Summary
The mTERF gene family encodes for nucleic acid binding proteins that are predicted to regulate organellar gene expression in eukaryotes. Despite the implication of this gene family in plant development and response to abiotic stresses, a precise molecular function was assigned to only a handful number of its c. 30 members in plants.
Using a reverse genetics approach in Arabidopsis thaliana and combining molecular and biochemical techniques, we revealed new functions for the chloroplast mTERF protein, MDA1.
We demonstrated that MDA1 associates in vivo with components of the plastid‐encoded RNA polymerase and transcriptional active chromosome complexes. MDA1 protein binds in vivo and in vitro with specificity to 27‐bp DNA sequences near the 5′‐end of psbE and ndhA chloroplast genes to stimulate their transcription, and additionally promotes the stabilization of the 5′‐ends of processed psbE and ndhA messenger (m)RNAs. Finally, we provided evidence that MDA1 function in gene transcription likely coordinates RNA folding and the action of chloroplast RNA‐binding proteins on mRNA stabilization.
Our results provide examples for the unexpected implication of DNA binding proteins and gene transcription in the regulation of mRNA stability in chloroplasts, blurring the boundaries between DNA and RNA metabolism in this organelle.
Keywords: DNA, gene expression, helical repeat protein, mTERF, plastid, RNA
Introduction
Owing to their endosymbiotic evolution, chloroplasts retained only c. 100 genes of their cyanobacterial ancestor genome (Sato et al., 1999) that encode messenger (m)RNAs of proteins involved in photosynthesis and a small fraction of the components of the chloroplast gene expression machinery (ribosomal proteins, rRNAs, tRNAs, plastid‐encoded RNA polymerase). Thus, the expression of chloroplast genes requires the import of hundreds of proteins that are encoded by nuclear genes (reviewed in Barkan, 2011). Consistent with their function in gene expression, many of these proteins bind to DNA or RNA in vivo. Some of these protein families are found only in eukaryotes and are specialized in the regulation of organellar genes (reviewed in Hammani et al., 2014). One such example is the mTERF (mitochondrial transcription termination factor) family. mTERF proteins are made of tandem repeats of a degenerate c. 31 amino acid motif that folds into three helices. These repeats stack to form a superhelix structure that is predicted to accommodate double‐stranded DNA in its central groove (Jimenez‐Menendez et al., 2010; Yakubovskaya et al., 2010). The mTERF family in metazoans includes four to five members that have preponderantly been implicated in DNA‐related functions in mitochondria like gene transcription or replication (reviewed in Roberti et al., 2009). By contrast, the family expanded to c. 30 members in higher plants (Babiychuk et al., 2011; Kleine, 2012; Zhao et al., 2014) and recent studies have suggested that this expansion has been accompanied by a functional diversification in RNA metabolism such as intron splicing and rRNA maturation in organelles (Hammani & Barkan, 2014; Hsu et al., 2014; Romani et al., 2015; Zhang et al., 2018). In addition to these studies, transcriptomic and physiological analyses conducted in Arabidopsis and crop species have highlighted the importance of mTERF genes for plant response to a variety of abiotic stresses (Zhao et al., 2014; Zhou et al., 2016; Xu et al., 2017; Robles et al., 2018; Nunez‐Delegido et al., 2019). Nevertheless, only a handful of mTERF genes have been characterized molecularly and biochemically in plants to better understand their functional diversification. To get deeper understanding of mTERF functions in plants, we conducted a reverse genetics approach with genes that had not been clearly characterized. Here, we describe new molecular and biochemical functions for the Arabidopsis mTERF protein, At4g14605, previously known as MDA1 (Robles et al., 2012) or mTERF5 which had been proposed to act as a positive regulator of psbE‐F‐L‐J genes transcription in Arabidopsis chloroplasts (Ding et al., 2019). We report the discovery of an additional site of action for MDA1 in the ndhA gene and evidence for a functional model in which MDA1 promotes the stabilization of the 5′‐ends of processed psbE and ndhA mRNAs besides their gene transcription. These findings provide examples for the unexpected implication of DNA binding proteins and gene transcription in the regulation of mRNA stability in chloroplasts.
Materials and Methods
Oligonucleotides used in this study are listed in Supporting Information Table S1.
Plant material
Arabidopsis thaliana ecotype Columbia (Col‐0) and Nicotania benthamiana were used in this study. The T‐DNA insertion mutant allele mda1‐2 (SAIL_425_E03) was obtained from the ABRC Stock Center. The hcf111‐1 allele was retrieved from a collection of EMS mutagenized Arabidopsis plants displaying high‐chlorophyll fluorescence (Meurer et al., 1996). Complemented mutants were obtained via Agrobacterium tumefaciens transformation of mda1‐2 homozygote plants. The binary vector (pGWB17) used for Agrobacterium‐transformation expressed the At4g14605 coding sequence fused with a 4xMyc C‐terminal tag under the control of the CaMV 35S promoter. Transgenic plants were selected on Murashige & Skoog (MS) plates containing 25 μg ml−1 hygromycin. All experiments were performed using 7‐d‐old plants grown in vitro (1 × MS, pH 5.7, 0.5% sucrose, 0.8% Agar; 16 h : 8 h, light : dark cycles; light intensity 65–85 μmol photons m−2 s−1) or 14‐d‐old plants grown on soil for immunoprecipitation combined with mass spectrometry experiments. The methods for measuring plant chlorophyll fluorescence are provided in Methods S1.
Protein extraction and immunoblotting
Total proteins were extracted in Tris pH 7.5, 10% glycerol, 1% NP40, 5 mM EDTA, 2 mM EGTA, 35 mM β‐mercaptoethanol, 1 × EDTA‐free protease inhibitor cocktail (Roche), resolved on SDS‐PAGE, and transferred onto PVDF membrane at 80 V for 1.5 h using wet transfer. Anti‐PsaD, ‐PetD antibodies were kindly donated by Alice Barkan (University of Oregon). Anti‐NdhL and ‐NdhB antibodies were kind donations of Toshiharu Shikanai (University of Kyoto) and anti‐RbcL antibodies were donated by Géraldine Bonnard (CNRS). Other antibodies against chloroplast proteins were purchased from Agrisera or PhytoAB and anti‐Myc antibodies (clone 9E10) from Sigma‐Aldrich.
Subcellular localization of MDA1
Nicotiana benthamiana leaves were infiltrated with A. tumefaciens GV3101 carrying pMDC83:MDA1 or pB7RWG2:RAP, each at OD600 = 0.5. Protoplasts were prepared as in Berglund et al. (2009) and examined under a Zeiss LSM 780 confocal microscope. Green fluorescent protein (GFP) was excited at 488 nm and emission was acquired between 493 and 556 nm. Red fluorescent protein (RFP) and chlorophyll were excited at 561 nm and emissions were acquired between 588–641 nm and 671–754 nm, respectively.
RNA extraction and analyses
Tissues were ground in liquid nitrogen and RNA was extracted with Trizol following manufacturer's protocol (Invitrogen). RNA was further extracted with phenol‐chloroform pH 4.3, and 5 μg Turbo DNase (Thermo Fisher, France) treated RNAs were used for Superscript IV reverse transcription with random hexamers. The resulting cDNA was diluted 20‐fold for quantitative (q)PCR reaction. ACT2 (AT3G18780) and TIP41 (AT1G13440) were used as reference genes. For RNA gel blotting, 5 μg (psbE operon genes) or 15 μg (ndhH operon genes) of RNA was fractionated on 1.2% agarose–1% formaldehyde gel and blotted as described previously (Barkan, 1998). Strand‐specific 60‐mer synthetic DNA oligonucleotides were used as probes (Table S1). For sRNA blotting, 5–10 μg low molecular weight (MW) RNAs enriched from total leaf RNA as in Lu et al. (2007) were blotted as described in Zhelyazkova et al. (2012). Results were visualized on an Amersham Typhoon imager and data quantification was performed with imagequant TL (GE Healthcare). Details about the tiling microarray plastid transcriptome and translatome analyses are provided in Methods S1.
RNA structure prediction
Secondary structures were predicted with the Mfold server (RNA folding form v.2.3 energies) at http://unafold.rna.albany.edu/ using default parameters and a folding temperature of 25°C.
Chloroplast isolation
Chloroplasts were purified by density gradient and differential centrifugations as described in (Kunst, 1998).
Chloroplast fractionation
Chloroplasts were lysed in 30 mM HEPES‐KOH pH 8, 10 mM MgOAc, 60 mM KOAc, 1 mM DTT, 1 × EDTA‐free protease inhibitor cocktail, 1 mM PMSF with or without the addition of 0.2 M Na2CO3, 1% NP‐40, 100 μg ml−1 RNase A, 250 U ml−1 RNase T1 or 50 U ml−1 DNase I (Thermo Fisher). Stromal and thylakoid proteins were separated by centrifugation at 20 000 g for 10 min at 4°C.
Transcription run‐on assay
The rate of transcription from 5 × 107 chloroplasts was analyzed as in Zubo et al. (2011). Results were visualized and signals quantified as described before.
Co‐immunoprecipitation (coIP)‐MS
Chloroplasts were cross‐linked in 300 mM Sorbitol, 30 mM HEPES‐KOH pH 8, 2 mM DSP for 1 h on ice. Chloroplasts were then resuspended in 20 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP‐40, 1 × EDTA‐free protease inhibitor cocktail, 1 mM PMSF, centrifuged for 20 min at 21 000 g, 4°C. Protein aliquots (2.5 mg) from the supernatant then were immunoprecipitated by the addition of 50 μl anti‐MYC Miltenyi magnetic beads and incubation for 30 min at 4°C on a rotator. Beads were washed and eluted as recommended by the manufacturer. Eluted proteins were digested with sequencing‐grade trypsin (Promega) and analyzed by nanoLC‐MS/MS at the ‘Plateforme Proteomic Strasbourg‐Esplanade’. Data were processed as described (Lange et al., 2019). A home‐made R package ipinquiry was used to identify significant MDA1 protein interactors by a statistical analysis on spectral counts using a negative binomial GLM model as described (Lange et al., 2019). The full list of protein interactants is provided in Table S4.
DIP‐qPCR
Chloroplasts were cross‐linked in 1 ml of 300 mM Sorbitol, 30 mM HEPES‐KOH pH 8, 1% formaldehyde during 30 min on ice. Cross‐linking was stopped by adding glycine to 125 mM and incubation on ice for 5 min. Chloroplasts were recovered by centrifugation and lysed in 1 ml DIP buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X100, 0.1% SDS, 0.1% Na‐deoxycholate, 1 × EDTA‐free protease inhibitor cocktail). Chloroplast DNA was sheared to 0.2–0.8 kb using a bioruptor on high settings, 30 s ON, 30 s OFF, 5 min, four times. For each IP, 2.5 mg proteins were used with 50 μl anti‐MYC Miltenyi magnetic beads and incubated on a rotator for 30 min at 4°C. Beads were washed four times with DIP buffer and eluted in 200 μl hot 100 mM NaHCO3, 1% SDS. Samples were reverse cross‐linked by adding 15 μl of 3 M NaCl, incubated overnight at 65°C and subsequently digested with 20 μg proteinase K, 10 mM EDTA at 37°C for 1 h. RNA was removed by adding 1 μl RNase A/T1 (Thermo Fisher) and incubation for 15 min at 37°C. DNA from the IP and supernatant fractions was extracted with phenol/chloroform and used at 1 : 50 dilution in qPCR reactions. Percentage recovery was calculated using the formula: 100 × 2−(Ct(IP) − Ct(sup)) (Saleh et al., 2008).
Recombinant MDA1 production
MDA1 coding sequence without the first 387 bp that are predicted to encode the chloroplast transit peptide was amplified by PCR on cDNA and cloned into BamHI and SalI in pMAL vector. The recombinant N‐terminal MBP fusion protein was expressed in Escherichia coli BL21 and purified as described in (Williams‐Carrier et al., 2008) except that the lysis buffer contained 250 mM NaCl and did not include CHAPS detergent.
In vitro binding assays
Synthetic DNA or RNA probes (Integrated DNA Technologies) were 5′ end‐labelled with [γ‐32P]‐ATP and T4 polynucleotide kinase and then purified by illustraTM Microspin G‐25 column filtration (GE Healthcare). dsDNA or RNA probes <60 bp were obtained by annealing two complementary oligonucleotide sequences. DNA probes >60 bp were amplified by PCR using gDNA as template, agarose gel purified and [γ‐32P]‐ATP 5′‐end‐labelled as described above. Except where otherwise indicated, rMDA‐binding reactions contained 50 mM Tris·HCl pH 7.5, 150 mM KCl, 4 mM DTT, 0.04 mg ml−1 BSA, 0.25 mM EDTA, 0.05 mg ml−1 poly(dI‐dC) competitor, 0.25% Tween‐20, 10% (v/v) glycerol and 15 pM radiolabelled probe. Poly(dI‐dC) was substituted by 10 units of RNasin (Promega) in RNA‐binding assays. Reactions were incubated for 30 min at 25°C and resolved on 5% native polyacrylamide gels in 1 × TBE. DNase I footprint assays were performed in similar binding conditions in 20‐μl volumes. First, binding reactions containing 5FAM‐end‐labelled psbE2 DNA probe in the presence or absence of rMDA (0.5 μM) were incubated for 30 min at 25°C. Then 10 μl of 0.015 U of DNase I in 0.5 mM CaCl2, 12.5 mM MgCl2 buffer subsequently was added and the reactions incubated for 5 min at 25°C. The reactions were brought to 25 mM EDTA, 0.125% SDS, 200 mM sodium acetate to stop cleavage and DNA fragments were purified by phenol/chloroform extraction and ethanol precipitation. Two hundred nanograms of DNA products for each reaction were mixed with 0.33 μl GeneScan™ 400HD ROX™ dye Size Standard and fractionated by automated fluorescent capillary electrophoresis on an ABI Prism 3130 XL Genetic Analyzer (Applied Biosystems). Product peaks were aligned with genemapper software to a sequencing ladder generated with a USB Thermo Sequenase Cycle Sequencing Kit.
Results
MDA1 encodes an mTERF‐repeat protein required for chloroplast biogenesis
At4g14605 encodes the chloroplast mTERF protein MDA1 that previously has been implicated in chloroplast development and abiotic stress responses in Arabidopsis, but whose molecular function had not been characterized (Robles et al., 2012). The MDA1 gene contains four exons and encodes a 493 amino acid protein harbouring eight mTERF tandem repeats and a predicted 43 amino acid chloroplast N‐terminal transit peptide (Fig. 1a). Homozygote mda1 mutant plants for the T‐DNA insertion line SAIL_425_E03 (mda1‐2) were obtained. The analysis of the T‐DNA flanking sequence tags in mda1‐2 revealed the presence of an inverted T‐DNA repeat arrangement inserted in the fourth exon of MDA1 gene between genomic positions +1133 and +1154. Disruption of MDA1 led to pale leaf pigmentation and a dwarf phenotype (Fig. 1b). Despite their severe phenotype, the mda1‐2 plants were fertile and produced siliques containing seeds. The introduction of a wild‐type (WT) copy of MDA1 gene into mda1‐2 fully restored the WT phenotype demonstrating that mda1‐2 phenotype resulted from MDA1 disruption. Reverse transcription (RT)‐PCR experiments conducted on cDNAs from WT, mda1‐2 and complemented plants using primers amplifying the full‐length MDA1 gene confirmed that the mda1‐2 mutant is a knockout (Fig. 1c).
Fig. 1.
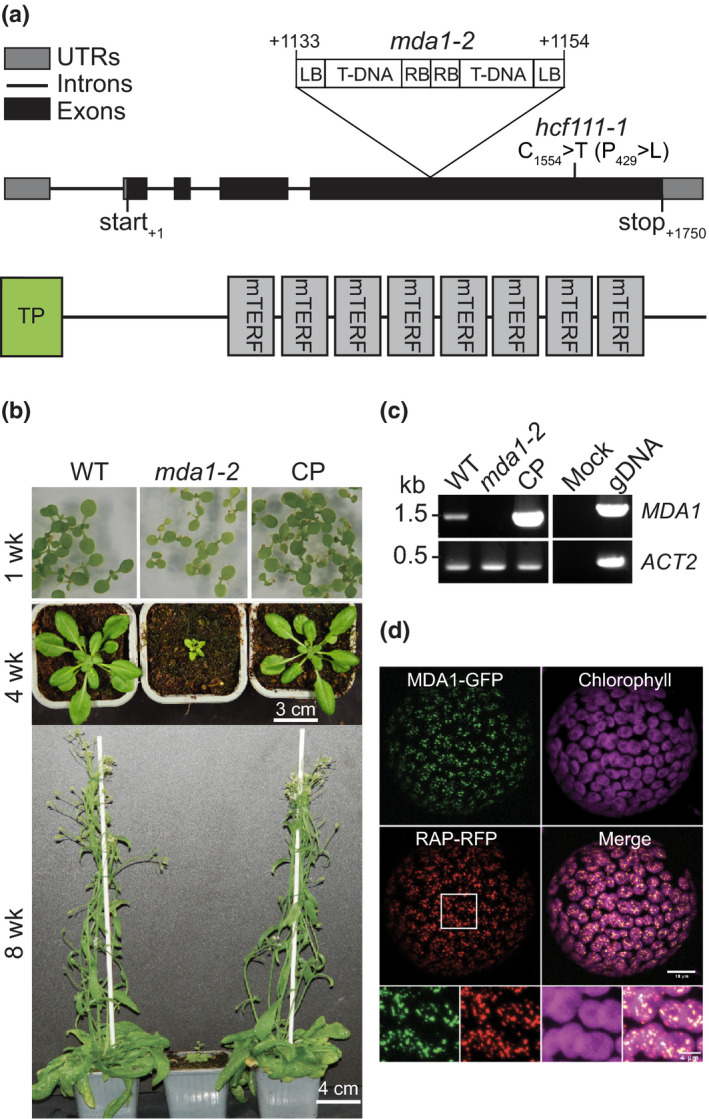
Characterization of mda1 mutant plants in Arabidopsis. (a) Schematic representations of the MDA1 gene and protein with the locations of mutant alleles used in this study. TP, putative chloroplast transit peptide. (b) Phenotypes of wild‐type (WT), mda1‐2 mutant and complemented (CP) plants grown in medium or soil at several developmental stages. wk, week. (c) Reverse transcription (RT)‐PCR analysis of mda1 gene expression in WT, mda1‐2 and CP plants. Genomic DNA (gDNA) template was used as a PCR positive control and ACTIN‐2 gene (ACT2) serves as the internal RT‐PCR control. (d) Transient coexpression assay of MDA1‐GFP and RAP‐RFP fusion proteins in tobacco protoplasts. Fluorescence images of green and red fluorescent protein (GFP, RFP) and chlorophyll as well as a merged image are shown. Magnified views of the region delineated by a white box are provided below.
A second mutant allele for MDA1 was retrieved independently from a collection of EMS mutagenized Arabidopsis plants displaying high chlorophyll fluorescence (Meurer et al., 1996) and was designated hcf111‐1. hcf111‐1 harbours a cytosine‐to‐thymine transition in the fourth exon of At4g15605 converting a proline‐to‐leucine residue in the last mTERF domain of MDA1 (Fig. 1a). hcf111‐1 plants grown on soil displayed a pale leaf and dwarf phenotype similar to mda1‐2 allele (Fig. S1a).
In order to establish the intracellular localization of MDA1, an MDA1‐GFP fusion protein was transiently expressed in tobacco leaves and leaf protoplasts were examined by confocal microscopy (Fig. 1d). The GFP fusion protein was detected in discrete foci within chloroplasts, which colocalize with the fluorescence signal of a coexpressed chloroplast nucleoid‐associated protein RAP, fused with RFP (Kleinknecht et al., 2014). Thus, these results demonstrate that MDA1 localizes to chloroplasts where it is associated with the nucleoids. Immunodetection on chloroplast subfractions isolated from complemented mutant plants expressing a 4xMyc epitope tagged version of MDA1 revealed that MDA1 was only detected in the membrane fraction consistent with the association of chloroplast nucleoids to membranes (Fig. S2; Sato et al., 2003). Altogether, these results indicate that MDA1 is a chloroplast protein and plays an important role in chloroplast biogenesis and plant development.
The accumulation of PSII and NDH subunits is impaired in mda1 mutants
In order to identify the defect in chloroplast biogenesis in mda1 more precisely, we performed immunoblot analyses on individual subunits of chloroplast protein complexes (Photosystem (PS) I and II, NADH dehydrogenase (NDH), Cytochrome b 6 f, ATP synthase, Rubisco and the ribosome) (Fig. 2). The immunoblot results showed that subunits of the PSII and NDH complexes were particularly decreased in mda1‐2 (c. 10% of WT level) and that their accumulation was fully restored in the complemented line. In addition, a moderate loss of the PSI subunit PsaD could be observed in mda1‐2.
Fig. 2.
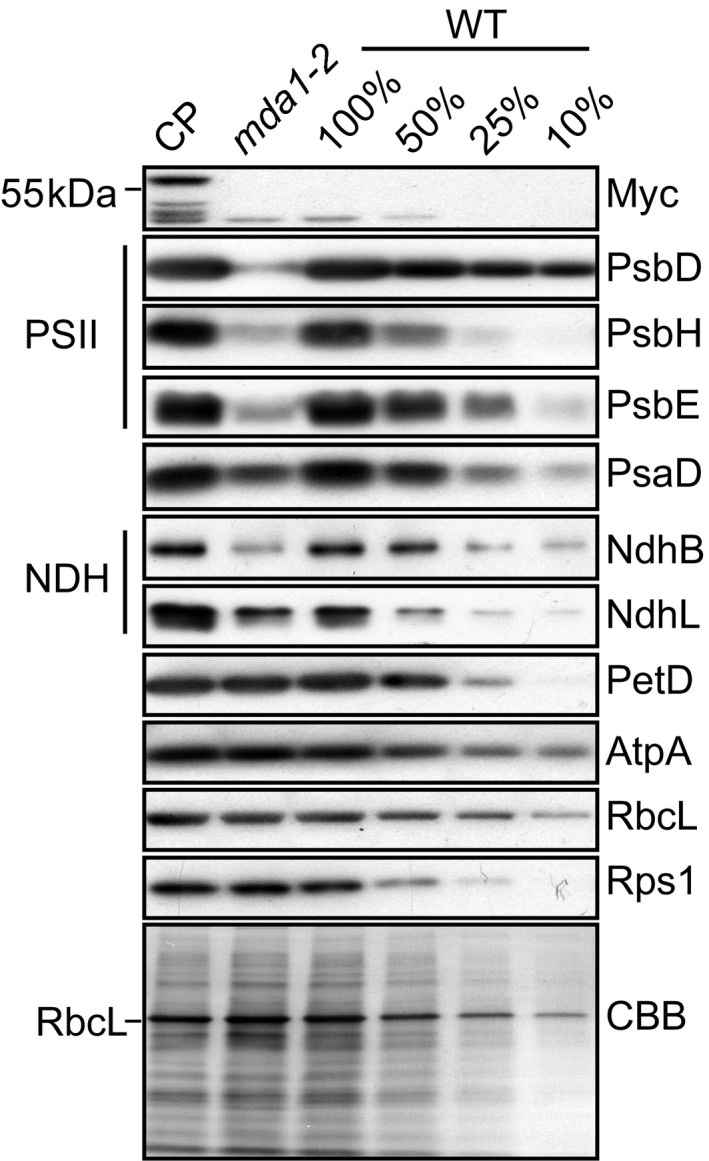
Immunodetection analyses of chloroplast proteins in Arabidopsis wild‐type (WT), mda1‐2 and complemented plants (CP). Replicate immunoblots of total leaf protein extract were probed with antibodies targeted against subunits of the chloroplast Photosystem II (PSII: PsbD, PsbH, PsbE), Photosystem I (PsaD), NADH dehydrogenase (NDH: NdhB, NdhL), cytochrome b 6 f (PetD), ATP synthase (AtpA), Rubisco (RbcL) and ribosome (Rps1) complexes. One of the replicate membranes was stained with Coomassie Blue (CBB) to show equal protein loading.
In order to confirm these results, the photosynthetic capacities of the two mutant alleles were measured by fluorometry along with the WT and complemented plants. Chlorophyll a fluorescence analyses demonstrated that the maximum quantum yield of PSII, expressed as F v/F m, was reduced < 0.5 in the mutants, which is indicative of primary defects in PSII (Meurer et al., 1996; Table S2). This can be explained in part by a three‐fold increased Fo level (Table S1). As observed in psbL, psbN and several other PSII mutants (Meurer et al., 1996; Swiatek et al., 2003; Torabi et al., 2014) the fluorescence dropped to 50% below the initial Fo level and then partially increased during induction again indicating primary defects in PSII. This caused a reduction of the PSII quantum yield (ΦPSII) to < 50% that of the WT and an increase in nonphotochemical quenching (NPQ). In order to estimate the rate‐limiting step in photosynthetic electron transport, PSI yield (ΦPSI), as well as donor and acceptor side limitation of PSI ([ΦPSI ND] and [ΦPSI NA], respectively) were measured (Table S2). Although ΦPSI was not severely changed, ΦPSI ND was twice as high and ΦPSI NA decreased several‐fold in the mutant as compared to the WT. This demonstrates that the electron flow towards PSI is rate‐limiting as can be expected by the reduced PSII activity. Overall, < 50% active P700 could be detected in mda1 mutants as compared to the WT. This indicates a partial loss of PSI in the mutants. Both mda1‐2 and hcf111‐1 behaved almost identically and complemented lines showed the WT phenotype indicating complete recovery.
Expression of chloroplast psbE and ndhH operons is impaired in the mda1 mutant
mTERF proteins are gene expression regulators in organelles (Kleine & Leister, 2015). To assess whether the loss of PSII and NDH proteins were caused by a defect in chloroplast gene expression, we assessed the accumulation of chloroplast gene transcripts by qRT‐PCR in mda1‐2 WT and complemented plants (Fig. 3). The results showed that the steady‐state levels of specific transcripts from PSII (psbE, psbF, psbL, psbJ) and NDH genes (ndhA, ndhI) were particularly diminished in mda1‐2 compared to the WT. The effect of the loss of MDA1 function on the expression of genes was fully mitigated in the complemented plants. These results correlated well with the specific loss of the PSII and NDH complex in mda1‐2 (Fig. 2). In addition, a chloroplast transcriptome‐wide analysis was conducted independently on the hcf111‐1 allele. This analysis measured the RNA steady‐state level of chloroplast genes in hcf111‐1 compared to the WT with their translation efficiency by tiling microarrays of the plastid ORFeome (Zoschke et al., 2013; Fig. S1b; Table S3). The abundance of psbE, psbF, psbL, psbJ and ndhA, ndhI transcripts was specifically affected in hcf111‐1 with a magnitude similar to what was observed for the mda1‐2 allele, although their translation was not impacted. Thus, we concluded that mutations in MDA1 gene compromise the expression of psbE, psbF, psbL, psbJ, ndhA and ndhI genes.
Fig. 3.
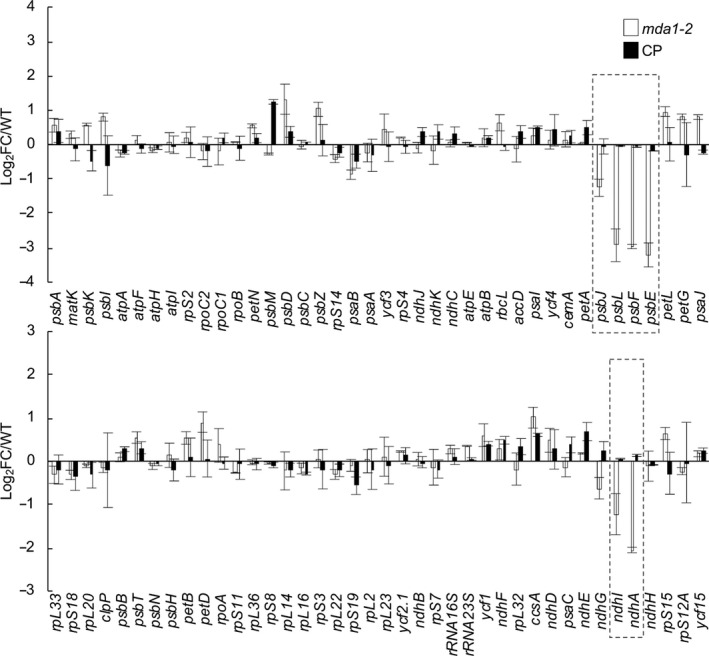
Steady‐state levels of chloroplast gene transcripts in Arabidopsis mda1. Transcript levels were determined by quantitative reverse transcription (qRT)‐PCR and are displayed as the log2 fold‐change (FC) between values obtained for the mutant or the complemented plants and the WT plants. Genes are ordered according to their genome positions. The nuclear ACT2 and TIP41 genes were used for data normalization. The values from two biological replicates performed each with technical triplicate were averaged per genotype and SEs are indicated.
Most chloroplast genes are organized in operon‐like structures and are cotranscribed as polycistronic messenger (m)RNA precursors whose post‐transcriptional maturation gives rise to a variety of overlapping mRNA isoforms (Barkan, 2011). The post‐transcriptional processing of these RNA precursors into mature mRNAs rely predominantly upon the cooperative actions of exoribonucleases and RNA‐binding proteins from the pentatricopeptide repeat (PPR) family that block their RNA degradation activity in vivo to stabilize the 5′ or 3′ ends of these processed mRNAs. The RNA fragments bound by these proteins usually accumulate as small RNA footprints (sRNAs) of c. 20–30 nt whose 5′ or 3′ ends coincide with those of the processed mRNAs these proteins stabilize in vivo (Ruwe & Schmitz‐Linneweber, 2012; Zhelyazkova et al., 2012; Ruwe et al., 2016).
Genes whose expression is affected in mda1 mutants are located in two independent transcriptional units (Fig. 4). To validate our findings and pinpoint which of the RNA isoforms from these gene clusters were missing in the mda1 mutant, RNA gel blot hybridization was performed (Fig. 4b,e). RNA blotting with probes for psbE, psbF, psbL and psbJ genes revealed a severe reduction of one prominent transcript of 1.1 kb in mda1‐2 which accumulated to WT level in CP plants, whereas the abundance of an additional 1.4 kb transcript was less affected in the mutant (Figs 4b, S3). The identical RNA hybridization patterns for the four genes indicates that the two transcripts correspond to tetracistronic psbE‐F‐L‐J mRNAs, consistent with previous observations (Westhoff et al., 1985; Xiong et al., 2020). The mapping of the transcript ends by circular RT‐PCR analysis (Figs 4c, S4) showed that the defective 1.1 kb transcript is expected to be a 5′‐end processed psbE‐F‐L‐J mRNA whose 5′‐end maps 2 nt downstream of the plastid‐encoded RNA polymerase (PEP) transcription initiation site (TSS); located at position −127 from psbE ATG (Allorent et al., 2013), and whose 3′‐end maps 95 nt downstream the psbJ stop codon. In agreement with the RNA blots, the frequency of the 1.1 kb psbE mRNA termini was particularly diminished in mda1 compared to the WT, but the reduction was more severe for the 5′‐ than the 3′‐end indicating that MDA1 acts primarily on the 5′‐end stability of the mature psbE‐F‐L‐J mRNA. Additional RNA blotting was conducted to map the 1.4 kb mRNA. Probes hybridizing to the 1.1 kb mRNA untranslated regions (UTRs) or immediately upstream the 5′‐UTR detected the 1.4 kb RNA form, whereas a probe hybridizing downstream the psbJ 3′‐UTR did not reveal any band. The RNA cross‐comparison of these hybridizations indicates that most 1.4 kb mRNAs differ from the 1.1 kb mRNAs by a longer 5′‐UTR. Circularized (c)RT‐PCR using a reverse primer located upstream the −125 psbE 5′‐end confirmed this and placed a scattered psbE 5′‐end in the −479 to −152 region, which suggested the existence of an alternative distal TSS (Fig. 4a,c).
Fig. 4.
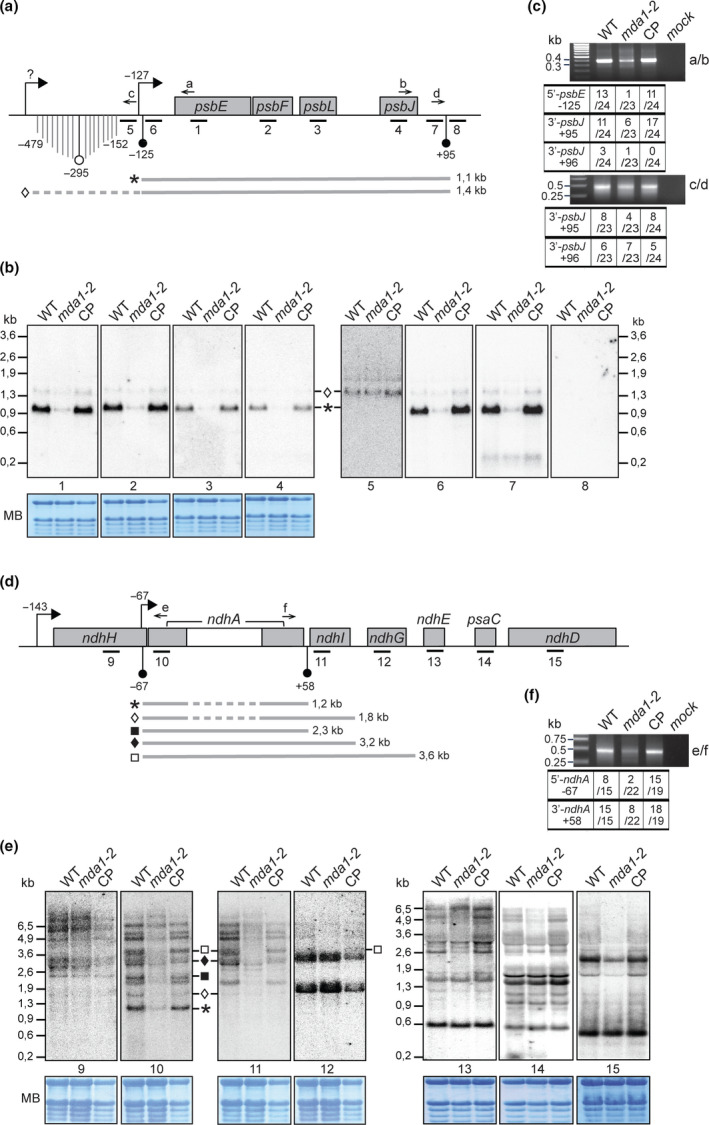
RNA gel blot analyses of transcripts of the Arabidopsis psbE and ndhH operons. (a) Genetic map of the psbE gene cluster indicating the positions of transcription initiation site (TSS) (right squared arrows) and mapped transcript termini (circle arrow tips). Positions are specified relative to gene start or stop codon. Black circle arrow tips indicate transcript termini whose positions coincide with the presence of an abundant sRNA in chloroplasts whose sequences are given in Supporting Information Fig. S4. (b) Replicate blots of wild‐type (WT), mda1‐2 and complemented plant (CP) RNA were hybridized with 60‐mer oligonucleotides strand‐specific probes whose positions are indicated beneath the map in (a). The methylene blue stained blots are shown to illustrate equal loading of rRNAs. Transcripts whose positions could be assigned from these results are diagrammed below the map and their length is given in kilobases (kb). (c) Mapping of transcript ends for psbE‐F‐J‐L genes in different genotypes by circularized reverse transcription (cRT)‐PCR. Primers used for PCR are indicated on the right and displayed on the map. The numbers of clones with the specified ends are indicated in the table. The RNA sequences of the predominant 5′ and 3′‐ends and the sRNAs are provided in Fig. S4. (d) Genetic map of the ndhA gene cluster. (e) RNA blots hybridized with strand specific probes. (f) Mapping of transcript ends for ndhA gene by cRT‐PCR.
Similar experiments were conducted to characterize ndhA and ndhI RNA accumulation in mda1 mutant. These genes belong to the ndhH gene cluster (Fig. 4d) that gives rise to overlapping RNAs whose positions have been partially mapped (Maria del Campo et al., 2006). The RNA blotting revealed that the abundance of distinct mRNAs containing ndhA and ndhI was specifically reduced in mda1 whereas mRNAs containing genes upstream or downstream were barely affected. We were able to assign positions for these disturbed transcripts based on their size, the probes to which they hybridized and the positions of chloroplast RNA termini in Arabidopsis that were mapped in this work or in a recent study (Castandet et al., 2019). Transcripts whose abundance is diminished in mda1 start with an ndhA 5′‐end. cRT‐PCR mapping of ndhA mRNA termini revealed that the frequency of the prominent ndhA 5′‐end that maps at position −67 in the WT was reduced in mda1 and increased in complemented plants overexpressing MDA1 compared to the WT (Figs 4f, S4). The ndhA RNA 3′‐end mapping at position +58 was reduced as well in mda1 but to a much lesser extent than the 5′‐end. These results argue that MDA1 contributes to the stabilization of the −67 ndhA 5′‐end in vivo. Interestingly, the position of this processed 5′‐end coincides with the existence of a recently identified TSS suggesting that the 5′‐end processing of ndhA mRNA is concomitant with transcription (Castandet et al., 2019).
Additional evidence that MDA1 promotes the post‐transcriptional stabilization of the 5′ ends of the processed psbE and ndhA mRNAs comes from the observation that chloroplast sRNAs sharing hallmarks of PPR footprints match the 5′‐ or 3′‐end of the mRNAs that require MDA1 for their accumulation in vivo (Ruwe & Schmitz‐Linneweber, 2012; Zhelyazkova et al., 2012; Ruwe et al., 2016) (Figs 4a,d, S4). RNA gel blots were performed to analyze the accumulation of these sRNAs in mda1 (Fig. 5) and it was found that mda1 specifically lacked the sRNAs from the psbE and ndhA 5′‐ends but not from their 3′ ends as compared to WT and complemented plants. Moreover, sRNAs mapping in other genomic locations (psaC and rbcL) accumulated normally in the mutant. The specific loss in mda1 of these two sRNAs that coincide with the 5′‐end of processed psbE and ndhA mRNAs demonstrates that post‐transcriptional RNA processing at these sites depends on the presence of MDA1. mTERF proteins are helical‐repeat proteins and share structural analogy to PPRs, and members of their family have been involved in RNA‐related functions in organelles (Hammani & Barkan, 2014; Hsu et al., 2014; Romani et al., 2015; Zhang et al., 2018). Thus, these sRNAs could be MDA1's footprints or MDA1 might promote the in vivo binding of PPR proteins to the 5′ ends of processed psbE and ndhA mRNAs.
Fig. 5.
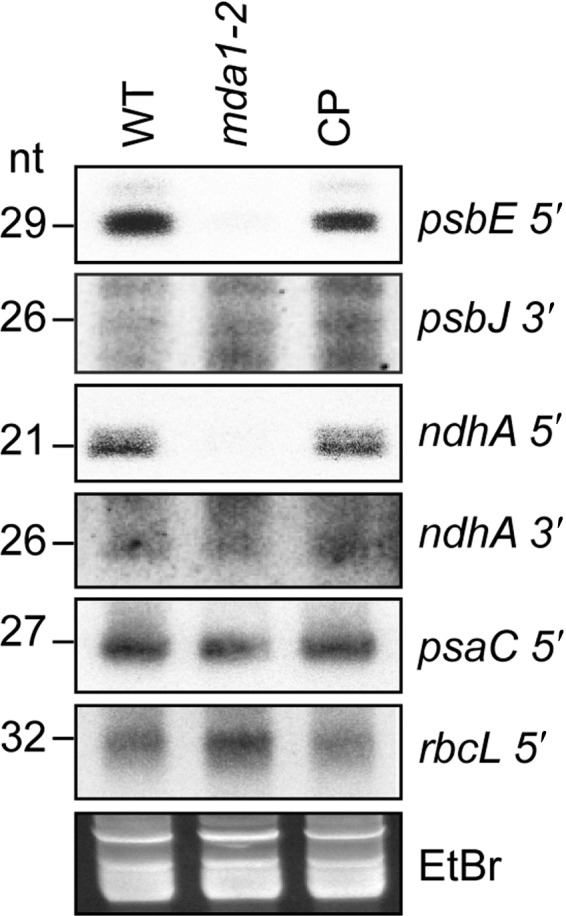
RNA gel blot analyses of chloroplast small (s)RNA accumulation in Arabidopsis mda1. Five µg of low‐molecular‐weight leaf RNA (or 10 μg for psbJ and ndhA 3′) of the indicated genotypes were fractionated in denaturing polyacrylamide gels and transferred to nylon membrane. Blots were hybridized with oligonucleotide probes complementary to sequences of sRNAs accumulating in the chloroplast regions listed on the right. The accumulation of two sRNAs matching the 5′‐end of psaC or rbcL messenger RNAs whose RNA abundance are unaffected in mda1 were monitored as internal controls. A portion of one of the gels stained by ethidium bromide (EtBr) is shown below to illustrate equal loading.
Altogether, the RNA blotting and end mapping results confirmed that MDA1 supports the in vivo accumulation of transcripts containing processed psbE and ndhA 5′‐ends. Nonetheless, the alteration of gene transcription could account for the decrease of the steady‐state level of these mRNAs in mda1. To determine whether transcriptional changes of these genes applied to mda1, chloroplast run‐on transcription assays were performed (Zubo et al., 2011). Genes whose transcript accumulation was defective in mda1 were selected as probes for hybridization with neosynthesized RNAs along with unaffected genes (Fig. 6a). Run‐on results and their quantification showed that the transcription rate of psbE and ndhA in mda1 was c. 44% and 68% of the WT, respectively, whereas transcription of ndhH or psaC was not affected compared to WT and complemented plants. To understand the contribution of this altered transcription to the severe reduction of the processed psbE‐F‐L‐J and ndhA mRNAs in mda1, their relative transcription rates were compared to the transcripts abundance measured by qRT‐PCR or RNA gel blots (Fig. 6b). RNA gel blots revealed that the loss of the 1.1 kb psbE‐F‐L‐J mRNA in mda1 was more severe than expected from qRT‐PCR analysis. This could be explained by qRT‐PCR not being strand‐specific and by its limitation to distinguish overlapping mRNAs. Although the transcription activity of psbE and ndhA genes in mda1 were c. 44% and 68% (respectively) of the WT, the mutant only accumulates c. 3.6% and 24% of their respective mRNAs. Therefore, the decrease of ndhA and psbE transcription in mda1 cannot in itself explain the more severe loss of the processed psbE and ndhA mRNAs in vivo. Altogether, the results indicate that MDA1 plays a role in promoting psbE and ndhA gene transcription as well as contributing to the post‐transcriptional stabilization of their 5′‐end processed mRNAs in vivo.
Fig. 6.
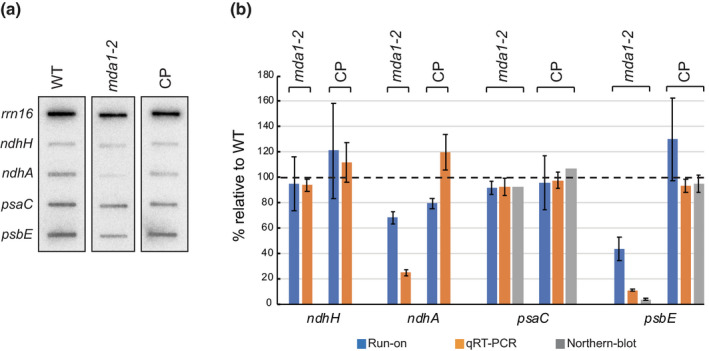
Run‐on transcription assay of chloroplast genes in Arabidopsis mda1. (a) Slot‐blot analysis of run‐on transcript samples from chloroplasts of the indicated genotypes hybridized to DNA fragments representing chloroplast rrn16, ndhH, ndhA, psaC and psbE genes. (b) Percentage changes relative to wild‐type (WT) of run‐on transcription rates, the mRNA steady‐state levels measured by transcription qRT‐PCR or /and RNA gel blot analyses. rrn16 was used as reference gene for the normalization of transcription rates. The quantification of mRNAs from RNA gel blots was performed on mature 1.1‐kb psbE‐F‐L‐J and 0.5‐kb monocistronic psaC mRNAs. Data are means of three independent experiments and SEs are indicated.
MDA1 is found in high MW complexes and associates with transcriptional active chromosome (TAC) components in vivo
In order to understand the basis for the association of MDA1 with chloroplast membranes (Fig. S2), membranes were isolated from the complemented plants expressing a 4xMyc tagged version of MDA1 and different treatments were applied to them. The release of MDA1 or the integral thylakoid membrane protein control, PsaD (Sane et al., 2005) to the soluble fraction was monitored by immunoblotting (Fig. 7a). Treatment with RNase or DNase had no effect on the membrane association of MDA1 indicating that MDA1 is not attached to the membranes via its association with DNA or RNA. However, treatments by sodium carbonate or the membrane solubilization agent NP40 detached the protein from the membranes, indicating that MDA1 is a peripheral protein as sodium carbonate treatment releases peripheral membrane proteins attached by hydrophobic interactions but not integral membrane proteins (Fujiki et al., 1982).
Fig. 7.
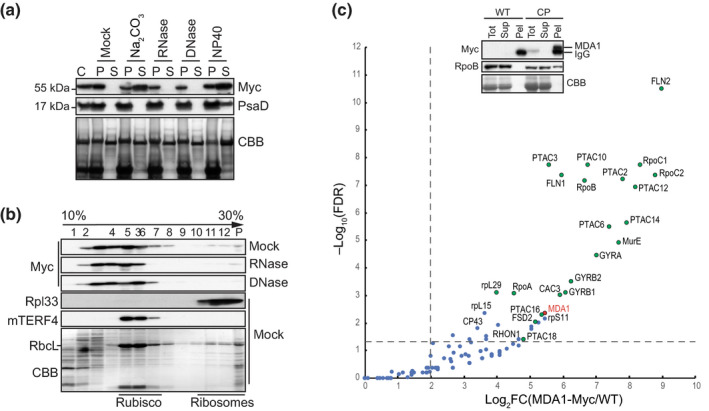
Arabidopsis MDA1 is found in high molecular weight (MW) complexes in vivo and associates with plastid‐encoded RNA polymerase (PEP) and transcriptional active chromosome (TAC) components. (a) MDA1 is a peripheral membrane protein whose membrane association is RNA and DNA‐independent. Chloroplast membrane fractions (C) were mock‐treated or 0.2 M Na2CO3, RNase, DNase, 1% NP40 treated before centrifugation. The presence of MDA1 in the pellet (P) or soluble fraction (S) after treatment was analyzed by immunoblotting with an antibody against the Myc epitope or the chloroplast integral membrane protein, PsaD as a control. The membrane stained with Coomassie Blue (CBB) is shown below. (b) Sucrose gradient fractionation of mock, RNase‐ and DNase‐treated chloroplast extracts. An equal volume of each fraction was analyzed on immunoblots using antibodies indicated at left. The chloroplast RNA intron splicing factor proteins, mTERF4 and the ribosomal protein Rpl33 are found in high MW complexes of c. 0.55 MDa and 1 MDa, respectively (Hammani & Barkan, 2014) and serve as size fractionation controls. (c) Chloroplast protein interactome of MDA1. MDA1 co‐immunoprecipitates (coIP) with components of the plastid TAC (Pfalz et al., 2006) including the subunits of the plastid encoded RNA polymerase. Solubilized chloroplasts from complemented mda1 plants expressing the 4Myc‐tagged MDA1 (CP) or wild‐type (WT) plants were used for immunoprecipitation with anti‐Myc antibody and coIP proteins were identified by MS analysis. Volcano plots show the enrichment of proteins copurified with 4Myc‐tagged MDA1 as compared with control IPs. IPs were performed on biological triplicate. The y‐ and x‐axes display the negative common logarithm of the adjusted false discovery rate (FDR) and fold‐changes, respectively. The dashed lines indicate the threshold above which proteins are significantly enriched (FDR < 0.05 and FC > 4). TAC components (Pfalz et al., 2006) are represented as green dots. The immunoblot validation of RpoB association is shown. Replicate immunoblots were probed with anti‐Myc or RpoB antibody. The full list of MDA1‐associated proteins is available in Supporting Information Table S4.
In order to analyze the association of MDA1 with high MW complexes in chloroplasts, solubilized chloroplasts were fractionated by sedimentation on sucrose gradients (Fig. 7b). MDA1 was detected mainly in macromolecular complexes ≤ 0.55 MDa (fractions 3 to 8) and to a lesser extent in complexes of c. 1 MDa (fractions 11 to P). DNase and RNase treatments of chloroplast extracts before sucrose gradient fractionation reduced the presence of MDA1 in fractions above 6 suggesting that some MDA1 proteins are found in large complexes containing chloroplast DNA and RNA.
In order to understand the protein composition of MDA1 complexes, coIP was performed on solubilized chloroplasts and proteins from immunoprecipitated fractions were identified by LC‐MS/MS (Fig. 7c). Out of 35 proteins that were significantly enriched in MDA1 IPs, 21 were components of the plastid TAC (Pfalz et al., 2006) including the four subunits of the plastid‐encoded RNA polymerase, PEP (RpoA, B, C1 and C2). Immunoblot analysis of anti‐Myc co‐immunoprecipitates confirmed that RpoB associates with MDA1 in chloroplast extract of the complemented plants (Fig. 7c). MDA1's protein partners support a function in chloroplast transcription. Altogether, these results in conjunction with mda1 transcriptomic analyses indicate that MDA1 associates in vivo with the TAC complex to promote transcription of specific genes.
MDA1 is a DNA binding protein that associates with psbE and ndhA genes in vivo
Consistent with their function in the regulation of organellar gene expression, several mTERF proteins have been described to bind DNA or RNA (reviewed in Kleine & Leister, 2015). To determine whether MDA1 binds to nucleic acids in vitro, recombinant and mature MDA1 (rMDA1) was expressed in E. coli and affinity‐purified (Fig. 8a). rMDA1 has a predicted MW of c. 51 kDa but eluted from a size exclusion chromatography column at a size of c. 100 kDa suggesting that it can form homodimers. rMDA1 containing fractions were pooled and the protein purity was confirmed by SDS‐PAGE analysis (Fig. 8b). To test the affinity of rMDA1 for nucleic acids, gel mobility shift (GMS) assays were performed in absence of competitors using a synthetic 43‐mer oligonucleotide probe in the form of ssDNA, dsDNA, ssRNA or dsRNA (Fig. 8c). rMDA1 virtually did not bind to dsRNA or ssDNA but showed clear binding to ssRNA and dsDNA with a more pronounced affinity for dsDNA. Thus, rMDA1 has the capacity to bind dsDNA and, to a lesser extent, ssRNA.
Fig. 8.
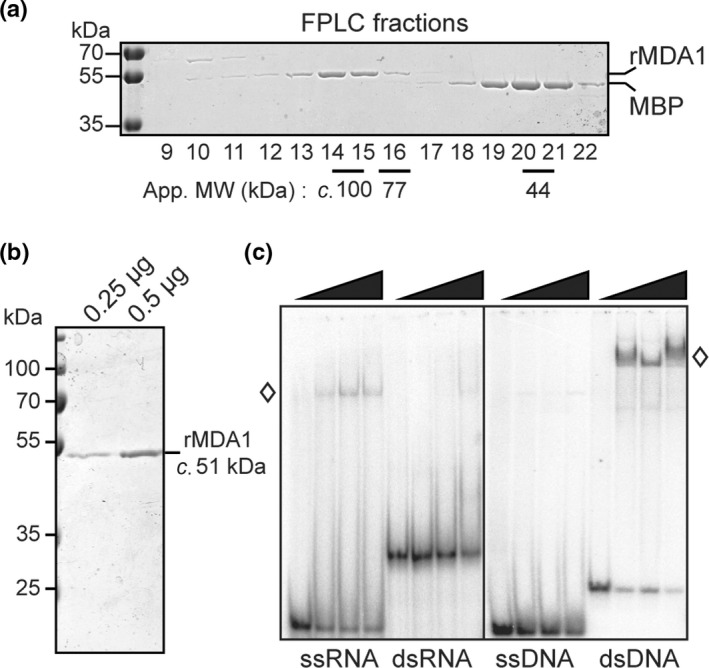
Recombinant Arabidopsis MDA1 binds ssRNA and dsDNA in vitro. (a) Purification of recombinant MDA1 (rMDA1). rMDA1 was expressed as maltose binding protein (MBP) fusion and purified by amylose affinity chromatography. After TEV protease cleavage, free rMDA1 and MBP were resolved by gel filtration (FPLC). Aliquots of column fractions were analyzed by SDS/PAGE and stained with Coomassie Blue. The elution positions of rMDA1 (c. 100 kDa) and the Conalbumine (77 kDa) and Ovalbumine (44 kDa) molecular weight (MW) markers are indicated below the gel. (b) SDS/PAGE analysis of the purity of the final rMDA1 used for in vitro assays. (c) Gel mobility shift assays showing ssRNA and dsDNA binding of rMDA1. Increasing amounts of rMDA1 (0, 50, 100 and 200 nM) were incubated with synthetic 43‐mer RNA or DNA oligonucleotides of the same random sequence in single‐ or double‐stranded forms and resolved on a native polyacrylamide gel. Free nucleic acids migrate to the bottom of the gel whereas rMDA1‐nucleic acid complexes are shifted up on the gel and are indicated by white diamonds.
In order to test whether these properties applied in vivo, RIP‐seq experiments on solubilized chloroplasts isolated from complemented plants were conducted but the results did not show substantial RNA enrichment in the immunoprecipitated samples when compared to the control (data not shown). This result indicates that MDA1 is not associated to RNA in vivo. We showed that MDA1 is required for the accumulation of transcripts whose processed forms are likely stabilized by PPR RNA‐binding protein caps (Figs 4, 5; Ruwe et al., 2016). To rule out the direct implication of MDA1 in this RNA stabilization process, additional GMS assays were performed using synthetic RNAs whose sequences correspond to the sRNAs matching processed 5′‐ends of ndhA and psbE mRNAs or 3′‐ends of ndhA and psbJ mRNAs (Fig. S4). In agreement with RIP‐seq results, rMDA1 did not bind to any of these sRNAs (Fig. S5). Based on these in vivo and in vitro data, we concluded that MDA1 is not the RNA‐binding protein stabilizing the termini of mature psbE‐F‐L‐J and ndhA containing mRNAs.
In a second step, we performed DIP‐qPCR on solubilized chloroplasts and DNA enrichment for several genes in the psbE and ndhH operons was analyzed in the immunoprecipitated samples (Fig. 9). The results showed specific and significant DNA recovery for psbE and ndhA genes with a more pronounced enrichment in regions near their putative promoter (Fig. 9b). Moreover, we observed that the degree of enrichment for psbE was higher than that of ndhA indicating that MDA1 has a higher affinity for psbE in vivo. Taken together, these results demonstrate that MDA1 binds specifically to DNA regions in psbE and ndhA genes whose transcript accumulation is impaired in mda1 mutants.
Fig. 9.
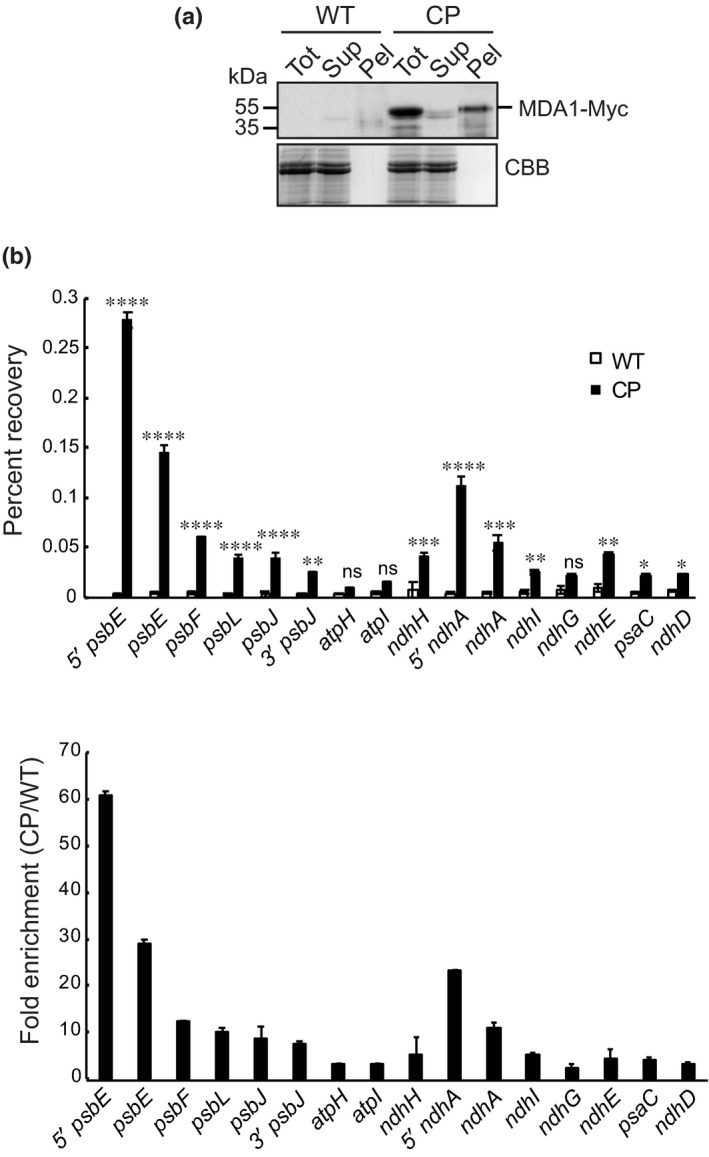
Arabidopsis chloroplast DNA immunoprecipitation quantitative PCR (DIP‐qPCR) analysis of MDA1 DNA binding activity in vivo. (a) Immunoprecipitation efficiency of 4Myc‐tagged MDA1 on chloroplasts extracted from complemented mda1 plants (CP). Wild‐type (WT) chloroplasts were used as input for negative experimental controls. A fraction of the input (Tot), supernatant (Sup) and pellet (Pel) samples were analyzed on immunoblots by probing with Myc antibodies. (b) Levels of immunoprecipitated (IP) DNA of various chloroplast DNA regions were calculated as percent recovery of the total input DNA. The significance of the variation between CP and control IPs were analyzed with a two‐way ANOVA, Tukey's test: *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.00005; ns, not significant. The DNA fold enrichment in CP IPs relative to WT is shown below. Data are means of three independent experiments and SEs are indicated.
MDA1 binds with specificity to 27‐nt DNA sequences located near psbE and ndhA promoter regions
In order to prove direct MDA1 binding to its in vivo target genes and define the DNA segments that are necessary for interaction, we assayed rMDA1 in vitro binding to overlapping DNA segments derived from 230‐ or 330‐bp regions covering (respectively) the psbE and ndhA putative promoters with regions near the 5′‐end of their ORFs (Fig. 10). rMDA1 bound with high affinity and specificity to a 100‐nt DNA segment, named E2 located upstream psbE (−125 to −26 bp from the ATG) and downstream the −127 PEP TSS (cf. E2 to E1 and E3) (Fig. 10a). To precisely map the protein binding site, a DNase I footprint assay was conducted using the E2 DNA fragment in the presence or absence of rMDA1 (Fig. S6). rMDA1 protected a 27‐bp DNA region from enzymatic cleavage mapping at position −96/−70 from the psbE ATG. MDA1 binding specificity to this genomic region was further tested in GMS assays using a 27‐bp DNA fragment corresponding to the DNase‐protected sequence (E4) or a DNA fragment of identical size mapping right downstream (E5). rMDA1 only bound the E4 segment demonstrating that the −96/−70 DNA region of the psbE gene is the putative MDA1 binding site.
Fig. 10.
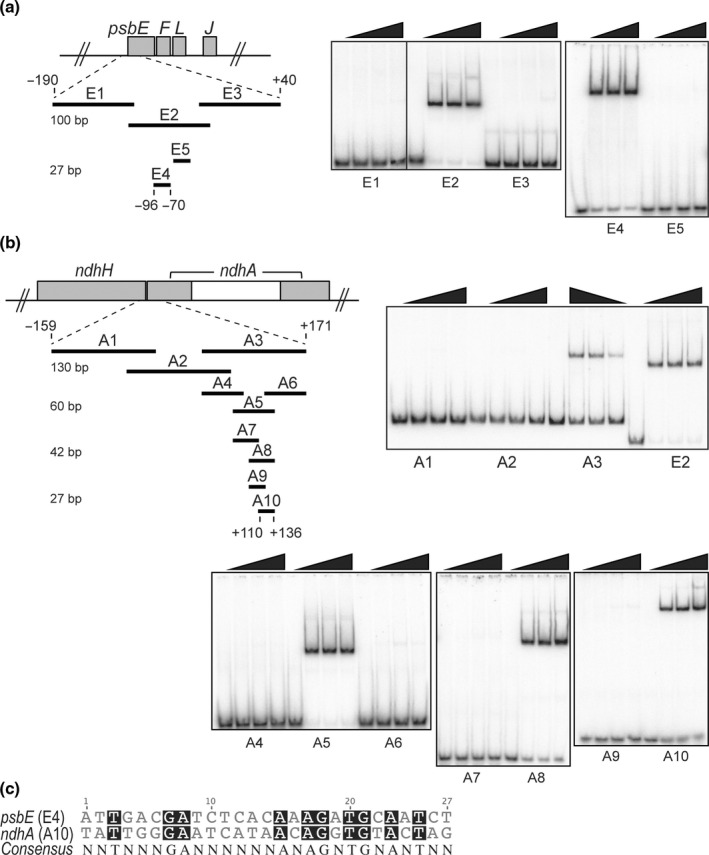
Preferential DNA binding of rMDA1 to regions near the 5′‐end of chloroplast psbE and ndhA Arabidopsis genes. (a) Gel mobility shift assays showing specific DNA binding of rMDA1 to a short DNA sequence upstream of psbE start codon. Different amounts of rMDA1 (0, 12.5, 25, 50 nM) were incubated with overlapping DNA segments of various sizes that span the −190/+40 genomic region from psbE start codon in the presence of poly(dI‐dC) competitor. The length of the DNA fragments (bp) is given on the left side. DNA fragment E2 was used for MDA1 DNase footprinting analysis shown in Supporting Information Fig S6. (b) Gel mobility shift assays showing specific DNA binding of rMDA1 to a DNA sequence downstream of ndhA start codon. Binding conditions were identical to those used in (a) and the DNA fragments covered a −159/+171 genomic region from the ndhA start codon. (c) Sequence alignment of the 27‐bp psbE and ndhA MDA1 binding sites. Conserved bases are shaded in black and the consensus sequence is given below.
MDA1 also acts on the ndhA locus in vivo. We showed that rMDA1 bound with specificity to a 130‐bp ndhA DNA segment (A3) mapping +42/+171 within ndhA ORF (compare probe A3 to A1 and A2; Fig. 10b). However, the affinity of rMDA1 for the ndhA3 binding site was weaker than for the psbE2 site as seen by the comparison of the amount of bound DNA at similar protein concentrations for each site. Contrary to psbE2, DNase footprinting performed with the ndhA3 fragment did not lead to significant cleavage protection probably due to the lower affinity of rMDA1 for the ndhA3 segment (data not shown). Thus, GMS assays were performed to delineate more precisely MDA1 binding sites using a series of shorter overlapping DNA fragments (from 60‐ to 27‐bp size) spanning the ndhA3 segment (probes A4 to A10). The results indicate that rMDA1 bound a 27‐bp region located +110/+136 within the ndhA ORF (probe A10). Altogether, the GMS assay results showed that MDA1 binds with specificity to two DNA sequences located near the 5′‐end of psbE and ndhA genes.
In agreement with MDA1 binding sites, our molecular analyses revealed that only a subset of chloroplast genes cotranscribed with psbE and ndhA is affected in mda1 mutants. We reasoned that the two MDA1 binding sites in psbE and ndhA genes should therefore contain sufficient nucleotide conservation to define DNA specificity among the entire chloroplast genome. The alignment between the 27‐bp psbE and ndhA binding sites revealed 10 conserved residues scattered along these sequences (Fig. 10c). This consensus sequence was matched against the entire Arabidopsis chloroplast genome to reveal three hits: the psbE4 and ndhA10 sites and an additional site in the ndhD gene. The DIP‐qPCR analysis did not reveal any MDA1 in vivo association with ndhD gene and the expression of ndhD was unaffected in mda1 mutants. These observations suggest that the two binding sites carry sufficient base conservation to define DNA specificity among the entire chloroplast genome but MDA1 might recognize additional elements to define its physiologically relevant gene targets in vivo.
Discussion
In this study, we have demonstrated novel aspects to the function of MDA1 in Arabidopsis, also known as mTERF5 (Ding et al., 2019). Similar to our results, Ding et al. showed that mTERF5/MDA1 is required for the accumulation of psbE‐F‐L‐J messenger (m)RNA in chloroplasts and positively regulates transcription at the psbE promoter. Consistent with our findings, they showed that mTERF5 bound in vitro and in vivo to a DNA sequence near the psbE promoter that maps to our MDA1 footprint, and demonstrated that this binding site contains a transcription pausing site. Finally, they revealed that mTERF5 associates in vivo with the plastid transcriptional active chromosome (TAC) component, pTAC6 and that this interaction allows the recruitment of the PEP complex near the psbE promoter to pause gene transcription in vivo. However, their functional model for mTERF5/MDA1 did not explain how the lack of transcription pausing at the psbE promoter causes the severe loss of the psbE‐F‐L‐J mRNA in the mterf5/mda1 mutant, particularly when considering that the transcription activity at psbE is reduced only partially in the mterf5/mda1 mutant compared to the wild‐type (WT) (Fig. 6). Our study showed that MDA1/mTERF5 had an additional in vivo gene target in the ndhH operon. We demonstrated that MDA1 was additionally required for the accumulation of specific ndhA‐I mRNAs and importantly, that the psbE and ndhA containing mRNAs that require MDA1 for their in vivo accumulation result from post‐transcriptional RNA processing most likely involving the participation of exoribonucleases with pentatricopeptide repeat (PPR) RNA‐binding proteins that protect and stabilize the processed mRNA ends. The analysis of the accumulation of the small RNA PPR footprints in mda1 indicated that MDA1 specifically promotes the in vivo binding of an RNA‐binding protein (presumably a PPR protein) to the 5′‐end of the processed psbE and ndhA mRNAs. In contrast to Ding et al., our MDA1 co‐immunoprecipitation (coIP) analysis revealed that MDA1 associates in vivo not only with pTAC6, but also with many TAC components, including the PEP core subunits. Finally, the relative quantification of the transcription activity at psbE and ndhA demonstrated that their reduction in mda1 could not justify in itself the more severe loss of their processed mRNAs in the mutant. Altogether, our results demonstrated that MDA1/mTERF5 functions not only in stimulating transcription of psbE and ndhA genes, but also in the stabilization of their post‐transcriptionally processed mRNAs. A potential functional model for MDA1 explaining its dual contribution to gene transcription and RNA stabilization in chloroplasts is discussed below.
MDA1: linking transcription and RNA stabilization of its target genes
MDA1 promotes both transcription of psbE and ndhA genes and the stabilization of their 5′‐end processed mRNAs. Our coIP results suggest that MDA1 interacts in vivo with the PEP transcription complex. Thus, the specific DNA binding of MDA1 near the psbE promoter most likely induces recruitment of components of the PEP complex to stimulate gene transcription locally. A puzzling question is how the DNA binding protein MDA1 additionally promotes the stabilization of the processed 5′‐end of psbE and ndhA RNAs. Answers to this question might come from the analysis of the genomic location of the MDA1 binding site in psbE (Fig. 11a). MDA1 binding site is placed 29 nucleotides downstream the position of the 5′‐end of processed psbE RNA and this binding site coincides with a transcriptional pausing site identified by Ding et al. whose in vivo activity requires MDA1 (Ding et al., 2019). The 29‐nt region directly upstream of this transcriptional pausing site matches the sequence of the sRNA that is bound in vivo by an unknown PPR‐like RNA‐binding protein that stabilizes the 5′‐end of processed psbE mRNA which is missing in mda1. The stability of psbE mRNA depends on the capacity of this RNA‐binding protein to bind its RNA target. Biochemical studies have shown that PPR proteins preferentially bind single‐stranded RNA whereas their capacity to invade RNA secondary structures is very limited (Williams‐Carrier et al., 2008; Hammani et al., 2011; McDermott et al., 2018). Therefore, in vivo mechanisms should exist to facilitate PPR binding to their RNA target sequences when these are folded in secondary structures (Jiang et al., 2019). A transcriptional pausing site strategically placed downstream of a PPR binding site offers an attractive mechanism by which to pause RNA elongation and preclude the formation of secondary structures that would otherwise be deleterious for PPR binding and RNA stabilization. We used the mfold server (Zuker, 2003) to predict RNA structures of neotranscribed psbE RNA segments including or excluding pausing at the MDA1 binding site (Fig. 11b). As expected, when transcription is paused at this site by MDA1, the 5′‐end of the psbE RNA and the PPR binding site do not fold into a stable RNA structure (dG > −5 kcal mol−1) and would allow PPR binding and the subsequent psbE mRNA stabilization. On the contrary, farther RNA elongation to the MDA1 binding site or the psbE start codon are predicted to allow the formation of stable RNA–RNA interactions (dG < −10 kcal mol−1) that occlude the PPR binding site and presumably compete with the PPR binding. Based on this observation and our experimental results, we hypothesize that MDA1 promotes psbE mRNA stability by pausing gene transcription to facilitate binding of a PPR protein to the RNA 5′‐end, which, in turn, stabilizes the mRNA in vivo.
Fig. 11.
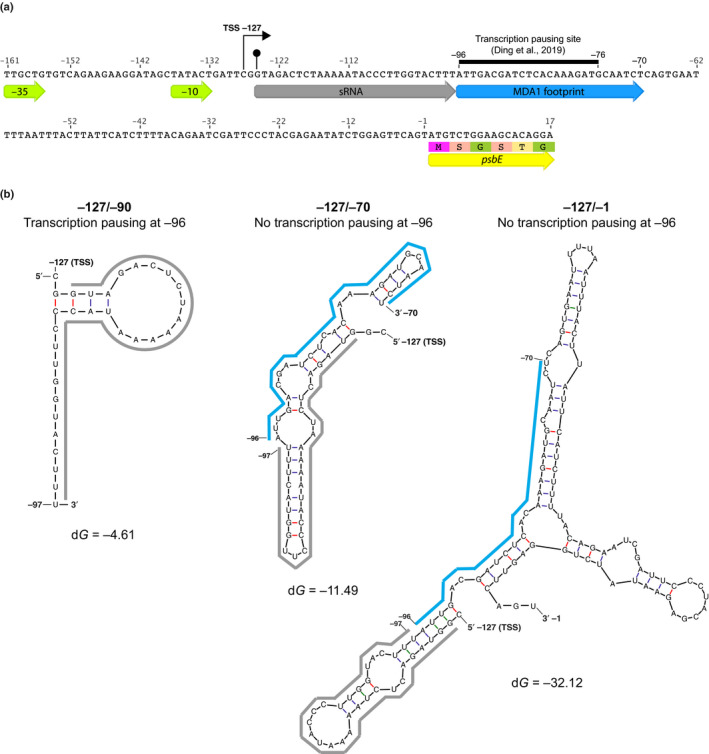
Potential mechanism of MDA1 action on psbE 5′ RNA stabilization in Arabidopsis. (a) Nucleotide sequence of the −161/+17 psbE genomic region. The genomic positions are given according to psbE start codon. The −35 and −10 consensus eubacterial plastid‐encoded RNA polymerase (PEP) promoter elements, transcription initiation site (TSS) (right squared arrow) and in vivo 5′‐end of processed psbE mRNA (circled arrow tip) are positioned on the map. The sequences of the 5′ psbE sRNA PPR footprint that is specifically lost in mda1 and MDA1 DNA binding site are underlined by grey and blue arrows, respectively. The MDA1 binding site contains a transcription pausing site (Ding et al., 2019). The 5′ region of the psbE coding sequence is underlined by a yellow arrow. (b) Mfold prediction of the most stable structure of neotranscribed RNA sequences from psbE TSS (−127) to the PEP pausing site (−96) or farther downstream to the MDA1 binding site (−70) or upstream the psbE start codon (−1). The sequences of the 5′ psbE sRNA and MDA1 footprint are underlined using the same color code as in (a). The calculated dG for each RNA structure is given in kcal mol–1. MDA1 DNA binding downstream of the PPR footprint sequence pauses gene transcription and would prevent the folding of the footprint into a secondary RNA structure that is deleterious for the PPR binding to the 5′ end of psbE mRNA and therefore, the post‐transcriptional stabilization of the processed psbE mRNA in vivo.
The MDA1 binding site in ndhA is located in the first exon and farther downstream from the gene promoter than for psbE. Although a transcriptional pausing site was not clearly identified in ndhA by Ding et al., the ndhA 5′ sRNA PPR footprint is specifically missing in mda1. This suggests that MDA1 might execute an analogous function at this site to cooperate with an RNA‐binding protein that stabilizes the 5′‐end of ndhA RNA in vivo.
Altogether, our results reveal the importance of DNA binding proteins in the regulation of post‐transcriptional processes in chloroplasts. Molecular and biochemical studies of the uncharacterized mTERF genes in plants will likely lead to the discovery of new regulatory mechanisms in chloroplast gene expression and a better understanding of the interplay between DNA and RNA metabolism in chloroplasts.
Author contributions
KH, RZ, L‐VM and RG designed the research; L‐VM, RG, KM, KH, AZ, JC, AA, NB and JM performed research; KH and L‐VM wrote the manuscript and RZ, RG and JM edited it.
Supporting information
Fig. S1 Phenotype and ribosome profiling analyses of the Arabidopsis mda1 mutant allele hcf111-1.
Fig. S2 MDA1 is associated to membranes in chloroplasts.
Fig. S3 Quantification of the abundance of the 1.4‐ and 1.1‐kb psbE‐F‐L‐J mRNAs in the different plant genotypes by RNA blot analyses.
Fig. S4 Genome mapping of the predominant in vivo 5′ and 3′ ends for the processed psbE‐F‐L‐J and ndhA mRNAs determined by cRT‐PCR in different genotypes.
Fig. S5 RNA gel mobility shift assays showing no binding of MDA1 to sRNAs.
Fig. S6 psbE DNase footprinting assays.
Methods S1 Photosynthetic capacity measurements and genome wide analyses of chloroplast transcriptome and translatome.
Table S1 List of oligonucleotides used in this study.
Table S2 Chlorophyll a fluorescence induction and light‐induced PSI absorbance changes.
Table S3 Ribosome profiling data of hcf111‐1.
Table S4 List of proteins identified by LC‐MS/MS in co‐immunopurification assays using MDA1 as bait.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
The authors thank Alice Barkan and Rosalind William‐Carrier (University of Oregon) for performing the RIP‐seq analysis. This study was supported by a grant from Agence Nationale de la Recherche (ANR‐16‐CE20‐0007) to KH, the DFG (ZO 302/5‐1) to RZ, and the SFB‐TRR175 to JM (A03) and RZ (A04).
References
- Allorent G, Courtois F, Chevalier F, Lerbs‐Mache S. 2013. Plastid gene expression during chloroplast differentiation and dedifferentiation into non‐photosynthetic plastids during seed formation. Plant Molecular Biology 82: 59–70. [DOI] [PubMed] [Google Scholar]
- Babiychuk E, Vandepoele K, Wissing J, Garcia‐Diaz M, De Rycke R, Akbari H, Joubes J, Beeckman T, Jansch L, Frentzen M et al 2011. Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proceedings of the National Academy of Sciences, USA 108: 6674–6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. 1998. Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Photosynthesis: Molecular Biology of Energy Capture 297: 38–57. [Google Scholar]
- Barkan A. 2011. Expression of plastid genes: organelle‐specific elaborations on a prokaryotic scaffold. Plant Physiology 155: 1520–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund AK, Pujol C, Duchene AM, Glaser E. 2009. Defining the determinants for dual targeting of amino acyl‐tRNA synthetases to mitochondria and chloroplasts. Journal of Molecular Biology 393: 803–814. [DOI] [PubMed] [Google Scholar]
- Castandet B, Germain A, Hotto AM, Stern DB. 2019. Systematic sequencing of chloroplast transcript termini from Arabidopsis thaliana reveals >200 transcription initiation sites and the extensive imprints of RNA‐binding proteins and secondary structures. Nucleic Acids Research 47: 11889–11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Zhang Y, Hu Z, Huang X, Zhang B, Lu Q, Wen X, Wang Y, Lu C. 2019. mTERF5 acts as a transcriptional pausing factor to positively regulate transcription of chloroplast psbEFLJ . Molecular Plant 12: 1259–1277. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. Journal of Cell Biology 93: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Barkan A. 2014. An mTERF domain protein functions in group II intron splicing in maize chloroplasts. Nucleic Acids Research 42: 5033–5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Bonnard G, Bouchoucha A, Gobert A, Pinker F, Salinas T, Giege P. 2014. Helical repeats modular proteins are major players for organelle gene expression. Biochimie 100C: 141–150. [DOI] [PubMed] [Google Scholar]
- Hammani K, des Francs‐Small CC, Takenaka M, Tanz SK, Okuda K, Shikanai T, Brennicke A, Small I. 2011. The pentatricopeptide repeat protein OTP87 is essential for RNA editing of nad7 and atp1 transcripts in Arabidopsis mitochondria. Journal of Biological Chemistry 286: 21361–21371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YW, Wang HJ, Hsieh MH, Hsieh HL, Jauh GY. 2014. Arabidopsis mTERF15 is required for mitochondrial nad2 intron 3 splicing and functional complex I activity. PLoS ONE 9: e112360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chai X, Manavski N, Williams‐Carrier R, He B, Brachmann A, Ji D, Ouyang M, Liu Y, Barkan A et al 2019. An RNA chaperone‐like protein plays critical roles in chloroplast mRNA stability and translation in Arabidopsis and Maize. Plant Cell 31: 1308–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez‐Menendez N, Fernandez‐Millan P, Rubio‐Cosials A, Arnan C, Montoya J, Jacobs HT, Bernado P, Coll M, Uson I, Sola M. 2010. Human mitochondrial mTERF wraps around DNA through a left‐handed superhelical tandem repeat. Nature Structural & Molecular Biology 17: 891–893. [DOI] [PubMed] [Google Scholar]
- Kleine T. 2012. Arabidopsis thaliana mTERF proteins: evolution and functional classification. Frontiers in Plant Science 3: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Leister D. 2015. Emerging functions of mammalian and plant mTERFs. Biochimica et Biophysica Acta 1847: 786–797. [DOI] [PubMed] [Google Scholar]
- Kleinknecht L, Wang F, Stube R, Philippar K, Nickelsen J, Bohne AV. 2014. RAP, the sole octotricopeptide repeat protein in Arabidopsis, is required for chloroplast 16S rRNA maturation. Plant Cell 26: 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L. 1998. Preparation of physiologically active chloroplasts from Arabidopsis. Methods in Molecular Biology 82: 43–48. [DOI] [PubMed] [Google Scholar]
- Lange H, Ndecky SYA, Gomez‐Diaz C, Pflieger D, Butel N, Zumsteg J, Kuhn L, Piermaria C, Chicher J, Christie M et al 2019. RST1 and RIPR connect the cytosolic RNA exosome to the Ski complex in Arabidopsis. Nature Communications 10: 3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Meyers BC, Green PJ. 2007. Construction of small RNA cDNA libraries for deep sequencing. Methods 43: 110–117. [DOI] [PubMed] [Google Scholar]
- Maria del Campo E, Sabater B, Martin M. 2006. Characterization of the 5′‐ and 3′‐ends of mRNAs of ndhH, ndhA and ndhI genes of the plastid ndhH‐D operon. Biochimie 88: 347–357. [DOI] [PubMed] [Google Scholar]
- McDermott JJ, Civic B, Barkan A. 2018. Effects of RNA structure and salt concentration on the affinity and kinetics of interactions between pentatricopeptide repeat proteins and their RNA ligands. PLoS ONE 13: e0209713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J, Meierhoff K, Westhoff P. 1996. Isolation of high‐chlorophyll‐fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198: 385–396. [DOI] [PubMed] [Google Scholar]
- Nunez‐Delegido E, Robles P, Ferrandez‐Ayela A, Quesada V. 2019. Functional analysis of mTERF5 and mTERF9 contribution to salt tolerance, plastid gene expression and retrograde signalling in Arabidopsis thaliana . Plant Biology 22: 459‐471. [DOI] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmuller R. 2006. pTAC2, ‐6, and ‐12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberti M, Polosa PL, Bruni F, Manzari C, Deceglie S, Gadaleta MN, Cantatore P. 2009. The MTERF family proteins: mitochondrial transcription regulators and beyond. Biochimica et Biophysica Acta 1787: 303–311. [DOI] [PubMed] [Google Scholar]
- Robles P, Micol JL, Quesada V. 2012. Arabidopsis MDA1, a nuclear‐encoded protein, functions in chloroplast development and abiotic stress responses. PLoS ONE 7: e42924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles P, Navarro‐Cartagena S, Ferrandez‐Ayela A, Nunez‐Delegido E, Quesada V. 2018. The characterization of Arabidopsis mterf6 mutants reveals a new role for mTERF6 in tolerance to abiotic stress. International Journal of Molecular Sciences 19: 2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani I, Manavski N, Morosetti A, Tadini L, Maier S, Kuhn K, Ruwe H, Schmitz‐Linneweber C, Wanner G, Leister D et al 2015. A member of the arabidopsis mitochondrial transcription termination factor family is required for maturation of chloroplast transfer RNAIle(GAU) . Plant Physiology 169: 627–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwe H, Schmitz‐Linneweber C. 2012. Short non‐coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins? Nucleic Acids Research 40: 3106–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwe H, Wang G, Gusewski S, Schmitz‐Linneweber C. 2016. Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle‐specific mRNA stabilization mechanisms. Nucleic Acids Research 44: 7406–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez‐Venegas R, Avramova Z. 2008. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nature Protocols 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Sane AP, Stein B, Westhoff P. 2005. The nuclear gene HCF107 encodes a membrane‐associated R‐TPR (RNA tetratricopeptide repeat)‐containing protein involved in expression of the plastidial psbH gene in Arabidopsis. The Plant Journal 42: 720–730. [DOI] [PubMed] [Google Scholar]
- Sato N, Terasawa K, Miyajima K, Kabeya Y. 2003. Organization, developmental dynamics, and evolution of plastid nucleoids. International Review of Cytology 232: 217–262. [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S. 1999. Complete structure of the chloroplast genome of Arabidopsis thaliana . DNA Research 6: 283–290. [DOI] [PubMed] [Google Scholar]
- Swiatek M, Regel RE, Meurer J, Wanner G, Pakrasi HB, Ohad I, Herrmann RG. 2003. Effects of selective inactivation of individual genes for low‐molecular‐mass subunits on the assembly of photosystem II, as revealed by chloroplast transformation: the psbEFLJoperon in Nicotiana tabacum . Molecular Genetics and Genomics 268: 699–710. [DOI] [PubMed] [Google Scholar]
- Torabi S, Umate P, Manavski N, Plochinger M, Kleinknecht L, Bogireddi H, Herrmann RG, Wanner G, Schroder WP, Meurer J. 2014. PsbN is required for assembly of the photosystem II reaction center in Nicotiana tabacum . Plant Cell 26: 1183–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff P, Alt J, Widger WR, Cramer WA, Herrmann RG. 1985. Localization of the gene for apocytochromeb‐559 on the plastid chromosome of spinach. Plant Molecular Biology 4: 103–110. [DOI] [PubMed] [Google Scholar]
- Williams‐Carrier R, Kroeger T, Barkan A. 2008. Sequence‐specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA 14: 1930–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong HB, Wang J, Huang C, Rochaix JD, Lin FM, Zhang JX, Ye LS, Shi XH, Yu QB, Yang ZN. 2020. mTERF8, a member of the mitochondrial transcription termination factor family, is involved in the transcription termination of chloroplast gene psbJ1 . Plant Physiology 182: 408–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Leister D, Kleine T. 2017. Arabidopsis thaliana mTERF10 and mTERF11, but Not mTERF12, are involved in the response to salt stress. Frontiers in Plant Science 8: 1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubovskaya E, Mejia E, Byrnes J, Hambardjieva E, Garcia‐Diaz M. 2010. Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell 141: 982–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cui YL, Zhang XL, Yu QB, Wang X, Yuan XB, Qin XM, He XF, Huang C, Yang ZN. 2018. A nuclear‐encoded protein, mTERF6, mediates transcription termination of rpoA polycistron for plastid‐encoded RNA polymerase‐dependent chloroplast gene expression and chloroplast development. Scientific Reports 8: 11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Cai M, Zhang X, Li Y, Zhang J, Zhao H, Kong F, Zheng Y, Qiu F. 2014. Genome‐wide identification, evolution and expression analysis of mTERF gene family in maize. PLoS ONE 9: e94126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P, Hammani K, Rojas M, Voelker R, Vargas‐Suarez M, Borner T, Barkan A. 2012. Protein‐mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Research 40: 3092–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Zhang L, Ullah A, Jin X, Yang X, Zhang X. 2016. Identification of multiple stress responsive genes by sequencing a normalized cDNA library from Sea‐Land Cotton (Gossypium barbadense L.). PLoS ONE 11: e0152927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Watkins KP, Barkan A. 2013. A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo . Plant Cell 25: 2265–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubo YO, Borner T, Liere K. 2011. Measurement of transcription rates in Arabidopsis chloroplasts. Methods in Molecular Biology 774: 171–182. [DOI] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Phenotype and ribosome profiling analyses of the Arabidopsis mda1 mutant allele hcf111-1.
Fig. S2 MDA1 is associated to membranes in chloroplasts.
Fig. S3 Quantification of the abundance of the 1.4‐ and 1.1‐kb psbE‐F‐L‐J mRNAs in the different plant genotypes by RNA blot analyses.
Fig. S4 Genome mapping of the predominant in vivo 5′ and 3′ ends for the processed psbE‐F‐L‐J and ndhA mRNAs determined by cRT‐PCR in different genotypes.
Fig. S5 RNA gel mobility shift assays showing no binding of MDA1 to sRNAs.
Fig. S6 psbE DNase footprinting assays.
Methods S1 Photosynthetic capacity measurements and genome wide analyses of chloroplast transcriptome and translatome.
Table S1 List of oligonucleotides used in this study.
Table S2 Chlorophyll a fluorescence induction and light‐induced PSI absorbance changes.
Table S3 Ribosome profiling data of hcf111‐1.
Table S4 List of proteins identified by LC‐MS/MS in co‐immunopurification assays using MDA1 as bait.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


