Abstract
Acetylcholine (ACh) signaling orchestrates mammalian movement, mental capacities, and inflammation. Dysregulated ACh signaling associates with many human mental disorders and neurodegeneration in an individual‐, sex‐, and tissue‐related manner. Moreover, aged patients under anticholinergic therapy show increased risk of dementia, but the underlying molecular mechanisms are incompletely understood. Here, we report that certain cholinergic‐targeting noncoding RNAs, named Cholino‐noncoding RNAs (ncRNAs), can modulate ACh signaling, agonistically or antagonistically, via distinct direct and indirect mechanisms and at different timescales. Cholino‐ncRNAs include both small microRNAs (miRNAs) and long noncoding RNAs (lncRNAs). The former may attenuate translation and/or induce destruction of target mRNAs that code for either ACh‐signaling proteins or transcription factors controlling the expression of cholinergic genes. lncRNAs may block miRNAs via ‘sponging’ events or by competitive binding to the cholinergic target mRNAs. Also, single nucleotide polymorphisms in either Cholino‐ncRNAs or in their recognition sites in the ACh‐signaling associated genes may modify ACh signaling‐regulated processes. Taken together, both inherited and acquired changes in the function of Cholino‐ncRNAs impact ACh‐related deficiencies, opening new venues for individual, sex‐related, and age‐specific oriented research, diagnosis, and therapeutics.
Keywords: acetylcholine, age, lncRNAs, miRNAs, noncoding RNA, sex, transcript regulation, transcription factors
![]()
Abbreviations
ACh, acetylcholine
AChE, acetylcholinesterase
BChE, butyrylcholinesterase
ChAT, choline acetyltransferase
CHRM, cholinergic receptor muscarinic
CHRNA, cholinergic receptor nicotinic α
CHT, choline transporter
CNS, central nerves system
COLQ, Acetylcholinesterase collagenic tail Q peptide
IL, interleukin
MRE, miRNA recognition element
ncRNA, noncoding RNA
PRIMA1, proline‐rich membrane anchor 1
PSG, pseudogene
VAChT, vesicular acetylcholine transporter
α7, nicotinic α7 ACh receptor
Acetylcholine (ACh) controls both peripheral and central nerves system (CNS) functions (Fig. 1A‐D), and impairments in the delicate balance between ACh production and elimination (here referred to as ‘the cholinergic tone’) may be detrimental to both functions. Numerous proteins take part in maintaining a stable cholinergic tone throughout the central and peripheral nervous systems (CNS, PNS) [1]. Specifically, the choline acetyltransferase (ChAT) protein synthesizes ACh, which is then packaged and transmitted to the synaptic cleft or to the bloodstream by the vesicular ACh transporter (VAChT) protein, produced from a gene embedded within the first intron of the ChAT gene. ACh signaling failure is observed in Alzheimer’s disease [2, 3, 4], amyotrophic lateral sclerosis (ALS) [5], and other neurodegenerative diseases [6, 7], whereas hyperactivity of ACh can lead to severe cardiac deficits [8, 9]. At its action sites, ACh can trigger the activation of multi‐subunit nicotinic ACh ionotropic receptor channels (CHRNs) that can be either homomeric or heteromeric and are composed of at least one α subunit and one β subunit [10] or metabotropic G protein‐coupled muscarinic receptors (GPCR CHRMs). Apart from the nicotinic α7 ACh receptor (α7) CHRN receptor isoform, which is a homomer of five subunits of the nicotinic receptor subunit α7, all CHRNs and cholinergic receptor muscarinic (CHRMs) are heteromeric, composed of five different subunits each. These ACh receptors are encoded by 16 CHRN and five CHRM subunit genes [11].
Fig. 1.
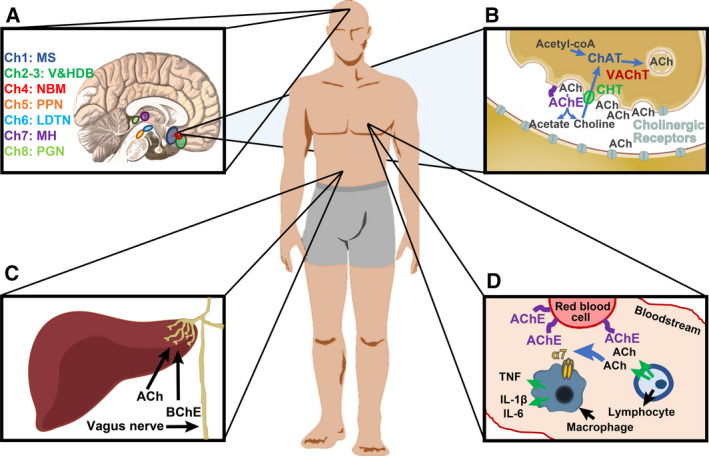
The cholinergic system. (A) The human brain includes eight cholinergic nuclei. Ch1 in the medial septum, Ch2 and Ch3 in the vertical and horizontal limbs of the diagonal band of Broca, Ch4 in the nucleus basalis of Meynert, Ch5 in the pedunculopontine nucleus, Ch6 in the laterodorsal tegmental nucleus, Ch7 in the medial habenula, and Ch8 in the para‐bigeminal nucleus [12]. (B) ChAT in the presynaptic cell synthesizes ACh from choline and acetyl‐CoA. VAChT packages ACh in vesicles, which are secreted to the cleft. There, ACh can activate pre (auto)‐ and postsynaptic cholinergic receptors (nicotinic or muscarinic). ACh in the cleft is hydrolyzed to acetate and choline by acetylcholinesterase (AChE) which is attached to the cellular membrane by proline‐rich membrane anchor 1 (PRIMA1, in the brain) or Acetylcholinesterase collagenic tail Q peptide (ColQ, in neuromuscular junctions). Choline transporters (CHT) transporters reuptake choline from the cleft to the presynaptic cell [1, 13]. (C) The vagus nerve reaches internal organs such as the liver, where it intercepts information and attenuates inflammation via ACh blockade of the NFkB pathway. In the liver, the main cholinesterase enzyme is butyrylcholinesterase (BChE) [13, 14]. (D) In the blood, ACh secreted by immune cells such as lymphocytes is intercepted by the α7 nicotinic receptor of other immune cells (e.g., macrophages), which reduces their inflammatory signal [TNF, interleukin (IL)‐1β, IL‐6]. Blood ACh can be hydrolyzed by AChE on the membrane of red blood cells [13].
Cholinergic receptors are located both in the postsynaptic or target cells, where they mediate cell‐to‐cell communication, and in the presynaptic (or secreting) cell, where they serve as indicators of ACh levels in the cleft or within the bloodstream. Correct ACh levels are maintained by a balance between the rate of synthesis and rate of degradation by the hydrolyzing enzymes AChE and BChE. The main nervous system cholinesterase is AChE, which is translated from several AChE mRNA splice variants [15]. The major AChE‐S splice variant is translated into a membrane‐bound tetramer, whereas the monomeric soluble splice variant (AChE‐R) is present at a far lower level and is induced under stressful conditions [16, 17, 18]. AChE‐S is anchored to the cell membrane via the structural protein PRIMA1 in the brain or the collagen‐related COLQ protein in neuromuscular junctions. BChE is a soluble tetramer, which is mainly expressed in the liver (https://www.proteinatlas.org/ENSG00000114200‐BCHE/tissue). After ACh breakdown, choline is retrieved to the cell through specific CHT [6].
Control processes sustaining balanced ACh levels involve direct communication linking internal brain cholinergic projections (Fig. 1A,B), brain–body messages (Fig. 1C), immune system functioning (Fig. 1D), and neuromuscular interaction. In the mammalian brain, cholinergic signals originate from eight distinct and autonomous nuclei (Fig. 1A). Cholinergic trajectories initiating in these areas innervate different brain regions and along with cholinergic interneurons contribute to the CNS cholinergic tone [12]. The main route of cholinergic brain–body communication involves the vagus nerve that innervates various peripheral tissues (including the heart, lung, liver, and abdomen). The afferent vagal input to the brain signals temperature, pain, touch, and stretch levels, and its efferent output are either paraganglionic or muscular [14]. ACh further plays a critical role in the innate immune system, where it mediates anti‐inflammatory reactions via its secretion and activation in many blood cells in the periphery (macrophages, monocytes, NK cells, granulocytes, B cells, diverse subtypes of T cells and others [13, 19, 20] (https://www.proteinatlas.org/ENSG00000175344‐CHRNA7/blood) and of microglia in the brain [21]).
Apart from its regulation of vital functions, ACh is an important effector of the sex‐specific, circadian, and age‐related variability between individuals [22, 23, 24, 25]. This reflects its capacity to modulate cognitive, behavioral, and immune defense features, and affects human health and well‐being in diverse ways. Notably, this immense phenotypic variability is difficult to explain by coding genes alone. Rather, a ‘cholinergic regulatome’ might be involved. This compound collection of genes may exert an important regulatory control over the cholinergic system, affecting its adaptation flexibility and its interindividual variability. The cholinergic regulatome maintains the ACh tone by regulating transcripts coding for the ACh‐synthesizing and ACh‐destructing enzymes, as well as transcription factors (TFs) and nucleases enhancing or silencing their expression, and upstream RNA controllers including short and long noncoding‐RNAs (ncRNAs). Together, these agents variably operate to control the expression of coding genes at the pre‐ and/or post‐transcription levels, as is briefly listed below.
Transcription factors are proteins that execute the first stage in the transcription–translation process of DNA. They may function as master regulators or as selective context‐dependent selectors of expression. The same TF can regulate different genes in different tissues or in the same tissue under diverse conditions [26]. TFs can enhance gene expression or silence it, and they may be recruited to the nucleus in response to a cellular event. Cholinergic TFs may hence execute the translation of an array of specific genes controlling the production and destruction of ACh‐related transcripts and proteins (Fig. 2A,B).
Fig. 2.
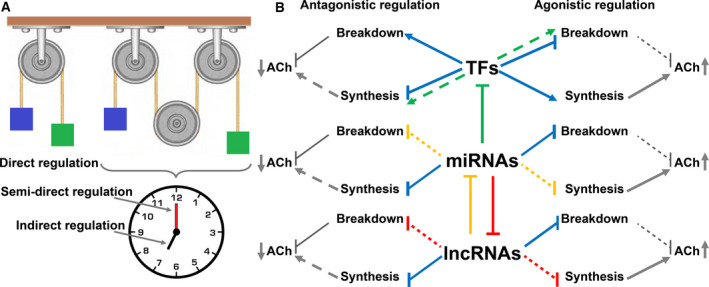
Types and forms of ncRNA regulation over the cholinergic tone. (A) Immediate regulation of cholinergic transcripts by ncRNA (one wheel) is referred to as ‘direct’, unlike the effect of lncRNAs over ACh signaling, which is mediated via other ncRNAs or TFs (three wheels). This may occur rapidly (the clock’s minutes hand), in which case it is referred to as ‘semidirect regulation’, or slowly (hours hand), to be defined ‘indirect regulation’. (B) TFs, miRNAs, and lncRNAs can each cause either agonistic (right hand side) or antagonistic (left hand side) regulation. Triangle arrows (→) indicate induction, and straight‐line arrows (Ⱶ) indicate suppression. Blue lines indicate innate features of the TF\miRNA\lncRNA, whereas the red, green, and yellow lines indicate complex systems in which a full line leads to the effect shown by the scattered lines. For example, when miRNAs repress enhancing TFs (green full line), they cease to induce genes involved in ACh breakdown (e.g., AChE, green scattered line to the right). This weakens the effect on ACh breakdown (leading to elevated ACh levels) [26, 27, 28, 29, 30, 31]. Note that each of the ncRNA types may affect the impact of the other types.
Both the properties and roles of ncRNAs are more diverse than those of TFs, and they can be divided into subgroups according to their length and function. Specifically, microRNAs (miRNAs) are, by far, the most intensively studied type of ncRNAs [32]. In their mature form, miRNAs are single‐stranded short RNAs (~ 20–23 bases). They can either be transcribed from a dedicated gene (intergenic miRNAs) [33] or be processed from a spliced‐out intron (intronic miRNAs) [34]. In spite of this general division and despite the fact that intronic miRNAs are normally coexpressed with their host genes, their expression can be uncoupled via alternative splicing or autonomous transcription [35, 36]. Further, intronic miRNAs can also control the expression of their host genes [37, 38] or cooperate with them to control cellular function [39]. miRNAs are processed into their final form by the Drosha and Dicer protein complexes and serve as guide RNAs leading to targeted mRNA degradation (e.g., via poly‐A shortening or cleavage) by the RNA‐induced silencing complex complex [27, 28] (Fig. 2B). In comparison, long noncoding RNAs (lncRNAs) are at least ~ 200 bases long and can originate from individual promoters, via splicing or from the minus strand of genes. lncRNAs may impact gene expression by silencing a segment of a specific chromosome, like the chromosome X lncRNA Xist [29]. Alternatively, they may take part in organizing nuclear paraspeckles, like NEAT1 [40]. Other lncRNAs are abundantly localized in the cytoplasm and operate as ‘sponges’ for short RNAs [30]. Thanks to their large number, certain lncRNAs play important roles in controlling cellular gene expression at large [31, 41] and ACh signaling in particular [42, 43], while others are being studied in different contexts.
Defining the ‘Cholino‐ncRNA’ landscape
To describe ncRNA regulation, one should first define the types and manners in which these regulatory processes take place. Largely, some ncRNAs can directly affect the transcript levels of the gene of interest (e.g., by preventing or inducing transcription or via binding to the mRNA; this type of regulation will be referred hereafter as direct regulation). Alternatively, ncRNAs’ effect on the gene of interest can be mediated via other ncRNAs (i.e., one type of ncRNA leads to under‐ or overexpression of another ncRNA that binds to the gene of interest) or via a coding gene (e.g., a ncRNA can affect transcript levels of a second gene, which indirectly affects the expression levels of the gene of interest). The two last manners are nondirect types of regulation, with the first (including ncRNAs mediators) being independent of translation and is, hence, more immediate; for the sake of terminology, this regulation mode will be referred hereafter as semidirect regulation, whereas the second nondirect regulatory pathway (consisting of protein products of genes other than the gene of interest) requires translation and is therefore slower. This type of control will be referred to as indirect regulation. Graphical representation of this classification is shown in Fig. 2A.
At another level, biological regulation can be classified by its final effect (i.e., does the regulation lead to an agonistic or an antagonistic effect). For ACh signaling processes, one may regard the signal intercepted by the accepting cell as the terminal stage. Hence, any regulation that reduces that signal (by lowering the amount of ACh released, by elevating the amount of ACh hydrolyzed in the intercellular space, or by reducing the amounts of receptors on the acceptor cell) may be regarded as antagonistic, whereas any regulation that inversely amplifies the intercepted signal will be regarded as agonistic (Fig. 2B). Each of these outcomes may involve TFs, miRNAs, or lncRNAs, as is briefly discussed below.
Direct regulation
At the basal state, the pre‐ and post‐transcriptional processes orchestrating ACh production and destruction (e.g., TFs and miRNAs) are kept balanced, maintaining a quiescent cholinergic tone (Fig. 3A). When the basal state is modified, prompt, rapid regulators may directly affect mRNAs whose expression directly impacts ACh signaling, for example, the ‘cholinergic genes’ include ChAT, VAChT, AChE‐S, AChE‐R, BChE, COLQ, CHT, PRIMA1, and the cholinergic receptors [25].Also, miRNAs can block translation and/or lead to degradation of their target transcripts, which carry complementary sequence motifs [32]. Therefore, miRNAs that control multiple cholinergic transcripts each may exert a pronounced agonistic or antagonistic impact via changing the cholinergic signals (e.g., by targeting AChE and attenuating ACh hydrolysis, they would operate as direct agonistic regulators; Fig. 3B). Inversely, miRNAs targeting ChAT, VAChT, and the cholinergic receptors may limit ACh synthesis, secretion, or interception, weakening the ACh signal and operating as direct antagonistic regulators (for miRNAs operating as direct regulators (agonistic or antagonistic), see Table S1). Other genes whose downregulation affects ACh signaling may add or delete activating or inhibitory cholinergic receptors to or from the postsynaptic membrane via endocytosis (e.g., Arrestin [44], Clathrin [45], Rab5, 11, 22, and Arf6 [46], and CA3 [47]). The complexity of direct RNA regulators of the cholinergic tone thus depends on and is amplified by the complexity of their affected protein targets.
Fig. 3.
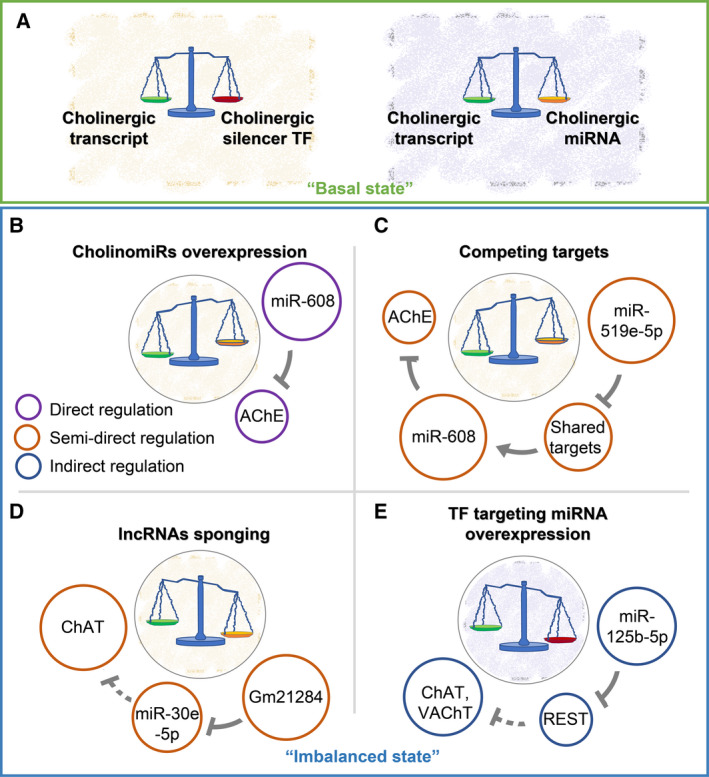
Variable ncRNA‐mediated routes control the cholinergic tone. ‘Basal state’ and ‘imbalanced state’ are noted by blue and green frames, respectively. (A) The ‘basal state’ indicates maintained balance between the cholinergic transcripts (green Libra’s basket) to their controlling TFs (red Libra’s basket) and miRNAs (orange Libra’s basket). (B‐E) The diameter of the colored circles indicates increased (larger) or reduced (smaller) expression B Overexpressed CholinomiRs (e.g., miR‐608) predict lower levels of their cholinergic targets (e.g., AChE) [48]. (C) Excess of a noncholinergic miRNA (e.g., miR‐519e‐5p) sharing targets with a cholinergic miRNA (e.g., miR‐608) leads to downregulation of the shared targets, elevated CholinomiR levels, and downregulated cholinergic targets [48, 49]. (D) Excess of sponging lncRNAs [including competing pseudogenes (PSGs)] decreases the levels of their miRNA targets, ascending the levels of the miRNAs’ cholinergic targets. For example, excess lncRNA Gm21284 downregulates miR‐30e‐5p, leading to elevated ChAT [50]. E. miRNAs targeting TFs modulate the targets of these TFs. For instance, miR‐125b‐5p excess (noted as relevant for men‐women brain differences in mental disease [51]) suppresses the silencing TF REST, consequently leading to upregulation of REST’s targets (e.g., ChAT, VAChT [49, 52]).
Semidirect regulation
In addition to the large numbers of cholinergic targets regulated by miRNAs targeting cholinergic transcripts (‘CholinomiRs’), these miRNA regulators often control noncholinergic transcripts as well. Therefore, transcripts sharing miRNA recognition motifs with cholinergic mRNAs would compete with them over the pool of those miRNAs. For example, the AChE‐targeting miR‐608 resides within an intron of the SEMA4G gene, which is widely expressed throughout the brain, abdomen, and immune blood cells (https://www.proteinatlas.org/ENSG00000095539‐SEMA4G). Notably, miR‐608 also targets the noncholinergic, anxiety‐related CDC42 and IL‐6 transcripts [48] (and its hosting gene itself; http://www.mirdb.org/cgi‐bin/target_detail.cgi?targetID=2034765). Therefore, other miRNAs that target CDC42 or IL‐6 can semidirectly affect AChE expression levels and the cholinergic tone by suppressing their noncholinergic shared targets, leading to higher levels of those free cholinergic‐targeting miRNAs and resulting in reduced cholinergic transcripts. Examples include miR‐519e‐5p, which is predicted to target both CDC42 and IL‐6, as well as 70 other miR‐608 targets [49]. Therefore, although intergenic miR‐519e‐5p (expressed throughout the brain and body tissues; https://ccb‐web.cs.uni‐saarland.de/imota/) does not target any cholinergic transcript, its increases may downregulate at least part of those 72 targets, ‘freeing’ miR‐608 chains and potentiating AChE downregulation (Fig. 3C).
lncRNAs as well may execute semidirect regulation over the cholinergic tone, for example, when they operate as sponges to miRNAs. Thus, lncRNA Gm21284 includes binding sites for the ChAT‐targeting miR‐30e‐5p and was shown to localize in the cytoplasm of rat brain cells [50] Overexpression of miR‐30e‐5p was accompanied by reduced ChAT mRNA levels, which was rescued by introducing higher levels of the lncRNA Gm21284. That Gm21284 may operate as a sponge to the ChAT‐targeting miR‐30e‐5p would prevent it from downregulating ChAT (Fig. 3D). Likewise, the lncRNA GAS5 operates as a sponge to miR‐96‐5p [53], which predictably targets ChAT mRNA. A recent report of a relatively slow but transient overexpression of ChAT and ACh [43] associates excess of GAS5 with lagged upregulation of both ChAT mRNA and protein levels. Finally, the paraspeckle‐regulating lncRNA NEAT1 targets miR‐132, a conserved miRNA that targets AChE [54, 55].
A subclass of lncRNAs includes PSGs, which do not code for proteins and many of which carry miRNA recognition elements (PSG+MRE). These PSGs compete with mRNAs over targeting miRNAs, specifically in the brain [56]. Knockdown of such PSG+MRE leads to specific elevation of the miRNAs targeting them and consequent downregulation of the mRNA targets of these miRNAs. For example, the PSG PGOHUM00000243565 (PSG565) carries miRNA recognition elements (MREs) for several miRNAs targeting cholinergic genes (AChE, BChE, VAChT). Knocking it down led to downregulation of these cholinergic transcripts that was proportional to the amount of shared MREs with the knocked‐down PSG+MRE [57] (for a list of cholinergic lncRNAs and PSGs and their target miRNAs, see Table S2).
Indirect regulation
Indirect (i.e., slow, lagged) regulation of ACh signaling may take two forms. First, it includes miRNAs controlling the expression levels of TFs controlling the production of cholinergic transcripts (Fig. 3E). The ChEA dataset [52] includes 62 TFs that regulate three or more cholinergic targets, including the cholinergic receptors. Several TFs, such as JARID2, only regulate agonistic cholinergic genes [ChAT, VAChT, cholinergic receptor nicotinic α4 (CHRNA4), CHRM3, and CHRM4]. Others, such as SRY, mainly control antagonistic genes (AChE, PRIMA1, COlQ) or agonistic genes that are expressed in the periphery (the non‐CNS receptors α3, β1, γ, δ, with M4 as the only CNS agonistic one). Yet, other TFs regulate cholinergic genes, which are expressed in distinct tissue types. For example, EGR1 regulates ChAT, VAChT, AChE PRIMA1, α2,4,6,7, β4, δ, ε, and M1,2,3,4 and is highly expressed throughout the brain and in blood immune cells (https://www.proteinatlas.org/ENSG00000120738‐EGR1 ). In comparison, AR regulates CHT, ChAT, VAChT, BChE, COLQ, PRIMA1, α2,3,5,7, β3, δ, and M3 and is highly expressed in the liver, the gall bladder, and the genitals (https://www.proteinatlas.org/ENSG00000169083‐AR/tissue ). This indicates that these two TFs may each control the basal expression level of cholinergic transcripts in their organs of expression (for a list of cholinergic TFs and their targets, see Table S3).
Transcription factors control the cholinergic tone in a tissue‐specific manner. Therefore, the impact of miRNAs that target those TFs can only affect the cholinergic tone in those tissues. For instance, several miRNAs such as miR‐16‐5p (highly expressed throughout the body) and miR‐155‐5p (expressed in most of the body tissues including the brain; https://ccb‐web.cs.uni‐saarland.de/imota/mit/) target each of the three ACh‐agonistic TFs MYC, c‐FOS, and JARID2 (these TFs are referred to as ‘agonistic’ since their target genes include at least two ACh synthesis genes and a couple of cholinergic receptors but no ACh breakdown genes; http://carolina.imis.athena‐innovation.gr/diana_tools/web/index.php?r=tarbasev8%2Findex). Likewise, TRIM28 is a TF that targets the α4 and β2 CHRNs but no other cholinergic transcript. This TF is highly expressed throughout the brain, suggesting that its downregulation may exert a specific antagonistic effect on the α4β2‐mediated cholinergic tone.
The above classification is inevitably incomplete, since certain miRNAs can operate both as direct and as indirect regulators. By targeting both a cholinergic mRNA and a TF that targets the very same gene, such miRNAs may downregulate the cholinergic tone both in the immediate and in the long term. For example, miR‐124, one of the most conserved and most abundantly expressed miRNAs in the mammalian brain [58], targets both the synaptic AChE transcript AChE‐S and the TF SOX9 that controls AChE transcription. Other examples include miR‐24, which targets BChE, the soluble variant of AChE (AChE‐R) and the TF HNF4A that regulates the transcription of both AChE and BChE [59]. Interestingly, HNF4A is exclusively expressed in the BChE‐expressing gastrointestinal, liver, gall bladder, pancreas, and kidney [6] (https://www.proteinatlas.org/ENSG00000101076‐HNF4A/tissue). Together, these reports suggest simultaneous, tissue‐specific miR‐24‐mediated direct and indirect regulation modes, both of which may exert context‐dependent agonistic effects.
Another example involves the internalization of CHT transporters via a Ca2+‐SNARE mechanism [60] removing CHT transporters from the presynaptic membrane. This would result in reduced reuptake of choline and consequent slowdown of ACh signaling and might lead to downregulation of the participant genes and an indirect antagonistic effect.
Reciprocity in the ncRNA‐cholinergic control
The vital properties of ACh signaling require maintenance of its responsiveness to modified conditions over day and night (Fig. 4A), for example, to ensure consistent surveillance over inflammatory states in central and peripheral tissues. This is ascertained via the vagus nerve, which mediates the capacity of the autonomic nervous system to regulate inflammation through the ‘cholinergic anti‐inflammatory pathway’, controlling ACh levels and its capacity to block inflammation via activating the nicotinic ACh receptor α7 on immune cells. Briefly, afferent fibers of the vagus nerve intercept inflammatory signals in the periphery [13]. In the brain, the muscarinic receptors M1 and M2 control ACh secretion by the efferent vagus in reaction to the afferent inflammatory signals [13]. ACh secreted from the vagus activates the α7 ACh receptors on macrophages, preventing NFkB from entering the nucleus. This ceases the inflammatory reaction of macrophages. When ACh binds to the α7 receptor in macrophages, the bound receptor physically interacts with Jak2 that, in turn, phosphorylates STAT3, inducing its translocation to the nucleus where it operates as a TF, and at the same time activating SOCS3 that inhibits further STAT3 [61, 62] (and, likewise, prevents IL‐6‐induced proinflammatory reaction via blockade of gp130 [63]). Intriguingly, transient expression of STAT3 has an anti‐inflammatory effect while its sustained expression leads to a proinflammatory effect via IL‐6 secretion [63, 64, 65]. Further, ACh‐bound α7 receptors recruit the intergenic miR‐124 that targets STAT3 and prevents its sustained expression [62, 65] (for regulation of miR‐124, see Ref. [66] and https://amp.pharm.mssm.edu/Harmonizome/gene/MIR124‐1). These two concomitant processes result in reduced secretion of inflammatory signals, which is intercepted by the afferent vagus that accordingly leads to attenuation of ACh secretion (Fig. 4B).
Fig. 4.
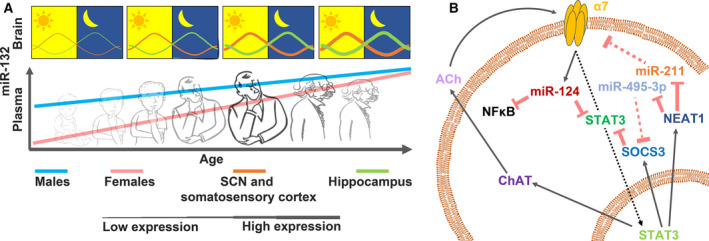
Reciprocity and changes in time and space in cholino‐ncRNAs regulation. (A) miR‐132 as an example of cholino‐ncRNA that changes between tissues and sexes, during daytime and along age. Upper bar: miR‐132 levels in the murine suprachiasmatic nucleus (SCN) and somatosensory cortex (orange line) and in the hippocampus (green line). Thick and thin lines indicate high and low miR‐132 levels, respectively. Lower bar: plasma miR‐132 levels in men and women along age [67, 68, 69, 70, 71]. (B) ACh activation of α7 receptors in macrophages may reciprocally change the cholinergic tone in an auto‐ or paracrine manner. Red arrows with flat heads: suppression. Scattered flat head arrows: Impaired suppression. ACh‐mediated activation of α7 receptors (via interaction with Jak2) induces STAT3 nuclear penetration (dotted arrow) and miR‐124 elevation. In the nucleus, STAT3 induces SOCS3, NEAT1, and ChAT elevation. Cytoplasmic ChAT elevates ACh secretion, and NEAT1 sponges miR‐211 and miR‐495‐3p, among others, reducing their blockade of the α7 NAChR and SOCS3 (respectively), whereas SOCS3 and miR‐124 suppress STAT3 [13, 19, 41, 52, 61, 62, 63, 64, 65, 72].
In addition to the above events, STAT3 also targets ChAT, whereas miR‐124 targets AChE. Since monocytes express ChAT [13, 19], and other immune cells express AChE (https://www.proteinatlas.org/ENSG00000087085‐ACHE/blood), this combination may elevate the cholinergic tone in the short term (by lowering AChE levels) and reduce it in the longer term (by preventing sustained, STAT3‐mediated ChAT expression), by enabling local control over the ACh tone. In human immune cells, miR‐211 targets the α7 subunit mRNA to restrict the cholinergic attenuation of inflammation [72]. Intriguingly, miR‐211 is sponged by the lncRNA NEAT1 [41], the transcription of which is promoted by STAT3 [52]. Moreover, NEAT1 sponges miR‐495‐3p that downregulates STAT3, yielding a feedback loop that can affect NEAT1 levels (Fig. 4B). In summary, the cholinergic tone in the immune system is strongly regulated by ncRNAs directly, semidirectly, and indirectly and in a reciprocal manner (for regulation of miR‐211 and miR‐495‐3p, see Ref. [73, 74] and https://amp.pharm.mssm.edu/Harmonizome/gene/MIR495, https://amp.pharm.mssm.edu/Harmonizome/gene/MIR211).
Another example of reciprocity involves the AChE‐Ca2+‐miR‐132 triad. miR‐132 targets AChE and reduces its synaptic amounts, elevating ACh levels and potentiating its capacity to bind cholinergic receptors [54]. The excess ACh can bind to muscarinic autoreceptors such as M1 and M3 [75, 76, 77], inducing calcium release from intracellular stores [78] and possibly interfering with REM sleep [79]. Since the promoter of the miR‐132 gene includes a calcium response element [54], ACh binding to the M1 and M3 autoreceptors may increase miR‐132 transcription. Thus, miR‐132 can cause a positive feedback loop maintaining a high cholinergic tone and affecting REM sleep. Likewise, new findings suggest that miR‐1010, a Drosophila intronic miRNA which resides in the SKIP gene, creates, with its hosting gene, a homeostatic feedback loop maintaining β2 nicotinic receptor levels. miR‐1010 targets the β2 transcript, thus reducing its transcription levels, whilst β2 receptors themselves initiate a cellular cascade resulting in the expression of miR‐1010 hosting gene [39]. The complex picture of miRNA regulation therefore affects various cholinergic receptors and enzymes, and the transcriptional regulators thereof.
Regulation in time and space
Apart from their capacity to alter the cholinergic tone, diverse ncRNAs are differentially expressed in men and women, throughout daytime, along age and between tissues, which reflects yet higher levels of complexity. This is compatible with the impact of the circadian clock on cholinergic‐related phenomena [80, 81], which may be partially due to cholinergic‐targeting ncRNAs. For instance, miR‐132 peaks during the day and decreases at night in the superchiasmatic nucleus [67] and in the somatosensory cortex [68] but presents an opposite rhythm (i.e., it peaks at night and nadirs at day) in the hippocampus [69] (Fig. 4A). Further, the lncRNA NEAT1 displays rhythmic expression in murine pituitary cells, and many other lncRNAs, including those shown to operate as sponges, show rhythmic expression patterns [82, 83]. Also, the cholinergic TF STAT3 that targets ChAT, BChE, COLQ, PRIMA1, CHRNβ1, CHRNβ4, and the M2 and M4 muscarinic receptors peaks in early morning hours and shows a circadian expression pattern [84, 85].
Notably, different pools of ncRNAs mainly target transcripts responsible to ACh synthesis or breakdown and present highly complex regulation patterns, in immediate and lagged terms. Moreover, the same ncRNA can simultaneously affect several transcripts. Since some ncRNAs are exclusively expressed in a certain tissue(s), a single ncRNA can yield an agonistic affect over the cholinergic tone in a specific tissue and at a certain time frame, while others function in a sustained manner. The complex patterns of those regulatory agents may account for the consequent complexity of the cholinergic tone. For instance, mir‐335‐5p targets both ChAT and REST (http://carolina.imis.athena‐innovation.gr/dianatools/web/index.php?r=tarbasev8%2Findex&miRNAs%5B%5D=hsa‐miR‐335‐5p&genes%5B%5D=&genes%5B%5D=CHAT&genes%5B%5D=REST&sources%5B%5D=1&sources%5B%5D=7&sources%5B%5D=9&publication_year=&prediction_score=&sort_field=&sort_type=&query=1). Mir‐335‐5p is highly expressed in the brain (https://ccb‐web.cs.uni‐saarland.de/tissueatlas/patterns). Since REST silences ChAT and VAChT, miR‐335‐5p can create an antagonistic effect in the brain at the short term (by targeting ChAT) and an agonistic effect in the long term (by targeting REST). In comparison, miR‐212 is highly expressed in the brain, liver, intestines and blood (https://ccb‐web.cs.uni‐saarland.de/tissueatlas/patterns), and targets the TF MYC, the soluble stress‐induced form of AChE (AChE‐R) and BChE [59]. MYC is a cholinergic agonistic TF (which induces transcription of CHT, VAChT, α1,7, β1,2,4, and M2,3,5) [52]. Altogether, miR‐212 may hence exert an agonistic role in the immediate term in the blood and liver (by targeting AChE‐R and BChE), whereas in the long term, it has an antagonistic effect in the brain, blood, and liver (where MYC is widely expressed; https://www.proteinatlas.org/ENSG00000136997‐MYC/tissue).
Further, age‐dependent changes in cholinergic‐regulating ncRNAs suggest either antagonistic pleiotropic effect or age‐beneficial one. That the cholinergic ncRNA expression pattern can change with age and in response to specific stressors may result in long term (and sometimes permanent) changes in the cholinergic tone. An example involves the cholinergic TF EGR1, which targets 15 cholinergic genes (including the two ACh synthesis and secretion genes and the two genes involved in ACh breakdown in the brain). EGR1 is highly coexpressed in the brain with ChAT, VAChT, and AChE (https://www.proteinatlas.org/ENSG00000120738‐EGR1/brain). Correlative changes in the levels of EGR1 and its AChE target occur in human Alzheimer’s disease brains, where both transcripts were shown to be normally expressed in the preclinical stages of the disease while being reduced in its later phases [86]. In rats, EGR1 levels change differently in diverse brain regions, in an age‐dependent manner and in reaction to fear, during learning and along age [87, 88, 89].
Sex‐related differences in these regulatory processes are of particular interest. In male, but not female rats, chronic adolescent stress is accompanied by elevated EGR1 levels, whereas stressed adolescent female, but not male, rats exhibited high CNS inflammation, suggesting sex‐ and tissue‐specific and stress‐induced differences in EGR1 expression [90]. That stress in female, but not male, rats was accompanied by elevated levels of IL‐6, IL‐1β, and NFkB may indicate that high EGR1 levels keep the cholinergic tone unimpaired in stressed males. Likewise, both NEAT1 and GAS5 show age‐correlated expression patterns in female humans [91], and NEAT1 expression elevates with age in murine hippocampi, in parallel to memory decline [92]. Furthermore, brain‐enriched miRNAs isolated from human plasma show sex‐ and age‐specific expression patterns. Those include miR‐132, with lower plasma levels in young females vs. males but with similarly increased levels in men and women older than 60 [70]. Likewise, miR‐132 expression levels ascend with age in murine hippocampi [71] (Fig. 4A).
Discussion
In the body of mammals, the cholinergic tone orchestrates significant shares of brain functioning, sustains balanced inflammatory reactions, and controls the communication with the periphery, among other functions. Despite the plethora of knowledge regarding the cholinergic system, much is left to discover about its ncRNA controllers. Specifically, the features creating the observed large interindividual variability are only partly deciphered. The cholinergic tone depends on a relatively small number of genes (three genes maintaining ACh synthesis, secretion, and reuptake, four controllers of ACh breakdown, and two dozen channels). Nevertheless, the ACh signals differ between tissues and change during the day, along lifetime and between males and females. As listed above, ncRNAs may be responsible for much of this variation.
In this review, we aimed to illuminate the important and complex role of ncRNAs in orchestrating the cholinergic tone, and to highlight the fact that there is still much to look forward to. For example, a rapidly growing body of evidence shows that among other agents, circular RNAs can also operate as sponges [42]. Additionally, transfer RNA fragments, a recently identified family of short ncRNAs that may operate as expression regulators via sequence‐specific transcript degradation, emerge as active regulators of multiple biological processes [93, 94]. Also, while we covered the different cholinergic‐related functions of the various ncRNA types, we ignored those genetic polymorphisms that alter their recognition elements in their target mRNAs, and which can further alter the regulatory effects of these ncRNAs. For example, miR‐132 and miR‐608 both target AChE. Two single nucleotide polymorphisms, one in miR‐608 itself [95] and one in the AChE MRE [48], can impair miR‐608 downregulation of AChE, leading to higher levels of AChE and thus affecting, semidirectly, the expression of miR‐132. Additionally, certain miRNAs may be targeted for destruction by those mRNAs carrying recognition elements complementary to their ‘seed’ sequences [96]. This inverse direction of regulation may add further complexity to the surveillance by ncRNAs over cholinergic signaling, while making this topic even more intriguing than it has been so far.
To conclude, further research is needed to explore the sex‐, age‐, and tissue‐specific interactions between cholino‐ncRNAs and the ever‐changing expression levels of their target transcripts. And yet, even on the basis of the existing knowledge summed up here, it seems that cholino‐ncRNAs can become a fruitful target for biomedical research, for identifying novel biomarkers and for developing new therapeutics. In particular, viewing the entire ncRNA landscape rather than a single cholino‐ncRNA, one is impressed by the intricacy and consequences of their actions as those are reflected in one specific pathway. Thus, observations of a seemingly biochemical pathway have led to the understanding that it actually reflects a complex pyramid of genes, RNAs, and proteins that keep interacting and cross‐interacting between them, culminating in a surprising balance, which is pivotal for human health and well‐being. Yet more specifically, this complexity indicates that to develop new diagnostic and/or therapeutic agents for treating a particular disease, one would gain substantially from exploring the ncRNAs that target the cholinergic balance in the diseased tissue, and doing that in men and women separately and in an age‐dependent manner.
Funding
The authors acknowledge support of this study by the European Research Council Advanced Award 321501 and the Israel Science Foundation Grant No.1016/18 (to HS) and the NOFAR Israel Innovations Authority Grant No. 65812 (to David S. Greenberg).
Supporting information
Table S1. miRNAs and their predicted (agonistic and antagonistic) cholinergic targets. miRNAs (blue) in ascending order. For every miRNA agonistic targets (receptors—turquoise, synthesis proteins—light green), the total number of agonistic targets (green), antagonistic targets (i.e., breakdown proteins—pink), and total number of antagonistic targets (red) are shown.
Table S2. lncRNAs and PSGs and their predicted miRNA targets. lncRNAs and PSGs in ascending alphabetical order. Every lncRNA\PSG has multiple targets (each of which in a different row) when all the rows of a specific lncRNA\PSG are colored in the same color (blue and green alternatively). Last column indicates the number of binding sites of the miRNA to the lncRNA\PSG. The table includes only lncRNAs\PSGs that are predicted to target at least three miRNAs with cholinergic targets and with each of the predicted miRNAs having at least five binding sites on the lncRNA\PSG.
Table S3. TFs and their (agonistic and antagonistic) cholinergic target genes. TFs (blue) in ascending alphabetical order. For every TF agonistic targets (synthesis proteins—light green, receptors—turquoise), the total number of agonistic targets (green), antagonistic targets (i.e. breakdown proteins—pink), and total number of antagonistic targets (red) are shown. CHRNB, cholinergic receptor nicotinic β.
Acknowledgements
The authors are grateful to Dr David S Greenberg, The Hebrew university of Jerusalem, for in‐depth comments on this text.
Edited by Qinghua Cui
References
- 1. Soreq H (2015) Checks and balances on cholinergic signaling in brain and body function. Trends Neurosci 38, 448–458. [DOI] [PubMed] [Google Scholar]
- 2. Mesulam M (2004) The cholinergic lesion of Alzheimer’s disease: pivotal factor or side show? Learn Mem 11, 43–49. [DOI] [PubMed] [Google Scholar]
- 3. Ulrich J, Meier‐Ruge W, Probst A, Meier E and Ipsen S (1990) Senile plaques: staining for acetylcholinesterase and A4 protein: a comparative study in the hippocampus and entorhinal cortex. Acta Neuropathol 80, 624–628. [DOI] [PubMed] [Google Scholar]
- 4. Sáez‐Valero J, Sberna G, McLean CA and Small DH (1999) Molecular isoform distribution and glycosylation of acetylcholinesterase are altered in brain and cerebrospinal fluid of patients with Alzheimer’s disease. J Neurochem 72, 1600–1608. [DOI] [PubMed] [Google Scholar]
- 5. Campanari ML, García‐Ayllón MS, Ciura S, Sáez‐Valero J and Kabashi E (2016) Neuromuscular junction impairment in amyotrophic lateral sclerosis: reassessing the role of acetylcholinesterase. Front Mol Neurosci 9, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith R, Chung H, Rundquist S, Maat‐Schieman MLC, Colgan L, Englund E, Liu Y‐J, Roos RAC, Faull RLM, Brundin P et al (2006) Cholinergic neuronal defect without cell loss in Huntington’s disease. Hum Mol Genet 15: 3119–3131. [DOI] [PubMed] [Google Scholar]
- 7. Bosboom JLW, Stoffers D and Wolters EC (2003) The role of acetylcholine and dopamine in dementia and psychosis in Parkinson’s disease. J Neural Transm Suppl 65, 185–195. [DOI] [PubMed] [Google Scholar]
- 8. Yanagisawa N, Morita H, Nakajima T, Okudera H, Shimizu M, Hirabayashi H, Nohara M, Midorikawa Y and Mimura S (1995) Sarin poisoning in Matsumoto, Japan. Lancet 346, 290–293. [DOI] [PubMed] [Google Scholar]
- 9. Rauh VA, Perera FP, Horton MK, Whyatt RM, Bansal R, Hao X, Liu J, Barr DB, Slotkin TA and Peterson BS (2012) Brain anomalies in children exposed prenatally to a common organophosphate pesticide. Proc Natl Acad Sci USA 109, 7871–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Giniatullin R, Nistri A and Yakel JL (2005) Desensitization of nicotinic ACh receptors: shaping cholinergic signaling. Trends Neurosci 28, 371–378. [DOI] [PubMed] [Google Scholar]
- 11. Brown DA (2019) Acetylcholine and cholinergic receptors. Brain Neurosci Adv 3, 239821281882050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dautan D, Bay HH, Bolam JP, Gerdjikov TV and Mena‐Segovia J (2016) Extrinsic sources of cholinergic innervation of the striatal complex: a whole‐brain mapping analysis. Front Neuroanat 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rosas‐Ballina M and Tracey KJ (2009) Cholinergic control of inflammation. J Intern Med 265, 663–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yuan H and Silberstein SD (2016) Vagus nerve and vagus nerve stimulation, a comprehensive review: Part I. Headache 56, 71–78. [DOI] [PubMed] [Google Scholar]
- 15. Soreq H and Seidman S (2001) Acetylcholinesterase — new roles for an old actor. Nat Rev Neurosci 2, 294–302. [DOI] [PubMed] [Google Scholar]
- 16. Meshorer E and Soreq H (2006) Virtues and woes of AChE alternative splicing in stress‐related neuropathologies. Trends Neurosci 29, 216–224. [DOI] [PubMed] [Google Scholar]
- 17. Kaufer D, Alon Friedman SS and Soreq H (1998) Acute stress facilitates long‐lasting changes in cholinergic gene expression. Nature 393, 373–377. [DOI] [PubMed] [Google Scholar]
- 18. Nadorp B and Soreq H (2014) Predicted overlapping microRNA regulators of acetylcholine packaging and degradation in neuroinflammation‐related disorders. Front Mol Neurosci 7, 00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hecker A, Lips KS, Pfeil U, Kummer W, Padberg W and Grau V (2006) Peripheral choline acetyltransferase is expressed by monocytes and upregulated during renal allograft rejection in rats. J Mol Neurosci 30, 23–24. [DOI] [PubMed] [Google Scholar]
- 20. Pavlov VA and Tracey KJ (2005) The cholinergic anti‐inflammatory pathway. Brain Behav Immun 19, 493–499. [DOI] [PubMed] [Google Scholar]
- 21. Schmitz TW, Soreq H, Judes Poirier X and Nathan Spreng XR (2020) Longitudinal basal forebrain degeneration interacts with TREM2/C3 biomarkers of inflammation in presymptomatic Alzheimer’s disease. J Neurosci 40, 1931–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vallianatou T, Shariatgorji M, Nilsson A, Fridjonsdottir E, Källback P, Schintu N, Svenningsson P and Andrén PE (2019) Molecular imaging identifies age‐related attenuation of acetylcholine in retrosplenial cortex in response to acetylcholinesterase inhibition. Neuropsychopharmacology 44, 2091–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takase K, Kimura F, Yagami T and Mitsushima D (2009) Sex‐specific 24‐h acetylcholine release profile in the medial prefrontal cortex: simultaneous measurement of spontaneous locomotor activity in behaving rats. Neuroscience 159, 7–15. [DOI] [PubMed] [Google Scholar]
- 24. Earnest DJ and Turek FW (1985) Neurochemical basis for the photic control of circadian rhythms and seasonal reproductive cycles: role for acetylcholine. Proc Natl Acad Sci USA 82, 4277–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lobentanzer S, Hanin G, Klein J and Soreq H (2019) Integrative transcriptomics reveals sexually dimorphic control of the cholinergic/neurokine interface in schizophrenia and bipolar disorder. Cell Rep 29, 764–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR and Weirauch MT (2018) The human transcription factors. Cell 172, 650–665. [DOI] [PubMed] [Google Scholar]
- 27. Selvarajan S, Vijayaraghavan J, Bobby Z and Ramalingam J (2019) Micro RNAs‐a review. J Evolution Med Dent Sci 8, 2918–2923. [Google Scholar]
- 28. Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richler C, Soreq H and Wahrman J (1992) X inactivation in mammalian testis is correlated with inactive X‐specific transcription. Nat Genet 2, 192–195. [DOI] [PubMed] [Google Scholar]
- 30. Ulitsky I (2018) Interactions between short and long noncoding RNAs. FEBS Lett 592, 2874–2883. [DOI] [PubMed] [Google Scholar]
- 31. Long Y, Wang X, Youmans DT and Cech TR (2017) How do lncRNAs regulate transcription? Sci Adv 3, eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bartel DP (2018) Metazoan MicroRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hwang JY, Kaneko N, Noh KM, Pontarelli F and Zukin RS (2014) The gene silencing transcription factor rest represses miR‐132 expression in hippocampal neurons destined to die. J Mol Biol 426, 3454–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ryan BM, McClary AC, Valeri N, Robinson D, Paone A, Bowman ED, Robles AI, Croce C and Harris CC (2012) rs4919510 in hsa‐mir‐608 is associated with outcome but not risk of colorectal cancer. PLoS ONE 7, e36306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramalingam P, Palanichamy JK, Singh A, Das P, Bhagat M, Kassab MA, Sinha S and Chattopadhyay P (2014) Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA 20, 76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bell ML, Buvoli M and Leinwand LA (2010) Uncoupling of expression of an intronic MicroRNA and its myosin host gene by exon skipping. Mol Cell Biol 30, 1937–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hübner M, Hinske C, Effinger D, Wu T, Thon N, Kreth F‐W and Kreth S (2018) Intronic miR‐744 inhibits glioblastoma migration by functionally antagonizing its host gene MAP2K4. Cancers 10, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paraboschi EM, Cardamone G, Rimoldi V, Duga S, Soldà G and Asselta R (2017) miR‐634 is a Pol III‐dependent intronic microRNA regulating alternative‐polyadenylated isoforms of its host gene PRKCA. Biochim Biophy Acta 1861, 1046–1056. [DOI] [PubMed] [Google Scholar]
- 39. Amourda C and Saunders TE (2020) The mirtron miR‐1010 functions in concert with its host gene SKIP to balance elevation of nAcRβ2. Sci Rep 10, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Bénard M, Fox AH et al (2014) NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell 25, 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xia D, Yao R, Zhou P, Wang C, Xia Y and Xu S (2020) LncRNA NEAT1 reversed the hindering effects of miR‐495‐3p/STAT3 axis and miR‐211/PI3K/AKT axis on sepsis‐relevant inflammation. Mol Immunol 117, 168–179. [DOI] [PubMed] [Google Scholar]
- 42. Maoz R, Garfinkel BP and Soreq H (2017) Alzheimer’s disease and ncRNAs. Adv Exp Med Biol 978, 337–361. [DOI] [PubMed] [Google Scholar]
- 43. Zhao HY, Zhang ST, Cheng X, Li HM, Zhang L, He H, Qin JB, Zhang WY, Sun Y and Jin GH (2019) Long non‐coding RNA GAS5 promotes PC12 cells differentiation into Tuj1‐positive neuron‐like cells and induces cell cycle arrest. Neural Regen Res 14, 2118–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee KB, Pals‐Rylaarsdam R, Benovic JL and Hosey MM (1998) Arrestin‐independent internalization of the M1, M3, and M4 subtypes of muscarinic cholinergic receptors. J Biol Chem 273, 12967–12972. [DOI] [PubMed] [Google Scholar]
- 45. Lambert L, Dubayle D, Fafouri A, Herzog E, Csaba Z, Dournaud P, el Mestikawy S and Bernard V (2018) Endocytosis of activated muscarinic m2 Receptor (m2R) in live mouse hippocampal neurons occurs via a clathrin‐dependent pathway. Front Cell Neurosci 12, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reiner C and Nathanson NM (2008) The internalization of the M2 and M4 muscarinic acetylcholine receptors involves distinct subsets of small G‐proteins. Life Sci 82, 718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Du A, Huang S, Zhao X, Feng K, Zhang S, Huang J, Miao X, Baggi F, Ostrom RS, Zhang Y et al (2017) Suppression of CHRN endocytosis by carbonic anhydrase CAR3 in the pathogenesis of myasthenia gravis. Autophagy 13, 1981–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hanin G, Shenhar‐Tsarfaty S, Yayon N, Yau YH, Bennett ER, Sklan EH, Rao DC, Rankinen T, Bouchard C, Geifman‐Shochat S et al (2014) Competing targets of microRNA‐608 affect anxiety and hypertension. Human Mol Genet 23, 4569–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen Y and Wang X (2020) miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 48, D127–D131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cheng X, Li H, Zhao H, Li W, Qin J and Jin G (2019) Function and mechanism of long non‐coding RNA Gm21284 in the development of hippocampal cholinergic neurons. Cell Biosci 72, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Camkurt MA, Karababa İF, Erdal ME, Kandemir SB, Fries GR, Bayazıt H, Ay ME, Kandemir H, Ay ÖI, Coşkun S et al (2020) MicroRNA dysregulation in manic and euthymic patients with bipolar disorder. J Affect Disord 261, 84–90. [DOI] [PubMed] [Google Scholar]
- 52. Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR and Ma'ayan A (2010) ChEA: transcription factor regulation inferred from integrating genome‐wide ChIP‐X experiments. Bioinformatics 26, 2438–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang W, Jia YJ, Yang Y‐L, Xue M, Zheng Z‐J, Wang L and Xue Y‐M (2020) LncRNA GAS5 exacerbates renal tubular epithelial fibrosis by acting as a competing endogenous RNA of miR‐96‐5p. Biomed Pharmacother 121, 109411. [DOI] [PubMed] [Google Scholar]
- 54. Shaked I, Meerson A, Wolf Y, Avni R, Greenberg D, Gilboa‐Geffen A and Soreq H (2009) MicroRNA‐132 potentiates cholinergic anti‐inflammatory signaling by targeting acetylcholinesterase. Immunity 31, 965–973. [DOI] [PubMed] [Google Scholar]
- 55. Zhou K, Zhang C, Yao H, Zhang X, Zhou Y, Che Y and Huang Y (2018) Knockdown of long non‐coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR‐132. Mol Cancer 17, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barbash S, Simchovitz A, Buchman AS, Bennett DA, Shifman S and Soreq H (2017) Neuronal‐expressed microRNA‐targeted pseudogenes compete with coding genes in the human brain. Translat Psychiatry 7, e1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Soreq H, Simchovitz A, Barbash S, Hanin G and Roitman M (2017) Microrna‐reacting pseudogenes control cholinergic signaling in brain neurons. Eur Neuropsychopharmacol 27, S417. [Google Scholar]
- 58. Makeyev EV, Zhang J, Carrasco MA and Maniatis T (2007) The MicroRNA miR‐124 promotes neuronal differentiation by triggering brain‐specific alternative pre‐mRNA splicing. Mol Cell 27, 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hanin G and Soreq H (2011) Cholinesterase‐targeting micrornas identified in silico affect specific biological processes. Front Mol Neurosci 4, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Parikh V, St. Peters M, Blakely RD and Sarter M (2013) The presynaptic choline transporter imposes limits on sustained cortical acetylcholine release and attention. J Neurosci 33, 2326–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Metz CN and Tracey KJ (2005) It takes nerve to dampen inflammation. Nat Immunol 6, 756–757. [DOI] [PubMed] [Google Scholar]
- 62. Ulloa L (2013) The cholinergic anti‐inflammatory pathway meets microRNA. Cell Res 23, 1249–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yasukawa H, Ohishi M, Mori H, Murakami M, Chinen T, Aki D, Hanada T, Takeda K, Akira S, Hoshijima M et al (2003) IL‐6 induces an anti‐inflammatory response in the absence of SOCS3 in macrophages. Nat Immunol 4, 551–556. [DOI] [PubMed] [Google Scholar]
- 64. Cevey ÁC, Penas FN, Alba Soto CD, Mirkin GA and Goren NB (2019) IL‐10/STAT3/SOCS3 axis is involved in the anti‐inflammatory effect of benznidazole. Front Immunol 10, 1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sun Y, Li Q, Gui H, Xu DP, Yang YL, Su DF and Liu X (2013) MicroRNA‐124 mediates the cholinergic anti‐inflammatory action through inhibiting the production of pro‐inflammatory cytokines. Cell Res 23, 1270–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Juźwik CA, S. Drake S, Zhang Y, Paradis‐Isler N, Sylvester A, Amar‐Zifkin A, Douglas C, Morquette B, Moore CS and Fournier AE (2019) microRNA dysregulation in neurodegenerative diseases: a systematic review. Prog Neurogibol 182, 101664. [DOI] [PubMed] [Google Scholar]
- 67. Cheng HYM, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S et al (2007) microRNA modulation of circadian‐clock period and entrainment. Neuron 54, 813–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Davis CJ, Clinton JM, Taishi P, Bohnet SG, Honn KA and Krueger JM (2011) MicroRNA 132 alters sleep and varies with time in brain. J Appl Physiol 111, 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Aten S, Hansen KF, Snider K, Wheaton K, Kalidindi A, Garcia A, Alzate‐Correa D, Hoyt KR and Obrietan K (2018) miR‐132 couples the circadian clock to daily rhythms of neuronal plasticity and cognition. Learn Mem 25, 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sheinerman K, Tsivinsky V, Mathur A, Kessler D, Shaz B and Umansky S (2018) Age‐ and sex‐dependent changes in levels of circulating brain‐enriched microRNAs during normal aging. Aging 10, 3017–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nudelman AS, DiRocco DP, Lambert TJ, Garelick MG, Le J, Nathanson NM and Storm DR (2010) Neuronal activity rapidly induces transcription of the CREB‐regulated microRNA‐132, in vivo . Hippocampus 20, 492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bekenstein U, Mishra N, Milikovsky DZ, Hanin G, Zelig D, Sheintuch L, Berson A, Greenberg DS, Friedman A and Soreq H (2017) Dynamic changes in murine forebrain miR‐211 expression associate with cholinergic imbalances and epileptiform activity. Proc Natl Acad Sci USA 114, E4996–E5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen H, Wang X, Bai J and He A (2017) Expression, regulation and function of miR‐495 in healthy and tumor tissues (Review). Oncol Lett 13, 2021–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chitnis NS, Pytel D, Bobrovnikova‐Marjon E, Pant D, Zheng H, Maas NL, Frederick B, Kushner JA, Chodosh LA, Koumenis C et al (2012) MiR‐211 is a prosurvival MicroRNA that regulates chop expression in a PERK‐dependent manner. Mol Cell 48, 353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nadal L, Garcia N, Hurtado E, Simó A, Tomàs M, Lanuza MA, Santafé M and Tomàs J (2016) Presynaptic muscarinic acetylcholine autoreceptors (M1, M2 and M4 subtypes), adenosine receptors (A1 and A2A) and tropomyosin‐related kinase B receptor (TrkB) modulate the developmental synapse elimination process at the neuromuscular junction. Mol Brain 9, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yokotani K, Okuma Y, Nakamura K and Osumi Y (1993) Release of endogenous acetylcholine from a vascularly perfused rat stomach in vitro; inhibition by M3 muscarinic autoreceptors and alpha‐2 adrenoceptors. J Pharmacol Exp Ther 266, 1190–1195. [PubMed] [Google Scholar]
- 77. Ohno‐Shosaku T, Matsui M, Fukudome Y, Shosaku J, Tsubokawa H, Taketo MM, Manabe T and Kano M (2003) Postsynaptic M1 and M3 receptors are responsible for the muscarinic enhancement of retrograde endocannabinoid signalling in the hippocampus. Eur J Neurosci 18, 109–116. [DOI] [PubMed] [Google Scholar]
- 78. Neher E, Marty A, Fukuda K, Kubo T and Numa S (1988) Intracellular calcium release mediated by two muscarinic receptor subtypes. FEBS Lett 240, 88–94. [DOI] [PubMed] [Google Scholar]
- 79. Yamada RG and Ueda HR (2020) Molecular mechanisms of REM sleep. Front Neurosci 13, 1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Elliott WJ (1998) Circadian variation in the timing of stroke onset: a meta‐analysis. Stroke 29, 992–996. [DOI] [PubMed] [Google Scholar]
- 81. Gibbs J, Ince L, Matthews L, Mei J, Bell T, Yang N, Saer B, Begley N, Poolman T, Pariollaud M et al (2014) An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat Med 20, 919–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Park J and Belden WJ (2018) Long non‐coding RNAs have age‐dependent diurnal expression that coincides with age‐related changes in genome‐wide facultative heterochromatin. BMC Genom 19, 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fan Z, Zhao M, Joshi PD, Li P, Zhang Y, Guo W, Xu Y, Wang H, Zhao Z and Yan J (2017) A class of circadian long non‐coding RNAs mark enhancers modulating long‐range circadian gene regulation. Nucleic Acids Res 45, 5720–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Moravcová S, Červená K, Pačesová D and Bendová Z (2016) Identification of STAT3 and STAT5 proteins in the rat suprachiasmatic nucleus and the Day/Night difference in astrocytic STAT3 phosphorylation in response to lipopolysaccharide. J Neurosci Res 94, 99–108. [DOI] [PubMed] [Google Scholar]
- 85. Moravcová S, Pačesová D, Melkes B, Kyclerová H, Spišská V, Novotný J and Bendová Z (2018) The day/night difference in the circadian clock’s response to acute lipopolysaccharide and the rhythmic Stat3 expression in the rat suprachiasmatic nucleus. PLoS ONE 13, e0199405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hu Y, Chen X, Huang S, Zhu Q, Yu S, Shen Y, Sluiter A, Verhaagen J, Zhao J, Swaab D et al (2019) Early growth response‐1 regulates acetylcholinesterase and its relation with the course of Alzheimer’s disease. Brain Pathol 29, 502–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Robinson‐Drummer PA, Chakraborty T, Heroux NA, Rosen JB and Stanton ME (2018) Age and experience dependent changes in Egr‐1 expression during the ontogeny of the context preexposure facilitation effect (CPFE). Neurobiol Learn Mem 150, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chakraborty T, Asok A, Stanton ME and Rosen JB (2016) Variants of contextual fear conditioning induce differential patterns of Egr‐1 activity within the young adult prefrontal cortex. Behaviour Brain Res 302, 122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Penner MR, Parrish RR, Hoang LT, Roth TL, Lubin FD and Barnes CA (2016) Age‐related changes in Egr 1 transcription and DNA methylation within the hippocampus. Hippocampus 26, 1008–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bekhbat M, Howell PA, Rowson SA, Kelly SD, Tansey MG and Neigh GN (2019) Chronic adolescent stress sex‐specifically alters central and peripheral neuro‐immune reactivity in rats. Brain Behav Immun 76, 248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Arshi A, Sharifi FS, Khorramian Ghahfarokhi M, Faghih Z, Doosti A, Ostovari S, Mahmoudi Maymand E and Ghahramani Seno MM (2018) Expression analysis of MALAT1, GAS5, SRA, and NEAT1 lncRNAs in breast cancer tissues from young women and women over 45 years of age. Mol Ther Nucleic Acids 12, 751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Butler AA, Johnston DR, Kaur S and Lubin FD (2019) Long noncoding RNA NEAT1 mediates neuronal histone methylation and age‐related memory impairment. Sci Signal 12, eaaw9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Jehn J, Treml J, Wulsch S, Ottum B, Erb V, Hewel C, Kooijmans RN, Wester L, Fast I and Rosenkranz D (2020) 5’ tRNA halves are highly expressed in the primate hippocampus and sequence‐specifically regulate gene expression. RNA 26, 1–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Grigoriev A and Karaiskos S (2016) Dynamics of tRNA fragments and their targets in aging mammalian brain. F1000Res 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhang AQ, Gu W, Zeng L, Zhang LY, Du DY, Zhang M, Hao J, Yue CL and Jiang J (2015) Genetic variants of microRNA sequences and susceptibility to sepsis in patients with major blunt trauma. Ann Surg 261, 189–196. [DOI] [PubMed] [Google Scholar]
- 96. Sheu‐Gruttadauria J, Pawlica P, Klum SM, Wang S, Yario TA, Schirle Oakdale NT, Steitz JA and MacRae IJ (2019) Structural basis for target‐directed MicroRNA degradation. Mol Cell 75, 1243–1255.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. miRNAs and their predicted (agonistic and antagonistic) cholinergic targets. miRNAs (blue) in ascending order. For every miRNA agonistic targets (receptors—turquoise, synthesis proteins—light green), the total number of agonistic targets (green), antagonistic targets (i.e., breakdown proteins—pink), and total number of antagonistic targets (red) are shown.
Table S2. lncRNAs and PSGs and their predicted miRNA targets. lncRNAs and PSGs in ascending alphabetical order. Every lncRNA\PSG has multiple targets (each of which in a different row) when all the rows of a specific lncRNA\PSG are colored in the same color (blue and green alternatively). Last column indicates the number of binding sites of the miRNA to the lncRNA\PSG. The table includes only lncRNAs\PSGs that are predicted to target at least three miRNAs with cholinergic targets and with each of the predicted miRNAs having at least five binding sites on the lncRNA\PSG.
Table S3. TFs and their (agonistic and antagonistic) cholinergic target genes. TFs (blue) in ascending alphabetical order. For every TF agonistic targets (synthesis proteins—light green, receptors—turquoise), the total number of agonistic targets (green), antagonistic targets (i.e. breakdown proteins—pink), and total number of antagonistic targets (red) are shown. CHRNB, cholinergic receptor nicotinic β.


