Abstract
Background
Maternal dietary exposures are considered to influence the development of infant allergies through changes in the composition of breast milk. Cohort studies have shown that ω3 polyunsaturated fatty acids (PUFAs) in breast milk may have a beneficial effect on the preventing of allergies in infants; however, the underlying mechanisms remain to be investigated. We investigated how the maternal intake of dietary ω3 PUFAs affects fatty acid profiles in the breast milk and their pups and reduced the incidence of allergic diseases in the pups.
Methods
Contact hypersensitivity (CHS) induced by 2,4‐dinitrofluorobenzene (DNFB) and fluorescein isothiocyanate was applied to the skin in pups reared by mother maintained with diets mainly containing ω3 or ω6 PUFAs. Skin inflammation, immune cell populations, and expression levels of immunomodulatory molecules in pups and/or human cell line were investigated by using flow cytometric, immunohistologic, and quantitative RT‐PCR analyses. ω3 PUFA metabolites in breast milk and infant's serum were evaluated by lipidomics analysis using LC‐MS/MS.
Results
We show that maternal intake of linseed oil, containing abundant ω3 α‐linolenic acid, resulted in the increased levels of ω3 docosapentaenoic acid (DPA) and its 14‐lipoxygenation products in the breast milk of mouse dams; these metabolites increased the expression of TNF‐related apoptosis‐inducing ligand (TRAIL) on plasmacytoid dendritic cells (pDCs) in their pups and thus inhibited infant CHS. Indeed, the administration of DPA‐derived 14‐lipoxygenation products to mouse pups ameliorated their DNFB CHS.
Conclusion
These findings suggest that an inhibitory mechanism in infant skin allergy is induced through maternal metabolism of dietary ω3 PUFAs in mice.
Keywords: 14‐lipoxygenation, breast milk, infant allergy, TRAIL, ω3 DPA
Maternal intake of α‐linolenic acid increased ω3 DPA and its 14‐lipoxygenation products in the breast milk of dams. 14‐lipoxygenation products of ω3 DPA increased TRAIL expression on plasmacytoid dendritic cells in the pups, which suppressed the IFN‐γ‐producing T cells and consequently inhibited infant allergic dermatitis.
Abbreviations
- 12‐LOX
12‐lipooxygenase
- ALA
α‐linolenic acid
- DPA
docosapentaenoic acid
- Elovl5
elongation of very long‐chain fatty acids protein 5
- EPA
eicosapentaenoic acid
- IFN‐γ
interferon‐γ
- TRAIL
TNF‐related apoptosis‐inducing ligand
1. INTRODUCTION
The numbers of infants with allergic diseases including allergic dermatitis, respiratory allergy, and food allergy have been increasing in recent years. Various genetic and environmental factors, including diet and commensal microorganisms, contribute to causing infant allergies.1, 2, 3 Accumulating clinical evidence indicates that maternal dietary exposures, due to their effects on breast milk composition, are key factors in the incidence of infant allergies.2 In addition, cohort studies in the Netherlands and Taiwan have shown that prolonged breastfeeding correlates with a reduction in the risk of allergic eczema in children.4, 5 In contrast, a Copenhagen cohort study yielded opposite results, where prolonged breastfeeding increased in the risk of infant allergic eczema.6 Together, these findings imply that the onset of infant allergy is not determined simply by the quantity or duration of infant intake of breast milk. Instead, infant allergies may be influenced by breast milk quality, which is determined through the metabolism of maternal dietary nutrients.7
Dietary oils, especially ω3 and ω6 essential polyunsaturated fatty acids (PUFAs), are associated with immunologic functions, including allergic responses in infants.2, 8, 9, 10, 11 The primary ω3 PUFA in linseed oil (also known as flaxseed oil) is α‐linolenic acid (ALA). After absorption into the body, ALA is converted to several ω3 PUFAs (eg, eicosapentaenoic acid [EPA], ω3 docosapentaenoic acid [DPA], and docosahexaenoic acid [DHA]) by several endogenous enzymes, including very long‐chain fatty acid elongase (Elovl) and desaturase.12 The ω3 PUFA‐derived metabolites have stronger bioactive and anti‐inflammatory functions than their parent compounds.13 We previously reported that maintaining mice on a diet containing 4% linseed oil ameliorated the incidence of allergic diarrhea and increased the amount of EPA in the intestinal compartments.14 Using lipidomics analysis, we identified 17,18‐epoxy eicosatetraenoic acid (17,18‐EpETE) as an anti‐allergic lipid metabolite abundantly produced in the intestine of mice fed linseed oil.14 Our subsequent study showed that 17,18‐EpETE prevents skin inflammation in mice and nonhuman primates by inhibiting neutrophil trafficking via GPR40.15
Cohort studies have shown that decreased levels of ω3 PUFAs and increased levels of ω6 PUFAs in breast milk and infant serum correlate with increased risk of allergy in infants.2 Consistent with those findings, maternal supplementation with fish oil, which contains abundant ω3 EPA and DHA, during pregnancy and lactation helped to prevent allergic eczema and food allergies in infants.9 In addition, recent studies revealed that human breast milk contains EPA‐ and DHA‐derived metabolites, which act as anti‐inflammatory mediators.16, 17, 18 Moreover, DPA is not only an intermediate between EPA and DHA but also a substrate for bioactive and anti‐inflammatory metabolites (eg, resolvin [Rv] sn‐3 DPA, protectinsn‐3 DPA, maresin [MaR] sn‐3 DPA).19, 20 However, how maternal PUFAs control various immunologic functions in the offspring remains to be elucidated.
In the current study, we extended our system to investigate the unresolved question of how increased levels of ω3 PUFAs in the maternal diet affect the fatty acid profiles of the breast milk of mouse dams and in their pups to reduce the incidence of allergic disease in the pups.
2. METHODS
2.1. Mice
BALB/c mice were purchased from Japan SLC and reared on a conventional diet (FR‐2; Funabashi Farm) or customized chow containing 4% soybean oil or 4% linseed oil (Oriental Yeast) under specific‐pathogen‐free conditions. The chow was stored refrigerated and was used during the period for optimal stability of α‐linolenic acid.14 For the cross‐fostering experiments, pups were cross‐fostered onto dams whose litters had been born within 48 h of the fostered pups. All animals were reared in the experimental animal facilities of the National Institutes of Biomedical Innovation, Health, and Nutrition, Japan. The experiments were approved by the Animal Care and Use Committees of the institute and were implemented in accordance with their guidelines.
2.2. Collection of milk samples from human mothers and diagnosis of allergic symptoms in their infants
Samples of human breast milk were collected within 1 month after childbirth from mothers (n = 34) at NTT Medical Center Tokyo. Sensitization using IgE as an indicator and allergic symptoms (atopic dermatitis and food allergy) in their infants (age, 1 year or younger) were diagnosed by a pediatrician. For the comparison of 14‐lipoxygenation products in breast milk, the infants were classified as having allergies or not according to these data. All experiments were approved by the Committee of NTT Medical Center Tokyo, The University of Tokyo, RIKEN, and NIBIOHN and were conducted in accordance with their guidelines (approval numbers: 14‐15, 26‐77‐1210, H27‐18, no. 71); informed consent was obtained from all participants.
2.3. DNFB‐ or FITC‐induced CHS in mice
Induction of 2,4‐dinitrofluorobenzene (DNFB)‐contact hypersensitivity (CHS) was performed as previously described,15, 21 with the modification for infant mice. At 2 weeks after birth, infant mice were anesthetized with isoflurane (Forane; AbbVie) and then sensitized through the application of 12.5 μL of 0.5% (wt/vol) DNFB (Nacalai Tesque) in 4:1 acetone:olive oil (vol/vol) to their shaved abdomens. Five days after sensitization, they again were anesthetized with isoflurane (AbbVie) and then challenged on both sides of each ear with 5 μL of 0.3% (wt/vol) DNFB. At 48 hours after DNFB challenge, the change in ear thickness measured by using a micrometer (Mitsutoyo) was compared with the ear thickness before elicitation with isoflurane (AbbVie), as shown in Supplemental Figure 1. Scratching behavior of pups was counted for 10 min by using video recording, following a previous report.22
Figure 1.
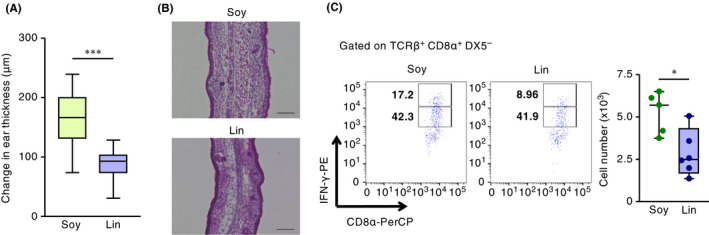
Maternal dietary intake of linseed oil ameliorates infant CHS. Pups were nursed for 2 weeks by dams maintained on diets containing 4% soybean oil (Soy) or linseed oil (Lin), after which infant mice were sensitized to DNFB. Five days after sensitization, infants were challenged with DNFB, and (A) ear swelling was calculated as the difference between ear thickness before and at 48 h after DNFB challenge. Horizontal lines indicate median values. P values were obtained by using the Mann‐Whitney U test (***P < .001). The graph shows data from individual infant mice (n = 48 [Soy] or 60 [Lin]); data were pooled from four representative independent experiments with reproducible results. (B) Frozen ear sections were stained with hematoxylin and eosin and analyzed microscopically at 48 h after DNFB challenge. Data are representative of three independent experiments with reproducible results. Bar, 100 μm. (C) Representative plots and numbers of IFN‐γhigh–producing CD8α+ T cells in infant skin at 48 h after DNFB challenge, as determined by using flow cytometry. The cell number shown is for the left (ie, challenged) ear of each pup. Horizontal lines indicate median values. P values were obtained by using the Mann‐Whitney U test (*P = .0173). Data from the left ear of individual mice are shown and are representative of three independent experiments with reproducible results
Induction of fluorescein isothiocyanate (FITC)‐CHS was performed as previously described,23 with modifications for infant mice. At 2 weeks after birth, infant mice were anesthetized with isoflurane (Forane; AbbVie), their ear thickness was measured by using a micrometer (Mitsutoyo), and their abdomens were shaved (day 0). Pups were sensitized through the application of 50 μL of 0.5% (wt/vol) FITC (Sigma) in 1:1 acetone:dibutyl phthalate (Sigma) (vol/vol) to their shaved abdomens on days 0 and 1. On day 5 after sensitization, pups again were anesthetized with isoflurane (AbbVie) and, for challenge, both sides of each ear were treated with 5 μL of 0.5% (wt/vol) FITC. At 24 h after FITC challenge, pups were anesthetized by using isoflurane (AbbVie); ear thickness was measured again and compared with that before CHS elicitation.
The protocol for intraperitoneal injection with lipids was previously described, with modifications for dams and infant mice.24 For lipid treatment, mouse dams were injected intraperitoneally with EPA, DPA, or DHA (1 mg/mouse in 200 μL of 5% ethanol in PBS; Cayman) or vehicle (200 μL of 5% ethanol in PBS) daily from day 14 of pregnancy until the pups were 20 days old, except on the day of parturition (day 0). Infant mice were injected intraperitoneally with EPA, DPA, or DHA (400 μg per pup in 100 μL of 4% ethanol in PBS; Cayman), a 14‐lipoxygenation product of DPA (the product synthesized from 9.5 μg of DPA per pup in 100 μL of 4% ethanol in PBS), or vehicle only (100 μL of 4% ethanol in PBS). Topical treatment with lipids was performed as previously described, with modifications for infant mice.15 Infant mice were treated topically with EPA, DPA, or DHA (10 μg per abdomen in 12.5 μL of 50% ethanol in distilled water, or 10 μg per each ear of pups in 10 μL of 50% ethanol in distilled water; Cayman), or vehicle only [12.5 μL (abdomen) or 10 μL (each ear) of 50% ethanol in distilled water] 30 min before DNFB sensitization and elicitation. For inhibition of TRAIL, the administration schedule and volume of anti‐mouse TRAIL monoclonal antibody (mAb) are based on previous reports,15, 25 with modifications for infant mice. Briefly, infant mice received purified anti‐mouse TRAIL mAb (100 μg/mouse; BioLegend, 109308) 30 min before DNFB sensitization and elicitation.
2.4. Immunohistochemistry
Frozen ear tissues were evaluated histologically as previously described.15 Briefly, ear specimens were embedded in optimal cutting temperature compound (Sakura Finetek) and were sectioned at 6 μm by using a cryostat (Leica Microsystems). Ear sections were stained with hematoxylin and eosin. The previously described whole‐mount staining protocol 21, 26 was modified as follows. The dorsal halves of the ears were incubated for 30 min at 37°C in 0.5 M ammonium thiocyanate. Subsequently, the dermal sheets were separated and fixed in acetone for 10 min at –20°C. To block nonspecific binding, the sheets were incubated for 30 min in PBS containing 2% newborn calf serum (NCS) at room temperature. The sheets then were incubated with purified anti‐mouse MHC class II mAb (BioLegend, 107602) overnight at 4°C in the dark, followed by incubation with mAb to rat IgG conjugated to Cy3 (Jackson ImmunoResearch Laboratories, 712‐165‐153) in PBS containing 2% NCS for 30 min at room temperature in the dark. The slides were mounted by using a ProLong Antifade Kit (Thermo Fisher Scientific) and observed under fluorescence microscopy (BIOREVO BZ‐9000; Keyence). The number and size of DC clusters were scored according to previously described criteria.21, 26 Samples were assessed histologically by two dermatopathologists, who were blinded to the treatment group.
2.5. Cell isolation and flow cytometry
Single‐cell suspensions were prepared from ear skin and spleen as previously described 15, 21 and modified as follows. The ear was divided into halves as the dorsal and ventral sides, and the cartilage was removed. The ear skin then was incubated for 90 min in RPMI containing 2% NCS and 2 mg/mL collagenase II (Wako). Cell suspensions were filtered by using a 100‐μm cell strainer.
Cells were treated with anti‐CD16/CD32 (TruStain fcX; BioLegend, 101320) to block nonspecific binding and subsequently with 7‐AAD (BioLegend, 420404); samples then were stained with the following mAbs: Alexa Fluor 488–conjugated anti‐mouse B220 (BD Biosciences, 557669) and MHC class II mAb (BioLegend, 107616) and anti‐human CD8α mAb (BioLegend, 300916); Alexa Fluor 647–conjugated anti‐mouse plasmacytoid dendritic cell antigen‐1 (PDCA‐1) (BioLegend, 127106) and TCRβ (BioLegend, 109218) mAbs; APC‐conjugated annexin V (BioLegend, 640919), anti‐mouse CD11c (BD Biosciences, 550261), PDCA‐1 (BioLegend, 127016), and TCRβ (BD Biosciences, 553174) mAbs; APC‐Cy7–conjugated anti‐CD11b (BioLegend, 101226) and CD45 (BioLegend, 103116) mAbs; BV421‐conjugated anti‐mouse CD11b (BioLegend, 101 236), CD4 (BioLegend, 100544), and CD45 (BioLegend, 103133) mAbs; FITC‐conjugated anti‐mouse B220 (BioLegend, 103206), DX5 (BioLegend, 108906), and Ly6G (BioLegend, 127606) mAbs; PE‐conjugated anti‐mouse CD11c (BD Biosciences, 557401), Langerin (eBioscience, 12‐2075‐82), and TRAIL (BioLegend, 109306) mAbs and anti‐human TRAIL mAb (BioLegend, 308206); PE‐Cy7–conjugated anti‐mouse CD11c (BioLegend, 117318), CD4 (BioLegend, 100528), F4/80 (BioLegend, 123114), Ly6C (BioLegend, 128018), and TCRβ (BioLegend, 109222) mAbs and anti‐human‐CD4 mAb (BioLegend, 317414); and PerCP‐conjugated CD11b (BioLegend, 101230) and CD8α (BioLegend, 100732) mAbs.
The intracellular staining was described previously 27 and modified as follows. For the intracellular cytokine staining, cells were incubated in the presence of phorbol 12‐myristate 13‐acetate (1.875 μg/mL; Sigma) and ionomycin (937.5 ng/mL; Sigma) in complete RPMI 1640 supplemented with brefeldin A (5.0 μg/mL; BioLegend) for 5 hours at 37°C. Cells were stained by using the Zombie‐NIR Fixable Viability Kit (BioLegend) and then treated with anti‐CD16/CD32 to block nonspecific binding, washed, and stained with antibodies to cell‐surface markers described above. Cells were then fixed and permeabilized by using an intracellular staining kit (BD Biosciences) or the Foxp3 staining kit (eBioscience). Subsequently, cells were stained with PE‐conjugated anti‐mouse IFN‐γ (BioLegend, 505808) and anti‐human IL‐4 mAb (BioLegend, 500704), eFluor 660–conjugated anti‐mouse indoleamine‐2,3‐dioxygenase mAb (eBioscience, 50‐9473), or PE‐conjugated anti‐mouse Foxp3 mAb (eBioscience, 12‐5773‐82) with anti‐CD16/CD32 to block nonspecific binding. Stained cells underwent flow cytometry (FACS Aria; BD Biosciences or MACSQuant; Miltenyi Biotec), and the data were analyzed by using FlowJo software (Tree Star).
2.6. Reverse transcription‐PCR analysis
A single‐cell suspension from the skin was obtained as described in Cell isolation and flow cytometry section. plasmacytoid dendritic cells (pDCs) and CD8+ T cells were sorted by using a FACSAria (BD Biosciences); sorted pDCs and CD8+ T cells were lysed in Sepasol‐RNA I Super G (Nacalai Tesque), and total RNA was extracted according to the manufacturer's protocol. Samples of abdominal mammary gland and liver tissue were collected from 12‐week‐old female BALB/c mice that had been lactating for 14 days, or samples of whole ears or epidermal sheets separated from the dorsal halves of the ears incubated for 30 min at 37°C in 0.5 M ammonium thiocyanate were collected from 3‐week‐old pups, and total RNA was extracted from samples by using the ReliaPrep RNA Tissue Miniprep System (Promega) according to the manufacturer's protocol. RNA was reverse‐transcribed by using the SuperScript VILO cDNA Synthesis Kit (Invitrogen); cDNA underwent real‐time reverse transcription‐PCR amplification by using the Universal ProbeLibrary probe (Roche) and primer sets specific for Actb (forward primer: 5′‐aaggccaaccgtgaaaagat‐3′, reverse primer: 5′‐gtggtacgaccagaggcatac‐3′, probe no. 56), ALox12 (forward primer: 5′‐gatcactgaagtggggctgt‐3′, reverse primer: 5′‐cacacatggtgaggaaatgg‐3′, probe no. 94), Cd86 (forward primer: 5′‐gaagccgaatcagcctagc‐3′, reverse primer: 5’‐ cagcgttactatcccgctct‐3′, probe no. 107), Cxcl1 (forward primer: 5′‐gactccagccacactccaac‐3′, reverse primer: 5′‐tgacagcgcagctcattg‐3′, probe no. 83), Cxcl2 (forward primer: 5′‐aaaatcatccaaaagatactgaacaa‐3′, reverse primer: 5′‐ctttggttcttccgttgagg‐3′, probe no. 26), Cxcl9 (forward primer: 5′‐cttttcctcttgggcatcat‐3′, reverse primer: 5′‐gcatcgtgcattccttatca‐3′, probe no. 1), Cxcl10 (forward primer: 5′‐gctgccgtcattttctgc‐3′, reverse primer: 5′‐tctcactggcccgtcatc‐3′, probe no. 3), Ccl8 (forward primer: 5′‐ttctttgcctgctgctcata‐3′, reverse primer: 5′‐gcaggtgactggagccttat‐3′, probe no. 26), Ccl17 (forward primer: 5′‐gctctgcttctggggacttt‐3′, reverse primer: 5′‐gaatggcccctttgaagtaa‐3′, probe no. 17), Dctrailr1 (forward primer: 5′‐tgtgaactttgctccacctgt‐3′, reverse primer: 5′‐cctatttggcactcgcattt‐3′, probe no. 83), Dctrailr2 (forward primer: 5′‐tttgctccacctgtgataaaga‐3′, reverse primer: 5′‐gttcggcactggcatttc‐3′, probe no. 49), Elovl2 (forward primer: 5′‐aacttgcagtgtcagaatctcg‐3′, reverse primer: 5′‐accacaagaccttggctacc‐3′, probe no. 2), Elovl5 (forward primer: 5′‐gtcctccatcccgtccat‐3′, reverse primer: 5′‐ctggatgattgtcagcacaaa‐3′, probe no. 31), Tcf4 (forward primer: 5′‐ccagcactgccgactaca‐3′, reverse primer: 5′‐ aagggtcgctgctgtgat‐3′, probe no. 69), Gzmb (forward primer: 5′‐ tgctgctcactgtgaaggaa‐3′, reverse primer: 5′‐ttaccatagggatgacttgctg‐3′, probe no. 2), Icosl (forward primer: 5′‐cgcaccatgcagctaaagt‐3′, reverse primer: 5′‐aaacatggagcttcttccaaac‐3′, probe no. 27), Il1ra (forward primer: 5′‐ctccttctcatccttctgtttca‐3′, reverse primer: 5’‐ggtcttctggttagtatcccagatt‐3′, probe no. 34), Il10 (forward primer: 5′‐cagagccacatgctcctaga‐3′, reverse primer: 5’‐tgtccagctggtcctttgtt‐3′, probe no. 41), Il17 (forward primer: 5′‐cagggagagcttcatctgtgt‐3′, reverse primer: 5’‐gctgagctttgagggatgat‐3′, probe no. 74), Il22 (forward primer: 5′‐tgacgaccagaacatccaga‐3′, reverse primer: 5’‐aatcgccttgatctctccac‐3′, probe no. 94), Pdl1 (forward primer: 5′‐ccatcctgttgttcctcattg‐3′, reverse primer: 5’‐tccacatctagcattctcacttg‐3′, probe no. 101), Tgfb1 (forward primer: 5′‐tggagcaacatgtggaactc‐3′, reverse primer: 5′‐gtcagcagccggttacca‐3′, probe no. 72), Tnfa (forward primer: 5′‐ctgtagcccacgtcgtagc‐3′, reverse primer: 5′‐ttgagatccatgccgttg‐3′, probe no. 25), Tnfrsf10b (forward primer: 5′‐ccctgagatctgccagtcat‐3′, reverse primer: 5’‐tttctctgggggtacaggaa‐3′, probe no. 103), Trail (forward primer: 5′‐gctcctgcaggctgtgtc‐3′, reverse primer: 5′‐ccaattttggagtaattgtcctg‐3′, probe no. 76), and Tslp (forward primer: 5′‐cagcttgtctcctgaaaatcg‐3′, reverse primer: 5’‐aaatgttttgtcggggagtg‐3′, probe no. 71). Real‐time reverse transcription‐PCR analysis was performed by using a Lightcycler II (Roche Diagnostics) to measure the expression levels of specific genes, previously described.15
2.7. Collection of milk from mouse dams
Murine breast milk was collected as described previously,28 with modifications. For both groups, dams were separated for 8 to 15 hours from pups by covering the infant mice with a mesh cage until breast milk was collected. Dams received oxytocin (dose, 2 IU in 200 μL saline intraperitoneally; Peptide Institute); 5 minutes later, milk began to be collected automatically (model WAT‐2006, Automated Milker for Rats and Mice; Muromachi Kikai) until at least 100 μL was obtained.
2.8. LC‐MS/MS lipidomics analysis
LC‐MS/MS lipidomics analysis was performed by using a high‐performance liquid chromatography system (Waters UPLC) with an Acquity UPLC BEH C18 column (Waters) and by using a linear ion‐trap quadrupole (QTRAP 5500; AB Sciex) 29 or hybrid (Orbitrap Elite; Thermo Scientific) mass spectrometer. MS/MS analysis was performed in anion mode; ω3 PUFAs and various lipid metabolites were quantified by using multiple reaction monitoring.29
2.9. Preparation of mixtures of DPA‐derived 14‐lipoxygenation products
The protocol for preparing mixtures of DPA‐derived 14‐lipoxygenation products was previously described,19 with modification as follows. Briefly, DPA (15 μM) was incubated with 12‐LOX (5.4 U/mL; Oxford Biomedical Research) in 0.05 M sodium phosphate buffer containing 0.02% Tween‐20 in the dark (pH 7.4, 37°C, 1 h). Methanol and chloroform were added to achieve a water:methanol:chloroform ratio of 1:1:2. The methanol‐chloroform fraction was collected and evaporated to dryness. The samples were reduced by using NaBH4 in methanol; 1 N HCl in water and chloroform was added to achieve a water:methanol:chloroform ratio of 1:1:2. The methanol‐chloroform fraction again was collected and evaporated to dryness. The mixtures of 14‐lipoxygenation products were dissolved in ethanol.
2.10. Culture of PMDC05 cell line
The human pDC line PMDC05 was established and kindly provided by M. Narita (Niigata University, Japan). PMDC05 cells were maintained in Iscove's modified Dulbecco's medium (IMDM; Invitrogen) supplemented with 10% fetal bovine serum (FBS), as previously described.30 PMDC05 cells (1 × 106/mL in IMDM containing 10% FBS) were cultured for 12 h with or without imiquimod (5 μg/μL; InvivoGen), DPA (1000 nM; Cayman), or the mixture of 14‐lipoxygenation products synthesized from 100 to 1000 nM DPA. Cell‐surface expression of TRAIL was evaluated through flow cytometry.
2.11. Co‐culture of PMDC05 cells and human PBMCs
PBMCs (Precision Bioservices; 2 × 105 cells in 300 µL of IMDM containing 10% FBS) were stimulated with plate‐bound anti‐human CD3 and CD28 antibodies (BioLegend, 317 302 and 302 902, respectively; 0.2 µg/mL) for 3 days. Prestimulated PBMCs were incubated for 3 days in the presence of PMDC05 cells (2 × 105 cells in a total of 500 µL of IMDM containing 10% FBS) with or without the 14‐lipoxygenation products from 1 μM DPA. Subsequent IL‐4 expression in T cells was evaluated by using flow cytometry.
2.12. Statistics
Statistical significance was evaluated by using the Kruskal‐Wallis test with Dunn's multiple‐comparison test to compare multiple groups; the Mann‐Whitney test or an unpaired t test was used to compare two groups (Prism; GraphPad Software). P values less than .05 were considered to be significant.
3. RESULTS
3.1. Increased levels of ω3 PUFAs in the maternal diet ameliorate infant Th1‐ and Th2‐type CHS
Initially, we wondered whether the maternal intake of linseed oil affected the development of skin inflammation in the progeny. We therefore maintained female mice on a diet containing 4% linseed oil or 4% soybean oil (as a control, frequently used in normal diets) for at least 2 months and then performed timed mating; when 2 weeks old, the pups of these dams underwent CHS testing (Figure S1). In this model of Th1‐type skin allergy, pups were sensitized through the application of the DNFB to their abdomens. Five days after sensitization, DNFB was applied to both sides of each ear; ear thickness was measured before and at 48 h after DNFB challenge (Figure S1). Pups whose mothers consumed the control chow (containing soybean oil) demonstrated marked skin inflammation and pronounced ear swelling, whereas skin inflammation was reduced in the offspring of dams that consumed linseed oil (Figure 1A,B). On the other hand, there was no difference in the number of scratching behaviors, as an itching indicator, between nonsensitized and sensitized pups applied DNFB to ear, as well as between soybean and linseed oil groups in the infant CHS (Figure S2). In addition, we used an FITC‐induced Th2‐type skin allergy model.23 Again, pups whose mothers consumed the control chow (containing soybean oil) demonstrated marked skin inflammation and pronounced ear swelling, whereas skin inflammation was reduced in the offspring of dams that consumed linseed oil (Figure S3A). These results suggest that increased levels of ω3 PUFAs in the maternal diet (ie, maternal intake of linseed oil) reduced both Th1‐ and Th2‐type allergic dermatitis in their infants.
3.2. Increased levels of ω3 PUFAs in the maternal diet decrease the number of IFN‐γhigh–producing CD8α+ T cells in the skin during infant CHS
During the sensitization phase of the murine DNFB‐induced CHS model, conventional dendritic cells (cDCs), epidermal Langerhans cells, and Langerin+ dermal DCs in the skin take up hapten and present it to T cells.31 In the elicitation phase, repeat exposure of cDCs to hapten leads to inflammation and consequent activation of IFN‐γ–producing CD8α+ T cells and other inflammatory cells (eg, neutrophils, M1‐ and M2‐type macrophages).31, 32, 33 It was suggested that IL‐17–producing T cells, which also have a potential to produce IL‐22,34 contributed to exacerbating DNFB CHS responses.33, 34 Moreover, keratinocytes produced chemokines including CXCL1, CXCL2, CXCL9, CXCL10, CCL8, and CCL17, which exacerbated CHS responses by inducing the accumulation of inflammatory cells in the skin,35 and IL‐1 receptor antagonist from keratinocytes ameliorated CHS responses.36 Thymic stromal lymphopoietin (TSLP) from epithelial cells and keratinocytes and DCs and mast cells were known to enhance Th2‐biased immune responses.37 In addition to their pathogenic immune cell functions, regulatory T (Treg) cells, Langerhans cells, and pDCs may help to prevent inflammatory reactions.32, 33, 38 Therefore, several mechanisms might be involved in the suppression of the DNFB‐induced CHS response in the linseed oil group, such as the inhibition of inflammatory cells, the induction of regulatory cells, or both.32, 33, 38
To examine these possibilities, we counted the IFN‐γ–producing CD8α+ T cells in infant skin. We noted that the numbers of dermal IFN‐γ–producing CD8α+ T cells, especially the IFN‐γhigh populations, were increased in the pups of the soybean oil group, whereas maternal intake of linseed oil reduced this cell count (Figure 1C). In addition to skin, we evaluated systemic IFN‐γ production in the spleen in the elicitation phase of DNFB CHS. There was no difference in CHS‐induced increasing of IFN‐γ production from splenic CD4 and CD8 T cells in pups of CHS between soybean and linseed oil group (Figure S4). Moreover, there was no difference in the mRNA levels of Il17, Il22, and Tslp in the ear skin of pups between soybean oil and linseed oil group (Figure S5). In contrast, the numbers of other inflammatory cells—including cDCs, Langerhans cells, M1‐ and M2‐type macrophages, neutrophils—in the pups’ skin did not differ between the two groups, although the number of Langerin+ dermal DCs was increased in the pups of the linseed group compared with the soybean group (Figure S6A‐C). In addition, although cDCs reportedly cluster together as an inducible skin‐associated lymphoid tissue during the elicitation phase and thus activate IFN‐γ–producing CD8α+ T cells,21, 26 neither the distribution, number, nor size of the clusters differed between our mice (Figure S6D,E). Moreover, there was no significance in the mRNA levels of Cxcl1, Cxcl2, Cxcl9, Cxcl10, Ccl8, Ccl17, and Il1ra in an epidermal sheet as keratinocytes between soybean and linseed oil groups (Figure S7). Together, these findings suggest that maternal intake of linseed oil mainly inhibited the induction of high IFN‐γ–producing CD8α+ T cells in the skin of infant mice.
3.3. Maternal intake of linseed oil increases TRAIL‐expressing pDCs in infant skin
Our current findings guided us to evaluate which types of cells responded to maternally ingested linseed oil to inhibit the induction of IFN‐γ–producing CD8α+ T cells in the progeny pups. To this end, we determined the number of dermal Treg cells in the pups because these cells are known to inhibit skin inflammation,39 but the cell counts did not differ between the soybean and linseed groups (Figure 2A).
Figure 2.
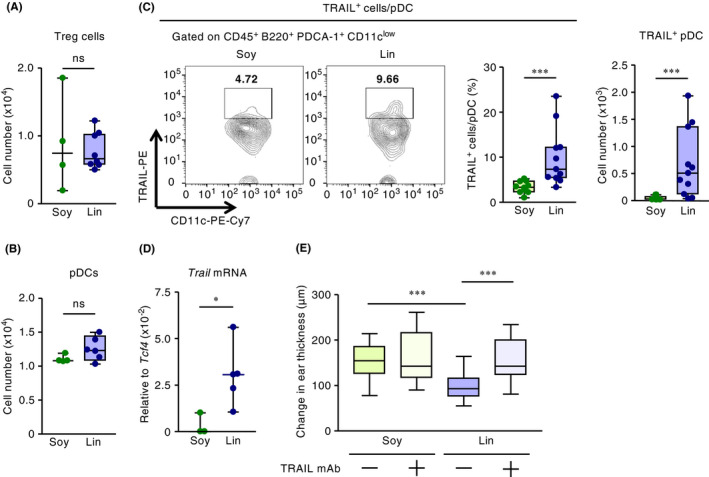
Maternal intake of linseed oil increases the expression of TRAIL on dermal pDCs in mouse pups and thus prevents infant CHS. After the induction of CHS, flow cytometric analyses were performed to enumerate (A) Foxp3+ regulatory T cells, (B) pDCs, and (C) TRAIL‐expressing pDCs in infant skin; this panel also includes a representative FACS plot and the proportion of TRAIL‐expressing pDCs among total pDCs. Data were pooled from two independent experiments with reproducible results (C). (D) The Trail mRNA expression level in pDCs in infant skin was determined by using real‐time RT‐PCR. Cell numbers shown are those for the left (ie, challenged) ear. Horizontal lines indicate median values. P values were obtained by using the Mann‐Whitney U test (*P < .05; ***P < .001; ns, not significant). (E) TRAIL expression–dependent regulation of infant CHS after maternal dietary intake of linseed oil was evaluated according to ear swelling in pups. To inhibit TRAIL function, infant mice were intraperitoneally injected with anti‐TRAIL mAb at 30 min before DNFB sensitization and challenge, and ear swelling was calculated as the difference in ear thickness before and at 48 h after DNFB challenge (n = 22 [Soy without anti‐TRAIL mAb], 20 [Soy with anti‐TRAIL mAb], 30 [Lin without anti‐TRAIL mAb], or 22 [Lin with anti‐TRAIL mAb]). Horizontal lines indicate median values. P values were obtained by using Dunn's multiple‐comparison test (***P < .001). Data were pooled from three independent experiments with reproducible results
We then looked at pDCs, which are reported to exert regulatory functions by producing immunomodulatory factors, including indoleamine 2,3‐dioxygenase (IDO), inducible T‐cell co‐stimulator ligand (ICOSL, which is encoded by Icosl), granzyme B (encoded by Gzmb), TNF‐α (encoded by Tnfa), and TNF‐related apoptosis‐inducing ligand (TRAIL, encoded by Trail).25, 40, 41, 42 Moreover, pDCs were reported to potentially express inhibitory molecules including programmed death ligand‐1 (PD‐L1), IL‐10, TGF‐β1, and CD86.43, 44 However, the number of pDCs in the skin did not differ significantly between the soybean and linseed groups (Figure 2B). In addition, mRNA levels of Tcf4, a transcription factor essential for the differentiation of pDCs,45, 46 in dermal pDCs were similar between groups (Figure S8B). Although the sorted fraction (CD45+ PDCA‐1+ B220+ CD11b– CD11clow) contains other cell types in addition to pDCs, their mRNA levels in pDC were exactly normalized by mRNA levels of Tcf4. In contrast to the similarities in cell numbers, cell‐surface expression of TRAIL and Trail mRNA transcript levels were increased in the skin pDCs of the linseed group compared with the control group during infant DNFB‐induced CHS (Figure 2B‐D). Furthermore, in the infant FITC‐induced CHS model, cell‐surface expression of TRAIL again was increased in the skin pDCs of the linseed group compared with the control group (Figure S3B). Unlike for TRAIL, the IDO+ proportion of pDCs and the mRNA expression levels of other regulatory molecules from pDCs, such as Icosl, Gzmb Tnfa, Pdl1, Il10, Tgfb1, and Cd86, did not differ between the soybean and linseed groups (Figure S8). Furthermore, the infant dermal TRAIL+ cells induced by maternal intake of linseed oil were mainly CD45+ PDCA‐1+ B220+ pDCs in the elicitation phase of CHS (Figure S9).
We next examined whether TRAIL expression on pDCs was a prerequisite for the inhibition of CHS in the linseed group. To test this issue, infant mice were treated with TRAIL‐specific neutralizing antibody during the development of CHS. The treatment canceled the anti‐inflammatory activity in the linseed group, resulting in the development of CHS, which was similar in severity to that in the soybean group (Figure 2E). These results collectively suggest that maternal intake of linseed oil increased the number of TRAIL‐expressing tolerogenic pDCs in the skin of pups and thus inhibited their CHS.
3.4. No effects on apoptosis by maternal intake of linseed oil
It was reported that TRAIL inhibited the cytokine (eg, IFN‐γ) production from activated T cells 47 and induced apoptosis in T cells 48 and keratinocytes.49 Therefore, we employed Annexin V staining to assess the effect of maternal intake of linseed oil on the induction of apoptosis in keratinocytes and CD8+ T cells in the skin of pups. There was no difference in the numbers of apoptotic cells in CD45– cells including keratinocytes and purified dermal CD8+ T cells between soybean and linseed oil groups (Figure S10A).
Since TRAIL receptors are composed of TRAIL receptor 2 (also known as death receptor 5, encoded by Tnfrsf10b), TRAIL receptor 3 (decoy TRAIL receptor 1, encoded by Dctrail1), and TRAIL receptor 4 (decoy TRAIL receptor 2, encoded by Dctrail2),50 we assessed which TRAIL receptors were expressed on keratinocytes and CD8+ T cells in pups in our CHS model. mRNA encoding Dctrail1, which is the decoy receptor, was highly expressed in keratinocytes compared to other TRAIL receptors (Figure S10B). Moreover, Dctrail1 mRNA was expressed in CD8+ T cells as highly as Tnfrsf10b mRNA in pups of CHS (Figure S10B). These findings suggest that different expression levels of each TRAIL receptor resulted in the specific inhibition of T‐cell activation without affecting apoptosis in keratinocytes and CD8+ T cells in our infant CHS.
3.5. Breast milk with increased levels of ω3 PUFAs ameliorated infant CHS
To test whether increased levels of ω3 PUFAs in the dams’ milk affected the development of DNFB‐induced CHS in mouse pups, we performed cross‐fostering experiments (Figure 3A). Skin inflammation in infant mice born to soybean oil–fed dams was ameliorated when these pups were nursed by mothers in the linseed group (Figure 3B). In agreement with this finding, the pups fostered to dams fed linseed oil showed increased proportion of TRAIL‐expressing pDCs in the skin (Figure 3C). Collectively, these results indicate that dams’ milk contained active lipid products derived from linseed oil that induced the production of TRAIL‐expressing pDCs and subsequently prevented infant CHS.
Figure 3.
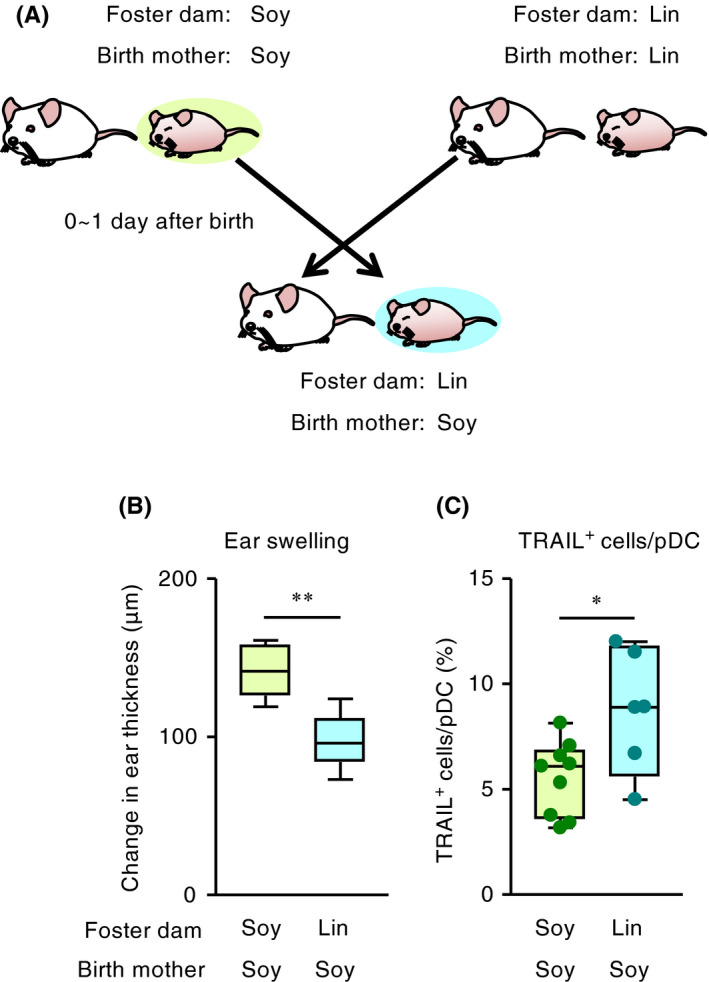
Breast milk mediates the inhibition of infant CHS and the induction of TRAIL on pDCs. (A) Experimental design for cross‐fostering. For each group of infant mice, cross‐fostering was established within 48 h after birth, and pups were fed for 2 weeks by foster dams. (B) Ear swelling in pups was evaluated at 48 h after DNFB challenge (n = 6 [birth mother, Soy; foster dam, Soy] or 11 [birth mother, Soy; foster dam, Lin]). Horizontal lines indicate median values. P values were obtained by using the Mann‐Whitney U test (**P < .01). Data are representative of two independent experiments with reproducible results. (C) The ratio of TRAIL expression on dermal pDCs in pups was evaluated at 48 h after DNFB challenge. Horizontal lines indicate median values. P values were obtained by using the Mann‐Whitney U test (*P < .05). Data are pooled from two independent experiments with reproducible results
3.6. Inhibition of infant CHS by ω3 DPA‐derived products in dams’ milk
We used lipidomics analysis involving liquid chromatography coupled with dual mass spectrometry (LC‐MS/MS) to quantify the levels of EPA, DPA, and DHA in dams’ milk and pups’ serum. The levels of these ω3 PUFAs were increased in the milk of the linseed group (Figure 4A‐C). Similarly, serum levels of EPA and DPA were increased in the pups of the linseed group, whereas the DHA level in the infant serum did not differ between the soybean and linseed groups (Figure 4D‐F). These results suggest that serum levels of ω3 PUFAs, especially EPA and DPA, in the infants were influenced by the maternal intake of these compounds.
Figure 4.
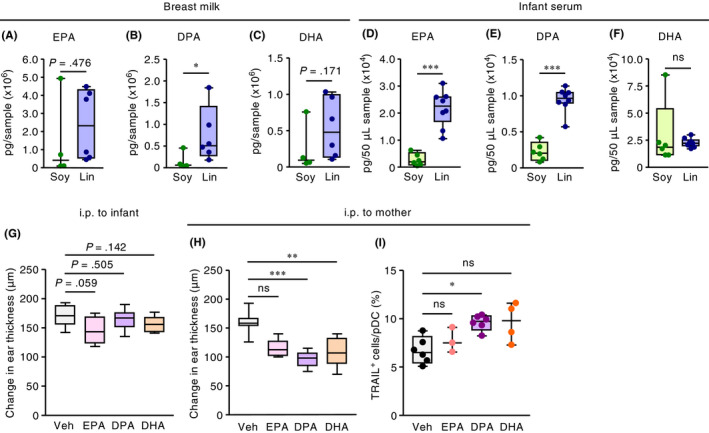
Increased levels of ω3 PUFAs regulate infant CHS and the induction of TRAIL expression on pDCs. (A to F) Three weeks after infants were born to mother mice maintained on diets containing 4% soybean (Soy) or linseed (Lin) oil, the amounts of EPA, DPA, and DHA in the (A to C) dams’ milk and (D to F) infants’ serum were quantified. Horizontal lines indicate median values. P values were obtained by using the Mann‐Whitney U test (*P < .05; **P < .01; ***P < .001; ns, not significant). Graphs show data from individual pups and dams. (G) Infant mice were intraperitoneally (i.p.) injected with EPA, DPA, DHA (400 μg/mouse daily), or vehicle (Veh) at 30 min and 1, 3, and 5 days before DNFB sensitization and at 30 min and 1 and 3 days before DNFB challenge; ear swelling was calculated as the difference between ear thickness before and at 48 h after DNFB challenge. Horizontal lines indicate median values. P values were obtained by using the Mann‐Whitney U test. (H, I) Mouse dams received intraperitoneal injection of EPA, DPA, DHA (1 mg/mouse daily), or Veh, and (H) ear swelling and (I) TRAIL expression on dermal pDCs of pups were evaluated at 48 h after DNFB challenge. Horizontal lines indicate median values. P values were obtained by using Dunn's multiple‐comparison test (**P < .01; ***P < .001; ns, not significant). The graph shows data from individual infant mice and is representative of three independent experiments with reproducible results
We next examined whether products derived from ω3 PUFAs were responsible for ameliorating skin inflammation in the pups. To this end, we intraperitoneally injected pups with EPA, DPA, or DHA; however, neither of these treatments reduced ear swelling due to CHS (Figure 4G). In contrast, intraperitoneal injection of ω3 PUFAs, especially DPA, into mother mice ameliorated skin inflammation in their pups, and this reduction in ear swelling was coincident with an increase in the proportion of TRAIL‐expressing pDCs in the pups’ skin (Figure 4H,I). Together, these results suggest that DPA‐derived metabolites generated in the dams were transferred to their infants, where they induced TRAIL‐expressing tolerogenic pDCs and consequently inhibited skin inflammation.
3.7. Lactating mammary gland preferentially generates 14‐lipoxygenation products of ω3 DPA
In the current study, we found that intraperitoneal administration of DPA to mouse dams—but not their progeny—inhibited infant allergy, thus suggesting that DPA metabolites generated in dams, presumably in mammary gland tissue, control the skin inflammation in their pups. Although our targeted, fully quantitative lipidomics analysis did not include DPA metabolites, the data regarding EPA‐ and DHA‐derived metabolites indicated that the levels of 12‐lipoxygenase (12‐LOX, encoded by Alox12) metabolites of EPA (ie, 12‐HEPE) and DHA (ie, 14‐HDoHE) were increased in the serum of pups from linseed‐fed dams (Figure S11A,B). These findings prompted us to suppose that a 12‐LOX‐mediated metabolite of DPA (14(S)‐hydroxy‐7Z,10Z,12E,16Z,19Z‐DPA; 14‐HDPA, a 14‐lipoxygenation product of DPA) might also be present in the serum of pups in the linseed group. Indeed, a virtual lipidomics analysis of DPA metabolites by using DHA metabolites as each standard revealed that 14‐HDPA was increased in the serum of infants in the linseed group, compared with other DPA metabolites (Figure S11C). To further confirm these findings, we synthesized DPA‐derived 14‐lipoxygenation products by incubating DPA with recombinant 12‐LOX. The fragments (eg, 207, 283, and 327 m/z) of the 345‐m/z precursor peak (approximate retention time, 25.1 min; likely monohydroxy DPA) of the product were consistent with previously reported fragments of 14‐HDPA (eg, 207, 283, and 327 m/z of the 345‐m/z precursor peak) 19 and identical to the fragments detected in the infant serum of the linseed group (Figure 5A). In addition, the fragments (eg, 143, 193, and 207 m/z) of the 361‐m/z precursor peak (approximate retention time, 20.5, 22.7, and 19.9 minutes, respectively; likely dihydroxy DPA) of the DPA‐derived 14‐lipoxygenation products were consistent with the previously reported fragments of MaR1n‐3 DPA, MaR2n‐3 DPA, and MaR3n‐3 DPA, respectively (Figure S12).19 The peak area of the 14‐lipoxygenation product predicted as 14‐HDPA (345 m/z precursor) in the infant serum was greater for the linseed group than the soybean group (Figure 5B). These results suggest that maternal intake of linseed oil increased the amounts of DPA‐derived 14‐lipoxygenation products (eg, 14‐HDPA), which then were transferred to infant mice.
Figure 5.
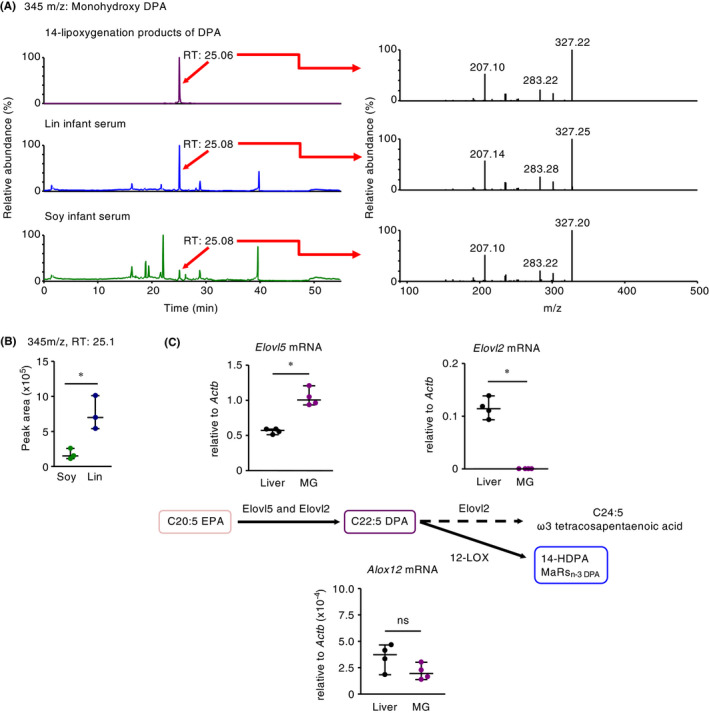
Maternal intake of linseed oil leads to the accumulation of 14‐lipoxygenation products of ω3 DPA in mouse pups due to the unique enzymatic environment in the mammary gland. (A) Representative chromatographs of the precursor ion at m/z 345 were obtained for the 14‐lipoxygenation products of DPA and of infant sera after maternal intake of linseed oil and soybean oil. Representative MS spectra (retention time [RT] 25.06‐25.08 min) from three independent experiments are shown. (B) The peak area of the precursor ion at m/z 345 was obtained for each 14‐lipoxygenation product of DPA in the sera of infants born to linseed oil– compared with soybean oil–fed dams. Horizontal lines indicate median values. P values were obtained by using the two‐tailed unpaired t test (*P < .05). (C) The mRNA expression levels of lipid metabolic genes (Elovl2, Elovl5, and Alox12) in the abdominal mammary glands (MG) and livers of 12‐week‐old dams, which had lactated for 2 weeks. Horizontal lines indicate median values. P values were obtained by using the Mann‐Whitney U test (*P < .05; ns, not significant). Data are representative of two independent experiments with reproducible results
Next, we investigated metabolic pathways in the mammary gland. For example, DPA is generated from EPA through the action of Elovl5 or Elovl2 51 and then metabolized to ω3 tetracosapentaenoic acid by Elovl2 or to 14‐lipoxygenation products including 14‐HDPA and MaRsn‐3 DPA by 12‐LOX.52, 53 Consistent with the production of DPA in the dams’ milk, mRNA expression of Elovl5 was significantly higher in the lactating mammary gland than in the liver (Figure 5C). In contrast, the mammary gland showed negligible expression of Elovl2 mRNA, whereas the liver expressed copious amounts of this transcript (Figure 5C). Furthermore, the expression levels of Alox12 mRNA were comparable between the mammary gland and the liver (Figure 5C), resulting in the 12‐LOX‐mediated metabolism of DPA to 14‐lipoxygenation products including 14‐HDPA and MaRsn‐3 DPA. These results suggest that lactating mammary gland tissue provides a unique environment in which DPA is generated preferentially from EPA by Elovl5 and is not elongated to ω3 tetracosapentaenoic acid by Elovl2 (Figure 5C).
3.8. Topical treatment with DPA reduces infant CHS
It was reported that the expression level of Elovl2 was negligible,54 and 12‐LOX was highly expressed in the skin,55 allowing us to hypothesize that the skin environment converted DPA to 14‐lipoxygenation products for the regulation of infant CHS. To test this hypothesis, we performed topical treatment with DPA on the ear swelling of pups in the CHS model. Unlike intraperitoneal injection, topical treatment of DPA to pups reduced ear swelling in DNFB CHS (Figure S13). These results suggested that topical DPA was converted to its 14‐lipoxygenation products for the regulation of infant CHS responses.
3.9. 14‐Lipoxygenation products of DPA directly induce the expression of TRAIL on pDCs to inhibit T‐cell activation for the reduction of infant CHS
We examined whether DPA‐derived 14‐lipoxygenation products were responsible for ameliorating skin inflammation in the pups. Intraperitoneal injection of these compounds into pups ameliorated skin inflammation, which was coincident with a decrease in the proportion of IFN‐γhigh CD8+ T cells in the skin (Figure 6A,B). These findings collectively suggest that DPA‐derived 14‐lipoxygenation products are the lipids responsible for controlling allergic inflammation in the skin of mouse pups.
Figure 6.
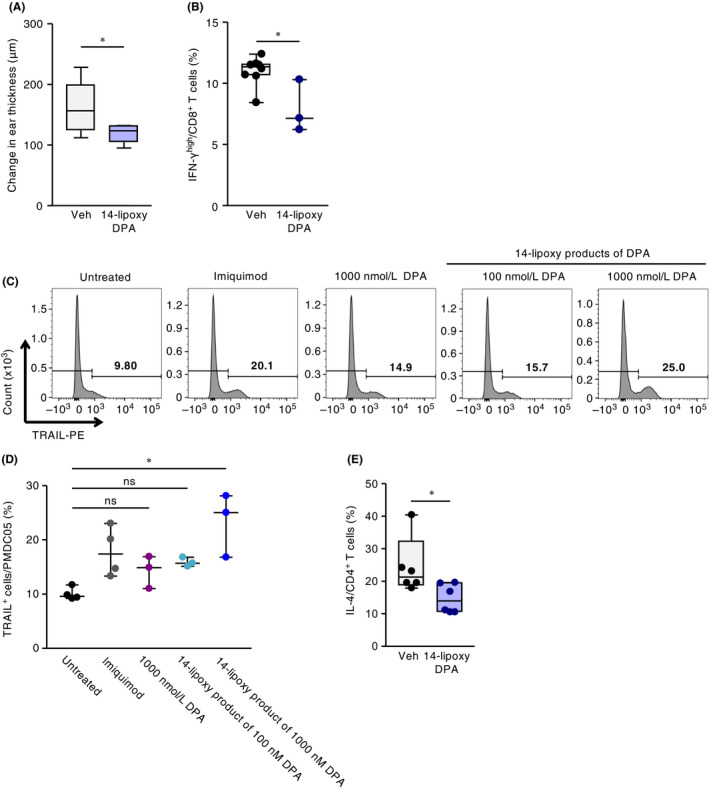
Exogenous 14‐lipoxygenation products of DPA suppressed infant CHS responses in vivo with the induction of TRAIL+ pDC and the suppression of T‐cell activation in vitro. Infant mice were intraperitoneally injected with 14‐lipoxygenation products of DPA (14‐lipoxy DPA) or vehicle (Veh) at 30 min and 1 and 3 days before DNFB sensitization and at 30 min and 1 and 3 days before DNFB challenge; (A) ear swelling was evaluated at 48 h after DNFB challenge. (B) The ratio of IFN‐γhigh–producing CD8+ T cells in infant skin at 48 h after DNFB challenge, as determined by using flow cytometry. Horizontal lines indicate median values. P values were obtained by using the Mann‐Whitney U test (*P < .05). Data were pooled for pups from three representative independent dams with reproducible results. (C, D) PMDC05 cells were treated with DPA, its 14‐lipoxygenation (14‐lipoxy) products, or imiquimod (5 μg/μL), and subsequent TRAIL expression was evaluated by using flow cytometry. (C) A representative histogram and (D) the proportion of TRAIL+ cells are shown. Horizontal lines indicate median values. P values were obtained by using Dunn's multiple‐comparison test (*P < .05). Data are pooled from two independent experiments with reproducible results. (E) PBMCs were stimulated with anti‐human CD3/CD28 antibody in the presence of PMDC05 cells with or without 1 μM of 14‐lipoxygenation product of DPA (14‐lipoxy DPA); subsequent IL‐4 expression in T cells was evaluated by using flow cytometry. Horizontal lines indicate median values. P values were obtained by using the Mann‐Whitney U test (*P < .05). Data are representative of two independent experiments with reproducible results
Next, we examined whether either DPA or its 14‐lipoxygenation products directly induced the expression of TRAIL on pDCs. We treated cells of the human pDC line PMDC05 with DPA or its 14‐lipoxygenation products including 14‐HDPA and MaRsn‐3 DPA in vitro. Flow cytometric analysis showed that treatment with 14‐lipoxygenation products of DPA—but not the parent compound itself—induced the expression of TRAIL on PMDC05 cells (Figure 6C,D).
We also evaluated the effects of increased TRAIL expression on pDCs induced by DPA‐derived 14‐lipoxygenation products on cytokine production from human PBMCs in vitro. Given that the PBMCs used in this experiment were biased to differentiate to IL‐4–producing cells, the activation of PBMCs with anti‐CD3/28 antibodies in the presence of PMDC05 induced IL‐4–producing T cells (Figure 6E). The increased TRAIL expression on PMDC05 induced by 14‐lipoxygenation products of DPA resulted in the suppression of IL‐4–producing T cells (Figure 6E). These results suggested that the induction of TRAIL expression on pDCs by DPA‐derived 14‐lipoxygenation products has the potential to suppress the activation of T cells.
3.10. Association of the levels of 14‐lipoxygenation DPA products in the breast milk with infant allergy in humans
We finally evaluated the relationship of our current findings with clinical phenomena. We investigated whether 14‐lipoxygenation DPA products could be detected in human breast milk by using LC‐MS/MS. Although 14‐HDPA, MaR1n‐3 DPA, MaR2n‐3 DPA, and MaR3n‐3 DPA were detected in the human milk (Figure S14A), the peak area of 14‐HDPA was larger than that of MaR1n‐3 DPA, MaR2n‐3 DPA, and MaR3n‐3 DPA (Figure S14B). We then compared the levels of 14‐lipoxygenation DPA products in the breast milk between nonallergic sensitized infants and allergic infants. The peak area of 14‐HDPA in the nonallergic group was larger than that of allergic one (Figure S15). On the contrary, there were no differences in MaR1n‐3 DPA, MaR2n‐3 DPA, and MaR3n‐3 DPA between allergic and nonallergic groups (Figure S15). Although we cannot exclude the possibility of the involvement of the other 14‐lipoxygenation DPA products (ie, MaR1n‐3 DPA, MaR2n‐3 DPA, and MaR3n‐3 DPA) in the control of the infant allergy, these results suggested that 14‐lipoxygenation DPA products (eg, 14‐HDPA) in the breast milk had a potential to prevent the onsets in infant allergies in humans.
4. DISCUSSION
Recent clinical studies have suggested that the PUFA quality of the maternal diet has the potential to affect immune responses, including allergic phenotypes, in infants.2, 8, 9, 10, 11 However, the mechanism through which maternal intake of ω3 or ω6 PUFAs regulates infant allergic disease remains to be clarified. In the current study, we showed the unique metabolic pathway of diet‐derived increased ω3 PUFAs in the lactating mammary glands of mouse dams modulated the incidence of CHS in their pups. Specifically, the environment in mammary gland tissue leads to the preferential accumulation of DPA and its 14‐lipoxygenation products (eg, 14‐HDPA) in the milk after maternal ingestion of linseed oil; in turn, these products reduce the incidence of skin inflammation in the infant pups through the induction of TRAIL expression on dermal pDCs, engaging them to promote a tolerogenic status (Figure S16). The lipid metabolic pathways in the lactating mammary gland differ from those in the liver (Figure 5C), suggesting a unique, coordinated system between mothers and infants that regulates a condition in infants through the metabolism of lipids in both the mother's diet (ie, dietary oils) and the infant's (ie, breast milk).
Previous meta‐analyses revealed discrepancies regarding the effects of ω3 PUFAs in the maternal diet and breast milk on the onset of infant allergy.56 In the current study, we showed that DPA metabolites generated by 12‐lipoxygenase might act as effector molecules to inhibit infant allergic symptoms through the breast milk in mice, although it remained to be undetermined whether 14‐lipoxygenated DPA detected in human milk was involved in the regulation of human infant allergy. Of note, a polymorphism of 12‐lipoxygenase in mammary gland tissue has been reported.57 In addition, accumulating evidence indicates that commensal bacteria participate in the generation of 14‐lipoxygenated lipids.58, 59 In this regard, a recent report suggested that the mammary gland contains commensal bacteria, which could affect the production of 14‐lipoxygenated lipids.60 Furthermore, the species and number of commensal bacteria are known to differ markedly between hosts. Therefore, interindividual differences in the production efficacy and types of ω3 PUFA‐derived metabolites including 14‐lipoxygenation products of DPA in breast milk are plausible.
We showed that milk from mouse dams fed linseed oil regulated CHS in pups (Figure 3). In a clinical study, EPA and DPA were predominant components of breast milk beginning 4 weeks after adding linseed oil to the mothers’ diet,61 similar to our current finding (Figure 4A‐C). Moreover, our study showed that EPA and DPA resulting from increased ω3 PUFAs in the maternal diet were translocated to the serum of pups (Figure 4D‐F). Intriguingly, intraperitoneal injection of exogenous DPA to mouse dams significantly reduced the infant CHS response (Figure 4I,J), but intraperitoneal injection of exogenous EPA or DPA to infant mice did not (Figure 4G). In addition, DHA suppresses CHS in adult mice,62 and many metabolites of EPA and DHA exert diverse biologic functions, including anti‐allergy and anti‐inflammation.13, 14, 21 However, because the ability to convert EPA to DPA or DHA is lower in infants than adults,63 EPA administered to infants may not sufficiently be converted to bioactive molecules for downregulation of CHS. In addition, because of the high demand for DHA during brain development, fatty acid‐binding proteins promptly carry DHA to the infant brain.64, 65 Therefore, DHA likely is used preferentially for brain development rather than for conversion into bioactive metabolites.
In contrast to intraperitoneal administration of DPA to pups, the injection of DPA into dams ameliorated the development of infant CHS, due to the generation of DPA‐derived 14‐lipoxygenation products, including 14‐HDPA and MaRsn‐3 DPA. Therefore, such 14‐lipoxygenation products might be generated preferentially in mouse mammary gland during lactation. Indeed, in lactating dams, the expression level of Elovl5 was higher and that of Elovl2 was much lower in the lactating mammary gland than in the liver (Figure 5C); this pattern creates an environment in mammary gland tissue where DPA is preferentially concentrated from EPA, without elongation to DHA. In humans, increasing ω3 PUFA intake consistently failed to significantly increase DHA levels in mammary gland.61
Moreover, topical treatment of DPA to pups reduced infant CHS response (Figure S13). This result implicated that skin environment efficiently converted DPA into its 14‐lipoxygenized products. Consistently, it was reported that 12‐LOX expression was detectable,55 and Elovl2 expression was low in the skin.54 Therefore, topical skin application of DPA might be effective for the regulation of infant CHS.
The development and functional changes of the mammary glands from pregnancy to lactation are regulated by transcriptional changes that are directed by estrogen, progesterone, and prolactin.66 In particular, lipid synthesis in lactating mammary gland is proposed to be activated through decreased progesterone concentrations and increased prolactin signaling, for the production of fat in breast milk.67 For example, miR‐150 suppressed the expression of Elovl5 in mammary gland throughout pregnancy, but Elovl5 expression increased and that of miR‐150 decreased once lactation began.66 Therefore, the biologic changes associated with lactation may create the unique environment that is necessary for the preferential concentration of DPA and its 14‐lipoxygenation products, which are generated by 12‐LOX, in mammary gland tissue.
According to a recent report, exogenous administration of DPA‐derived products reduced neutrophil‐endothelial cell interaction and therefore ameliorated inflammation in colitis model.20 Our current findings show that intraperitoneal injection of exogenous DPA to mouse dams led to the generation of 14‐lipoxygenation products, which ameliorated the infant CHS response and increased TRAIL expression by dermal pDCs in pups (Figure 4I,J). We also showed that injecting pups with 14‐lipoxygenation products ameliorated the infant CHS response (Figure 6A,B). Moreover, we found that the increased TRAIL expression on pDCs was sufficient to inhibit cytokine production from human PBMCs in vitro (Figure 6C‐E). In a previous study, TRAIL inhibited the activation of Th1 and Th2 responses in CD4+ T cells and the activation of CD8+ T cells.47 Consistent with this report, in our current study, maternal intake of linseed oil led to the induction of TRAIL expression on pDCs in pups and thus reduced infant CHS responses in murine models of both Th1‐ and Th2‐type allergies (Figures 1 and 2 and Figure S3).
Maternal intake of linseed oil decreased the number of dermal IFN‐γhigh–producing CD8+ T cells in their infants and thus inhibited Th1‐type CHS in the pups (Figure 1C). Since it was reported IFN‐γhigh and IFN‐γlow populations were present in PBMC T cells with functional differences,68 IFN‐γ is known to be a key factor in the exacerbation of contact hypersensitivity,33 and therefore, our data indicate that the decreased numbers of IFN‐γhigh population plausibly play a critical role in the amelioration of infant contact hypersensitivity by maternal intake of linseed oil. Moreover, CD8+ T cells exposed to a secondary antigen reportedly induce high production of IFN‐γ through IL‐2.69 Although IL‐2 incubation increases the levels of TRAIL receptor expressed on CD8+ T cells,70 CD8+ T cells exposed to a secondary hapten—in contrast to their response to primary exposure—may highly express IFN‐γ and TRAIL receptors during the elicitation phase of infant CHS. TRAIL is reported to inhibit the activation of CD8+ T cells or to induce their apoptosis,47, 48 and our current findings revealed that maternal intake of linseed oil decreased the number of IFN‐γ–producing CD8+ T cells in the skin of pups. On the other hand, maternal intake of linseed oil did not affect the apoptosis in the skin CD8+ T cells and keratinocytes in the pups. This finding was explained by a fact that they highly expressed Dctrail1, a decoy receptor for TRAIL in both CD8+ T cells and keratinocytes (Figure S10A). In addition, Tnfrsf10b mRNA was highly expressed in CD8+ T cells, but not keratinocytes, as much as Dctrail1 mRNA (Figure S10B). These results suggested that Tnfrsf10b and Dctrail1 signals coordinated to inhibit the activation of CD8+ T cells without increasing apoptosis.
Maternal intake of linseed oil did not affect IFN‐γ‐production from CD4+ T cells and CD8+ T cells in the spleen of pups in CHS (Figure S4). Consistently, it was reported that, unlike liver pDCs, splenic pDCs had no effect on the induction of oral tolerance to CHS responses.35 It is likely that the effect of maternal intake of linseed oil on the TRAIL expression on pDCs was different among organs. Therefore, the expression of TRAIL by dermal pDCs, which is induced by maternal consumption of linseed oil, may inhibit IFN‐γhigh–producing CD8+ T cells during the elicitation phase of CHS in mouse pups. Given that during allergic contact dermatitis, CD8+ T cells produce IFN‐γ, which induces epidermal spongiosis, with increased hyaluronan production and decreased E‐cadherin expression in keratinocytes,33, 71 TRAIL potentially may reduce epidermal spongiosis during infant CHS by inhibiting IFN‐γ–producing CD8+ T cells. As an additional possibility, although TRAIL demonstrated an anti‐inflammatory function in vitro by inducing interleukin‐1 receptor antagonist (IL‐1Ra) expression in keratinocytes,72 and IL‐1Ra diminishes the CHS response,37 our results suggested that the inhibitory effect of infant CHS responses by maternal intake of linseed oil was independent on IL‐1Ra (Figure S5). As transcriptional regulators in vitro, NGFI‐A‐binding protein 2 (NAB2) upregulates TRAIL expression in pDCs, natural killer cells, and T cells, whereas early growth response 1 (EGR1) downregulates TRAIL expression in natural killer cells.73, 74 Therefore, although the intracellular signals for TRAIL induction in pDCs have not yet been elucidated, TRAIL expression on dermal pDCs in infants might be increased through the upregulation of NAB2 or downregulation of EGR1 due to maternal intake of linseed oil.
Among 14‐lipoxygenation DPA products, 14‐HDPA in human breast milk was much detected in the group of nonallergic sensitized infants compared with that of allergic infants (Figure S15). These results might mean that 14‐lipoxygenation DPA products reduce the onsets of infant allergy rather than their sensitization of that, consistent with murine results.
In conclusion, we have shown that the synthesis of DPA and its metabolism into 14‐lipoxygenation products occurred preferentially in the mammary glands of mice and that these compounds were transferred to pups through the dams’ milk. This transfer helps to prevent allergic responses in the pups by increasing the number of TRAIL‐expressing dermal pDCs and their TRAIL expression level, consequently leading to decreased activation of T cells. These findings support a new mechanism of dietary regulation of allergic diseases through mother‐infant nutritional interaction in murine model and suggest a promising strategy for preventing infant allergies.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTION
SH conceived and designed the study, performed experiments, analyzed data, and wrote the manuscript. TN, HS, TH, SO, and KK performed immunologic analyses and contributed to discussions. KS, JA, YA, JI, TT, and MA performed lipidomics analyses and helped to set up the LC‐MS/MS analyses. NS and ES collected human milk samples. KH, AM, and HK provided helpful suggestions and discussion. JK conceived, designed, and supervised experiments; provided reagents; and contributed to manuscript preparation.
Supporting information
ACKNOWLEDGEMENTS
We thank M. Narita (Niigata University) for providing the human plasmacytoid dendritic cell line (PMDC05 cells). We also thank our laboratory members for helpful discussions and support.
Hirata S‐I, Nagatake T, Sawane K, et al. Maternal ω3 docosapentaenoic acid inhibits infant allergic dermatitis through TRAIL‐expressing plasmacytoid dendritic cells in mice. Allergy. 2020;75:1935–1951. 10.1111/all.14217
Funding information
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) and the Japan Society for the Promotion of Science (JSPS) KAKENHI (nos. 18H02150, 18H02674, and 17K09604 [to JK], JP18K17997 [to KH], 15638619 [to KK and JK], and 15H05897, 15H05898, and 15H04648 [to MA]); the Japan Agency for Medical Research and Development (nos. 17fk0108223h0002, 17fk0108207h0002, 17ek0210078h0002, 17ak0101068h0001, 17gm1010006s0101, 18ck0106243h0003, and 19ek0410062h0001 [to JK], and 17ek0410032s0102 [to HK and JK], and JP19K07617 [to TN]); the Ministry of Health and Welfare of Japan (to JK); The Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries, and Food Industry (to MA and JK); the Grant for Joint Research Project of the Institute of Medical Science, the University of Tokyo (to JK); Astellas Foundation for Research on Metabolic Disorders (to JK); Nipponham Foundation for the Future of Food (to JK); The Canon Foundation (to JK); and ONO Medical Research Foundation (to JK). The authors have no conflicting financial interests.
REFERENCES
- 1. Cook‐Mills JM. Maternal influences over offspring allergic responses. Curr Allergy Asthma Rep. 2015;15:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Munblit D, Boyle RJ, Warner JO. Factors affecting breast milk composition and potential consequences for development of the allergic phenotype. Clin Exp Allergy. 2015;45:583‐601. [DOI] [PubMed] [Google Scholar]
- 3. Minniti F, Comberiati P, Munblit D, et al. Breast‐milk characteristics protecting against allergy. Endocr Metab Immune Disord Drug Targets. 2014;14:9‐15. [DOI] [PubMed] [Google Scholar]
- 4. Snijders BEP, Thijs C, Dagnelie PC, et al. Breast‐feeding duration and infant atopic manifestations, by maternal allergic status, in the first 2 years of life (KOALA study). J Pediatr. 2007;151(347‐351):351. [DOI] [PubMed] [Google Scholar]
- 5. Chiu C‐Y, Liao S‐L, Su K‐W, et al. Exclusive or Partial breastfeeding for 6 months is associated with reduced milk sensitization and risk of eczema in early childhood: the PATCH birth cohort study. Medicine (Baltimore). 2016;95:e3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Giwercman C, Halkjaer LB, Jensen SM, Bønnelykke K, Lauritzen L, Bisgaard H. Increased risk of eczema but reduced risk of early wheezy disorder from exclusive breast‐feeding in high‐risk infants. J Allergy Clin Immunol. 2010;125:866‐871. [DOI] [PubMed] [Google Scholar]
- 7. Bravi F, Wiens F, Decarli A, Dal Pont A, Agostoni C, Ferraroni M. Impact of maternal nutrition on breast‐milk composition: a systematic review. Am J Clin Nutr. 2016;104:646‐662. [DOI] [PubMed] [Google Scholar]
- 8. Dunstan JA, Mori TA, Barden A, et al. Fish oil supplementation in pregnancy modifies neonatal allergen‐specific immune responses and clinical outcomes in infants at high risk of atopy: a randomized, controlled trial. J Allergy Clin Immunol. 2003;112:1178‐1184. [DOI] [PubMed] [Google Scholar]
- 9. Furuhjelm C, Warstedt K, Fagerås M, et al. Allergic disease in infants up to 2 years of age in relation to plasma omega‐3 fatty acids and maternal fish oil supplementation in pregnancy and lactation. Pediatr Allergy Immunol. 2011;22:505‐514. [DOI] [PubMed] [Google Scholar]
- 10. Rogers LK, Valentine CJ, Pennell M, et al. Maternal docosahexaenoic acid supplementation decreases lung inflammation in hyperoxia‐exposed newborn mice. J Nutr. 2011;141:214‐222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Valentine CJ. Maternal dietary DHA supplementation to improve inflammatory outcomes in the preterm infant. Adv Nutr. 2012;3:370‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baker EJ, Miles EA, Burdge GC, Yaqoob P, Calder PC. Metabolism and functional effects of plant‐derived omega‐3 fatty acids in humans. Prog Lipid Res. 2016;64:30‐56. [DOI] [PubMed] [Google Scholar]
- 13. Serhan CN. Treating inflammation and infection in the 21st century: new hints from decoding resolution mediators and mechanisms. FASEB J. 2017;31:1273‐1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kunisawa J, Arita M, Hayasaka T, et al. Dietary ω3 fatty acid exerts anti‐allergic effect through the conversion to 17,18‐epoxyeicosatetraenoic acid in the gut. Sci Rep. 2015;5:srep09750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nagatake T, Shiogama Y, Inoue A, et al. The 17,18‐epoxyeicosatetraenoic acid‐G protein‐coupled receptor 40 axis ameliorates contact hypersensitivity by inhibiting neutrophil mobility in mice and cynomolgus macaques. J Allergy Clin Immunol. 2018;142(470‐484):e12. [DOI] [PubMed] [Google Scholar]
- 16. Arnardottir H, Orr SK, Dalli J, Serhan CN. Human milk proresolving mediators stimulate resolution of acute inflammation. Mucosal Immunol. 2016;9:757‐766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weiss GA, Troxler H, Klinke G, Rogler D, Braegger C, Hersberger M. High levels of anti‐inflammatory and pro‐resolving lipid mediators lipoxins and resolvins and declining docosahexaenoic acid levels in human milk during the first month of lactation. Lipids Health Dis. 2013;12:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson DT, Palac HL, Baillif V, et al. Long chain fatty acids and related pro‐inflammatory, specialized pro‐resolving lipid mediators and their intermediates in preterm human milk during the first month of lactation. Prostaglandins Leukot Essent Fatty Acids. 2017;121:1‐6. [DOI] [PubMed] [Google Scholar]
- 19. Dalli J, Colas RA, Serhan CN. Serhan CN. Novel n‐3 immunoresolvents: structures and actions. Sci Rep. 2013;3:srep01940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gobbetti T, Dalli J, Colas RA, et al. Protectin D1n–3 DPA and resolvin D5n–3 DPA are effectors of intestinal protection. Proc Natl Acad Sci USA. 2017;114:3963‐3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sawada YU, Honda T, Hanakawa S, et al. Resolvin E1 inhibits dendritic cell migration in the skin and attenuates contact hypersensitivity responses. J Exp Med. 2015;212:1921‐1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takamori A, Nambu A, Sato K, et al. IL‐31 is crucial for induction of pruritus, but not inflammation, in contact hypersensitivity. Sci Rep. 2018;8:6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nara H, Komatsu M, Tekeda Y, Araki A, Akhter N, Asao H. IL‐21 Attenuates FITC‐Induced Contact Hypersensitivity Response via Regulation of Dendritic Cell Function. J Invest Dermatol. 2018;138:2174‐2184. [DOI] [PubMed] [Google Scholar]
- 24. Trépanier M‐O, Lim J, Lai TKY, et al. Intraperitoneal administration of docosahexaenoic acid for 14days increases serum unesterified DHA and seizure latency in the maximal pentylenetetrazol model. Epilepsy Behav. 2014;33:138‐143. [DOI] [PubMed] [Google Scholar]
- 25. Taieb J, Chaput N, Ménard C, et al. A novel dendritic cell subset involved in tumor immunosurveillance. Nat Med. 2006;12:214‐219. [DOI] [PubMed] [Google Scholar]
- 26. Natsuaki Y, Egawa G, Nakamizo S, et al. Perivascular leukocyte clusters are essential for efficient activation of effector T cells in the skin. Nat Immunol. 2014;15:1064‐1069. [DOI] [PubMed] [Google Scholar]
- 27. Nielsen MM, Lovato P, MacLeod AS, et al. IL‐1β–dependent activation of dendritic epidermal T cells in contact hypersensitivity. J Immunol. 2014;192:2975‐2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DePeters EJ, Hovey RC. Methods for collecting milk from mice. J Mammary Gland Biol Neoplasia. 2009;14:397‐400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arita M. Mediator lipidomics in acute inflammation and resolution. J Biochem (Tokyo). 2012;152:313‐319. [DOI] [PubMed] [Google Scholar]
- 30. Narita M, Watanabe N, Yamahira A, et al. A leukemic plasmacytoid dendritic cell line, PMDC05, with the ability to secrete IFN‐alpha by stimulation via Toll‐like receptors and present antigens to naïve T cells. Leuk Res. 2009;33:1224‐1232. [DOI] [PubMed] [Google Scholar]
- 31. Ono S, Kabashima K. Novel insights into the role of immune cells in skin and inducible skin‐associated lymphoid tissue (iSALT). Allergo J Int. 2015;24:170‐179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Christensen AD, Haase C. Immunological mechanisms of contact hypersensitivity in mice. APMIS Acta Pathol Microbiol Immunol Scand. 2012;120:1‐27. [DOI] [PubMed] [Google Scholar]
- 33. Honda T, Egawa G, Grabbe S, Kabashima K. Update of immune events in the murine contact hypersensitivity model: toward the understanding of allergic contact dermatitis. J Invest Dermatol. 2013;133:303‐315. [DOI] [PubMed] [Google Scholar]
- 34. Eyerich S, Traidl‐Hoffmann C, Behrendt H, et al. Novel key cytokines in allergy: IL‐17, IL‐22. Allergol Sel. 2017;1:71‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suzuki K, Meguro K, Nakagomi D, Nakajima H. Roles of alternatively activated M2 macrophages in allergic contact dermatitis. Allergol Int. 2017;66:392‐397. [DOI] [PubMed] [Google Scholar]
- 36. Kondo S, Pastore S, Fujisawa H, et al. Interleukin‐1 Receptor Antagonist Suppresses Contact Hypersensitivity. J Invest Dermatol. 1995;105:334‐338. [DOI] [PubMed] [Google Scholar]
- 37. Ziegler SF, Roan F, Bell BD, Stoklasek TA, Kitajima M, Han H. The biology of thymic stromal lymphopoietin (TSLP). Adv Pharmacol. 2013;66:129‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goubier A, Dubois B, Gheit H, et al. Plasmacytoid dendritic cells mediate oral tolerance. Immunity 2008;29:464‐475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ring S, Schäfer SC, Mahnke K, Lehr H‐A, Enk AH. CD4+ CD25+ regulatory T cells suppress contact hypersensitivity reactions by blocking influx of effector T cells into inflamed tissue. Eur J Immunol. 2006;36:2981‐2992. [DOI] [PubMed] [Google Scholar]
- 40. Hardy AW, Graham DR, Shearer GM, Herbeuval J‐P. HIV turns plasmacytoid dendritic cells (pDC) into TRAIL‐expressing killer pDC and down‐regulates HIV coreceptors by Toll‐like receptor 7‐induced IFN‐alpha. Proc Natl Acad Sci USA. 2007;104:17453‐17458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kingham TP, Chaudhry UI, Plitas G, Katz SC, Raab J, DeMatteo RP. Murine liver plasmacytoid dendritic cells become potent immunostimulatory cells after Flt‐3 ligand expansion. Hepatology 2007;45:445‐454. [DOI] [PubMed] [Google Scholar]
- 42. Jahrsdörfer B, Vollmer A, Blackwell SE, et al. Granzyme B produced by human plasmacytoid dendritic cells suppresses T‐cell expansion. Blood 2010;115:1156‐1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Contractor N, Louten J, Kim L, Biron CA, Kelsall BL. Cutting edge: Peyer's patch plasmacytoid dendritic cells (pDCs) produce low levels of type I interferons: possible role for IL‐10, TGFbeta, and prostaglandin E2 in conditioning a unique mucosal pDC phenotype. J Immunol. 2007;179:2690‐2694. [DOI] [PubMed] [Google Scholar]
- 44. Mathan TSMM, Figdor CG, Buschow SI. Human plasmacytoid dendritic cells: from molecules to intercellular communication network. Front Immunol. 2013;4:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cisse B, Caton ML, Lehner M, et al. Transcription factor E2–2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 2008;135:37‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ghosh HS, Cisse B, Bunin A, Lewis KL, Reizis B. Continuous expression of the transcription factor e2–2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity 2010;33:905‐916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lehnert C, Weiswange M, Jeremias I, et al. TRAIL‐receptor costimulation inhibits proximal TCR signaling and suppresses human T cell activation and proliferation. J Immunol. 1950;2014(193):4021‐4031. [DOI] [PubMed] [Google Scholar]
- 48. Green DR, Ferguson T, Zitvogel L, Kroemer G. Immunogenic and tolerogenic cell death. Nat Rev Immunol. 2009;9:nri2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wehrli P, Viard I, Bullani R, Tschopp J, French LE. Death receptors in cutaneous biology and disease. J Invest Dermatol. 2000;115:141‐148. [DOI] [PubMed] [Google Scholar]
- 50. Falschlehner C, Emmerich CH, Gerlach B, Walczak H. TRAIL signalling: decisions between life and death. Int J Biochem Cell Biol. 2007;39:1462‐1475. [DOI] [PubMed] [Google Scholar]
- 51. Gregory MK, Gibson RA, Cook‐Johnson RJ, Cleland LG, James MJ. Elongase reactions as control points in long‐chain polyunsaturated fatty acid synthesis. PLoS ONE 2011;6:e29662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yeung J, Hawley M, Holinstat M. The expansive role of oxylipins on platelet biology. J Mol Med. 2017;95:575‐588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gregory MK, Cleland LG, James MJ. Molecular basis for differential elongation of omega‐3 docosapentaenoic acid by the rat Elovl5 and Elovl2. J Lipid Res. 2013;54:2851‐2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Y, Botolin D, Christian B, Busik J, Xu J, Jump DB. Tissue‐specific, nutritional, and developmental regulation of rat fatty acid elongases. J Lipid Res. 2005;46:706‐715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mashima R, Okuyama T. The role of lipoxygenases in pathophysiology; new insights and future perspectives. Redox Biol. 2015;6:297‐310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Waidyatillake NT, Dharmage SC, Allen KJ, et al. Association of breast milk fatty acids with allergic disease outcomes‐A systematic review. Allergy 2018;73:295‐312. [DOI] [PubMed] [Google Scholar]
- 57. Prasad VVTS, Kolli P, Moganti D. Association of a functional polymorphism (Gln261Arg) in 12‐lipoxygenase with breast cancer. Exp Ther Med. 2011;2:317‐323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chiang N, Fredman G, Bäckhed F, et al. Infection regulates pro‐resolving mediators that lower antibiotic requirements. Nature 2012;484:524‐528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hirata S‐I, Kunisawa J. Gut microbiome, metabolome, and allergic diseases. Allergol Int. 2017;66:523‐528. [DOI] [PubMed] [Google Scholar]
- 60. Shively CA, Register TC, Appt SE, et al. Consumption of Mediterranean versus western diet leads to distinct mammary gland microbiome populations. Cell Rep. 2018;25(47‐56):e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Francois CA, Connor SL, Bolewicz LC, Connor WE. Supplementing lactating women with flaxseed oil does not increase docosahexaenoic acid in their milk. Am J Clin Nutr. 2003;77:226‐233. [DOI] [PubMed] [Google Scholar]
- 62. Tomobe YI, Morizawa K, Tsuchida M, Hibino H, Nakano Y, Tanaka Y. Dietary docosahexaenoic acid suppresses inflammation and immunoresponses in contact hypersensitivity reaction in mice. Lipids 2000;35:61‐69. [DOI] [PubMed] [Google Scholar]
- 63. Shek LP, Chong M‐F, Lim JY, Soh S‐E, Chong Y‐S. Chong Y‐S. Role of dietary long‐chain polyunsaturated fatty acids in infant allergies and respiratory diseases. Clin Dev Immunol. 2012;2012:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weiser MJ, Butt CM, Mohajeri MH. Docosahexaenoic acid and cognition throughout the lifespan. Nutrients. 2016;8(2):99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Elsherbiny M, Goruk S, Monckton E, et al. Long‐term effect of docosahexaenoic acid feeding on lipid composition and brain fatty acid‐binding protein expression in rats. Nutrients. 2015;7:8802‐8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Heinz RE, Rudolph MC, Ramanathan P, et al. Constitutive expression of microRNA‐150 in mammary epithelium suppresses secretory activation and impairs de novo lipogenesis. Dev Camb Engl. 2016;143:4236‐4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mohammad MA, Haymond MW. Regulation of lipid synthesis genes and milk fat production in human mammary epithelial cells during secretory activation. Am J Physiol Endocrinol Metab. 2013;305:E700‐716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Orlando V, La Manna MP, Goletti D, et al. Human CD4 T‐cells with a naive phenotype produce multiple cytokines during mycobacterium tuberculosis infection and correlate with active disease. Front Immunol. 2018;9:1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sa Q, Woodward J, Suzuki Y. IL‐2 produced by CD8+ immune T cells can augment their IFN‐γ production independently from their proliferation in the secondary response to an intracellular pathogen. J Immunol. 2013;190:2199‐2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mirandola P, Ponti C, Gobbi G, et al. Activated human NK and CD8+ T cells express both TNF‐related apoptosis‐inducing ligand (TRAIL) and TRAIL receptors but are resistant to TRAIL‐mediated cytotoxicity. Blood 2004;104:2418‐2424. [DOI] [PubMed] [Google Scholar]
- 71. Ohtani T, Memezawa AI, Okuyama R, et al. Increased hyaluronan production and decreased E‐cadherin expression by cytokine‐stimulated keratinocytes lead to spongiosis formation. J Invest Dermatol. 2009;129:1412‐1420. [DOI] [PubMed] [Google Scholar]
- 72. Vassina E, Leverkus M, Yousefi S, Braathen LR, Simon H‐U, Simon D. Increased expression and a potential anti‐inflammatory role of TRAIL in atopic dermatitis. J Invest Dermatol. 2005;125:746‐752. [DOI] [PubMed] [Google Scholar]
- 73. Balzarolo M, Karrich JJ, Engels S, Blom B, Medema JP, Wolkers MC. The transcriptional regulator NAB2 reveals a two‐step induction of TRAIL in activated plasmacytoid DCs. Eur J Immunol. 2012;42:3019‐3027. [DOI] [PubMed] [Google Scholar]
- 74. Balzarolo M, Watzl C, Medema JP, Wolkers MC. NAB2 and EGR‐1 exert opposite roles in regulating TRAIL expression in human Natural Killer cells. Immunol Lett. 2013;151:61‐67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


