological processes, such as the generation of cyclic guanosine monophosphate, inactivation of certain enzymes to cause DNA damage, and inhibition or induction of apoptotic cell death (Hess et al., 2005; Martínez et al., 2011).
The ADH5 of insects was first found in several Drosophila species with 70% identity to the corresponding human form and was highly selective for formaldehyde rather than ethanol, its enzymatic property being compatible with the constitutive nature of the homolog in vertebrates (Danielsson et al., 1994). In the past two decades, little research on ADH5 has been done in insects. The expression and activity of ADH5 was significantly changed as a metabolizing enzyme in response to toxic industrial additives and heavy metals in larvae of Chironomus riparius (Park & Kwak, 2009a, 2009b). Then, ADH5 was confirmed to play a critical role in degradation of odorants and xenobiotics as a biotransformation enzyme within antennae of Cydia pomonella (Huang et al., 2016). In recent years, thanks to the rapid development of transcriptome, more and more ADH5 was found and suggested to participate in the metamorphosis and pheromone chemosensory system in a lot of insects. The transcriptome displayed ADH5 present in wing discs during the larva‐to‐pupa metamorphosis and might be involved in chitin biosynthesis of Bombyx mori (Ou et al., 2014). The transcriptomic analysis of the female sex pheromone glands showed ADH5 could be involved in sex pheromone biosynthesis and degradation pathways in diamondback moth, black cutworm and three noctuid moths (Vogel et al., 2010; Gu et al., 2013; Li et al., 2015; He et al., 2017).
Helicoverpa armigera (Lepidoptera: Noctuidae) seriously impacts the output of many agricultural and economic crops. Meanwhile, these plants could synthesize a variety of toxic substances to defend against this herbivory pest. 2‐tridecanone (2‐TD) is an important secondary substance in Solanaceae plants. Its concentration in leaves can be as high as 0.39%, which can induce excessive expression of various insect cytochrome P450 genes including CYP6B6 (Yu et al., 2002; Liu et al., 2006). CYP6B6 is important for the detoxification of exogenous toxic substances, growth, and developmental process in cotton bollworm (Zhao et al., 2016a). In previous research, we identified a 2‐TD responsive region in the CYP6B6 promoter, designated as the HE1 element, using transient transfection and mobility shift assays (Li et al., 2014). Using the yeast one‐hybrid method, we subsequently identified candidate regulators that bind to the CYP6B6 HE1 element, and one of these was found to be a previously uncharacterized ADH5 of H. armigera (HaADH5) based on the results of National Center for Biotechnology Information Basic Local Alignment Search Tool (NCBI BLAST) analysis.
The functions of insect ADH5 in growth and developmental processes have not been clearly confirmed in the available literature. To investigate whether HaADH5 regulates the expression of CYP6B6 upon treatment with the toxic substance 2‐TD, we cloned the HaADH5 complementary DNA (cDNA) sequence using rapid amplification of cDNA ends (RACE) and also tested the interaction between HaADH5 protein and the CYP6B6 HE1 element through an in vitro electrophoretic mobility shift assay and transcription activity validation in yeast. Using real‐time quantitative polymerase chain reaction (PCR) and Western blotting, HaADH5 and CYP6B6 expression were also detected in the midgut of 6th instar larvae exposed to 2‐TD. The present findings demonstrate the broader range of ADH5 function as a regulatory factor affecting the expression of a gene involved in detoxification, metabolism, growth, and development in cotton bollworm.
Materials and methods
Insects
The cotton bollworm population used in this study (a laboratory population) was initially collected from the Anningqu town of Urumqi in Xinjiang Autonomous Region, China, and reared on an artificial diet in a growth room maintained at 26 ± 1 °C, 70%–80% relative humidity, with a photoperiod of 16 : 8 h (L : D). The population was never exposed to any pesticides. The composition of the artificial diet was as follows: corn flour 300 g, soybean powder 100 g, yeast extract powder 100 g, citric acid 2.5 g, vitamin C 10 g, sorbic acid 1.5 g, vitamin B 1.5 g, erythromycin 0.05 g, propionic acid 5 mL, vitamin E 1.5 g, water 2.5 L.
Plasmids, antisera, and bacterial and yeast strains
The pMD18‐T, pET32a, and pGADT7 vectors were purchased from Takara Bio, EMD Biosciences, and Clontech, respectively. The polyclonal antisera of anti‐CYP6B6, anti‐β‐Actin, and anti‐HaADH5 were prepared by our research group (Huang et al., 2015; Wei et al., 2015). Competent cells of Escherichia coli strains DH5α and Transetta were purchased from TransGen Biotech. The yeast strains Y1HGold (p4r‐AbAi) and Y1HGold (p4m‐AbAi) were constructed by our research group as previously reported (Zhao et al., 2016b).
HaADH5 sequencing by RACE
Total RNA was extracted from the midgut of 6th instar larvae using the TansZol Up Plus RNA Kit (TransGen Biotech, Beijing, China) then reverse transcribed into RACE‐Ready cDNA using the SMARTer RACE 5′/3′ Kit (Clontech Laboratories, Mountain View, CA, USA).
Based on the nucleotide sequence determined by yeast one‐hybrid screening, we designed primers for RACE amplification of HaADH5 by DNAMAN software (Table 1). The outer PCR reaction conditions were 94 °C for 5 min, followed by 94 °C for 30 s, 68 °C for 30 s, and 72 °C for 3 min (25 cycles). The outer PCR product was diluted 50‐fold then used as the template for the inner PCR reaction. The inner PCR conditions were 94 °C for 5 min, followed by 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 3 min (30 cycles). After the inner PCR amplification, the product was recycled, ligated into pMD18‐T vector, and transformed into E. coli DH5α. Positive clones were identified by bacterial solution PCR detection and sent for sequencing (Sangon Biotech, Shanghai, China). Using DNAMAN software, the 5′ and 3′ end sequences were assembled to obtain the cDNA sequence of HaADH5, and its open reading frame (ORF) was predicted using ORF Finder tool in the NCBI database.
Table 1.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Purpose |
|---|---|---|
| 5′‐outer | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | HaADH5 5′ RACE outer PCR |
| 5′‐P1 | TCATCCAACGCCACACAAGG | |
| 5′‐inner | CTAATACGACTCACTATAGGGC | HaADH5 5′ RACE inner PCR |
| 5′‐P2 | GCAGGTTGTAGGATAGGTGCCTTGGG | |
| 3′‐outer | TACCGTCGTTCCACTAGTGATTT | HaADH5 3′ RACE outer PCR |
| 3′‐P1 | CTATCATCGATCCCAACGACAAAA | |
| 3′‐inner | CGCGGATCCTCCACTAGTGATTTCACTATAGG | HaADH5 3′ RACE inner PCR |
| 3′‐P2 | TTGTCAGCCCAAAGCAGGT | |
| HaADH5‐F | GAGGTACCATGGTGAAGGCACGGAAA, Kpn Ι | HaADH5 ORF amplification for ligation into pET32a |
| HaADH5‐R | GCGTCGACTTATAATTTAACAACAGCCT, Sal Ι | |
| HaADH5‐yF | CCATATGATGGTGAAGGCACGGAAATA, Nde I | HaADH5 ORF amplification for ligation into pGADT7 |
| HaADH5‐yR | CCTCGAGTTATAATTTAACAACAGCCTTTC, Xho I | |
| β‐Actin‐QF | CACACCTTCTACAACGAGCTG | qPCR of target and reference genes |
| β‐Actin‐QR | GAGGATCTTCATGAGGTAGTCG | |
| Tubulin‐QF | TCCAACTCACACACTCGCT | |
| Tubulin‐QR | GGAAGCAGATGTCGTATAATG | |
| HaADH5‐QF | GTCTCCGCGGCGCTCAAA | |
| HaADH5‐QR | TCATCCAACGCCACACAAGG | |
| CYP6B6‐QF | TTCAAACTTATACCATGTCCACAAT | |
| CYP6B6‐QR | CCAATTGACGGAGCTCTAGAATCA |
PCR, polymerase chain reaction; RACE, rapid amplification of complementary DNA ends; ORF, open reading frame.
The conserved amino acid of HaADH5 was analyzed by Protein BLAST in the NCBI database. The phylogenetic tree of HaADH5 was constructed by neighbor‐joining analyses using MEGA software with 1000 replicates of bootstrap values. The secondary structure of HaADH5 was predicted including signal peptide, transmembrane domain and subcellular localization by SignalP, TMHMM and TargetP tools of CBS Prediction Servers software, then its tertiary structure was predicted using SWISS‐MODEL software and Protein Data Bank (PDB) database.
Expression and purification of HaADH5 protein in E. coli
The HaADH5 ORF was amplified using the following reaction conditions: 94 °C for 5 min; 35 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 1 min; and 72 °C for 7 min. The purified PCR product was digested with Kpn I and Not I and ligated into the pET32a vector linearized with the same enzymes, followed by transformation into E. coli DH5α competent cells (Cycle Pure Kit, Gel Extraction Kit, Plasmid Mini Kit, OMEGA Bio‐tek, Norcross, GA, USA). After double enzyme digestion to ensure the insertion, pET32a‐HaADH5 was transformed into the E. coli Transetta strain for protein expression. The Transetta (pET32a‐HaADH5) strain was incubated in lysogeny broth (LB) medium at 37 °C until the optical density at 600 nm reached 0.5. Isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) was added into the culture broth to a final concentration of 0.5 mmol/L. The culture was subsequently incubated at 20 °C for 4 h and collected by centrifugation at 7104 g at 4 °C (Ai et al., 2017).
For purification of the His‐HaADH5 protein, cell pellets from 1 L of induced culture broth were completely suspended in phosphate‐buffered saline (PBS, pH 7.0) by gently vortexing. The cell suspension was sonicated for 15 min with 5 s bursts alternated with chilling on ice to decrease viscosity, followed by centrifugation at 15 984 g for 10 min at 4 °C. The supernatant was directly loaded onto a 2 mL column of Ni2+‐NTA His Bind Resins (Novagen, Madison, WI, USA) equilibrated with Ni2+‐NTA Binding Buffer. The settled resin was washed with wash buffer (300 mmol/L NaCl, 50 mmol/L NaH2PO4, pH 8.0) containing 5 mmol/L imidazole then eluted with a linear gradient of imidazole (10, 10, 20, 20, 50, 100, 200, 200 mmol/L). All protein purification steps were performed at 4 °C. All collected outflows were separated by 12% SDS‐PAGE and stained with Coomassie blue.
The concentration and purity of His‐HaADH5 protein was tested using the Pierce Bicinchoninic Acid (BCA) Protein Assay Kit (Thermo Scientific, Waltham, , MA, USA) and Western blotting. The protein was transferred onto a nitrocellulose (NC) membrane using the electrode diverting method. The NC membrane incubation steps were as follows: blocking buffer (PBS containing 3% defatted milk powder) for 1 h at 37 °C, anti‐His‐tag mouse monoclonal antibody in blocking buffer for 2 h at 37 °C, and peroxidase‐conjugated goat anti‐mouse immunoglobulin G (IgG) (H+L) for 2 h at 37 °C. After each incubation step, the membrane was washed with PBS three times. Finally, the membrane was stained with chemiluminescent substrate for 10 min using the BeyoECL Plus kit (Beyotime, Shanghai, China). The membrane was viewed using an ImageQuant LAS4000 imager (Fujifilm, Japan).
In vitro detection of HaADH5 interaction with the CYP6B6 promoter
The CYP6B6 HE1 element was prepared as the DNA probe for the electrophoretic mobility shift assay (EMSA). The negative control contained only HE1 probe without HaADH5 protein. For specific competition, excess unlabeled HE1 DNA was added. For non‐specific competition, a 300 bp non‐correlated sequence was added and denoted as bHLH that was a basic helix‐loop‐helix gene from Chenopodium glaucum. The EMSA/Gel‐shift kit (Beyotime) was used for the reaction. Protein‐bound probes were separated from free probes on 6% (w/v) non‐denaturing PAGE in Tris‐borate ethylenediaminetetraacetic acid buffer. All DNA bands were transferred onto Amersham Hybond‐N+ Membrane (GE Healthcare, Waukesha, WI, USA) using the electrode diverting method. DNA probe labeling and subsequent color detection were performed using the DIG High Prime DNA Labeling and Detection Starter Kit ΙΙ (Roche, Basel, Switzerland) according to the manufacturer's protocol. Finally, the membrane was detected using an ImageQuant LAS4000 imager.
Validation of HaADH5 interaction with the CYP6B6 promoter in yeast
The reaction conditions for amplification of the HaADH5 ORF were 94 °C for 5 min; 35 cycles of 94 °C for 30 s, 59 °C for 30 s, and 72 °C for 1 min; and 72 °C for 7 min. The purified PCR product was digested with Nde I and Xho I then ligated into pGADT7 vector linearized with the same enzymes. The pGADT7‐HaADH5 plasmid was transformed into the Y1HGold (p4r‐AbAi) strain with the 2‐TD responsive region of the CYP6B6 promoter and into a mutant Y1HGold (p4m‐AbAi) strain. The two strains were subsequently coated onto SD/‐Leu medium with or without 100 ng/mL Aureobasidin A (AbA). In this analysis, if HaADH5 interacted with the 2‐TD responsive region of CYP6B6, the AbAr reporter gene would be active in the Y1HGold (p4r‐AbAi) strain but not active in the Y1HGold (p4m‐AbAi) strain.
HaADH5 and CYP6B6 expression in the midgut of H. armigera exposed to 2‐TD
The newly molted 6th instar larvae, after molting for 1 day, were treated by 4 h of starvation treatment, then the larvae were exposed to the artificial diet mixed with 2‐TD (Sigma‐Aldrich, St Louis, MO, USA) (99% purity) 0 or 10 mg/g (w:w) for 6, 12, 20, 30, and 48 h. Each treatment contained 100 larvae. The tested larvae were starved for 30 min to defecate after the different feeding times. The midgut tissue was isolated from the experimental larvae on ice, cleaned in sterile water, then frozen in liquid nitrogen. Each sample contained three midgut tissues, and each group contained three samples.
Total RNA was extracted using the TansZol Up Plus RNA Kit, followed by cDNA synthesis using Reverse Transcriptase Moloney Murine Leukemia Virus (RNase H−) (TaKaRa, Kusatsu, Japan). The reaction setup and quantitative PCR (qPCR) cycling conditions were based on the protocol for the QuantiNova SYBR Green PCR kit (Qiagen, Germantown, MD, USA). The β‐Actin and Tubulin genes were used to normalize the expression levels of the target genes among samples (Vandesompele et al., 2002; Chandra et al., 2014; Shakeel et al., 2015). qPCR was performed in triplicate for each cDNA sample. All of the primer sets are listed in Table 1. The relative expression levels of the target genes were calculated using the method. Using GraphPad Prism 5, one‐way analysis of variance was used to compare the results for samples in the same group, and a paired t‐test was used to compare samples collected at the same time.
Total protein from the midgut was extracted using the TCA/acetone method. The quantity was determined using the Pierce BCA Protein Assay Kit, and 3 μg total protein was separated by 12% SDS‐PAGE and transferred onto a nitrocellulose membrane using the electrode diverting method. According to the predicted sizes of CYP6B6 (58 kD), β‐Actin (42 kD), and HaADH5 (37 kD), the membrane was divided and tested using anti‐CYP6B6 rabbit, anti‐β‐Actin mouse, and anti‐HaADH5 mouse polyclonal antiserum, respectively. The membrane was subsequently incubated with peroxidase‐conjugated goat anti‐mouse or anti‐rabbit IgG and stained with a chemiluminescent substrate. The Western blotting results were quantified using Integrated Density (IntDen) measurement by grayscale analyses, with the β‐Actin IntDen value used for normalization of the target protein. The relative HaADH5 and CYP6B6 IntDen values were analyzed by a paired t‐test for samples from different groups collected at the same time.
Results
HaADH5 gene sequence analysis
The HaADH5 cDNA sequence was amplified and assembled by 5′‐ and 3′‐RACE to obtain the ORF. The 1128 bp HaADH5 cDNA consisted of 38 bp 5′‐UTR, 1005 bp ORF (from 39 to 1043), and 85 bp 3′‐UTR (containing the polyadenylation signal ATTAAA). The ORF encoded a mature protein of 334 amino acids with a predicted molecular weight of 36.67 kD and a theoretical isoelectric point of 6.91. The HaADH5 protein was conserved with most of the key residues for its function: 26 conserved residues important for interaction with nicotinamide adenine dinucleotide phosphate (P48, M129, T133, G154, G157, A158, V159, A178, G179, K183, Y198, N221, V222, C243, G244, S245, I246, S247, Y249, F276, L277, V278, I321, L322, G324, and N326) and five conserved residues important for substrate interaction (Y49, A52, C243, Y249, and W279) (Fig. 1).
Figure 1.

The nucleotide and amino acid sequences of HaADH5. Conserved sites in HaADH5 important for nicotinamide adenine dinucleotide phosphate (NADP) and substrate interaction are underlined and shaded, respectively. The dotted line indicates the poly(A) signal in the nucleotide sequence.
A phylogenetic tree was constructed to assess the relationship of HaADH5 with published ADHs from Homo, Drosophila, and Bombyx species (Fig. 2). HaADH5 belonged to the class III of MDR‐ADHs and tightly clustered together with the published ADH5 in H. armigera (GenBank accession number: AKD01727.1), with which it had 82% amino acid identity. The presently identified HaADH5 has a disulfide bond (C142, C217) and five phosphorylation sites (Y49, Y57, Y61, S241, and S245), and it does not have an N‐glycosylation site, signal peptide, or transmembrane helices. HaADH5 structure prediction indicated homodimerization based on the homologous leukotriene B4 12‐hydroxydehydrogenase/15‐oxo‐prostaglandin 13‐reductase (LTB412‐HD/PGR, PDB ID: 1v3u), which had 49.85% sequence identity. Its predicted structure contains 14 helices, 15 beta strands, five beta bulges, three beta sheets, 28 beta turns, three gamma turns forming three beta hairpins, seven helix‐helix interactions, and five beta‐alpha‐beta units (Fig. 3). HaADH5 homodimerization involves 11 hydrogen bonds and 146 non‐bonded contacts of the dimer interface through 24 residues (E14, P17, K18, R19, A52, S55, R56, F237, V242, C243, G244, S245, S248, L266, V267, Q270, L271, K272, I273, E274, G275, F276, L277, and R280).
Figure 2.
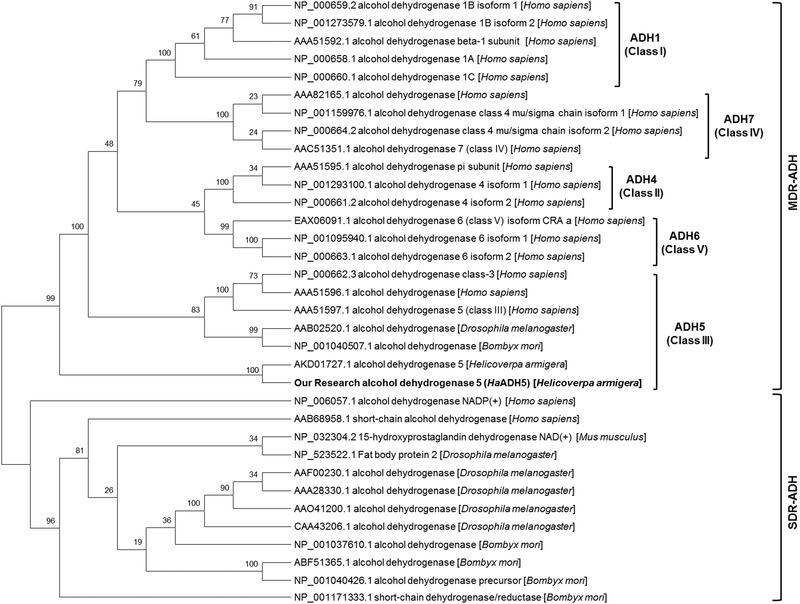
Neighbor‐joining tree of the HaADH5 and published alcohol dehydrogenases (ADHs) from Homo sapiens, Drosophila melanogaster, Bombyx mori, and Helicoverpa armigera. MDR‐ADH: medium‐chain dehydrogenase/reductase family; SDR‐ADH: short‐chain dehydrogenase/reductase family.
Figure 3.
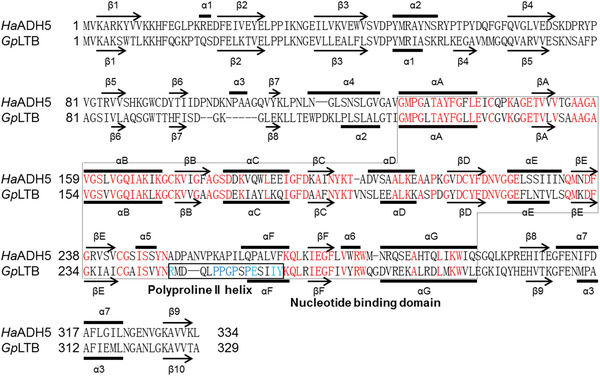
Structure‐based sequence alignment of HaADH5 and GpLTB. GpLTB, guinea pig LTB412‐HD/PGR, Protein Data Bank identification code 1v3u. Secondary structural elements of HaADH5 and GpLTB are indicated by arrows (β‐strand) and bold lines (α‐helix). The nucleotide‐binding domain and the left‐handed polyproline II helix region are surrounded by a thin dotted line and a thick solid line, respectively.
His‐HaADH5 synthesis and purification
A highly expressed band over 45 kD was observed in the Transetta (pET32a‐HaADH5) strain with 0.5 mmol/L IPTG induction at 20 °C for 4 h (released into the supernatant after gentle sonication); this band corresponded to the predicted size of the His‐HaADH5 fusion protein of 53.5 kD (Fig. A). The purification of His‐HaADH5 was performed by Ni2+ affinity chromatography at 4 °C. SDS‐PAGE showed that His‐HaADH5 was well purified with only one band after the two steps with 200 mmol/L imidazole buffer (Fig. B). The amount of the fusion protein estimated by the protein BCA assay was 443.1 μg per 1 L LB induced culture, suggesting sufficient protein purification for activity determination. Testing of the purified fusion protein with anti‐His tag monoclonal antibody (Fig. C) showed specific binding of anti‐His tag to His‐HaADH5, the size of which was slightly less than 55 kD.
Figure 4.
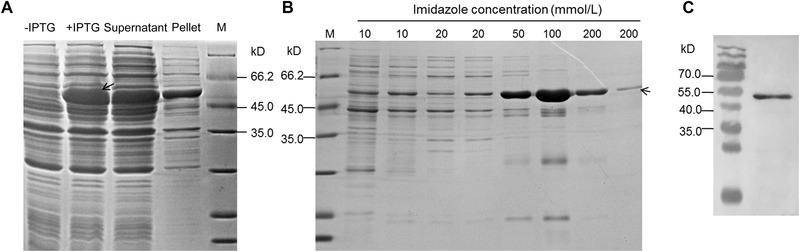
The expression and purification of the His‐HaADH5 fusion protein. (A) Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) for the protein products of the Transetta (pET32a‐HaADH5) strain with isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) induction. (B) SDS‐PAGE tracking the His‐HaADH5 protein purification progress. (C) Western blotting of the purified fusion protein with anti‐His tag mouse monoclonal antibody.
Interaction between HaADH5 and the CYP6B6 promoter in vitro and in yeast
We investigated whether HaADH5 protein could bind the HE1 element of the CYP6B6 promoter using both in vitro and in vivo analyses (Fig. 5). Prokaryotically expressed HaADH5 was used for the binding reaction with the HE1 probe by EMSA. Both 1 μg and 2 μg HaADH5 protein led to a band shift in the positive test with HE1 probe (Fig. A). To confirm the DNA‐binding activity of HaADH5, competition analyses were performed using unlabeled HE1 and an unrelated bHLH DNA fragment. The addition of excess unlabeled HE1 (lane 3) abolished the band shift; in contrast, the band shift was retained when the unrelated bHLH fragment was added (lane 4) (Fig. B). The results in Figure A and B show that HaADH5 could bind to the HE1 element of CYP6B6 in vitro. Transcription activity validation in yeast was also performed to test the interaction between HaADH5 and the 2‐TD responsive region of the CYP6B6 promoter (Fig. C). The pGADT7‐HaADH5 plasmid was transformed into two reporter yeast strains, Y1HGold (p4r‐AbAi) and Y1HGold (p4m‐AbAi), and these cells were coated onto SD/‐Leu and SD/‐Leu/AbA media. The transformed cells exhibiting normal growth on the SD/‐Leu medium demonstrated that the whole process of transformation was feasible. For transformed Y1HGold (p4r‐AbAi), there were four colonies on the SD/‐Leu/AbA medium, whereas no growth was observed for the Y1HGold (p4m‐AbAi) strain. Thus, on +AbA medium, the AbA gene was activated in the Y1HGold (p4r‐AbAi) strain but not in the Y1HGold (p4m‐AbAi) strain, indicating that HaADH5 interacts with the CYP6B6 promoter in yeast. Collectively, these results strongly suggest that HaADH5 can bind to the HE1 element (2‐TD responsive region) of the CYP6B6 promoter in a sequence‐specific manner.
Figure 5.
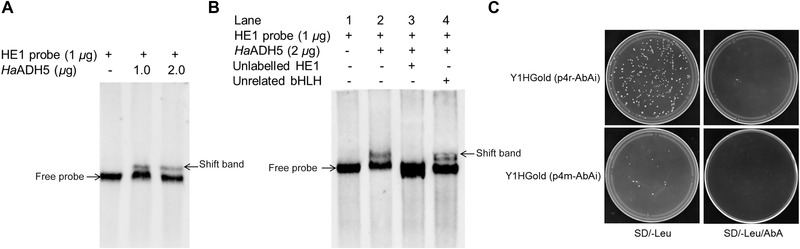
Analysis of the interaction between HaADH5 and the HE1 fragment of the CYP6B6 promoter. (A) Interaction of the HE1 fragment and different amounts of HaADH5 protein. (B) Electrophoretic mobility shift assay (EMSA) competition analysis of HaADH5 and the HE1 fragment. Lane 1: negative control without protein; Lane 2: positive control with 2 μg protein; Lane 3: competition reaction with 2 μg protein and 200‐fold unlabeled HE1 fragment; Lane 4: non‐competition reaction with 2 μg protein and 200‐fold unrelated bHLH fragment. (C) The interaction between HaADH5 and the 2‐tridecanone (2‐TD) responsive region of the CYP6B6 promoter in yeast.
HaADH5 and CYP6B6 expression in the midgut of larvae exposed to 2‐TD
To further investigate whether HaADH5 can respond to 2‐TD, we detected HaADH5 expression in the midgut of 6th instar larvae treated with 10 mg/g 2‐TD. Under treatment, HaADH5 transcript levels significantly increased to the maximum level at 6 h (P < 0.001) and rapidly returned to the normal level at 12 h (P > 0.05). The normal level was maintained until 30 h then decreased significantly to the minimum level at 48 h in the treatment group (P < 0.001). In light of the finding that HaADH5 could bind to the CYP6B6 promoter, CYP6B6 expression was also monitored in response to 2‐TD treatment. Its relative messenger RNA (mRNA) transcript levels were significantly increased at 6 h (P < 0.001) and reached the maximum level at 12 h (P < 0.001) under treatment. After a reduction to the normal level at 20 h, CYP6B6 transcript levels decreased to the minimum level at 48 h (P < 0.05). Both transcripts of HaADH5 and CYP6B6 had significantly reduced expression at 48 h under 2‐TD treatment (Fig. A).
Figure 6.
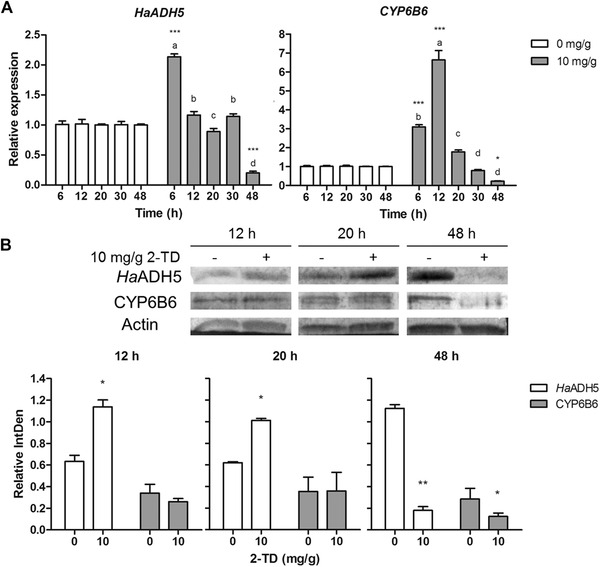
The effect of 2‐tridecanone (2‐TD) on the expression of HaADH5 and CYP6B6 in the midgut of 6th instar larvae. (A) Relative expression of the target genes as indicated by messenger RNA transcript levels. (B) Relative amounts of protein. Different letters above the bars indicate significant differences between different time point samples under the same treatment condition (P < 0.05, one‐way analysis of variance test). Asterisks indicate significant differences between the treatment and the control at the same time point (* P < 0.05, ** P < 0.01, *** P < 0.001, paired t‐test).
Analysis of the relative amount of HaADH5 protein indicated a significant increase at 12 and 20 h in the treatment group compared to the control group (P < 0.05) and a significant decrease at 48 h (P < 0.01). The relative amount of CYP6B6 protein did not change at 12 h and 20 h in the treatment group, but it was significantly reduced at 48 h (P < 0.05). The relative amounts of HaADH5 and CYP6B6 protein were clearly reduced after 2‐TD treatment for 48 h (Fig. B). Correlation analysis was subsequently performed for the relative amounts of HaADH5 and CYP6B6 protein with 2‐TD treatment. The results indicated that the two proteins were positively correlated (y = 0.1931x + 0.0981, R = 0.851, df = 3) in the 10 mg/g treatment group.
Discussion
Insects and plants have complex and sophisticated mechanisms to adapt to each other during coevolution. To escape or survive from attacks by herbivorous insect, plants not only are equipped with physical carriers but also synthesize a kind of toxic defensive compound. On the other hand, herbivorous pests enhance the biological activity of certain detoxifying and metabolic enzymes to degrade these toxic substances for normal growth. Wild tomatoes rely on high concentration of 2‐TD to resist the feeding of several pests, such as H. armigera, Aphis gossypii, Manduca sexta and Leptinotarsa decemlineata (Gonçalves et al., 1998; Lv et al., 2012). 2‐TD can stimulate ecdysone 20‐monooxygenase activity in Spodoptera frugiperda and induce an enhanced level of tolerance to the carbamate insecticide carbaryl in Heliothis zea (Kennedy, 1984; Yu, 1995). Our published research showed that 2‐TD treatment changed expression of CYP6B6 in cotton bollworm and identified the 2‐TD responsive element present in the CYP6B6 promoter (Li et al., 2014). And 2‐TD was confirmed to suppress the 20‐hydroxyecdysone (20E) titer and affect larval weight, pupation rate and adult emergence in cotton bollworm (Zhang et al., 2016).
ADH5 is a central player in formaldehyde detoxification and S‐nitrosoglutathione reduction to regulate oxidative stress, neuronal development, cardiovascular health, immune system balance, carcinogenesis, and single nucleotide polymorphisms (Lima et al., 2009; Leung et al., 2013; Blonder et al., 2014). In this paper, we cloned a new HaADH5 and found its expression significantly increased in a short time from the larval midgut of 10 mg/g 2‐TD treated cotton bollworm. This result consists of the main function of insect ADH5 to work in the metabolism of endogenous compounds and toxic xenobiotics (Park & Kwak, 2009a; Huang et al., 2016). In addition, ADH5 was suggested to be involved in metamorphosis process and mating stage in many Lepidoptera species (Li et al., 2015; He et al., 2017). Although ADH5 is not a typical transcription factor, previous studies showed that ADH5 also localizes to the nucleus and has a nucleotide‐binding domain and a left‐handed polyproline II helix region (Höög et al., 2001; Hori et al., 2004). In our study, the predicted structure of HaADH5 was similar to that of LTB412‐HD/PGR in terms of the presence of a nucleotide‐binding domain. There were four prolines and a helix structure in the polyproline II helix region of LTB412‐HD/PGR, corresponding to a proline percentage of 25%. Although the amino acids in this particular region were not conserved between HaADH5 and LTB412‐HD/PGR, HaADH5 was also found to have a helix structure and four prolines, corresponding to 22.2% of the total amino acids in this region. Therefore, we suggest HaADH5 may contribute to CYP6B6 expression in response to 2‐TD treatment to balance juvenile and ecdysis hormones and regulate the metamorphosis and development of H. armigera.
In this study, we found that HaADH5 can bind to the 2‐TD responsive element of the CYP6B6 promoter both EMSA in vitro and transcription activity validation in yeast. In addition to the new HaADH5, there is another published ADH5 in H. armigera (GenBank accession number: AKD01727.1). We also investigated whether the published ADH5 protein could bind to the CYP6B6 promoter using the same methods. Both analyses indicated that it does not interact with the CYP6B6 HE1 element (Fig. S1). Structure prediction of the published ADH5 protein indicated that it has 15 helices, 14 beta strands, 30 beta turns, and four gamma turns; however, it does not have a helix in the region homologous to the polyproline II helix of LTB412‐HD/PGR, and it only has three prolines in this region, accounting for 16.6% of the total amino acids (data not shown). According to these results, the ability of the presently described HaADH5 to bind and potentially regulate CYP6B6 does not apply to the published ADH5 protein. This dissimilarity is also present among the three isoforms of the transcription factor‐like forkhead box A (FOXA) protein of Homo sapiens (FOXA1, FOXA2, and FOXA3), which each bind to slightly different nucleotide sequences (Tsai et al., 2006).
After finding that HaADH5 could interact with the 2‐TD responsive region of the CYP6B6 promoter, we wanted to know how HaADH5 regulates the expression of CYP6B6 in 6th larval midgut. Before this, we detected the HaADH5 mRNA in all larval stages from the 1st to prepupa and four major tissues of 6th instar larvae including fat body, midgut, integument, and head. The result shows its mRNA existed in all tested samples, and the highest amount was in the prepupa and fat body (Fig. S2). The mRNA and protein of ADH5 were highly expressed during various developmental stages from egg to adult in C. riparius, and in all the tested adult tissues including antennae, thoraxes, abdomens, legs and wings of C. pomonella (Park & Kwak, 2009a; Huang et al., 2016). Moreover, the prepupa is a transition stage from larva to pupa; at this stage, tremendous changes in the concentration of 20‐hydroxyecdysone and juvenile hormone regulate numerous differentially expressed genes for metamorphosis (Zhao et al., 2006). The insect fat body is a central site for the organization of metabolic activity for growth and metamorphosis, and it contains many cell types that metamorphose upon hormone induction. The result of spatial and temporal expression further indicated HaADH5 has an effect on the metamorphosis and development of H. armigera.
Then, we detected the expression of HaADH5 and CYP6B6 in 6th instar larvae in response to 2‐TD treatment. HaADH5 mRNA transcript levels increased the highest at 6 h and gradually decreased to the lowest at 48 h; similarly, CYP6B6 transcripts peaked at 12 h and reached a minimum level at 48 h. There was a temporal order in the increase of transcription between HaADH5 and CYP6B6. The HaADH5 protein were significantly increased and CYP6B6 protein did not change at 12 h and 20 h, while the two proteins were significantly reduced at 48 h and positively correlated in the treatment group. According to these results, we propose that upon 2‐TD treatment, overexpressed HaADH5 can bind to the HE1 element to positively activate CYP6B6 expression in a short timeframe.
Considering the influences of 2‐TD treatment, functions of ADH5 and the ability of HaADH5 to bind to the CYP6B6 promoter, we further propose the hypothesis pathway about HaADH5 regulates CYP6B6 expression to control molting and metamorphosis when larvae are exposed to 2‐TD in H. armigera (Fig. 7). First, kinds of precursors of formaldehyde are produced from cytoplasmic and nuclear sources by toxic 2‐TD, and the expression of HaADH5 is increased at the same time, then the catalyzed reactions of formaldehyde metabolism are enhanced and overexpress HaADH5 actively or passively enter the nucleus. The HaADH5 maybe form a dimerization transcript factor with an unknown protein and bind with the HE1 element of CYP6B6 DNA. Subsequently, the transcription of CYP6B6 is activated and increased in the nucleus, the overexpressed CYP6B6 is released to cytoplasm and directly affect the downstream molting and metamorphosis processes. Surely, this hypothesis warrants further investigation using other methods in our next steps.
Figure 7.
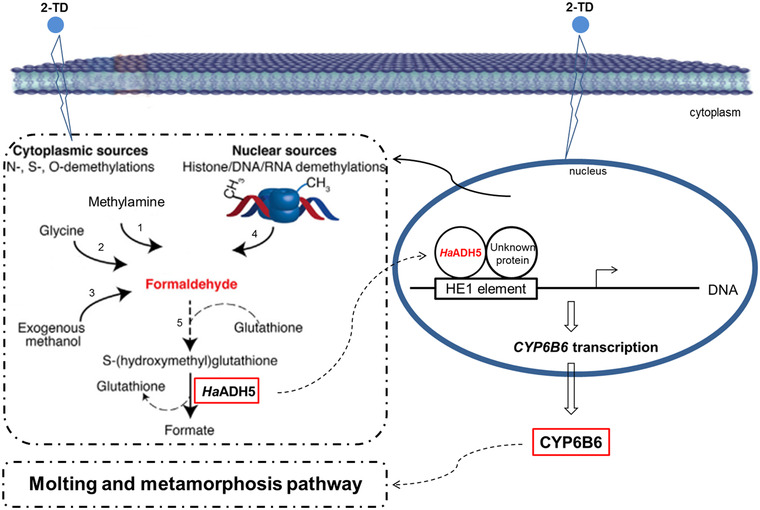
The hypothesis pathway of HaADH5 regulates CYP6B6 expression to control growth and development in Helicoverpa armigera after 2‐tridecanone (2‐TD) treatment. The red font and box display the content of compound or protein was increased in the processes. 1: Semicarbazide‐sentivite amine oxidase; 2: myeloperoxidase; 3: alcohol dehydrogenase 1 (ADH1) and catalase; 4: protein and nucleic acid demethylases; 5: spontaneous reaction.
In conclusion, the present findings show that HaADH5 could bind with the HE1 element of CYP6B6 promoter and activate CYP6B6 transcription in response to 2‐TD treatment. Therefore, HaADH5 could potentially be used as a molecular marker to study detoxification mechanisms triggered by exogenous substances and also as a molecular target to control the growth and development of cotton bollworm.
Disclosure
The authors have no conflict of interest, including specific financial interests and relationships and affiliations relevant to the subject of this manuscript.
Supporting information
Fig. S1. The interaction between the published ADH5 and HE1 fragment of CYP6B6 promoter.
Fig. S2. The temporal and spatial expression profile of the HaADH5 in H. armigera.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31471781), Natural Science Foundation of Xinjiang Region in China (2016D01C042) and Tianshan Cedar Project in 2017 (2017xs20).
References
- Ai, X.Y. , Zhao, J. , Wang, Y.M. , Wei, Y.J. and Liu, X.N. (2017) Optimization of prokaryotic expression of soluble alcohol dehydrogenase from Helicoverpa armigera . Biotechnology, 27(1), 65–70. [Google Scholar]
- Anand, P. , Hausladen, A. , Wang, Y.J. , Zhang, G.F. , Stomberski, C. , Brunengraber, H. et al (2014) Identification of S‐nitroso‐CoA reductases that regulate protein S‐nitrosylation. Proceedings of the National Academy of Science USA, 111(52), 18572–18577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blonder, J.P. , Mutka, S.C. , Sun, X.C. , Qiu, J. , Green, L.H. , Mehra, N.K. et al (2014) Pharmacologic inhibition of S‐nitrosoglutathione reductase protects against experimental asthma in BALB/c mice through attenuation of both bronchoconstriction and inflammation. Pulmonary Medicine, 14, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos, B.G. , Wit, N. , Meiser, J. , Dingler, F.A. , Pietzke, M. , Mulderrig, L. et al (2017) Mammals divert endogenous genotoxic formaldehyde into one‐carbon metabolism. Nature, 548, 549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra Sharath, G. , Asokan, R. , Manamohan, M. , Kumar, K.N. and Sita, T. (2014) Evaluation of reference genes for quantitative real‐time PCR normalization in cotton bollworm, Helicoverna armigera . Molecular Biology (Mosk), 48, 927–938. [PubMed] [Google Scholar]
- Chase, J.R. , Poolman, M.G. and Fell, D.A. (2009) Contribution of NADH increases to ethanol's inhibition of retinol oxidation by human ADH isoforms. Alcoholism: Clinical and Experimental Research, 33, 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti, A. , Franzini, M. , Scataglini, I. and Pompella, A. (2014) Mechanisms and targets of the modulatory action of S‐nitrosoglutathione (GSNO) on inflammatory cytokines expression. Archives of Biochemistry and Biophysics, 562, 80–91. [DOI] [PubMed] [Google Scholar]
- Danielsson, O. , Atrian, S. , Luque, T. , Hjelmqvist, L. , Gonzàlez, D.R. and Jörnvall, H. (1994) Fundamental molecular differences between alcohol dehydrogenase classes. Proceedings of the National Academy of Sciences USA, 91, 4980–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltour, L. , Foglio, M.H. and Duester, G. (1999) Metabolic deficiencies in alcohol dehydrogenase Adh1, Adh3, and Adh4 null mutant mice. Journal of Biological Chemistry, 274, 16796–16801. [DOI] [PubMed] [Google Scholar]
- Fernández, M.R. , Biosca, J.A. and Parés, X. (2003) S‐nitrosoglutathione reductase activity of human and yeast glutathione‐dependent formaldehyde dehydrogenase and its nuclear and cytoplasmic localisation. Cellular and Molecular Life Sciences, 60, 1013–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves, M.I.F. , Maluf, W.R. , Gomes, L.A.A. and Barbosa, L.V. (1998) Variation of 2‐tridecanone level in tomato plant leaflets and resistance to two mite species (Tetranychus sp.). Euphytica, 104, 33–38. [Google Scholar]
- Gu, S.H. , Wu, K.M. , Guo, Y.Y. , Pickett, J.A. , Field, L.M. , Zhou, J.J. et al (2013) Identification of genes expressed in the sex pheromone gland of the black cutworm Agrotis ipsilon with putative roles in sex pheromone biosynthesis and transport. BMC Genomics, 14:636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, P. , Zhang, Y.F. , Hong, D.Y. , Wang, J. , Wang, X.L. , Zuo, L.H. et al (2017) A reference gene set for sex pheromone biosynthesis and degradation genes from the diamondback moth, Plutella xylostella, based on genome and transcriptome digital gene expression analyses. BMC Genomics, 18: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, D.T. , Matsumoto, A. , Kim, S.O. , Marshall, H.E. and Stamler, J.S. (2005) Protein S‐nitrosylation: Purview and parameters. Nature Reviews Molecular Cell Biology, 6, 150–166. [DOI] [PubMed] [Google Scholar]
- Höög, J.O. , Hedberg, J.J. , Strömberg, P. and Svensson, S. (2001) Mammalian alcohol dehydrogenase‐functional and structural implications. Journal Biomedical Science, 8, 71–76. [DOI] [PubMed] [Google Scholar]
- Hori, T. , Yokomizo, T. , Ago, H. , Sugahara, M. , Ueno, G. , Yamamoto, M. et al (2004) Structural basis of leukotriene B4 12‐hydroxydehydrogenase/15‐Oxo‐prostaglandin 13‐reductase catalytic mechanism and a possible Src homology 3 domain binding loop. The Journal of Biological Chemistry, 279, 22615–22623. [DOI] [PubMed] [Google Scholar]
- Huang, L.N. , Cheng, T.T. , Wang, X.H. , Wei, Y.J. , Zhao, J. , Li, J.Y. et al (2015) The cloning, expression and preparation of polyclonal antibody of β‐actin gene from Helicoverpa armigera . Biotechnology Bulletin, 31(12), 138–145. [Google Scholar]
- Huang, X.L. , Liu, L. , Su, X.J. and Feng, J.N. (2016) Identification of biotransformation enzymes in the antennae of codling moth Cydia pomonella . Gene, 580, 73–79. [DOI] [PubMed] [Google Scholar]
- Jensen, D.E. , Belka, G.K. and DuBois, G.C. (1998) S‐Nitrosoglutathione is a substrate for rat alcohol dehydrogenase class III isoenzyme. Biochemical Journal, 331, 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, R. , Holmquist, B. , Hempel, J. , Vallee, B.L. and Jörnvall, H. (1988) Class III human liver alcohol dehydrogenase: a novel structural type equidistantly related to the class I and class II enzymes. Biochemistry, 27, 1132–1140. [DOI] [PubMed] [Google Scholar]
- Kawanishi, M. , Matsuda, T. and Yag, T. (2014) Genotoxicity of formaldehyde: molecular basis of DNA damage and mutation. Frontiers in Environmental Science, 2, 1–8. [Google Scholar]
- Kennedy, G.G. (1984) 2‐tridecanone, tomatoes and Heliothis zea: potential incompatibility of plant antibiosis with insecticidal control. Entomologia Experimentalis et Applicata, 35, 305–311. [Google Scholar]
- Koivusalo, M. , Baumann, M. and Uotila, L. (1989) Evidence for the identity of glutathione‐dependent formaldehyde dehydrogenase and class III alcohol dehydrogenase. FEBS Letters, 257, 105–109. [DOI] [PubMed] [Google Scholar]
- Li, F. , Liu, X.N. , Zhu, Y. , Ma, J. , Liu, N. and Yang, J.H. (2014) Identification of the 2‐tridecanone responsive region in the promoter of cytochrome P450 CYP6B6 of the cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). Bulletin of Entomological Research, 104, 801–808. [DOI] [PubMed] [Google Scholar]
- Li, Z.Q. , Zhang, S. , Luo, J.Y. , Wang, C.Y. , Lv, L.M. , Dong, S.L. et al (2015) Transcriptome comparison of the sex pheromone glands from two sibling Helicoverpa species with opposite sex pheromone components. Scientific Reports, 10.1038/srep09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima, B , Lam, G.K.W. , Xie, L. , Diesen, D.L. , Villamizar, N. , Nienaber, J. et al (2009) Endogenous S‐nitrosothiols protect against myocardial injury. Proceedings of the National Academy of Sciences USA, 106, 6297–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.N. , Liang, P. , Gao, X.W. and Shi, X.Y. (2006) Induction of the cytochrome P450 activity by plant allelochemicals in the cotton bollworm, Helicoverpa armigera (Hübner). Pesticide Biochemistry and Physiology, 84, 127–134. [Google Scholar]
- Leung, J. , Wei, W. and Liu, L. (2013) S‐nitrosoglutathione reductase deficiency increases mutagenesis from alkylation in mouse liver. Carcinogenesis, 34, 984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, X.G. , Kranzler, H.R. , Zuo, L.J. , Zhang, H.P. , Wang, S. and Gelernter, J. (2008) ADH7 variation modulates extraversion and conscientiousness in substance dependent subjects. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 147, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, M. , Sun, H.H. , Wang, L.H. and Gao, X.W. (2012) Effects of secondary metabolites on activities of glutathione S‐transferases, carboxylesterase in aphid. Chinese Agricultural Science Bulletin, 28, 253–256. [Google Scholar]
- Martínez, R.A. , Cadenas, S. and Lamas, S. (2011) Nitric oxide signaling: classical, less classical, and nonclassical mechanisms. Free Radical Biology and Medicine, 51, 17–29. [DOI] [PubMed] [Google Scholar]
- Oscar, B.M. and Marinkovic, K. (2003) Alcoholism and the brain: an overview. Alcohol Research and Health, 27, 125–133. [PMC free article] [PubMed] [Google Scholar]
- Östberg, L.J. , Persson, B. and Höög, J.O. (2016) Computational studies of human class V alcohol dehydrogenase‐the odd sibling. BMC Biochemistry, 17: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, J. , Deng, H.M. , Zheng, S.C. , Huang, L.H. , Feng, Q.L. and Liu, L. (2014) Transcriptomic analysis of developmental features of Bombyx mori wing disc during metamorphosis. BMC Genomics, 15(1): 820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, K. and Kwak, I.S. (2009a) Alcohol dehydrogenase gene expression in Chironomus riparius exposed to di(2‐ethylhexyl) phthalate. Comparative Biochemistry and Physiology, 150, 361–367. [DOI] [PubMed] [Google Scholar]
- Park, K. and Kwak, I.S. (2009b) Characterization and expression of Chironomus riparius alcohol dehydrogenase gene under heavy metal stress. Environmental Health and Toxicology, 24, 107–117. [Google Scholar]
- Pontel, L.B. , Rosado, I.V. , Burgos, B.G. , Garaycoechea, J.I. , Yu, R. , Arends, M.J. et al (2015) Endogenous formaldehyde is a hematopoietic stem cell genotoxin and metabolic carcinogen. Molecular Cell, 60, 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchandani, V.A. , Bosron, W.F. and Li, T.K. (2001) Research advances in ethanol metabolism. Pathologie Biologie, 49, 676–682. [DOI] [PubMed] [Google Scholar]
- Shakeel, M. , Zhu, X. , Kang, T.H. , Wan, H. and Li, J.H. (2015) Selection and evaluation of reference genes for quantitative gene expression studies in cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). Journal of Asia‐Pacific Entomology, 18, 123–130. [Google Scholar]
- Smith, B.C. and Marletta, M.A. (2012) Mechanisms of S‐nitrosothiol formation and selectivity in nitric oxide signaling. Current Opinion in Chemical Biology, 16, 498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts, A.S. and Appling, D.R. (2010) Compartmentalization of mammalian folate‐mediated one‐carbon metabolism. Annual Review of Nutrition, 30, 57–81. [DOI] [PubMed] [Google Scholar]
- Tsai, K.L. , Huang, C.Y. , Chang, C.H. , Sun, Y.J. , Chuang, W.J. and Hsiao, C.D. (2006) Crystal structure of the human FOXK1a‐DNA complex and its implications on the diverse binding specificity of winged helix/forkhead proteins. The Journal of Biological Chemistry, 281(25), 17400–17409. [DOI] [PubMed] [Google Scholar]
- Vandesompele, J. , De Preter, K. , Pattyn, F. , Poppe, B. , Van Roy, N. , De Paepe, A. et al (2002) Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3(7), research0034.1‐0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, H. , Heidel, A.J. , Heckel, D.G. and Groot, A.T. (2010) Transcriptome analysis of the sex pheromone gland of the noctuid moth Heliothis virescens . BMC Genomics, 11, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, W. , Li, B. , Hanes, M.A. , Kakar, S. , Chen, X. and Liu, L.M. (2010) S‐nitrosylation from GSNOR deficiency impairs DNA repair and promotes hepatocarcinogenesis. Science Translational Medicine, 2(19): 19ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y.J. , Zhang, L , Cheng, T.T. , Ma, J. and Liu, X.N. (2015) Preparation of the polyclonal antibody against P450 CYP6B6 from Helicoverpa armigera . Life Science Research, 19(6), 497–500. [Google Scholar]
- Yang, Z.N. , Bosron, W.F. and Hurley, T.D. (1997) Structure of human chi chi alcohol dehydrogenase: a glutathione‐dependent formaldehyde dehydrogenase. Journal of Molecular Biology, 265, 330–343. [DOI] [PubMed] [Google Scholar]
- Yu, C.H. , Gao, X.W. and Zheng, B.Z. (2002) Induction of the cytochrome P450 by 2‐tridecanone in Helicoverpa armigera . Acta Entomologica Sinica, 45, 1–7. [Google Scholar]
- Yu, S.J. (1995) Allelochemical stimulation of ecdysone 20‐monooxygenase in fall armyworm larvae. Archives Insect Biochemistry and Physiology, 28, 365–375. [Google Scholar]
- Zhang, L. , Lu, Y. , Xiang, M. , Shang, Q.L. and Gao, X.W. (2016) The retardant effect of 2‐tridecanone, mediated by cytochrome P450, on the development of cotton bollworm, Helicoverpa armigera . BMC Genomics, 17, 954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Liu, N. , Ma, J. , Huang, L.N. and Liu, X.N. (2016a) Effect of silencing CYP6B6 of Helicoverpa armigera (Lepidoptera: Noctuidae) on its growth, development, and insecticide tolerance. Journal of Economic Entomology, 109(6), 2506–2516. [DOI] [PubMed] [Google Scholar]
- Zhao, J. , Liu, X.N. , Li, F. , Zhuang, S.Z. , Huang, L.N. , Ma, J. et al (2016b) Yeast one‐hybrid screening the potential regulator of CYP6B6 overexpression of Helicoverpa armigera under 2‐tridecanone stress. Bulletin of Entomological Research, 106, 182–190. [DOI] [PubMed] [Google Scholar]
- Zhao, X.F. , He, H.J. , Dong, D.J. and Wang, J.X. (2006) Identification of differentially expressed proteins during larval molting of Helicoverpa armigera . Journal of Proteome Research, 5, 164–169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The interaction between the published ADH5 and HE1 fragment of CYP6B6 promoter.
Fig. S2. The temporal and spatial expression profile of the HaADH5 in H. armigera.


