Abstract
Aims
Methylation profiling (MP) is increasingly incorporated in the diagnostic process of central nervous system (CNS) tumours at our centres in The Netherlands and Scandinavia. We aimed to identify the benefits and challenges of MP as a support tool for CNS tumour diagnostics.
Methods
About 502 CNS tumour samples were analysed using (850 k) MP. Profiles were matched with the DKFZ/Heidelberg CNS Tumour Classifier. For each case, the final pathological diagnosis was compared to the diagnosis before MP.
Results
In 54.4% (273/502) of all analysed cases, the suggested methylation class (calibrated score ≥0.9) corresponded with the initial pathological diagnosis. The diagnosis of 24.5% of these cases (67/273) was more refined after incorporation of the MP result. In 9.8% of cases (49/502), the MP result led to a new diagnosis, resulting in an altered WHO grade in 71.4% of these cases (35/49). In 1% of cases (5/502), the suggested class based on MP was initially disregarded/interpreted as misleading, but in retrospect, the MP result predicted the right diagnosis for three of these cases. In six cases, the suggested class was interpreted as ‘discrepant but noncontributory’. The remaining 33.7% of cases (169/502) had a calibrated score <0.9, including 7.8% (39/502) for which no class indication was given at all (calibrated score <0.3).
Conclusions
MP is a powerful tool to confirm and fine‐tune the pathological diagnosis of CNS tumours, and to avoid misdiagnoses. However, it is crucial to interpret the results in the context of clinical, radiological, histopathological and other molecular information.
Keywords: central nervous system tumours, diagnostics, methylation profiling
Abbreviations
- CEP6
centromere 6
- Chr
chromosome
- CNS
central nervous system
- CNV
copy‐number variation
- CTNNB1
beta‐catenin 1
- DDx
differential diagnosis
- DK
Denmark
- DKFZ
Deutsches Krebsforschungszentrum/German Cancer Research Centre
- Dx
diagnosis
- FF
fresh‐frozen
- FFPE
formalin‐fixed paraffin‐embedded
- FISH
fluorescent in‐situ hybridization
- MP
methylation profiling
- NL
Netherlands
- OUH
Odense University Hospital
- PMC
Princess Máxima Centre for Pediatric Oncology
- PXA
pleomorphic xanthoastrocytoma
- RELA
V‐rel avian reticuloendotheliosis viral oncogene homolog A
- SHH
sonic hedgehog
- SNP
single‐nucleotide polymorphism
- UMCU
University Medical Centre Utrecht
- WHO
World Health Organization
- WNT
wingless/integrated
Introduction
Diagnostics of benign to highly malignant central nervous system (CNS) tumours is often complex. Some tumours can be treated by surgery alone, whereas others require multimodal treatment and the use of diagnostic, prognostic and predictive biomarkers. Based on histology, there is significant interobserver variation in the diagnostics of particular CNS tumours [1]. Molecular analyses can help to reduce this interobserver variation, especially now that there is increasing knowledge about the characteristic molecular features of many CNS tumour entities [2].
Mapping of epigenetic alterations in CNS tumours has recently been shown to offer promising perspectives. The diagnostic potential of DNA methylation profiling (MP) by assessing the methylation status of 850,000 CpG sites across the entire human genome has been explored by the German Cancer Research Centre (DKFZ) and Heidelberg University [3, 4, 5]. They have shown that methylation profiles of CNS tumours share common features and that these profiles differ between (histological) tumour entities. Hereby, they conceived the concept of brain tumour DNA ‘fingerprinting’ by MP for CNS tumour classification. A DNA methylation‐based classifier tool is publicly available for CNS tumour classification through a webpage (www.molecularneuropathology.org, henceforth referred to as ‘the Classifier’). Methylation profiles can be uploaded to the Classifier and matched to a database containing a reference cohort, initially of 2801 samples covering 82 CNS tumour classes and nine control tissue classes, which has been updated with many more samples ever since [5]. The automated analysis results in a calibrated score, representing the degree of match between the methylation profile of the tumour of interest and predefined methylation classes.
Recent studies using the Classifier have shown that most WHO CNS tumour entities can be precisely identified [6, 7]. Especially for paediatric tumours, the approach is promising. Namely, medulloblastomas, which often have low mutation rates and absence of frequently recurring hotspot mutations, can be separated into several subgroups that are clinically important [8]. Furthermore, DNA MP of ependymomas has distinguished nine distinct subgroups [9] with one subgroup, ependymoma, RELA fusion‐positive, already recognized as a separate entity in the updated WHO 2016 classification. In fact, groups of ependymomas categorized by methylation profiles are clinically more homogenous than grouping based on histology using WHO classification and grading [9.
The copy‐number variation (CNV) profile, obtained along with the Classifier score, provides additional information of high value in specific differential diagnostics settings [6]. The diagnostic relevance of CNVs is emphasized, for example, in the cIMPACT‐NOW update 3, introducing the combined gain of complete chromosome (Chr) 7 and loss of complete Chr 10 as a diagnostic criterion for a diagnosis of ‘molecular glioblastoma’[10]. Additionally, the DNA methylation‐based Classifier tool gives information on the methylation status of the MGMT promoter: a variable that is relevant for therapy and prognosis in patients with glioblastoma [11].
The aim of this study was to assess the diagnostic impact of DNA methylation‐based classification in CNS tumour diagnostics and to discuss both benefits and pitfalls encountered during the implementation of the tool at our respective centres. Monitoring this is, from our perspective, an important part of introducing new methods for (CNS) tumour diagnostics.
Materials and methods
DNA methylation‐based tumour classification has been increasingly incorporated in the diagnostic process at our centres. In The Netherlands (NL), MP is performed at the University Medical Centre Utrecht (UMCU) for (challenging) in‐hospital CNS tumour cases, for all CNS tumours of paediatric patients from the Princess Máxima Centre for Paediatric Oncology (PMC) and for referred cases with a challenging diagnosis from other Dutch centres. In Odense, Denmark (DK), MP was performed on selected, often challenging in‐hospital CNS tumour cases (including some paediatric cases), as well as referred cases from other centres in Denmark, Sweden, Norway and Finland.
Patients
About 502 CNS tumour samples analysed between October 2016 and April 2018 were included, of which 279 samples were from adults and 223 from children. The following exclusion criteria were applied: diagnosis other than primary CNS tumour, no suggested histological diagnosis before DNA MP, missing clinical information, cases analysed in research setting and duplicate cases (e.g. repeated tests on the same material or if more than one sample from a tumour was available, only the result with the best calibrated score was kept for analysis). The total number of unique patients was 480: 20 of these patients were included with two samples and one patient with three samples. In case of samples from one patient from different locations/components (n = 3) or different points in time (e.g. initial and recurrent disease, n = 18), both entries were included for statistical analysis.
Reasons to perform MP were categorized in four major groups: (i) ‘Routine/gain experience with tool’ (e.g. all paediatric cases from the PMC were routinely subjected to MP); (ii) ‘Challenging diagnosis’ (including cases with unusual/nonspecific histology or unclear molecular findings); (iii) ‘Subclassification’ (especially in case of ependymomas or medulloblastomas); (iv) ‘Revision/re‐evaluation’. This last category included a mix of cases with an unexpected clinical course (e.g. unusual recurrent disease or long‐term survival, metastasis outside of CNS), unresolved tumour cases and cases analysed specifically to find possible therapeutic options. A list of the (first differential) diagnosis of all analysed tumours is given in Table S1.
Sample preparation
Most (n = 448) samples were isolated from formalin‐fixed paraffin‐embedded (FFPE) tissue and 17 from fresh‐frozen (FF) tissue. In 36 cases, DNA was isolated at different hospitals (generally also from FFPE tissue) and sent to our centres for MP. One sample was formalin‐fixed agar‐embedded. Whole unstained slides were used or areas with the highest tumour cell content were macrodissected for DNA isolation. Estimated tumour cell content was highly variable (Figure 1 B). DNA was isolated using NorDiag Arrow using the DiaSorin DNA extraction kit (NL) or GeneRead DNA FFPE Kit (Qiagen, Hilden, Germany) (DK) according to the respective manufacturer’s instructions. DNA concentration was measured using the Qubit 2.0 fluorometer and ranged from 0.1 to 875.5 ng/μl. Per sample, we aimed to use 500 ng (DK) or 200 ng (NL) of DNA. Bisulphite conversion was performed with EZ DNA Methylation™ Kit (Zymo Research, Irvine, CA, USA).
Figure 1.
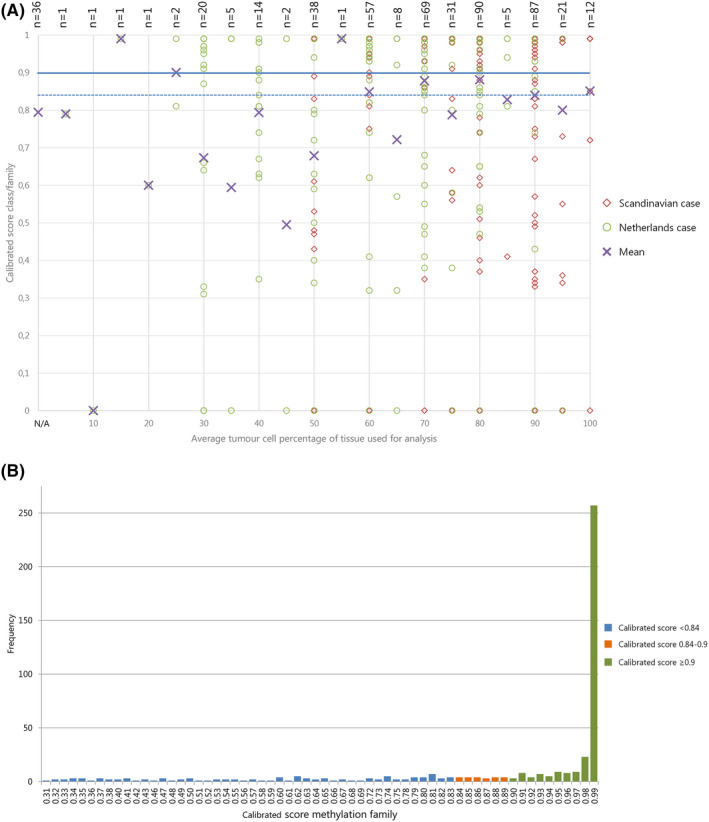
(A) Effect of estimated tumour cell percentage on calibrated score: Scatter plot of average tumour cell percentage (x‐axis) versus calibrated score for MP class family (Y‐axis). Cases from NL: green circles; cases from Scandinavia: red diamonds; purple crosses: mean. Horizontal lines: blue solid ‐ threshold of calibrated score ≥0.9; blue dashed ‐possible alternative threshold for calibrated score at ≥0.84 as suggested by [6] Cases with ‘no match <0.3’ (no calibrated score provided) were given the value 0 to be able to visualize them in this plot. The mean calibrated score of samples for which no tumour cell percentage was available is plotted at the bottom of the x‐axis, marked with ‘N/A’. NB. Symbols are often superimposed; labels at the top show the number of plotted cases. (B) Distribution of cases by calibrated scores for methylation class family: bar chart showing frequency of cases (Y‐axis) with specified calibrated score (X‐axis) for 468 cases with MP result. Valid matches with calibrated score ≥0.9 presented in green; no match cases with calibrated scores 0.84‐<0.9 in orange, remaining no match cases with calibrated scores 0.31‐<0.84 in blue. Data on no match cases with scores < 0.3 are not shown.
Methylation profile analysis
All methylation data were generated using the Illumina® MethylationEPIC (850 k) BeadChip platform as described by Capper et al. (2018) [5]. All FF samples were processed along with FFPE samples on the same EPIC chip. The profiles, contained in paired IDAT files, were matched with the Classifier using the current version at the time of original upload (mostly v11b2, v11b4 since January 2018; www.molecularneuropathology.org). The Classifier will provide, if possible, a match to a methylation ‘family’ and if applicable to a ‘subclass’ within that family, each accompanied by a calibrated score (ranging from 0.3 to 0.99). This score gives an indication of the degree of match between the methylation profile of the sample and the methylation profiles of samples in the reference database. The Classifier is unable to assign a methylation class when the calibrated score falls <0.3 and no exact calibrated score is given between 0.0 and 0.3. Samples were not generally reclassified after the Classifier version update during the preparation of this manuscript, except for 23 cases from NL for which the calibrated score for the subclass was missing due to temporary problems with the online access to the Classifier tool at the time of original upload. Reclassification of these 23 cases did not lead to significant changes in calibrated scores or assigned methylation classes.
Data collection and statistical analysis
The variables that were collected in the database for all cases are presented in Table 1. Tumours were graded according to the WHO Classification of Tumours of the CNS that was valid at the time of first diagnosis (mostly 4th revised edition of 2016). This study includes 84 tumour samples dating from 2007 to 2015, 11 tumour samples dating from 2002 to 2006 and one tumour sample from 1994. For these samples, the initial diagnosis was based on the WHO classifications from 2007, 2000 and 1993 respectively. Integration of the MP result into the final diagnosis, and (if applicable) reconsideration of the histopathology in case of a discrepant methylation class, was done by the pathologist requesting the test (hereafter referred to as the ‘original pathologist’). Next, all tumours were categorized into eight groups comparable to the categorization used by Capper et al. [5]. The ‘no match’ cases were subdivided into three groups based on the calibrated score for the methylation class family: <0.3; 0.3 to <0.7 and 0.7 to <0.9. Detailed information on these categories is given in Supplement 1. For each case with a changed diagnosis, the consequence for the WHO grade (upgraded, downgraded or unchanged) was evaluated.
Table 1.
Collected variables per case for database
| Collected variables per case |
|---|
| Tissue type |
| Original pathological diagnosis prior to MP |
| Differential diagnosis prior to MP |
| WHO grade of (first differential) diagnosis prior to MP |
| Reason to perform methylation profiling |
| Tumour location |
| Age at time of diagnosis |
| Gender |
| Tumour cell percentage |
| DNA concentration (ng/µl) |
| DNA amount added to assay (ng/45 µl) |
| Year of tissue block |
| Year of array analysis |
| Highest scoring methylation class or family |
| Highest scoring methylation subclass |
| Calibrated scores for family and subclass |
| Final pathological diagnosis post MP |
| WHO grade of the final pathological diagnosis |
Statistical analyses (mainly descriptive such as frequencies, mean and standard deviation, t‐tests, Fisher’s exact test and scatter plots) were performed using IBM SPSS version 25.
Results
Cohort characteristics
Of 502 included samples (Table 2), 223 were paediatric (mean age 8.7 years) and 279 were from adult patients (mean age 50.9 years); 55.6% (279/502) samples were from males, and 44.4% (223/502) samples from female patients. Most samples were obtained at initial presentation of the disease (88.2%, 443/502), while fewer were from recurrent disease (11.6%, 58/502) or a metastasis (0.2%, 1/502). The main reason to perform MP was ‘routine/gain experience with tool’ (67.3%, 338/502). The other cases were analysed because of a challenging (differential) diagnosis (17.7%, 89/502), specifically to be able to subclassify the tumour (8.6%, 43/502, of which 40 were paediatric) or as a part of revision/re‐evaluation during the course of the disease (6.4%, 32/502).
Table 2.
Cohort characteristics of 502 cases included for analysis
| Adult (n = 279) | Paediatric (n = 223) | Combined (n = 502) | ||||
|---|---|---|---|---|---|---|
| Freq | % | Freq | % | Freq | % | |
| Region | ||||||
| Scandinavia | 148 | 53.0 | 38 | 17.0 | 186 | 37.1 |
| Netherlands | 131 | 47.0 | 185 | 83.0 | 316 | 62.9 |
| Gender | ||||||
| M | 150 | 53.8 | 129 | 57.8 | 279 | 55.6 |
| F | 129 | 46.2 | 94 | 42.2 | 223 | 44.4 |
| Disease stage | ||||||
| Initial | 241 | 86.4 | 202 | 90.6 | 443 | 88.2 |
| Recurrent | 37 | 13.3 | 21 | 9.4 | 58 | 11.6 |
| Metastasis | 1 | 0.4 | 0 | 0 | 1 | 0.2 |
| Reason to perform MP | ||||||
| Routine/gain experience with tool | 194 | 69.5 | 144 | 64.6 | 338 | 67.3 |
| Challenging diagnosis | 60 | 21.5 | 29 | 13.0 | 89 | 17.7 |
| Subclassification | 3 | 1.1 | 40 | 17.9 | 43 | 8.6 |
| Revision/re‐evaluation | 22 | 7.9 | 10 | 4.5 | 32 | 6.4 |
| Mean | SD | Mean | SD | |||
| Age | 50.83 | 16.917 | 8.69 | 5.545 | ||
Freq, frequency; SD, standard deviation.
Assay performance
Using a calibrated score with a cut‐off of ≥0.9 (as suggested by Capper et al. [5]), the Classifier was able to find a match with a methylation class family/subclass in 66.3% of cases (333/502 analysed samples) (Figure 1 A, and subclasses in Figure S1). When using the cut‐off of ≥0.84 (possible alternative cut‐off value, as more recently suggested by Capper et al. [6]), this percentage rises to 70.9% (356/502) (Figure S2). Despite the slightly different approach in DNA extraction and added DNA amount in NL and DK (max. 200 and 500 respectively), there was no significant difference in the percentage of cases for which a match (cut‐off ≥0.9) was obtained: 68.4% (216/316) for NL and 62.4% (116/186) for DK (P = 0.173). A match was obtained in 71.7% (160/223) of paediatric cases (cut‐off ≥0.9) and in 61.6% of adult cases (172/279). Assay performance was similar for cases at initial presentation vs. cases of recurrent disease, with a match made at a calibrated score of ≥0.9 in 66.1% (293/443) vs. 65.5% (39/58) of cases, respectively, and in 70.9% (314/443) vs. 70.7% (41/58) of cases, respectively, when using the cut‐off of ≥0.84. The case presenting as metastatic disease of a primary CNS tumour (n = 1) was excluded from this analysis. For the 18 patients from whom both material from initial and recurrent disease were analysed, the effect of the MP result was various, including, for example, no match (<0.9) for both samples (six patients); establishing new diagnosis (≥0.9) for one (five patients) or both samples (one patient) or confirmation (and refinement) of diagnosis (≥0.9) for both samples (six patients). Out of nine patients with a calibrated score ≥0.9 for both samples, five patients had identical final and initial diagnoses. In one case, there was indication of disease progression with diffuse astrocytoma, IDH‐mutant (grade II) diagnosed on the initial sample and glioblastoma, IDH‐mutant diagnosed on the recurrent sample. In another case, glioblastoma IDH‐wildtype, subtype midline was diagnosed on the recurrent sample, after an initial diagnosis of low‐grade diffuse astrocytoma/glioma, IDH‐wildtype (grade II). Finally, in one case, the MP results and CNV profiles of initial (final diagnosis pilocytic astrocytoma) and ‘recurrent’ sample (final diagnosis glioblastoma, IDH‐wildtype) contributed to identifying the latter as a second primary tumour.
Tissue block age was computed by subtracting the year the tissue block was made from the year of MP analysis. Tissue block age ranged from 0 to 23 years, with a mean of 1.22 years (SD 2.82). For cases, for which a match to a methylation class could be made at cut‐off ≥0.9, tissue age ranged from 0 to 13 years, with a mean of 1.08 years (SD 2.45) and for no match cases (cut‐off <0.9), the tissue age ranged from 0 to 23 with a mean of 1.47 years (SD 3.43) (P = 0.147). For all cases combined (n = 502) (including cases with calibrated scores <0.3 given the value ‘0’), the determination coefficient R2 was 0.012, showing that 1.2% of the variance in calibrated score for the class family can be explained by tissue block age, with a P‐value of 0.015 (Figure S3). Excluding the cases that did not reach the threshold of 0.3 for the calibrated score (n = 463), the R 2 was 0.006 (P = 0.104).
In our series, the use of FF (although few in number) or FFPE material did not significantly affect the assay performance. A match with a calibrated score of ≥0.9 was obtained in 64.7% (11/17) of FF cases and in 66.5% of FFPE cases (298/448) (P = 0.531). The cases in which DNA was isolated elsewhere (n = 36) and the case for which the tissue was embedded in agar (n = 1) were left out of this analysis.
In 7.8% of cases (39/502), the calibrated score of the Classifier was too low (<0.3) and no match to a methylation class was found. There was no clear technical explanation why the performance of the assay was insufficient for these cases. For example, tumour cell percentage ranged from 10 to 100% (mean 62.6; SD 23.3, Figure 1 B). Also, DNA concentration (range 0.2–49 ng/μl, mean 12 ng/μl, SD 12.9 ng/μl) and total amount of DNA added (range 11–500 ng, mean 193.6 ng, SD 114 ng) were not particularly low for these cases. Similarly, the age of the tissue blocks used for analysis did not strikingly differ from those for which a match was made at a calibrated score >0.3. For these cases, tissue block age ranged from 0 to 16 years (mean 2.03 years; SD 3.67 years). The diagnoses prior to MP were also not particularly ‘rare’ (listed in Table S2) and there was no indication that germline mutations could be responsible for the low score in these cases.
Impact of suggested methylation class on diagnosis
Confirmation of diagnosis/confirmation and refinement (≥0.9)
In 54.4% of cases (273/502), the outcome of MP matched the initial histological diagnosis: in 41.0% (206/502) of the analysed cases, the suggested methylation class corresponded exactly with the initial pathological diagnosis (based on histology with or without molecular analysis) and the diagnosis of 13.3% of cases (67/502) was more refined after incorporation of the MP result (Figure 2). For example, cases with the histological diagnosis medulloblastoma could be assigned a subgroup (WNT‐activated, SHH‐activated and non‐WNT/non‐SHH) (n = 24). For ependymomas, particular groups such as ependymoma with RELA fusion (n = 2) or myxopapillary ependymoma (n = 2) could be specified. There were a few cases in which the diagnosis of myxopapillary ependymoma was suggested by the Classifier, while (according to the reporting pathologist) histologically no myxopapillary features could be identified upon revision (n = 4). These ependymoma cases were categorized as ‘confirmation of diagnosis (≥0.9)’. Of note, according to the DKFZ/Heidelberg methylation class description (www.molecularneuropathology.org), this class encompasses both ependymomas with myxopapillary features and to a lesser extent ependymomas with classical or rarely tanycytic histology. The classifier effect ‘confirmation and refinement’ was more common in the paediatric part of the cohort. This can be explained by the much larger proportion of medulloblastomas and ependymomas in the paediatric cohort, that is, tumours for which subclassification is clinically relevant and nowadays warranted [8, 9].
Figure 2.
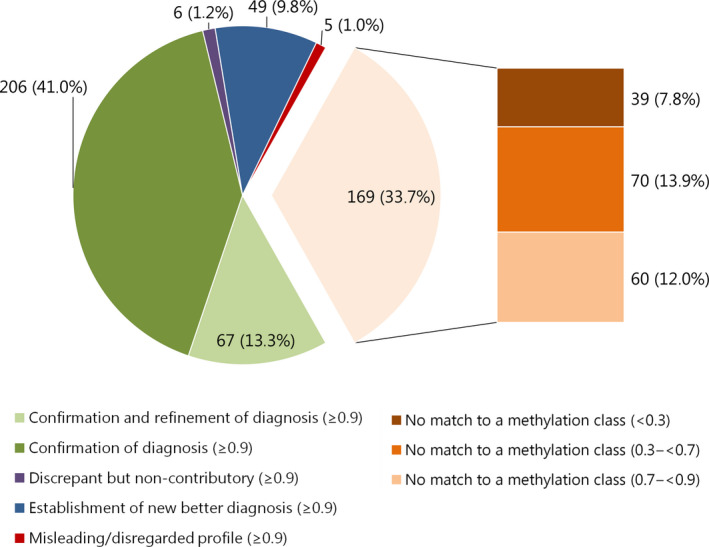
Effect of methylation profiling on diagnosis signed out to the clinicians for 502 cases with a calibrated score ≥0.9: light orange pie section represents all ‘no match’ cases combined. These are subdivided into calibrated scores <0.3; 0.3 to <0.7 and 0.7 to <0.9 in the bar to the right of this pie section. Labels represent: n (%) of 502 cases total.
Establishment of new diagnosis (≥0.9)
In 9.8% of cases (49/502), the MP result led to a new integrated diagnosis to that of the original pathologist (Figure 3 A). Of the cases in which a new diagnosis was established after MP, 11 cases had a lower WHO grade, 24 cases had a higher WHO grade and in 14 cases, the WHO grade was unchanged (Figure 3 B). A change in WHO grade occurred in challenging cases (n = 7), cases analysed during revision/re‐evaluation (n = 7) and in cases analysed as a part of routine diagnostic work‐up (n = 20). The effects of the Classifier result were similar for the paediatric cases compared to the adult cases. For example, a new diagnosis was established in 8.5% of paediatric cases (19/223) vs. 11.1% of adult cases (31/279) (P = 0.451) (Figure S4A,B).
Figure 3.
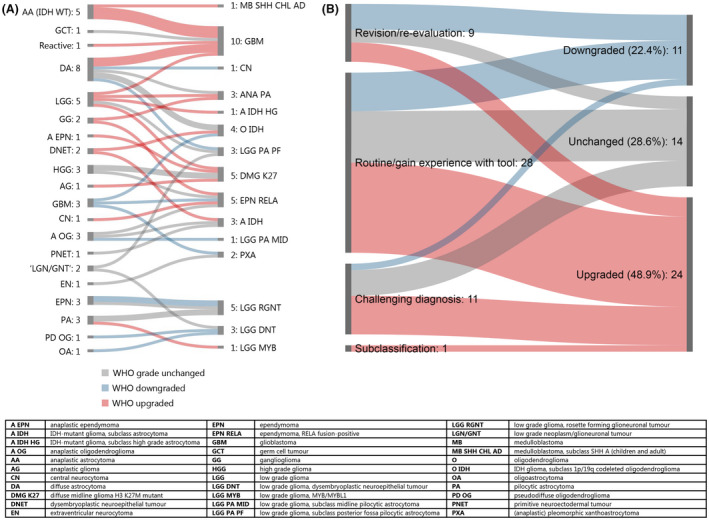
New diagnoses after methylation profiling: (A) Overview of initial (left) and new diagnoses after MP (right) in cases classified as ‘Establishing new better diagnosis (≥0.9)’ (n = 49). (B) WHO grade effects in cases with establishment of new diagnosis: Difference between WHO grade original diagnosis and final diagnosis for cases categorized as ‘Establishment of new better diagnosis (≥0.9)’ (n = 49), subdivided by reasons to perform MP. Green shades: downgraded, blue shades: unchanged, red shades: upgraded.
‘Misleading/disregarded profile’ (≥0.9)
In very few cases with a calibrated score ≥0.9 (1%, 5/502), the suggested class based on MP was not adopted by the original pathologist, because the MP outcome did not match the preferred histological diagnosis and/or was considered not to fit the clinical or radiological context (Table S3). An example of a case in this category is described in detail under ‘Special illustrative cases’. Importantly, thorough review of the follow‐up of these cases showed that in three of these cases, the MP result actually predicted clinical behaviour better than the diagnosis signed out by the original pathologist.
Discrepant but noncontributory (≥0.9)
Six cases were categorized as ‘discrepant but noncontributory (≥0.9)’ (Table S4). Five of these cases were classified as control tissue, which did not match the presence of tumour cells upon histology. Interestingly, three of these cases had relatively high estimated tumour cell contents (60–80%) and adequate quantities of DNA were available in four cases (200 ng). It is unclear why these cases failed to classify into a specific tumour entity. The sixth case concerns a tumour initially diagnosed as ganglioglioma with a differential diagnosis of pleomorphic xanthoastrocytoma (PXA), which was classified as PXA. The Classifier description for this class states that ‘tumours in this class may also show a ganglion cell‐like differentiation and may then histologically appear as anaplastic ganglioglioma’. The reporting pathologist interpreted this result as ‘not excluding the diagnosis of ganglioglioma’ and the final report stated a preferred diagnosis of ganglioglioma. Thus, the MP result was not strictly misleading but was not completely followed either, which is why the case was categorized as discrepant but noncontributory.
No match cases (0.3 to <0.7 and 0.7 to <0.9)
Twenty‐six per cent of all cases (130/502) were assigned to a methylation class with calibrated scores between 0.3 and <0.7 (n = 70) or 0.7 and <0.9 (n = 60). For some cases, the methylation class and accompanying CNV plot could still be helpful for the diagnosis: for example, establishing a new diagnosis in 8.3% (5/60) of cases with calibrated score of 0.7 to <0.9 and in 7.1% (5/70) of cases with calibrated score of 0.3 to <0.7 (Figure 4 A,B). Also, for a large proportion of cases, the histological diagnosis could be confirmed or confirmed and refined: 51.7% (31/60) and 5% (3/60) of cases with calibrated score 0.7 to <0.9, respectively, and 35.7% (25/70) and 10% (7/70) of cases with calibrated score 0.3 to <0.7 respectively. As expected, the percentage of cases for which the methylation class would have been potentially misleading was high in these groups: 20.0% (12/60) of cases with calibrated score 0.7 to <0.9 and 31.4% (22/70) of cases with calibrated score 0.3 to <0.7.
Figure 4.
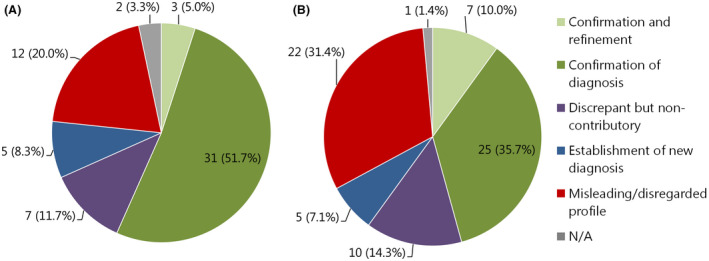
(A) Effect of methylation profiling on final pathological diagnosis of 60 cases with calibrated score of 0.7 to <0.9 for the methylation class family: N/A refers to unresolved cases. Labels represent: n (%) of 60 cases total. (B) Effect of methylation profiling on final pathological diagnosis of 70 cases with calibrated score of 0.3 to <0.7 for the methylation class family: N/A refers to unresolved cases. Labels represent: n (%) of 70 cases total.
Special illustrative cases
Case 1‐ ‘Misleading/disregarded profile’
In this 15‐year‐old girl who presented with progressive headache, nausea, blurred vision and diplopia, imaging revealed a tumour in the pineal region (Figure 5 A). The differential diagnosis at that time included germinoma, astrocytoma and pineocytoma/pineoblastoma. Histopathological evaluation of the biopsies revealed a small blue round cell tumour with brisk mitotic activity and a few dispersed Homer Wright‐like rosettes (Figure 5 B). Immunohistochemically, the tumour cells were positive for synaptophysin and showed a MIB‐1 labelling index of 60%. Combined with the information on its location, the tumour was histologically diagnosed as pineoblastoma. Surprisingly, MP suggested ‘medulloblastoma, WNT’ with an almost perfect calibrated score (0.99), with loss/monosomy of complete Chr 6 in the CNV plot (Figure 5 C). Additionally, next‐generation sequencing (NGS) revealed a CTNNB1 mutation (p.(Ser33Phe)). The CNV plot generated with single‐nucleotide polymorphism (SNP) array analysis was identical to that based on MP, and fluorescent in‐situ hybridization (FISH) and SNP data combined suggested polysomy with (indeed) relative loss of Chr 6 (Figure S5). The combination of CTNNB1 mutation and monosomy of Chr 6 is typically found in WNT‐activated medulloblastomas. One may speculate that occasionally otherwise prototypical medulloblastomas can occur in extraordinary locations such as the pineal region, for example, because of ectopic location of progenitor cells that usually are confined to the posterior fossa. For the time being, however, because of the tumour location (and after ruling out additional tumours in the posterior fossa), the diagnosis suggested by MP analysis was considered to be unfitting and the final integrated diagnosis was ‘pineoblastoma (WNT‐activated), WHO grade IV’. It is presently unknown if WNT activation has the same favourable prognostic meaning in pineoblastomas as in medulloblastomas.
Figure 5.
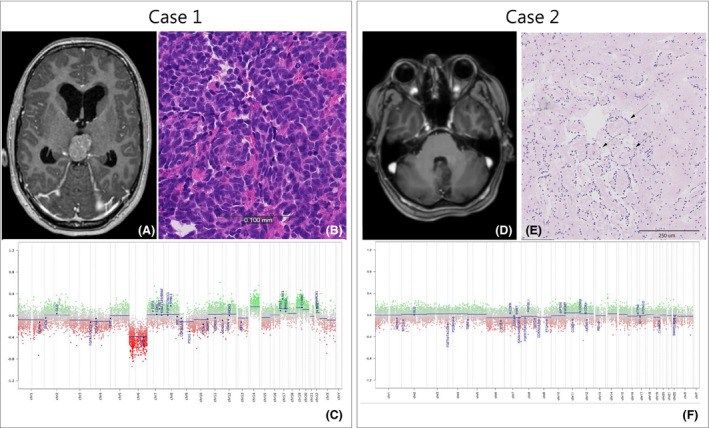
Case 1. (A) MR image, T1‐weighted after IV Gadolinium‐based contrast administration: tumour in the pineal region of a 15‐year‐old girl. (B) H&E stain 10×. (C) MP CNV plot, showing loss of chromosome 6. Case 2. (D) MR image, T1‐weighted after IV Gadolinium showing a tumour in cerebellum with close relation to the cerebral aqueduct and brainstem. (E) H&E stains 10×, arrows indicating focal perivascular pseudorosettes. (F) CNV plot showing a flat baseline with no indication of chromosomal changes in this tumour. Scale bars indicate (B) 100 μm; (E) 250 μm.
Case 2 ‐ Establishment of new diagnosis
A 42‐year‐old woman presented with a tumour in the cerebellum close to the brainstem (Figure 5 D). The histology (5‐mm biopsy) showed a tumour with low cellularity composed of small monomorphic tumour cells focally arranged in perivascular pseudorosettes (Figure 5 E). Immunohistochemistry showed positive staining for GFAP, Olig2 and ATRX and a low MIB‐1 labelling index (<1%). IDH1‐R132H staining was negative. Sequencing identified PIK3CA and NF1 mutations. The findings indicated a low‐grade glioma, possibly an ependymoma, and irradiation was considered. A match to the methylation class low‐grade glioma, rosette‐forming glioneuronal tumour and a flat CNV plot was obtained by MP (Figure 5 F). Additional synaptophysin immunostaining was performed and immunoreactivity was found in the pericapillary area of perivascular pseudorosettes as expected for this entity. This diagnosis was also supported by the coexisting mutations in PIK3CA and NF1, which is in line with the recent study on rosette‐forming glioneuronal tumours by Sievers et al. [12].With a WHO grade I diagnosis instead of a WHO grade II diagnosis, the oncologists decided not to irradiate the tumour and thereby to avoid potential radiation‐related side effects.
Discussion
DNA methylation‐based tumour classification of CNS tumours was implemented in our clinical diagnostic practices using the Classifier developed by Capper et al. (2018). 502 CNS tumours were analysed for a variety of reasons, for example, as a part of routine tumour work‐up, because of a challenging diagnosis, because it was requested by an external lab or to learn about the tool. A match to a specific methylation class with a calibrated score ≥0.9 was reached for 66.3% (333/502) of the tumour samples. Capper et al. reported a match to a methylation class in 88% of the analysed tumours in a clinical validation study with more than 1100 CNS tumours [6]. Our results are more in line with the data from a few of the five external centres, also reported by Capper et al., which had implemented MP in a clinicopathological setting. Combined, these centres analysed 401 cases, and for the individual centres, the percentage of match to a methylation class varied between 58 and 95% (78% match at a calibrated score of ≥0.9 for all five centres combined). Of note, the results presented in this study are based on the analysis of samples in a daily diagnostic setting, including, for example, suboptimal biopsy material. Although the majority of included cases were from our own neurosurgery departments, referred cases with challenging diagnoses were also included, most likely creating a slight bias towards more difficult cases. This possibly resulted in a higher percentage of cases that were difficult to classify, although Karimi et al. (2019) reported a clinically significant contribution of the MP result to the final diagnosis in 84% of a cohort of 55 challenging cases [13]. It remains unclear in what setting the cases analysed by the individual centres presented by Capper et al. were selected, and this might explain the better performance of the Classifier at some of these centres.
A new diagnosis was established in 9.8% (49/502) of the cases in favour of the diagnosis indicated by MP. This was based on a match to a specific DNA methylation class with a calibrated score ≥0.9, sometimes in combination with aberrations as seen in the CNV profile generated based on MP and/or available immunohistochemical findings or NGS results. In comparison, Capper et al. reclassified 12% of the cases (for individual centres, this varied between 6 and 25%). The IDH‐wildtype astrocytic gliomas, WHO grade II‐III were most frequently reclassified (28.6%, 14/49): most of these as glioblastoma (n = 8). The fact that a number of IDH‐wildtype astrocytic gliomas, WHO grade II‐III were reclassified as glioblastoma is in accordance with the new recommendations from cIMPACT‐NOW update 3, which states that such tumours without prototypical histological features of glioblastoma but with combined gain of complete Chr 7 and loss of complete Chr 10 and/or a high copy amplification of EGFR and/or a TERT promoter mutation should nowadays be considered as molecular glioblastoma, IDH‐wildtype [10]. Thereby, the CNV profile that accompanies the Classifier result can thus be very helpful in the diagnostic process. Of note, further refinement of diagnoses can now also be provided for meningiomas using the Meningioma classifier [14] and for medulloblastomas group 3/4 using the Medulloblastoma classifier [8].
We found that in 1% (5/502) of the cases, the result of the MP was initially disregarded/considered misleading, and the diagnosis before MP was maintained by the original pathologist (e.g. Case 1, Figure 5). However, thorough revision of these cases learned that the MP result for three of these cases was actually right. Since the start of the implementation of the Classifier tool in our centres, new insights have emerged and new molecular diagnostic criteria (e.g. c‐IMPACT‐NOW update 3 [10]) have been introduced in the field of CNS tumour diagnostics. Based on current insight on IDH‐wildtype astrocytic tumours, WHO grade II‐III, one of the misleading cases (low‐grade glioma, IDH‐wildtype) would not have been classified as misleading but instead, the MP result would have confirmed the diagnosis of a diffuse astrocytic glioma, IDH‐wildtype, with molecular features of glioblastoma, WHO grade IV (Table S3). In two of the other misleading cases (anaplastic astrocytoma, IDH‐wildtype and diffuse astrocytoma, IDH‐wildtype), later tumour recurrences displayed the histological high‐grade features that were initially missing, primarily preventing the neuropathologists from following the MP result when evaluating the primary tumour. Regarding the last misleading case (rosette‐forming glioneuronal tumour), later molecular analyses did not support the histopathological diagnosis given. In retrospect, these cases would have been categorized in the category ‘Establishment of new diagnosis’. Thus, these cases illustrate the significance of the learning curve of those involved in the interpretation of MP results and that categorization of cases in the predefined categories is not ‘static’ but may change as knowledge and experience expand.
The identification of cases that do not achieve a match at a calibrated score of ≥0.9 (33.7%, 169/502 in this study) is important, as they contribute to the ongoing improvement and refinement of the DNA methylation‐based Classifier tool. Investigation of underlying molecular alterations and identification of unifying features helps segregate these cases and may help to define novel entities [15, 16].
Among our 502 cases, 223 samples were from paediatric patients. It has previously been suggested that the performance of the Classifier is poorer for samples from paediatric patients than for samples from adult patients, because more rare tumours tend to occur in the paediatric cohort [6]. This does not hold true for our cohort (71.7% match made at ≥0.9). An explanation for this might be that most of the tumour samples from paediatric cases in our series consisted of tumour types that are well represented in the reference cohort used for the development of the Classifier (e.g. medulloblastoma, ependymoma and pilocytic astrocytoma). Similar to previously reported data by Pickles et al. (2019) [17], MP results especially contributed to (molecular) subtyping/refinement of the diagnosis for a large portion of the paediatric cohort (39%, 93/223). In addition to its role in routine diagnosis, MP may also prove to be an invaluable tool in the identification of new paediatric brain tumour classes [18].
We have looked at several technical parameters that could influence the performance of the MP assay, such as tumour cell percentage, sample fixation (FF/FFPE), DNA concentration, DNA amount and age of the tissue block. However, none of these parameters seemed to have a clear effect on the likelihood that a match could be made with a methylation class family/subclass. The regression analysis between tissue block age and assay performance revealed that the tissue block age had a weak/negligible effect (1.2% variance, P = 0.012) on the calibrated score for the class family. These results indicate a very good assay performance even on old tissue blocks (>5–10 years). However, our results may be biased as the majority of samples included in our study were less than 5 years old. Additional comparative studies exclusively including old tissue blocks of different age intervals are needed to get a better idea of any tissue age‐dependent effects on the assay performance.
The correlation between tumour cell percentage and array performance was not straightforward. For the cases with ‘no match <0.3’, tumour cell percentage ranged from 10 to 100% and there were several cases with good assay performance (that is, match made at calibrated score ≥0.9) with low estimated tumour cell percentage (as low as 15%). Of note, the estimation of tumour cell percentage is notoriously imprecise [19]. Nonetheless, intuitively, it appears important to analyse tumour DNA of vital tumour and to minimalize DNA isolation from normal tissue as well as from necrotic areas, as these might result in suboptimal or erroneous classification (e.g. too low calibrated score or classification as normal/control tissue). Therefore, in our routine diagnostics, we aim to macrodissect vital tumour tissue to obtain preferably >30% tumour cells.
Although the number of assays performed with FF material was small, we did not observe a significant difference in assay performance when using FF or FFPE as input material with regard to the percentage of cases for which a match was made with a good calibrated score (FF: 64.7%; FFPE: 66.5%). It should be noted that the recommended restore step was performed for all FFPE samples. In addition, the DKFZ/Heidelberg Classifier was built on MP data sets obtained from DNA isolated from FFPE patient samples. However, because DNA extracted from FF material is generally of higher concentration and superior quality compared to what can be obtained from FFPE material, the former may be preferred (especially in cases where FFPE material is very old).
In this study, the overall fraction of cases with a match at a calibrated score ≥0.9 was comparable between The Netherlands and Scandinavia, with 68% and 62% match respectively. This suggests that MP is a robust approach, withstanding slightly different approaches in purification procedures and variation in DNA amounts. The original instructions for the performance of the EPIC array by Illumina state that the assay requires an input of ≥250 ng of genomic DNA. We did not attempt to define a lower cut‐off of DNA amount in this study, but in our experience, even samples with low DNA quantities of questionable quality from ‘old’ material can be successfully classified. Yet, to facilitate the interpretation of the MP results, four technical parameters deemed relevant for assay performance are valuable to include in the pathology reports: quantity of DNA input, estimated tumour cell percentage, quality of bisulphite conversion and percentage of detected CpG sites.
We have not investigated the possible treatment consequences of each case after DNA MP. Also, we do not have clinical follow‐up of all the cases that were included, to allow consideration of clinical behaviour of the tumour. It remains to be elucidated whether MP results with a good calibrated score (≥0.9) might overrule the histopathological diagnosis, such as in cases like the ‘ganglioglioma’ classified as pleomorphic xanthoastrocytoma as shown in Table S4. Furthermore, in our experience, MP results with a calibrated score <0.9 can also be useful, though the percentage of ‘misleading profiles’ increased significantly the lower the calibrated score (2.2% when using ≥0.84 cut‐off; 20% for cases with calibrated score 0.7 to <0.9 and 32.4% for cases with calibrated score 0.3 to <0.7). Further exploration of the best cut‐off value for the calibrated score and how to interpret the data in cases with a (somewhat) lower score are warranted.
No specific data on the turn‐around time have been included in this study, as many cases were analysed retrospectively. In addition, workflows have been optimized and adjusted to the expected sample load since the start of this study, making it difficult to provide exact data on turn‐around times. Now, 3 years after the introduction of the tool, we have a turn‐around time of a maximum of 2 weeks (array processed once per week). Only in the instance that analysis has to be repeated due to technical problems (registered for 12 out of 502 cases in this database, 2–3% of current cases) or if there are left over cases but too few to fill an additional chip, this may be longer. Our clinicians are aware of this workflow and know they can anticipate the MP result about 2 weeks after MP testing has been ordered.
To conclude, DNA MP is a very powerful tool to support the clinical diagnosis of CNS tumours, especially in cases where morphological and genetic features are inconclusive. The fact that the assay provides different levels of information (tumour classification with family and subclass, CNV’s and single gene promoter methylation status) makes it extra valuable. However, to avoid misdiagnoses and to achieve therapeutic management decisions, it is crucial to interpret the results of MP in the context of clinical, radiological, histopathological and other molecular information. For transparent integration of the results obtained with MP in the clinical diagnostic process, we suggest to add these results to ‘layer 4’ of the integrated diagnostics approach as proposed by the International Society of Neuropathology‐Haarlem consensus guidelines [20] and more recently by the International Collaboration on Cancer Reporting (ICCR) guidelines as well ((http://www.iccr‐cancer.org/datasets/published‐datasets/central‐nervous‐system). This would facilitate continuous multidisciplinary evaluation of the molecular and epigenetic information and adjustment of the interpretation as knowledge evolves.
Ethical approval
The Danish part of the study was approved by the Regional Committee on Health Research Ethics for Southern Denmark (Project‐ID S‐20150148) as well as the Danish Data Protection Agency (file number: 16/11065). The use of tissue was not prohibited by any patient according to the Danish Tissue Application Register. For the Dutch part of the study, anonymous or coded use of clinical data is part of the standard treatment agreement with patients in our centres.
Author contributions
The database was built by NB, LPA, JKP and HB. FDV also assisted in the setup of the database (NL). LPA performed statistical analyses and created visual representations of the outcomes. LPA and JPK created illustrations for case examples. LPA, JKP and HB wrote the manuscript, with input from BWK, PW and WDL. DS, BPU, MG, TB, ST, EA, BK, WTD, PW and FDV provided cases. All authors read and approved the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Fig S1 .Distribution of cases by calibrated scores for methylation subclass:Frequencies of calibrated score for methylation subclass of 280 cases for which subclass was specified.Cut‐off value for the MP subclass was set at ≥0.50 by Capper et al. (5).
Fig S2 . Effect of methylation profiling on diagnosis for 502 cases with a calibrated score ≥0.84 (possible alternative cut‐off value for calibrated score, suggested by Capper et al.(6)): Categorization of the cases based on the effect of methylation profiling on the diagnosis signed out to the clinicians. Light orange pie section represents all ‘no match’ cases combined. These are subdivided in calibrated scores <0.3; 0.3‐<0.7 and 0.7‐<0.84 in the bar to the right of this pie section. Labels represent: n (%) of 502 cases total..
Fig S3 . (A) Distribution of calibrated scores for methylation class family by tissue block age of FFPE samples (n=448): purple circles: values for individual samples (some symbols superimposed); red crosses: mean calibrated for methylation class family. Horizontal lines ‐ blue solid: threshold of calibrated score ≥0.9; blue dashed: possible alternative threshold for calibrated score at ≥0.84 as suggested by (6).Cases with ‘no match <0.3’ (no calibrated score provided) were given the value 0 to visualize them in this plot.
Fig S4. Effect of methylation profiling on final pathological diagnosis of specific subgroups. (A) Paediatric (n=223); (B) Adult (n=279); (C) Subclassification (n=43); (D) Challenging diagnosis (n=89).
Fig S5. Case 1. (A) FISH CMYC break‐apart probe (red and green signals) showing 4‐6 signal pairs per nucleus, indicative for polysomy; (B) FISH centromere 6 probe (yellow) showing 2‐3 signals per nucleus, suggesting relative loss of Chr 6; (C) SNP array suggesting loss of chromosome 6; (D) SNP array adjusted for copy number neutral loss of chromosome 6.
Data S1 . Supplementary data.
Acknowledgement
We acknowledge the excellent laboratory work performed by the technicians Rene Sørensen, Anmar Omara Kafel and Tobias Teken Christensen, from the PCR laboratory, at the Department of Pathology, Odense University Hospital, Denmark, as well as by Erwin van der Biezen, Carmen de Voijs and Willem Hoefakker from the molecular laboratory at the Department of Pathology, UMC Utrecht, The Netherlands.
Priesterbach‐Ackley L. P., Boldt H. B., Petersen J. K., Bervoets N., Scheie D., Ulhøi B. P., Gardberg M., Brännström T., Torp S. H., Aronica E., Küsters B., den Dunnen W. F. A., de Vos F. Y. F. L., Wesseling P., de Leng W. W. J. and Kristensen B. W. (2020) Neuropathology and Applied Neurobiology 46, 478–492 Brain tumour diagnostics using a DNA methylation‐based classifier as a diagnostic support tool
References
- 1. Kros JM, Gorlia T, Kouwenhoven MC, Zheng P, Collins VP, Figarella‐branger D et al Panel review of anaplastic oligodendroglioma from European organization for research and treatment of cancer trial 26951: assessment of consensus in diagnosis, influence of 1p/19q Loss, and correlations with outcome. J Neuropathol Exp Neurol 2007; 66: 545–51 [DOI] [PubMed] [Google Scholar]
- 2. Kristensen BW, Priesterbach‐Ackley LP, Petersen JK, Wesseling P. Molecular pathology of tumors of the central nervous system. Ann Oncol 2019; 30: 1265–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP et al Identification of a CpG Island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010; 17: 510–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Danielsson A, Nemes S, Tisell M, Lannering B, Nordborg C, Sabel M et al MethPed: a DNA methylation classifier tool for the identification of pediatric brain tumor subtypes. Clin Epigenetics 2015; 7: 62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D et al DNA‐methylation‐based classification of central nervous system tumours. Nature. 2018; 555: 469–474. 10.1038/nature26000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Capper D, Stichel D, Sahm F, Jones DTW, Schrimpf D, Sill M et al Practical implementation of DNA methylation and copy‐number‐based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathol 2018; 136: 181–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jaunmuktane Z, Capper D, Jones DTW, Schrimpf D, Sill M, Dutt M et al Methylation array profiling of adult brain tumours: diagnostic outcomes in a large, single centre. Acta Neuropathol Commun 2019; 8: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sharma T, Schwalbe EC, Williamson D, Sill M, Hovestadt V, Mynarek M et al Second‐generation molecular subgrouping of medulloblastoma: an international meta‐analysis of Group 3 and Group 4 subtypes. Acta Neuropathol 2019; 138: 309–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pajtler KW, Witt H, Sill M, Jones DTW, Hovestadt V, Kratochwil F et al Molecular classification of ependymal tumors across All CNS compartments, histopathological grades, and age groups. Cancer Cell 2015; 27: 728–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brat DJ, Aldape K, Colman H, Holland EC, Louis DN, Jenkins RB et al cIMPACT‐NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH‐wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol 2018; 136: 805–810. 10.1007/s00401-018-1913-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hegi ME, Diserens A‐C, Gorlia T, Hamou M‐F, de Tribolet N, Weller M et al MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N Engl J Med 2005; 352: 997–1003 [DOI] [PubMed] [Google Scholar]
- 12. Sievers P, Appay R, Schrimpf D, Stichel D, Reuss DE, Wefers AK. Rosette‐forming glioneuronal tumors share a distinct DNA methylation profile and mutations in FGFR1, with recurrent comutation of PIK3CA and NF1. Acta Neuropathol 2019; 138: 497–504 [DOI] [PubMed] [Google Scholar]
- 13. Karimi S, Zuccato JA, Mamatjan Y, Mansouri S, Suppiah S, Nassiri F et al The central nervous system tumor methylation classifier changes neuro‐oncology practice for challenging brain tumor diagnoses and directly impacts patient care. Clin Epigenetics 2019; 11: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sahm F, Schrimpf D, Stichel D, Jones DTW, Hielscher T, Schefzyk S et al DNA methylation‐based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol 2017; 18: 682–94 [DOI] [PubMed] [Google Scholar]
- 15. Koelsche C, Mynarek M, Schrimpf D, Bertero L, Serrano J, Sahm F. Primary intracranial spindle cell sarcoma with rhabdomyosarcoma ‐ like features share a highly distinct methylation profile and DICER1 mutations. Acta Neuropathol 2018; 136: 327–37 [DOI] [PubMed] [Google Scholar]
- 16. Reinhardt A, Stichel D, Schrimpf D, Sahm F, Korshunov A. Anaplastic astrocytoma with piloid features, a novel molecular class of IDH wildtype glioma with recurrent MAPK pathway, CDKN2A/B and ATRX alterations. Acta Neuropathol 2018; 136: 273–91 [DOI] [PubMed] [Google Scholar]
- 17. Pickles JC, Fairchild AR, Stone TJ, Brownlee L, Merve A, Yasin SA et al DNA methylation‐based profiling for paediatric CNS tumour diagnosis and treatment : a population‐based study. Lancet Child Adolesc Heal 2019; 4642: 1–10 [DOI] [PubMed] [Google Scholar]
- 18. Perez E, Capper D. DNA‐methylation‐based classification of paediatric brain tumours. Neuropathol Appl Neurobiol 2020. https://doi.org/1010.1111/nan.12598. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 19. Smits AJJ, Kummer JA, De Bruin PC, Bol M, Van Den Tweel JG, Seldenrijk KA et al The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod Pathol 2014; 27: 168–74 [DOI] [PubMed] [Google Scholar]
- 20. Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, Von Deimling A et al International Society of Neuropathology‐Haarlem Consensus Guidelines for Nervous system tumor classification and grading. Brain Pathol 2014; 24: 429–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1 .Distribution of cases by calibrated scores for methylation subclass:Frequencies of calibrated score for methylation subclass of 280 cases for which subclass was specified.Cut‐off value for the MP subclass was set at ≥0.50 by Capper et al. (5).
Fig S2 . Effect of methylation profiling on diagnosis for 502 cases with a calibrated score ≥0.84 (possible alternative cut‐off value for calibrated score, suggested by Capper et al.(6)): Categorization of the cases based on the effect of methylation profiling on the diagnosis signed out to the clinicians. Light orange pie section represents all ‘no match’ cases combined. These are subdivided in calibrated scores <0.3; 0.3‐<0.7 and 0.7‐<0.84 in the bar to the right of this pie section. Labels represent: n (%) of 502 cases total..
Fig S3 . (A) Distribution of calibrated scores for methylation class family by tissue block age of FFPE samples (n=448): purple circles: values for individual samples (some symbols superimposed); red crosses: mean calibrated for methylation class family. Horizontal lines ‐ blue solid: threshold of calibrated score ≥0.9; blue dashed: possible alternative threshold for calibrated score at ≥0.84 as suggested by (6).Cases with ‘no match <0.3’ (no calibrated score provided) were given the value 0 to visualize them in this plot.
Fig S4. Effect of methylation profiling on final pathological diagnosis of specific subgroups. (A) Paediatric (n=223); (B) Adult (n=279); (C) Subclassification (n=43); (D) Challenging diagnosis (n=89).
Fig S5. Case 1. (A) FISH CMYC break‐apart probe (red and green signals) showing 4‐6 signal pairs per nucleus, indicative for polysomy; (B) FISH centromere 6 probe (yellow) showing 2‐3 signals per nucleus, suggesting relative loss of Chr 6; (C) SNP array suggesting loss of chromosome 6; (D) SNP array adjusted for copy number neutral loss of chromosome 6.
Data S1 . Supplementary data.


