SUMMARY
Receptor‐like kinases (RLKs) represent the largest group of cell surface receptors in plants. The monophyletic leucine‐rich repeat (LRR)‐RLK subfamily II is considered to contain the somatic embryogenesis receptor kinases (SERKs) and NSP‐interacting kinases known to be involved in developmental processes and cellular immunity in plants. There are only a few published studies on the phylogenetics of LRR‐RLKII; unfortunately these suffer from poor taxon/gene sampling. Hence, it is not clear how many and what main clades this family contains, let alone what structure–function relationships exist. We used 1342 protein sequences annotated as ‘SERK’ and ‘SERK‐like’ plus related sequences in order to estimate phylogeny within the LRR‐RLKII clade, using the nematode protein kinase Pelle as an outgroup. We reconstruct five main clades (LRR‐RLKII 1–5), in each of which the main pattern of land plant relationships re‐occurs, confirming previous hypotheses that duplication events happened in this gene subfamily prior to divergence among land plant lineages. We show that domain structures and intron–exon boundaries within the five clades are well conserved in evolution. Furthermore, phylogenetic patterns based on the separate LRR and kinase parts of LRR‐RLKs are incongruent: whereas the LRR part supports a LRR‐RLKII 2/3 sister group relationship, the kinase part supports clades 1/2. We infer that the kinase part includes few ‘radical’ amino acid changes compared with the LRR part. Finally, our results confirm that amino acids involved in each LRR‐RLKII–receptor complex interaction are located at N‐capping residues, and that the short amino acid motifs of this interaction domain are highly conserved throughout evolution within the five LRR‐RLKII clades.
Keywords: LRR‐RLKII, SERK, kinase, leucine‐rich repeat receptor, phylogeny
Significance Statement
In the present study, based on 1342 amino acid sequences, five main LRR‐RLKII clades (1–5) were identified, each of which represents mixed functions. Furthermore, our results confirm that short amino acids motifs and single residues, involved in each LRR‐RLKII–receptor complex interaction, are highly conserved in the different clades.
INTRODUCTION
If plant cells are to sense signals from their environment as well communicate with each other they must perceive and process information through so‐called cell surface receptors. For instance, during plant development and growth, as well as during cell specification, proper organisation and communication among cells is of obvious importance. Whereas plant hormones and transcription factors have long been understood to be of greatest importance in such regulation, cell surface receptors are now also considered potentially crucial (e.g. De Smet et al., 2009). Receptor‐like kinases (RLKs), for which the structural basis of ligand perception and signal activation was reviewed by Hohmann et al. (2017) and Chakraborty et al. (2019), represent the largest group of cell surface receptors in plants, and as such are considered the largest plant gene family, with more than 600 members and representing about 2.5% of protein‐coding genes in Arabidopsis (Shiu and Bleecker, 2001; Torii, 2004; Aan den Toorn et al., 2015). Based on Arabidopsis thaliana kinase sequence comparisons using neighbour joining (Shiu and Bleecker, 2001) RLKs were found to be monophyletic, and appear to form one of the clades in the kinase superfamily. Up to 50 different kinase clades or subfamilies were found within the RLKs by Shiu et al. (2004), based on only Arabidopsis and Oryza sequence comparisons, and confirming the classification of Shiu and Bleecker (2001). One of the largest RLK subfamilies is the leucine‐rich repeat (LRR)‐RLKs, which are considered to comprise 235 out of the 610 known RLKs (Aan den Toorn et al., 2015) and which combine an extracellular LRR with an intracellular kinase domain, whose activity is known to process the gathered information. Shiu et al. (2004) showed the LRR‐RLK subfamily to be distributed among 23 clusters, but because they used neighbour joining and did not include bootstraps in their analysis monophyly of these clusters remains unsettled. Sakamoto et al. (2012), based on ‘approximately‐maximum likelihood’ analysis (FastTree; Price et al., 2009) of amino acid kinase sequences from only Arabidopsis and Oryza, concluded that LRR‐RLKs occur in 15 clades (their Figure 1). Most LRR‐RLK clades appear to be fairly well supported, based on Shimodaira–Hasegawa tests for ‘local support’ (see the supplementary data in Sakamoto et al. (2012)), but as CLUSTAL was used for the alignment, and given the nature of FastTree, we are not sure how robust this pattern actually is. The same study (Sakamoto et al., 2012) used LRR‐RLKII amino acid sequences in a separate phylogenetic analysis, but it is not clear what this tree is rooted on and hence their claim that there are three well‐supported clades, named NIK (NSP‐interacting kinase), SERK (somatic embryogenesis receptor kinase) and LRRIIc (based on annotation of the Arabidopsis members in each cluster), is difficult to interpret.
Another important class of surface receptors other than RLKs is formed by the receptor‐like proteins (RLPs), which always include a LRR but lack a cytoplasmic kinase domain (Wang et al., 2010); an example is the extracellular‐like SERK (ELS) proteins (Schmidt et al., 2009), a name that appears not to have been used in the literature so far. Functionally, most RLKs and RLPs are involved in either plant development or plant immunity (He et al., 2018). In addition, there are RLKs that possess neither an extracellular region nor a transmembrane domain and are called receptor‐like cytoplasmic kinases (Sakamoto et al., 2012); these known to be involved in plant immunity.
For all RLKs, ligand–receptor bonding is usually required in order for the kinase to work properly. In plants, most reported RLKs have serine/threonine kinase specificity in ligand–receptor bonding (Butenko et al., 2009) whereas animal RLKs have tyrosine specificity (Shiu and Bleeker, 2001). As indicated above, Shiu and Bleecker (2001) made the first attempt to classify the different RLKs into clade‐based groups, using a kinase sequence‐based phylogenetic tree as a comparative pattern. According to their study, land plant RLKs (including from mosses, ferns, conifers and flowering plants) are monophyletic and constitute a sister group to other monophyletic groups including Raf serine/threonine kinases, related to retroviral genes and the animal receptor tyrosine kinase (RTK) family. The authors concluded that in Arabidopsis 24% of RLK genes have an intracellular kinase region only, and belong to either ‘unique subfamilies’ or are related to ‘kinases with a receptor topology’ (Shiu and Bleecker, 2001) but did not indicate whether they are monophyletic. The authors hypothesised that fusion of the kinase domain with different extracellular structures has led to the current land plant RLK gene family. Furthermore, the same authors proposed that, based on the observed expansion and distribution pattern of RLKs through plant chromosomes, the current length and structure of RLK genes were probably already in place before the diversification of land plants (Shiu and Bleecker, 2001). This scenario has been supported by later studies (e.g. Sakamoto et al., 2012; Liu et al., 2017).
For the LRR‐RLKs, a division into 13 main monophyletic subfamilies (LRR‐RLK I to LRR‐RLK XIII) was proposed by Liu et al. (2017) based on maximum‐likelihood phylogenetic analysis of kinase domain amino acid sequences and on subsequent evolutionary reconstruction of gene structure. Among these 13 subfamilies, additional subdivisions are recognised in subfamilies VI, VII and XIII (i.e. VI‐1, VI‐2, VII‐1, VII‐2, XIII‐1 and XIII‐2), with plant RLL‐RLKs containing 19 clades in total. Using gene expression analysis, Chae et al. (2009) showed experimentally that the LRR‐RLK subfamilies are not correlated with function but each comprise proteins with mixed responses and expression patterns.
As outlined above, the monophyletic LRR‐RLK subfamily II (according to Sakamoto et al., 2012; Liu et al., 2017; Li et al., 2018) contains three distinct and well‐supported LRR‐RLKII clades, named NIK, SERK and LRRIIc (with unassigned function), as claimed by Sakamoto et al. (2012), who also showed that these have tissue‐specific expression patterns. The LRR‐RLKII SERK clade is considered to contain SERKs, known to be involved in both developmental processes (stomatal patterning, root meristem development, floral organ abscission, plant growth, xylem differentiation and male gametophyte development) and as cellular immunity in plants (Li, 2010; He et al., 2018). In this process, members of the LRR‐RLK subfamily II take part in the first phase of the immune process in plants as the elicitors. They detect conserved protein structures of micro‐organisms, so‐called microbe‐associated molecular patterns, such as the 22‐amino‐acid conserved bacterial flagellin (flg22) protein (Newman et al., 2013). Several RLKs, for instance flagellin‐sensing 2 (FLS2), Botrytis‐induced kinase 1 (BIK1), elongation factor‐Tu receptor (EFR), DAMP peptide receptor 1 (AtPEPR1) and BAK1 (brassinosteroid insensitive 1‐associated receptor kinase 1), interacting with RLK1–3 (BIR1–3), activate plant immune systems after pathogen attacks by forming heterodimers with the kinase domain of the SERK proteins by a phosphorylation event between the SERK protein and the receptor (Wang et al., 2010; Roux et al., 2011; Halter et al., 2014; Tang et al., 2015; He et al., 2018). Previous surveys indicated that whereas BAK1 (which is considered by Nam and Li (2002) to be SERK3) and SERK4 are involved in plant immunity, SERK1 and SERK2 appear not to have this function (Albrecht et al., 2008).
Moreover, NIK genes are closely related to SERK genes but are involved in interaction with nuclear shuttle proteins (NSPs) of geminiviruses during viral infection, thereby highlighting the role of NIKs in disease resistance. Although Hecht et al. (2001) showed that the presence of an SPP motif, which is enriched by serine (S) and proline (P), is a unique and specific motif in SERK proteins that can be used as a criterion to distinguish them from other subfamily members like NIKs, Nolan et al. (2011) showed that the SPP motif is actually not present in SERK4 and SERK5. They proposed that these two types of protein (SERKs and NIKs) cannot be separated structurally or functionally and are both involved in development and defence mechanisms of plants.
Plant RLK proteins have confusing nomenclature in public databases and in the literature (e.g. Schmidt et al., 1997; Chinchilla et al., 2007; Albrecht et al., 2008; Schmidt et al., 2009; Li, 2010), with different names such as BAK1, SERK or RKS (for ‘receptor kinase‐like SERK’) having been submitted to NCBI, for example, and some never published in peer‐reviewed studies. SERK1 (BAK1‐LIKE1) and SERK4 (BKK1) can heterodimerize with BRI1 to regulate plant growth (Gou et al., 2012). Whereas Schmidt et al. (2009) labelled plant RLKs as ‘RKS 0–16’ based on their associated domain structures as found in A. thaliana, other authors have used ‘SERK’ (types 1–5) for plant RLK proteins based on phylogenetic and functional studies of the proteins (e.g. Albrecht et al., 2008; Wang et al., 2010; Liebrand et al., 2014; Aan Den Toorn et al., 2015) – although the latter two studies were limited by low LRR‐RLK taxonomic sampling and the use of neighbour joining. ‘BAK1’ has been used as a synonym for SERK 3 proteins (Chinchilla et al., 2007; Albrecht et al., 2008; Li, 2010). Apart from addressing the question of the extent to which these terms are congruent or synonymous, the main aim of our study is to refine phylogenetic patterns among members of the LRR‐RLKII clade, using all available homologous data to date and exploring phylogenetic contrasts between extracellular LRR parts versus that inferred from the kinase. We elucidate trends in SERK structural and functional evolution and try and identify amino acid sites that may be involved in active sites in the SERK proteins, both extra‐ and intracellular.
RESULTS
Phylogenetic analysis of plant SERK proteins
Our final amino acid multiple sequence alignment (MSA) included 1342 sequences of proteins annotated as ‘SERK’ or ‘SERK like’; it comprised 1319 alignment positions and showed contrasting patterns of sequence conservation (Data S1 in the online Supporting Information). Overall alignment quality was good and we found that MAFFT gave reproducible, consistent results (not shown). Moreover, the ASaturA analysis did not indicate apparent saturation in amino acid sites among the sequences. As the Ext (‘extracellular’) matrix comprises the extracellular LRR part of SERK and SERK‐like proteins, rooting for Ext was inferred from the full matrix‐based tree (see Experimental Procedures) as no outgroup sequence is known for LRR.
As can be deduced from Figures S2 and S3, the Ext partition (Data S2), comprising the LRR and transdomain regions, is slightly less conserved than the intracellular (Int) partition (Data S3), comprising the kinase domains. Using PAUP* (Swofford, 2002), we estimated 58% of the characters in Ext to be ‘parsimony informative’, 16% ‘uninformative’ and 26% ‘constant’, whereas in Int these proportions are 54%, 14% and 32%, respectively.
We reconstructed a maximum‐likelihood tree based on the entire MSA of 1319 characters using IQ‐TREE (Nguyen et al., 2015), which selected the JTT model (Jones et al., 1992), with four categories of Gamma rate distribution modelling, as best‐fitting. Using Trichinella Pelle as outgroup in our IQ‐TREE ML tree topology, we found five main clades, four of which had bootstrap support of 100%, and one with 92%. We label these clades LRR‐RLKII 1–5 in order to reflect the general structure and function of these proteins, irrespective of their multiple specific functions (see Figure 1a). Each clade comprised multiple types of ‘RKS type similarities’ and ‘SERK’ or ‘NIK’ annotated sequences; the occurrence and distribution of all types across the five main clades found are summarised in Table 1.
Figure 1.
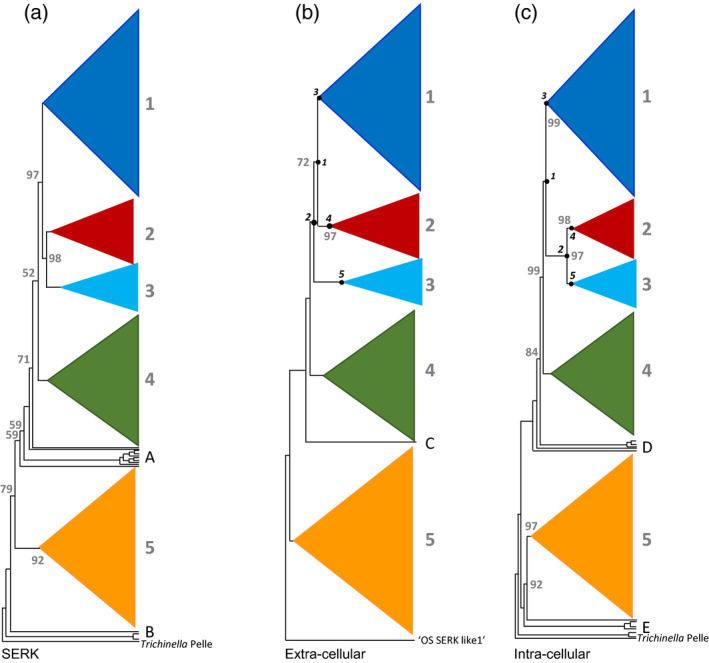
Maximum likelihood phylogenetic trees (cladogram style) of 1342 land plant leucine‐rich repeat receptor‐like kinase II (LRR‐RLKII) amino acid sequences, with five main clades (collapsed), labelled here as 1–5, and using the nematode Trichinella Pelle kinase sequence as the outgroup.
Trees are based on (a) the combined intracellular (‘Int’) + extracellular (‘Ext’) sequence matrix, (b) the extracellular LRR domain (matrix ‘Ext’, see text) rooted by inference from (a), and (c) the intracellular kinase domain (matrix ‘Int’). Bootstrap support values of less than 100 are indicated at the main nodes. Numbers in circles indicate nodes used in apomorphy reconstruction (see text). Terminals not belonging to any of the five main clades are: (A) Marchantia polymorpha [receptor kinase‐like SERK (RKS)2, RKS13, RKS0], Closterium ehrenbergii somatic embryogenesis receptor kinase (SERK), Physcomitrella patens (RKS8, RKS0), Selaginella moellendorffii, Adiantum capillus‐veneris; (B) Oryza sativa (SERK‐like 1); (C) M. polymorpha (RKS2); (D) P. patens (RKS8, RKS0), S. moellendorffii, A. capillus‐veneris; (E) Closterium ehrenbergii SERK, S. moellendorffii, M. polymorpha (RKS13, RKS0).
Table 1.
Occurrence and distribution of sequences annotated as similar to RKS (receptor kinase‐like SERK) and SERK (somatic embryogenesis receptor kinase) across the main leucine‐rich repeat receptor‐like kinase II (LRR‐RLKII) clades found in this study. RKS types are designated according to similarity to types in Schmidt et al. (2009). The SERK and NSP interacting kinase (NIK) types are based on NCBI annotation files
| LRR‐RLKII clade | No. of terminals | Similar to RKS types | SERK types | Non‐SERK proteins |
|---|---|---|---|---|
| 1 | 405 | 1, 4, 5, 7, 9, 11, 14 | 1, 2 | ‘NIK1’, ‘NIK2’ |
| 2 | 132 | 1, 5, 7, 9, 14 | – | ‘NIK3’ |
| 3 | 102 | 5 | 1, 2 | ‘Poap SERK‐like 1’, ‘Poap SERK like 2’ |
| 4 | 280 | 2, 3, 6 | 1, 2 | ‘AtLRRRLK1, AtLRRRLK2, ‘Ta SERK‐like’, ‘OS SERK‐like 1’ |
| 5 | 404 | 0, 8, 10, 12, 13, 16 | 1, 2, 3, 4, 5, BAK1 |
In almost all clades we see land plant and Angiosperm Phylogeny Group relationships reflected [i.e. in Newick notation (Marchantia (Physcomitrella (Amborella (monocots (lower Eudicots (Asterids, Rosids); Figures 2c and 3]. This indicates that gene duplication and subsequent clade proliferation would have occurred before the split among land plants, as also outlined by Shiu and Bleecker (2001) and previous studies (e.g. Liu et al., 2017). Moreover, as indicated in Figure 3(a), ELS proteins, lacking a cellular kinase component, are located in clade 5. We found that NIK and other non‐SERK proteins from the LRR‐RLKII family do not form a separate clade, as has been suggested in previous studies (Sakamoto et al., 2012; Aan Den Toorn et al., 2015), but are distributed among four out of five clades in this study (Table 1, Figure S5).
Figure 2.
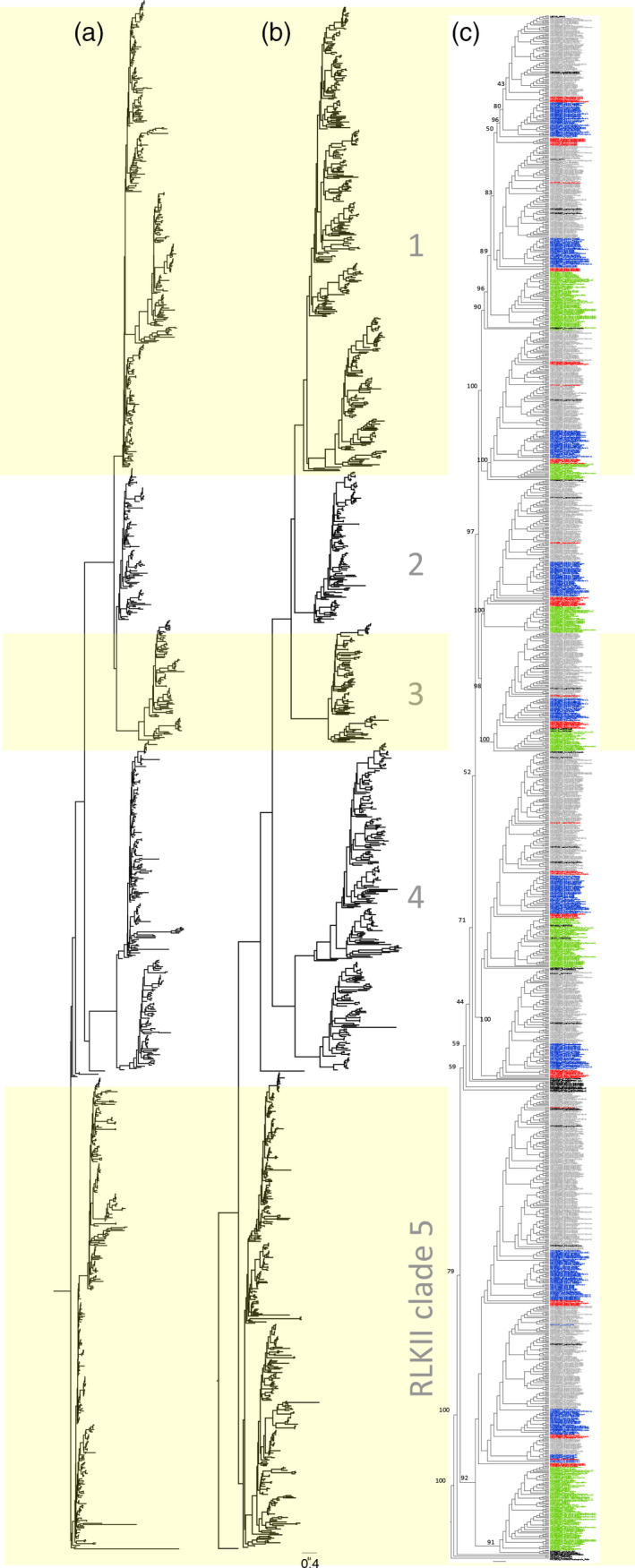
Maximum likelihood phylogenetic trees of 1342 terminals.
Maximum likelihood phylogenetic trees of 1342 terminals based on (a) the receptor‐like kinase II (RLKII) kinase domain (intracellular, ‘Int’, matrix) rooted by Trichinella Pelle, and (b) the extracellular partition (‘Ext’ matrix) rooted by inference from the full matrix‐based tree, both scaled to the actual branch lengths (see scale bar) and (c), in cladogram style, the full matrix, with bootstrap support values indicated at main nodes. Clade labelling is as in Figure 1.
Figure 3.
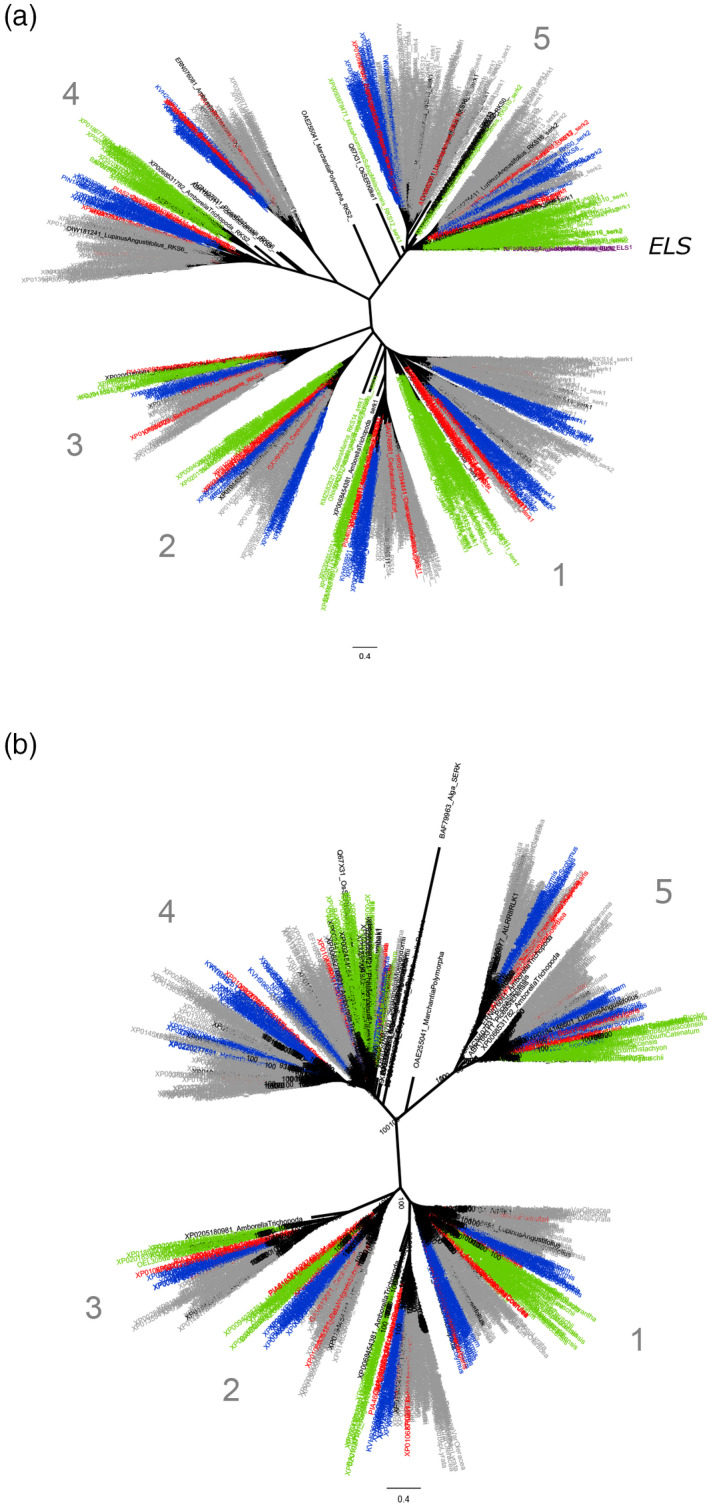
As in Figure 2 but presented as unrooted trees.
(a) The tree based on the extracellular part and (b) that based on the internal kinase part. Asterids, rosids, lower eudicots and monocots are marked with blue, grey, red and green, respectively. Clade labelling is as in Figure 1. The placement of extracellular‐like somatic embryogenesis receptor kinase (ELS) receptors (lacking a cellular kinase component, see text) is indicated.
Trees based on Ext and Int partitions of our MSA showed topological differences (Figure 1): in the Ext topology clades 1 and 2 are sister groups, followed by clade 3. In Int, however, clades 2 and 3 are sister and 1 is sister to them. The Int‐based tree did not differ in topology from the Ext + Int‐based tree (Figure 1).
While clade 1 comprises seven different RKS type similarities, clade 3 predominantly consists of sequences with RKS type 5 similarity (see Table 1). Almost all sequences in clade 2 are determined as NIK3, while clade 1 is a set of NIK1, NIK2 and SERK proteins (Table 1). The LRR‐RLK clades 4 and 5 comprise three and six RKS type similarities, respectively. It appears that RKS type similarity and SERK type distribution over five clades are not dependent on whether Ext or Int partition was used. The known SERK types in clades 1, 3 and 4 are limited to types 1 and 2, whereas in clade 5 all types of SERKs (1–5) are present.
From Table 1, we infer that the relationship between SERK type and RKS type similarity is complicated and contains several incongruences. The BAK1 sequences, which refer to BRI‐I associated kinase, were actually considered as SERK3 in previous studies (e.g. Li, 2010; Roux et al., 2011). All detected BAK1s are in clade 5, and their RKS type similarities are 10, 12, 13 or 16.
In Figure 2, the Ext‐based and Int‐based trees are shown with actual branch lengths in amino acid changes per site. In order to investigate the possibility of paralogue‐specific rate changes following gene duplications, we calculated the number of amino acid substitutions of each of the five clades, divided by its number of terminals, for the Ext‐ and Int‐partitions separately (Table 2), and applied a G‐test (expected/observed goodness of fit) in order to test for significance of differences, both among clades and among parts. According to our results, the main clades do not differ significantly. However, as indicated in Figure 2, the largest difference appears to occur in LRR‐RLKII clade 4 of the Ext‐based tree.
Table 2.
G‐test of branch lengths among the main leucine‐rich repeat receptor‐like kinase II (LRR‐RLKII) clades and in extracellular (‘Ext’) versus intracellular (‘Int’) partitions of somatic embryogenesis receptor kinase (SERK) sequences. Values indicate the total number of amino acids in the particular clade, divided by the number of terminals it comprises
| Main LRR‐RLKII clade | 1 | 2 | 3 | 4 | 5 | E | Χ2 df=4 |
|---|---|---|---|---|---|---|---|
| O | O | O | O | O | ΣO/5 | 4.107 | |
| Ext | 8148/405 = 20.1 | 2323/132 = 17.6 | 2239/102 = 21.9 | 7633/280 = 27.3 | 6186/404 = 15.3 | 102.2/5 | |
| (O – E)2/E | 0.004 | 0.384 | 0.110 | 2.334 | 1.275 | ||
| Int | 4961/405 = 12.2 | 1588/132 = 12.0 | 1838/102 = 18.0 | 3441/280 = 12.3 | 3456/404 = 8.6 | 63.1/5 | 3.633 |
| (O – E)2/E | 0.013 | 0.029 | 2.314 | 0.007 | 1.270 | ||
| O clade total | 32.3 | 29.6 | 39.9 | 39.6 | 23.9 | 165.3/5 |
O, observed; E, expected.
Motif structure in five main clades
Figure 4 summarises the motif structure of proteins for each clade based on the MSA of the Ext + Int matrix as visualized by Mesquite v.3.10 (Maddison and Maddison, 2016) and the sequence logos generated by WebLogo (Crooks et al., 2004). All structures have a signal sequence at the N‐terminal of their protein. As indicated in Figure S3, a conserved motif with unknown function (LSPxY/FExAL) is present in clades 2 and 3 but not in the others. Immediately following this motif, clades 2 and 5 have two leucine zipper domains formed by two leucines each, with six other amino acids in between. Clades 1 and 4 have only one leucine zipper domain, whereas clade 3 does not have this motif (Figure 4). A conserved cysteine pair with four to seven spacer amino acids in between is present in all five clades and is located in between the leucine zipper domains and the LRR domain. The region also consists of leucine zipper domains and the conserved cysteine pair, called N‐capping residues (Aan den Toorn et al., 2015). Additionally, based on the MSA, we found a region enriched by several serines and prolines in some terminals of clade 5 but not in other clades (Figure S7). This motif was designated the ‘SPP (serine–proline–proline) motif’ in Aan den Toorn et al. (2015) but is called here the ‘hinge domain’ as it probably provides the flexible hinge activity of the LRR‐RLKII proteins (Schmidt et al., 1997). In all terminals of clades 1, 2, 3 and 4, and in some terminals of clade 5, another conserved cysteine pair was detected at the end of the LRR motif. We find that the extracellular part of LRR‐RLKII proteins is terminated by a transmembrane motif, and detect a semi‐conserved motif (xGxL/TK/RxF/YxxxEL/Tx) with unknown function at the end of the transmembrane motif in all clades (Figure 4).
Figure 4.
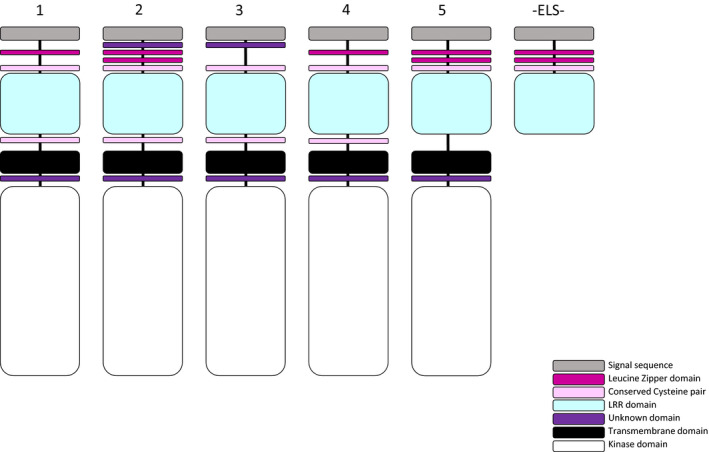
Motif structure of leucine‐rich repeat receptor‐like kinase II (LRR‐RLKII) proteins for each main clade inferred here, as well as for extracellular‐like somatic embryogenesis receptor kinase (ELS).
Structures are based on observations in the WebLogo analyses of all available sequences in this study, displayed in Figures S1–S3.
As is indicated in Figures S1 and S2, the intracellular structure of all terminals is conserved across all clades and appears to be initiated by a kinase domain and ends with a semi‐conserved C‐terminal sequence (xxE/YLSG/xP/xR).
From the DNA perspective, looking at the exon boundaries of the detected domains using ARAPORT (Arabidopsis Information Portal; https://www.araport.org) and NCBI (https://www.ncbi.nlm.nih.gov/) within selected annotated sequences of each five clades revealed that there are 11–12 exons. As is illustrated in Figure 5, unlike the variable size of introns, the size and the order of exons are highly conserved among the different main clades (Table S3). The first exon harbours a signal peptide domain followed by an exon with an N‐capping residue including a leucine zipper domain and a conserved cysteine pair. Next, there are four exons with a total of five LRR domains, with 24 amino acids each, followed by two exons with one LRR domain, including the other conserved cysteine pair or SPP domain, and one transmembrane domain. Overall, the first eight domains are located in the extracellular part of the proteins. The last three or two detected exons are associated with the intracellular part of the protein, recognized by PFAM (https://pfam.xfam.org) as a kinase domain. Sequences 9, 11 and 12 (Figure 5, located in clade 5, do not have a second conserved cysteine pair. The three kinase domains in sequence 7 (A. thaliana, clade 4), have been merged into two exons. Moreover, as illustrated in Figure 5, the exon–intron boundaries of ELS proteins (sequences 11 and 12) are very similar to clade 5 proteins apart from the first exon of ELS which is a combination of the first and second exons of clade 5 proteins.
Figure 5.
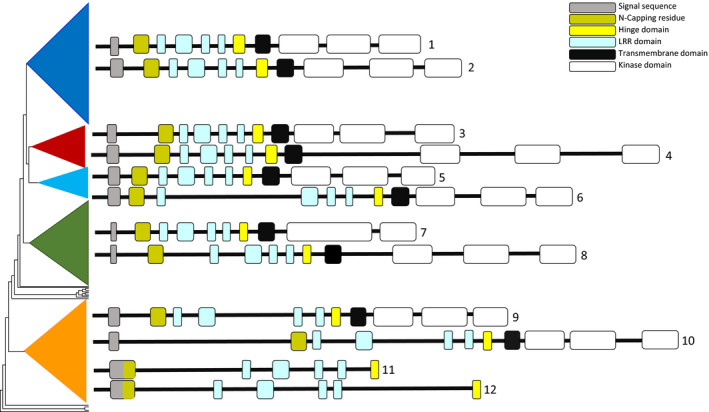
Exon and intron structure of leucine‐rich repeat receptor‐like kinase (LRR‐RLK) proteins.
Exon and intron boundaries for LRR‐RLK genes, numbered 1–12, of the main clades inferred here, including extracellular‐like somatic embryogenesis receptor kinase (ELS) genes (nos 11 and 12, indicated by missing kinase domains). Structures represent individual sequences of six selected Arabidopsis thaliana and six selected Oryza sativa accessions (see Table S3 for GenBank accession numbers).
Apomorphy detection for clades 2 and 3
Detection of synapomorphic amino acid substitutions shows that in the Ext‐based tree the number of apomorphies between nodes 2 and 1 (Figures 1b and 6a) is 25, seven of which are considered as radical changes according to calculated Grantham scores (Grantham, 1974). Among the characters with radical changes, none of them have unique changes [i.e. with consistency index (CI) = 1]. Similarly, from node 2 to node 5 (Figures 1b and 6a) we counted 68 apomorphies, 17 of which can be considered as radical changes, with a single one, G116 > W, having CI = 1 (Table S2), indicating it is a unique change in the Ext‐based tree. Additionally, 8 of 34 detected apomorphies for nodes 1 to 3 and 8 out of 39 for nodes 1 to 4 (Figures 1b and 6a) appear radical (Table 3), and none of them are unique.
Figure 6.
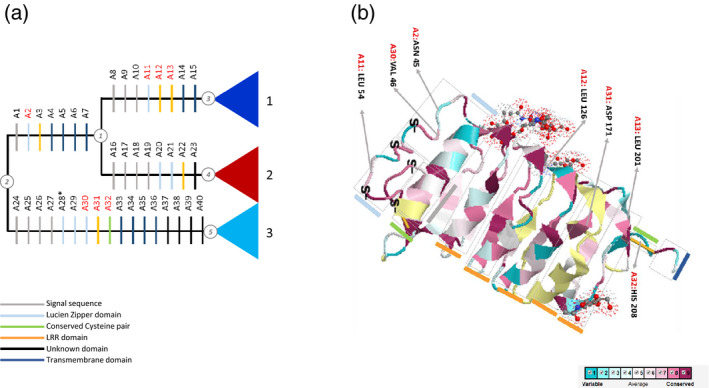
Apomorphies associated with the main leucine‐rich repeat receptor‐like kinase II (LRR‐RLKII) clades 1, 2 and 3.
(a) ‘Radical’ amino acid character optimisation on nodes 1 and 4 (see Figure 6b) of the extracellular (Ext) tree topology. Each individual apomorphy is indicated by numbering and colouring, the latter being related to the type of motif to which the detected apomorphy belongs. Unique apomorphies (consistency index = 1) are marked by a red asterisk.
(b) Conservation pattern of the external part of protein sequences aligned to a SERK1 ectodomain structure model (PDB 4LSC). Circled numbers indicates nodes used in apomorphy reconstruction.
Table 3.
Apomorphy detection in extracellular and intracellular trees for leucine‐rich repeat receptor‐like kinase clades 2 and 3 (see Figure 6a for the extracellular tree and Figure 7a for the intracellular tree)
| Branch | Apomorphies | |||
|---|---|---|---|---|
| Total no. (†) | Cons. a | Non‐cons. b | Rad. c (‡) | |
| Extracellular tree: | ||||
| Node 1–node 3 | 34 (0) | 11 | 15 | 8 (0) |
| Node 1–node 4 | 39 (1) | 17 | 14 | 8 (0) |
| Node 2–node 1 | 25 (4) | 10 | 8 | 7 (0) |
| Node 2–node 5 | 68 (5) | 29 | 22 | 17 (1) |
| Intracellular tree: | ||||
| Node 1–node 2 | 33 (7) | 11 | 10 | 12 (3) |
| Node 1–node 3 | 30 (7) | 8 | 13 | 9 (1) |
| Node 2–node 4 | 15 (1) | 7 | 2 | 6 (1) |
| Node 2–node 5 | 44 (1) | 22 | 14 | 8 (0) |
Conservative changes.
Non‐conservative changes.
Radical changes based on the Grantham score (see text). (†) indicates the number of characters with consistency index (CI) = 1 while (‡) indicates the number of radical changes with CI = 1.
In the Int‐based tree, 30, 33, 15 and 44 substitutions were recorded, respectively, between nodes 1 and 3, nodes 1 and 2, nodes 2 and 4 and nodes 2 and 5 (Figures 1c and 7a). Out of all reconstructed apomorphies, we detected 12 radical changes (with three of them, A574 > N, A646 > D and N653 > I, having CI = 1) between nodes 1 and 2, and nine between nodes 1 and 3 (with one of them, P645 > W, having CI = 1). We found six radical changes (one of them, N567 > L, having CI = 1) between nodes 2 and 4 and eight between nodes 2 and 5 (without unique changes; see Tables 3 and S3). All in all, roughly a quarter of all amino acid substitutions along these branches were radical, both in the extracellular and intracellular parts of the LRR‐RLKII protein.
Figure 7.
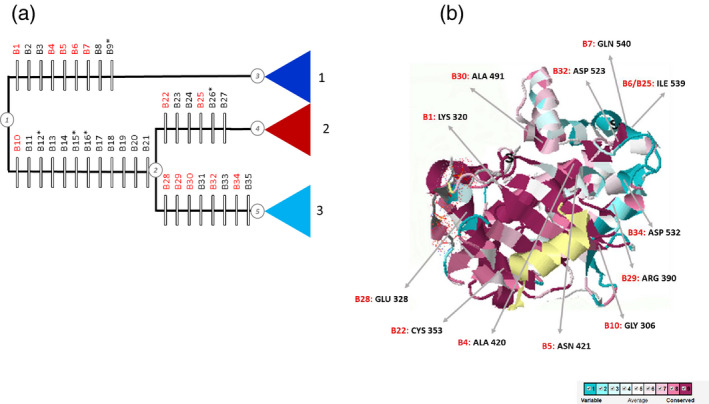
Apomorphies associated with the main leucine‐rich repeat receptor‐like kinase II (LRR‐RLKII) clades 1, 2 and 3 inferred here.
(a) Radical apomorphies for clades 1, 2 and 3, based on the intracellular (Int) tree topology. Each block represents an inferred apomorphy, with black indicating absent apomorphies and red apomorphies present in the protein model. Unique apomorphy (consistency index = 1) are marked by an asterisk.
(b) Conservation pattern of the internal part of LRR‐RLKII protein sequences aligned to an available SERK1 kinase domain with PDB ID 3TL8. Circled numbers indicates nodes used in apomorphy reconstruction.
Furthermore, when plotting the CIs for the above‐outlined apomorphies against their corresponding Grantham score changes (Figure S4) no direct relationship between these two criteria appears to exist; most of the changes had a CI of less than 0.4 and a Grantham score of less than 150, indicating that the majority of the changes are not specific and not unique to just one clade but occur independently several times in different clades. For the unique changes, on the other hand, around 23% concerned radical changes.
Conservation of the 3D structure of the extra‐ and intracellular domains of plant LRR‐RLKII proteins
Figure 6(b) illustrates the conservation pattern of the aligned LRR‐RKLII Ext‐matrix over the ectodomain of a ‘SERK1 co‐receptor’ protein 3D structure (PDB ID 4LSC). Several sulphide bridges are observed in the N‐terminal of the protein. Of 40 detected apomorphies with radical changes associated with the first three clades (Figure 6a), seven are present in the sequence of PDB ID 4LSC (indicated in Figure 6b). Most detected apomorphies are located at signal and transmembrane sequences (12 and 10 apomorphies, respectively), followed by leucine zipper domains (seven apomorphies). Others are located at the LRR domain (five apomorphies), the second conserved cysteine pair (one apomorphy) and the second unknown domain (five apomorphies) (Figure 6a).
Apomorphies associated with the intracellular part of LRR‐RLKII are indicated in Figure 7, which depicts the PDB structure of a SERK protein (BAK1) (PDB ID 3TL8). Thirty‐five radical apomorphies are located in the kinase domain (Figure 7a), of which 12 characters available in the sequence of 3TL8 protein are indicated in the kinase protein structural model in Figure 7(b).
DISCUSSION
In this paper we have studied the phylogenetic relationships within the LRR‐RLKII subfamily using currently available protein sequences in GenBank annotated as ‘SERK’, ‘NIK’ or ‘SERK‐like’ proteins, as well as based on pBLAST searches using these and other related sequences as queries. We inferred five well‐supported main clades which we labelled here as LRR‐RLKII clades 1–5. For two main clades, sister group relationships differed depending on whether the extracellular part or the cytoplasmic kinase part was used to build the tree. We detected apomorphies for these incongruences and evaluated them in light of the 3D structure. At the DNA level, for each main clade we determined the motif structure and exon–intron boundaries.
Proper rooting of the tree was a challenging part of our phylogenetic analyses as it was not directly clear what the sister group of SERK proteins is. Sakamoto et al. (2012) showed that the SERK clade is a sister group of NIK and other members of LRRII in tomato and Arabidopsis. Therefore, previous phylogenetic studies (Nolan et al., 2011; Liebrand et al., 2014; Aan den Toorn et al., 2015; Zhou et al., 2016) have considered non‐SERK LRR‐II and LRR‐RLK BRL1 (including NIK1, NIK2 and the SERK of the alga Closterium) as outgroups. In contrast with these studies, we found that the non‐SERK proteins are placed in different main clades which could imply that these studies used incorrect rooting. As land plant RLKs have been previously considered a sister group of the animal RLK family (Shiu and Bleecker, 2001) we selected the nematode Trichinella Pelle (which we detected as the most similar animal sequence to SERK proteins using BLAST) as the outgroup.
The pattern of main angiosperm clade relationships (i.e. monocots (asterids, rosids)) can be observed six times in our main tree, indicating that at least five duplication events happened in the gene subfamily prior to the divergence among angiosperm lineages. The presence of these genes in the land plants but not in the green algae (which do not have a LRR domain) can be considered as evidence for an ancient merger of LRR and kinase domains after the divergence of land plants from green algae, as also hypothesised by Liu et al. (2017).
We present amino acid sequence motif structures for each of five main clades based on 1342 available sequences. In contrast with Hecht et al. (2001), who used a SPP motif as a criterion for distinguishing the SERK proteins from other LRR‐RLK proteins, we could not find such structure in all clades, only in some terminals in clade 5 (Figure S7). Therefore, we cannot use it to distinguish SERK proteins from non‐SERK ones in the LRR‐RLKII subfamily.
From a DNA perspective, our results confirm the findings of previous studies (Shiu et al., 2004; Wei et al., 2015; Liu et al., 2017) showing that domain structures and intron–exon boundaries of all proteins were well conserved in evolution. The number of exons is 11, but in some terminals two kinase exons have merged into one exon (e.g. Figure 5, A. thaliana in clade 4). In agreement with other studies (e.g. Nolan et al., 2011), we confirmed the existence of five LRR domains using PFAM, a 3D structure model of SERK proteins (PDB) as well as available annotation files in public datasets. However, we could not detect any conserved amino acid structure, the existence of which was suggested by Liu et al. (2017). Based on extracellular LRR domains, clade 1 (comprising sequences similar to RKS types 1, 4, 5, 7, 9, 11 and 14, SERK1,2 and NIK1,2) and clade 2 (comprising sequences similar to RKS types 1, 5, 7, 9 and 14 and NIK3) are sister groups, but based on the intracellular kinase domain clades 2 and 3 are sister groups. These different placements are well supported and could point to possible independent phylogenetic histories between the extra‐ and intracellular parts of the LRR‐RLKII proteins. But that would imply there having been a ‘switch’ between the extra‐ and intracellular domains following the split between clade 4 versus clades 1, 2 and 3. This would entail a new connection between the extra‐ and intracellular domains from clades 1 or 2, ‘abandoning’ the extracellular domain of clade 3. It is possible that this could have occurred through an ancient recombination event, but more data are needed to investigate this. Our G‐test results indicate no significant differences in the number of amino acid substitutions among clades 1–5. The largest observed/expected difference occurs in clade 5 of the Ext‐based tree, which is also apparent from Figure 2. If this reflects actual rate differences, these could have evolved following the second gene duplication, leaving the newly formed paralogue (clade 4) to be under low functional constraints. For only three out of the 282 included sequences in this clade could we detect sequence similarity to RKS types, perhaps indicating that function in clade 4 in Ext‐based tree has evolved in a different way from that in the other clades.
Our phylogenetic study indicates that each main clade is associated with a range of RKS type similarities, apart from clade 3 where only sequence similarity to RKS type 5 is encountered (for comparison see Figure S6). Clades 1, 2 and 5 contain multiple RKS type similarities which may be caused by the fact that the first 100 sequences found in our RKS type‐based pBLASTs are not as abundant in the plant kingdom or are more similar between clades 1 and 2 than between the other clades. Although SERK3, ‐4 and ‐5 appear to be confined to clade 5, SERK types 1 and 2 are distributed in all clades apart from clade 2. Additionally, clade 2 only contains NIK3 proteins. Since the function of other RKS types has not yet been characterised we cannot functionally annotate each of our main clades.
Tracing the apomorphies in trees based on the extracellular part of the proteins showed that most of the detected ‘radical’ (according to Grantham distance) amino acid changes occur in signal and transmembrane domains. Moreover, the number of apomorphy characters with radical changes is lower (24) in the extracellular tree than in the cytoplasmic kinase‐based tree (35; Table 3). It is possible that positive selection on these residues is involved; however, we do not have the underlying codon sequence available in order to (model and) test this. Shiu et al. (2004) found indications of positive selection in extracellular domains as measured by K a and K s values, as did Zhang et al. (2006) for the 14 A. thaliana LRR‐RLKII genes known at the time. Liu et al. (2017) checked possible positive selection on LRR‐RLK genes in each subfamily and found possible candidates, claiming that positive selection may have driven the evolution of LRR‐RLKs.
In terms of protein structure, alignment of amino acid sequences to the available 3D structure model (PDB 4LSC, SERK1) reconstructed by Santiago et al. (2013) shows structural variation in the extracellular LRR part of the SERK protein. Figure 6(b) shows that it forms six loops (parallel beta sheets), the last five of which are LRR repeats, with the first being an ‘unknown’ domain at the N‐terminal of the protein. The leucine residues at the inner side of the LRR protein are known as the ‘hydrophobic core’ of the molecules (Morita et al., 2016). The disulphate bridges represent the first conserved cysteine pair where hydrogen bond interactions of the 4LSC protein with a conserved sequence of CxxFHxTCN occur (Santiago et al., 2013). We see that all these reported amino acids except for the last asparagine are conserved (CTWFHVTC) over clade 5 (comprising sequences with similarity to RKS types 0, 8, 10, 12, 13 and 16 and BAK1) but not in other clades (Figure 8). This conservation suggests an evolutionarily conserved interaction with receptors like BRI1, the brassinosteroid receptor and possibly other heterodimerizing receptors. The interaction of SERK proteins with BRI1, FLS2, PSKR (phytosulphokine receptor) and HAESA receptors which are responsible for plant growth, plant immunity and floral abscission have been studied previously (Santiago et al., 2013, 2016; Sun et al., 2013; Wang et al., 2015). According to Santiago et al. (2013), the N‐terminal cap of SERK1 folds on top of the BRI1 steroid‐binding pocket, where it establishes contact with the BRI1 island domain with RI1LRR25 and with the hormone itself. Here, the phenylalanine residue in the cysteine motif (which appears disordered in the isolated SERK1 structure) makes a stacking interaction with the C ring of the hormone, while the neighbouring histidine establishes hydrogen bonds with the 2a,3a‐diol moiety of brassinolide (Santiago et al., 2013). According to the authors, the brassinolide ligands can be considered as a molecular ‘glue’ between the BRI1 receptor and the ‘BAK1 co‐receptor’. Additionally, other amino acids including cysteine, threonine and asparagine in the N‐capping residue (see Figure 8, sequence 4LSC) were found to interact with BRI1 hormones (Santiago et al., 2013). Morita et al. (2016) showed that LRR23–LRR25 and LRR21 and LRR22 of BRI1 are heterodimerized with BAK1. The interaction surface with BRI1 occupies only about 10% of the total accessible surface area of SERK1. The amino acids in and around the conserved motif CTWFHVTC, which contacts with HAESA, FLS2 and PSKR receptors based on previous studies (Sun et al., 2013; Wang et al., 2015; Santiago et al., 2016), are highlighted in Figure 8. Although the CTWFHVTC motif has been detected in all studied SERK proteins of clade 5, the interaction between SERK and different receptors occurs via different amino acids. Phenylalanine is the only amino acid that contacts with all mentioned receptors. Studies by Santiago et al. (2013, 2016), Sun et al. (2013) and Wang et al. (2015) discovered that several amino acids located in the LRR1–4 motifs of SERK proteins are in contact with different receptors. Each SERK–receptor complex has its particular interaction position (indicated in Figure 8); among all of them only arginine, tyrosine and phenylalanine are conserved among the four studied interactions. According to our WebLogo analyses of the structure of the extracellular part of SERK proteins in Figure 8, all amino acids involved in the receptor–SERK protein complex in clade 5 are conserved throughout the clade.
Figure 8.
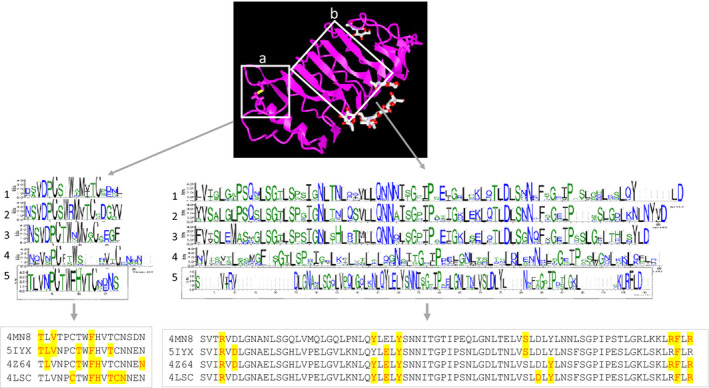
The amino acids involved in various somatic embryogenesis receptor kinase (SERK) complex interactions.
The amino acids involved in SERK (ID 4MN8)–FLS2, SERK (ID 5IYX)–HAESA, SERK (ID 4Z64)–PSKR and SERK (ID 4LSC)–BRI1 complex interactions, located in (a) the N‐capping residue and (b) the leucine‐rich repeat domains of associated SERK proteins; WebLogo frequency plots indicate the conservation pattern across entire clades.
In Figure 4 we summarised the modifications taking place in LRR‐RLKII proteins within the different main clades, based on the defined domains of the different receptors. Whereas the overall structure of the receptor kinase molecules remains conserved, specific elements are conserved within each clade, separating them from other clades. These elements are all positioned at the extracellular domain of the receptors, and are therefore likely to play a specific role in extracellular interactions with other protein and peptide sequences (see Hohmann et al., 2017). Since the SERK receptors are known to be involved in a multitude of intracellular signal transduction cascades with numerous defined heterodimerizing receptors (and probably many more undefined proteins and peptides), the specific elements conserved in the different clades could represent the basis for this multitude of interactions. Another interesting observation is that SERKs are very similar to proteins defined as ELS, which are almost identical in sequence to clade 5 proteins (see Figures 4 and 5). The transmembrane and intracellular domains of these different gene products are missing, and therefore it is assumed that these ELS proteins will float freely in the intracellular space, competing with SERK for binding to its target proteins and peptides. Local concentrations of ELS expression can be very high in specific organs or developmental stages (E. Schmidt, unpublished data, Wageningen, The Netherlands), which would suggest that SERK activity could be severely restricted there.
To summarise, we found five main clades for the LRR‐RLKII group of plant receptor kinases throughout the plant kingdom. These receptors are involved in many aspects of signal transduction, and in this study we have defined conserved motifs which modulate the multitude of intercellular signals.
EXPERIMENTAL PROCEDURES
Sequence mining and compilation
The A. thaliana RKS proteins (0–16) originally reported by Schmidt et al. (2009) and shown in Figure S6 were used as query sequences for pBLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) searches, enabling compilation of the amino acid sequences in this study. We performed extensive pBLAST searches, using default settings, in order to harvest from GenBank as thoroughly as possible and in order to avoid mis‐spellings and mis‐namings in sequence names. The first 100 hits for each specific RKS were selected as sequences with similarity to that A. thaliana RKS type. As a result we compiled 1528 sequences and consider these to represent all publically available ‘SERK’ and ‘non‐SERK’ sequences associated with the LRR‐RLKII subfamily to date. The set comprised 317 asterids, 897 rosids, 39 lower eudicots, 259 monocots and 16 magnoliids; in order to represent land plants, we included Amborella trichopoda, Picea sitchensis, Adiantum capillus‐veneris, Selaginella moellendorffii, Physcomitrella patens and Marchantia polymorpha. We also included the alga Closterium ehrenbergii and three animal kinase sequences, two from the nematode Trichuris suis and a kinase Pelle protein from the nematode Trichinella pseudospiralis as outgroup candidates. We added RLK sequences annotated ‘NIK1’, ‘NIK2’ and ‘LRRII non‐SERKs’ which were used as ‘outliers’ in Aan den Toorn et al. (2015), following the suggestion by Sakamoto et al. (2012) that there are three LRR‐RLKII clades. Moreover, we included three ‘extracellular like SERK receptor’ sequences (labelled ‘ELS protein’ by Schmidt et al., 2009), which have the LRR domains only (see Table S1).
As outlined above, several names including ‘SERK’ (types 1–5), ‘RKS’ (0–16), ‘NIK’ and ‘BAK1’ have been used by authors for these sequences. We therefore explored overlap and redundancy in RLK classification and nomenclature, summarizing these over each main clade found in this study (see below and Table 1). The SERK and NIK types were assigned as given in the GenBank (National Centre for Biotechnology Information) accession information (Table S1).
Sequence alignment and phylogenetic analysis
We performed MSA using a Linux version of MAFFT v.7 (Katoh and Standley, 2013) with ‘Auto’ settings in effect. The resulting MSA was inspected visually and adjusted manually when needed using Mesquite v.3.10 (Maddison and Maddison, 2016) but no columns/residues were removed. However, redundant sequences (with identical names or sequences) and sequences that did not align properly were discarded from the MSA. We also removed some terminals which only contain a LRR or kinase domain. As indicated above, we used the serine/threonine‐protein kinase Pelle of the nematode T. pseudospiralis as the outgroup sequence. In order to do this we had to exclude parts of this sequence so as to maintain homology with the ingroup MSA. Positions 1–156 from the Pelle sequence were excluded, corresponding to the LRR part (not homologous to plant RLK LRRs) of the protein. As a final matrix, 1342 terminals were kept for further analyses; these are listed in Table S1. The possibility of amino acid saturation in our MSA was checked for with ASaturA (Van de Peer et al., 2002), using the JTT substitution matrix (Jones et al., 1992). According to the results of motif searching using PFAM 32.0 (Finn et al., 2016; https://pfam.xfam.org/), we divided the MSA into two parts: one (named ‘Ext’, with alignment positions 1–573) containing the extracellular domains up to the transmembrane part and a second part (named ‘Int’, with alignment positions 574–1319) containing the intracellular kinase, with position 574 detected as the first position of a kinase by PFAM.
Final MSAs were subjected to phylogenetic reconstruction using a Linux version of IQ‐TREE (Nguyen et al., 2015), which is maximum likelihood‐based and includes automatic amino acid substitution model selection (ModelFinder) as well as 1000 replicates of ‘ultra fast’ bootstrapping (UFBoot). Tree searches were performed on both the full alignment and separate Ext and Int matrices. Resulting trees were visualized using Figtree v.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
To see if there is any significant difference between substitution rate of different main clades we used the IQ‐TREE ML tree topology and calculated the number of amino acid substitutions (parsimony branch lengths) of each main clade (see below) divided by the number of terminals in that clade, and for Int and Ext partitions separately, using PAUP* 4.0 (Swofford, 2002). Subsequently, we applied a G‐test to test for the significance of differences in the number of amino acid substitutions among main clades and between the Ext and Int partitions.
Amino acid conservation and protein structure analysis
We used a Linux version of WebLogo 3 (Crooks et al., 2004) to check for and visualise the amino acid residue conservation patterns in the MSA for separate main clades. Furthermore, for each main clade we selected one A. thaliana and one Oryza sativa sequence (see Table S3) as well as one A. thaliana and one O. sativa ELS sequence and compared their exon and intron boundaries as presented in ARAPORT (https://www.araport.org) and NCBI (https://www.ncbi.nlm.nih.gov/).
Apomorphy detection for the clades is incongruent between the Ext‐ and Int‐based trees
In order to identify synapomorphic amino acid substitutions for each main clade, which could probably lead us to functionally relevant residues, we used the ML tree topology and optimised the amino acid MSA onto it using PAUP* 4.0 (Swofford, 2002) for Linux, with accelerated transformation (ACCTRAN). An apomorphy list (from ‘DescribeTree’) was compiled and, in addition, the CIs for individual changes were recorded in order to measure the fit of these sites to the tree (Farris, 1989), and hence to establish whether they represent unique changes.
To evaluate the evolutionary distance between two amino acids, we calculated Grantham scores (Grantham, 1974) between original and replaced amino acids for each node using a python script (available upon request). These scores range from 5 to 215 and are based on side chain atomic composition, polarity and volume properties of all amino acids (Grantham, 1974). Higher Grantham scores therefore show more physico‐chemical and hence functional distance between two amino acids (Grantham, 1974). Amino acid substitutions involving Grantham scores of 5–60, 60–100 and more than 100 have been considered ‘conservative’, ‘non‐conservative’ and ‘radical’, respectively (Abkevich, 2004; Balasubramanian et al., 2005). Grantham scores were plotted against the CI to see if there is any correlation between the level of homoplasy and the level of Grantham similarity between replaced amino acids.
Finally, we selected two SERK 3D protein structures from the PDB (Berman et al., 2000; http://www.rcsb.org) including the extracellular part (PDB 4LSC) and intracellular part (PDB 3TL8) of the protein, determined using X‐ray diffraction by Santiago et al. (2013) and Cheng et al. (2011), respectively. We mapped the MSA of Ext and Int onto the corresponding selected 3D protein structures using the ConSurf server at https://consurf.tau.ac.il (Ashkenazy et al., 2010, 2016; Celniker et al., 2013), in order to calculate the evolutionary conservation of amino acid positions in relation to the SERK structure (Berman et al., 2000; http://www.rcsb.org). We used ‘empirical Bayesian method’, to compute the evolutionary rate.
AUTHOR CONTRIBUTIONS
All authors analysed the selected datasets, performed the computational experiments, contributed to the development and experimental design and interpreted the results as well as writing of the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTERESTS
The authors declare that they have no conflict of interests.
Supporting information
Table S1. Accession number, organism, SERK and NIK types and LRR‐RLKII clade membership of sequences used in this analysis. SERK and NIK types are based on NCBI annotation files.
Table S2. Amino acid changes in the defined nodes in Figure 1(b,c) and their corresponding calculated Grantham score.
Table S3. Exon sizes in selected Arabidopsis thaliana and Oryza sativa HDR genes.
Figure S1. WebLogo of main LRR‐RLKII clades 1–5 (see text) based on the Int + Ext‐matrix.
Figure S2. WebLogo of main LRR‐RLKII clades 1–5 (see text) based on the Ext‐matrix.
Figure S3. WebLogo of main LRR‐RLKII clades 1–5 (see text) based on the Int‐matrix.
Figure S4. Relationship between Grantham score and calculated consistency index of amino acid substitutions leading to LRR‐RLKII clades 1 and 2 (see text).
Figure S5. IQ‐TREE maximum likelihood tree based on the Int + Ext sequence matrix (see text).
Figure S6. Unrooted IQ‐TREE maximum likelihood tree based on RKS protein sequences of Arabidopsis thaliana from Schmidt et al. (2009), patented as US2009126041 (A1), ES2468190 (T3), NZ550620 (A), BRPI0312721 (A2), with bootstrap values shown at nodes.
Figure S7. ‘Hinge motifs’, including the second conserved ‘cysteine pair/SPP motif’, for representatives of LRR‐RLKII clades 1–5 (see text), along with their WebLogo frequency plots.
Data S1. Alignment of the full LRR‐RLKII sequences (‘Ext + Int’ matrix) as a FASTA file.
Data S2. Alignment of the extracellular part of LRR‐RLKII sequences (the ‘Ext matrix’) as a FASTA file.
Data S3. Alignment of the intracellular part of LRR‐RLKII sequences (the ‘Int matrix’) as a FASTA file.
Acknowledgements
We thank our two reviewers for their constructive comments on the manuscript.
DATA AVAILABILITY STATEMENT
All relevant data can be found within the manuscript and its supporting materials.
REFERENCES
- Aan Den Toorn, M. , Albrecht, C. and De Vries, S. (2015) On the origin of SERKs: bioinformatics analysis of the somatic embryogenesis receptor kinases. Mol. Plant, 8, 762–782. [DOI] [PubMed] [Google Scholar]
- Abkevich, V. (2004) Analysis of missense variation in human BRCA1 in the context of interspecific sequence variation. J. Med. Genet. 41, 492–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht, C. , Russinova, E. , Kemmerling, B. , Kwaaitaal, M. and de Vries, S.C. (2008) Arabidopsis somatic embryogenesis receptor kinase proteins serve brassinosteroid‐dependent and independent signaling pathways. Plant Physiol. 148, 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy, H. , Erez, E. , Martz, E. , Pupko, T. and Ben‐Tal, N. (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy, H. , Abadi, S. , Martz, E. , Chay, O. , Mayrose, I. , Pupko, T. and Ben‐Tal, N. (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian, S. , Xia, Y. , Freinkman, E. and Gerstein, M. (2005) Sequence variation in G‐protein‐coupled receptors: analysis of single nucleotide polymorphisms. Nucleic Acids Res. 33, 1710–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M. , Westbrook, J. , Feng, Z. , Gilliland, G. , Bhat, T.N. , Weissig, H. , Shindyalov, I.N. and Bourne, P.E. (2000) The protein data bank. Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko, M.A. , Vie, A.K. , Brembu, T. , Aalen, R.B. and Bones, A.M. (2009) Plant peptides in signalling, Looking for new partners. Trends Plant Sci. 14, 255–263. [DOI] [PubMed] [Google Scholar]
- Celniker, G. , Nimrod, G. , Ashkenazy, H. , Glaser, F. , Martz, E. , Mayrose, I. , Pupko, T. and Ben‐Tal, N. (2013) ConSurf, using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 53, 199–206. [Google Scholar]
- Chae, L. , Sudat, S. , Dudoit, S. , Zhu, T. and Luan, S. (2009) Diverse transcriptional programs associated with environmental stress and hormones in the Arabidopsis receptor‐like kinase gene family. Mol. Plant, 2, 84–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, S. , Nguyen, B. , Wasti, S.D. and Guozhou, X. (2019) Plant Leucine‐Rich Repeat Receptor Kinase (LRR‐RK): structure, ligand perception, and activation mechanism. Molecules, 24, 3081 10.3390/molecules24173081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, W. , Munkvold, K.R. , Gao, H. , Mathieu, J. , Schwizer, S. , Wang, S. , Martin, G.B. and Chai, J. (2011) Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase‐interacting domains in a type III effector. Cell Host Microbe, 10, 616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla, D. , Zipfel, C. , Robatzek, S. , Kemmerling, B. , Nürnberger, T. , Jones, J.D.G. , Felix, G. and Boller, T. (2007) A flagellin‐induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature, 448, 497–500. [DOI] [PubMed] [Google Scholar]
- Crooks, G.E. , Hon, G. , Chandonia, J.M. and Brenner, S.E. (2004) WebLogo, a sequence logo generator. Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet, I. , Voß, U. , Jürgens, G. and Beeckman, T. (2009) Receptor‐like kinases shape the plant. Nat. Cell Biol. 11, 1166–1173. [DOI] [PubMed] [Google Scholar]
- Farris, J.S. (1989) The retention index and rescaled consistency index. Cladistics, 5, 417–419. [DOI] [PubMed] [Google Scholar]
- Finn, R.D. , Coggill, P. , Eberhardt, R.Y. et al (2016) The Pfam protein families database, towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou, X. , Yin, H. , He, K. , Du, J. , Yi, J. , Xu, S. , Lin, H. , Clouse, S.D. and Li, J. (2012) Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet, 8, e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham, R. (1974) Amino acid difference formula to help explain protein evolution. Science, 185, 862–864. [DOI] [PubMed] [Google Scholar]
- Halter, T. , Imkampe, J. , Mazzotta, S. et al (2014) The Leucine‐rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr Biol. 24, 134–143. [DOI] [PubMed] [Google Scholar]
- He, Y. , Zhou, J. , Shan, L. and Meng, X. (2018) Plant cell surface receptor‐mediated signaling – a common theme amid diversity. J Cell Sci. 131, jcs209353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht, V. , Vielle‐Calzada, J.P. , Hartog, M.V. , Schmidt, E.D.L. , Boutilier, K. , Grossniklaus, U. and de Vries, S.C. (2001) The Arabidopsis somatic embryogenesis receptor kinase 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127, 803–16. [PMC free article] [PubMed] [Google Scholar]
- Hohmann, U. , Lau, K. and Hothorn, M. (2017) The structural basis of ligand perception and signal activation by receptor kinases. Annu. Rev. Plant Biol. 68, 109–137. [DOI] [PubMed] [Google Scholar]
- Jones, D.T. , Taylor, W.R. and Thornton, J.M. (1992) The rapid generation of mutation data matrices from protein sequences. Bioinformatics, 8, 275–282. [DOI] [PubMed] [Google Scholar]
- Katoh, K. and Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7, Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. (2010) Multi‐tasking of somatic embryogenesis receptor‐like protein kinases. Curr. Opin. Plant Biol. 13, 509–514. [DOI] [PubMed] [Google Scholar]
- Li, X. , Salman, A. , Guo, C. , Yu, J. , Cao, S. , Gao, X. , Li, W. , Li, H. and Guo, Y. (2018) Identification and characterization of LRR‐RLK family genes in potato reveal their involvement in peptide signaling of cell fate decisions and biotic/abiotic stress responses. Cells, 7, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand, T.W.H. , van den Burg, H.A. and Joosten, M.H.A.J. (2014) Two for all, Receptor‐associated kinases SOBIR1 and BAK1. Trends Plant Sci. 19, 123–132. [DOI] [PubMed] [Google Scholar]
- Liu, P.L. , Du, L. , Huang, Y. , Gao, S.M. and Yu, M. (2017) Origin and diversification of Leucine‐rich repeat receptor‐like protein kinase (LRR‐RLK) genes in plants. BMC Evol. Biol. 17, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison, W.P. and Maddison, D.R. (2016) Mesquite, A modular system for evolutionary analysis. Version 3.10. http://mesquiteproject.org
- Morita, J. , Kato, K. , Nakane, T. , Kondo, Y. , Fukuda, H. , Nishimasu, H. , Ishitani, R. and Nurekib, O. (2016) Crystal structure of the plant receptor‐like kinase TDR in complex with the TDIF peptide. Nat. Commun. 7, 12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, K.H. and Li, J. (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell, 110(2), 203–212. [DOI] [PubMed] [Google Scholar]
- Newman, M.A. , Sundelin, T. , Nielsen, J.T. and Erbs, G. (2013) MAMP (microbe‐associated molecular pattern) triggered immunity in plants. Front Plant Sci. 4, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, L.T. , Schmidt, H.A. , Von Haeseler, A. and Minh, B.Q. (2015) IQ‐TREE, A fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan, K.E. , Kurdyukov, S. and Rose, R.J. (2011) Characterisation of the legume SERK‐NIK gene superfamily including splice variants, Implications for development and defence. BMC Plant Biol. 11, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, M.N. , Dehal, P.S. and Arkin, A.P. (2009) Fasttree, computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, M. , Schwessinger, B. , Albrecht, C. , Chinchilla, D. , Jones, A. , Holton, N. , Malinovsky, F.G. , Tor, M. , de Vries, S. and Zipfe, C. (2011) The Arabidopsis Leucine‐rich repeat receptor–like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell, 23, 2440–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, T. , Deguchi, M. , Brustolini, O.J.B. , Santos, A.A. , Silva, F.F. and Fontes, E.P.B. (2012) The tomato RLK superfamily, phylogeny and functional predictions about the role of the LRRII‐RLK subfamily in antiviral defense. BMC Plant Biol. 12, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago, J. , Henzler, C. and Hothorn, M. (2013) Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co‐receptor kinases. Science, 341, 889–892. [DOI] [PubMed] [Google Scholar]
- Santiago, J. , Brandt, B. , Wildhagen, M. , Hohmann, U. , Hothorn, L.A. , Butenko, M.A. and Hothorn, M. (2016) Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife, 5, e15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, E.L. , Guzzo, F. , Toonen, M.A.J. and de Vries, S.C. (1997) A Leucine‐rich repeat containing receptor‐like kinase marks somatic plant cells competent to form embryos. Development, 124, 2049–2062. [DOI] [PubMed] [Google Scholar]
- Schmidt, E.D.L. , De Boer, A.D. and Van der Kop, D.A.M. (2009) US2009126041 (A1) – Regeneration. https//worldwide.espacenet.com/publicationDetails/biblio?FT=D&date=20090514&DB=&locale=en_EP&CC=US&NR=2009126041A1&KC=A1&ND=4
- Shiu, S.H. and Bleecker, A.B. (2001) Receptor‐like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA, 98, 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.H. , Karlowski, W.M. , Pan, R. , Tzeng, Y.H. , Mayer, K.F.X. and Li, W.H. (2004) Comparative analysis of the receptor‐like kinase family in Arabidopsis and Rice. Plant Cell, 16, 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. , Li, L. , Macho, A.P. , Han, Z. , Hu, Z. , Zipfel, C. , Zhou, J.M. and Chai, J. (2013) Structural basis for flg22‐induced activation of the Arabidopsis FLS2‐BAK1 immune complex. Science, 342, 624–628. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2002) Phylogenetic Analysis Using Parsimony. Sunderland, Massachusetts: Sinauer Associates. [Google Scholar]
- Tang, J. , Han, Z. , Sun, Y. , Zhang, H. , Gong, X. and Chai, J. (2015) Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res. 25, 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii, K.U. (2004) Leucine‐rich repeat receptor kinases in plants, structure, function and signal transduction pathways. Int. Rev. Cytol. 234, 1–46. [DOI] [PubMed] [Google Scholar]
- Van de Peer, Y. , Frickey, T. , Taylor, J.S. and Meyer, A. (2002) Dealing with saturation at the amino acid level: a case study based on anciently duplicated zebrafish genes. Gene, 295, 205–211. [DOI] [PubMed] [Google Scholar]
- Wang, G. , Fiers, M. , Ellendorff, U. , Wang, Z. , de Wit, P.J.G.M. , Angenent, G.C. and Thomma, B.P.H.J. (2010) The diverse roles of extracellular leucine‐rich repeat‐containing receptor‐like proteins in plants. CRC Crit. Rev. Plant Sci. 29, 285–299. [Google Scholar]
- Wang, J. , Li, H. , Han, Z. , Zhang, H. , Wang, T. , Lin, G. , Chang, J. , Yang, W. and Chai, J. (2015) Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature, 525, 265–268. [DOI] [PubMed] [Google Scholar]
- Wei, Z. , Wang, J. , Yang, S. and Song, Y. (2015) Identification and expression analysis of the LRR‐RLK gene family in tomato (Solanum lycopersicum) Heinz. Genome, 58, 121–34. [DOI] [PubMed] [Google Scholar]
- Zhang, X.S. , Choi, J.H. , Heinz, J. and Chetty, C.S. (2006) Domain‐Specific positive selection contributes to the evolution of Arabidopsis Leucine‐rich repeat receptor‐like kinase (LRR RLK) genes. J. Mol. Evol. 63, 612–621. [DOI] [PubMed] [Google Scholar]
- Zhou, F. , Guo, Y. and Qiu, L.J. (2016) Genome‐wide identification and evolutionary analysis of Leucine‐rich repeat receptor‐like protein kinase genes in soybean. BMC Plant Biol. 16, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Accession number, organism, SERK and NIK types and LRR‐RLKII clade membership of sequences used in this analysis. SERK and NIK types are based on NCBI annotation files.
Table S2. Amino acid changes in the defined nodes in Figure 1(b,c) and their corresponding calculated Grantham score.
Table S3. Exon sizes in selected Arabidopsis thaliana and Oryza sativa HDR genes.
Figure S1. WebLogo of main LRR‐RLKII clades 1–5 (see text) based on the Int + Ext‐matrix.
Figure S2. WebLogo of main LRR‐RLKII clades 1–5 (see text) based on the Ext‐matrix.
Figure S3. WebLogo of main LRR‐RLKII clades 1–5 (see text) based on the Int‐matrix.
Figure S4. Relationship between Grantham score and calculated consistency index of amino acid substitutions leading to LRR‐RLKII clades 1 and 2 (see text).
Figure S5. IQ‐TREE maximum likelihood tree based on the Int + Ext sequence matrix (see text).
Figure S6. Unrooted IQ‐TREE maximum likelihood tree based on RKS protein sequences of Arabidopsis thaliana from Schmidt et al. (2009), patented as US2009126041 (A1), ES2468190 (T3), NZ550620 (A), BRPI0312721 (A2), with bootstrap values shown at nodes.
Figure S7. ‘Hinge motifs’, including the second conserved ‘cysteine pair/SPP motif’, for representatives of LRR‐RLKII clades 1–5 (see text), along with their WebLogo frequency plots.
Data S1. Alignment of the full LRR‐RLKII sequences (‘Ext + Int’ matrix) as a FASTA file.
Data S2. Alignment of the extracellular part of LRR‐RLKII sequences (the ‘Ext matrix’) as a FASTA file.
Data S3. Alignment of the intracellular part of LRR‐RLKII sequences (the ‘Int matrix’) as a FASTA file.
Data Availability Statement
All relevant data can be found within the manuscript and its supporting materials.


